Abstract
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT), nucleocapsid protein (NC), genomic RNA, and the growing DNA strand all influence the copying of the HIV-1 RNA genome into DNA. A detailed understanding of these activities is required to understand the process of reverse transcription. HIV-1 viral DNA is initiated from a tRNA3Lys primer bound to the viral genome at the primer binding site. The U3 and R regions of the RNA genome are the first sequences to be copied. The TAR hairpin, a structure found within the R region of the viral genome, is the site of increased RT pausing, RNase H activity, and RT dissociation. Template RNA was digested approximately 17 bases behind the site where polymerase paused at the base of TAR. In most template RNAs, this was the only cleavage made by the RT responsible for initiating polymerization. If the RT that initiated DNA synthesis dissociated from the base of the TAR hairpin and an RT rebound at the end of the primer, there was competition between the polymerase and RNase H activities. After the complete heteroduplex was formed, there were additional RNase H cleavages that did not involve polymerization. Levels of NC that prevented TAR DNA self-priming did not protect genomic RNA from RNase H digestion. RNase H digestion of the 100-bp heteroduplex produced a 14-base RNA from the 5′ end of the RNA that remained annealed to the 3′ end of the minus-strand strong-stop DNA only if NC was present in the reaction.
The polymerase and RNase H domains of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) are components of a single protein which may help to link minus-strand DNA synthesis to the degradation of genomic RNA. The crystal structure of RT suggests that the polymerase and RNase H domains can simultaneously contact a nucleic acid substrate (8), which suggests that, with an RNA-DNA substrate, the polymerase and RNase H active sites can be simultaneously engaged. Moreover, the crystal structures suggest that the initial RNase H cleavage sites should be about 17 or 18 bases behind the polymerase binding site, which agrees with the biochemical analyses (reviewed in reference 3). However, DNA synthesis and RNA degradation are not rigorously coupled; there appears to be relatively little RNase H degradation during active polymerization in vitro (6). Furthermore, both murine leukemia virus and HIV-1 can replicate in vivo by complementation between RNase H-defective and polymerase-defective RTs (24; our unpublished observations). Pausing of the polymerase, at either secondary structures, homopolymeric G or C sequences, or sites where chain terminators are incorporated, leads to RNase H cleavage 17 to 20 bases downstream of the polymerase active site (13, 23, 25, 26). Experiments following the binding of a single RT show that the RT responsible for polymerization can also cleave the RNA template, but only when RT pauses (6, 10). Before the structure of RT was known, it was suggested that RNase H activation might involve a conformational switch that stops polymerization and promotes dissociation of RT (6, 10). The available structural information for HIV-1 RT suggests that this is unlikely. What, then, is the explanation for the inability of the RNase H of HIV-1 RT to cleave when there is active polymerization, and how does this inability affect the behavior of RT when it copies the HIV-1 genome?
We have begun an investigation of these questions by asking how HIV-1 RT behaves when it copies the 5′ end of the HIV-1 genome. The rate of RT polymerization can be affected by secondary structures present in the template. For example, the TAR hairpin, an imperfectly base-paired 57-base hairpin present within the R region of the HIV-1 genome that binds the Tat protein and activates transcription (3), can cause RT to pause during DNA synthesis (13). The minus-strand DNA that is created when RT copies the template RNA from the end of the primer binding site to the end of R is called minus-strand strong-stop DNA (−sssDNA). The TAR sequence allows −sssDNA to adopt a secondary structure similar to the TAR RNA hairpin, and this DNA hairpin promotes self-priming. There does not appear to be self-priming in vivo; in vitro self-priming can be prevented by the nucleocapsid protein (NC) (9, 11, 15, 16, 22). We used a model DNA substrate based on HIV-1 to show that a 17-base oligonucleotide complementary to the 3′ end of −sssDNA can prevent self-priming in the presence of NC and developed a model of how NC, RT, and the secondary structure of TAR collaborate to allow RT to copy the 5′ end of the genome and prepare the nascent DNA for strand transfer (9). Several structures with different thermodynamic stabilities arise during the copying of the 5′ end of the HIV-1 genome. Once −sssDNA synthesis is complete, RNase H degrades genomic RNA. The degradation of the RNA must be sufficient to allow strand transfer but not so extensive that −sssDNA can form a secondary structure that is the complement of the TAR hairpin. If the problem is described in energetic terms, the stability of the residual RNA-DNA duplex must be sufficient to prevent −sssDNA from forming a hairpin but not so stable that it interferes with strand transfer. In in vitro assays, NC is required to prevent hairpin formation, which suggests that NC affects the processing of the RNA by RNase H, the stability of the RNA-DNA hybrid, or both. The experiments presented here were designed to monitor the processing and fate of the RNA template during −sssDNA synthesis and to measure the effects of NC on the polymerase and RNase H activities of HIV-1 RT.
MATERIALS AND METHODS
Preparation of RT and nucleic acid substrates.
Wild-type HIV-1 RT (p66/p51) was expressed in Escherichia coli and purified as described previously (2). Wild-type HIV-1 NC (p7 Zn2+ holoenzyme) was generously provided by Robert Gorelick, Louis Henderson, and Larry Arthur. NC was reconstituted to a final concentration of 30 μM from lyophilized powder in 1× RT binding buffer (50 mM Tris Cl [pH 8.0], 80 mM KCl, 1 mM dithiothreitol, 1 mg of bovine serum albumin/ml, 20% glycerol) and stored in 4-μl aliquots within 150-μl tubes at −80°C. Fresh aliquots of NC were thawed immediately prior to use. Oligonucleotides were purchased from Life Technologies, Inc. (Rockville, Md.). HIV-1 genomic sequences were subcloned from the pNL4-3 clone (GenBank GI322415) as described previously (9). Unlabeled and internally labeled RNAs were synthesized with the Megashortscript T7 kit (Ambion, Austin, Tex.) according to the manufacturer's instructions, using a PCR-generated DNA fragment as the template for RNA synthesis. RNA was purified by electrophoresis on a 5% denaturing polyacrylamide gel, excised, eluted by soaking overnight in 10 mM Tris Cl (pH 8.0) and 15 μg of proteinase K/ml, precipitated, and resuspended in 10 mM Tris Cl (pH 8.0). RNA was quantitated using a UV spectrophotometer. Quantitation of the relative intensities of bands on specified gels was accomplished using a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and ImageQuant software (Molecular Dynamics). DNA oligomers were 32P labeled using [γ-32P]ATP (Amersham Pharmacia, Piscataway, N.J.) and T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.).
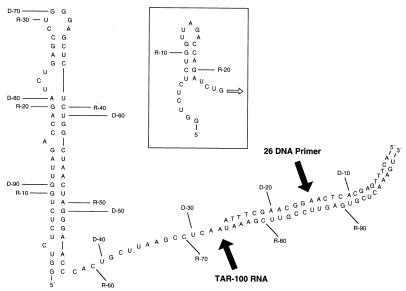
TAR-100 RNA.
The 100-base sequence at the 5′ end of the pNL4-3 RNA genome (GenBank accession no. GI322415; positions 455 to 554) was designated TAR-100 (see Fig. 1). This RNA sequence encompassed the TAR hairpin and most of the poly(A) hairpin (5′-GGUCUCUCUGGUUAGACCAGAUCUG AGCCUGGGAGCUCUCUGGCUAACUAGGGAACCCACUGCUUAAG CCUCAAUAAAGCUUGCCUUGAGUGCUCAAAGU-3′). A 26-base DNA primer complementary to the 3′ end of TAR-100 RNA was designated 26 DNA primer (5′-ACTTTGAGCACTCAAGGCAAGCTTTA-3′).
FIG. 1.
Sequence and structure of the 100-base RNA template TAR-100. An RNA template consisting of 100 bases at the 5′ end of the HIV-1 RNA pNL4-3 genome annealed at its 3′ end to a 26-mer DNA primer was predicted to form the TAR hairpin (57-base structure at left) and a secondary hairpin (inset) if base-pairing of the TAR hairpin were disrupted. The DNA primer (26 DNA Primer) is shown annealed to the RNA. The RNA is numbered according to whether the label was at the 5′ end of the DNA primer (prefix D-) or at the 5′ end of the RNA (prefix R-).
5′ 32P-end-labeled RNA.
