Abstract
Immature retrovirus particles contain radially arranged Gag polyproteins in which the N termini lie at the membrane and the C termini extend toward the particle's center. We related image features to the polyprotein domain structure by combining mutagenesis with cryoelectron microscopy and image analysis. The matrix (MA) domain appears as a thin layer tightly associated with the inner face of the viral membrane, separated from the capsid (CA) layer by a low-density region corresponding to its C terminus. Deletion of the entire p6 domain has no effect on the width or spacing of the density layers, suggesting that p6 is not ordered in immature human immunodeficiency virus type 1 (HIV-1). In vitro assembly of a recombinant Gag polyprotein containing only capsid (CA) and nucleocapsid (NC) domains results in the formation of nonenveloped spherical particles which display two layers with density matching that of the CA-NC portion of immature HIV-1 Gag particles. Authentic, immature HIV-1 displays additional surface features and an increased density between the lipid bilayers which reflect the presence of gp41. The other internal features match those of virus-like particles.
The genome of the human immunodeficiency virus (HIV) contains three open reading frames common to all retroviruses (28, 45): gag—the group-specific antigen frame which encodes the precursor (Gag) of the major structural proteins of the virus interior (matrix [MA], capsid [CA], nucleocapsid [NC] domains and protein p6 in HIV), pol—which encodes the enzymatic activities of the virus (protease, reverse transcriptase, RNase H, and the integrase), and the env frame—which encodes the precursor (gp160) of the viral envelope proteins (gp41 [TM] and gp120 [SU]). HIV is a complex retrovirus (5) and hence encodes a further six regulatory proteins which enhance and control the replication of the virus (6, 7). The pol open reading frame overlaps that of gag and is expressed via a frame-shifting event that produces a Gag-Pol protein (45).
Virus assembly begins at the cell surface with the clustering of roughly 2,000 Gag proteins, 200 Gag-Pol proteins, and the two strands of genomic RNA. Gag and Gag-Pol proteins are bound to the inner surface of the membrane by covalently linked myristate at their N termini and a charged surface region. The budding particle includes envelope protein complexes (TM-SU) if they are present. After budding, the protease cleaves the Gag and Gag-Pol proteins to produce the proteins of the mature, infectious virion (43). This maturation process changes the arrangement of the structural components inside the virion: the radially arranged Gag molecules are dismantled, and a conical core structure is assembled in the center of the particle (45).
Mutagenesis and expression studies have shown the remarkable robustness of particle assembly. Expression of the Gag protein alone in mammalian and insect cells leads to budding of virus-like particles (VLPs) or Gag particles which are very similar in morphology to immature HIV (9, 20, 39–41). Mutated Gag proteins from which large regions have been deleted remain capable of directing budding (e.g., see reference 11).
Early work on HIV and other retroviruses was based on the assumption of icosahedral symmetry for the interpretation of images (17–19, 23, 26, 31, 32, 34). Indeed, surface views and glancing sections were interpreted as consistent with a triangulation number (T = 7 laevo) for HIV (18). Recent work using cryoelectron microscopy (cEM) has demonstrated that the retrovirus particle is neither icosahedral nor consistent in size and shape (13, 47, 48, 51). This absence of regularity cripples the traditional methods of structural analysis which have been so successful in more regular systems (3). Components display local order but are assembled in various ways to produce the plethora of sizes and shapes which characterizes the assembled particle (13). Approaching retrovirus structure requires setting aside the familiar tools of structural analysis and focusing on the characterization of the regular elements of the structure within the variation that is the hallmark of the complete particle. The result of such an analysis will be parameters of the local organization that can be combined with other information, such as that provided by X-ray crystallography, to generate testable models of the interactions which drive particle formation. It will not produce a single, unique three-dimensional structure for the virion, since a single structure cannot represent the array of structures observed in preparations. While this approach is more difficult (47) and may be unsatisfactory to some (33), it is the only one that accurately reflects observations.
