Abstract
Background
Moyamoya disease (MMD) signifies a cerebrovascular disorder with obscure origin and a more rapid and severe progression in children than adults. This investigation aims to uncover age-associated distinctions through proteomic and metabolomic profiling to gain insights into the underlying mechanisms of MMD.
Methods
Twelve MMD patients—six children and six adults—along with six healthy controls (HC), participated, each providing a 10 mL blood sample. Serum proteomic and metabolomic analyses were conducted using ultra-performance liquid chromatography and high-resolution mass spectrometry, complemented by bioinformatics to identify differential biomolecules and their interactions. Pathway implications were ascertained using GO and KEGG enrichment analysis.
Results
Notable proteomic and metabolomic discrepancies were observed between pediatric and adult MMD subjects. A total of 235 and 216 proteins varied in adult and pediatric cases compared to HCs, with 73 proteins shared. In addition, 129 and 74 anionic, plus 96 and 104 cationic metabolites, were differentially expressed in the pediatric and adult groups, respectively, with 34 anionic and 28 cationic metabolites in common. Age-specific biomolecules further characterized these distinctions. Enrichment analysis pinpointed immunity and inflammation pathways, with vitamin digestion and absorption highlighted as pivotal in pediatric MMD.
Conclusion
This study unveils distinct metabolic and proteomic patterns within pediatric and adult MMD patients. The critical role of the vitamin digestion and absorption pathway in the pathogenesis of pediatric MMD offers novel insight into disease mechanisms.
Keywords: proteomics, metabolomics, pediatric patients, moyamoya disease, integration analysis
Graphical Abstract
Introduction
Moyamoya disease (MMD) is a chronic cerebrovascular disease characterized by the progressive narrowing of intracranial arteries, with the origin segment occurring in the middle and anterior cerebral arteries, and it leads to the development of a non-typical vascular network at the skull base. This often results in ischemia, stroke, and other clinical manifestations.1 Affected vessels are prone to rupture or occlusion, which can lead to permanent neurological deficits and death if interventions are not applied.
Although MMD was previously predominantly diagnosed in Asian populations, it has now been observed in various ethnic groups worldwide, including European and North American populations.2 The disease has a bimodal age of onset, with patients typically presenting in early childhood (between the ages of 5 and 9 years) or mid-life (around the age of 40).3 The clinical presentation varies with age, with hemorrhagic cerebrovascular events being less common in the pediatric population. Conversely, hemorrhagic and ischemic manifestations occur equally in adult- and childhood-onset disease.4,5 Studies have shown that the disease prognosis is worse in younger children, especially those who are younger than age four.6
Currently, drugs have not proven effective in stopping or reversing the progression of MMD. Aggressive surgical revascularization lowers the risk of cerebral infarction, reduces the frequency of transient ischemic attacks (TIAs), and improves the activities of daily living (ADL) and neurocognitive function in patients who undergo the procedure. Furthermore, indirect revascularization is suitable for young individuals.7 The application of spatial coefficient of variation in arterial spin labeling MRI could aid in predicting surgical revascularization outcomes for pediatric patients with MMD.8 However, owing to rapid disease progression in pediatric patients, preoperative infarction is common.9 The etiology of MMD, as well as the significant variations in symptom manifestation and disease prognosis between adult and pediatric patients, are poorly understood. Recent research has proposed RNF213 as a significant genetic factor in the etiology of both MMD and Moyamoya Syndrome (MMS).10 There has been a lack of comprehensive investigations of the molecular differences in pediatric patients with MMD. It is crucial to explore these molecular changes to determine the underlying mechanisms responsible for the varying clinical characteristics and prognosis of MMD in children.
Recent advances in omics technologies, including proteomics and metabolomics, have provided valuable tools for comprehensive analysis of biological samples and identifying key proteins and metabolites involved in molecular changes associated with various diseases. Araki et al found that the expression of fibrinogen, a protein involved in blood clotting, was higher in the cerebrospinal fluid of adult patients with MMD than in healthy controls.11 Metabolomic studies conducted on patients with MMD revealed that certain amino acids, such as phenylalanine and tyrosine, were present at lower levels in the serum of adult patients with MMD than in healthy control subjects.12 Our previous studies demonstrated that the proinflammatory and immunosuppressive abilities of adult MMD patients were significantly enhanced, while the anti-inflammatory and immunomodulatory abilities were significantly decreased.13 Current studies on MMD primarily concentrate on adult patients with MMD, However, the multi-omics understanding of pediatric MMD is lacking. The development of liquid chromatography-mass spectrometry (LC-MS) has paved the way for multi-omic progress, allowing high-throughput quantitative data of biomolecules to be rapidly acquired, thus providing a better understanding of the underlying mechanisms of diseases.14
Our study aimed to comprehensively investigate how the proteome and metabolome are involved in MMD through serum sample analysis. By utilizing proteomic and metabolomic data obtained using LC-MS-based methods, combined with univariate and multivariate analyses, as well as pathway analysis, we identified specific protein and metabolic alterations that could potentially correlate with MMD. Our findings have significant implications for creating more precise and unbiased diagnostic tools for MMD and enhancing our knowledge of the underlying pathophysiology of MMD.
Materials and Methods
Study Design and Blood Samples
At the Fifth Medical Center of the PLA General Hospital, blood samples were collected from 18 participants, consisting of six adult and six pediatric patients with MMD, as well as six adult healthy controls (HCs). Flow chart illustrating the participant selection process for MMD patients and HC in the study (Figure S1). The MMD patients included in this study were diagnosed in accordance with both the Research Committee on MMD (Spontaneous Occlusion of Circle of Willis) of the Ministry of Health, Labor Welfare, Japan, and the Japan Stroke Society’s Guideline Committee 2021.9 All 12 patients with MMD recovered and were discharged from the hospital. None of the MMD cases included in this study were deemed critical. Patient details are shown in Table S1.
Patients and Samples
Fasting blood samples were obtained from six adult patients with MMD, six pediatric patients with MMD, and six HC controls. The samples were collected in ethylenediaminetetraacetic acid-treated vacuum blood collection tubes, and the coagulation layers were incubated at 37 °C for 1 h. The supernatant was then centrifuged at 1597.5 x g for 5 min. The upper clear liquid was removed after centrifuging at 12000 rpm, 4°C for 10 min. The clear liquid (0.5 mL) was then placed in a 1.5-mL centrifuge tube, flash frozen in liquid nitrogen for 15 min, and stored in a refrigerator at −80°C until subsequent assays. This study complied with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of the Fifth Medical Center of the General Hospital of the People’s Liberation Army. Informed consents were obtained from the legal guardians and relatives of the study participants prior to study commencement. The adult patients and healthy controls also provided informed consent.
Proteomics
Protein Extraction and Peptide Preparation
Protein extraction and peptide preparation were performed according to laboratory protocols. Initially, each sample was lysed with DB lysis buffer (8 M Urea, 100 mM TEAB, pH 8.5) and centrifuged at 12000 rpm for 20 min at 4 °C. In addition, a pool sample for building the spectral library for DIA protein identification was created by mixing 20–30 μL of serum from each sample. For data-dependent acquisition (DDA) protein identification, the proteins in the pooled sample were divided into two duplicates, one of which was processed using the ProteoMinerTM Protein Enrichment Kit (Bio-Rad) to remove high-abundance proteins. These duplicates were treated with 2 mM dithiothreitol (DTT) for 1 h at 56 °C, followed by alkylation with iodoacetic acid for 1 h at room temperature in the dark. After precipitation with acetone, the protein precipitate was dissolved using 0.1 M triethylammonium bicarbonate (pH 8.5) and 8 M urea buffer. The supernatant of each sample, containing precisely 0.1 mg of protein, was digested with Trypsin Gold (Promega) at 37 °C for 16 h. Subsequently, a desalination method was used to remove the high urea content, and the sample was dried by vacuum centrifugation.
Library Construction
To fractionate low-abundance pool sample peptides, a C18 column (Waters BEH C18, 4.6×250 mm, 5 m) was used in Rigol L3000 HPLC, operating at 1 mL/min. The column oven was maintained at 50 °C, while the elution was carried out using mobile phases A (2% acetonitrile with ammonium hydroxide adjusted to pH 10.0) and B (98% acetonitrile with ammonium hydroxide adjusted to pH 10.0) in a gradient fashion. The solvent gradients were set at 3% B for 5 min, 3–8% B for 0.1 min, 8–18% B for 11.9 min, 18–32% B for 11 min, 32–45% B for 7 min, 45–80% B for 3 min, 80% B for 5 min, 80–85% B for 0.1 min, and 5% B for 6.9 min. The eluates were monitored at 214 nm, collected in tubes every minute, and then merged into four fractions. To analyze the peptides of high-abundance and low-abundance protein fractions, samples were vacuum dried and reconstituted using 0.1% (v/v) formic acid (FA) in water, with 0.2 L of standard peptides (iRT kit, Biognosys).
To construct the transition library, DDA mode shotgun proteomic analyses were performed using an EASY-nLCTM 1200 UHPLC system equipped with an Orbitrap Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific). Low- and high-abundance peptides from the four fractions were reconstituted with 0.1% FA and injected into a homemade C18 NanoTrap column (2 cm × 100 μm, 3 μm). They were then separated on a homemade analytical column (15 cm × 150 μm, 1.9 μm), with a 120 min linear gradient of eluent B (0.1% FA in 80% ACN) in eluent A (0.1% FA in H2O) at a flow rate of 600 nL/min. The solvent gradient details were 5–10% B for 2 min, 10–40% B for 105 min, 40–50% B for 5 min, 50–90% B for 3 min, and 90–100% B for 5 min.
The Q-Exactive HF-X mass spectrometer was set to positive polarity mode and a spray voltage of 2.3 kV, and a capillary temperature of 320°C was used. Full MS scans ranging from 350 m/z to 1500 m/z were taken at a resolution of 60,000× (at 200 m/z) with an AGC target value of 3×106 and a maximum ion injection time of 20 ms. The 40 most abundant precursor ions from this scan were fragmented based on a HCD fragment analysis at a resolution of 15,000× (at 200 m/z), with an AGC target value of 5×104 and a maximum ion injection time of 45 ms. The normalized collision energy was set at 27%, with an intensity threshold of 2.2×104 and a dynamic exclusion parameter of 40s.
