Abstract
Background: Juvenile myoclonic epilepsy (JME) is a common adolescent epilepsy characterized by myoclonic, generalized tonic–clonic, and sometimes absence seizures. Prognosis varies, with many patients experiencing relapse despite pharmacological treatment. Recent advances in imaging and artificial intelligence suggest that combining microstructural brain changes with traditional clinical variables can enhance potential prognostic biomarkers identification. Methods: A retrospective study was conducted on patients with JME at the Severance Hospital, analyzing clinical variables and magnetic resonance imaging (MRI) data. Machine learning models were developed to predict prognosis using clinical and radiological features. Results: The study utilized six machine learning models, with the XGBoost model demonstrating the highest predictive accuracy (AUROC 0.700). Combining clinical and MRI data outperformed models using either type of data alone. The key features identified through a Shapley additive explanation analysis included the volumes of the left cerebellum white matter, right thalamus, and left globus pallidus. Conclusions: This study demonstrated that integrating clinical and radiological data enhances the predictive accuracy of JME prognosis. Combining these neuroanatomical features with clinical variables provided a robust prediction of JME prognosis, highlighting the importance of integrating multimodal data for accurate prognosis.
Keywords: juvenile myoclonic epilepsy, prognosis, machine learning, artificial intelligence, magnetic resonance imaging
1. Introduction
Juvenile myoclonic epilepsy (JME) is a prevalent adolescent epilepsy, accounting for approximately 10% of all epilepsies [1,2]. Characterized by recurrent myoclonic seizures, primarily in the shoulders and limbs, JME typically begins during puberty and is often accompanied by generalized onset motor tonic–clonic seizures and generalized onset non-motor (absence) seizures [3,4]. Classified under idiopathic generalized epilepsy (IGE), the prognosis for JME with pharmacological treatment is relatively favorable, with approximately 60% of the patients achieving five years or more of seizure freedom on medication, and approximately 25% maintaining remission without medication [5,6]. However, many patients relapse after stopping medication, necessitating lifelong treatment. Meta-analyses suggest that approximately 35% of the patients exhibit drug-refractory epilepsy [7,8].
Clinical factors influencing the prognosis with antiseizure medication include female gender, a younger age of onset, a history of absence seizures, praxis-induced seizures, childhood absence epilepsy, comorbid psychiatric disorders, family history, epileptiform asymmetries on Electroencephalogram (EEG), and the absence of photoparoxysmal response [9,10]. While brain magnetic resonance imaging (MRI) in patients with JME appears visually normal, recent advancements in quantitative and functional MRI analysis reveal differences in structural and functional connectivity compared to healthy controls, along with widespread neocortical thinning as the disease progresses [11,12].
Despite the progress in imaging techniques, studies analyzing quantitative imaging features for prognosis in JME remain scarce. Recent efforts have utilized machine learning and deep learning to develop diagnostic and prognostic models for various neurological diseases, including epilepsy [13,14]. Some studies have combined clinical information and brain MRI images to predict drug response, but these models often underperform due to the insufficient integration of comprehensive variables [15,16].
To address this gap, our study develops and validates artificial intelligence models that combine clinical and imaging variables to predict the prognosis of drug treatment response in JME.
2. Materials and Methods
2.1. Data and Participants
This study was approved by the institutional review board (no. 4-2021-1196), and the requirement for informed consent was waived. A retrospective review of consecutive patients who presented with seizures and visited the epilepsy clinic of the Severance Hospital (a tertiary care center) between January 2000 and May 2021 was conducted [17]. The patients diagnosed with JME by epilepsy specialists were included. The exclusion criteria were as follows: (1) patients with less than a 3-year follow-up, and (2) participants who underwent a 2D protocol MRI.
The initial diagnosis of JME was confirmed by reviewing the medical records of the institution’s neurologists. The diagnosis was based on the clinical and EEG features set by the International League against Epilepsy. The patients with JME experienced both generalized onset motor myoclonic and generalized onset motor tonic–clonic seizures. Additionally, the EEG findings consistently demonstrated generalized polyspikes or spike-and-wave complexes in all the patients with JME. The MRI readings were confirmed to be normal by board-certified neuroradiologists.
