Abstract
Virus infection induces an antiviral response that is predominantly associated with the synthesis and secretion of soluble interferon. Here, we report that herpes simplex virus type 1 virions induce an interferon-independent antiviral state in human embryonic lung cells that prevents plaquing of a variety of viruses. Microarray analysis of 19,000 human expressed sequence tags revealed induction of a limited set of host genes, the majority of which are also induced by interferon. Genes implicated in controlling the intracellular spread of virus and eliminating virally infected cells were among those induced. Induction of the cellular response occurred in the absence of de novo cellular protein synthesis and required viral penetration. In addition, this response was only seen when viral gene expression was inhibited, suggesting that a newly synthesized viral protein(s) may function as an inhibitor of this response.
Mammalian cells respond to virus infection by launching a transcription program that generates an intracellular antiviral state. In many but not all cases, cells undergoing this response also synthesize and secrete alpha/beta interferon (IFN-α/β) (49), which renders neighboring uninfected cells resistant to virus infection. IFNs are pleiotropic cytokines that mediate antiviral and antiproliferative responses and modulate the immune system (42). IFN-α and -β and IFN-γ signal through distinct, yet related, pathways in a rapid and direct manner. Binding of IFN-α/β to its cell surface receptor induces the tyrosine kinases Tyk2 and JAK1 to phosphorylate STAT-1 and STAT-2, enabling these proteins to bind p48 and form the IFN-stimulated gene factor 3 (ISGF3) complex. This complex translocates to the nucleus, where it binds to the IFN-stimulated response element (ISRE) and activates transcription. Many IFN-stimulated genes (ISGs) encode proteins that contribute to the antiviral state. For example, the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) phosphorylates eIF-2α, resulting in inhibition of protein synthesis, and activated 2′→5′ oligoadenylate synthetase (OAS) produces 2-5A, which in turn activates RNase L, resulting in mRNA degradation (42).
ISGs can also be directly activated by dsRNA or virus infection in the absence of IFN (2, 44). These responses presumably act to limit virus replication in the first cells that are infected in a tissue or organism. IFN, dsRNA, and virus infection each utilize a different signaling pathway for induction of mRNA from an ISG coding for a protein with a molecular weight of 56,000 (ISG 56K) in human fibrosarcoma cells (11, 16, 50). The degree of overlap between these signaling pathways has yet to be precisely defined; however, they all appear to converge on the ISRE. Several viruses stimulate the formation of alternative ISRE-binding transcription complexes that are distinct from the ISGF3 induced by IFN. For example, Sendai virus induces a novel transcriptional activator complex composed of the IFN regulatory factor proteins IRF-3 and IRF-7, along with several transcriptional coactivator proteins, that binds the ISRE of the ISG 15K gene (50). Similarly, measles virus induces the C-X-C chemokine IFN-inducible protein 10 (IP-10) through the same ISRE as IFN-α, but with a different transcription factor (29). Human cytomegalovirus (HCMV) induces IFN-responsive RNAs in the absence of viral and cellular protein synthesis following binding of viral glycoprotein B (gB) to an unknown cell surface receptor (4, 53, 54). HCMV-induced activation of the ISG 54K gene is STAT independent and is mediated by a novel transcriptional activator complex that contains IRF3 (28).
Here, we studied the transcriptional response of human cells to infection with herpes simplex virus type 1 (HSV-1). HSV-1 is a large enveloped DNA virus composed of an icosahedral capsid surrounded by an amorphous tegument that contains proteins that become available to the virus immediately following penetration of the host cell (37). During the lytic cycle, HSV genes are expressed in a tightly regulated temporal cascade beginning with transcription of the immediate-early (IE) genes. The IE genes are activated by the virion-associated transactivator, VP16, through a specific sequence motif within their promoters (33). HSV-1 encodes five IE proteins: ICP-0, -4, -22, -27, and -47. The first four are nuclear regulators that activate expression of the viral early and late genes (37), while ICP47 blocks a host antigen presentation pathway (52).
We have previously reported the construction and characterization of KM110, an HSV-1 mutant bearing lesions that eliminate the transactivation functions of VP16 and ICP0 (26). KM110 is incapable of launching the lytic program of viral gene expression in most cell types, and human embryonic lung (HEL) fibroblasts survive infection with KM110, with no evidence of viral gene expression. Here we use the KM110 isolate to show that the HSV particle induces an IFN-independent antiviral state that protects cells from infection by several RNA and DNA viruses. The antiviral state is induced in the absence of viral gene expression. Microarray analysis of 19,000 human expressed sequence tags (ESTs) revealed induction of a limited set of host genes, many of which are also induced by IFN. Wild-type HSV-1 also induced the same set of cellular genes, but only when viral gene expression was inhibited. Thus, the HSV particle induces an IFN-independent cellular antiviral response that is subsequently disarmed following the onset of viral gene expression.
MATERIALS AND METHODS
Viruses and cells.
HEL, U2OS, and Vero cells, obtained from the American Type Culture Collection, were maintained in Dulbecco's minimal essential medium (DMEM) supplemented with 10% (HEL and U2OS) or 5% (Vero) fetal bovine serum (FBS). Vesicular stomatitis virus (VSV) and the HSV-1 strains KOS, d22lacZ (ICP22−) (23), N38 (ICP47−) (46), and ΔICP6 (ICP6−) (15) were propagated on Vero cells. HSV-1 strains n212 (ICP0−) (7), dlX3.1 (ICP0−) (39), V422 (VP16−) (21), and KM110 (VP16− ICP0−) (26) were propagated on U2OS cells in the presence of 3 mM hexamethylene bisacetamide (Sigma, St. Louis, Mo.). HSV-1 mutants bearing lesions in essential genes were grown on their respective complementing cell lines as follows: 5dl1.2 (ICP27−) was grown on V27 cells (35), d120 (ICP4−) was grown on E5 cells (10), K082 (gB−) (6) was grown on VB38 cells (Vero cells containing the HSV-1 gB gene under the control of its own promoter using histidinol as a selection marker; kindly provided by D. Johnson, Oregon Health Sciences University, Portland), and F-gDβ (gD− gI−) and F-US6kan (gD−) were grown on VD60 cells (18, 22). UV inactivation of HSV-1 was performed with a UV Stratalinker 2400 (Stratagene) for a period of 1 min. The treatment reduced viral titers by a factor of ∼104 (data not shown). KOS virions were purified by banding on a dextran gradient as previously described (26).