MS Analysis: DIA Mode
To analyze the peptides in each serum sample, they were mixed with standard peptides and injected into a system consisting of an EASY-nLCTM 1200 UHPLC system and an Orbitrap Q-Exactive HF-X mass spectrometer set to operate in DIA mode. The liquid conditions were identical to those used for the DDA model. During DIA acquisition, the MS1 resolution was set to 60,000× (at 200 m/z), and the MS2 resolution was set to 30,000× (at 200 m/z), with an m/z range of 350–1500 separated into 30 acquisition windows. The full-scan AGC target was 3 × 106, with an injection time of 50 ms. The DIA settings included a normalized collision energy (NCE) of 27%, a target value of 1 × 106, and an automatic maximum injection time to allow for continuous operation of the MS in parallel ion filling and detection mode.
Proteome Data Analysis
The Proteome Discoverer (PD 2.2, Thermo Fisher Scientific) platform, Biognosys Spectronaut v.9.0, and R statistical framework were used for data analysis and visualization of the DIA data. Biognosys Spectronaut v.9 was used for MS2-based label-free quantification of raw DIA data. The analysis was performed using the method described by Bruder et al15 The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to analyze protein families and pathways. The enrichment pipeline16 was used to perform the KEGG enrichment analysis.
Metabolomics
To analyze the serum samples, 100 μL of each sample was resuspended in 80% pre-chilled methanol by vortexing, and it was then incubated on ice for 5 min. The samples were centrifuged at 15,000×g and 4 °C for 20 min. After the supernatant was properly diluted to a final concentration containing 53% methanol with LC-MS grade water, it was transferred and centrifuged again at 15,000×g and 4 °C for 20 min.15 The LC-MS/MS analysis was performed using a Hypersil Gold column (100 × 2.1 mm, 1.9 μm) (ThermoFisher, Germany), coupled with an Orbitrap Q-ExactiveTM HF-X mass spectrometer (Thermo Fisher, Germany). Samples were loaded onto the column with a linear gradient for 17 min at a flow rate of 0.2 mL/min. Raw UHPLC-MS/MS data were analyzed using the Compound Discoverer software (version 3.1, Thermo Fisher), R (version 3.4.3), Python (version 2.7.6), and CentOS (version 6.6) to perform peak alignment, peak picking, and quantitation of each metabolite, followed by normalization of the peak intensities to the total spectral intensity. The peaks were further matched to mzCloud (https://www.mzcloud.org/), mzVault, and MassList databases to obtain accurate and quantitative results.
Metabolome Data Analysis
Metabolites were annotated using the KEGG database. To process the metabolomic data, we used metaX software16 for principal component analysis (PCA). Univariate analysis (t-test) was used to determine statistical significance (p-value). Metabolites with VIP > 1 and P < 0.05, and fold change ≥ 2.0 or FC ≤ 0.5, were identified as differential metabolites. To filter metabolites of interest, we utilized volcano plots to plot log2 (fold change) and log10 (P-value) of the metabolites with the ggplot2 package in R. We normalized the data using z-scores for the intensity areas of differential metabolites, followed by plotting the cluster heatmaps using the Pheatmap package in R. To analyze the correlation between differential metabolites, we employed cor () in the R language (method = Pearson) and established statistically significant correlations using the cor. Mtest () in the R language at P < 0.05. Correlation plots were generated using the Corrplot package in the R software. We performed enrichment analysis of the metabolic pathways for the differential metabolites by checking whether the ratios satisfied x/n > y/N. When metabolic pathways met this criterion, they were considered enriched, and statistically significant enrichment of metabolic pathways was determined.
Statistical Analysis
We used R (version 3.5.2) for all statistical analyses of metabolomic and proteomic data. Data were first transformed to Log2 and normalized to the median. Using the Limma R package, we employed the lmFit and eBay functions to perform differential expression (DE) analysis of proteins and metabolites in the disease groups. In Limma’s design, we used multiple comparisons with all samples to estimate the variance of the expression of each protein or metabolite. We then compared the two groups of interest to pinpoint the direct differential expression of proteins or metabolites in adult MMD and HC, and in child MMD and adult HC. We used the KEGG database (https://www.kegg.jp) to annotate the pathways. Furthermore, we utilized the Pathview online tool (https://Pathview/uncc.edu/overview) to develop the KEGG pathways. The igraph R package was used to generate a network diagram of the DE proteins and metabolites. In the network diagram, the nodes represent DE proteins between adult MMD and HC, and child MMD and HC.
Results
Proteomic Analysis of Adult MMD, Child MMD, and Adult HC
Quantitative proteomic analysis was performed of serum from patients with MMD using ultra-performance liquid chromatography and data-independent acquisition mass spectrometry (UHPLC-DIA-MS) to examine tryptic peptides. Overall, 932 proteins from 18 samples were detected and quantified by UHPLC-DIA-MS.
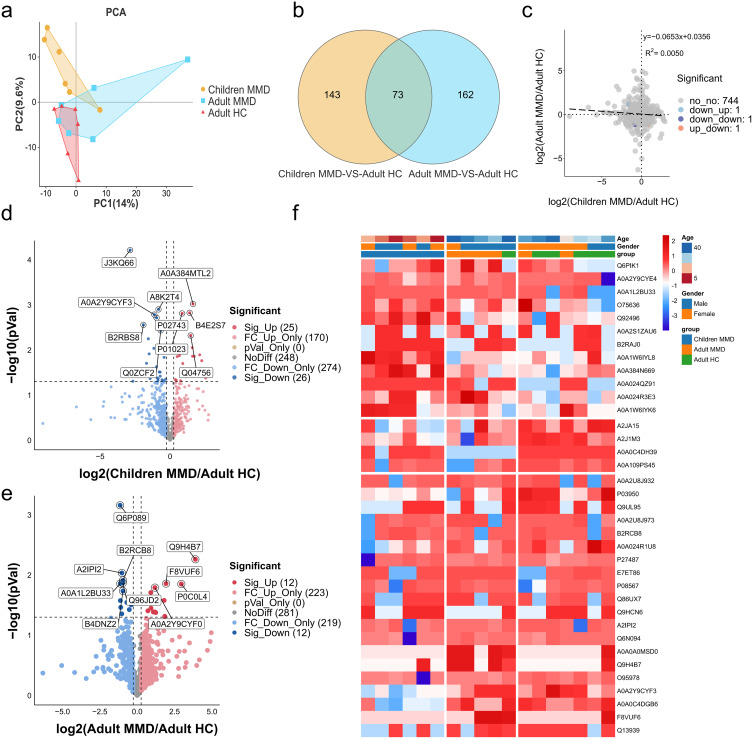
We performed a principal component analysis (PCA) to analyze the relationship between proteins in adults with MMD, adult HCs, and children with MMD, which identified the protein interactions and differential expression levels (Figure 1a). A total of 235 and 216 proteins showed differential expression between adult MMD and HCs, and pediatric MMD and adult HC, respectively. Of these, 73 were expressed in both adult and pediatric MMD, whereas 143 and 162 were specific to pediatric and adult MMD, respectively. This indicates that protein expression varies among patients at different ages with MMD (Figure 1b). Omics association analysis revealed that certain proteins presented consistent upregulation or downregulation trends between children and adult patients with moyamoya disease (Figure 1c). Figure 1d and e shows the protein fold-change differences between adult MMD and HC and between pediatric MMD and adult HC. A0A2Y9CYF0 was decreased in children with MMD but increased in adults with MMD, whereas P01023 exhibited the opposite trend. Figure 1f presents the proteomic data and confirms the PCA results. Hierarchical clustering analysis demonstrated unique clustering patterns among adult MMD, pediatric MMD, and adult HC, revealing the co-regulation of various proteins.
Figure 1.
Differential protein expression levels among adults with MMD, children with MMD, and adult HCs. (a). PCA of proteomic data of adult and child MMD, as well as adult HC. (b). Venn diagram summarizing the differential and overlapping proteins between adult MMD, child MMD, and adult HCs. (c). Protein fold-changes in adult MMD and child MMD. (d). Volcano plot summarizing the differential and overlapping proteins between child MMD and adult HCs. (e). Volcano plot summarizing the differential and overlapping proteins between adult MMD and adult HCs. (f). Heatmap of DE protein levels for adult MMD, child MMD, and adult HCs. DE, differential expression.
The most upregulated protein (14.86-fold change, P = 0.005) in adult MMD compared to adult HC was Q9H4B7 (serum amyloid A4 protein), an acute-phase protein activated and produced during chronic inflammatory processes.17 The most downregulated protein (0.45-fold change, P = 0.001) in adult MMD compared to adult HC was Q6P089, also known as NUBPL. The expression of several proteins involved in immune and inflammatory responses were significantly altered, including P0C0L4 (FC = 7.74, P = 0.014), F8VUF6 (FC = 3.81, P = 0.014), B1AHL2 (FC = 3.59, P = 0.048), A0A1L2BU33 (FC = 0.46, P = 0.014), and O95978 (FC = 0.47, P = 0.034) (Table 1).
Table 1.