Unlabeled RNA was 5′ dephosphorylated using shrimp alkaline phosphatase (Roche Biochemicals, Indianapolis, Ind.) and labeled in 1× RT binding buffer using T4 polynucleotide kinase (New England Biolabs) in the presence of RNasin (1 U/μl; Promega, Madison, Wis.). Labeled RNA was fractionated by electrophoresis on a 5% denaturing polyacrylamide gel, and the full-length 5′-labeled product was eluted and purified as described above.
32P-RNA RT-NC assays using internally labeled or 5′-end-labeled RNA.
32P-labeled RNA was incubated with a fivefold excess of unlabeled primer in 1× RT binding buffer supplemented with RNasin (1 U/μl) at 37°C for 30 min. RT start solution was added to give a final concentration of 80 μM for each deoxynucleoside triphosphate and 6 mM MgCl2, and the mixture was divided in half. One half received NC protein sufficient to coat the primer and template strands at the indicated levels (assuming seven bases/NC), and the second half received an equivalent volume of 1× RT binding buffer. After incubation with NC for 4 min at 37°C, the indicated amount of RT was added. For assays performed in the presence of heparin, polymerization was allowed to proceed for 5 s, after which heparin (heparin sodium; Amersham Pharmacia) was added to a final concentration of 0.8 mg/ml. Aliquots (4 μl) were removed at the appropriate times into an equal volume of formamide gel loading buffer (9) and heated to 75°C for 3 min. Samples were loaded on a 12% denaturing polyacrylamide gel containing 0.05% sodium dodecyl sulfate, fractionated by electrophoresis, dried under vacuum, and exposed to a PhosphorImager screen (Molecular Dynamics).
In some experiments, the 4-μl aliquot removed at each time point was stopped by addition of heparin (1 μl of a 4-mg/ml solution) and 1 μl of E. coli RNase H (2 U/μl; Promega). The reactions were incubated with RNase H for 15 min at 37°C in order to digest the RNA contained in an RNA-DNA heteroduplex. After the incubation was complete, formamide gel loading buffer was added and the samples were electrophoresed as described above.
32P-DNA RT-NC assays.
32P-DNA primer was incubated with a fivefold excess of 32P-labeled template RNA in 1× RT binding buffer supplemented with RNasin (1 U/μl) at 37°C for 30 min. RT start solution was added, and then a sufficient amount of unlabeled primer was added to bring the total primer amount to a fivefold excess over the RNA, and the mixture was divided in half as described above. After incubation with NC for 4 min at 37°C, the indicated amount of RT was added. For assays performed in the presence of heparin, polymerization was allowed to proceed for 5 s, and then heparin was added to a final concentration of 0.8 mg/ml, as described above. Aliquots were transferred at the appropriate times into DNA stop solution (400 mM EDTA [pH 8.0], 100 μg of RNase A/ml). Reactions were incubated for 30 min at 65°C to digest the 32P-labeled template RNA, followed by the addition of an equal volume of formamide gel loading buffer. The reactions were heated to 75°C for 3 min. Samples were fractionated on a 12% denaturing polyacrylamide gel containing 0.05% sodium dodecyl sulfate, electrophoresed at 70 W, dried under vacuum, and exposed to a PhosphorImager screen (Molecular Dynamics).
Digestion of TAR-100 RNA in the intact 100-base heteroduplex by RT.
Internally labeled TAR-100 RNA was mixed with a 1.5-fold excess of unlabeled complementary 100-base DNA (called −TAR DNA) in 1× RT binding buffer. The mixture was heated to 65°C for 3 min and cooled to 37°C. RT start solution (10×) was then added, and the reaction was divided in half. NC was added to half the reactions in an amount sufficient for twofold coating of the heteroduplex and allowed to bind for 3 min at 37°C. The second half received an equivalent volume of 1× RT binding buffer. A 0.12 molar ratio of RT was added to the annealed template-primer mixtures, aliquots were removed at the indicated times and mixed with formamide loading buffer, and the samples were electrophoresed as described above.
Self-priming of −TAR DNA after RT-dependent RNase H digestion of TAR-100 RNA in the 100-base heteroduplex.
A threefold excess of internally labeled TAR-100 RNA was mixed with 32P-labeled complementary −TAR DNA in 1× RT binding buffer. The mixture was heated to 65°C for 3 min and cooled to 37°C. RT start solution (10×) was then added, and the reaction was divided in half as described above. A 0.25 molar ratio of RT was added to the annealed template-primer mixtures, and aliquots were removed at the indicated times into DNA stop solution. Reactions were incubated for 30 min at 65°C to digest the 32P-labeled template RNA. After digestion of the RNA, an equal volume of formamide gel loading buffer was added, and the samples were electrophoresed as described above.
RNA secondary-structure prediction.
RNA structures were predicted using RNA Structure 3.0 (17).
RESULTS
Predicted hairpins in TAR-100 RNA.
The computer program RNA Structure 3.0 (17) was used to predict secondary structures formed by TAR-100 RNA (Fig. 1). The bases were numbered to correspond to either the size of the DNA copied from the RNA template (D) or the size of the 5′-end-labeled RNA (R) (Fig. 1). As expected, the 5′ 57 bases of the RNA formed the TAR hairpin (1, 18). Further analysis demonstrated that when base pairing within the TAR hairpin was disrupted, a small secondary hairpin could be formed near the 5′ end of the RNA (Fig. 1, inset).
HIV-1 RT pauses at the base of hairpins in the TAR region.
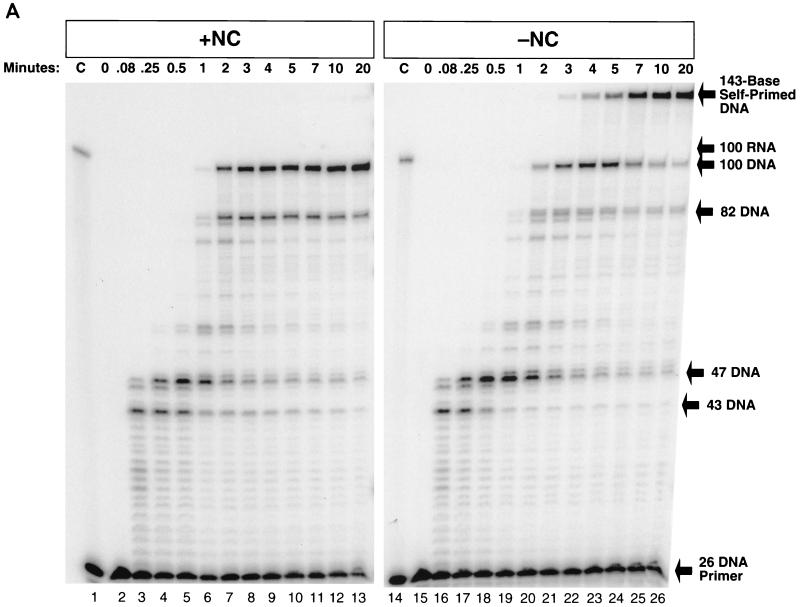
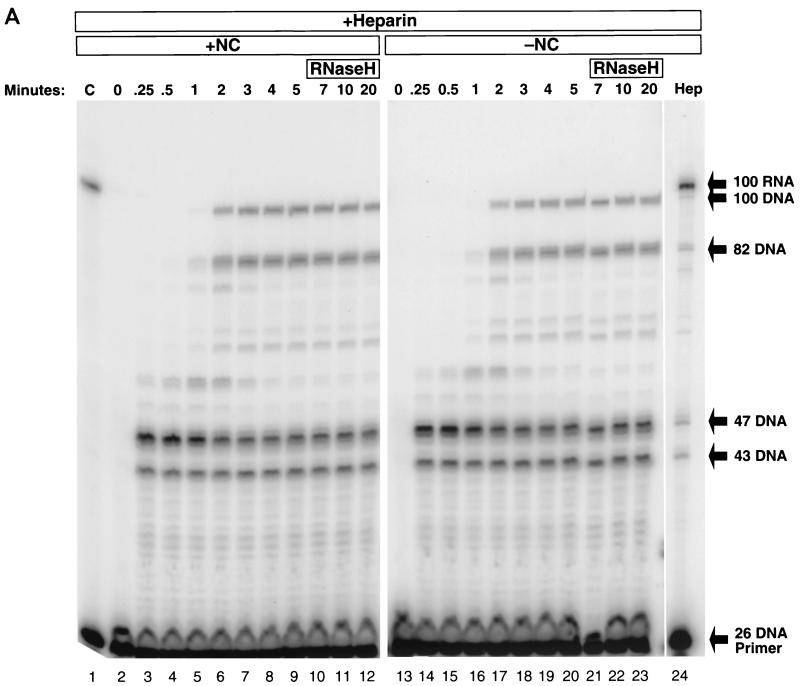
Secondary structures within the HIV-1 R region impede the progress of RT. In Fig. 2A, a 5′-end-labeled 26-base DNA primer (26 DNA primer [Fig. 1]) was annealed to TAR-100 RNA. The annealed mixture was divided in half and mixed either with a fourfold coating level of NC (Fig. 2A, lanes 2 to 13) or with buffer alone (Fig. 2A, lanes 15 to 26). The RNA, which was labeled, was degraded at the end of the experiment by addition of RNase A to the reactions. Previous experiments indicated that two- to fourfold coating levels of NC were optimal for inhibiting self-priming synthesis of the complementary DNA strand, and allowing high levels of polymerization (9). In lanes 2 to 13, in which the primer was extended by RT in the presence of NC, there were strong pauses in the first 30 s (Fig. 2A, lanes 2 to 5). After 5 s (0.08 min), RT paused at the base of the TAR hairpin, when the DNA product was 43 bases long (D-43). The melting of the first three bases of the TAR hairpin, promoted by the mismatch near the base of the RNA hairpin (Fig. 1), allowed the addition of the next four bases, followed by a second strong pause when the DNA was 47 bases long (D-47). This second pause was caused by a longer annealed region that was less easily traversed. The pause at D-47 was more pronounced, and persisted longer, than the pause at D-43. DNA synthesis continued into the TAR hairpin, with low levels of pausing after passing the mismatch in the RNA hairpin when the DNA was 57 bases long. Once the base pairing in the TAR RNA hairpin was disrupted by synthesis of the complementary DNA strand, the remaining single-stranded RNA at the 5′ end of the template could refold, forming a second, smaller hairpin that briefly blocked polymerization at D-82 (Fig. 2A, lanes 6 to 13; Fig. 1 inset). After the pause at D-82, the full-length 100-base product was formed, appearing in significant amounts by 2 min (lane 7). After 20 min, the 100-base DNA (which we call −TAR DNA) was the major product, constituting 68% of the total extended products.