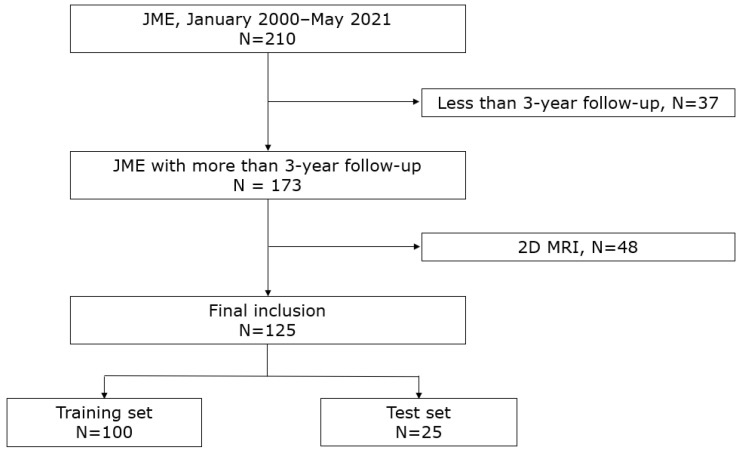
The clinical variables collected from the patients included age, sex, age at onset, the duration of disease, treatment history of seizures, number of antiseizure medications attempted prior to the visit, a family history of epilepsy, a history of febrile seizures, and a history of absence seizures. The patients who were seizure-free for 2 years or longer at any time after diagnosis were considered to have a favorable outcome. We randomly divided the entire subjects into training and test sets in an 8:2 ratio and used them for training and testing the artificial intelligence model. The study flow is shown in Figure 1.
Figure 1.
Flow chart depicting the study subjects.
2.2. Image Acquisition and Processing
All the MRI scans were performed using a 3T MRI system with an eight-channel sensitivity-encoding head coil (Achieva or Ingenia, Philips Healthcare; TrioTim, Siemens). The 3D T1-weighted images with isotropic voxels were obtained using a spoiled gradient echo sequence. For preprocessing, the T1-weighted images were resampled to a uniform spatial resolution of 1 × 1 × 1 mm3, followed by the N4 bias field correction and z-score normalization of the images. All the images were submitted to imaging experts for visual analysis. Structural MRI data were processed using the FreeSurfer 6.0.0 software. The processing pipeline included several key steps: Raw Digital Imaging and Communications in Medicine (DICOM) files were converted to Neuroimaging Informatics Technology Initiative (NIfTI) format and imported into the FreeSurfer environment. The automated segmentation of the brain was performed using the recon-all pipeline, which included motion correction, skull stripping, and the segmentation of cortical and subcortical regions [18,19,20]. The resulting segmentations were visually inspected for accuracy, and manual corrections were applied as necessary. Following segmentation, the volumetric measurements of various brain regions were obtained from the automatically generated aseg.stats file. Key 29 regions of interest (ROI), including bilateral thalamus, bilateral caudate, bilateral putamen, bilateral globus pallidus, bilateral hippocampus, bilateral amygdala, bilateral nucleus accumbens, bilateral ventral diencephalon, bilateral choroid plexus, bilateral cerebellum (cortex and white matter), optic chiasm, brain stem, and corpus callosum (anterior, mid-anterior, central, mid-posterior, and posterior), were extracted and compiled into a comprehensive dataset for the subsequent statistical analysis. This process ensured the consistent and accurate measurement of brain region volumes across all the subjects.
2.3. Machine Learning Models
Machine learning models were developed using a dataset composed of clinical variables from the patients with JME and volume data from the various regions of interest (ROI). The dataset was divided into training and test sets to train and evaluate the models.
We employed six machine learning algorithms (decision tree, random forest, XGBoost, LightGBM, support vector machine (SVM), and artificial neural network (ANN)) to predict post-surgery delirium. Each model was trained on the training set, with hyperparameters tuned using grid search with cross-validation on the training data. The models’ performances were evaluated on the testing set based on accuracy and the area under the receiver operating characteristic curve (AUROC).
A decision tree classifier was implemented using the scikit-learn library, with tree depth restricted to prevent overfitting, and the maximum depth set through cross-validation. The random forest model, also implemented in scikit-learn, consisted of an ensemble of decision trees, with parameters such as the number of trees (n_estimators) and the maximum depth of each tree (max_depth) optimized via cross-validation. XGBoost was used to train a gradient boosting model that minimizes a regularized objective function, with hyperparameters including n_estimators, max_depth, learning_rate, and subsample rates tuned to optimize performance. LightGBM, another gradient boosting framework, was chosen for its efficiency with large datasets, with parameters such as num_leaves, max_depth, learning_rate, and n_estimators tuned to find the optimal settings. The SVM model classified the data by finding the optimal hyperplane that maximizes the margin between classes. A kernel function was applied to handle non-linear relationships in the data. An ANN model with input, hidden, and output layers was used. Neurons applied weighted sums and non-linear activation functions, and the model was trained using backpropagation and gradient descent to minimize the loss function.