The simplest level of organization in the retrovirus particle is a radial one. The Gag protein in the immature particle appears to be arranged with its amino terminus (MA domain) at the viral envelope and the carboxy terminus (NC domain) within the center of the particle where it interacts with the genomic RNA. This organization is seen in the VLPs which bud from insect cells upon expression of the Gag protein. A well-established radial organization can serve as a framework for addressing specific questions. For example, we were able to use fine immunocytochemical mapping to demonstrate that actin interacts specifically with the NC domain of the immature HIV particle (48) rather than the MA domain (35) as suggested by occasional images. This paper describes the use of cEM in combination with profile analysis and mutagenesis to explore the radial organization of immature HIV-1. The mutagenesis and cEM approaches complement each other. cEM provides a faithful representation of the density in the particle when corrected for the imaging effects of defocus phase contrast. Fortunately, the phase-contrast transfer function (CTF) is well understood (10), and its correction has become routine (8, 29, 30) so that quantitative evaluation of the particle density is possible. Mutagenesis is a particularly powerful tool of analysis for the HIV system because such a large array of mutants have been characterized and assembly of VLPs is surprisingly tolerant to significant modifications of the Gag protein.
Here we probe the radial organization of the immature particle by examining VLPs formed by deleted versions of Gag, particles assembled in vitro without membranes, and the authentic immature HIV particle. Our results resolve the MA domain underneath the membrane, provide evidence for the TM density within the bilayer, and demonstrate that p6 is arranged irregularly in the Gag particle and the virion.
MATERIALS AND METHODS
Preparation of Gag particles.
Gag particles were produced by expression of HIV-1 Gag (pNL4-3) (2) with the recombinant baculoviruses AcNPVgag12myr (for expression of wild-type Gag, see reference 39), AcNPVgag13myr (for HIV-1 GagΔp6 expression, see Gagamb438 in reference 4), and AcNPVgag dl41-143myr (for HIV-1 GagΔMA expression, see reference 16) in BTI-TN-5B1-4 cells (known as High 5 cells). Immature HIV-1 particles (pNL4-3) (2) were produced from infected MT-4 cells treated with an inhibitor of the viral protease (0.05 μg/ml) (48). VLPs and immature HIV-1 were purified (49) and prepared for cEM (13) as described previously.
In vitro assembly.
Recombinant HIV-1GagΔMAΔp6 protein was purified from Escherichia coli (24) and stored in 500 mM NaCl and 50 mM Tris (pH 8.0) at a final concentration of 1 mg/ml. The assembly reaction was initiated by the addition of 15 μM 73-mer DNA oligonucleotide, and it was performed with a dialysis bag dialyzing for 60 min at 4°C against 100 mM NaCl and 50 mM Tris (pH 8.0). Assembled particles were collected by centrifugation for 10 min at 4°C in an Eppendorf centrifuge and were resuspended in 100 mM NaCl and 50 mM Tris (pH 8.0).
Microscopy and image analysis.
cEM was performed as described previously using a Philips CM200FEG electron microscope operated at 200 kV (13). The radial density distribution in the virus particles was calculated by using the Fourier-Bessel expansion method that was described previously (13). The profiles from individual particles were averaged by aligning the density corresponding to the two leaflets of the membrane of the bilayer. Profiles of HIV-1 GagΔMA were aligned using the position of the two peaks corresponding to the CA protein layer, and the radial density profiles were derived from measurements of well-ordered areas. The program for performing the radial density measurements (13) is written in FORTRAN using standard subroutines (37) and is available upon request. Each averaged profile combines up to 3,600 (360° × 10 samples) measurements. The defocus was determined from the positions of the local minima in a radially averaged power spectrum of the individual particles. Contrast transfer function (CTF) correction was performed by division of the transform of the image with the appropriate CTF as described previously (8). The radial density distribution of CTF-corrected images is an average of 50 measurements at different positions of the particle. The individual profiles were aligned by placing the paired leaflets of the lipid bilayers into coincidence.
RESULTS
Wild-type HIV-1 Gag particles visualized by cEM.