Plaque reduction assay.
HEL cells were seeded in 12-well dishes such that monolayers were completely confluent the next day. Monolayers were then mock infected or infected with the indicated virus at a multiplicity of infection (MOI) of 5 in serum-free DMEM for 1 h followed by replacement with DMEM containing 5% FBS. Universal IFN-α (Research Diagnostics, Inc.) was added to mock-infected samples at 1,000 U/ml. Twenty-four hours later, monolayers were inoculated with approximately 100 PFU of VSV, followed by replacement with DMEM containing 0.5% methylcellulose. Monolayers were fixed and stained 24 h later.
RNA extraction and Northern blot analysis.
Total cellular RNA was extracted from 100-mm-diameter dishes of infected cells by using Trizol (Gibco BRL) according to the manufacturer's instructions. Where indicated, cycloheximide (100 μg/ml) was added 1 h prior to infection and maintained continuously. Aliquots (5 μg) were subjected to electrophoresis as previously described (26). Membranes were hybridized to a 32P-labeled probe generated by random priming in ExpressHyb buffer (Clonetech) as specified by the manufacturer. The ISG 56K and stress 70 chaperone probes were derived from IMAGE Consortium clones 325364 and 27801, respectively.
DNA microarrays.
DNA microarrays comprising about 19,000 human EST clones were printed at the Microarray Centre (Ontario Cancer Institute, Toronto, Ontario, Canada) on CMT-GAPS aminosaline-coated glass slides (Corning, N.Y.) with a 32-pin contact arrayer (SDDC II; Engineering Services, Inc.). The genes were arrayed in duplicate on two slides, each bearing 9,500 clones spotted in duplicate. Detailed information on the layout of the microarrays can be found on the website of the Microarray Centre (http://www.oci.utoronto.ca/services/microarray).
Microarray analysis.
Total cellular RNA was harvested from 107 cells with 7.5 ml of Trizol. For each microarray, 10 μg of total RNA was reverse transcribed with 400 U of SuperScript II (Gibco, Life Technologies) in a total reaction volume of 40 μl. The reverse transcription was primed with an AncT primer (T20VN; Sigma Genosys) and performed in the presence of dATP, dGTP, and dTTP (Pharmacia; final concentration of 168 μM each); dCTP (Pharmacia; final concentration of 50 μM); and Cy3-dCTP or Cy5-dCTP (NEN; final concentration of 50 μM). Twenty units of RNasin (Promega) was added to each reaction mixture. The mixture (minus the enzyme) was heated at 65°C for 5 min and then at 42°C for 5 min. The reverse transcriptase was added, and the reaction mixture was incubated at 42°C for 2 h. The reverse transcription was stopped with 6.25 mM EDTA, and the RNA template was degraded by the addition of 0.5 N NaOH, followed by incubation at 65°C for 20 min. The mixture was neutralized by the addition of 0.5 M acetic acid, and the labeled cDNA was precipitated by adding 1 volume of isopropanol and incubating on ice for 30 min. After rinsing with 70% ethanol, the labeled cDNA was resuspended in 3 μl of DNase-free, RNase-free water (Sigma). In order to eliminate labeling biases, two pairs of slides were hybridized for each pair of samples: one pair in which the control RNA was labeled with Cy3 and the experiment RNA was labeled with Cy5 and one pair in which the control RNA was labeled with Cy5 and the experiment RNA was labeled with Cy3.
For hybridization, 3 μl of purified Cy3-labeled cDNA and 3 μl of purified Cy5-labeled cDNA were added to 75 μl of DIG Easy hybridization buffer (Boehringer Mannheim). Two microliters of yeast tRNA (Sigma; 10 mg/ml) and 2 μl of single-stranded salmon sperm DNA (Sigma; 10 mg/ml) were added to the hybridization mixture, and the solution was heated at 65°C for 2 min. This solution was then carefully pipetted between two microarrays (parts 1 and 2) placed face to face. The slides were incubated at 37°C in a humid hybridization chamber for 8 to 12 h. Before scanning, the slides were washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (three times for 15 min at 50°C), rinsed in 0.1× SSC (three times for 5 min each at room temperature), and dried by centrifugation. The arrays were read on a laser confocal scanner (ScanArray 4000; GSI Lumonics), and the images obtained were quantified by using the QuantArray 2.0 software (GSI Lumonics). Normalization of the raw data and analysis of the data sets were performed with an algorithm developed in house (A. B. Goryachev et al., unpublished data).
Production and quantitation of glycoprotein-deficient viruses.
Vero cells (2 × 107) were inoculated with KOS, F-gDβ, F-US6kan, or K082 at an MOI of 5. Two days later, cells were harvested and then spun at 1,400 × g for 7 min, and the pellets were resuspended in 1 ml of serum-free DMEM. Following three freeze-thaw cycles and sonication, samples were respun to pellet cellular debris, and the supernatant was harvested. The titers of the resulting virus stocks were determined on Vero cell monolayers. Titers of F-gDβ, F-US6kan, and K082 were reduced by a factor of ∼104 compared to KOS (data not shown). In order to standardize the number of viral particles used in subsequent experiments, particles were counted in the presence of a fixed amount of 90-nm-diameter polystyrene latex particles (Dow Diagnostics) by using a Philips model 410 transmission electron microscope. The volume of F-gDβ, F-US6kan, or K082 virus stock used was adjusted accordingly in order to inoculate cells with the same number of viral particles calculated for a specific MOI for KOS.
RESULTS
HSV-1 virions induce an antiviral state in the absence of de novo viral gene expression.