Shapley additive explanations (SHAP) were adopted to verify the explainability of the AI models [21,22]. These values highlight the key factors in predicting JME prognosis, providing insights into model patterns.
3. Results
3.1. Clinical Characteristics
The clinical variables are summarized in Table 1. Among the 125 patients, 85 were seizure-free for over 2 years (favorable prognosis), whereas 40 were not (poor prognosis). In the favorable prognosis group, the proportion of males was higher compared with the poor prognosis group (60.0% vs. 40.0%, p = 0.036). There were no significant differences between the two groups regarding the age, onset age, and epilepsy duration. Similarly, there were no differences in the family history, febrile seizure history, or the presence of absence seizures between the groups. Regarding treatment, there were no differences in the proportion of treated patients, the number of antiseizure medications used, or the use of valproic acid, lamotrigine, levetiracetam, and topiramate. Additionally, the follow-up duration was comparable between the two groups.
Table 1.
Demographic and clinical characteristics of the patients in the study.
| Favorable Prognosis (n = 85) |
Poor Prognosis (n = 40) |
p-Value | |
|---|---|---|---|
| Age (years) | 23.5 ± 8.7 | 23.2 ± 7.7 | 0.843 |
| Male sex, n (%) | 51 (60.0) | 16 (40.0) | 0.036 |
| Onset age (years) | 15.2 ± 4.5 | 15.2 ± 4.8 | 0.991 |
| Epilepsy duration (years) | 8.3 ± 9.4 | 8.0 ± 8.2 | 0.888 |
| Family history, n (%) | 16 (18.8) | 4 (10.0) | 0.209 |
| Febrile seizure history *, n (%) | 9 (10.6) | 5 (12.5) | 0.767 |
| Absence seizure, n (%) | 31 (36.5) | 14 (35.0) | 0.873 |
| Treated, n (%) | 44 (51.8) | 24 (60.0) | 0.388 |
| Number of ASMs | 2 (1–3) | 2 (1–3) | 0.577 |
| VPA, n (%) | 65 (76.5) | 30 (75.0) | 0.857 |
| LTG, n (%) | 39 (45.9) | 21 (52.5) | 0.490 |
| LEV, n (%) | 40 (47.1) | 24 (60.0) | 0.177 |
| TPM, n (%) | 17 (20.0) | 6 (15.0) | 0.501 |
| Follow-up duration (years) | 13.9 ± 6.7 | 11.1 ± 7.2 | 0.178 |
* Fisher’s exact test was used. ASM, antiseizure medication; VPA, valproic acid; LTG, lamotrigine; LEV, levetiracetam; TPM, topiramate.
3.2. Radiological Characteristics
The radiological characteristics are summarized in Table 2. Comparing the volume data of the regions of interest (ROI) between the two groups demonstrated that the volumes of the left amygdala (1739.9 ± 263.4 vs. 1601.7 ± 358.2, p = 0.017) and the right hippocampus (4396.6 ± 417.7 vs. 4128.8 ± 825.7, p = 0.017) were significantly smaller in the poor prognosis group. No significant differences were observed between the groups in the volumes of the other bilateral subcortical structures, the brain stem, the corpus callosum, or the total intracranial volume.
Table 2.
Radiologic features and their association with the prognosis of JME.