Top 10 DE Proteins
| Adult MMD vs adult HC | Child MMD vs adult HC | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein ID | FC | P-value | log2FC | Up/Down | Protein ID | FC | P-value | log2FC | Up/Down |
| Q9H4B7 | 14.86 | 0.005 | 3.89 | up | P05556 | 6.17 | 0.032 | 2.63 | up |
| P0C0L4 | 7.74 | 0.014 | 2.95 | up | B2RAJ0 | 4.27 | 0.027 | 2.09 | up |
| F8VUF6 | 3.81 | 0.014 | 1.93 | up | A0A2S1ZAU6 | 4.07 | 0.011 | 2.03 | up |
| B1AHL2 | 3.59 | 0.048 | 1.85 | up | A0A1W6IYK6 | 3.68 | 0.038 | 1.88 | up |
| A0A0A0MSD0 | 3.49 | 0.026 | 1.8 | up | Q9BYJ0 | 3.56 | 0.002 | 1.83 | up |
| Q6P089 | 0.45 | 0.001 | −1.14 | down | Q6MZX9 | 0.06 | 0.038 | −4.13 | down |
| A0A1L2BU33 | 0.46 | 0.014 | −1.13 | down | Q7Z379 | 0.09 | 0.023 | −3.47 | down |
| A8K061 | 0.46 | 0.027 | −1.13 | down | A0A0C4DGB6 | 0.25 | 0.001 | −2.02 | down |
| O95978 | 0.47 | 0.034 | −1.1 | down | Q9UL95 | 0.28 | 0.042 | −1.84 | down |
| A0A2Y9CYE4 | 0.49 | 0.044 | −1.03 | down | B2RBS8 | 0.31 | 0.037 | −1.67 | down |
P05556, or Integrin Beta 1 (ITGB1), was the most significantly upregulated protein in children with MMD compared to that in adult HC, with a 6.17-fold change (P = 0.032). Compared to that in HC, the expression of Q6MZX9 was the most downregulated in adult MMD, with a 0.06-fold decrease (P = 0.038). Protein levels were significantly altered in MMD patients, with B2RAJ0, A0A2S1ZAU6, A0A1W6IYK6, and Q9BYJ0 expression increasing by 3.56–4.27-fold (P < 0.05). Conversely, Q7Z379, A0A0C4DGB6, Q9UL95, and B2RBS8 expression decreased, with 0.09 to 0.31-fold changes (P < 0.05) (Table 1).
Metabolomics of Adult MMD, Child MMD, and Adult HC
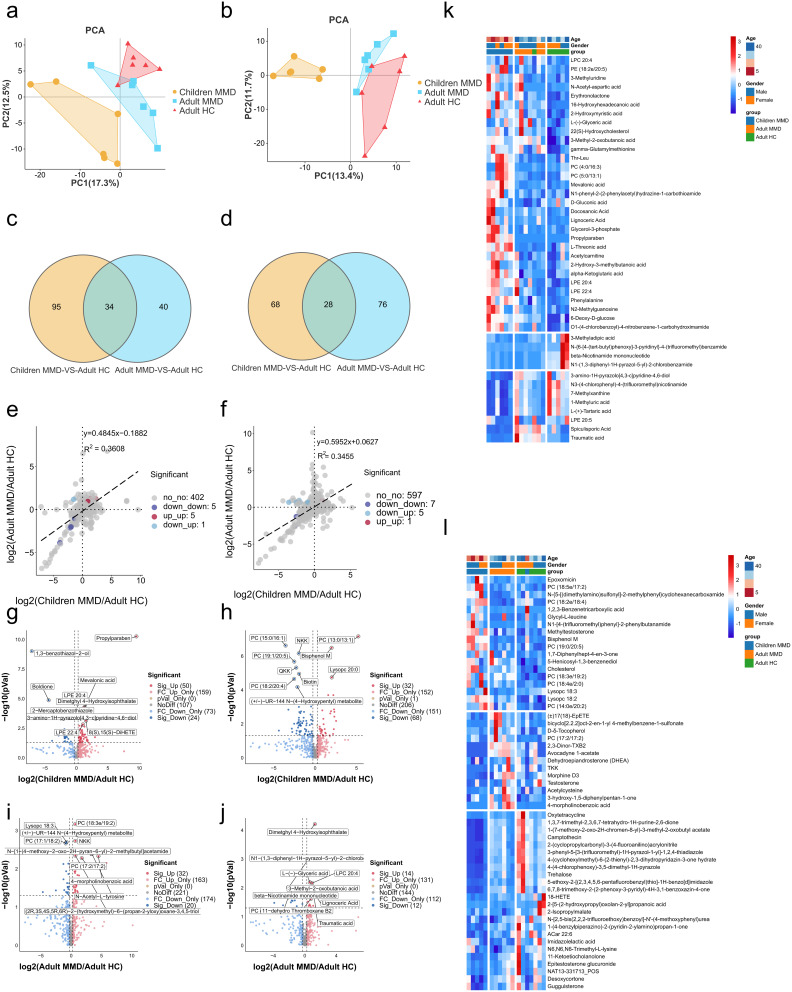
LC-MS was used in conjunction with UHPLC-MRM-MS for non-targeted metabolomics. Similar to the proteomics data, metabolomics data were classified using PCA, which showed significant differences in anionic and cationic metabolites between adult HC and adult/child patients with MMD at the global metabolomics level (Figure 2a and b). The expression of 129 anionic metabolites varied between child MMD and adult HC, and 74 varied between adult MMD and adult HCs. The expression of 34 metabolites were altered in adult versus child MMD, whereas 95 and 40 were uniquely associated with child or adult MMD, respectively (Figure 2c). Additionally, 96 and 104 cationic metabolites had varied expression levels between childhood MMD and adult HCs, and adult MMD and HCs, respectively. The expression of 28 metabolites were altered in both adult and child MMD, whereas 68 and 76 metabolites were uniquely associated with child and adult MMD, respectively (Figure 2d). The metabolic profiles of patients with MMD vary by age group, with both commonalities and variations observed. Figure 2e and f illustrate the fold-changes in anionic and cationic metabolite expression between adult MMD and adult HC, as well as between pediatric MMD and adult HCs. Some anionic metabolites showed similar changes in expression in pediatric and adult patients with MMD. The expression of beta-nicotinamide mononucleotide, 3-methyladipic acid, LPC 20:5, N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl) benzamide, and N1-(1,3-diphenyl-1H-pyrazol-5-yl)-2-chlorobenzamide was downregulated, while that of 6-deoxy-d-glucose, dimethyl 4-hydroxyisophthalate, lignoceric acid, gamma-glutamylmethionine, and LPC 20:4 was upregulated. Traumatic acid regulation showed opposite trends in pediatric and adult MMD (downregulated and upregulated, respectively) (Figure 2e).
Figure 2.
Differential metabolite expression levels between adult HCs and adults and children with MMD. The PCA of anionic and cationic metabolic data between adult HCs and adults with MMD (a) and children with MMD (b). Venn diagram illustrating differences in and overlap of anionic and cationic metabolites between adult HCs and adults with MMD (c) and children with MMD (d). Metabolic fold-changes in anionic and cationic metabolites in adults with MMD, and children with MMD (e), and between adults and children (f). Volcano plot summarizing the shared and distinct anionic and cationic metabolites between children with MMD and adult HCs (g) and between adult with MMD and adult HCs (h). Volcano plot summarizing the shared and distinct anionic and cationic metabolites between adults with MMD and adult HCs (i and j). Heatmap of DE metabolites levels between adult HCs and adults with MMD (k) and children with MMD (l). DE, differential expression.
Adult and pediatric MMD exhibited downregulated expression of several cationic metabolites, such as 4-(cyclohexylmethyl)-6-(2-thienyl)-2,3-dihydropyridazin-3-one hydrate, 2-Isopropylmalate, 5-ethoxy-2-[(2,3,4,5,6-pentafluorobenzyl)thio]-1H-benzo[d]imidazole, 2-[5-(2-hydroxypropyl)oxolan-2-yl]propanoic acid, ethyl 2-{[2-(4-methoxyphenoxy)-5-nitrobenzoyl]amino}acetate, PC (18:5e/2:0), and N-[methyl(oxo)phenyl-lambda-sulfanylidene]-N’-phenylurea, and upregulated expression of SM (d14:0/20:0). Notably, the expression of (±)-UR-144 N-(4-Hydroxypentyl) and PC (18:2/20:4), were downregulated in pediatric MMD patients but upregulated in adult MMD patients (Figure 2f). Figure 2g and h show that the expression of 66 anionic and 86 cationic metabolites exhibited significant differences (P < 0.05) in pediatric MMD vs adult HC, whereas Figure 2i and j display 39 anionic and 21 cationic metabolites with significantly different expression (P < 0.05) between adult MMD and adult HC. Figure 2k and l illustrate the differential metabolites data, supporting the PCA results. Hierarchical clustering revealed unique cluster patterns between anionic and cationic metabolites in adult MMD patients, pediatric MMD patients, and adult HCs. Moreover, this analysis revealed the co-regulation of a wide range of metabolites.
Propylparaben and 1.3-benzothiazol-2-ol showed the most significant upregulated and downregulated expression, respectively, in pediatric MMD patients compared to adult HCs. Propylparaben expression increased by 746.92-fold, whereas 1.3-benzothiazol-2-ol expression decreased by 0.01-fold (Table 2). However, its role in MMD pathogenesis remains unclear. The cationic metabolites PC (13:0/13:1) and avocadyne 1-acetate exhibited the highest upregulated and downregulated expression in both pediatric and adult MMD, with 37.32-fold and 0.05-fold changes, respectively (Table 2).
Table 2.