FIG. 2.
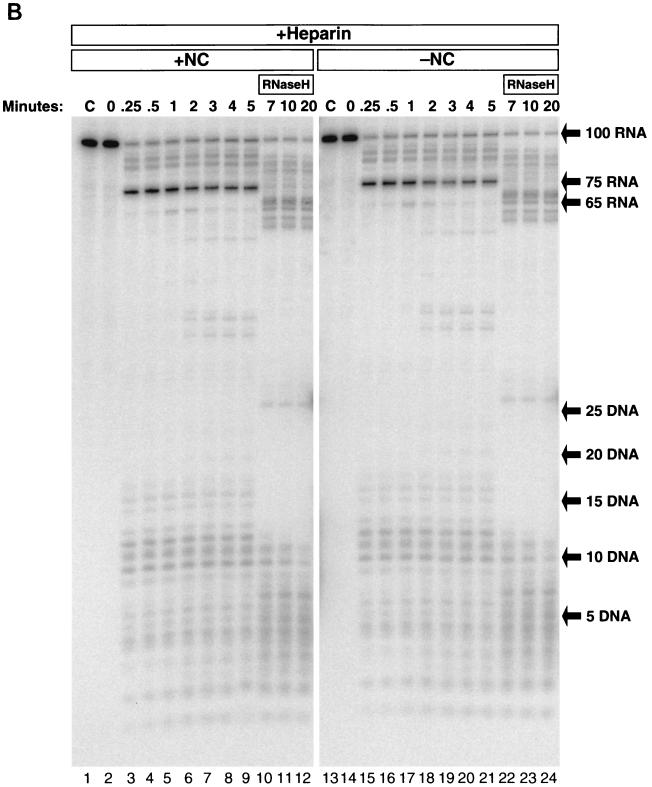
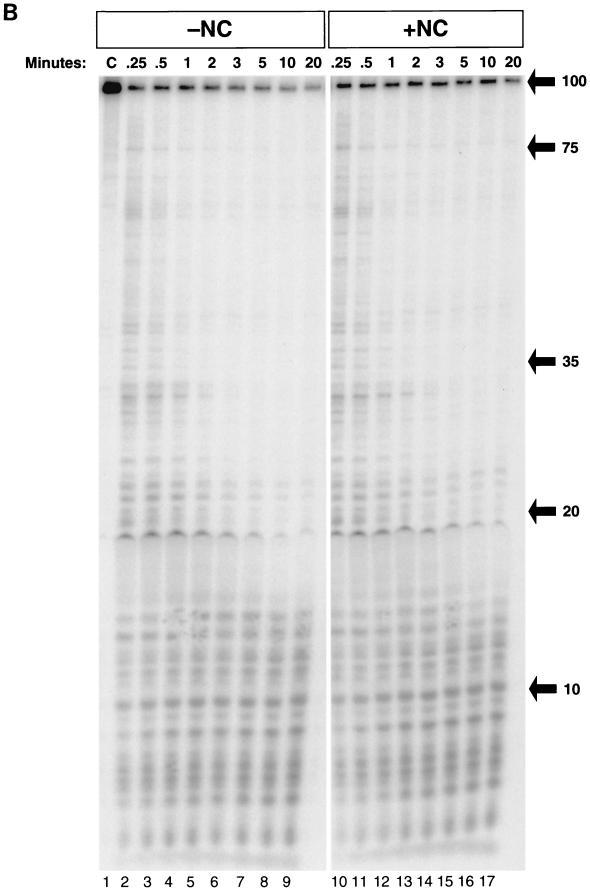
Self-priming of −TAR DNA and the fate of the template RNA in the presence or absence of NC. (A) 32P-labeled 26 DNA primer was annealed to TAR-100 RNA and extended by RT in the presence (lanes 2 to 13) or absence (lanes 15 to 26) of NC. Reactions were initiated by the addition of RT (see Materials and Methods). Aliquots of the reactions were added to DNA stop solution at the time points indicated above the gel. The internally labeled RNA was digested by adding RNase A to DNA stop solution, except in lanes 1 and 14 (C, control). In lanes 1 and 14, an aliquot of the reaction was incubated in the absence of RT for the 20-min reaction time course and loaded onto a 12% denaturing polyacrylamide gel to demonstrate that the RNA remained intact and the primer was not extended in the absence of RT. The lengths (in bases) of the prominent pause products, as well as the full-length DNA and RNA and the self-primed DNA, are indicated on the right. (B) The fate of the internally labeled TAR-100 RNA was monitored as DNA synthesis proceeded. Unlabeled 26 DNA primer was annealed to TAR-100 RNA and extended by RT in the presence (lanes 2 to 13) or absence (lanes 15 to 26) of NC. Aliquots of the reactions were added to DNA stop solution at the time points indicated above the gel. In lanes 1 and 14, an aliquot of the reaction was incubated in the absence of RT for the 20-min reaction time course and loaded onto a 12% denaturing polyacrylamide gel to demonstrate that the RNA remained intact in the absence of RT. Lane 27 contained the 5′-end-labeled 26-base DNA oligomer digested with DNase I as a marker. The migration of DNA markers and RNA fragments (in bases) is shown on the right. (C) The 5′-end-labeled RNA was visualized as DNA synthesis proceeded, in a reaction similar to that shown in panel B. The migration of DNA markers and RNA fragments (in bases) is shown on the right. The sizes of the RNAs were determined by overexposing the autoradiographs and counting down from the full-length (100-mer) RNA.