| Favorable Prognosis (n = 85) |
Poor Prognosis (n = 40) |
p-Value | |
|---|---|---|---|
| Left | |||
| Thalamus | 8054.0 ± 844.4 | 7748.2 ± 1393.7 | 0.131 |
| Caudate | 3569.1 ± 433.8 | 3458.2 ± 643.6 | 0.259 |
| Putamen | 5075.5 ± 619.2 | 4902.2 ± 895.1 | 0.211 |
| Pallidum | 2073.8 ± 244.5 | 2017.6 ± 353.9 | 0.303 |
| Hippocampus | 4180.6 ± 415.2 | 4027.1 ± 630.2 | 0.107 |
| Amygdala | 1739.9 ± 263.4 | 1601.7 ± 358.2 | 0.017 |
| Nucleus accumbens | 511.4 ± 98.5 | 286.6 ± 133.8 | 0.246 |
| Ventral diencephalon | 4175.7 ± 457.7 | 4003.7 ± 621.1 | 0.084 |
| Choroid plexus | 437.0 ± 163.7 | 427.5 ± 159.2 | 0.765 |
| Cerebellum–cortex | 56,595.1 ± 5734.4 | 54,113.8 ± 8452.0 | 0.057 |
| Cerebellum–white matter | 14,800.9 ± 1846.9 | 14,133.2 ± 2293.1 | 0.084 |
| Right | |||
| Thalamus | 7574.4 ± 775.1 | 7219.0 ± 1275.0 | 0.056 |
| Caudate | 3637.9 ± 444.1 | 3544.3 ± 567.1 | 0.318 |
| Putamen | 5130.4 ± 617.7 | 4985.8 ± 809.3 | 0.273 |
| Pallidum | 1977.6 ± 238.8 | 1942.0 ± 284.8 | 0.467 |
| Hippocampus | 4396.6 ± 417.7 | 4128.8 ± 825.7 | 0.017 |
| Amygdala | 1848.7 ± 279.3 | 1744.6 ± 361.0 | 0.080 |
| Nucleus accumbens | 577.2 ± 105.1 | 554.5 ± 117.2 | 0.280 |
| Ventral diencephalon | 4173.3 ± 444.1 | 4017.0 ± 562.6 | 0.095 |
| Choroid plexus | 430.6 ± 154.9 | 422.9 ± 188.5 | 0.809 |
| Cerebellum–cortex | 56,266.6 ± 5838.5 | 53,800.4 ± 8392.4 | 0.059 |
| Cerebellum–white matter | 14,242.4 ± 2001.1 | 13,540.7 ± 2200.4 | 0.079 |
| Midline | |||
| Brainstem | 21,182.7 ± 2266.9 | 20,565.8 ± 3631.8 | 0.248 |
| Optic-chiasm | 154.3 ± 58.7 | 140.1 ± 60.8 | 0.213 |
| Corpus callosum | |||
| Anterior | 862.7 ± 141.4 | 836.8 ± 158.5 | 0.360 |
| Mid-anterior | 669.4 ± 181.2 | 666.1 ± 171.8 | 0.922 |
| Central | 688.8 ± 172.1 | 660.2 ± 171.5 | 0.388 |
| Mid-posterior | 552.8 ± 103.9 | 560.6 ± 128.1 | 0.720 |
| Posterior | 990.4 ± 178.0 | 982.0 ± 205.2 | 0.815 |
| Total intracranial volume | 1,581,418.5 ± 175,060.8 | 1,502,151.6 ± 250,957.1 | 0.077 |
The presented values represent the volume of each brain region, with the units in mm3.
3.3. Performances of Machine Learning Models
Table 3 shows the performance of the six machine learning models—logistic regression, random forest, XGBoost, LightGBM, SVM, and ANN—developed using the training set and validated on an independent test set. Among these, XGBoost achieved the highest performance (AUROC, 0.700), followed by LightGBM (AUROC, 0.618), random forest (AUROC, 0.7317), SVM (AUROC, 0.500), logistic regression (AUROC, 0.431), and ANN (AUROC, 0.425).
Table 3.
Performances of machine learning models on the test set.
| Models | Accuracy | Precision | Recall | F1-Score | AUROC |
|---|---|---|---|---|---|
| Logistic Regression | 0.600 | 0.560 | 0.600 | 0.565 | 0.431 |
| Random Forest | 0.680 | 0.664 | 0.680 | 0.652 | 0.580 |
| XGBoost | 0.680 | 0.816 | 0.680 | 0.712 | 0.700 |
| Light GBM | 0.560 | 0.486 | 0.560 | 0.505 | 0.618 |
| SVM | 0.640 | 0.410 | 0.640 | 0.500 | 0.500 |
| ANN | 0.600 | 0.400 | 0.600 | 0.480 | 0.425 |
AUROC, area under the receiver operating characteristic curve; XGBoost, extreme gradient boosting; LightGBM, light gradient boosting machine; SVM, support vector machine; ANN, artificial neural network.