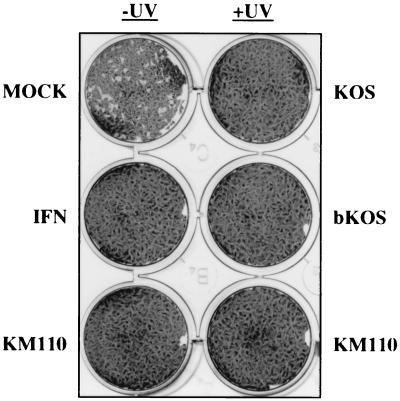
The HSV-1 mutant KM110 bears mutations that inactivate the transactivation functions of VP16 and ICP0 and therefore cannot launch the lytic program of viral gene expression (26). HEL fibroblasts infected with KM110 display no evidence of viral gene expression and survive for at least 10 days in culture after virus inoculation. We asked if cells infected previously with KM110 displayed altered susceptibility to subsequent virus infection. HEL monolayers were either mock infected or infected with 5 PFU of KM110 per cell and then superinfected 24 h later with ca. 100 PFU of wild-type HSV-1 KOS, VSV, or vaccinia virus per monolayer. All three superinfecting viruses produced the expected number of plaques on mock-infected monolayers, but no plaques were observed on HEL monolayers that had been previously infected with KM110 (Fig. 1 [only the data obtained with VSV are shown]). KM110 retained the ability to block VSV plaque formation even when its genome was inactivated by irradiation with UV light, confirming that development of resistance does not require expression of the KM110 genome. UV-inactivated wild-type HSV-1 strain KOS also blocked plaque formation by all three viruses, showing that the antiviral effect is not specific to the KM110 mutant (Fig. 1). KOS virions retained UV-resistant antiviral activity following purification by banding on a dextran gradient, indicating that the effect is induced by virions rather than a soluble factor present in the virus inoculum (Fig. 1). HEL cells did not develop resistance to VSV following exposure to medium harvested from KM110-infected cells (data not shown), arguing that HSV-1 virions do not induce the production of functional levels of IFN or other factors capable of inducing an antiviral state. Taken in combination, these data suggest that HSV-1 virions are capable of inducing a nonspecific, IFN-independent antiviral state in the absence of de novo viral gene expression.
FIG. 1.
Induction of an antiviral state by HSV-1 KM110 and UV-inactivated wild-type KOS. HEL monolayers were mock infected (mock, IFN) or infected with KM110 (with [+] or without [−] UV inactivation) or UV-inactivated KOS at an MOI of 5. IFN-α was added at 1,000 U/ml following the infection. The next day, ∼100 PFU of VSV was added to each well, and monolayers were stained 24 h later. A similar inhibition of plaquing was observed for HSV-1 KOS and vaccinia virus (data not shown).
HSV-1 virions induce expression of host genes involved in antiviral defense.
The foregoing data suggested that HSV-1 virions induce a host antiviral defense mechanism in the absence of viral gene expression. We therefore asked if HSV-1 virions induce expression of specific cellular genes, using DNA microarrays that comprise over 19,000 unique human genes or ESTs. Duplicate cultures of HEL cells were mock infected, infected with virus (KM110, KOS, or UV-inactivated KOS), or treated with IFN-α. Total cellular RNA isolated 24 h later was used to generate cDNA for DNA microarray analysis. Approximately 10,000 of the 19,000 genes represented on the microarrays were expressed at levels enabling detection and quantification with statistical confidence (A. B. Goryachev et al., unpublished data). Genes whose expression levels changed more than a factor of 2 (up or down) in at least one of two experiments between the infected or IFN-treated and mock-infected cells were identified (Table 1). Both KM110 and UV-inactivated KOS increased the levels of expression of a small set of cellular genes (33 and 32, respectively). The two sets were highly related, with 27 genes common to both. Strikingly, 20 of these 27 shared genes were also induced by IFN-α. Most of the genes thus identified that were not common to all of the IFN, KM110, and UV-inactivated KOS data sets had induction ratios close to the cutoff for inclusion (and/or scored as positive in only one of the duplicates).
TABLE 1.
Microarray analysis of IFN-treated and HSV-1-infected HEL cells
| Accession no. | Gene or gene product and description | Relative change in level of expression
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| KOS
|
UV-treated KOS
|
KM110
|
IFN-α
|
||||||
| Set A | Set B | Set A | Set B | Set A | Set B | Set A | Set B | ||
| AA193601 | MX1, myxovirus (influenza virus) resistance 1, homolog of murine (IFN-inducible protein p78) | 20.06 | 7.68 | 10.08 | 7.40 | 16.55 | 7.44 | ||
| R82716 | ISG15, IFN-stimulated protein, 15 kDa | 18.33 | 16.53 | 20.46 | 11.83 | 16.55 | 13.88 | ||
| N63988 | EST | 13.69 | 9.1 | 6.25 | 6.21 | ||||
| W52254 | IFIT1, IFN-induced protein 56 | 9.83 | 12.05 | 4.93 | 7.72 | 6.16 | 4.57 | ||
| AA152305 | SCYB10, small inducible cytokine subfamily B (Cys-X-Cys), member 10 | 7.63 | 2.81 | 2.42 | 2.58 | ||||
| W49782 | MX1, myxovirus (influenza virus) resistance 1, homolog of murine (IFN-inducible protein p78) | 6.45 | 4.59 | 3.78 | 3.55 | —a | 5.17 | ||
| AA035361 | STAT1, signal transducer and activator of transcription 1, 91 kDa | 6.33 | 4.13 | 3.94 | 3.48 | 4.37 | 3.08 | ||
| T95113 | Homo sapiens cig5 mRNA, partial sequence | 5.57 | 2.18 | 4 | 1.61 | 2.01 | 1.4 | ||
| R76719 | EST, weakly similar to GEF-2 protein (H. sapiens) | 5.38 | 4.76 | 6.53 | 4.27 | 8.2 | 6.78 | ||