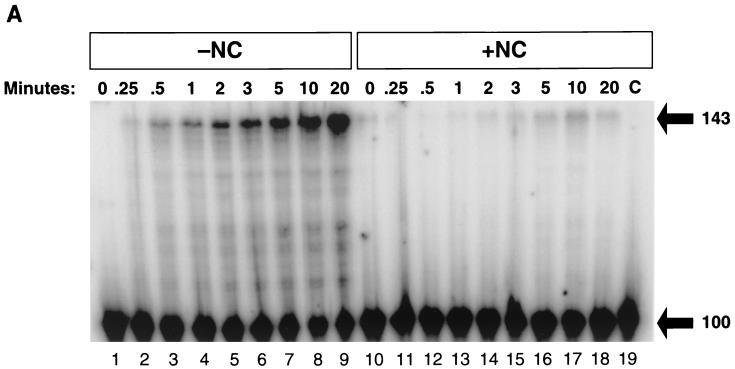
The pause sites seen in the absence of NC (Fig. 2A, lanes 15 to 26) were similar to those seen in the presence of NC (lanes 2 to 13), with pronounced pausing at D-43, D-47, and D-82. There was a clear and reproducible difference in the amount of pausing at D-82: almost threefold more product remained after 20 min in the presence of NC. The increase in pausing at D-82 was likely caused by an NC-dependent increase in the formation of the secondary hairpin (Fig. 1 inset). In the absence of NC, the full-length 100-base −TAR DNA could be detected after 1 min, and the level was maximal after 4 to 5 min and subsequently declined. In the absence of NC, there was self-priming of the 100-base DNA at the TAR hairpin which resulted in the disappearance of the 100-mer and the appearance of a 143-base-long product. By 3 min, the 143-base self-primed DNA product began to appear; at 20 min, this DNA comprised 62% of the total product extended by RT. A comparison of the left and right panels of Fig. 2A shows that RT can complete the synthesis of the 100-mer DNA more quickly in the presence of NC.
RT pausing at secondary structures correlates with RNase H activity.
Digestion of internally labeled TAR-100 RNA during DNA synthesis is shown in Fig. 2B in the presence (lanes 2 to 13) and absence (lanes 15 to 26) of NC. The reactions shown in Fig. 2B were similar to those shown in Fig. 2A except that the DNA was not labeled and the labeled RNA was not degraded by RNase A. The gels were run with DNA size markers. The sizes of the RNA fragments were determined by overexposing the gels and counting down from the full-length RNA (100 bases). In reactions performed in the absence of NC, two cleavages occurred in the first 5 s in the region of the RNA annealed to the 26-base DNA primer, approximately 17 and 8 bases from the 3′ end of the DNA primer (Fig. 2B, lanes 3 and 16). These cleavages were made by RT before the primer was extended. Other cleavages were dependent on DNA synthesis. Quantitation of the bands in lanes 3 and 16 revealed that after 5 s of incubation with RT, at least 75% of the template RNA was digested in the region where the DNA primer was annealed, before polymerization was initiated. Because 15% of the template remained uncut at 5 s but was digested at later times, it appears that 90% of the total template was digested before polymerization began. The first polymerization-dependent RNase H cleavage generated a 75-base RNA. This RNA appeared when the polymerase domain of RT was paused at the base of TAR (D-43) (Fig. 2A) and corresponds to an RNase H cleavage approximately 17 bases behind the stalled polymerase active site. If no additional cleavages were made before RT reached the base of the TAR hairpin, a 25-base-long RNA fragment corresponding to the 3′ end of the RNA would be produced. No band corresponding to this RNA can be seen on the gel. This suggests that all the template RNAs that were copied by RT were digested opposite the DNA primer before polymerization began. Thus, RNase H digestion appears to be much faster than polymerization when RT first binds to this template-primer. The 75-base fragment disappeared after 5 min, and a shorter fragment of approximately 65 bases was formed which persisted over the entire 20-min time course. The 65-base RNA was neither used as a primer and extended by RT nor digested by RNase H, suggesting that it was single stranded. How was this single-stranded RNA generated? A simple interpretation is that in some cases, RNase H digestion occurred sufficiently close to the end of the DNA to permit the dissociation of the uncopied 65-base RNA template from the DNA primer. Smaller amounts of single-stranded template RNA were released from the duplex at the secondary RNA hairpin, where pausing at the hairpin (producing the 82-base-long DNA band shown in Fig. 2A, lanes 7 to 13) resulted in low levels of RNase H cleavage 37 bases from the 5′ end of the RNA (Fig. 2B, lanes 7 to 9). RNase H digestion of the RNA template generated a 32- or 33-base single-stranded RNA product (Fig. 2B, lanes 8 to 13). Over the course of the reaction, short RNA fragments (less than 20 bases long) were generated by the various cleavage reactions; these can be seen near the bottom of the gel. The data in Fig. 2B show that NC has no measurable effect on the pattern of RNase H digestion.
A 14-base RNA is the shortest 5′ end product.
Comparison of the digestion patterns of the internally labeled and 5′-end-labeled TAR-100 RNA allows unambiguous interpretation of how the 5′ end of the RNA is processed by RNase H. As expected, copying the 5′-end-labeled TAR-100 RNA in either the presence or the absence of NC was similar to what was observed with internally labeled RNA (Fig. 2B; data not shown). Figure 2C shows the digestion of the 5′-end-labeled RNA. The appearance of the cleavage at R-75 (while RT was paused at the base of the TAR hairpin) and the cleavage at R-37, as well as the inability of RT to digest the 65- and 32-base templates, was similar to what was seen with the internally labeled RNA (Fig. 2B and C). However, the experiment with 5′-end-labeled RNA showed that the shortest RNA fragment generated by RT at the 5′ end of the genome migrated on a sequencing gel at a position suggesting it was 14 bases long.
Secondary structures promote both RT pausing and RT dissociation.
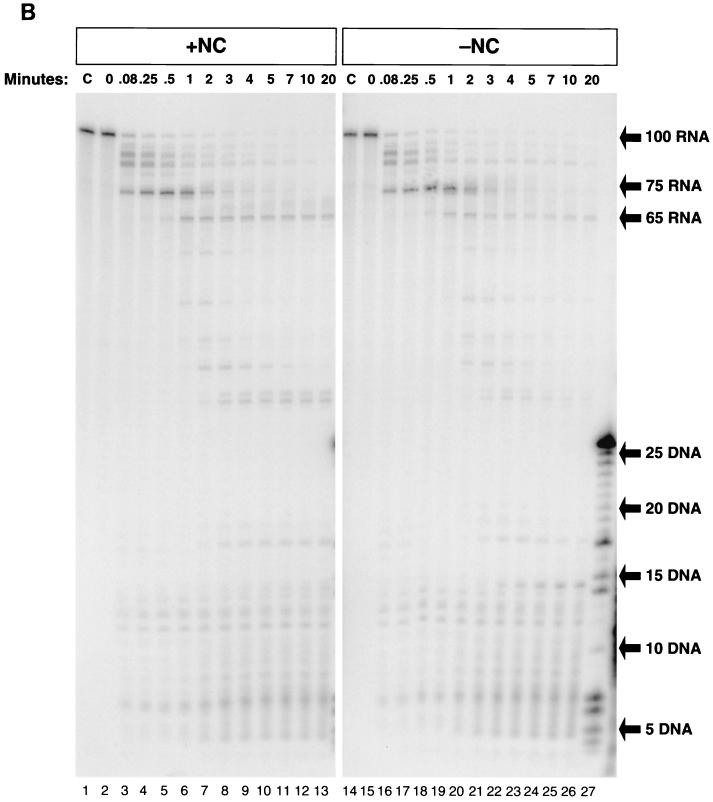
The experiments shown in Fig. 2 were performed using conditions that allowed RT to dissociate from the RNA-DNA heteroduplex and then reassociate, potentially undergoing multiple rounds of DNA synthesis and/or RNA cleavage on either the same or different template-primers. Heparin, which binds to and sequesters RT, was included in the experiments shown in Fig. 3 to limit the reaction to a single RT binding event. Heparin was added 5 s after the addition of RT, before significant polymerization occurred (Fig. 2A and B, lanes 3). Figure 3A shows the results of DNA synthesis in the presence of heparin. It is clear that the presence or absence of NC had little effect on the ability of RT to polymerize in the presence of heparin. The pattern of the bands in lanes 2 to 12 was identical to that of those in lanes 13 to 23; however, as seen before, the 100-mer DNA is synthesized more rapidly in the presence of NC. Without heparin, in the absence of NC, the maximal amount of 100-base full-length product was formed after 3 min. However, in the presence of heparin, the full-length product (100-mer or longer) is only 9% of the extended products, compared with the more than 60% full-length product formed in the absence of heparin after 20 min (Fig. 2A). Although the pattern of pause sites remained the same as in the absence of heparin (Fig. 2A), the relative intensities of the bands at D-43, D-47, and D-82 were increased substantially at the 20-min time point in the presence of heparin (Fig. 3A). The percentage of the 43-base-long DNA product that remained unextended at the 20-min time point increased from less than 1 without heparin (Fig. 2A) to 18 in the presence of heparin (Fig. 3A). The amount of the 47-mer product was 4% of the total extended DNA without heparin (Fig. 2A) and 30% with heparin (Fig. 3A). When corrected for the loss of 18% of the extendable template at the first pause site, the percentage of templates that failed to be extended past 47 bases was actually 37 in the presence of heparin. In the absence of heparin, no correction was required, because over the course of the incubation almost all of the product was extended beyond 47 bases. In the presence of heparin, 14% of the total extended product was halted at the pause at 82, and 9% was extended to 100 bases, indicating that approximately 60% of the template that was extended to 82 bases could not be extended further. In the presence of heparin, only 9% of the extended template was copied to 100 bases in a single RT binding event, compared with the 62 to 68% completion rate when RT was allowed to rebind. Comparing the products formed during a single round of RT synthesis with the products formed when multiple rounds of DNA synthesis were permitted shows that RT pausing at secondary structures in the RNA genome can lead to a substantial dissociation of RT. This effect is illustrated more clearly by the time course of the pause at D-47 shown in Fig. 2A and 3A. In the first 30 s, the 47-base DNA accumulated as the RT paused at the base of the TAR hairpin, both in the presence (Fig. 3A) and absence (Fig. 2A) of heparin. In the presence of heparin, over the next 2 min, the RT that remained bound resumed polymerization and the amount of 47-base DNA decreased. However, the RT that dissociated during the pause and was sequestered by heparin was unable to rebind, and these templates were not extended further, leaving significant amounts of the 47-base DNA at the 4-min time point and beyond. In the absence of heparin (Fig. 2A), most of the 47-base DNA was extended by RT, although a small portion of the RNA template was digested by RNase H to such an extent that the RNA dissociated from the nascent DNA, preventing further DNA synthesis.