Top 10 DE Anionic & Cationic Metabolites in Childhood MMD Vs Adult HC
| Compound | FC | log2FC | P-value | VIP | Up/Down | |
|---|---|---|---|---|---|---|
| Anionic metabolites | ||||||
| Propylparaben | 746.92 | 9.54 | 0.000 | 2.33 | up | |
| 11-trans Leukotriene C4 | 8.79 | 3.14 | 0.012 | 1.76 | up | |
| FAHFA (18:0/3:0) | 7.77 | 2.96 | 0.018 | 1.72 | up | |
| Lignoceric Acid | 4.93 | 2.30 | 0.004 | 1.97 | up | |
| Docosanoic Acid | 3.48 | 1.80 | 0.033 | 1.61 | up | |
| 1,3-benzothiazol-2-ol | 0.01 | −7.10 | 0.000 | 2.27 | down | |
| 2-Mercaptobenzothiazole | 0.01 | −6.45 | 0.000 | 2.27 | down | |
| Boldione | 0.05 | −4.42 | 0.000 | 2.26 | down | |
| N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide | 0.07 | −3.87 | 0.037 | 1.59 | down | |
| 3-(3-Methoxyphenyl)propionic acid | 0.14 | −2.83 | 0.013 | 1.50 | down | |
| Cationic metabolites | ||||||
| PC (13:0/13:1) | 37.32 | 5.22 | 0.000 | 2.34 | up | |
| 2-(2-amino-3-methylbutanamido)-3-phenylpropanoic acid | 5.98 | 2.58 | 0.001 | 2.01 | up | |
| Proline-hydroxyproline | 5.80 | 2.54 | 0.007 | 1.87 | up | |
| N-(3-chloro-2-methylphenyl)-N’-(3-methoxypropyl)thiourea | 5.03 | 2.33 | 0.004 | 1.87 | up | |
| Dulcitol | 5.02 | 2.33 | 0.002 | 2.17 | up | |
| Avocadyne 1-acetate | 0.05 | −4.37 | 0.025 | 1.45 | down | |
| PC (15:0/16:1) | 0.08 | −3.56 | 0.000 | 2.08 | down | |
| 6-Keto-prostaglandin f1alpha | 0.11 | −3.22 | 0.001 | 1.71 | down | |
| Terephthalic Acid | 0.14 | −2.81 | 0.019 | 1.43 | down | |
| 2-[5-(2-hydroxypropyl)oxolan-2-yl]propanoic acid | 0.15 | −2.76 | 0.030 | 1.64 | down | |
In adult MMD and HC, the most notable changes in anionic metabolite expression were observed for indole-3-acrylic acid (2.55-fold change) and N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide (0.07-fold decrease) (Table 3). Cationic metabolite levels differed significantly between adult MMD and HCs, specifically for 2-[3-(4-pyridyl)-1H-1,2,4-triazol-5-yl] pyridine and PC (20:5e/17:2). The former exhibited 56.64-fold upregulated expression, whereas the latter demonstrated 0.04-fold downregulated expression (Table 3).
Table 3.
Top 10 DE Anionic & Cationic Metabolite in Adult MMD Vs Adult HC
| Compound | FC | log2FC | P-value | VIP | Up/Down | |
|---|---|---|---|---|---|---|
| Anionic metabolites | ||||||
| Indole-3-acrylic acid | 2.55 | 1.35 | 0.049 | 1.66 | up | |
| Dimethyl 4-Hydroxyisophthalate | 2.43 | 1.28 | 0.000 | 2.81 | up | |
| Lignoceric Acid | 2.34 | 1.23 | 0.016 | 1.51 | up | |
| Traumatic acid | 2.27 | 1.18 | 0.020 | 1.56 | up | |
| Capric acid | 2.10 | 1.07 | 0.042 | 2.00 | up | |
| N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide | 0.07 | −3.87 | 0.033 | 2.23 | down | |
| (±)18-HEPE | 0.15 | −2.76 | 0.040 | 1.76 | down | |
| beta-Nicotinamide mononucleotide | 0.23 | −2.13 | 0.010 | 2.35 | down | |
| PC (4:0/18:5) | 0.23 | −2.09 | 0.021 | 1.50 | down | |
| N1-(1,3-diphenyl-1H-pyrazol-5-yl)-2-chlorobenzamide | 0.25 | −2.02 | 0.009 | 2.36 | down | |
| Cationic metabolites | ||||||
| 2-[3-(4-pyridyl)-1H-1,2,4-triazol-5-yl]pyridine | 56.64 | 5.82 | 0.007 | 2.43 | up | |
| 4-morpholinobenzoic acid | 22.59 | 4.50 | 0.005 | 2.49 | up | |
| N-Acetyl-L-tyrosine | 10.71 | 3.42 | 0.004 | 2.45 | up | |
| N-[1-(4-methoxy-2-oxo-2H-pyran-6-yl)-2-methylbutyl]acetamide | 10.20 | 3.35 | 0.004 | 2.57 | up | |
| bicyclo[2.2.2]oct-2-en-1-yl 4-methylbenzene-1-sulfonate | 3.63 | 1.86 | 0.009 | 1.98 | up | |
| PC (20:5e/17:2) | 0.04 | −4.68 | 0.041 | 2.06 | down | |
| 2-[5-(2-hydroxypropyl)oxolan-2-yl]propanoic acid | 0.17 | −2.52 | 0.037 | 1.98 | down | |
| 5-ethoxy-2-[(2,3,4,5,6-pentafluorobenzyl)thio]-1H-benzo[d]imidazole | 0.41 | −1.28 | 0.007 | 1.51 | down | |
| PC (18:5e/2:0) | 0.45 | −1.15 | 0.036 | 1.78 | down | |
| PC (17:1/18:2) | 0.53 | −0.91 | 0.002 | 2.45 | down | |
Differential Protein Pathway Analysis
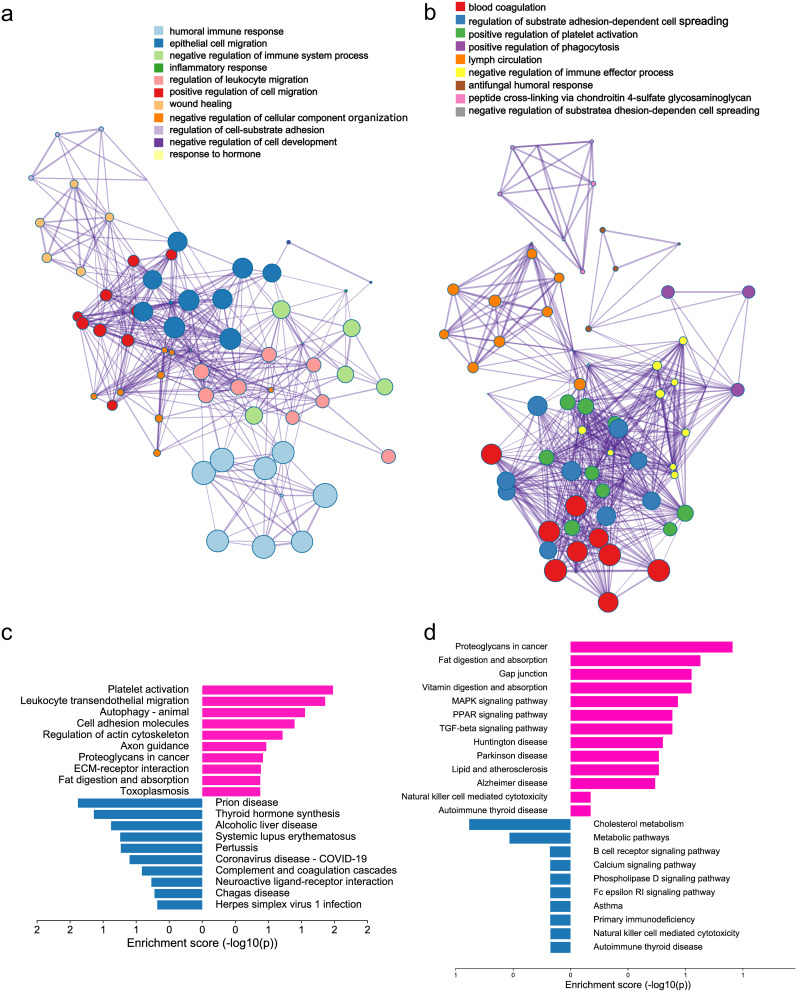
Gene Ontology (GO) pathway enrichment network clustering analysis was conducted on differentially expressed proteins in child MMD and adult HC using an online tool (http://metascape.org/gp). The Benjamini-Hochberg correction method was applied to correct for multiple comparisons (false discovery rate < 0.05). The analysis identified 11 significantly enriched GO pathways (Figure 3a), including four related to immune and inflammatory responses: the humoral immune response, negative regulation of immune system processes, inflammatory response, and regulation of leukocyte migration. This further confirmed the presence of immunosuppression in childhood MMD. Using the same approach, we identified nine enriched GO pathways in adult MMD patients versus adult HC. Three of these are related to the immune and inflammatory responses: positive regulation of platelet activation, positive regulation of phagocytosis, and negative regulation of immune effector processes (Figure 3b). These findings further confirm the presence of pro-inflammatory and immune suppression in adult MMD. The enriched KEGG signaling pathways were analyzed using the ClusterProfiler package in R software (version 3.18.0) to demonstrate the biological actions of DE proteins in children with MMD and adult HC. The pathways with the highest upregulation and downregulation in children with MMD compared to adult HC were platelet activation and prion disease, respectively (Figure 3c). The most significantly upregulated and downregulated pathways in adult MMD patients compared to those in adult HC were leukocyte transendothelial migration and thyroid hormone synthesis, respectively (Figure 3d). These findings indicate that abnormalities in energy metabolism and immune system regulation may occur in individuals with MMD.
Figure 3.
Differential protein pathway analysis. (a) Diagram showing GO enrichment analysis clustered networks of differentially expressed proteins in pediatric MMD vs adult HC (b). Clustered network diagram GO enrichment analysis of differentially expressed proteins in adult MMD vs adult HCs. (c) KEGG pathway enrichment analysis of differentially expressed proteins in pediatric MMD vs adult HCs. (d) Enrichment analysis of KEGG pathways for proteins showing differential expression in adult MMD vs adult HCs. Red: upregulation, Blue: downregulation.