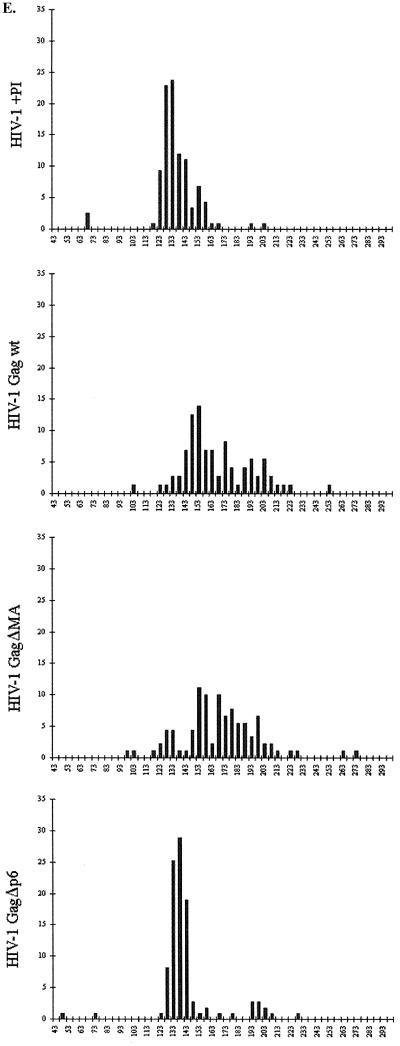
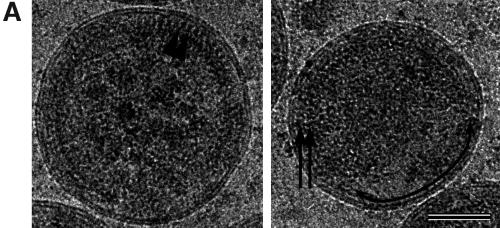
We have previously proposed a model for the arrangement of Gag polyproteins in immature HIV-1 particles (13). Here we provide experimental evidence for the proposed arrangement and present further details of the structural organization of immature HIV particles. Wild-type VLPs were expressed in H5 cells after infection with recombinant baculovirus. Particles in the cell culture supernatants were harvested 20 to 24 h after infection, concentrated by centrifugation through a cushion of sucrose, and gently resuspended in buffer. Analysis of stained sodium dodecyl sulfate-polyacrylamide gels and Western blots showed that the Gag protein was the single most abundant protein in the preparation (data not shown). cEM demonstrated that a large number of particles had been released from baculovirus-infected cells (data not shown). The most abundant particles found in the cell culture supernatant were the spherical HIV-1 Gag particles. The Gag particles' size and shape were very heterogeneous as reported previously (13, 16), ranging in diameter from 120 to 250 nm (Fig. 1E) and often showing a distorted profile rather than a spherical one. The Gag particles were easily distinguished from the elongated baculovirus particles that remained intact under our gentle conditions of purification.
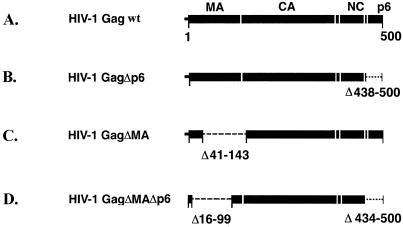
FIG. 1.
Wild-type HIV-1 Gag and Gag deletion mutants. Constructs A through C were expressed in H5 cells by infection with recombinant baculovirus. Construct D was expressed in E. coli and assembled in vitro in the presence of oligonucleotides. (E) The histograms show the effect of the deletions on the particle size. The x axis indicates the particle diameter in nanometers, and the y axis indicates the frequency of occurrence in the population as a percentage.
Observation of individual Gag particles at high magnification revealed further details (Fig. 2A). No consistent external features were seen on the surface of the viral membrane, reflecting the absence of viral glycoproteins. A regular, azimuthal variation of contrast was visible between the two leaflets of the membrane, matching the periodic density variation of the underlying protein layer. The bulk of the internal density was separated from the membrane by a 40-Å-wide, low-density space. Thin tethers run through this region below the membrane and connect the internal structural proteins with the inner leaflet of the membrane (Fig. 2A). The spacing between neighboring connections was 33 ± 5 Å (n = 75) and did not vary between particles. The internal density was subdivided into two major densities separated by a 15-Å-wide low-density region (Fig. 2A). The outer domain typically displayed a rod-like shape. This appearance was not maintained throughout the entire circumference of the particle. In some regions, the straight tip of the rod appeared more globular and the distance and angle between neighboring rods increased accordingly. This variation in appearance could reflect conformational variation between regions. The inner Gag domains always appeared to be tightly packed.
FIG. 2.