| R09187 | EST | 5.18 | 2.63 | 4.87 | 1.82 | 2.6 | 2.29 | ||
| AA035024 | MX2, myxovirus (influenza virus) resistance 2, homolog of murine | 4.68 | 2.03 | 2.9 | 1.92 | ||||
| T92814 | EST, homolog to H. sapiens cig41 | 4.5 | 2.91 | 3.41 | 3.07 | 2.54 | 2.28 | ||
| H49854 | IFITM1, IFN-induced transmembrane protein 1 (9–27) | 4.43 | 4.49 | 5.69 | 4.41 | 6.69 | 5.38 | ||
| AA046075 | EST | 3.9 | 2.91 | 2.38 | 1.92 | 2.33 | 2.15 | ||
| R59506 | EST, homologue to H. sapiens cyclin-E binding protein | 3.85 | 2.13 | 2.92 | 1.64 | ||||
| AA043535 | IMAGE-487558 | 3.31 | 1.98 | 2.34 | 2.14 | 2.46 | 2.15 | ||
| W47619 | OAS3, 2′→5′ oligoadenylate synthetase 3 | 2.99 | 2.13 | 2.73 | 2.26 | 2.13 | 2.46 | ||
| N63887 | EST, weakly similar to TYKi protein (Mus musculus) | 2.81 | 1.75 | 2.21 | 1.67 | ||||
| N43008 | SP100, nuclear antigen Sp100 | 2.81 | 1.91 | 2.23 | 1.77 | 2.38 | 2.38 | ||
| T74462 | ABCB2, ATP-binding cassette, subfamily B (MDR/TAP), member 2 | 2.77 | 2.03 | 2.44 | 1.4 | 2.21 | 1.67 | ||
| R34567 | OAS2, 2′→5′ oligoadenylate synthetase 2 | 2.59 | 1.93 | 2.3 | 1.71 | 2.21 | 2.45 | ||
| H70440 | GS3686, hypothetical protein, expressed in osteoblast | 2.48 | 2.06 | 3.29 | 1.41 | 2.28 | 1.95 | ||
| T86070 | EST | 2.42 | 1.88 | 2.08 | 1.66 | ||||
| R23341 | B2M, β-2-microglobulin | 2.3 | 1.91 | 3.09 | 1.94 | 2.83 | 2.29 | ||
| W94862 | ADAR, adenosine deaminase, RNA specific | 2.29 | 1.94 | 2 | 1.95 | ||||
| AA028065 | BRF2, butyrate response factor 2 (EGF-response factor 2) | 2.25 | 1.93 | ||||||
| W47350 | RARRES3, retinoic acid receptor responder (tazarotene induced) 3 | 2.13 | 1.97 | 2.03 | 1.95 | ||||
| T80567 | KIAA072, KIAA0725 protein | 2.02 | 1.74 | 2.31 | 1.5 | ||||
| AA001748 | EST, weakly similar to angiopoietin Y1 (H. sapiens) | 2.01 | 2.35 | ||||||
| AA040506 | IMAGE-485950 | 2 | 1.75 | ||||||
| AA040032 | KIAA0015, KIAA0015 gene product | 0.55 | 0.36 | 1.99 | 2.37 | 2.04 | 2.18 | 3.37 | 3.59 |
| W63787 | PHGDH, β-phosphoglycerate dehydrogenase | 0.42 | 0.5 | ||||||
| R13977 | EST | 0.37 | 0.43 | 0.52 | 0.47 | ||||
| R80595 | SOD2, superoxide dismutase 2, mitochondrial | 3.7 | 1.81 | ||||||
| H60340 | IMAGE-207631 | 1.78 | 2.08 | ||||||
| H17813 | TOP2A, topoisomerase II alpha | 0.48 | 0.48 | ||||||
| R40483 | Homo sapiens clone 24877 mRNA sequence | 3.31 | 1.85 | ||||||
| N34456 | NT5, 5′ nucleotidase (CD73) | 2.19 | 1.83 | ||||||
| AA040062 | EST, weakly similar to yeast antiviral protein SK12 and ATP-dependent DNA helicase | 3.65 | 1.48 | 2.84 | 2.14 | 2 | 1.59 | ||
| N58703 | IMAGE-247579 | 2.1 | 1.51 | 2.15 | 1.89 | ||||
| W88741 | EST | 5.41 | 44.08 | ||||||
| N30287 | EST, weakly similar to Wiscott-Aldrich syndrome protein homolog | 2.32 | 2.58 | ||||||
| W67568 | HLA-A, MHC class I, A | 0.43 | 0.18 | 2.64 | 3.21 | ||||
| AA053162 | HLA-B, MHC class I, B | 2.42 | 2.32 | ||||||
| W00378 | HLA-C, MHC class I, C | 2.22 | 2.43 | ||||||
—, Intensity of the spot on the array was below local background.
Infection with the wild-type KOS virus had a more dramatic effect on cellular mRNA levels. However, only two of the genes whose expression was changed by infection with KM110 or UV-inactivated KOS or after treatment with IFN-α were also altered after KOS infection. In both cases, the level of expression was decreased by KOS, but increased by the other treatments (Table 1). A comprehensive analysis of the effects of wild-type HSV-1 on cellular gene expression will be presented elsewhere.
We drew two broad conclusions from the microarray data. First, KM110 and UV-inactivated KOS increase the expression of remarkably similar sets of cellular genes, which overlap extensively with those induced by IFN-α. Some of the proteins encoded by the genes that are common to all three sets act to limit intracellular virus replication (e.g., MX1, OAS, and PML) (42), and others serve as secreted proinflammatory chemokines (e.g., SCYB10 [also known as IP-10] and ISG15) (1). Second, wild-type HSV-1 does not induce the expression of any of these genes, implying that induction of IFN-responsive genes occurs only when viral gene expression is inhibited.
The transcriptional activation function of VP16 prevents induction.
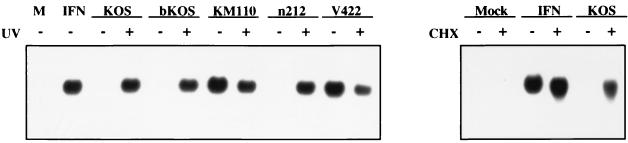
Transcriptionally inactive HSV-1 (KM110 and UV-inactivated KOS) induced IFN-responsive genes, but transcriptionally competent virus did not. One possibility is that HSV-1 produces one or more gene products shortly after infection that block the cellular response to the infecting virion. To further investigate this possibility and to validate the results of the microarray analysis, we monitored the accumulation of ISG 56K RNA as an indicator of viral gene induction by using Northern blot analysis (Fig. 2). ISG 56K, which encodes a 56-kDa IFN-inducible protein, is one of the transcripts most strongly induced by KM110 (Table 1). Northern blot analysis confirmed that ISG 56K message is strongly induced by KM110 and UV-inactivated KOS, but does not accumulate following infection with wild-type KOS. However, KOS strongly induced ISG 56K mRNA when the infection was carried out in the presence of cycloheximide, confirming that wild-type virus is competent for induction when viral protein synthesis is blocked and demonstrating that the response does not require cellular protein synthesis.