Differential Metabolite Pathway Analysis
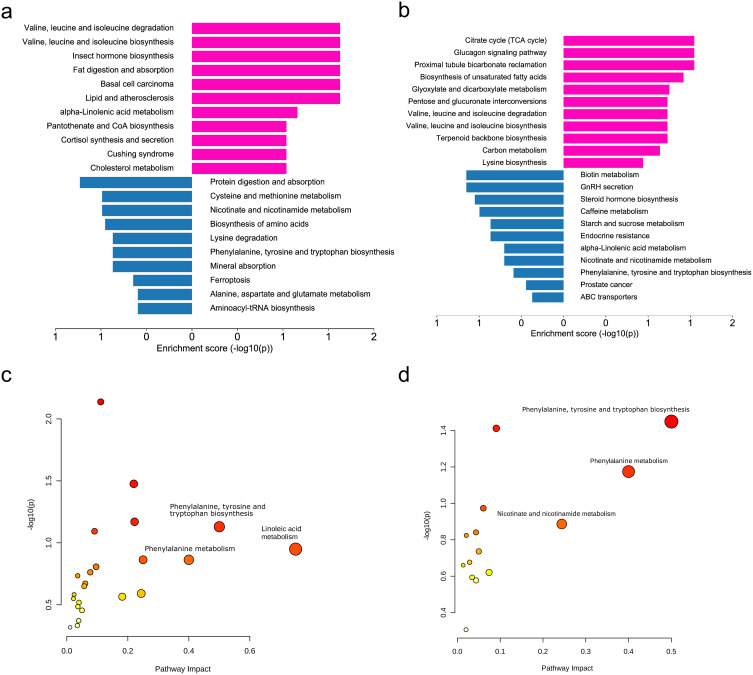
KEGG-enriched signaling pathways were analyzed using the R ClusterProfiler package to identify the primary biological effects of DE metabolites in children with MMD compared to those in adult HC (Figure 4a and b). The citrate cycle was found to be significantly upregulated, while biotin metabolism was prominently downregulated in children with MMD compared to adult HC. Valine, leucine, and isoleucine degradation were the most significantly upregulated pathways in adult MMD vs adult HCs, whereas protein digestion and absorption pathways were downregulated. Cholesterol metabolism was upregulated in children with MMD but downregulated in adults with MMD. These findings are important for understanding and managing MMD in different age groups. Integrative analysis of the differential metabolites and proteins was conducted using an online tool (https://genap.metaboanalyst.ca/) (Figure 4c and d). Phenylalanine, tyrosine, and tryptophan biosynthesis, along with phenylalanine, nicotinate, and nicotinamide metabolism, have a more significant impact on children with MMD than on healthy adults. These findings suggest a potential connection between these pathways and the development of MMD.
Figure 4.
Differential metabolite pathway analysis. (a) KEGG pathway enrichment analysis of differentially expressed proteins in pediatric MMD vs adult HCs. (b) KEGG enrichment analysis of pathways for proteins showing differential expression in adult MMD vs adult HCs. (c) Integrative analysis of differential metabolites and proteins in pediatric MMD vs adult HCs at the pathway level. (d) Integrative analysis of differential metabolites and proteins in adult MMD vs adult HCs at the pathway level. Red: upregulation, Blue: downregulation.
Correlation Analysis of DE Proteins and Metabolites
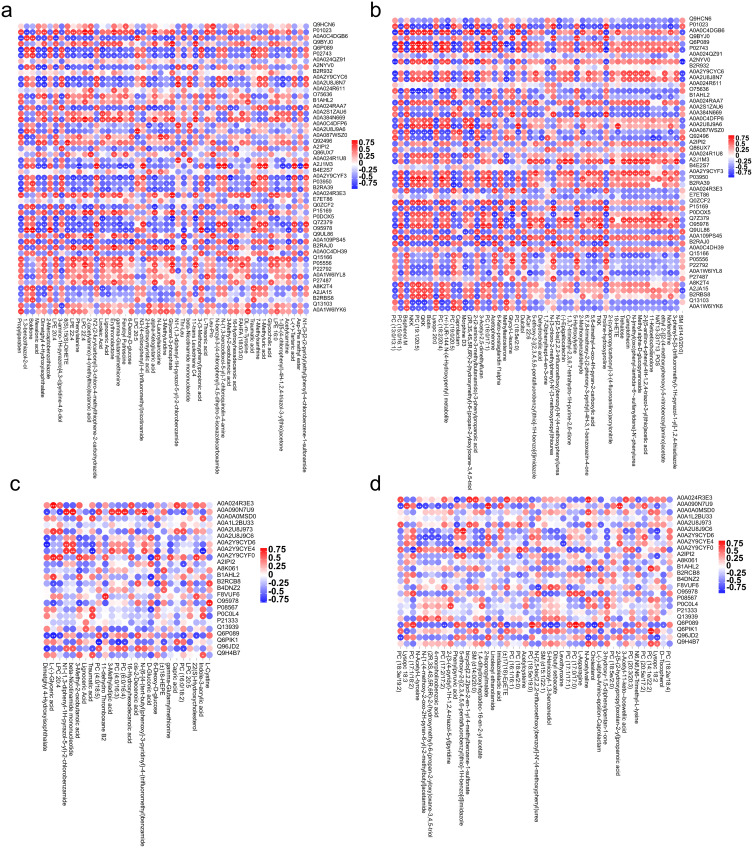
Pearson’s correlation coefficient analysis was conducted to determine the correlation between the identified differential proteins and the negative or positive ion metabolites from the metabolomic analysis. The following pairs were highly correlated: LPC 20:5 and Q6MZX9 (CC = 0.93, P < 0.001); LPC 22:4 and P01023 (CC = 0.92, P < 0.001); Boldione and Q9UL95 (CC = 0.92, P < 0.001); Boldione and O95978 (CC = 0.91, P < 0.001); and Erythronolactone and B2RAJ0 (CC = 0.91, P < 0.001) (Figure 5a). The top five protein and cationic metabolite correlations in MMD children vs adult HC were: PC (19:1/20:5) and Q6P089 (CC = 0.96, P < 0.001), N-(3-chloro-2-methylphenyl)-N’-(3-methoxypropyl)thiourea and B2RAJ0 (CC = 0.95, P < 0.001), PC (18:2/20:4) and P03950 (CC = 0.94, P < 0.001), (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-(propan-2-yloxy)oxane-3,4,5-triol and A0A2U8J9A6 (CC = 0.93, P < 0.001), and TKK and O95978 (CC = 0.93, P < 0.001) (Figure 5b). There were strong correlations between specific protein and anion metabolite pairs differentially expressed in Adult MMD vs HC, including L-(-)-Glyceric acid and A0A024R3E3 (CC = 0.86, P < 0.001), N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide and A0A090N7U9 (CC = 0.83, P = 0.001), beta-Nicotinamide mononucleotide and A0A090N7U9 (CC = 0.82, P = 0.001), L-Cystine and O95978 (CC = 0.80, P = 0.001), and N1-(1,3-diphenyl-1H-pyrazol-5-yl)-2-chlorobenzamide and A0A090N7U9 (CC = 0.80, P = 0.001) (Figure 5c). The top five correlated proteins and cationic metabolites in adult MMD vs adult HC were PC (17:0/17:0) and Q6P089 (CC = 0.92, P < 0.001), PC (14:1e/22:2) and Q6PIK1 (CC = 0.89, P < 0.001), 5-ethoxy-2-[(2,3,4,5,6-pentafluorobenzyl)thio]-1H-benzo[d]imidazole and A0A2U8J9C6 (CC = 0.87, P < 0.001), PC (18:5e/19:0) and Q6P089 (CC = 0.87, P < 0.001), 2-isopropylmalate and A0A090N7U9 (CC = 0.87, P < 0.001) (Figure 5d).
Figure 5.
Correlation analysis of DE proteins and metabolites. (a) Correlation analysis of DE proteins and anionic metabolites in pediatric MMD vs adult HCs. (b) Correlation analysis of DE proteins and cationic metabolites in pediatric MMD vs adult HCs. (c) Correlation analysis of DE proteins and cationic metabolites in adult MMD vs adult HCs. (d) Correlation analysis of DE proteins and cationic metabolites in adult MMD vs adult HCs.
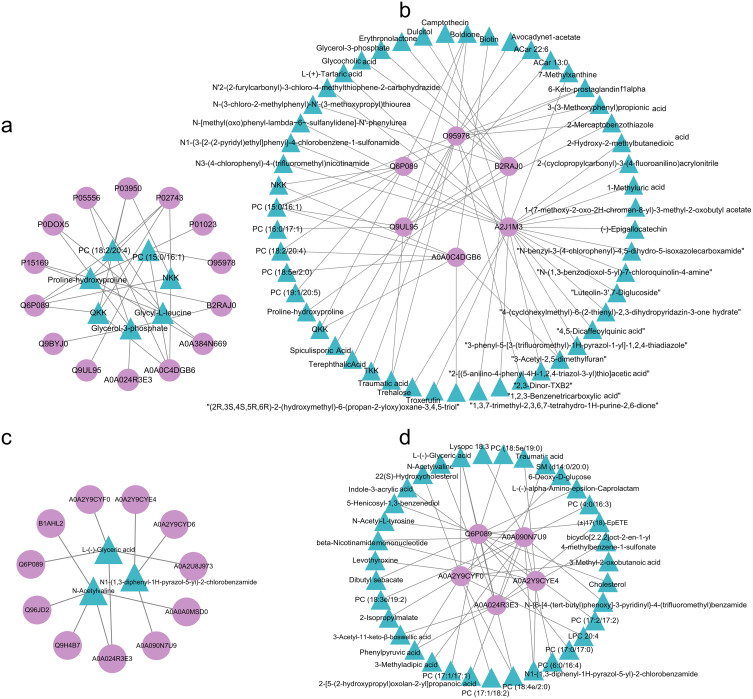
We used the Cytoscape software to identify the core metabolites and proteins involved in both childhood and adult MMD through differential network analysis. The core metabolites of childhood MMD included PC (18:2/20:4), PC (15:0/16:1), NKK, Glycy-L-leucine, Glycerol-3-phosphate, QKK, and proline hydroxyproline (Figure 6a). The core proteins found in children with MMD included O95978, Q6P089, Q9UL95, A0A0C4DGB6, B2RAJ0, and A2J1M3 (Figure 6b). Moreover, adult MMD presented only three core metabolites, namely, L-(-)-l-glyceric acid, N-acetylvaline, and N1-(1,3-diphenyl-1H-pyrazol-5-yl)-2-chlorobenzamide (Figure 6c), and five core proteins, namely, A0A024R3E3, A0A2Y9CYE4, A0A2Y9CYF0, A0A090N7U9, and Q6P089 (Figure 6d).