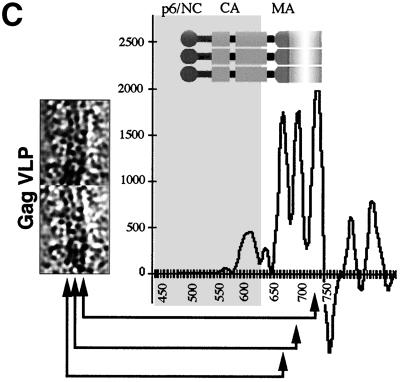
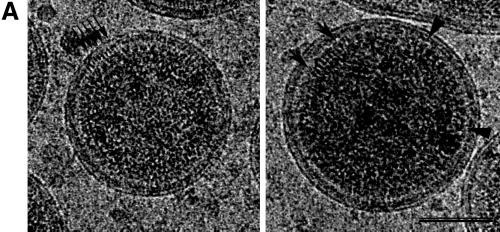
Wild-type HIV-1 Gag particles in cEM. (A) cEM of wild-type Gag particles shows thin tethers (arrows, left panel) spanning the low density submembrane space. The density at the lower radius is subdivided into two major bands (arrows, right panel). Arrowheads mark the CA rods. Areas of white correspond to higher density and hence mass in all of the images presented. Scale bar, 50 nm. (B) The average of 10 radial density distributions of wild-type HIV-1 Gag particles aligned to the position of the membrane reflects the presence of radially arranged Gag domains. The inset shows the assignment of individual peaks of density to the domains of the HIV-1 Gag polyprotein. The plot shows density (y axis) as a function of distance from the particle center in angstroms (x axis). (C) CTF-corrected images of two typical regions of a VLP membrane and the corresponding average radial density distribution from 50 regions of the membranes of five Gag particles. The internal density is masked (gray area) in this image. The interpretation of the CTF-corrected density of the particle (left) is shown by the arrows matching its features with the corresponding radial density profile and a schematic of the arrangement of the proteins. The peaks corresponding to the MA domain and the inner leaflet of the membrane were not resolved in the uncorrected image (B) and are represented by a single, broad peak of density in the same radial position (C).
We employed image processing to analyze the radial organization of the particles in greater detail. cEM images of particles were digitized, and their radial density distribution was calculated (Fig. 2B). Simple averaging of the projected radial density of particles of different diameters would result in a blurring of features. A Fourier-Bessel method (13) was used to overcome this problem by averaging the three-dimensional radial density distributions from different radii of the particles after aligning them on the position of the membrane. The average particle profile is shown in Fig. 2B. The result confirms our previously reported observation of separately folded domains in a radially arranged Gag polyprotein. The viral membrane itself shows up as two closely juxtaposed density peaks separated by a distance of ∼50 Å. The density peak at the inner leaflet of the membrane is broader than that of the outer, reflecting the presence of additional mass, presumably contributed by the MA domain of the Gag polyprotein. The remainder of the Gag polyprotein is located further inside and is separated from the viral membrane by a ∼40 Å-wide (70-Å peak-to-peak distance) region of low density. The adjacent density is formed by the broad protein layer, composed of rod-like subunits (see Fig. 2A). The profile reveals two peaks of density in this position. These paired peaks are characteristic of the CA protein, which contains two subdomains. The peaks are separated by 39 ± 4 Å (n = 10) and contribute to a CA-derived protein layer of 85 ± 6 Å (n = 10) width. Another density peak at a lower radial position indicates the location of the innermost protein layer, presumably created by the NC and p6 domains and the p6 domain. This most-intense density peak is separated from the neighboring density by a distance of 66 ± 4 Å (n = 10, peak-to-peak spacing), leaving a gap of 15 Å.
cEM revealed thin tethers crossing from the inner face of the particle membrane to the internal layers of density. This suggested that a portion of the viral polyprotein was anchored to the lipid bilayer of the particle; however, the nature of the arrangement near the membrane was not apparent from the original, uncorrected images. The contrast in a cEM image arises primarily from defocus phase contrast. This contrast allows the visualization of features in images of unstained material; however, it does so by distorting the relative densities of features in the particle. A complete interpretation of the density in the particle requires that the defocus and hence the CTF (10) be determined so that the densities can be corrected computationally (8). Naturally, this correction cannot restore the information at the nodes of the CTF, and so multiple defocuses must be examined to avoid misinterpretation.