FIG. 2.
Northern blot analysis of ISG 56K RNA. HEL cells were mock infected (M, IFN) or infected with the indicated virus (with [+] or without [−] UV inactivation) at an MOI of 5. Where indicated, cycloheximide (CHX) was added at 100 μg/ml 1 h prior to infection and maintained throughout. At 24 h postinfection, RNA was extracted and analyzed for ISG 56K RNA levels by Northern blot hybridization.
The genetic basis for the ability of untreated KM110 to induce the transcriptional response was determined. KM110 bears two separate mutations: the V422 lesion truncates the C-terminal acidic transcriptional activation domain of VP16 after residue 422 (21), and the n212 mutation truncates the IE protein ICP0 after residue 212 (7). As shown in Fig. 2, a virus bearing only the V422 mutation triggered induction of ISG 56K RNA as efficiently as did KM110. In contrast, the n212 mutant failed to induce this transcript. Therefore, truncation of the transcriptional activation domain of VP16 is associated with the induction of the ISGs.
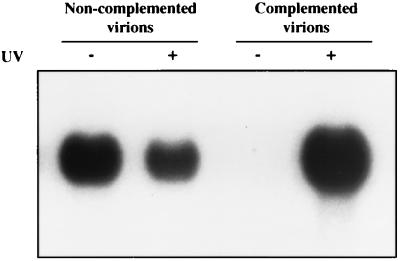
VP16 is a component of the infecting virion that acts during the very earliest stages of infection to stimulate transcription of the five viral IE genes (33). It also serves an essential structural role in virion assembly and egress (25, 51). The V422 mutation abolishes the transcriptional activity of VP16, but leaves its structural functions intact (21). The V422 mutation might therefore unmask inducing activity, because V422 virions, which are devoid of transcriptionally competent VP16, are less able to synthesize a possible inhibitor of induction. If so, then one would predict that V422 virions would be unable to induce ISG gene expression when loaded with wild-type VP16. We generated a V422 virus stock harboring wild-type VP16 by passaging the virus on 16-8 cells that provide wild-type VP16 in trans. The resulting complemented virions were then used to infect HEL cells. Unlike noncomplemented virions, the complemented virions failed to induce the ISG 56K RNA (Fig. 3). However, inducing activity was restored when the genome of the complemented virus was inactivated with UV irradiation. The differences in intensity between V422 and UV-inactivated V422 seen in Fig. 2 and 3 are not consistent between individual experiments and thus are not significant. These data demonstrate that the V422 mutant induces ISG 56K RNA, because V422 virions lack wild-type VP16.
FIG. 3.
Induction of ISG 56K RNA by V422 is suppressed by loading wild-type VP16 into mutant virions. HEL cells were infected with V422 grown on either U2OS cells (noncomplemented virions) or 16-8 cells (Vero cells that provide VP16 in trans [complemented virions]) at an MOI of 5 (with [+] or without [−] UV inactivation). At 24 h postinfection, RNA was extracted and analyzed for ISG 56K RNA levels by Northern blot hybridization.
Induction requires viral entry.
HCMV virions trigger expression of host IFN-inducible genes, and soluble HCMV glycoprotein B (gB) is apparently sufficient to induce this effect (4, 54). Presumably, gB located in the envelope of the infecting HCMV virion binds to a cell surface receptor and activates intracellular signaling events. These data imply that HCMV need not enter the host cell in order to induce cellular gene expression. We asked if entry is required for HSV-1 to induce the expression of ISG 56K mRNA. To accomplish this, we examined the phenotypes of several HSV-1 mutants that are competent to bind to the cell surface, but are unable to penetrate the plasma membrane.
HSV entry is a multistep process that requires many viral envelope glycoproteins (34). gC (and to a lesser extent, gB) binds to heparin sulfate proteoglycans, providing the initial attachment to the cell surface. gD then interacts with several cell surface receptors, and the virion envelope fuses with the host plasma membrane by using gB, gD, gH, and gL. Viral isolates bearing null mutations in the genes encoding the glycoproteins required for membrane fusion must be propagated on complementing cells that provide the missing glycoprotein in trans. The complemented virions that result are capable of one round of productive infection on noncomplementing cells, producing noninfectious (noncomplemented) virions that are competent to bind to the cell surface, but are unable to penetrate (6, 14, 18, 38). Noncomplemented virions lacking gD and gI (F-gDβ), gD (F-US6kan), or gB (KO82), which are unable to enter cells, did not induce ISG 56K RNA, even after UV inactivation; in contrast, the corresponding complemented virions, which are able to enter cells, showed efficient induction when they were UV inactivated (Fig. 4). The simplest interpretation of this result is that induction requires viral entry into host cells.
FIG. 4.
Viral penetration is required for induction of ISG 56K RNA. HEL cells were infected with HSV-1 KOS (MOI 5) or the indicated glycoprotein mutants (with [+] or without [−] UV inactivation). Mutants grown on their respective complementing cell line were used to infect monolayers at an MOI of 5 PFU/cell. Mutants grown for one round on noncomplementing Vero cells produce viral particles, the titers of which cannot be determined due to their inability to penetrate. Thus, viral particles were counted with a transmission electron microscope (see Materials and Methods), and the inoculum was adjusted so that a dose of viral particles equivalent to an MOI of 5 PFU of KOS per cell was used. At 24 h postinfection, RNA was extracted and analyzed for ISG 56K RNA levels by Northern blot hybridization.
Evidence for a potential virus-encoded inhibitor of the antiviral response.