Figure 6.
Correlation network analysis of DE proteins and metabolites. Core metabolites (a) and proteins (b) in childhood MMD; and core metabolites (c) and proteins (d) in adult MMD. Triangle: metabolites; circle: proteins.
Analysis of Shared KEGG Pathways Between DE Proteins and DE Metabolites
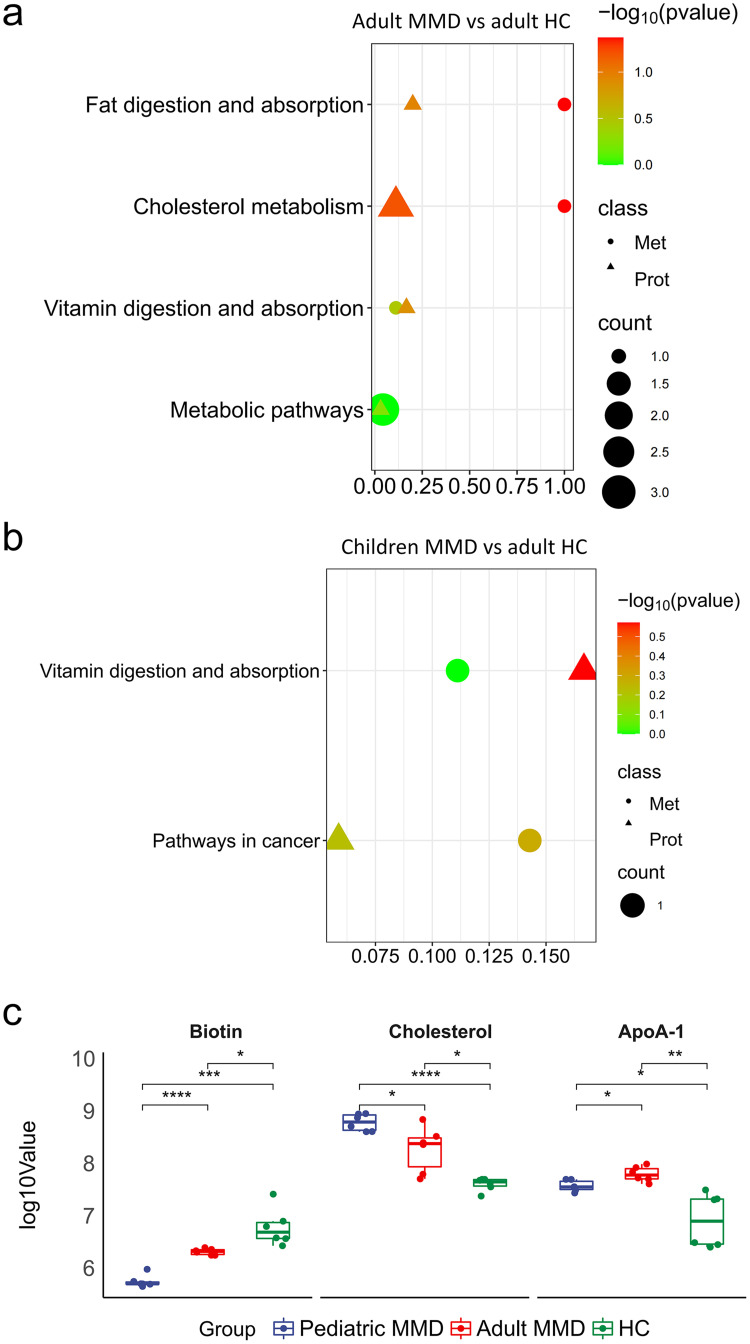
The KEGG pathway database was used to analyze the integration of differential proteins and metabolites, as well as the main biochemical and signal transduction pathways involved. Our study found that vitamin digestion and absorption were the primary pathways altered in children with MMD compared to adult HC, and adult MMD compared to adult HC (Figure 7a and b). PathView analysis identified biotin, cholesterol, and ApoA-1 as key molecules in the vitamin digestion and absorption pathways in patients with MMD. These molecules play important roles in the metabolism of vitamins and lipids, which are impaired in MMD (Figure S2). Abundance analysis of biotin, cholesterol, and ApoA-1 showed that biotin levels were significantly different among the groups (P < 0.05), indicating their importance in the development of MMD (Figure 7c). Biotins had a more significant role in the occurrence of MMD in children. Furthermore, ApoA-1 expression was also found to be significantly different in children with MMD vs adult HC, and in adult MMD vs adult HC; thus, it was deemed to be important in the onset of MMD. Abundance analysis demonstrated that biotin and ApoA-1 could serve as potential biomarkers for the diagnosis and treatment of MMD.
Figure 7.
Analysis of shared KEGG pathway between DE proteins and metabolites. Shared KEGG pathways between DE proteins and metabolites in childhood MMD vs adult HC (a) and adult MMD vs adult HC (b). Variations in the levels of metabolites and proteins involved in the digestion and absorption pathways of vitamins (c).
Discussion
We identified key disease pathways in both pediatric and adult MMD sera compared with sex-matched adult controls and created a protein-metabolite resource for MMD researchers. We verified the known disease mechanisms in both pediatric and adult patients with MMD, including alterations in numerous inflammatory and immune proteins that are well-established pathological features of MMD.
In children with moyamoya disease (MMD), propylparaben expression increased by 746.92-fold when compared to healthy controls (HC), suggesting a strong pro-inflammatory.18 Conversely, anti-inflammatory and antioxidant metabolites, including 1.3-benzothiazol-2-ol19 and avocadyne 1-acetate,20 were markedly downregulated. The presence of elevated PC (13:0/13:1) levels was observed in pediatric MMD for the first time, with its role in MMD pathology being uncertain. Nonetheless, 1.3-benzothiazol-2-ol is recognized for its biological activities, potentially implicating its deficiency in inflammation and disease processes.21 Additionally, increased PC (13:0/13:1) has been linked to inflammation and viral infections such as hepatitis B.22 Avocado-derived avocadyne 1-acetate exhibits anti-inflammatory, antioxidant, and anticancer potential.23 Intestinal synthesis of indole-3-acrylic acid shows promise in delaying atherosclerosis progression,24 and benzamide treatment can mitigate inflammation and ischemia-reperfusion injury.25 The involvement of these factors in MMD pathophysiology warrants further exploration. Likewise, the therapeutic applications of pyridine compounds26 and the role of PC (20:5e/17:2)27 in cardiovascular health, due to effects on cholesterol metabolism, should be investigated.
Upregulation of P05556I (integrin β1) in non-alcoholic steatohepatitis (NASH) mouse models suggests involvement in promoting monocyte adhesion and hepatitis. Similarly, integrin β1 is upregulated in pediatric MMD, potentially contributing to disease progression.28 Q6MZX9, akin to immunoglobulin alpha-2 heavy chain and vital to IgA function, presents an unknown role in MMD, yet emphasizes the need for research to unveil its biological significance. IgA is crucial for mucosal immunity and managing infections, autoimmune conditions, allergies, and gut diseases.29 Decreased expression of A8K2T4 (C7), a complement system component, in both adult and pediatric MMD patients may predispose to bacterial infections and autoimmune disorders.30 Q6P089, encoded by NUBPL, is essential for mitochondrial respiratory chain complex I integrity. NUBPL deficiency can escalate ROS production and mitochondrial dysfunction.31 ITGB1 participates in cell adhesion, signal transmission, and immune regulation.32 A significant downregulation of Q6MZX9 in pediatric MMD further implicates the role of inflammation in its pathogenesis. These findings contrast with adult MMD cases; hence, targeted research is essential to discern age-related effects in the disease’s progression and manifestations.
Lignoceric acid, N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide and 2-[5-(2-hydroxypropyl)oxolan-2-yl]propanoic acid are among the top ten differential metabolites in children and adults with MMD, of which lignoceric acid expression was upregulated, and N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide and 2-[5-(2-hydroxypropyl)oxolan-2-yl]propanoic acid were downregulated. Lignoceric acid is a circulating very-long chain saturated fatty acid, which has a negative correlation with cardiovascular disease (CVD) in anti-atherosclerotic activity, as atherosclerosis is a pathological indicator of early cardiovascular disease.33 Further research is necessary to fully understand the intricate relationship between lignoceric acid and MMD, as this protective mechanism against atherosclerosis conflicts with the vascular damage mechanisms underlying MMD.
N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide, also known as GSK583, is a novel, selective inhibitor of the receptor-interacting protein kinase 2 (RIP2), which has been shown to have potential therapeutic applications in the treatment of inflammatory diseases.34 We speculate that a decrease in N-{6-[4-(tert-butyl)phenoxy]-3-pyridinyl}-4-(trifluoromethyl)benzamide levels may compromise the inhibitory action of RIP2 on MMD, which could result in pro-inflammatory responses. 2-[5-(2-Hydroxypropyl) oxylan-2-yl] propanoic acid is a type of propanoic acid (PA) that reduces intestinal cholesterol absorption and aortic atherosclerotic lesion areas. Additionally, it increases the number of regulatory T cells and the level of interleukin (IL)-10 in the intestinal microenvironment, which inhibits the expression of the major intestinal cholesterol transporter Niemann Pick C1 like 1 (Npc1l1).35 Although these metabolites have been widely investigated in arteriosclerosis, this is the first report on changes in these metabolites in MMD. According to the previous only literature on non-targeted metabolomics of moyamoya disease using plasma samples,36 MMD patients were found to have higher levels of phenol, 2-hydroxybutyric acid, L-isoleucine, L-serine, glycerol, pelargonic acid, L-methionine, myristic acid, pyroglutamic acid, palmitic acid, palmitoleic acid, stearic acid, octadecanamide, monoglyceride (MG) (16:0/0:0/0:0), and MG (0:0/18:0/0:0), as well as lower levels of L-alanine, L-valine, urea, succinic acid, L-phenylalanine, L-threonine, L-tyrosine, edetic acid, and oleamide, when compared to the levels of HCs. The previous research results were not replicated well in our study. Upon careful comparison, we found that the previous study utilized a non-targeted gas chromatography-mass spectrometry (GC-MS) approach, while we utilized a liquid chromatography tandem mass spectrometry approach (LC-MS/MS). Although both GC-MS and LC-MS/MS can be used to analyze metabolites in biological samples, differences in their principles, detection targets, and sensitivity can lead to different results. LC-MS/MS has high selectivity, sensitivity, and resolution, which can detect very low levels of specific molecules in small samples; and its detection method is not limited to volatile molecules. As a result, it is more reliable in producing results.37 In addition, there is a metabolomics study on patients with MMD using cerebrospinal fluid samples.38 They used nuclear magnetic resonance (NMR) analysis to complete the analysis of cerebrospinal fluid, and their results did not replicate our results or those obtained through GC-MS analysis. This further indicates that different analytical techniques may focus on different analytes.