Figure 2C shows a characteristic view of the wild-type VLP membrane after CTF correction of the image and the radial density distribution across the membrane. The presence of three individual layers of density is now clearly discernible. We assigned the two external layers to the outer and inner leaflets of the particle membrane and speculated that the submembrane density may be contributed by the globular portion of the MA domain of Gag. Careful examination of a large number of images showed that the same arrangement was retained over most of the VLP membrane. It was noted that a thin space of low density is present between the individual layers of density. Hence, the measurement of the corrected radial density distribution reveals three separate peaks of density in the position of the membrane (Fig. 2C). The peaks of the bilayer leaflets were ∼26 Å apart. The inner density layer is ∼22 Å below the peak of the inner leaflet.
A deletion in the MA domain affects the VLP membrane and the submembrane space.
We assigned the density immediately apposed to the inner surface of the membrane to the MA domain and assigned the peaks at lower radius to CA and the NC-p6 domain. Previously we suggested that this 40-Å-wide region of low density below the membrane represents the C-terminal residues of MA (47, 48). We tested this hypothesis by expressing the deletion mutant HIV-1GagΔMA, which lacks the carboxy-terminal two-thirds of the MA domain and the 11 amino-terminal residues of CA. We chose this construct because it is known to lead to efficient particle release (16). Many other constructs affect particle release or stability or result in redirecting budding to inappropriate domains (11).
Infection of H5 cells with the corresponding recombinant baculovirus resulted in efficient release of HIV-1 GagΔMA particles into the cell culture supernatant. Particles were harvested 20 to 24 h after infection, concentrated by centrifugation through a cushion of sucrose, and gently resuspended in buffer. The HIV-1 GagΔMA particles migrated to a density of 1.166 g/cm3 on a sucrose density gradient, similar to the density of authentic retrovirus particles. Analysis of stained sodium dodecyl sulfate-polyacrylamide gels and Western blots showed that the Gag polyprotein was the single most abundant protein in the preparation (data not shown). Examination of the pelleted material by cEM revealed the presence of enveloped VLPs in the cell culture supernatant. Many of the VLPs were disrupted, indicating that the deletion in MA reduced particle stability (data not shown). In contrast, most baculovirus particles remained intact and undistorted by the preparation for cEM. Wild-type Gag particles and HIV-1GagΔMA particle diameters were measured and plotted in a histogram (Fig. 1E). The diameters of wild-type and mutant VLPs ranged from ∼100 nm to more than 250 nm (Fig. 1E), with an average particle diameter of 169 ± 29 nm (wild-type HIV-1 Gag, n = 97) and 170 ± 33 nm (HIV-1GagΔMA, n = 107), respectively. HIV-1GagΔMA particles showed a size heterogeneity similar to that of wild-type Gag particles. Sixty-six percent of wild-type HIV-1 Gag VLPs and 72% of HIV-1GagΔMA VLPs deviated from the average diameter by more than 10 nm (Fig. 1E).
The HIV-1GagΔMA particles displayed an uneven mass distribution (Fig. 3A) in which the bulk of the mutant Gag polyproteins were found in one hemisphere of the particle. This hemisphere contained regular arrays of Gag polyproteins, similar to those observed in wild-type particles (Fig. 3A). Particles always showed areas of unstructured density with an apparent size which depended on the orientation of the particle in the water layer. Careful examination of structured areas within mutant VLPs demonstrated that the lipid bilayer and the space below were clearly affected by the deletion in the MA domain. The regular internal-density variation detected between the two leaflets of wild-type membranes was not observed in HIV-1GagΔMA particles; instead, the membranes were smooth and featureless. The inner leaflet of the HIV-1GagΔMA VLPs also appeared thinner than that of the wild type, probably reflecting the removal of the MA domain from the inner face of the lipid bilayer. Further, the 40-Å gap beneath the wild-type VLP membrane was much narrower in the HIV-1GagΔMA mutant so that the internal density was brought closer to the lipid bilayer.
FIG. 3.
