Abstract
It is becoming increasingly clear that any human immunodeficiency virus (HIV) vaccine should induce a strong CD8+ response. Additional desirable elements are multispecificity and a focus on conserved epitopes. The use of multiple conserved epitopes arranged in an artificial gene (or EpiGene) is a potential means to achieve these goals. To test this concept in a relevant disease model we sought to identify multiple simian immunodeficiency virus (SIV)-derived CD8+ epitopes bound by a single nonhuman primate major histocompatibility complex (MHC) class I molecule. We had previously identified the peptide binding motif of Mamu-A*012, a common rhesus macaque MHC class I molecule that presents the immunodominant SIV gag-derived cytotoxic T lymphocyte (CTL) epitope Gag_CM9 (CTPYDINQM). Herein, we scanned SIV proteins for the presence of Mamu-A*01 motifs. The binding capacity of 221 motif-positive peptides was determined using purified Mamu-A*01 molecules. Thirty-seven peptides bound with apparent Kd values of 500 nM or lower, with 21 peptides binding better than the Gag_CM9 peptide. Peripheral blood mononuclear cells from SIV-infected Mamu-A*01+ macaques recognized 14 of these peptides in ELISPOT, CTL, or tetramer analyses. This study reveals an unprecedented complexity and diversity of anti-SIV CTL responses. Furthermore, it represents an important step toward the design of a multiepitope vaccine for SIV and HIV.
With more than 30 million human immunodeficiency virus (HIV)-infected individuals (World Health Organization [WHO] web site http://hivinsite.ucsf.edu/social/un/2098.371d.html estimates), there can be few other more pressing biomedical priorities than to produce an effective vaccine for HIV. Given the important role that CD8+ lymphocytes play in controlling viral replication (11, 32, 43, 49, 58), it is critical that this vaccine stimulate strong cytotoxic T-lymphocyte (CTL) responses. Simian immunodeficiency virus (SIV) infection of macaques provides the best nonhuman primate model to determine whether the generation of virus-specific CTLs can alter the course of disease after infection (33, 65). The nucleotide sequences of the SIVs are closely related to those of HIV-1 and -2 (12, 24). SIV and HIV have similar tropisms for CD4 (16, 36), and infection with SIV causes an AIDS-like disease in the majority of infected macaques by 1 year postinoculation (35). Since macaques and humans have very similar immune systems (10, 31, 63, 76), SIV infection of macaques is also an excellent model to study the immunology of HIV infection of humans.
SIV infection of macaques is currently the only cost-effective animal model to test vaccine efficacy in vivo. Several vaccine studies in macaques have already suggested that a strong immune response to SIV can be generated in appropriately immunized monkeys (15, 19, 29, 42, 46, 47) and that this response can, in some cases, protect against the development of AIDS. In particular, cell-mediated responses to SIV appear to represent a crucial component of vaccine protective efficacy. CD8+ lymphocytes recognize pathogen-infected cells, are involved in the host's defensive response to intracellular pathogens (34), and may play an important role in the containment of the AIDS virus in infected individuals (74). This is especially evident during the first few weeks postinfection (8, 39, 53, 57) and during most phases of disease by mechanisms which include killing of infected cells and suppression of replication (69, 75). It has recently been shown that depletion of CD8+ cells using monoclonal antibodies (MAbs) resulted in increases in virus loads in SIV-infected animals (32, 43, 58). Besides this role in containment of disease, CTLs may also be involved in providing protection from infection with HIV (17, 54, 55). Thus, these observations collectively provide the rationale to explore whether CTLs can protect from AIDS virus infection in an animal model.
Currently, a single useful major histocompatibility complex (MHC) class I molecule (Mamu-A*01) in the rhesus macaque has been well characterized. This allele is present in approximately 25% of rhesus macaques of Indian descent (38, 73), and tetramers and ELISPOT assays for the single Mamu-A*01-restricted CTL epitope Gag_CM9 (CTPYDINQM; p11C, C→M) have been developed (2, 3, 28, 41). However, thus far only a limited number of SIV-derived, Mamu-A*01-restricted epitopes have been defined (2, 4, 22, 25, 44). Therefore, we wanted to examine whether additional Mamu-A*01-restricted CTL epitopes derived from other regions of SIV could be identified. Vaccination with multiple epitopes is likely of importance since escape from CTL induced against a single epitope is possible (9, 23, 26, 45, 51, 64). CTL against epitopes in different proteins may also have very different effects on reducing viral burden. Finally, definition of multiple epitopes will allow more precise characterization and quantitation of immune responses against SIV, either during the course of natural infection or following immunization with experimental vaccines.
MATERIALS AND METHODS
Motif scanning of SIV proteins and peptide synthesis.
The Mamu-A*01 peptide binding motif is defined by the requirement for proline (P) in position 3 (2). Live-cell binding assays indicated that in addition to the requirement for P in position 3, Mamu-A*01 preferentially bound peptides bearing a small residue in position 2 (A, V, S, T, or P) and hydrophobic (A, L, I, V, and M) or aromatic (F, W, and Y) residues at the C terminus.
This motif was utilized to scan the SIVmac251 sequence to identify potential Mamu-A*01 binding peptides between 8 and 11 residues in length, and 111 peptides were identified. Additionally, 50 9-mer and 50 10-mer sequences were selected by removing the restriction for small residues in position 2, for a total of 211 peptides. The corresponding peptides were then synthesized as crude material by Chiron Mimotopes (San Diego, Calif.). Lyophilized material was resuspended at 20 mg/ml in 100% dimethyl sulfoxide and then diluted to required concentrations in phosphate-buffered saline (PBS).
Radiolabeled probe peptides and peptides subsequently determined to bind Mamu-A*01 with high affinity (500 nM or less) were resynthesized at Epimmune on a larger scale using standard tert-butoxycarbonyl or 9-fluorenylmethoxy carbonyl solid-phase methods, as previously described (56). These were purified to >95% homogeneity by reverse-phase high-pressure liquid chromatography, and composition was ascertained by amino acid analysis, sequencing, and/or mass spectrometry analysis.
Mamu-A*01 purification.
721.221 cells transfected with the Mamu-A*01 cDNA were utilized as the source of Mamu-A*01 molecules. Cells were maintained in vitro by culture in RPMI 1640 medium (Flow Laboratories, McLean, Va.) supplemented with 2 mM l-glutamine (Gibco, Grand Island, N.Y.), 100 U (100 μg/ml) of penicillin-streptomycin solution (Gibco), and 10% heat-inactivated fetal calf serum (FCS; Hazleton Biologics) and grown for large-scale cultures in roller bottle apparatuses.
Mamu-A*01 was purified from cell lysates as previously described (62). Briefly, cells were lysed at a concentration of 108 cells/ml in 50 mM Tris-HCl (pH 8.5) containing 1% NP-40 (Fluka Biochemika, Buchs, Switzerland), 150 mM NaCl, 5 mM EDTA, and 2 mM phenylmethylsulfonyl fluoride. Lysates were then passaged through 0.45-μm filters and cleared of nuclei and debris by centrifugation at 10,000 × g for 20 min, and MHC molecules were purified by affinity chromatography.
For affinity purification, columns of inactivated Sepharose CL4B and protein A-Sepharose were used as precolumns. Mamu-A*01 was captured by repeated passage over protein A-Sepharose beads conjugated with the anti-HLA(A,B,C) antibody W6/32 as previously described (2). After two to four passages, the W6/32 column was washed with 10 column volumes of 10 mM Tris-HCl (pH 8.0) with 1% NP-40, 2 column volumes of PBS, and 2 column volumes of PBS containing 0.4% n-octylglucoside. Finally, Mamu-A*01 molecules were eluted with 50 mM diethylamine in 0.15 M NaCl containing 0.4% n-octyglucoside (pH 11.5). A 1/25 volume of 2.0 M Tris (pH 6.8) was added to the eluate to reduce the pH to ∼8.0. The eluate was then concentrated by centrifugation in Centriprep 30 concentrators at 2,000 rpm (Amicon, Beverly, Mass.). Protein purity, concentration, and effectiveness of depletion steps were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Mamu-A*01 binding assay.
Quantitative assays for the binding of peptides to soluble Mamu-A*01 molecules on the basis of the inhibition of binding of a radiolabeled standard probe peptide to detergent-solubilized MHC molecules were performed utilizing the protocol previously described for the binding of peptides to HLA class I molecules (62). Briefly, 1 to 10 nM radiolabeled probe peptide, iodinated by the chloramine T method (27), was coincubated at room temperature with various amounts of purified Mamu-A*01 in the presence of 1 μM human β2-microglobulin (Scripps Laboratories, San Diego, Calif.) and a cocktail of protease inhibitors. Following a 2-day incubation, the percent of MHC bound radioactivity was determined by size exclusion gel filtration chromatography on a TSK 2000 column.
A position 1 C→A analog of the SIV Gag 181-190 peptide (ATPYDINQML) was used as the radiolabeled probe. In the case of competitive assays, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide was calculated. Peptides were initially tested at one or two high doses. The 50% inhibitory concentration (IC50) of peptides yielding positive inhibition was then determined in subsequent experiments, in which two to six further dilutions were tested, as necessary. Since under the conditions used the concentration of label is less than that of MHC and the IC50 is equal to the MHC concentration, the measured IC50 values are reasonable approximations of the true Kd values. Each competitor peptide was tested in two to four completely independent experiments. As a positive control, in each experiment the unlabeled version of the radiolabeled probe was tested.
IFN-γ ELISPOT assay.