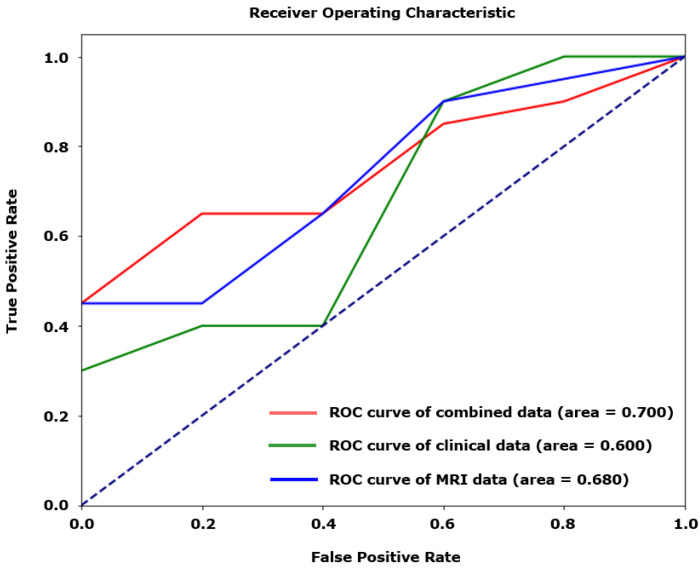
When the best-performing XGBoost model was developed using only clinical data, AUROC was 0.0.600. Using only the MRI data, AUROC was 0.680. Combining both the clinical and MRI data improved AUROC to 0.700 (Figure 2).
Figure 2.
Receiver operating characteristic (ROC) curves for models using clinical, radiological, and combined variables to predict poor prognosis of JME.
3.4. Feature Importances
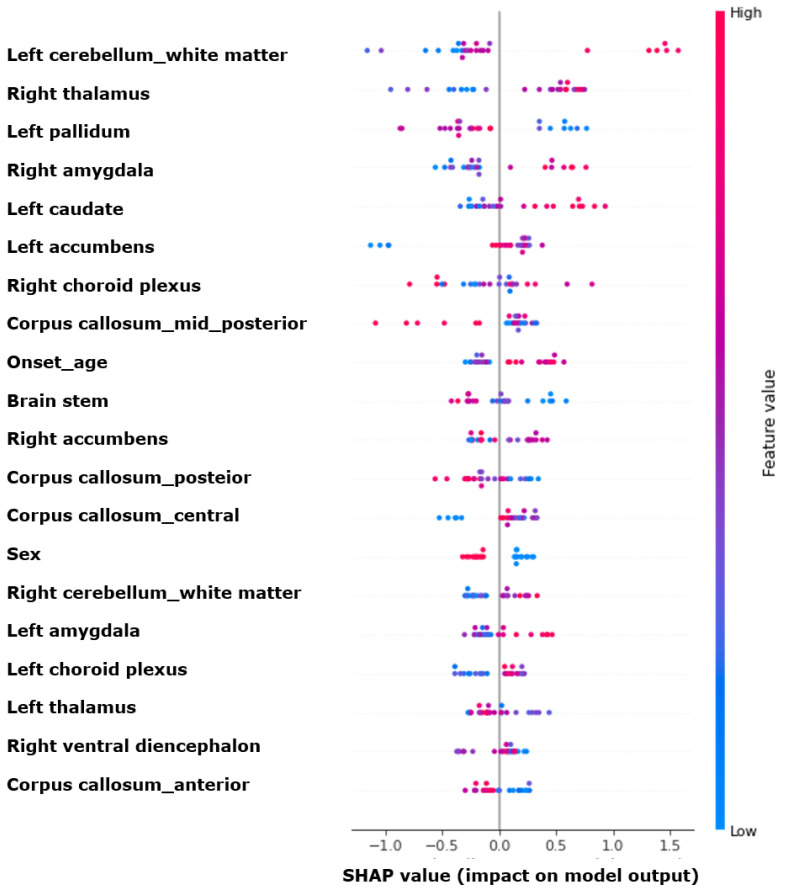
Figure 3 illustrates the feature importance using SHAP values. Among the top ten features, the five most significant MRI variables were as follows: left cerebellum white matter, right thalamus, left globus pallidus, right amygdala, and left caudate. Following these are the left nucleus accumbens, right choroid plexus, corpus callosum mid-posterior, onset age, and brain stem.
Figure 3.
SHAP value summary plot for the XGBoost model. SHAP, Shapley additive explanations; XGBoost, extreme gradient boosting.
4. Discussion
The key findings of this study are as follows:
The volumes of the left amygdala and right hippocampus, along with male gender, were associated with prognosis;The XGBoost model demonstrated a performance of approximately 0.700 in predicting prognosis, with higher performance observed when combining the clinical and radiological variables;
The cerebellum, thalamus, and globus pallidus were crucial for the machine learning model’s prediction of prognosis.
In our clinical variables, male gender was associated with a favorable prognosis, consistent with previous studies on the prognosis of JME [6,23]. This may be due to the broader range of antiseizure medications available to males compared to females of childbearing age. Additionally, this study identified the amygdala and hippocampus as the structures associated with prognosis. These structures have shown differences in comparisons between the patients with JME and healthy controls, and are involved in emotion and cognition, which may influence prognosis in JME.