FIG. 3.
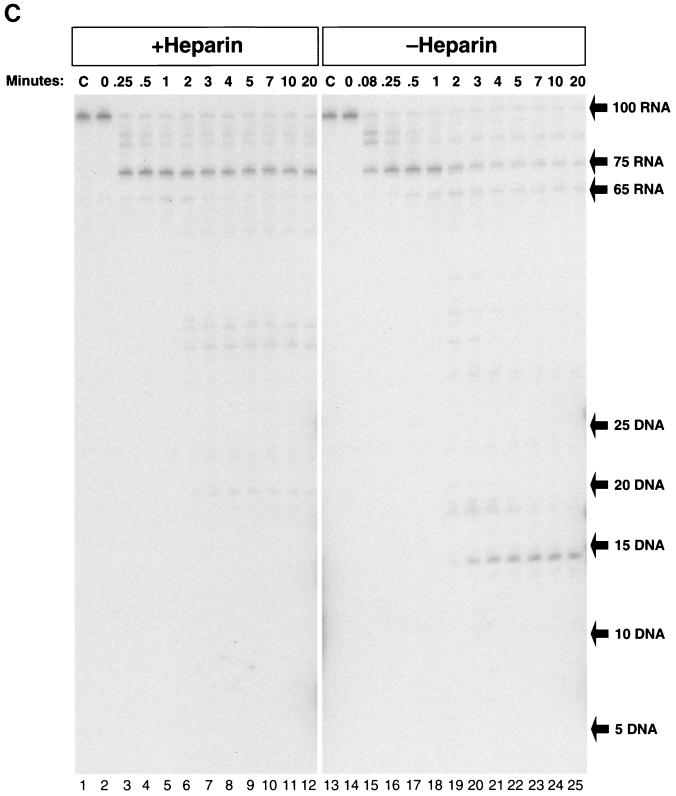
A single round of RT synthesis does not allow self-priming or release of single-stranded template RNA and reveals increased RT dissociation at pause sites. (A) 32P-labeled 26 DNA primer was annealed to internally labeled TAR-100 RNA and extended by RT in the presence (lanes 2 to 12) or absence (lanes 13 to 23) of NC. In lane 1, an aliquot of the reaction was incubated in the absence of RT as described in the legend to Fig. 2B to show that the RNA remained intact. Reactions were initiated by the addition of RT, and after 5 s, heparin was added, as described in Materials and Methods. Aliquots were removed at each time point as for Fig. 2A, except that E. coli RNase H was added immediately after the 5-min aliquot was removed. In lane 24, heparin was added before HIV-1 RT, which blocked almost all the polymerase activity. (B) The fate of the internally labeled TAR-100 RNA was monitored as DNA synthesis proceeded. Unlabeled 26 DNA primer was annealed to TAR-100 RNA and extended by RT in the presence (lanes 2 to 12) or absence (lanes 14 to 24) of NC. Aliquots of the reactions were added to DNA stop solution at the time points indicated at the top of the figure and loaded onto a 12% denaturing polyacrylamide gel. As for Fig. 4A, E. coli RNase H was added to the reaction after the 5-min time point. The migration of DNA markers (in bases) and the RNA fragments are shown on the right. Lanes 1 and 13 (marked C) are control lanes showing untreated TAR-100 RNA. (C) 5′-end-labeled TAR-100 RNA was annealed to unlabeled 26 DNA primer in the presence (lanes 2 to 12) or the absence (lanes 14 to 25) of heparin. Reactions were initiated by the addition of RT as described in the legend to Fig. 2A, and heparin was added after 5 s of incubation. Aliquots were removed at the indicated times. The migration of DNA markers and RNA fragments is indicated to the right of the figure. Lanes 1 and 13 are control lanes (marked C) showing untreated TAR-100 RNA.
It is important to determine the fate of RT after it reaches the end of the template, because both digestion of the template RNA and subsequent strand transfer events are affected by the ability of RT to recognize the end of the genome and the growing DNA strand. Does RT remain bound after completing synthesis and continue to digest the RNA template, or does it dissociate? This question can be answered based on the results shown in Fig. 3A and B. Because heparin sequestered any RT that dissociated in the experiments shown in Fig. 3A, only the RT that remained bound to the completed heteroduplex could digest the RNA template. Self-priming of the completed DNA requires RNase H activity. If RT remained bound to the end of the completed substrate and continued digesting the template RNA, it could do so with a bind-and-slide mechanism. It appears that this does not occur, however, because there was no 143-base self-primed product formed in the presence of heparin in Fig. 3A. Alternatively, RT could remain bound to the end of the completed TAR-100 RNA/−TAR DNA heteroduplex but lack the ability to slide to make additional RNase H cleavages and therefore be unable to self-prime because of insufficient digestion of the template RNA. To test whether insufficient RNA digestion was responsible for the lack of self-priming, we added E. coli RNase H to the experiments shown in Fig. 3A after 5 min of polymerization. Although the duplex RNA was digested (Fig. 3B), no self-priming occurred, indicating that RT did not remain bound to the end of the template in a state that would allow additional DNA synthesis after the −TAR DNA was completed.
E. coli RNase H activity was not blocked by heparin.
The specificity of heparin for RT was demonstrated by the fact that although heparin efficiently sequestered RT, it did not block E. coli RNase H activity. The levels of heparin that were added to the experiments were sufficient to block most of the RT polymerase activity (Fig. 3A, lane 24), and as expected, heparin also eliminated the RNase H activity of RT, as discussed above. However, E. coli RNase H activity was not blocked by heparin (Fig. 3B, lanes 10 to 12 and 22 to 24). The protein concentration of the E. coli RNase H was too low to be measured by standard methodologies, suggesting that the binding capacity of the heparin was not overcome by an excess of E. coli RNase H. Instead, it appears that much less heparin is required to sequester RT activity than is required to block E. coli RNase H.
Rebinding of RT is required for RNase H to cleave the RNA close enough to the end of the DNA to release RNA fragments.