Ninety-six-well flat-bottomed plates (U-Cytech-BV, Amsterdam, The Netherlands) were coated with 5 μg of anti-gamma interferon (IFN-γ) MAb MD-1 (U-Cytech-BV) overnight at 4°C. The plates were then washed 10 times with PBST (PBS [Gibco-BRL] containing 0.05% Tween 20 [Sigma Chemical, St. Louis, Mo.]), and then the plates were blocked with 2% PBSA (PBS containing 2% bovine serum albumin [BSA; Sigma Chemical]) for 1 h at 37°C. The 2% PBSA was discarded from the plates, and freshly isolated peripheral blood mononuclear cells (PBMC) were added. Cells were resuspended in RPMI 1640 (Mediatech) supplemented with penicillin, streptomycin, and 5% fetal bovine serum (FBS; Biocell) (R05). The R05 also contained either 5 μg of concanavalin A (Sigma Chemical) per ml, 1 to 10 μM various Mamu-A*01-bound peptides, 1 to 10 μM irrelevant SIV envelope peptide E→V (ELGDYKLV), or no peptide. Input cell numbers were 2.0 × 105 peripheral blood lymphocytes in 100 μl/well in triplicate wells.
Cells were then incubated for 16 h at 37°C in 5% CO2, after which the cells were removed from the plates by shaking and 200 μl of ice-cold deionized water was added per well to lyse the remaining PBMC. Plates were incubated on ice for 15 min and then washed 20 times with PBST. Next, 1 μg of rabbit anti-IFN-γ polyclonal biotinylated detector antibody solution (U-Cytech-BV) per well was added, and the plates were incubated for 1 h at 37°C. The plates were washed 10 times with PBST, after which 50 μl of a gold-labeled anti-biotin immunoglobulin G solution (U-Cytech BV) was added. The plates were incubated for 1 h at 37°C and washed 10 times with PBST. Thirty microliters of activator mix (U-Cytech BV) per well was added, and the plates were developed for about 30 min. The activator mix consists of a silver salt solution that precipitates at the sites of gold clusters (from the gold-labeled antibiotin solution), visualizing the sites where the IFN-γ was secreted. When black spots appeared in the wells under an inverted microscope, the wells were washed with distilled water to stop development and then air dried.
Wells were imaged with IP Lab Spectrum 3.23 software using a Hamamatsu C4880 series camera attached to a Nikon TE 300 inverted microscope. Spots were counted manually. A spot-forming cell (SFC) was defined as a large black spot with a fuzzy border (37). To determine significance levels, a baseline for each peptide was first established using the average and standard deviation of the number of SFCs in three independent assays as performed on Mamu-A*01+ but SIV-naive animals. A threshold significance value corresponding to this average plus two standard deviations was then determined. In our analysis of samples from SIV-infected Mamu-A*01+ animals, a response was considered positive if the number of SFCs exceeded the threshold significance level for that specific peptide.
Generation of in vitro-cultured CTL effector cells.
CTL cultures were established from EDTA-treated peripheral blood samples as previously described (2). Briefly, Ficoll-Hypaque-separated PBMC were stimulated 1:1 with 5 × 106 γ-irradiated (3,000 rad) autologous B lymphoblastoid cell line cells (B-LCLs) pulsed with the appropriate peptide (5 μM) in R10 medium. Cultures were supplemented with R10 containing 20 U of recombinant interleukin-2 (rIL-2), a gift from Hoffman-LaRoche (Nutley, N.J.), per ml. On day 7, viable cells were restimulated and again expanded in the presence of rIL-2. CTL activity was assessed after 14 days of culture in a standard 51Cr release assay.
CTL analysis.
SIV-specific CTL activity was assessed using a standard 51Cr release assay (2). 51Cr-labeled Mamu-A*01+ B-LCL targets were pulsed with SIV peptides or an irrelevant influenza virus NP peptide (SNEGSYFF). Target cells (5 × 103) were incubated for 5 h with CTL effectors at effector-to-target cell ratios ranging from 20:1 to 50:1. CTL activity was calculated from the counts per minute present in harvested supernatants using the formula % specific release = (experimental release − spontaneous release)/(maximal release − spontaneous release) × 100. The reported percent specific lysis represents the 51Cr released from the Mamu-A*01 peptide-pulsed targets minus the 51Cr released from target cells pulsed with the irrelevant influenza virus NP peptide (SNEGSYFF). Spontaneous release was always less than 20% of maximal release.
Mamu-A*01 tetramers.
Soluble tetrameric Mamu-A*01 MHC class I/SIV Gag_CM9 peptide complexes were constructed as previously described (3, 5).
Tetramer staining.
Fresh unstimulated PBMC (106) were washed twice in fluorescence-activated cell sorting (FACS) buffer (PBS [Gibco] with 2% FCS [BioCell]) in a 96-well U-bottomed plate. In a 100-μl volume, cells were stained in the dark for 40 min at room temperature with the tetramer (1 μg/ml for in vitro cultures, 5 μg/ml for fresh PBMC), anti-rhesus CD3 fluorescein isothiocyanate (FITC) MAb (10 μl; BioSource), and anti-CD8α-PerCP antibody (3 μl; Becton Dickinson). Cells were washed four times with FACS buffer and fixed by adding 450 μl of 2% paraformaldehyde (PFA). Sample data were acquired on a Becton Dickinson FACSCalibur instrument and analyzed using CellQuest software (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). Background tetramer staining of fresh, unstimulated PBMC from naive Mamu-A*01+ animals was routinely less than 0.08%.
Intracellular IFN-γ staining.
A total of 2 × 105 cells from in vitro-stimulated CTL cultures were incubated at 37°C for 1 h with phorbol myristate acetate-ionomycin (50 ng/ml and 1 μg/ml, respectively), 5 μM Gag-CM9 peptide, or a control influenza virus peptide (SNEGSYFF) in the presence of Mamu-A*01+ B-LCL (105) as antigen-presenting cells (APC). Cells were then treated with 10 μg of brefeldin A per ml to inhibit protein trafficking and incubated a further 4 to 5 h at 37°C. Cells were then washed twice with FACS buffer (PBS plus 2% FCS) and stained with CD8α-PerCP and Mamu-A*01-phycoerythrin (PE) tetramers. After fixation with PFA overnight, cells were washed twice with FACS buffer and treated with 150 μl of permeabilization buffer (0.1% saponin in FACS buffer) for 5 min at room temperature. Cells were washed once more with 0.1% saponin and then incubated in the dark for 50 min with 1 μl of anti-human IFN-γ-FITC MAb (Pharmingen; clone 4S.B3; catalog no. 18904A). Finally, cells were washed four times with 0.1% saponin buffer, and a 100-μl cell suspension was fixed with 450 μl of 2% PFA.
Animals, viruses, and infections.
Rhesus macaques used in this study were identified as Mamu-A*01+ by PCR-SSP and direct sequencing as previously described (38). All rhesus macaques used in this study were Mamu-A*01+ with the exception of animal 95003. Rhesus macaques 96078 and 96087 are naive macaques. Animals 94004 and 96031 were vaccinated 10 weeks previously with a DNA-modified vaccinia virus Ankara (MVA) regimen expressing the Gag_CM9 peptide (3). Animal 95024 was infected intravenously with 40 50% tissue culture infectious doses of a heterogeneous SIV stock (originally provided by R. C. Desrosiers, Harvard University and New England Regional Primate Research Center). The stock was amplified by growth on rhesus PBMC with a final passage on CEMx174 cells to increase titers (50, 68). Rhesus macaques 95114, 95115, 96031, and 95003 were infected intrarectally with a molecularly cloned virus, SIVmac239. This stock was amplified on rhesus PBMC only. SIV-infected animals were cared for according to an experimental protocol approved by the University of Wisconsin Research Animal Resource Committee.
RESULTS
Identification of 37 SIV-derived peptides which bind to Mamu-A*01.
To explore whether multiple CTL epitopes in Mamu-A*01+ rhesus macaques could be identified, we used the previously defined motif for Mamu-A*01 to scan all SIV proteins (2). A total of 211 peptides were identified which were analyzed using in vitro peptide-binding experiments utilizing purified Mamu-A*01 molecules. Each potential binder was used to outcompete the radiolabeled probe peptide in our peptide binding assay. Under the stoichometric conditions used in the assay, IC50 is a reasonable approximation of Kd. It was found that 37 peptides bound with an IC50 of less than 500 nM (Table 1). The 500 nM affinity threshold has previously been shown to be associated with recognition in vivo in both murine and human systems (59, 60, 70, 72). Seventeen of the peptides identified herein bound Mamu-A*01 with IC50 values of 50 nM or less and therefore would be classified as high-affinity binders. The remaining 20 peptides bound in the 51 to 500 nM range and would be classified as intermediate binders (56). It is noteworthy that 21 peptides bound with greater affinity than the known Gag_CM9 epitope. Interestingly, no potential Mamu-A*01-restricted peptides that bound with IC50 values of less than 500 nM were identified in Nef or Vpr.
TABLE 1.