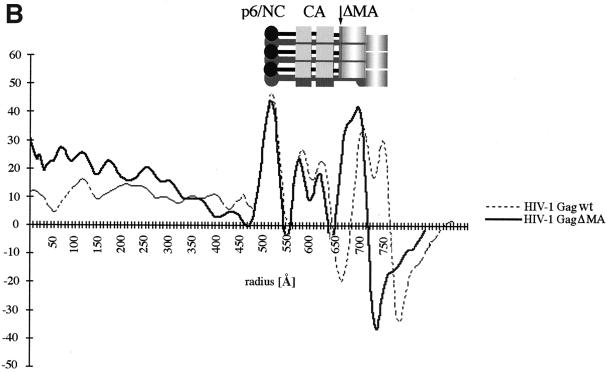
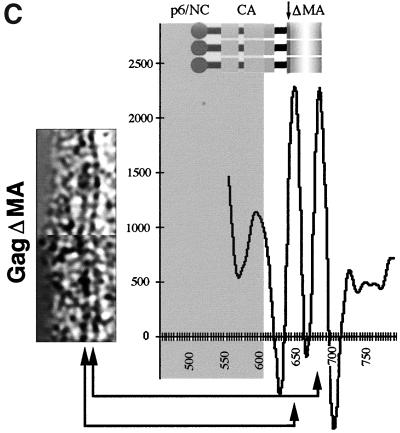
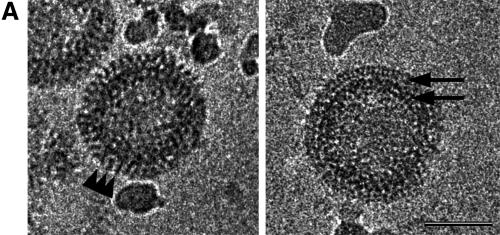
HIV-1 GagΔMA particles visualized by cEM. (A) A cEM of an HIV Gag particle with a deletion of residues 41 to 143. The deletion of MA sequences results in the thinning of the submembrane space and a reduced inner leaflet thickness and affects the regular variation in density within the membrane. Frequently the internal organization is disrupted, and the regular array of Gag molecules is only preserved in limited regions of the circumference. The arc within the membrane (right) shows the presence of a vacant region of the particle. The arrowheads (left) mark CA-derived rods of Gag density while the arrows (right) mark radial CA and NC density. Size bar, 50 nm. (B) The radial density distribution of HIV-1 GagΔMA demonstrates the effect of the matrix deletion. The mutation results in an approximation of regular internal density and the particle membrane. The superimposition with the profile of wild-type Gag particles illustrates the effect of the deletion. The plot shows density (y axis) as a function of distance from the particle center in angstroms (x axis). (C) CTF-corrected image of the membrane region of an HIV-1 GagΔMA particle and the corresponding region of the average of the radial density distribution from 50 particles with the central region (gray) masked.
These changes in the membrane region and the region beneath the membrane were also reflected in the radial density profiles of HIV-1GagΔMA particles (Fig. 3B). Measurements were taken from clearly structured portions of the particle, since inclusion of unstructured regions blurred the average profile. The deletion had no significant effect on the general structural features of the internal protein layers in HIV-1GagΔMA particles so that individual profiles could be aligned. The comparison of wild-type and HIV-1GagΔMA radial density profiles revealed the dramatic effect of the deletion on the membrane-associated regions. A gap of ∼15 Å (rather than the 40 Å seen in wild-type VLPs) separates the inner leaflet from the first internal protein layer. This spacing varied somewhat between individual HIV-1GagΔMA particles, suggesting that an intact matrix protein fixes the position of the Gag polyprotein more precisely relative to the inner face of the viral membrane.
The analysis of the wild-type VLP membrane had shown that CTF correction of the images revealed a layer beneath the membrane (Fig. 2C). We hypothesized that this density was created by the globular head of the MA protein and should therefore be affected by the deletion in HIV-1GagΔMA particles. This hypothesis was confirmed by the CTF-corrected images of the mutant VLP membrane, which contained two, rather than three, density layers (Fig. 3C). The remaining radial density peaks in the profile superimpose well. The outer and inner leaflets of the bilayer retain the same spacing of ∼26 Å as in the wild-type VLP membrane (Fig. 3C).
Deletion of the p6 domain has no apparent effect on VLP structure.