In the experiments shown in Fig. 3B, RT-dependent RNase H digestion of the internally labeled RNA that was annealed to the 26-base primer occurred near positions R-84 and R-91, identical to what was described above. A large amount of R-75 accumulated by 15 s, as was described above, but in the presence of heparin, most of the R-75 did not undergo further processing to yield the 65-base single-stranded RNA unless E. coli RNase H was introduced into the reaction (after 5 min). A small amount of the 65-base RNA was produced at between 1 and 2 min but disappeared by 3 min, showing that this RNA remained annealed to DNA and could be used as a template. This finding clearly indicates that the RNase H cleavages that lead to the dissociation of the uncopied portion of the RNA template require rebinding of RT and that the polymerizing RT does not extensively digest RNA at this site. A second example of this type of pausing during polymerization and cleavage is seen, albeit less distinctly, at the site of secondary hairpin cleavage. The initial RNase H cleavages of the RNA resulted in a 37- to 39-base RNA fragment, which was digested to a 26-mer only after E. coli RNase H was added. Thus, in both cases, processing of the RNA that resulted in the release of uncopied, single-stranded template RNA involved exogenous RNase H activity.
Degradation of the RNA template requires the rebinding of RT.
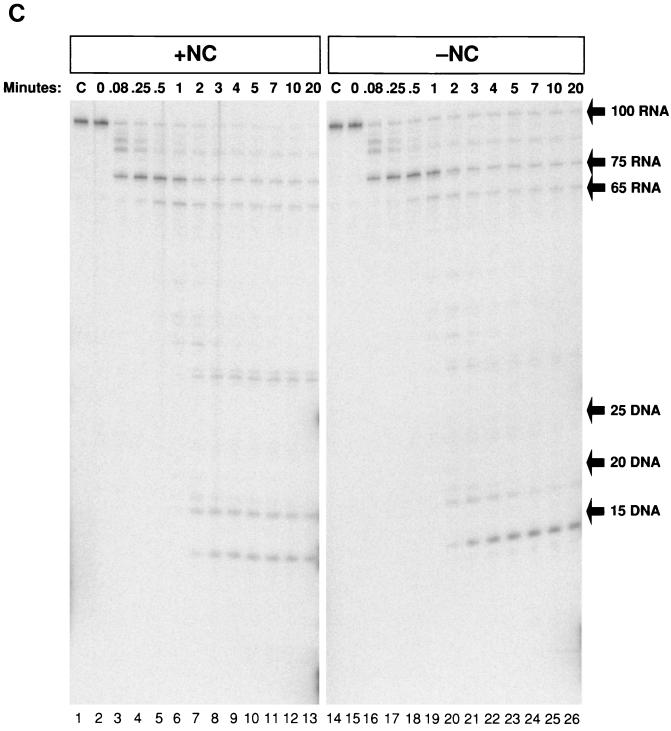
The inclusion of heparin in the RT assays allowed us to determine which RNase H cleavages were made by the RT involved in the first round of polymerization and which were made by RT molecules that were rebound. In Fig. 3C, 5′-end-labeled TAR-100 RNA was used as a template for DNA synthesis in the presence or absence of heparin. Figure 3C (lanes 2 to 12) shows the four cleavages that were made in the RNA, as discussed above. The first two cleavages were made in the region bound to the primer, before polymerization began, approximately 17 and 8 bases behind the 3′ end of the DNA primer. After initiation of polymerization, there was only a single major cleavage made in the RNA, which generated a 75-base RNA fragment, most of which (70%) was still present after 20 min. In the absence of heparin (lanes 14 to 25), additional cleavages were made in the 75-base RNA, resulting in a final product that was 14 bases long, similar to what was seen in the experiments shown in Fig. 2C. Therefore, although the cleavage in the template RNA at R-75 was made by the polymerizing RT, the cleavage at the end of the genome that generated the 14-base fragment at the 5′ end was dependent on the ability of RT to rebind. This finding is also consistent with the idea, suggested by the experiments in which E. coli RNase H was added to the reactions (Fig. 3A and B), that RT dissociates from the completed heteroduplex upon reaching the end of the RNA template.
FIG. 5.
The 5′ 14-base RNA remains annealed to −sssDNA only in the presence of NC. 26 DNA primer was annealed to 5′-end-labeled TAR-100 RNA and extended by RT in the presence (lanes 2 to 13) or absence (lanes 15 to 26) of NC. At the indicated times, aliquots of the reactions were added to a mixture of heparin (to sequester RT) and E. coli RNase H (to digest RNA annealed to DNA). After incubation with E. coli RNase H, the reactions were stopped by the addition of formamide gel loading buffer, and the samples were loaded onto a 12% denaturing polyacrylamide gel and fractionated by electrophoresis. The sizes of DNA and RNA are shown on the right of the figure. Lanes 1 and 14 are control lanes (marked C) showing untreated TAR-100 RNA.
The RNase H activity of RT cleaves almost every template RNA at the base of the TAR hairpin.
The total amount of RNA in the 100-mer band shown in Fig. 3C, lane 2, was quantitated and compared with that in the bands shown in lane 4. At 0.5 min, the RNA digestion pattern shown in Fig. 3C, lane 4, shows that most of the RNA (75%) was cleaved at R-75 after RT paused at the base of the TAR hairpin. The remaining 25% was either uncut or cleaved at R-84 and R-91 before polymerization began. Therefore, at 0.5 min, essentially all of the RNA that was copied to the base of the hairpin underwent digestion at R-75, approximately 17 bases behind the hairpin.
The RNase H activity of RT extensively degrades the RNA in the 100-bp RNA-DNA heteroduplex.
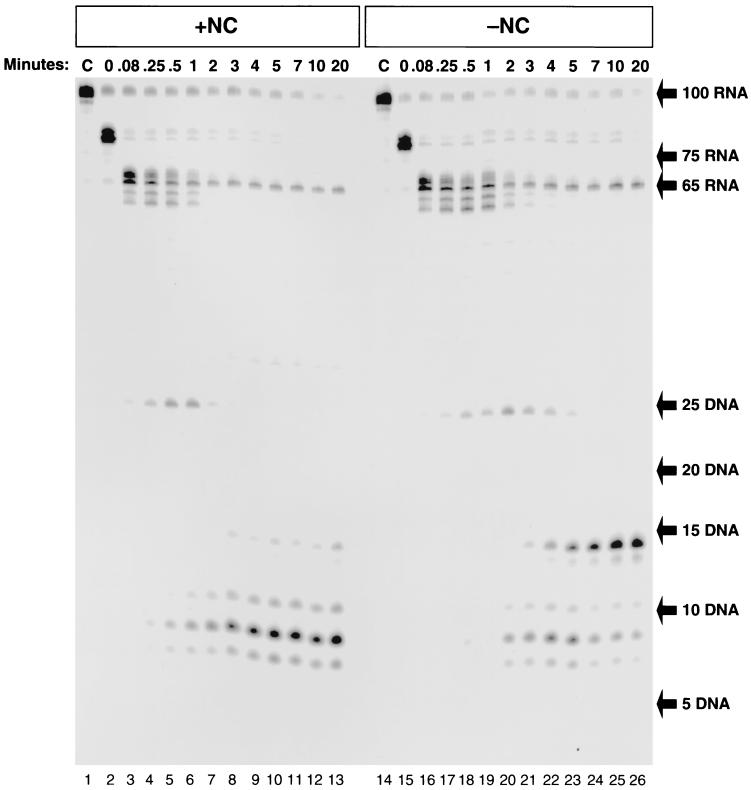
Figure 4 shows the results of an experiment in which RT was added to the 100-bp TAR-100 RNA/−TAR DNA heteroduplex in either the presence or absence of NC. In Fig. 4A, the DNA was 32P labeled. In the absence of NC (lanes 1 to 9), the DNA was able to self-prime, yielding the expected 143-base product. Self-priming was greatly reduced by the addition of NC (lanes 10 to 18). The experiment shown in Fig. 4B was performed under the same conditions as that shown in Fig. 4A, except that the RNA instead of the DNA was labeled. After only 0.25 min of digestion with RT, there was a ladder of RNA products ranging from 1 to 99 bases. By 1 or 2 min, the majority of the digested RNA was 35 bases or smaller. After 3 min, the RNA was almost completely digested into fragments of 20 bases or smaller. Using a fully double-stranded substrate containing 5′-end-labeled RNA, we found that the shortest product produced from the 5′ end of the RNA migrates at the position expected for a 14-mer (data not shown). The rapid appearance of a ladder of RNA fragments indicates that the RNase H activity of RT was able to bind and cleave apparently at random rather than generating a series of fragments by stepwise degradation from one end of the substrate. However, the RNase cleavage can produce specific products, for example, the 14-mer from the 5′ end of the RNA (Fig. 3C).