The top ten proteins in children and adults with MMD were not shared between the two groups. In adult MMD, the expression of Q9H4B7, encoded by TUBB1, and Q6P089, encoded by IGH@, exhibited the highest degrees of upregulation and downregulation among all the proteins, respectively. TUBB1 expression is positively correlated with gamma delta T cells, but is negatively correlated with helper follicular T cells infiltration.39 Furthermore, downregulated IGH@ expression has been associated with altered B cell development and function. Studies have shown that changes in IGH@ expression can affect B cell receptor signaling and development of B cell lymphomas. In addition, IGH@ plays a crucial role in the elimination of pathogens and long-term regulation of immunity.40 These findings suggest that the etiology and advancement of MMD may vary between pediatric and adult populations via distinct mechanisms.
Previous studies have shown that multiple inflammatory signaling pathways are involved in the pathogenesis of MMD, including the nuclear factor-κB (NF-κB),41 transforming growth factor-β (TGF-β),42 and mitogen-activated protein kinase (MAPK) pathways.43 Our GO enrichment results identified four pathways related to inflammation and immunity, including humoral immune response, negative regulation of immune system processes, inflammatory response, and regulation of leukocyte migration, which were involved in the occurrence and progression of childhood MMD. Only three pathways, including positive regulation of platelet activation, positive regulation of phagocytosis, and negative regulation of immune effector processes, were involved in adult MMD. These newly identified functional pathways exhibited no overlap, highlighting notable distinctions in the proteins that undergo changes during disease development in pediatric and adult populations. KEGG enrichment analysis results underscored the role of the previously identified pathological pathways in adult MMD, such as MAPK and TGF-β. Moreover, we identified novel pathways relevant to pediatric and adult MMD. Consistent with the findings of Geng et al36, the metabolite KEGG enrichment analysis revealed phenylalanine, tyrosine, and tryptophan biosynthesis, and phenylalanine metabolism had the greatest impact on the pathways in both children and adults with MMD. Our findings revealed new pathways in childhood MMD, such as the citrate cycle (TCA cycle) and biotin metabolism. The role of metabolism in the TCA cycle in immune cells is not limited to adenosine triphosphate production. It has been increasingly recognized as a source of the synthesis of metabolites that function as signaling molecules in the immune system.44 Biotin deficiency is associated with high blood sugar levels and insulin resistance in both animals and humans. Biotin supplementation reduces coronary artery thickening and stroke incidence in rats.45 Although there have been relevant studies on the role of these pathways in promoting inflammation and preventing arteriosclerosis, further research is needed to elucidate the exact role of these mechanisms in MMD.
To obtain valuable insights into MMD pathophysiology, we performed an integrated study combining proteomic and metabolomic analyses. Correlation network analysis succinctly illustrated the most important nodes in children and adults with MMD. Many nodal metabolites such as NKK and QKK were discovered for the first time in children with MMD. No overlapping metabolite nodes were found in adults and children with MMD, suggesting a significant difference in metabolism between children and adults with MMD. An overlapping node protein, Q6P089, was identified in adults and children with MMD, which has been found to participate in the immune response by affecting B cell function.40 In the shared KEGG pathway analysis, we identified vitamin digestion and absorption as the core pathways of MMD in children and adults. Further analysis of the abundance of metabolites and proteins involved in this pathway revealed significant differences in the abundance of biotin between the three groups. Biotin deficiency was more pronounced in children than in adults with MMD. Therefore, we speculate that biotin deficiency is related to faster disease progression and that biotin supplementation therapy may slow the rapid progression of childhood MMD. In addition, we found that ApoA-1 is important in the development of MMD. Similar to our research, previous studies46 showed that serum proteomics of DIA in patients with moyamoya disease showed that apolipoprotein C-I (ApoC1), apolipoprotein D (ApoD), and apolipoprotein A-IV (ApoA4) were reduced.
Limitations
While we are the first to integrate proteomic and metabolomic data to study MMD in children, our study has certain limitations. The small sample size necessitates caution in data interpretation and warrants future studies with expanded cohorts to corroborate our findings. Our sample is regionally specific, drawn from a single Chinese center; thus, the results may reflect regional traits. Furthermore, the lack of an age, sex, race, weight, and height-matched pediatric control group limited our choice to adult patient comparisons, introducing potential biases due to age-related differences. Although multiple comparisons with adult controls provided valuable insights, they inherently restrict the generalizability and reliability of our findings. Future research should aim to include more representative control groups to overcome this constraint. External variables, such as dietary and environmental factors, could influence plasma metabolite profiles, introducing additional layers of complexity to our analysis. Validation of our results with larger, independent datasets and experimental models is essential to substantiate our conclusions.
Conclusion
This study unveils distinct metabolic and proteomic patterns within pediatric and adult MMD patients. The critical role of the vitamin digestion and absorption pathway in the pathogenesis of pediatric MMD offers novel insight into disease mechanisms.
Acknowledgments
We thank the individuals who contributed to the study or manuscript preparation but did not fulfill all the criteria of authorship.
Funding Statement
This study was supported by grants from the National Natural Science Foundation of China (grant numbers: 82171280 and 82201451).
Abbreviations
ADL, activities of daily living; AGC, automatic gain control; adult MMD, adults with moyamoya disease; BCAA, branched-chain amino acids; CVD, cardiovascular disease; child MMD, children with moyamoya disease; DDA, data-dependent acquisition; DE, differentially expressed; DEP, differentially expressed proteins; DTT, dithiothreitol; FA, formic acid; FAO, fatty acid oxidation; GO, Gene Ontology; HC, healthy control; HCD, higher energy collisional dissociation; HMDB, Human Metabolome Database; HRMS, high-resolution mass spectrometry; ICA, internal carotid artery; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC-MS, liquid chromatography-mass spectrometry; MMD, moyamoya disease; rpm, rotations per minute; NAD, nicotinamide adenine dinucleotide; NCE, normalized collision energy; NMR, nuclear magnetic resonance; PA, propanoic acid; PCA, principal component analysis; ROS, reactive oxygen species; TCA, citrate cycle; TIA, transient ischemic attack; TXM, thromboxane metabolites; UHPLC, ultra-high-performance liquid chromatography; UHPLC-DIA-MS, ultra-performance liquid chromatography and data-independent acquisition mass spectrometry.
Data Sharing Statement
Our team offers proteomic results via a user-friendly online repository (https://www.iprox.org/; DOI: IPX0006333000) as a publicly available resource, enabling researchers to access moyamoya disease patient sera. Metabolomics results were generated and stored at the Fifth Medical Center of the People’s Liberation Army of China (PLA) General Hospital. Data supporting the results of this study may be obtained from the corresponding authors.
Ethical Approval and Consent to Participate
This study was approved by the Ethics Committee of the Fifth Medical Center of the PLA General Hospital (Approval number: ky-2020-9-22) and complied with the Declaration of Helsinki. Informed consents were obtained from the legal guardians and relatives of the study participants prior to study commencement. The adult patients and healthy controls also provided informed consent.