SIV-derived peptides that bind to Mamu-A*01 with IC50 values below 500 nMa
| Peptide no. or virus | Protein and amino acids | Peptide | Sequence | IC50 (nM) | Reference |
|---|---|---|---|---|---|
| 1 | Env 235–242 | Env_CL8 | CAPPGYAL | 1.9 | |
| 2 | Pol 143–152 | Pol_LV10 | LGPHYTPKIV | 3.0 | |
| 3 | Pol 51–61 | Pol_EA11 | EAPQFPHGSSA | 3.7 | |
| 4 | Env 235–243 | Env_CL9 | CAPPGYALL | 5.5 | |
| 5 | Env 729–738 | Env_ST10 | SPPSYFQTHT | 8.2 | |
| 6 | Pol 621–629 | Pol_SV9621 | STPPLVRLV | 8.7 | |
| 7 | Pol 588–596 | Pol_QV9 | QVPKFHLPV | 9.1 | |
| 8 | Env 622–630 | Env_TL9 | TVPWPNASL | 10 | |
| 9 | Vpx 102–111 | Vpx_GL10 | GPPPPPPPGL | 23 | |
| 10 | Pol 474–483 | Pol_IL10 | IYPGIKTKHL | 23 | |
| 11 | Pol 621–628 | Pol_SL8 | STPPLVRL | 26 | |
| 12 | Pol 147–155 | Pol_YI9 | YTPKIVGGI | 26 | |
| 13 | Gag 372–379 | Gag_LF8 | LAPVPIPF | 29 | |
| 14 | Rev 87–96 | Rev_DL10 | DPPTNTPEAL | 39 | |
| 15 | Pol 359–368 | Pol_GM10 | GSPAIFQYTM | 48 | |
| 16 | Gag 372–380 | Gag_LA9 | LAPVPIPFA | 50 | |
| 17 | Vpx 39–48 | Vpx_HV10 | HLPRELIFQV | 50 | |
| 18 | Env 763–771 | Env_SI9 | SWPWQIEYI | 67 | |
| 19 | Pol 957–964 | Pol_MI8 | MTPAERLI | 77 | |
| 20 | Pol 34–43 | Pol_QF10 | QMPRQTGGFF | 78 | |
| 21 | Pol 359–367 | Pol_GT9 | GSPAIFQYT | 82 | |
| 22 | Gag 181–189 | Gag_CM9 | CTPYDINQM | 86 | |
| 23 | Vif 75–82 | Vif_LL8 | LTPERGWL | 90 | |
| 24 | Gag 170–177 | Gag_VL8 | VVPGFQAL | 99 | |
| 25 | Vif 100–107 | Vif_VI8 | VTPDYADI | 103 | |
| 26 | Vif 144–152 | Vif_QA9 | QVPSLQYLA | 141 | |
| 27 | Tat 28–35 | Tat_TL8 | TTPESANL | 148 | |
| 28 | Vif 14–22 | Vif_RW9 | RIPERLERW | 170 | |
| 29 | Env 133–140 | Env_AV8 | AAPTSAPV | 175 | |
| 30 | Gag 254–262 | Gag_QI9 | QNPIPVGNI | 196 | |
| 31 | Env 431–439 | Env_YI9 | YVPCHIRQI | 210 | |
| 32 | Env 728–736 | Env_ST9 | SSPPSYFQT | 220 | |
| 33 | Vpx 8–18 | Vpx_II11 | IPPGNSGEETI | 241 | |
| 34 | Gag 340–349 | Gag_VT10 | VNPTLEEMLT | 267 | |
| 35 | Env 504–512 | Env_IT9 | ITPIGLAPT | 286 | |
| 36 | Gag 149–157 | Gag_LW9 | LSPRTLNAW | 355 | |
| 37 | Pol 692–700 | Pol_SV9692 | SGPKTNIIV | 366 | |
| SIV | Env 235–243 | Env_CL9 | CAPPGYALL | 25 | |
| SIV | Pol 621–629 | Pol_SV9 | STPPLVRLV | 22 | |
| SIV | Env 622–630 | Env_TL9 | TVPWPNETL | 25 | |
| SIV | Gag 181–189 | Gag_CM9 | CTPYDINQM | 2, 44 | |
| SHIV | Env 431–439 | Env_YI9 | YAPPISGQI | 22 |
Previously defined Mamu-A*01-restricted SIV/SHIV CTL epitopes are shown in boldface.
Elispot identifies 14 Mamu-A*01-bound peptides in SIV-infected macaques.
We then analyzed whether the 37 selected peptides (IC50 <500 nM) were actually recognized in vivo by fresh PBMC derived from SIV-infected Mamu-A*01+ animals (Table 2). Two naive, uninfected Mamu-A*01+ animals (96078 and 96087) were initially tested in Elispot assays. None of the 37 peptides induced significant responses at either 1 or 10 μM peptide concentrations in either of these control animals (data not shown).
TABLE 2.
MHC type, vaccination, and infection status of animals used to detect multiple CTL epitopesa
| Animal no. | MHC type | Vaccination | Infection |
|---|---|---|---|
| 96078 | A*01 | No | No |
| 96087 | A*01 | No | No |
| 96031 | A*01 | Yes, 2 wk prior to infection | Yes, SIVmac239 i.r. |
| 94004 | A*01 | Yes, 10 wk previously | No |
| 95024 | A*01 | No | Yes, SIVmac biological isolate i.v. |
| 95114 | A*01 | No | Yes, SIVmac239 i.r. |
| 95115 | A*01 | No | Yes, SIVmac239 i.r. |
| 95003 | — | No | Yes, SIVmac239 i.r. |
Animals were vaccinated with DNA-MVA containing the Gag_CM9 peptide (3). Infection with SIVmac239 or SIVmac biological isolate was done intravenously (i.v.) or intrarectally (i.r.).
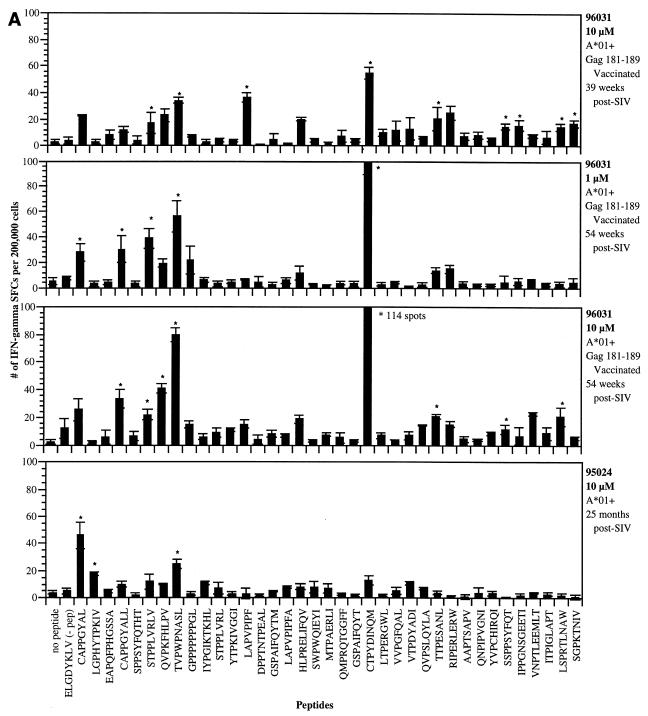
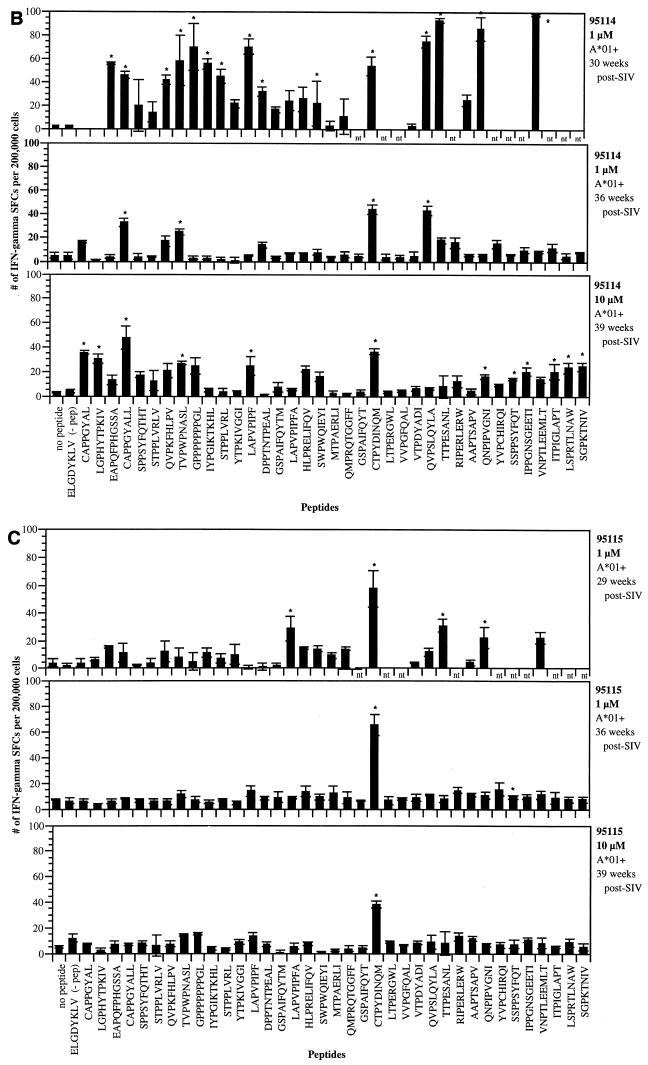
Using IFN-γ ELISPOT analysis of fresh PBMC derived from four SIV-infected Mamu-A*01+ macaques, we were able to demonstrate that 14 of these newly defined peptides, in addition to the previously identified Gag_CM9 epitope (2, 44), were well recognized (Fig. 1). The number of SFCs detected against each peptide in these animals ranged from 11 to 114 per 200,000 PBMC plated. While considerable variability existed from animal to animal with respect to peptides that were recognized, with few exceptions replicate assays conducted on PBMC from each animal gave reproducible responses. Stimulation with 12 of these peptides gave positive responses in animal 96031 (Fig. 1A). While this animal demonstrated a very broad immune response, the strongest response was to the Gag_CM9 peptide against which this animal had been previously vaccinated (Table 2). When PBMC from animal 96031 were stimulated with a lower concentration of peptide (1 μM), while many of the epitopes still induced equally strong responses, some of the weakly responding epitopes were no longer stimulatory (Fig. 1A). Unlike animal 96031, animal 95024 (25 months post-SIV infection) responded to only a few of the peptides (Fig. 1A). Interestingly, three new peptides (Env_CL8, Pol_LV10, and Env_TL9) gave better responses than the Gag_CM9 epitope. Animal 95114 also demonstrated a very broad Mamu-A*01-restricted immune response after SIV infection (Fig. 1B). In this animal, a total of 22 peptides were recognized. In the first assay conducted on this animal, eight of the peptides gave SFC values greater than that for the Gag_CM9 epitope. Finally, in animal 95115, while a few low-responding peptides were detected in the initial assay, with the exception of responses against the Gag_CM9 epitope, these responses appeared to subside over time (Fig. 1C). A summary of the ELISPOT responses in the four SIV-infected animals is presented in Table 3. In total, 14 peptides which gave significant ELISPOT responses in at least two independent assays were considered positive. Since the Env_CL8 and Env_CL9 peptides overlap, we are considering this to represent a single positive response.