FIG. 4.
RNase H digestion of the 100-base RNA-DNA heteroduplex is rapid and nonspecific. (A) Internally labeled TAR-100 RNA was annealed to a complementary 5′-end-labeled 100-base DNA in the absence (lanes 1 to 9) or presence (lanes 10 to 18) of NC. RT was added, and aliquots of the reactions were added to DNA stop solution at the indicated times and visualized on a 12% denaturing polyacrylamide gel. The migration of the 100-base starting material and the 143-base self-primed DNA product is indicated on the right. (B) Reactions were identical to those described for Fig. 4A except that the 100-base DNA was unlabeled. The internally labeled RNA digestion products were fractionated on a 12% denaturing polyacrylamide gel. The sizes of DNA markers are shown on the right of the figure.
The 5′ 14-base RNA remains annealed to the 3′ end of −TAR DNA only in the presence of NC.
Digestion of the RNA after the synthesis of the −TAR DNA was complete resulted in a 14-base RNA product at the 5′ end of the RNA, which was generated in similar amounts in the presence and absence of NC. In the experiment shown in Fig. 5, we used E. coli RNase H to determine whether the 14-base RNA remained annealed to −TAR DNA in the presence or absence of NC. In an experiment similar to that shown in Fig. 2C, 5′-end-labeled TAR-100 RNA was copied from 26 DNA primer. Heparin was added to aliquots of the reactions at the indicated times. Heparin bound to RT and sequestered both the polymerase and RNase H activities of the enzyme. E. coli RNase H was included in the heparin stop mixture, which resulted in the digestion of any RNA fragments that remained annealed to the −TAR DNA strand at that time point. The digestion pattern in the presence of NC (Fig. 5, lanes 2 to 13) was similar to that in the absence of NC (lanes 15 to 26), except in the case of the 14-base RNA. In the presence of NC, E. coli RNase H digested almost all of the 14-base RNA to 8 bases after 20 min of polymerization, showing that the RNA was annealed to the −TAR DNA. In contrast, after 20 min without NC, almost all of the 14-base RNA was undigested, showing that it had dissociated from the −TAR DNA.
Immediately after the completion of −TAR DNA synthesis, the RNA was mostly intact and was digested by RT to a 14-mer over time in a series of rebinding events. As a result, for time points up to 5 min, most of the 5′ RNA was longer than 14 bases; therefore, the RNA remained annealed and was digested in the absence of NC. However, after 20 min in the absence of NC (lane 26), the 14-base RNA was the primary product produced by RT from the 5′ end, and this RNA could not be digested by E. coli RNase H, showing that the 14-base RNA produced by RT dissociated from −TAR DNA in the absence of NC.
DISCUSSION
A thorough characterization of the interplay among RT, NC, and the DNA and RNA elements in the 5′ portion of the HIV-1 genome is needed to understand how the virus efficiently converts its single-stranded RNA genome into double-stranded DNA. The results presented here allow us to develop a specific model for the roles of RT, NC, and nucleic acid in copying the 5′ 74 bases of the HIV-1 RNA genome into minus-strand DNA. The cleavages made by the RNase H of RT in the region where the 26 DNA primer is bound to the template RNA are not included in this model (Fig. 6) because these cleavages are specific to our model substrate and occur before polymerization begins. After initiating polymerization, RT progresses until the polymerase contacts the base of the TAR RNA hairpin, where RT stalls, permitting RNase H cleavage approximately 17 bases behind the polymerase active site. The pause increases the likelihood of RT dissociation, resulting in the release of 50% of the RT. Dissociation and rebinding increase the RNase H activity relative to the polymerase activity, resulting in RNase H cleavage at the −8 position relative to the polymerase active site. The −8 cleavage decreases the stability of the annealed heteroduplex, thereby increasing the likelihood that the incomplete minus-strand will be released from the template RNA. Rebinding of RT can also result in continued extension of the growing DNA strand.
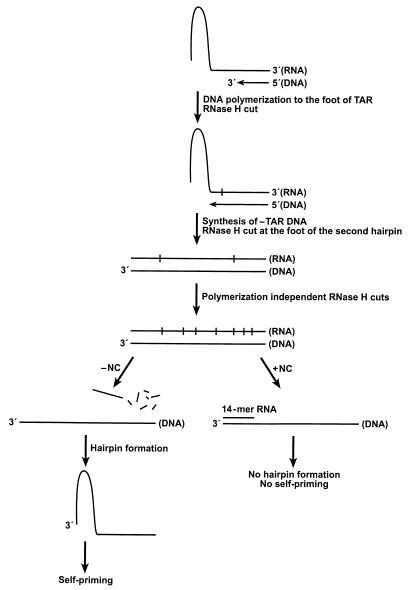
FIG. 6.
The 26-mer DNA primer annealed to the 3′ end of the 100-mer TAR RNA is extended to the base of the TAR hairpin where RT pauses. RNase H cleaves the RNA once. RT can copy the template to the 5′ end. RT falls off the end of the fully double-stranded RNA-DNA duplex. RT can rebind and degrade the RNA strand, leaving a 14-mer from the 5′ end of the RNA template. In the absence of NC, the 14-mer can fall off, allowing hairpin formation and self-priming. In the presence of NC, the 14-mer remains annealed, preventing hairpin formation and self-priming.
As DNA polymerization continues to copy the RNA into the TAR hairpin, the RNA base pairing within the hairpin is disrupted. At this point, there is a brief opportunity for the uncopied 5′ end of the RNA to refold, forming a smaller, secondary hairpin (Fig. 1). Formation of the secondary hairpin is enhanced by NC. The secondary RNA hairpin, because of its lower stability, causes only modest RT pausing, with attendant RNase H digestion and template RNA dissociation, compared with pausing at the TAR RNA hairpin. Complete copying of the RNA template produces a blunt-ended heteroduplex, with the majority of the RNA in the heteroduplex remaining intact, except for a single cleavage 17 bases behind the base of the TAR hairpin. In most cases, RT dissociates from the end of the RNA template before RNase H activity cleaves the RNA at the 5′ end. Multiple RT rebinding events result in extensive RNase H digestion of the template RNA, leaving fragments shorter than 20 bases. In the complete heteroduplex, the 5′ end of the genomic RNA is efficiently cleaved to produce a product that migrates at the position of a 14-mer DNA. NC inhibits dissociation of the 14-base RNA from the newly synthesized −TAR DNA, preventing self-priming at the DNA TAR hairpin, in a fashion similar to what we have shown previously with DNA oligomers (9). In the absence of NC, the kinetic advantage of hairpin formation can overcome the inherent stability of the 14-base heteroduplex, resulting in the dissociation of the 14-base RNA. Dissociation of the 14-base RNA allows either self-priming or low-specificity strand transfer. NC blocks the dissociation of the 14-base RNA from −sssDNA, which prevents hairpin formation (Fig. 6). The 14-base RNA would remain annealed to −sssDNA unless it is displaced by a region of greater complementarity, which occurs in the highly specific first-strand transfer reaction.
RT pauses as a consequence of the secondary structure of the RNA template (5, 14, 20, 25). The 57-base RNA TAR hairpin is sufficiently stable to impede the progress of RT, stalling its forward progress and allowing RNase H digestion. Clearly, the extent of RNase H digestion is inversely correlated with polymerization, as has been reported by other investigators (6, 10, 25). It may be that RNase H activity requires pausing during polymerization and/or dissociation and reassociation of polymerizing RT. Interestingly, in our experiments RT did not cleave the RNA downstream of the second pause site (D-47 at the base of TAR). This pause site was only 4 bases downstream of the first pause at D-43, where the RNA was cleaved at R-75. This result was somewhat unexpected, because the second pause was stronger than the first; however, either the structure of the heteroduplex or the cleavage at R-75 (or both) may have interfered with the additional RNase H cleavage (26).