Disclosure
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Suzuki J. Cerebrovascular “moyamoya” disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol. 1969;20(3):288–299. doi: 10.1001/archneur.1969.00480090076012 [DOI] [PubMed] [Google Scholar]
- 2.Kuroda S, Houkin K. Moyamoya disease: current concepts and future perspectives. Lancet Neurol. 2008;7(11):1056–1066. doi: 10.1016/S1474-4422(08)70240-0 [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Zheng L, Feng L. Epidemiology, diagnosis and treatment of moyamoya disease. Exp Ther Med. 2019;17(3):1977–1984. doi: 10.3892/etm.2019.7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzman R, Lee M, Achrol A, et al. Clinical outcome after 450 revascularization procedures for moyamoya disease. Clinical Article J Neurosurg. 2009;111(5):927–935. doi: 10.3171/2009.4.JNS081649 [DOI] [PubMed] [Google Scholar]
- 5.Scott RM, Smith JL, Robertson RL, et al. Long-term outcome in children with moyamoya syndrome after cranial revascularization by pial synangiosis. J Neurosurg. 2004;100(2 Suppl Pediatrics):142–149. doi: 10.3171/ped.2004.100.2.0142 [DOI] [PubMed] [Google Scholar]
- 6.Papavasiliou A, Bazigou-Fotopoulou H, Ikeda H. Familial Moyamoya disease in two European children. J Child Neurol. 2007;22(12):1371–1376. doi: 10.1177/0883073807307101 [DOI] [PubMed] [Google Scholar]
- 7.Fiaschi P, Scala M, Piatelli G, et al. Limits and pitfalls of indirect revascularization in moyamoya disease and syndrome. Neurosurg Rev. 2021;44(4):1877–1887. doi: 10.1007/s10143-020-01393-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tortora D, Scavetta C, Rebella G, et al. Spatial coefficient of variation applied to arterial spin labeling MRI may contribute to predict surgical revascularization outcomes in pediatric moyamoya vasculopathy. Neuroradiology. 2020;62(8):1003–1015. doi: 10.1007/s00234-020-02446-4 [DOI] [PubMed] [Google Scholar]
- 9.Fujimura M, TOMINAGA T, KURODA S, et al. 2021 Japanese Guidelines for the Management of Moyamoya Disease: guidelines from the Research Committee on Moyamoya Disease and Japan Stroke Society. Neurol Med Chir. 2022;62(4):165–170. doi: 10.2176/jns-nmc.2021-0382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunet T, Zott B, Lieftüchter V, et al. De novo variants in RNF213 are associated with a clinical spectrum ranging from Leigh syndrome to early-onset stroke. Genet Med. 2024;26(2):101013. doi: 10.1016/j.gim.2023.101013 [DOI] [PubMed] [Google Scholar]
- 11.Araki Y, Yoshikawa K, Okamoto S, et al. Identification of novel biomarker candidates by proteomic analysis of cerebrospinal fluid from patients with moyamoya disease using SELDI-TOF-MS. BMC Neurol. 2010;10:112. doi: 10.1186/1471-2377-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X. Targeted metabolomics analysis of serum amino acid profiles in patients with Moyamoya disease. Amino Acids. 2022;54(1):137–146. doi: 10.1007/s00726-021-03100-w [DOI] [PubMed] [Google Scholar]
- 13.Guo Q, Wang Q-N, Li J, et al. Proteomic and metabolomic characterizations of moyamoya disease patient sera. Brain Behav. 2023;13(12):e3328. doi: 10.1002/brb3.3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacMullan MA, Dunn ZS, Graham N, et al. Quantitative Proteomics and Metabolomics Reveal Biomarkers of Disease as Potential Immunotherapy Targets and Indicators of Therapeutic Efficacy. Theranostics. 2019;9(25):7872–7888. doi: 10.7150/thno.37373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruderer R, Bernhardt OM, Gandhi T, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to Acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14(5):1400–1410. doi: 10.1074/mcp.M114.044305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malle E, De Beer FC. Human serum amyloid A (SAA) protein: a prominent acute-phase reactant for clinical practice. Eur J Clin Invest. 1996;26(6):427–435. doi: 10.1046/j.1365-2362.1996.159291.x [DOI] [PubMed] [Google Scholar]
- 18.Watkins DJ, Ferguson KK, Anzalota Del Toro LV, et al. Associations between urinary phenol and paraben concentrations and markers of oxidative stress and inflammation among pregnant women in Puerto Rico. Int J Hyg Environ Health. 2015;218(2):212–219. doi: 10.1016/j.ijheh.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamal A, Syed MA, Mohammed SM. Therapeutic potential of benzothiazoles: a patent review. Expert Opin Ther Pat. 2015;25(3):335–349. doi: 10.1517/13543776.2014.999764 [DOI] [PubMed] [Google Scholar]
- 20.Tcheng M, Minden MD, Spagnuolo PA. Avocado-derived avocadyne is a potent inhibitor of fatty acid oxidation. J Food Biochem. 2022;46(3):e13895. doi: 10.1111/jfbc.13895 [DOI] [PubMed] [Google Scholar]
- 21.Tariq S, Kamboj P, Amir M. Therapeutic advancement of benzothiazole derivatives in the last decennial period. Arch Pharm. 2019;352(1):e1800170. doi: 10.1002/ardp.201800170 [DOI] [PubMed] [Google Scholar]
- 22.Saito T, Sugimoto M, Okumoto K, et al. Serum metabolome profiles characterized by patients with hepatocellular carcinoma associated with hepatitis B and C. World J Gastroenterol. 2016;22(27):6224–6234. doi: 10.3748/wjg.v22.i27.6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhuyan DJ. The Odyssey of Bioactive Compounds in Avocado (Persea americana) and Their Health Benefits. Antioxidants. 2019;8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang HL, Chiu -C-C, Lo C, et al. Circulating gut microbiota-related metabolites influence endothelium plaque lesion formation in ApoE knockout rats. PLoS One. 2022;17(5):e0264934. doi: 10.1371/journal.pone.0264934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ducrocq S, Benjelloun N, Plotkine M, et al. Poly(ADP-ribose) synthase inhibition reduces ischemic injury and inflammation in neonatal rat brain. J Neurochem. 2000;74(6):2504–2511. doi: 10.1046/j.1471-4159.2000.0742504.x [DOI] [PubMed] [Google Scholar]
- 26.Alrooqi M, Khan S, Alhumaydhi FA, et al. A Therapeutic Journey of Pyridine-based Heterocyclic Compounds as Potent Anticancer Agents: a Review (From 2017 to 2021). Anticancer Agents Med Chem. 2022;22(15):2775–2787. doi: 10.2174/1871520622666220324102849 [DOI] [PubMed] [Google Scholar]
- 27.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44(6):1071–1079. doi: 10.1194/jlr.R300004-JLR200 [DOI] [PubMed] [Google Scholar]
- 28.Guo Q, Furuta K, Lucien F, et al. Integrin β(1)-enriched extracellular vesicles mediate monocyte adhesion and promote liver inflammation in murine NASH. J Hepatol. 2019;71(6):1193–1205. doi: 10.1016/j.jhep.2019.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugino H, Sawada Y, Nakamura M. IgA Vasculitis: etiology, Treatment, Biomarkers and Epigenetic Changes. Int J Mol Sci. 2021;22(14):7538. doi: 10.3390/ijms22147538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corvini M, Randolph C, Aronin SI. Complement C7 deficiency presenting as recurrent aseptic meningitis. Ann Allergy Asthma Immunol. 2004;93(2):200–205. doi: 10.1016/S1081-1206(10)61476-7 [DOI] [PubMed] [Google Scholar]
- 31.Keeney PM, Xie J, Capaldi RA, et al. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J Neurosci. 2006;26(19):5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu W, Sun H, Zhang M, et al. ITGB1 as a prognostic biomarker correlated with immune suppression in gastric cancer. Cancer Med. 2023;12(2):1520–1531. doi: 10.1002/cam4.5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M, Zuo L-S-Y, Sun T-Y, et al. Circulating Very-Long-Chain Saturated Fatty Acids Were Inversely Associated with Cardiovascular Health: a Prospective Cohort Study and Meta-Analysis. Nutrients. 2020;12(9). doi: 10.3390/nu12092709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tigno-Aranjuez JT, Benderitter P, Rombouts F, et al. In vivo inhibition of RIPK2 kinase alleviates inflammatory disease. J Biol Chem. 2014;289(43):29651–29664. doi: 10.1074/jbc.M114.591388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haghikia A, Zimmermann F, Schumann P, et al. Propionate attenuates atherosclerosis by immune-dependent regulation of intestinal cholesterol metabolism. Eur Heart J. 2022;43(6):518–533. doi: 10.1093/eurheartj/ehab644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng C, Cui C, Guo Y, et al. Metabolomic Profiling Revealed Potential Biomarkers in Patients With Moyamoya Disease. Front Neurosci. 2020;14:308. doi: 10.3389/fnins.2020.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Núñez O, Gallart-Ayala H, Martins CPB, et al. State-of-the-art in fast liquid chromatography-mass spectrometry for bio-analytical applications. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:3–21. doi: 10.1016/j.jchromb.2012.12.031 [DOI] [PubMed] [Google Scholar]
- 38.Jeon JP, Yun T, Jin X, et al. 1H-NMR-based metabolomic analysis of cerebrospinal fluid from adult bilateral moyamoya disease: comparison with unilateral moyamoya disease and atherosclerotic stenosis. Medicine. 2015;94(17):e629. doi: 10.1097/MD.0000000000000629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Gong M, Xiong Z, et al. ADAM19 and TUBB1 Correlate with Tumor Infiltrating Immune Cells and Predicts Prognosis in Osteosarcoma. Comb Chem High Throughput Screen. 2023;26(1):135–148. doi: 10.2174/1386207325666220406112305 [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Li F. [Molecular Characteristics and Clinical Significance of IGH Gene Rearrangement in B-Cell Lymphoma--Review]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2022;30(4):1291–1295. doi: 10.19746/j.cnki.issn.1009-2137.2022.04.052 [DOI] [PubMed] [Google Scholar]
- 41.Takeda M, Tezuka T, Kim M, et al. Moyamoya disease patient mutations in the RING domain of RNF213 reduce its ubiquitin ligase activity and enhance NFκB activation and apoptosis in an AAA+ domain-dependent manner. Biochem Biophys Res Commun. 2020;525(3):668–674. doi: 10.1016/j.bbrc.2020.02.024 [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Tang M, Li H, et al. TGFβ1 as a Predictive Biomarker for Collateral Formation Within Ischemic Moyamoya Disease. Front Neurol. 2022;13:899470. doi: 10.3389/fneur.2022.899470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chelleri C, Scala M, De Marco P, et al. Case report: revascularization failure in NF1-related moyamoya syndrome after selumetinib: a possible pathophysiological correlation? Front Pediatr. 2023;11:1051026. doi: 10.3389/fped.2023.1051026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams NC, O’Neill LAJ. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front Immunol. 2018;9:141. doi: 10.3389/fimmu.2018.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riveron-Negrete L, Fernandez-Mejia C. Pharmacological Effects of Biotin in Animals. Mini Rev Med Chem. 2017;17(6):529–540. doi: 10.2174/1389557516666160923132611 [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Ji C, Han Q, et al. Data-Independent Acquisition-Based Serum Proteomic Profiling of Adult Moyamoya Disease Patients Reveals the Potential Pathogenesis of Vascular Changes. J Mol Neurosci. 2022;72(12):2473–2485. doi: 10.1007/s12031-022-02092-w [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Our team offers proteomic results via a user-friendly online repository (https://www.iprox.org/; DOI: IPX0006333000) as a publicly available resource, enabling researchers to access moyamoya disease patient sera. Metabolomics results were generated and stored at the Fifth Medical Center of the People’s Liberation Army of China (PLA) General Hospital. Data supporting the results of this study may be obtained from the corresponding authors.