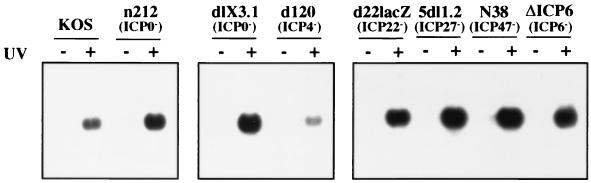
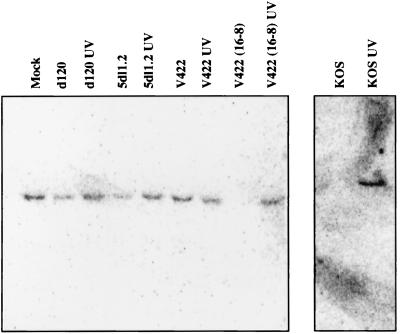
We attempted to determine if a viral gene product(s) is responsible for blocking the antiviral response during HSV infection by surveying the phenotypes of selected mutant viruses. The IE protein ICP4 is the major HSV transcriptional regulator and is stringently required for expression of the viral early and late genes (37). Previous work showed that the ICP4 null mutant, d120, synthesizes only ICP0, ICP22, ICP27, ICP47, and ICP6, albeit at exaggerated levels (10). The d120 mutant failed to efficiently induce ISG 56K RNA (unless the virus was first UV inactivated) suggesting that ICP4 is not required to block the response (Fig. 5). The simplest interpretation of this finding is that one or more of the other IE proteins and/or ICP6 normally acts to block induction (although it remains possible that overproduction of these proteins contributes to the d120 phenotype). Noteably, ISG 56K RNA was not induced in cells infected with any of a panel of viral mutants bearing lesions that individually inactivate each of these proteins (Fig. 5). The difference in intensity of signal between UV-inactivated viruses is not reproducible and thus is not significant. One interpretation of these results is that HSV-1 encodes two or more proteins that are each sufficient to block the response. Another is that induction is not detected when viral gene expression is allowed to proceed, because HSV-induced delayed shutoff of the cellular gene precludes or masks the response. Consistent with this interpretation, both KOS and complemented V422 caused a large decline in the levels of mRNA derived from the cellular gene encoding the 60-kDa stress 70 protein chaperone by 24 h postinfection (Fig. 6). However, stress 70 mRNA levels did not decline following infection with d120 or 5dl1.2 (Fig. 6), indicating that these isolates do not globally shut off cellular gene expression. Taken together, these data suggest that wild-type infection may indeed preclude induction of an antiviral response through a general host shutdown mechanism. This explanation, however, seems insufficient to explain the lack of a response during infection with a number of IE mutant viruses, lending support to the idea that the virus may in fact produce one or more specific inhibitors.
FIG. 5.
HSV-1 may disarm the antiviral response by synthesizing an inhibitor. Viral mutants bearing mutations that inactivate individual IE genes were used to infect HEL cells (with [+] or without [−] UV inactivation) at an MOI of 5. At 24 h postinfection, RNA was extracted and analyzed for ISG 56K RNA levels by Northern blot hybridization.
FIG. 6.
Northern blot of cellular stress 70 protein chaperone mRNA following infection with various HSV-1 recombinants. Wild-type and mutant HSV-1 viruses were used to infect HEL cells (with or without UV inactivation) at an MOI of 5. At 24 h postinfection, RNA was extracted and analyzed for the cellular stress 70 protein chaperone mRNA by Northern blot hybridization.
Induction by HSV-1 virions can be uncoupled from IFN signaling.
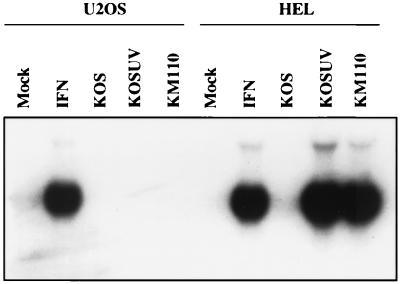
We have shown previously that although HEL cells fail to support growth of KM110, this virus replicates efficiently on the human osteosarcoma cell line U2OS (26). In order to determine if HSV virions induce a response in U2OS cells similar to that seen in HEL cells, we monitored ISG 56K RNA induction in U2OS cells treated with IFN-α or infected with KOS, UV-inactivated KOS, or KM110. ISG 56K RNA was not induced by any of the viruses in U2OS cells. However, ISG 56K RNA was induced in U2OS cells after treatment with IFN, demonstrating that these cells have a functional IFN signaling cascade (Fig. 7). Entirely analogous results were obtained for the mRNA encoding the C-X-C chemokine IP-10, which was induced by IFN and HSV-1 virions in HEL cells, but only by IFN in U2OS cells (data not shown). These data argue that HSV-1 virions do not trigger expression of IFN response genes by engaging the IFN receptor and demonstrate that our virion preparations lack detectable IFN activity. They also suggest a possible correlation between the permissiveness of a given cell line for KM110 and the appearance of an IE cellular transcription response.
FIG. 7.
HSV-1 does not induce ISG 56K RNA in the U2OS cell line. HEL and U2OS monolayers were mock infected (mock, IFN) or infected with KOS (with or without UV inactivation) or KM110 at an MOI of 5. IFN-α was added at 1,000 U/ml following infection. At 24 h postinfection, RNA was extracted and analyzed for ISG 56K RNA levels by Northern blot hybridization.
DISCUSSION
Treatment of cells with IFN rapidly induces an antiviral state (42). Here, we show that HSV-1 virus particles that are incapable of gene expression produce a similar effect. Induction of the antiviral state by HSV-1 is inhibited by viral gene expression and occurs in an IFN-independent fashion. The HSV-induced antiviral state is linked to enhanced expression of a specific set of cellular genes, many of which are also induced by IFN. Some of these genes, such as those coding for MX1/2, OAS2/3, and β-2-microglobulin, are known to limit intracellular virus replication (42). MX proteins are dynamin superfamily GTPases that interfere with viral replication at many levels. The OAS pathway activates RNase L and leads to degradation of viral mRNAs. β-2-Microglobulin is required for expression of major histocompatibility complex (MHC) class I molecules, which are critical for recognition and lysis of virally infected cells by T cells. Other induced cellular genes, such as ISG15 and IP-10, serve as proinflammatory cytokines (1); IP-10 has been implicated as an important mediator of Th1 dominant immune responses (48). Induction appears to require viral penetration, but does not occur when viral gene expression is permitted, implying that HSV encodes one or more gene products that normally act to disarm the response. A recent report by Preston and colleagues found that HSV-1 induces expression of four IFN-inducible genes if viral gene expression is blocked, in a process that does not require cellular protein synthesis (31).