This study develops a novel prognostic prediction model for JME based on clinical and radiological variables. Recent efforts have concentrated on developing various AI models that utilize clinical variables for diagnosis and treatment in clinical settings [16,24]. Our research group’s previous studies have investigated the use of radiological variables in diagnosing and classifying JME and other generalized epilepsies. [25]. These studies demonstrated that microstructural changes in the brain MRI of patients with JME could enhance diagnosis and classification. This study further shows that combining clinical and radiological variables yields superior predictive performance compared to using either type of variable alone. Thus, it is crucial to use multimodal data in AI models to enhance prognostic prediction capabilities. It is expected that this model will assist physicians, especially those who are not epileptologists, in diagnosing and explaining the prognosis of epilepsy patients based on clinical and MRI data. Increasing the generalizability of these models through diverse datasets is essential for improving the application of AI in epilepsy treatment.
In our study, the SHAP value analysis of the most efficient XGBoost model revealed that the cerebellum and thalamus were the two most important variables for predicting prognosis. While cerebellar volume reduction has been associated with the use of antiseizure medications (ASMs) such as phenytoin, none of our patients were treated with phenytoin. Additionally, there were no significant differences between the groups in terms of the number of ASMs used or the duration of epilepsy [26]. Previous studies have demonstrated that patients with JME show a reduced cerebellar white matter volume compared to healthy controls [27,28]. This finding aligns with the theory that an altered cerebello-thalamo-cortical network contributes to JME pathogenesis, potentially influencing treatment response and prognosis. Moreover, the thalamus undergoes microstructural changes in JME, with volume alterations in specific regions being well documented [29,30]. A previous study has also suggested the potential involvement of altered thalamo-frontal connectivity with the widespread network in seizure regulation [31,32]. In our study, the thalamus was similarly identified as a crucial feature in determining prognosis, reinforcing its importance in the disease’s underlying mechanisms and clinical outcomes.
This study has several limitations. First, being a single-center study, it has a small sample size, and external validation will be crucial in future research to verify reproducibility. Second, the multimodal approach was limited, utilizing only the volume analysis of T1 3D MRI among the various radiological variables, and EEG signals were not used in the model development. Third, the study included a heterogeneous sample with varied treatment exposures and disease durations. Future research with larger samples and stricter inclusion criteria may provide more robust results.
This study has several notable strengths. First, it presents the novel prognosis of JME by combining clinical and radiological variables, demonstrating that radiological variables significantly enhance the prognostic prediction capability alongside clinical variables. We identified models with appropriate performance levels using various machine-learning techniques. Second, the study confirmed that both clinical and radiological variables, previously identified as relevant to JME in earlier studies, showed differences according to prognosis. The importance of these variables in the machine learning models was also related to the mechanisms of JME, providing clinical explainability.
5. Conclusions
This study demonstrated the clinical utility of a machine learning model combining clinical and radiological variables to predict the prognosis of JME. It highlighted the potential of AI-supported care using explainable machine learning models in the clinical management of epilepsy.
Author Contributions
K.M.K.: Conception and design, acquisition of data, data analysis and interpretation, and manuscript writing. B.K.C.: Data analysis and interpretation. W.-S.H., S.C., M.K.C. and K.H.: Revision of manuscript critically for important intellectual content. W.-J.K.: Conception and design, data analysis and interpretation, supervision, and final approval of the version to be submitted. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the Institutional Review Board of Severance Hospital, Yonsei University (No. 4-2021-1196) approved on 20 October 2021.
Informed Consent Statement
Patient consent was waived due to the retrospective nature and use of de-identified data, with approval from the Institutional Review Board of Severance Hospital, Yonsei University (No. 4-2021-1196).
Data Availability Statement
Anonymized data relevant to this study will be shared upon request with a qualified investigator pending appropriate Institutional Review Board approvals.
Conflicts of Interest
All the authors have no conflicts to disclose.