The RNA genome of HIV-1 is a dynamic and malleable structure, as evidenced by the behavior of the TAR hairpin during polymerization. The RNA-RNA base pairing in the TAR hairpin was disrupted by the polymerase as DNA synthesis continued into the hairpin, allowing the 5′ portion of the original hairpin to refold, forming a smaller, secondary hairpin. Judging by the amount of RT pausing, the formation of the secondary hairpin is enhanced by NC. NC could enhance hairpin formation in two ways. First, NC could increase the rate of annealing of complementary sequences within the hairpin; second, NC can slow the rate of polymerization of RT (9, 21), giving the secondary hairpin more time to form. Although secondary structure appears to be a major determinant of RT pausing, the extent of pausing, dissociation, and RNase H activity is also influenced by other factors (7, 19).
In addition to interacting with the genomic RNA, NC has profound effects on the nascent DNA. After copying the TAR sequence, RT reaches the end of the template RNA. For the first-strand transfer, the growing DNA strand must be transferred to the R region at the 3′ end of the genome. However, the newly synthesized DNA strand can self-prime, short-circuiting the process of copying the genome. NC has the ability to block self-priming (9, 11). We previously showed that a 17-base DNA oligomer annealed to the 3′ end of −sssDNA is sufficient to prevent self-priming (9). Significantly, the blunt-ended nature of the completed RNA-DNA heteroduplex appears to limit digestion of the RNA oligomer bound to the 3′ end of −TAR DNA, resulting in a final product that is 14 bases long. The size of the shortest RNA generated by RT-dependent RNase H activity in the present study was not affected by the addition of NC. Previously, we suggested that NC might prevent self-priming by protecting the RNA from digestion and/or by maintaining longer annealed RNA-DNA hybrids (9). Although NC can protect RNA against digestion, it is clear from the results presented here that an NC concentration sufficient to block self-priming did not significantly inhibit the digestion of the genomic RNA. Using a model substrate, we showed that NC prevents formation of a −sssDNA hairpin by preferentially maintaining the annealing of the 5′ 14-base genomic RNA fragment even though hairpin formation is kinetically favored, in agreement with our previous findings using DNA oligomers (9). Stabilization of the annealing of this 14-base RNA by NC prevents the self-priming of −sssDNA.
Although the coupling of DNA polymerization and RNase H digestion is possible, it appears, at least in our in vitro model system, that these activities, for the most part, are mutually exclusive. Before extension of the DNA primer, nearly all the template RNA was cleaved either 17 or 8 bases behind the polymerase domain, which is positioned at the 3′ end of the DNA primer. This result indicates that RNase H activity is more rapid than polymerization when RT first binds to a DNA primer on an RNA template, a result that has been reported by other investigators (12). RNase H digestion by the polymerizing RT occurred only during pauses in polymerization, when the polymerizing RT was stalled at secondary structures. RT molecules that were not involved in polymerization were responsible for the majority of the RNase H activity, extensively degrading the RNA template after polymerization was largely complete. Binding of RT to completed RNA-DNA heteroduplexes appeared to be random and resulted in rapid digestion of most of the template RNA into oligomers that were 20 bases or shorter, in the presence or absence of NC. No preferential pattern of cleavages could be seen either in the presence or absence of NC; however, a specific 14-base product was derived from the 5′ end of the RNA. Because the heteroduplex substrate and RT were added in approximately equal amounts in the experiments presented here, each RT had to cleave each template multiple times to fully digest the RNA. There are two mechanisms by which RT could make multiple cleavages in the RNA: through multiple, separate binding events (where each binding event produces either a single cleavage or a very limited number of cleavages) or through a bind-and-slide mechanism (where many cleavages are made per binding event). Extensive digestion of the RNA template required that RT dissociate and rebind. The data do not, however, rule out the possibility that under certain conditions RT may cleave the RNA in the heteroduplex by using a bind-and-slide mechanism.
The rapid, random digestion of heteroduplex RNA, combined with the propensity of RT to dissociate at secondary structures, could present a problem for completion of DNA synthesis. Extensive digestion of the template RNA following dissociation of RT, even when the substrate and RT are present at a 1:1 molar ratio, can result in the release of the RNA template from the growing DNA strand. Similar results have been reported previously by other investigators under comparable assay conditions (4, 25). If the heteroduplex dissociates and single-stranded RNA is released, synthesis of the minus-strand DNA will be interrupted. Synthesis of the DNA strand will continue only if it either reanneals to the RNA template from which it just dissociated or is transferred to another RNA template. Although it is clear that pausing at hairpins can result in a substantial amount of RT dissociation, studies of viral DNA synthesis suggest that this effect does not pose a significant problem in vivo. Presumably, the virus must have a means by which it controls the production of an incomplete substrate. NC may facilitate polymerization through regions with secondary structure by helping to maintain the integrity of the RNA template-nascent DNA heteroduplex and may also facilitate the transfer of any released minus-strand DNA to the second copy of genomic RNA that is present in the virion.
ACKNOWLEDGMENTS
We thank Pat Clark and Peter Frank for purifying HIV-1 RT, Hilda Marusiodis for help with the manuscript, and Anne Arthur for expert editorial assistance. We are also grateful to Larry Arthur, Louis Henderson, and Robert Gorelick for generously providing NC and thank Edward Arnold and Steve Harrison for providing access to unpublished structures of HIV-1 RT in complex with RNA-DNA heteroduplexes.
Research was sponsored by the National Cancer Institute, DHHS, under contract with ABL, and by the National Institute of General Medical Sciences.
REFERENCES
- 1.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 2.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 4.DeStefano J J, Bambara R A, Fay P J. The mechanism of human immunodeficiency virus reverse transcriptase-catalyzed strand transfer from internal regions of heteropolymeric RNA templates. J Biol Chem. 1994;269:161–168. [PubMed] [Google Scholar]
- 5.DeStefano J J, Buiser R G, Mallaber L M, Fay P J, Bambara R A. Parameters that influence processive synthesis and site-specific termination by human immunodeficiency virus reverse transcriptase on RNA and DNA templates. Biochim Biophys Acta. 1992;1131:270–280. doi: 10.1016/0167-4781(92)90025-u. [DOI] [PubMed] [Google Scholar]
- 6.DeStefano J J, Buiser R G, Mallaber L M, Myers T W, Bambara R A, Fay P J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991;266:7423–7431. [PubMed] [Google Scholar]
- 7.DeStefano J J, Mallaber L M, Fay P J, Bambara R A. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 1993;21:4330–4338. doi: 10.1093/nar/21.18.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding J, Das K, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Hughes S H, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll M D, Hughes S H. HIV-1 nucleocapsid protein can prevent self-priming of minus-strand strong-stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J Virol. 2000;74:8785–8792. doi: 10.1128/jvi.74.19.8785-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudding L R, Nkabinde N C, Mizrahi V. Analysis of the RNA- and DNA-dependent DNA polymerase activities of point mutants of HIV-1 reverse transcriptase lacking ribonuclease H activity. Biochemistry. 1991;30:10498–10506. doi: 10.1021/bi00107a019. [DOI] [PubMed] [Google Scholar]
- 11.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J K, Palaniappan C, Wu W, Fay P J, Bambara R A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 14.Klarmann G J, Schauber C A, Preston B D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem. 1993;268:9793–9802. [PubMed] [Google Scholar]
- 15.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J L, Kleiman L, Parniak M A, Weinberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependency of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 18.Muesing M A, Smith D H, Capon D J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 19.Palaniappan C, Fuentes G M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 20.Pathak V K, Temin H M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992;66:3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raja A, DeStefano J J. Kinetic analysis of the effect of HIV nucleocapsid protein (NCp) on internal strand transfer reactions. Biochemistry. 1999;38:5178–5184. doi: 10.1021/bi9828019. [DOI] [PubMed] [Google Scholar]
- 22.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 23.Suo Z, Johnson K A. Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1997;36:12468–12476. doi: 10.1021/bi971218+. [DOI] [PubMed] [Google Scholar]
- 24.Telesnitsky A, Goff S P. Two defective forms of reverse transcriptase can complement to restore viral infectivity. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkmann S, Wohrl B M, Tisdale M, Moelling K. Enzymatic analysis of two HIV-1 reverse transcriptase mutants with mutations in carboxyl-terminal amino acid residues conserved among retroviral ribonucleases H. J Biol Chem. 1993;268:2674–2683. [PubMed] [Google Scholar]
- 26.Wohrl B M, Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990;29:10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]