Differential display and microarray analysis showed previously that the related herpesvirus HCMV induces IFN-responsive RNAs in primary human fibroblasts (53, 54). The HSV-induced response described in this report is similar to that induced by HCMV in that induction does not require viral gene expression or cellular protein synthesis. However, the response to HSV is evident only when viral gene expression is blocked, while HCMV induces IFN response genes even when viral gene expression is allowed to proceed. In addition, purified HCMV gB suffices to induce the response (4), implying that binding of HCMV virions to the cell surface is sufficient, while our data strongly argue that HSV-1 must penetrate the plasma membrane in order to induce. Several aspects of the HSV-induced cellular response are common to other viral systems. Adenovirus capsids induce the expression of multiple chemokines, including IP-10 (3, 19, 27), in the absence of viral gene expression, while the human immunodeficiency and Epstein-Barr viruses induce a cellular response following virus attachment (5, 36, 43). Attachment, penetration, and limited viral transcription suffice for induction of the chemokine RANTES during infection with measles virus (32).
While the signaling pathway used in the HSV-induced response remains to be identified, it apparently does not involve signaling through the IFN receptors: U2OS cells respond to IFN, yet fail to show expression of ISG 56K mRNA upon infection with either KM110 or UV-inactivated KOS. A similar conclusion using cell lines mutated for Tyk2, JAK1, or STAT1 was recently reported (31). Our data indicate that induction requires viral penetration of the host plasma membrane, but occurs in the absence of viral transcription. A number of IFN-responsive genes, including ISG 56K, can be induced directly by dsRNA in the absence of IFN (2, 44). However, dsRNA is unlikely to be involved in the response to HSV, because viral transcription is not required. Our data therefore suggest that HSV activates a novel intracellular sensor that detects a very early step during virus infection. Possible inducing events include fusion of the viral envelope with the host plasma membrane, introduction of viral tegument proteins into the cytoplasm, or changes in the cytoskeleton, because HSV capsids are transported to the nucleus via microtubules (41) and the HSV-1 tegument protein VP22 exhibits the properties of a microtubule-associated protein (12). Alternatively, it is possible that delivery of viral DNA into the nucleus triggers the host response. Further studies are required to distinguish between these possibilities. The availability of cell lines such as U2OS that are defective in this signaling pathway should facilitate these studies.
Induction of the antiviral response occurs only when viral gene expression is blocked, suggesting that a newly made gene product may function as an inhibitor. The IE protein ICP4 is a prominent HSV transcriptional regulator that is essential for expression of viral early and late genes. Inasmuch as an ICP4 null mutant failed to efficiently induce ISG 56K in the absence of UV inactivation, we concluded that any viral inhibitor must be an IE gene product. However, mutants bearing lesions that individually inactivate each IE protein failed to induce ISG 56K. Therefore, if a viral inhibitor does exist, then HSV-1 likely encodes two or more proteins that are each sufficient to block the response. This apparent redundancy of inhibitors may indicate that disarming the cellular antiviral response is of great importance to the virus. We have recently shown that the IE protein ICP0 contributes to the relative resistance of HSV-1 to IFN (24), indicating that ICP0 is capable of overcoming an already established antiviral state. Thus, ICP0 is a likely candidate for one of the putative inhibitors.
The potential biological significance of the cellular response to HSV particles is many fold. The efficiency of the cellular response in a given cell type may influence the decision of whether incoming viral genomes enter the lytic cycle or remain quiescent. It will be interesting to learn if a similar virion-induced response occurs during infection of neurons and influences the entry into latency. The response likely enhances the ability of HSV to induce antiviral immunity in vivo and may partly explain the self-limiting nature of HSV infections in the intact human host. The response has potentially broad implications for gene therapy, which requires efficient transfer of the therapeutic gene to the desired location and the sustained expression of that gene (47). HSV has been identified as a potentially ideal vector for gene delivery, because the viral genome can accept insertions of multiple therapeutic genes and HSV can be targeted to the nervous system (9, 13). However, current HSV vectors have been designed to preclude expression of the viral IE proteins in order to eliminate cytotoxicity (17, 40). Our results predict that such vectors will trigger the host antiviral response in the same fashion as KM110. Such a response would likely severely limit the duration of transgene expression in vivo, through immune-mediated clearance of the infected cells. In support of this hypothesis, replication-defective recombinant adenoviral vectors induce cytotoxic T lymphocytes capable of lysing infected cells (19). In addition, the IFN-induced antiviral state blocks transcription of both viral and heterologous promoters located in the HSV genome (30), suggesting that the virion-induced antiviral state would contribute to extinction of transgene expression. Consistent with this possibility, HSV vectors that establish genome quiescence in the same fashion as KM110 support only very low levels of expression of heterologous transgenes (17, 40).
Viruses both induce and evade host antiviral responses (8, 20, 45). Our data point to the existence of a novel IFN-independent intracellular mechanism for detecting virus infection. Deciphering the mechanisms by which HSV induces and disarms this system will enhance our understanding of the basic biology of virus-host interactions and aid in the rational design of useful viral vectors for gene therapy.
ACKNOWLEDGMENTS
We thank David Johnson for viral mutants lacking glycoproteins and for valuable advice and discussions. We are grateful to Rob Maranchuk and Holly Saffran for excellent technical assistance; Richard Sherburne for help with electron microscopy; and Bryan MacNeil, Eric Ho, and Brian Li for assistance with the microarray analysis.
This research was supported by a grant from the Medical Research Council to J.R.S. and an NRC/NSERC/MRC grant to A.M.E. A.M.E. is an MRC Scientist, K.L.M. holds postdoctoral fellowships from the MRC and the Alberta Heritage Foundation for Medical Research, and A.B.G. holds a postdoctoral fellowship from the National Science and Engineering Research Council of Canada.