Funding Statement
This study was supported by a new faculty research seed money grant from Yonsei University College of Medicine for 2024 (2024-32-0050). The sponsors had no role in the study design; the collection, analysis, and interpretation of the data; the writing of the report; or the decision to submit this article for publication.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hauser W.A. The prevalence and incidence of convulsive disorders in children. Epilepsia. 1994;35((Suppl. 2)):S1–S6. doi: 10.1111/j.1528-1157.1994.tb05932.x. [DOI] [PubMed] [Google Scholar]
- 2.Grünewald R.A., Panayiotopoulos C.P. Juvenile myoclonic epilepsy: A review. Arch. Neurol. 1993;50:594–598. doi: 10.1001/archneur.1993.00540060034013. [DOI] [PubMed] [Google Scholar]
- 3.Proposal for classification of epilepsies and epileptic syndromes Commission on classification and terminology of the international league against epilepsy. Epilepsia. 1985;26:268–278. [PubMed] [Google Scholar]
- 4.Baykan B., Wolf P. Juvenile myoclonic epilepsy as a spectrum disorder: A focused review. Seizure. 2017;49:36–41. doi: 10.1016/j.seizure.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Baykan B., Martínez-Juárez I.E., Altindag E.A., Camfield C.S., Camfield P.R. Lifetime prognosis of juvenile myoclonic epilepsy. Epilepsy Behav. 2013;28((Suppl. 1)):S18–S24. doi: 10.1016/j.yebeh.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Giuliano L., Mainieri G., Aguglia U., Bilo L., Durante V., Ermio C., Galimberti C.A., La Neve A., Monti G., Ranzato F., et al. Long-term prognosis of juvenile myoclonic epilepsy: A systematic review searching for sex differences. Seizure. 2021;86:41–48. doi: 10.1016/j.seizure.2021.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Fayad C., Saad K., Kahwagi G.J., Hallit S., Griffin D., Abou-Khalil R., El-Hayek E. A systematic review and meta-analysis of factors related to first line drugs refractoriness in patients with juvenile myoclonic epilepsy (jme) PLoS ONE. 2024;19:e0300930. doi: 10.1371/journal.pone.0300930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevelink R., Koeleman B.P.C., Sander J.W., Jansen F.E., Braun K.P.J. Refractory juvenile myoclonic epilepsy: A meta-analysis of prevalence and risk factors. Eur. J. Neurol. 2019;26:856–864. doi: 10.1111/ene.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guaranha M.S., Filho G.M., Lin K., Guilhoto L.M., Caboclo L.O., Yacubian E.M. Prognosis of juvenile myoclonic epilepsy is related to endophenotypes. Seizure. 2011;20:42–48. doi: 10.1016/j.seizure.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Rubboli G., Beier C.P., Selmer K.K., Syvertsen M., Shakeshaft A., Collingwood A., Hall A., Andrade D.M., Fong C.Y., Gesche J., et al. Variation in prognosis and treatment outcome in juvenile myoclonic epilepsy: A biology of juvenile myoclonic epilepsy consortium proposal for a practical definition and stratified medicine classifications. Brain Commun. 2023;5:fcad182. doi: 10.1093/braincomms/fcad182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domin M., Bartels S., Geithner J., Wang Z.I., Runge U., Grothe M., Langner S., von Podewils F. Juvenile myoclonic epilepsy shows potential structural white matter abnormalities: A tbss study. Front. Neurol. 2018;9:509. doi: 10.3389/fneur.2018.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J.H., Suh S.i., Park S.Y., Seo W.K., Koh I., Koh S.B., Seol H.Y. Microstructural white matter abnormality and frontal cognitive dysfunctions in juvenile myoclonic epilepsy. Epilepsia. 2012;53:1371–1378. doi: 10.1111/j.1528-1167.2012.03544.x. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y., Chen Z., Yang Y., Zhang J., Wang Y. Development and validation of an interpretable machine learning model for predicting post-stroke epilepsy. Epilepsy Res. 2024;205:107397. doi: 10.1016/j.eplepsyres.2024.107397. [DOI] [PubMed] [Google Scholar]
- 14.Han K., Liu C., Friedman D. Artificial intelligence/machine learning for epilepsy and seizure diagnosis. Epilepsy Behav. 2024;155:109736. doi: 10.1016/j.yebeh.2024.109736. [DOI] [PubMed] [Google Scholar]
- 15.Kaushik M., Mahajan S., Machahary N., Thakran S., Chopra S., Tomar R.V., Kushwaha S.S., Agarwal R., Sharma S., Kukreti R., et al. Predicting efficacy of antiseizure medication treatment with machine learning algorithms in north indian population. Epilepsy Res. 2024;205:107404. doi: 10.1016/j.eplepsyres.2024.107404. [DOI] [PubMed] [Google Scholar]