FIG. 1.
Detection of IFN-γ production by PBMC using the ELISPOT assay. PBMC from various Mamu-A*01+ SIV-infected animals were tested with the Mamu-A*01 peptides in 16-h ELISPOT assays. (A) Animal 96031 (Gag_CM9 vaccinated) and animal 95024. (B) Animal 95114. (C) Animal 95115. PBMC were plated in 96-well plates at 2 × 105 cells/well and stimulated with various peptides (1 to 10 μM concentration). Mean values and standard deviations from triplicate wells were averaged for each assay, and SFCs were enumerated as described in Materials and Methods. Asterisks indicate statistically significant responses (see the text). Responses to concanavalin A were always greater than 200 SFCs per 2 × 105 cells. The ELGDYKLV peptide represents an irrelevant SIV Env peptide (negative control).
TABLE 3.
Positive responses detected against 14 Mamu-A*01-bound peptides by ELISPOT in SIV-infected Mamu-A*01+ macaquesa
| Peptide no. | Peptide name | Sequence | Response to peptide at concn (μM)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 96031
|
95024
|
95114
|
95115
|
|||||||||
| 10 | 1 | 10 | 10 | 1 | 1 | 10 | 1 | 1 | 10 | |||
| 1 | Env CL8 | CAPPGYAL | − | + | − | + | − | − | + | − | − | − |
| 2 | Pol LV10 | LGPHYTPKIV | − | − | − | + | − | − | + | − | − | − |
| 3 | Pol_EA11 | EAPQFPHGSSA | − | − | − | − | + | − | − | − | − | − |
| 4 | Env CL9 | CAPPGYALL | − | + | + | − | + | + | + | − | − | − |
| 5 | Env_ST10 | SPPSYFQTHT | − | − | − | − | − | − | − | − | − | − |
| 6 | Pol_SV9621 | STPPLVRLV | + | + | + | − | − | − | − | − | − | − |
| 7 | Pol_SV9 | QVPKFHLPV | − | − | + | − | + | − | − | − | − | − |
| 8 | Env_TL9 | TVPWPNASL | + | + | + | + | + | + | + | − | − | − |
| 9 | Vpx_GL10 | GPPPPPPPGL | − | − | − | − | + | − | − | − | − | − |
| 10 | Pol_IL10 | IYPGIKTKHL | − | − | − | − | + | − | − | − | − | − |
| 11 | Pol_SL8 | STPPLVRL | − | − | − | − | + | − | − | − | − | − |
| 12 | Pol_YI9 | YTPKIVGGI | − | − | − | − | − | − | − | − | − | − |
| 13 | Gag_LF8 | LAPVPIPF | + | − | − | − | + | − | + | − | − | − |
| 14 | Rev_DL10 | DPPTNTPEAL | − | − | − | − | + | − | − | − | − | − |
| 15 | Pol_GM10 | GSPAIFQYTM | − | − | − | − | − | − | − | − | − | − |
| 16 | Gag_LA9 | LAPVPIPFA | − | − | − | − | − | − | − | + | − | − |
| 17 | Vpx_HV10 | HLPRELIFQV | − | − | − | − | − | − | − | − | − | − |
| 18 | Env_SI9 | SWPWQIEYI | − | − | − | − | + | − | − | − | − | − |
| 19 | Pol_MI8 | MTPAERLI | − | − | − | − | − | − | − | − | − | − |
| 20 | Pol_QF10 | QMPRQTGGFF | − | − | − | − | − | − | − | − | − | − |
| 21 | Pol_GT9 | GSPAIFQYT | − | − | − | − | nt | − | − | nt | − | − |
| 22 | Gag_CM9 | CTPYDINQM | + | + | + | − | + | + | + | + | + | + |
| 23 | Vif_LL8 | LTPERGWL | − | − | − | − | nt | − | − | nt | − | − |
| 24 | Gag_VL8 | VVPGFQAL | − | − | − | − | nt | − | − | nt | − | − |
| 25 | Vif_VI8 | VTPDYADI | − | − | − | − | − | − | − | − | − | − |
| 26 | Vif_QA9 | QVPSLQYLA | − | − | − | − | + | + | − | − | − | − |
| 27 | Tat_TL8 | TTPESANL | + | − | + | − | + | − | − | + | − | − |
| 28 | Vif_RW9 | RIPERLERW | − | − | − | − | nt | − | − | nt | − | − |
| 29 | Env_AV8 | AAPTSAPV | − | − | − | − | − | − | − | − | − | − |
| 30 | Gag_QI9 | QNPIPVGNI | − | − | − | − | + | − | + | + | − | − |
| 31 | Env_YI9 | YVPCHIRQI | − | − | − | − | nt | − | − | nt | − | − |
| 32 | Env_ST9 | SSPPSYFQT | + | − | + | − | nt | − | + | nt | + | − |
| 33 | Vpx_II11 | IPPGNSGEETI | + | − | − | − | nt | − | + | nt | − | − |
| 34 | Gag_VT10 | VNPTLEEMLT | − | − | − | − | + | − | − | − | − | − |
| 35 | Env_IT9 | ITPIGLAPT | − | − | − | − | nt | − | + | nt | − | − |
| 36 | Gag_LW9 | LSPRTLNAW | + | − | + | − | nt | − | + | nt | − | − |
| 37 | Pol_SV9692 | SGPKTNIIV | + | − | − | − | nt | − | + | nt | − | − |
Since the Env_CL8 and Env_CL9 peptides overlap, we consider this a single positive response. Mamu-A*01-bound SIV-derived peptides were considered positive responders (shown in boldface) if peptides yielded two or more significant responses (+) by ELISPOT in the four SIV-infected animals tested. Each column of data is from a separate experiment. Not all peptides were tested in each animal (nt). Peptides were tested at 1 and 10 μM concentrations.
To rule out the possibility that the reactivity against these different peptides is actually the result of cross-reactivity of T cells generated against the Gag_CM9 peptide, we carried out ELISPOT assays using the complete set of peptides in a Mamu-A*01+ animal (94004) that had been vaccinated only against the Gag_CM9 peptide. This animal had previously shown good reactivity against this peptide, with up to 20% of this animal's CD3 CD8α lymphocytes being positive for tetramer staining against the Gag_CM9 epitope 1 week following its first MVA (3). Analysis of Gag_CM9-reactive lymphocytes from this animal 10 weeks after receiving MVA revealed 91 SFCs per 200,000 cells (data not shown). No reactivity was seen against any of the other peptides, indicating that the responses in the SIV-infected animals were likely not a result of cross-reactivity to the Gag_CM9 peptide. To investigate whether SIV infection alone was responsible for these reactivities, we used all 37 peptides in an ELISPOT assay in an SIV-infected Mamu-A*01-negative animal (95003). None of the peptides were recognized in this animal (data not shown).
To determine whether the peptide-specific production of IFN-γ was an MHC class I-restricted response, a replicate ELISPOT assay (200,000 cells/well, 10 μM peptide) was conducted using CD8+-depleted PBMC from animal 95114. In this assay, positive responses were no longer detected from peptides which had induced positive responses in bulk PBMC (data not shown), confirming the role of CD8+ T cells in mediating these responses.
Activity of newly identified epitopes in recall CTL assays from SIV-infected animals.
We then used 51Cr release CTL assays to determine whether these peptides could recall in vitro memory CTL activity from SIV-infected animals. When tested in a chronically SIV-infected Mamu-A*01+ macaque (95024), several of the peptides recalled good CTL activity after a 2-week culture period (Table 4). Additional cultures were also initiated from two other SIV-infected Mamu-A*01+ animals (95114 and 95115). As with ELISPOT, not all peptides that induced a CTL response in a particular animal did so in all animals. Seventeen peptides were reproducibly considered positive by 51Cr release assays under the criteria listed in Table 4. Fourteen of these peptides were recognized in more than one animal, while the three remaining peptides were reproducibly detected only in a single animal.
TABLE 4.