REFERENCES
- 1.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay S K, Leonard G T, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 3.Borgland S L, Bowen G P, Wong N C W, Libermann T A, Muruve D A. Adenovirus vector-induced expression of the C-X-C chemokine IP-10 is mediated through capsid-dependent activation of NF-κB. J Virol. 2000;74:3941–3947. doi: 10.1128/jvi.74.9.3941-3947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle K A, Pietropaolo R L, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briant L, Signoret N, Gaubin M, Robert-Hebmann V, Zhang X, Murali R, Greene M I, Piatier-Tonneau D, Devaux C. Transduction of activation signal that follows HIV-1 binding to CD4 and CD4 dimerization involves the immunoglobulin CDR3-like region in domain 1 of CD4. J Biol Chem. 1997;272:19441–19450. doi: 10.1074/jbc.272.31.19441. [DOI] [PubMed] [Google Scholar]
- 6.Cai W, Person S, Warner S C, Zhou J, DeLuca N A. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cebulla C M, Miller D M, Sedmak D D. Viral inhibition of interferon signal transduction. Intervirology. 1999;42:325–330. doi: 10.1159/000053968. [DOI] [PubMed] [Google Scholar]
- 9.Coffin R S, Latchmann D S. Herpes simplex virus based vectors. In: Latchmann D S, editor. Genetic manipulation of the nervous system. London, United Kingdom: Academic Press; 1995. pp. 99–112. [Google Scholar]
- 10.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Der S D, Zhou A, Williams B R, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott G, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fink D J, DeLuca N A, Goins W F, Glorioso J C. Gene transfer to neurons using herpes simplex virus vectors. Annu Rev Neurosci. 1996;19:265–287. doi: 10.1146/annurev.ne.19.030196.001405. [DOI] [PubMed] [Google Scholar]
- 14.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein D J, Weller S K. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology. 1988;166:41–51. doi: 10.1016/0042-6822(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Peters K L, Sen G C. Induction of the human protein p56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 17.Jamieson D R S, Robinson L H, Daksis J I, Nicholl M J, Preston C M. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus Vmw65 mutants. J Gen Virol. 1995;76:1417–1431. doi: 10.1099/0022-1317-76-6-1417. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kafri T, Morgan D, Krahl T, Sarvetnick N, Verma I. Cellular immune response to adenoviral vector infected cells does not require de novo viral gene expression: implications for gene therapy. Proc Natl Acad Sci USA. 1998;95:11377–11382. doi: 10.1073/pnas.95.19.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalvakolanu D V. Virus interception of cytokine-regulated pathways. Trends Microbiol. 1999;7:166–171. doi: 10.1016/s0966-842x(99)01476-6. [DOI] [PubMed] [Google Scholar]
- 21.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 22.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long M C, Leong V, Schaffer P A, Spencer C A, Rice S A. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J Virol. 1999;73:5593–5604. doi: 10.1128/jvi.73.7.5593-5604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossman K L, Saffran H A, Smiley J R. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J Virol. 2000;74:2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mossman K L, Sherburne R, Lavery C, Duncan J, Smiley J R. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J Virol. 2000;74:6287–6299. doi: 10.1128/jvi.74.14.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mossman K L, Smiley J R. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J Virol. 1999;73:9726–9733. doi: 10.1128/jvi.73.12.9726-9733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muruve D A, Barnes M J, Stillman I E, Libermann T A. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum Gene Ther. 1999;10:965–976. doi: 10.1089/10430349950018364. [DOI] [PubMed] [Google Scholar]
- 28.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nazar A S, Cheng G, Shin H S, Brothers P N, Dhib-Jalbut S, Shin M L, Vanguri P. Induction of IP-10 chemokine promoter by measles virus: comparison with interferon-g shows the use of the same response element but with differential DNA-protein binding profiles. J Neuroimmunol. 1997;77:116–127. doi: 10.1016/s0165-5728(97)00070-2. [DOI] [PubMed] [Google Scholar]
- 30.Nicholl M J, Preston C M. Inhibition of herpes simplex virus type 1 immediate-early gene expression by alpha interferon is not VP16 specific. J Virol. 1996;70:6336–6339. doi: 10.1128/jvi.70.9.6336-6339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholl M J, Robinson L H, Preston C M. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol. 2000;81:2215–2218. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- 32.Noe K H, Cenciarelli C, Moyer S A, Rota P A, Shin M L. Requirements for measles virus induction of RANTES chemokine in human astrocytoma-derived U373 cells. J Virol. 1999;73:3117–3124. doi: 10.1128/jvi.73.4.3117-3124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 34.Rajcani J, Vojvodova A. The role of herpes simplex virus glycoproteins in the virus replication cycle. Acta Virol. 1998;42:103–118. [PubMed] [Google Scholar]
- 35.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts M L, Luxembourg A T, Cooper N R. Epstein-Barr virus binding to CD21, the virus receptor, activates resting B cells via an intracellular pathway that is linked to B cell infection. J Gen Virol. 1996;77:3077–3085. doi: 10.1099/0022-1317-77-12-3077. [DOI] [PubMed] [Google Scholar]
- 37.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1043–1107. [Google Scholar]
- 38.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 43.Sugano N, Chen W, Roberts M L, Cooper N R. Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-kappaB induction. J Exp Med. 1997;186:731–737. doi: 10.1084/jem.186.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiwari R K, Kusari J, Sen G C. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 46.Umene K. Conversion of a fraction of the unique sequences to part of the inverted repeats in the S component of the herpes simplex virus type 1 genome. J Gen Virol. 1986;67:1035–1048. doi: 10.1099/0022-1317-67-6-1035. [DOI] [PubMed] [Google Scholar]
- 47.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 48.Ward S G, Bacon K, Westwick J. Chemokines and T-lymphocytes: more than just an attraction. Immunity. 1998;9:1–11. doi: 10.1016/s1074-7613(00)80583-x. [DOI] [PubMed] [Google Scholar]
- 49.Wathelet M G, Berr P M, Huez G A. Regulation of gene expression by cytokines and virus in human cells lacking the type-1 interferon locus. Eur J Biochem. 1992;206:901–910. doi: 10.1111/j.1432-1033.1992.tb16999.x. [DOI] [PubMed] [Google Scholar]
- 50.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 51.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R., II Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.York I, Roop C, Andrews D W, Riddel S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8− T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 53.Zhu H, Cong J, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu H, Cong J, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]