- 16.Hakeem H., Feng W., Chen Z., Choong J., Brodie M.J., Fong S.L., Lim K.S., Wu J., Wang X., Lawn N., et al. Development and validation of a deep learning model for predicting treatment response in patients with newly diagnosed epilepsy. JAMA Neurol. 2022;79:986–996. doi: 10.1001/jamaneurol.2022.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye B.S., Cho Y.-J., Jang S.H., Lee M.K., Lee B.I., Heo K. The localizing and lateralizing value of auras in lesional partial epilepsy patients. Yonsei Med. J. 2012;53:477–485. doi: 10.3349/ymj.2012.53.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ségonne F., Dale A.M., Busa E., Glessner M., Salat D., Hahn H.K., Fischl B. A hybrid approach to the skull stripping problem in mri. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 19.Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- 20.Ségonne F., Pacheco J., Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans. Med. Imaging. 2007;26:518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- 21.Giuste F., Shi W., Zhu Y., Naren T., Isgut M., Sha Y., Tong L., Gupte M., Wang M.D. Explainable artificial intelligence methods in combating pandemics: A systematic review. IEEE Rev. Biomed. Eng. 2023;16:5–21. doi: 10.1109/RBME.2022.3185953. [DOI] [PubMed] [Google Scholar]
- 22.Cho S., Joo B., Park M., Ahn S.J., Suh S.H., Park Y.W., Ahn S.S., Lee S.K. A radiomics-based model for potentially more accurate identification of subtypes of breast cancer brain metastases. Yonsei Med. J. 2023;64:573–580. doi: 10.3349/ymj.2023.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shakeshaft A., Panjwani N., McDowall R., Crudgington H., Peña Ceballos J., Andrade D.M., Beier C.P., Fong C.Y., Gesche J., Greenberg D.A., et al. Trait impulsivity in juvenile myoclonic epilepsy. Ann. Clin. Transl. Neurol. 2021;8:138–152. doi: 10.1002/acn3.51255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z., Jiang D., Zhao X., Yang J., Liang D., Wang H., Zhao C., Liao J. Predicting drug treatment outcomes in childrens with tuberous sclerosis complex-related epilepsy: A clinical radiomics study. AJNR Am. J. Neuroradiol. 2023;44:853–860. doi: 10.3174/ajnr.A7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K.M., Hwang H., Sohn B., Park K., Han K., Ahn S.S., Lee W., Chu M.K., Heo K., Lee S.K. Development and validation of mri-based radiomics models for diagnosing juvenile myoclonic epilepsy. Korean J. Radiol. 2022;23:1281–1289. doi: 10.3348/kjr.2022.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang S., An H., Shin D.W., Lee H.W. Retention rates and successful treatment with antiseizure medications in newly-diagnosed epilepsy patients. Yonsei Med. J. 2024;65:89–97. doi: 10.3349/ymj.2022.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H.-J., Lee D.A., Park K.M. Altered cerebellar volumes and intrinsic cerebellar network in juvenile myoclonic epilepsy. Acta Neurol. Scand. 2023;2023:7907887. doi: 10.1155/2023/7907887. [DOI] [Google Scholar]
- 28.Kim J.H., Kim J.B., Suh S.-i. Alteration of cerebello-thalamocortical spontaneous low-frequency oscillations in juvenile myoclonic epilepsy. Acta Neurol. Scand. 2019;140:252–258. doi: 10.1111/ane.13138. [DOI] [PubMed] [Google Scholar]
- 29.Mory S.B., Betting L.E., Fernandes P.T., Lopes-Cendes I., Guerreiro M.M., Guerreiro C.A., Cendes F., Li L.M. Structural abnormalities of the thalamus in juvenile myoclonic epilepsy. Epilepsy Behav. 2011;21:407–411. doi: 10.1016/j.yebeh.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Betting L.E., Mory S.B., Lopes-Cendes I., Li L.M., Guerreiro M.M., Guerreiro C.A.M., Cendes F. Mri reveals structural abnormalities in patients with idiopathic generalized epilepsy. Neurology. 2006;67:848–852. doi: 10.1212/01.wnl.0000233886.55203.bd. [DOI] [PubMed] [Google Scholar]
- 31.O’Muircheartaigh J., Vollmar C., Barker G.J., Kumari V., Symms M.R., Thompson P., Duncan J.S., Koepp M.J., Richardson M.P. Abnormal thalamocortical structural and functional connectivity in juvenile myoclonic epilepsy. Brain. 2012;135:3635–3644. doi: 10.1093/brain/aws296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauffin H., Landtblom A.-M., Vigren P., Frick A., Engström M., McAllister A., Karlsson T. Similar profile and magnitude of cognitive impairments in focal and generalized epilepsy: A pilot study. Front. Neurol. 2022;12:746381. doi: 10.3389/fneur.2021.746381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data relevant to this study will be shared upon request with a qualified investigator pending appropriate Institutional Review Board approvals.