Recall CTL responses detected against 16 Mamu-A*01-bound peptides by CTL assays in SIV-infected Mamu-A*01+ macaquesa
| Peptide no. | Peptide name | Sequence | Positive responses/ no. of tests
|
||
|---|---|---|---|---|---|
| 95024 | 95114 | 95115 | |||
| 1 | Env_CL8 | CAPPGYAL | 2/3 | 0/1 | 1/1 |
| 2 | Pol_LV10 | LGPHYTPKIV | 1/3 | 1/1 | 1/1 |
| 3 | Pol_EA11 | EAPQFPHGSSA | 0/3 | 0/1 | 1/1 |
| 4 | Env_CL9 | CAPPGYALL | 3/4 | 0/1 | 1/1 |
| 5 | Env_ST10 | SPPSYFQTHT | 1/3 | 1/1 | 1/1 |
| 6 | Pol_SV9621 | STPPLVRLV | 0/3 | 0/1 | 1/1 |
| 7 | Pol_QV9 | QVPKFHLPV | 1/3 | 0/1 | 1/1 |
| 8 | Env_TL9 | TVPWPNASL | 0/3 | 0/1 | 1/1 |
| 9 | Vpx_GL10 | GPPPPPPPGL | 0/3 | 0/1 | 0/1 |
| 10 | Pol_IL10 | IYPGIKTKHL | 0/3 | 1/1 | nt |
| 11 | Pol_SL8 | STPPLVRL | 0/3 | 1/1 | nt |
| 12 | Pol_YI9 | YTPKIVGGI | 2/3 | 0/1 | nt |
| 13 | Gag_LF8 | LAPVPIPF | 3/3 | 1/1 | nt |
| 14 | Rev_DL10 | DPPTNTPEAL | 0/3 | 0/1 | nt |
| 15 | Pol_GM10 | GSPAIFQYTM | 3/4 | 1/1 | nt |
| 16 | Gag_LA9 | LAPVPIPFA | 3/4 | 1/1 | nt |
| 17 | Vpx_HV10 | HLPRELIFQV | 0/3 | 0/1 | nt |
| 18 | Env_SI9 | SWPWQIEYI | 1/3 | 0/1 | nt |
| 19 | Pol_MI8 | MTPAERLI | 2/2 | 1/1 | 0/1 |
| 20 | Pol_QF10 | QMPRQTGGFF | 1/2 | 1/1 | 1/1 |
| 21 | Pol_GT9 | GSPAIFQYT | 0/2 | 0/1 | 0/1 |
| 22 | Gag_CM9 | CTPYDINQM | 0/2b | 1/1 | 1/1 |
| 23 | Vif_LL8 | LTPERGWL | 0/2 | 0/1 | 0/1 |
| 24 | Gag_VL8 | VVPGFQAL | 0/2 | 0/1 | 0/1 |
| 25 | Vif_VI8 | VTPDYADI | 1/2 | 1/1 | 0/1 |
| 26 | Vif_QA9 | QVPSLQYLA | 2/2 | 1/1 | 0/1 |
| 27 | Tat_TL8 | TTPESANL | 1/2 | 1/1 | 0/1 |
| 28 | Vif_RW9 | RIPERLERW | 0/2 | 0/1 | 0/1 |
| 29 | Env_AV8 | AAPTSAPV | 2/2 | nt | nt |
| 30 | Gag_QI9 | QNPIPVGNI | 1/2 | nt | nt |
| 31 | Env_YI9 | YVPCHIRQI | 0/2 | nt | nt |
| 32 | Env_ST9 | SSPPSYFQT | 0/2 | nt | nt |
| 33 | Vpx_II11 | IPPGNSGEETI | 0/2 | nt | nt |
| 34 | Gag_VT10 | VNPTLEEMLT | 2/2 | nt | nt |
| 35 | Env_IT9 | ITPIGLAPT | 0/2 | nt | nt |
| 36 | Gag_LW9 | LSPRTLNAW | 0/2 | nt | nt |
| 37 | Pol_SV9692 | SGPKTNIIV | 0/2 | nt | nt |
Since the Env_CL8 and Env_CL9 peptides overlap, we consider this a single positive response. Two-week in vitro-stimulated PBMC from three SIV-infected Mamu-A*01+ macaques were tested against each of the 37 identified peptides. The number of positive CTL responses per total number of assays tested for each peptide is listed. 51Cr release assay responses were considered positive if the percentage of specific lysis was >15% over that of an irrelevant peptide. Peptides were considered positive responders (shown in boldface) if 50% or more of the assays (were two or more assays were conducted) in a given animal were positive or if two or more assays from any of the three animals tested combined were positive. Effector-to-target cell ratios in the 51Cr release assays ranged from 20:1 to 50:1. Not all peptides were tested in all animals (nt).
The CTPYDINQM CTL epitope had escaped at the time of the assays in 95024.
Compared to the 14 peptides positively identified by the ELISPOT assays, recall in vitro memory CTL activity was detected against 7 of these peptides. Of the remaining seven ELISPOT-positive peptides, three yielded a positive CTL response in a single CTL assay (Table 4). Unfortunately, replicate CTL assays were not conducted to confirm these responses. In addition, nine peptides that were not consistently recognized in ELISPOT demonstrated positive recall memory CTL activity. Therefore, recall memory CTL activity was detectable against a significant number of the peptides which had yielded positive responses by ELISPOT.
Tetramer analysis of antigen-specific CD8+ cells.
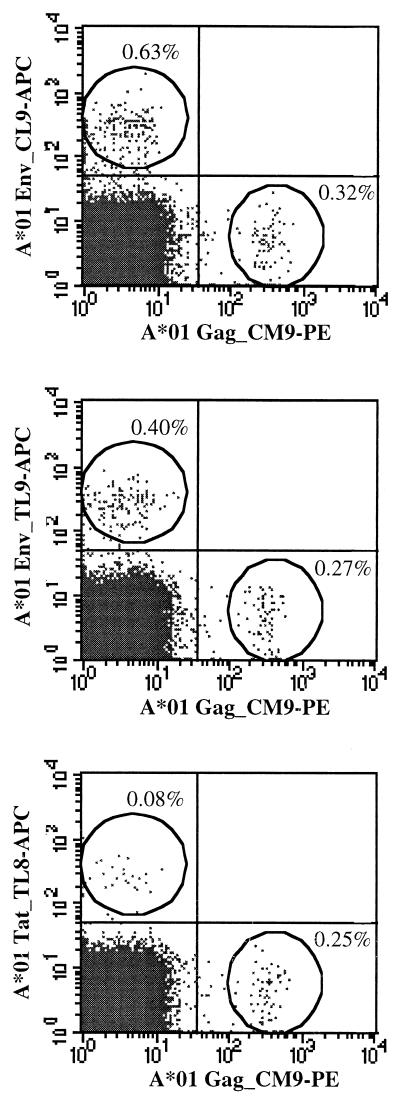
We next determined whether we could detect antigen-specific CD8+ responses against four of the newly defined peptides in fresh PBMC using Mamu-A*01 tetramers refolded with each of these peptides (Table 5). Responses were examined in three of the SIV-infected macaques (96031, 95114, and 95115) which had demonstrated positive ELISPOT responses to these peptides, as well as in a naive Mamu-A*01+ animal (96078). The Gag_CM9 tetramer detected levels of antigen-specific CD3 CD8α T lymphocytes ranging from 0.35 to 2.68% in the SIV-infected animals. Positive responses detected using the other four tetramers, however, were lower and ranged between 0.14 and 0.48%. The Env_TL9 (TVPWPNASL) and Env_CL8 (CAPPGYAL) tetramers detected good responses in all three animals, while responses with the Env_CL9 (CAPPGYALL) and Tat_TL8 (TTPESANL) tetramers varied considerably among animals. In animal 95114, staining with the Env_CL8 (0.48%) and Env_TL9 (0.40%) tetramers was actually higher than that for the Gag_CM9 tetramer (0.35%), although responses to these three peptides in ELISPOT varied between assays (Fig. 1B). Overall, the levels of tetramer staining correlated well with the levels of IFN-γ-producing SFCs detected against each peptide in the ELISPOT assays. After in vitro stimulation of PBMC from 95114 with either the Gag_CM9, Env_CL9, or Tat_TL8 peptide, the frequency of tetramer-staining cells specific for the corresponding peptides increased substantially (data not shown).
TABLE 5.
Tetramers detect responses against five epitopes in fresh PBMC of SIV-infected macaquesa
| Peptide no. | Peptide name | Sequence | % of T lymphocytes
|
|||
|---|---|---|---|---|---|---|
| 96031 (SIV+) | 95114 (SIV+) | 95115 (SIV+) | 96078 (naive) | |||
| 1 | Env_CL8 | CAPPGYAL | 0.41 | 0.48 | 0.24 | 0.02 |
| 4 | Env_CL9 | CAPPGYALL | 0.14 | 0.31 | 0.05 | 0.01 |
| 8 | Env_TL9 | TVPWPNASL | 0.34 | 0.40 | 0.25 | 0.02 |
| 22 | Gag_CM9 | CTPYDINQM | 2.68 | 0.35 | 1.49 | 0.06 |
| 27 | Tat_TL8 | TTPESANL | 0.26 | 0.15 | 0.06 | 0.04 |
Fresh PBMC from three SIV-infected Mamu-A*01+ animals were stained with Mamu-A*01 tetramers refolded with five of the peptides recognized by ELISPOT assays. PBMC from a naive Mamu-A*01+ animal (96078) served as a control. The frequencies of tetramer-staining cells are presented as a percentage of CD3 CD8α T lymphocytes. Significant responses are shown in boldface. Values represent levels detected in a single experiment. Responses were reproducible in two additional tetramer stains (data not shown). Background levels of tetramer staining in fresh PBMC from a naive Mamu-A*01+ animal (96078) were below 0.06%.
To determine whether any of these tetramers were cross-reacting and staining the same population of lymphocytes, we conducted double stains using the Gag_CM9-PE tetramer and three of the other four APC-labeled Mamu-A*01 tetramers. PBMC from animal 95114 were selected because this animal elaborated responses to each of these peptides. In each case, the Gag_CM9-PE tetramer stained a distinctly separate population of CD3 CD8α T lymphocytes than the other APC-labeled Mamu-A*01 tetramers (Fig. 2). The levels of tetramer-positive cells differ from those listed in Table 5 because the stains were conducted at different times postinfection. To confirm these results, we combined tetramer staining with intracellular staining for IFN-γ to measure the ability of different populations of cells to respond after stimulation with specific peptides through the production of IFN-γ. To accomplish this, we used a CTL line cocultured with the Gag_CM9 and Gag_LA9 (LAPVPIPFA) peptides. As expected, a subset of cells from this culture stained positive with the Gag_CM9 tetramer. Stimulation of these cells with the Gag_LA9 peptide induced a separate Gag_CM9 tetramer-negative population of cells to produce IFN-γ (data not shown). Similarly, while stimulation of a Gag_CM9-specific in vitro CTL line with the Gag_CM9 peptide induced 97% of the culture to produce IFN-γ, stimulation with six other Mamu-A*01-bound peptides induced less than 0.3% of this culture to produce IFN-γ (data not shown). Thus, lymphocytes reactive with the Gag_CM9 epitope do not recognize other Mamu-A*01-bound peptides.
FIG. 2.
Mamu-A*01 tetramers refolded with individual peptides stain unique populations of lymphocytes. The specificity of the tetramers was assessed through double staining of fresh PBMC (animal 95114) with the Gag_CM9-PE-labeled tetramer and one of three APC-labeled Mamu-A*01 tetramers. Each of the tetramers stained a unique population of CD3+ CD8+ T lymphocytes, with no cells staining with more than one tetramer. Background levels of tetramer staining in fresh PBMC from a naive Mamu-A*01+ animal (96078) were below 0.06%.
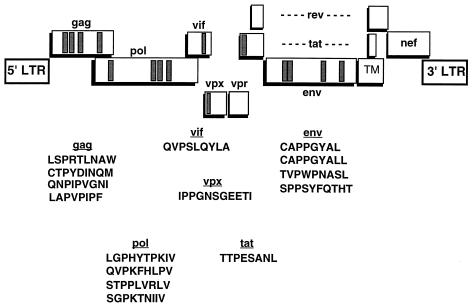
In summary, ELISPOT, CTL assays, and tetramer staining were able to identify a total of 14 Mamu-A*01-restricted SIV CTL epitopes (Table 6). Although none of the peptides that bound to Mamu-A*01 were found in Nef or Vpr, the positive peptides were distributed relatively uniformly throughout the remaining SIV proteins (Fig. 3).
TABLE 6.
Summary of ELISPOT, CTL, and tetramer responses against 37 Mamu-A*01-bound SIV-derived peptidesa
| Peptide no. | Peptide name | Sequence | ELISPOT | CTL | Tetramer |
|---|---|---|---|---|---|
| 1 | Env_CL8 | CAPPGYAL | + | + | + |
| 2 | Pol_LV10 | LGPHYTPKIV | + | + | |
| 3 | Pol_EA11 | EAPQFPHGSSA | − | − | |
| 4 | Env_CL9 | CAPPGYALL | + | + | + |
| 5 | Env_ST10 | SPPSYFQTHT | − | + | |
| 6 | Pol_SV9621 | STPPLVRLV | + | − | |
| 7 | Pol_QV9 | QVPKFHLPV | + | + | |
| 8 | Env_TL9 | TVPWPNASL | + | − | + |
| 9 | Vpx_GL10 | GPPPPPPPGL | − | − | |
| 10 | Pol_IL10 | IYPGIKTKHL | − | − | |
| 11 | Pol_SL8 | STPPLVRL | − | − | |
| 12 | Pol_YI9 | YTPKIVGGI | − | + | |
| 13 | Gag_LF8 | LAPVPIPF | + | + | |
| 14 | Rev_DL10 | DPPTNTPEAL | − | − | |
| 15 | Pol_GM10 | GSPAIFQYTM | − | + | |
| 16 | Gag_LA9 | LAPVPIPFA | − | + | |
| 17 | Vpx_HV10 | HLPRELIFQV | − | − | |
| 18 | Env_SI9 | SWPWQIEYI | − | − | |
| 19 | Pol_MI8 | MTPAERLI | − | + | |
| 20 | Pol_QF10 | QMPRQTGGFF | − | + | |
| 21 | Pol_GT9 | GSPAIFQYT | − | − | |
| 22 | Gag_CM9 | CTPYDINQM | + | + | + |
| 23 | Vif_LL8 | LTPERGWL | − | − | |
| 24 | Gag_VL8 | VVPGFQAL | − | − | |
| 25 | Vif_VI8 | VTPDYADI | − | + | |
| 26 | Vif_QA9 | QVPSLQYLA | + | + | |
| 27 | Tat_TL8 | TTPESANL | + | + | + |
| 28 | Vif_RW9 | RIPERLERW | − | − | |
| 29 | Env_AV8 | AAPTSAPV | − | + | |
| 30 | Gag_QI9 | QNPIPVGNI | + | − | |
| 31 | Env_YI9 | YVPCHIRQI | − | − | |
| 32 | Env_ST9 | SSPPSYFQT | + | − | |
| 33 | Vpx_II11 | IPPGNSGEETI | + | − | |
| 34 | Gag_VT10 | VNPTLEEMLT | − | + | |
| 35 | Env_IT9 | ITPIGLAPT | − | − | |
| 36 | Gag_LW9 | LSPRTLNAW | + | − | |
| 37 | Pol_SV9692 | SGPKTNIIV | + | − |
Since the Env_CL8 and Env_CL9 peptides overlap, we consider this a single positive response. Positive responses (+) in ELISPOT, 51Cr release assays, and tetramer stains are indicated. Fourteen Mamu-A*01-bound SIV-derived peptides were considered positive responders (bold) based on results from ELISPOT assays and confirmed in the majority of cases by CTL assays and tetramer staining.
FIG. 3.
Locations of Mamu-A*01-bound peptides recognized in SIV-infected rhesus macaques. Boxes within each of the proteins correspond to the position of the Mamu-A*01-restricted CTL epitopes which were identified by ELISPOT or CTL assays.
DISCUSSION
Herein, we report the identification of 14 SIV-derived CTL epitopes restricted by the same rhesus macaque MHC class I molecule, Mamu-A*01. These results have significance for our basic understanding of the phenomenon of immunodominance (78) as well as for vaccine development. The description of 14 epitopes bound by Mamu-A*01 and recognized by CD8+ lymphocytes from SIV-infected macaques demonstrates that the CTL repertoire against SIV is very broad (in the sense of multiple antigens being recognized) and multispecific (in the sense of multiple epitopes being recognized within the same antigen). Given that an animal can simultaneously recognize up to 15 different peptides bound by a single MHC class I molecule (animal 95114), the repertoire of SIV-derived epitopes could well exceed 100 different specificities in heterozygous individuals, in which up to six different MHC class I molecules are expressed. It is possible that this unsuspected and unprecedented breadth of repertoire is unusual and restricted to the Mamu-A*01 MHC class I molecule in the rhesus macaque. Clues from the literature, however, suggest that this might not be the case. In HIV-infected individuals, responses have been described against six different epitopes bound by HLA-B*5101 (67). Similarly, up to five different HLA-B*3501-bound peptides can be recognized by HLA-B*3501 individuals (61, 66), and 11 different HLA-A*2402-restricted HIV-1 CTL epitopes have been described (30). Furthermore, CTL responses against as many as 13 different peptides have already been described in HIV-infected patients (14) and against as many as five peptides in an SIV-infected macaque (23). Data hinting at the possibility that the CTL response is indeed broad and multispecific for other pathogens have also recently been obtained for human hepatitis B virus (48, 52), Epstein-Barr virus (14), and malaria (20) infections. In this context, it is of great interest to note that only chimpanzees that made broad, polyspecific CTL responses to multiple hepatitis C virus-derived epitopes were able to clear the virus (13). Similarly, the polyclonality of the anti-HIV CD8+ response in HIV-infected patients correlated with the levels of CD4 counts (14). Together, these studies suggest that a broad CTL response may be effective at controlling virus replication.
The accumulated evidence suggests that in humans and primates in general, immunodominance is not as strict as originally portrayed. Rather, a complex pattern of multispecific responses, in which T cells specific for many different epitopes coexist, is starting to emerge. Depending on timing, disease course, and the particular individual studied, the immunodominance of particular peptides may fluctuate. For example, it is interesting that in animals 95024 and 95114, the Gag_CM9 epitope was not the most immunodominant, and many other peptides were better recognized. In one of these animals (95024), this was likely due to escape in the Gag_CM9 epitope which had acquired a position 182 T→A mutation at the time of the assay (data not shown). Additionally, tetramer staining showed that lymphocytes from animal 95114 bearing T-cell receptors specific for the Env_CL8 and Env_TL9 epitopes were present at higher frequencies than the Gag_CM9-reactive lymphocytes (Table 5). These data suggest that the previously described Gag_CM9 epitope may not be the most immunodominant epitope in all SIV-infected Mamu-A*01+ animals. This current study, because of the identification of a large number of well-defined epitopes, will enable future studies to address the immunological basis of these different patterns of immunodominance and their potential significance in terms of disease pathogenesis.
Two other groups have independently confirmed that three of the epitopes described herein are recognized in SIV-infected, Mamu-A*01+ animals. The Env_CL9 (CAPPYGALL) and Env_TL9 (TVPWPNETL) (note the A→E and S→T substitutions in the SIVsmE660 peptide) epitopes have been shown to be recognized when pulsed onto Mamu-A*01/721.221 transfectants in SIVsmE660-infected rhesus macaques (25). Similarly, the Pol_SV9 (STPPLVRLV) epitope was recognized by CTL from an SIVmac251-infected Mamu-A*01+ rhesus macaque (22). Interestingly, tetramer analysis revealed that there were far fewer CD8+ lymphocytes recognizing this CTL epitope than the Gag_CM9 epitope (22), suggesting that this new epitope is subdominant. Thus, two other groups have validated the results of our approach to epitope discovery in SIV-infected rhesus macaques.
Vaccination with multiple CTL epitopes encoded by experimental minigenes increases the cell surface density of peptide-MHC class I complexes, inducing a more robust epitope-specific primary CD8+ response compared to vaccination with the entire proteins (6, 18). Additional advantages of the multiple-epitope approach include the potential for focusing immune responses against conserved epitopes and for generating responses simultaneously against multiple epitopes. In this context, it is noteworthy that several of the newly described CTL epitopes are found in many different conserved regions of the virus (data not shown). Simultaneous vaccination against a broad range of CTL epitopes may also reduce the possibility that escape will occur by limiting the amount of early virus replication.
The rhesus macaque is the only cost-effective animal model to address the design of a CTL-based, multiepitope vaccine against HIV. While HLA transgenic mice can be vaccinated with epitope constructs encoding HIV peptides (1, 77), they cannot be challenged with a virus similar to HIV. The definition of 10 new Mamu-A*01 epitopes, in addition to the four previously defined Mamu-A*01 epitopes (2, 22, 25, 44), is an important step in analyzing whether vaccination with multiepitope vaccines can induce protective immunity against HIV in humans. In practical terms, this study underlines the power of a rational approach to epitope identification based on the combined use of motif analysis, in vitro binding assays utilizing purified MHC molecules, and functional ELISPOT, CTL, and tetramer analyses. While similar approaches to define additional CTL epitopes in influenza virus, hepatitis B virus, and HIV have been successful in both mice and humans (7, 40, 52, 71), this is the first validation of this approach in SIV-infected rhesus macaques. It should, however, be noted that our approach might not identify all CTL epitopes, especially those bound at lower affinity. We have now determined motifs for four other rhesus MHC class I molecules (21, 23) and are in the process of applying a similar approach to define additional CTL epitopes bound by each of these newly defined MHC class I molecules.
In conclusion, besides shedding new light on the degree of complexity involved in anti-SIV-specific CD8 responses, this study represents an important step toward facilitating the testing of the multiepitope approach in the SIV model of HIV infection in humans.
ACKNOWLEDGMENTS
T.M.A. and B.R.M. contributed equally to this work.
We thank Rafi Ahmed for helpful discussions and the Immunology and Virology Core Laboratory at WRPRC for infection and monitoring of rhesus macaques. We also thank Lettie Smith for help in preparation of the manuscript.
D.I.W. is an Elizabeth Glaser scientist. This work was supported by grants R44 AI38081, R01 AI41913, R01 AI46366, and RR00167 and a Cremer Scholarship from the Department of Pathology, UW Madison (B.R.M.).
REFERENCES
- 1.Alexander J, Oseroff C, Sidney J, Wentworth P, Keogh E, Hermanson G, Chisari F V, Kubo R T, Grey H M, Sette A. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J Immunol. 1997;159:4753–4761. [PubMed] [Google Scholar]
- 2.Allen T M, Sidney J, Delguercio M F, Glickman R L, Lensmeyer G L, Wiebe D A, Demars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 3.Allen T M, Vogel T U, Fuller D H, Mothe B R, Steffen S, Boyson J E, Shipley T, Fuller J, Hanke T, Sette A, Altman J D, Moss B, McMichael A J, Watkins D I. Induction of AIDS virus-specific CTL activity in fresh, unstimulated PBL from rhesus macaques vaccinated with a DNA prime/MVA boost regimen. J Immunol. 2000;164:4968–4978. doi: 10.4049/jimmunol.164.9.4968. [DOI] [PubMed] [Google Scholar]
- 4.Allen T M, Watkins D I. SIV and SHIV CTL epitopes identified in macaques, p. IV 8–13 ( http://hiv-web.lanl.gov/immunology/) In: Korber B, editor. HIV molecular immunology database 1998. Los Alamos, N. Mex: Los Alamos National Laboratory; 1998. [Google Scholar]
- 5.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 6.Anton L C, Yewdell J W, Bennink J R. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J Immunol. 1997;158:2535–2542. [PubMed] [Google Scholar]
- 7.Bertoni R, Sidney J, Fowler P, Chestnut R W, Chisari F V, Sette A. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J Clin Investig. 1997;100:503–513. doi: 10.1172/JCI119559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrow P, Lewicki H, Wei X P, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B A, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 10.Boyson J E, Shufflebotham C, Cadavid L F, Urvater J A, Knapp L A, Hughes A L, Watkins D I. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- 11.Brodie S J, Lewinsohn D A, Patterson B K, Jiyamapa D, Krieger J, Corey L, Greenberg P D, Riddell S R. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat Med. 1999;5:34–41. doi: 10.1038/4716. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti L, Guyader M, Alizon M, Daniel M D, Desrosiers R C, Tiollais P, Sonigo P. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature. 1987;328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- 13.Cooper S, Erickson A L, Adams E J, Kansopon J, Weiner A J, Chien D Y, Houghton M, Parham P, Walker C M. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 14.Dalod M, Dupuis M, Deschemin J C, Sicard D, Salmon D, Delfraissy J F, Venet A, Sinet M, Guillet J G. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8+ responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 16.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 17.De Maria A, Cirillo C, Moretta L. Occurrence of human immunodeficiency virus type 1 (HIV-1)-specific cytolytic T cell activity in apparently uninfected children born to HIV-1-infected mothers. J Infect Dis. 1994;170:1296–1299. doi: 10.1093/infdis/170.5.1296. [DOI] [PubMed] [Google Scholar]
- 18.Deng Y P, Yewdell J W, Eisenlohr L C, Bennink J R. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J Immunol. 1997;158:1507–1515. [PubMed] [Google Scholar]
- 19.Desrosiers R C, Wyand M S, Kodama T, Ringler D J, Arthur L O, Sehgal P K, Letvin N L, King N W, Daniel M D. Vaccine protection against simian immunodeficiency virus infection. Proc Natl Acad Sci USA. 1989;86:6353–6357. doi: 10.1073/pnas.86.16.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolan D L, Hoffman S L, Southwood S, Wentworth P A, Sidney J, Chesnut R W, Keogh E, Appella E, Nutman T B, Lal A A, Gordon D M, Oloo A, Sette A. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. 1997;7:97–112. doi: 10.1016/s1074-7613(00)80513-0. [DOI] [PubMed] [Google Scholar]
- 21.Dzuris J L, Sidney J, Appella E, Chesnut R W, Watkins D I, Sette A. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J Immunol. 2000;164:283–291. doi: 10.4049/jimmunol.164.1.283. [DOI] [PubMed] [Google Scholar]
- 22.Egan M A, Kuroda M J, Schmitz J E, Charini W A, Lord C I, Forman M A, Letvin N L. Use of major histocompatibility complex class I/peptide/β2M tetramers to quantitate CD8+ cytotoxic T lymphocytes specific for dominant and nondominant viral epitopes in simian-human immunodeficiency virus-infected rhesus monkeys. J Virol. 1999;73:5466–5472. doi: 10.1128/jvi.73.7.5466-5472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans D T, O'Connor D H, Jing P, Dzuris J L, Sydney J, da Silva J, Allen T M, Horton H, Venham J E, Rudersdorf R A, Pauza C D, Bontrop R E, DeMars R, Sette A, Hughes A L, Watkins D I. Virus-specific CTL responses select for amino acid variation in SIV Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 24.Franchini G, Gurgo C, Guo H G, Gallo R C, Collalti E, Fargnoli K A, Hall L F, Wong-Staal F, Reitz M S., Jr Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328:539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- 25.Furchner M, Erickson A L, Allen T M, Watkins D I, Sette A, Johnson P R, Walker C M. The simian immunodeficiency virus envelope glycoprotein contains two epitopes presented by the Mamu-A*01 class I molecule. J Virol. 1999;73:8035–8039. doi: 10.1128/jvi.73.10.8035-8039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goulder P J R, Phillips R E, Colbert R A, McAdam S, Ogg G, Nowak M A, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowlandjones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 27.Greenwood F, Hunter W, Glover J. The preparation of 131I-labeled human growth hormone of high specific radioactivity. Biochem J. 1963;89:114. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanke T, Blanchard T J, Neumann V C, Boyson J E, Sweeney P, Allen T M, Watkins D I, Smith G L, McMichael A. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques using a DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J Virol. 1999;73:7524–7532. doi: 10.1128/jvi.73.9.7524-7532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch V M, Goldstein S, Hynes N A, Elkins W R, London W T, Zack P M, Montefiori D, Johnson P R. Prolonged clinical latency and survival of macaques given a whole inactivated simian immunodeficiency virus vaccine. J Infect Dis. 1994;170:51–59. doi: 10.1093/infdis/170.1.51. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda-Moore Y, Tomiyama H, Ibe M, Oka S, Miwa K, Kaneko Y, Takiguchi M. Identification of a novel HLA-A24-restricted cytotoxic T-lymphocyte epitope derived from HIV-1 Gag protein. AIDS. 1998;12:2073–2074. doi: 10.1097/00002030-199815000-00020. [DOI] [PubMed] [Google Scholar]
- 31.Jaeger E E, Bontrop R E, Lanchbury J S. Nucleotide sequences, polymorphism and gene deletion of T cell receptor beta-chain constant regions of Pan troglodytes and Macaca mulatta. J Immunol. 1993;151:5301–5309. [PubMed] [Google Scholar]
- 32.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L Q, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson R P. Macaque models for AIDS vaccine development. Curr Opin Immunol. 1996;8:554–560. doi: 10.1016/s0952-7915(96)80046-x. [DOI] [PubMed] [Google Scholar]
- 34.Kagi D, Ledermann B, Burki K, Zinkernagel R M, Hengartner H. Molecular mechanisms of lymphocyte-mediated cytotoxicity and their role in immunological protection and pathogenesis in vivo. Annu Rev Immunol. 1996;14:207–232. doi: 10.1146/annurev.immunol.14.1.207. [DOI] [PubMed] [Google Scholar]
- 35.King N W, Chalifoux L V, Ringler D J, Wyand M S, Sehgal P K, Daniel M D, Letvin N L, Desrosiers R C, Blake B J, Hunt R D. Comparative biology of natural and experimental SIVmac infection in macaque monkeys: a review. J Med Primatol. 1990;19:109–118. [PubMed] [Google Scholar]
- 36.Klatzmann D, Barre-Sinoussi F, Nugeyre M T, Danquet C, Vilmer E, Griscelli C, Brun-Veziret F, Rouzioux C, Gluckman J C, Chermann J C, Montagnier L. Selective tropism of lymphadenopathy associated virus (LAV) for helper-inducer T lymphocytes. Science. 1984;225:59–63. doi: 10.1126/science.6328660. [DOI] [PubMed] [Google Scholar]
- 37.Klinman D M. ELISPOT assay to detect cytokine-secreting murine and human cells. Curr Protocols Immunol. 1994;6.19:1–8. doi: 10.1002/0471142735.im0619s83. [DOI] [PubMed] [Google Scholar]
- 38.Knapp L A, Lehmann E, Piekarczyk M S, Urvater J A, Watkins D I. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens. 1997;50:657–661. doi: 10.1111/j.1399-0039.1997.tb02927.x. [DOI] [PubMed] [Google Scholar]
- 39.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundu S K, Dupuis M, Sette A, Celis E, Dorner F, Eibl M, Merigan T C. Role of preimmunization virus sequences in cellular immunity in HIV-infected patients during HIV type 1 MN recombinant gp160 immunization. AIDS Res Hum Retroviruses. 1998;14:1669–1678. doi: 10.1089/aid.1998.14.1669. [DOI] [PubMed] [Google Scholar]
- 41.Kuroda M J, Schmitz J E, Barouch D H, Craiu A, Allen T M, Sette A, Watkins D I, Forman M A, Letvin N L. Analysis of gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehner T, Wang Y, Cranage M, Bergmeier L A, Mitchell E, Tao L, Hall G, Dennis M, Cook N, Brookes R, Klavinskis L, Jones I, Doyle C, Ward R. Protective mucosal immunity elicited by targeted iliac lymph node immunization with a subunit SIV envelope and core vaccine in macaques. Nat Med. 1996;2:767–775. doi: 10.1038/nm0796-767. [DOI] [PubMed] [Google Scholar]
- 43.Matano T, Shibata R, Siemon C, Connors M, Lane H C, Martin M A. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller M D, Yamamoto H, Hughes A L, Watkins D I, Letvin N L. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol. 1991;147:320–329. [PubMed] [Google Scholar]
- 45.Mortara L, Letourneur F, Gras-Masse H, Venet A, Guillet J G, Bourgault-Villada I. Selection of virus variants and emergence of virus escape mutants after immunization with an epitope vaccine. J Virol. 1998;72:1403–1410. doi: 10.1128/jvi.72.2.1403-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossman S P, Bex F, Berglund P, Arthos J, O'Neil S P, Riley D, Maul D H, Bruck C, Momin P, Burny A, Fultz P N, Mullins J I, Liljestrom P, Hoover E A. Protection against lethal simian immunodeficiency virus SIVsmmPBj14 disease by a recombinant Semliki Forest virus gp160 vaccine and by a gp120 subunit vaccine. J Virol. 1996;70:1953–1960. doi: 10.1128/jvi.70.3.1953-1960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphey-Corb M, Martin L N, Davison-Fairburn B, Montelaro R C, Miller M, West M, Ohkawa S, Baskin G B, Zhang J Y, Putney S D, Allison A C, Eppstein D A. A formalin-inactivated whole SIV vaccine confers protection in macaques. Science. 1989;246:1293–1297. doi: 10.1126/science.2555923. [DOI] [PubMed] [Google Scholar]
- 48.Nayersina R, Fowler P, Guilhot S, Missale G, Cerny A, Schlicht H J, Vitiello A, Chestnut R, Person J L, Redeker A G, Chisari F V. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J Immunol. 1993;150:4659–4671. [PubMed] [Google Scholar]
- 49.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y Z, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 50.Pauza C D, Emau P, Salvato M S, Trivedi P, MacKenzie D, Malkovsky M, Uno H, Schultz K T. Pathogenesis of SIVmac251 after atraumatic inoculation of the rectal mucosa in rhesus monkeys. J Med Primatol. 1993;22:154–161. [PubMed] [Google Scholar]
- 51.Price D A, Goulder P J, Klenerman P, Sewell A K, Easterbrook P J, Troop M, Bangham C R, Phillips R E. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehermann B, Fowler P, Sidney J, Person J, Redeker A, Brown M, Moss B, Sette A, Chisari F V. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J Exp Med. 1995;181:1047–1058. doi: 10.1084/jem.181.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, Takiguchi M, Schultz T, McMichael A, Whittle H. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 55.Rowland-Jones S L, Dong T, Fowke K R, Kimani J, Krausa P, Newell H, Blanchard T, Ariyoshi K, Oyugi J, Ngugi E, Bwayo J, MacDonald K S, McMichael A J, Plummer F A. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Investig. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruppert J, Sidney J, Celis E, Kubo R T, Grey H M, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 57.Safrit J T, Andrews C A, Zhu T, Ho D D, Koup R A. Characterization of human immunodeficiency virus type 1-specific cytotoxic T lymphocyte clones isolated during acute seroconversion: recognition of autologous virus sequences within a conserved immunodominant epitope. J Exp Med. 1994;179:463–472. doi: 10.1084/jem.179.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8(+) lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 59.Sette A, Sidney J, del Guercio M F, Southwood S, Ruppert J, Dahlberg C, Grey H M, Kubo R T. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol Immunol. 1994;31:813–822. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 60.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast W M, Melief C J, Oseroff C, Yuan L, Ruppert J, Sidney J, del Guercio M F, Southwood S, Kubo R T, Chesnut R W, Grey H M, Chisari F V. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 61.Shiga H, Shioda T, Tomiyama H, Takamiya Y, Oka S, Kimura S, Yamaguchi Y, Gojoubori T, Rammensee H G, Miwa K, Takiguchi M. Identification of multiple HIV-1 cytotoxic T-cell epitopes presented by human leukocyte antigen B35 molecules. AIDS. 1996;10:1075–1083. [PubMed] [Google Scholar]
- 62.Sidney J, Southwood S, Oseroff C, del Guercio M-F, Sette A, Grey H M. Measurement of MHC/peptide interactions by gel filtration. Curr Protocols Immunol. 1998;1998:18.3.1–18.3.19. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- 63.Slierendregt B L, van Noort J T, Bakas R M, Otting N, Jonker M, Bontrop R E. Evolutionary stability of transspecies major histocompatibility complex class II DRB lineages in humans and rhesus monkeys. Hum Immunol. 1992;35:29–39. doi: 10.1016/0198-8859(92)90092-2. [DOI] [PubMed] [Google Scholar]
- 64.Soudeyns H, Paolucci S, Chappey C, Daucher M B, Graziosi C, Vaccarezza M, Cohen O J, Fauci A S, Pantaleo G. Selective pressure exerted by immunodominant HIV-1-specific cytotoxic T lymphocyte responses during primary infection drives genetic variation restricted to the cognate epitope. Eur J Immunol. 1999;29:3629–3635. doi: 10.1002/(SICI)1521-4141(199911)29:11<3629::AID-IMMU3629>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 65.Stott J, Almond N. Assessing animal models of AIDS. Nat Med. 1995;1:295–297. doi: 10.1038/nm0495-295. [DOI] [PubMed] [Google Scholar]
- 66.Tomiyama H, Miwa K, Shiga H, Moore Y I, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Evidence of presentation of multiple HIV-1 cytotoxic T lymphocyte epitopes by HLA-B*3501 molecules that are associated with the accelerated progression of AIDS. J Immunol. 1997;158:5026–5034. [PubMed] [Google Scholar]
- 67.Tomiyama H, Sakaguchi T, Miwa K, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum Immunol. 1999;60:177–186. doi: 10.1016/s0198-8859(98)00113-x. [DOI] [PubMed] [Google Scholar]
- 68.Trivedi P, Meyer K K, Streblow D N, Preuninger B L, Schultz K T, Pauza C D. Selective amplification of simian immunodeficiency virus genotypes after intrarectal inoculation of rhesus monkeys. J Virol. 1994;68:7649–7653. doi: 10.1128/jvi.68.11.7649-7653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsubota H, Lord C I, Watkins D I, Morimoto C, Letvin N L. A cytotoxic T lymphocyte inhibits acquired immunodeficiency syndrome virus replication in peripheral blood lymphocytes. J Exp Med. 1989;169:1421–1434. doi: 10.1084/jem.169.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Most R G, Murali-Krishna K, Whitton J L, Oseroff C, Alexander J, Southwood S, Sidney J, Chesnut R W, Sette A, Ahmed R. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology. 1998;240:158–167. doi: 10.1006/viro.1997.8934. [DOI] [PubMed] [Google Scholar]
- 71.Vitiello A, Marchesini D, Furze J, Sherman L A, Chestnut R W. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vitiello A, Yuan L, Chestnut R W, Sidney J, Southwood S, Farness P, Jackson M R, Peterson P A, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 73.Vogel T, Norley S, Beer B, Kurth R. Rapid screening for Mamu-A1-positive rhesus macaques using a SIVmac Gag peptide-specific cytotoxic T-lymphocyte assay. Immunology. 1995;84:482–487. [PMC free article] [PubMed] [Google Scholar]
- 74.Walker B D, Plata F. Cytotoxic T lymphocytes against HIV. AIDS. 1990;4:177–184. doi: 10.1097/00002030-199003000-00001. [DOI] [PubMed] [Google Scholar]
- 75.Walker C M, Moody D J, Stites D P, Levy J A. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 76.Watkins D I. MHC of nonhuman primates. Curr Top Microbiol Immunol. 1994;188:145–159. doi: 10.1007/978-3-642-78536-8_8. [DOI] [PubMed] [Google Scholar]
- 77.Woodberry T, Gardner J, Mateo L, Eisen D, Medveczky J, Ramshaw I A, Thomson S A, French R A, Elliot S L, Firat H, Lemonnier F A, Suhrbier A. Immunogenicity of a human immunodeficiency virus (HIV) polytope vaccine containing multiple HLA A2 HIV CD8+ cytotoxic T-cell epitopes. J Virol. 1999;73:5320–5325. doi: 10.1128/jvi.73.7.5320-5325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yewdell J W, Bennink J R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]