Images and radial density measurements of wild-type particles displayed two prominent internal layers in wild-type VLPs. We assigned the innermost layer to the NC and p6 domains; however, the relative contributions of the NC and p6 domain to the density were unclear. We expressed the Gag deletion mutant HIV-1GagΔp6 that lacks the entire p6 domain (Fig. 1) to address this question. Infection of H5 cells with the recombinant baculovirus resulted in the efficient release of mutant Gag particles into the cell culture supernatant as revealed by Western blot and cEM analysis of particle preparations (data not shown). The mutant VLPs were clearly less heterogeneous than wild-type particles, having a much narrower distribution of particle diameters (Fig. 1E). Only 25% of HIV-1GagΔp6 particles deviated by more than 10 nm from their average value (140 ± 23 nm; n = 111), whereas more than 66% of wild-type Gag particles fell outside this range. The better-defined particle diameter may indicate a better control of size during assembly of HIV-1GagΔp6 polyproteins. Most HIV-1 GagΔp6 particles appeared truly spherical (Fig. 4). Fewer of the mutant particles were distorted or disrupted than in wild-type VLPs.
FIG. 4.
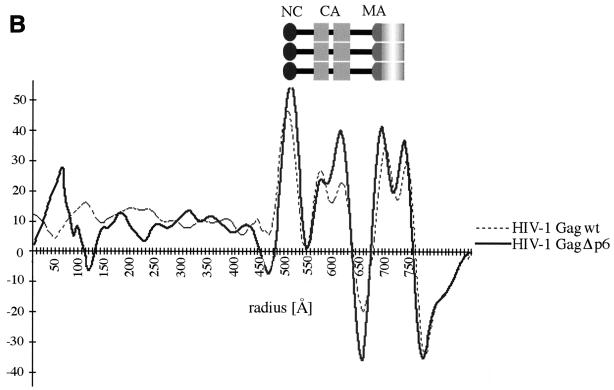
HIV-1 GagΔp6 particles in cEM. (A) cEM of HIV-1 GagΔp6 particles shows no effect of the p6 deletion on the internal layers of density. Mutant particles display the same details as wild-type Gag particles. The arrows (left) mark the tethers. The arrows (right) indicate a region of typical rod density for comparison with a region of altered rod density marked by arrowheads. Bar, 50 nm. (B) Radial density distribution of HIV GagΔp6 particles. The superimposition with the radial density distribution of wild-type VLPs demonstrates that the C-terminal p6 domain does not contribute to the averaged innermost density of the Gag protein layer. The plot shows density (y axis) as a function of distance from the particle center in angstroms (x axis). Positive and negative density values correspond to areas of black/white in panel A, respectively.
The structural details in HIV-1 GagΔp6 particles match those of wild-type VLPs (Fig. 4A). A regular variation of density was noticed between the two leaflets of the membrane that matched the periodic density variation of the underlying protein layer seen in the wild type. The deletion of the C-terminal p6 domain had no apparent effect on the overall organization of the mutant particles, nor did it alter the spacing of protein layers inside the particle. Thin connections were visible crossing the space below the membrane and establishing linkages between the inner leaflet of the membrane and the CA-derived protein layer further inside (Fig. 4A, left). Neighboring connections were separated by 33 ± 5 Å (n = 54), which exactly matches the spacing seen in wild-type VLPs (33 ± 5 Å, n = 75).
Intriguingly, the similarity of the wild-type and mutant particles extended to the innermost density layer, which we originally ascribed to the NC-p6 domain. This indicates that the p6 domain of the Gag polyprotein does not contribute ordered density to the wild-type Gag particle. This observation is confirmed by the comparison of the average radial density profiles of wild-type and HIV-1 GagΔp6 particles. Figure 4B demonstrates that the deletion of the entire p6 domain had no significant effect on the organization of internal layers, the spacing between protein layers, or the width of the Gag domains in the Gag particles. The fact that the innermost portion of the average density layer was not altered demonstrates that it only reflects contributions from the NC domain of the Gag polyprotein.
Organization of immature HIV-1 particles in the absence of a membrane.
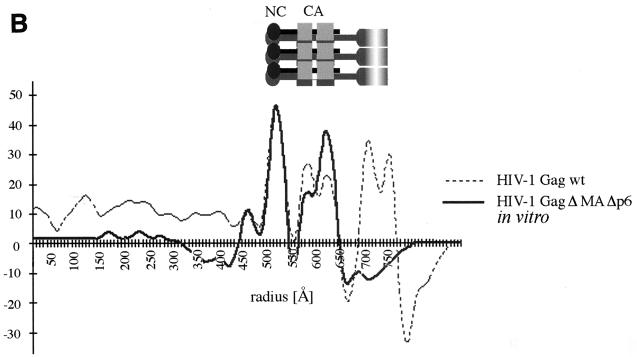
We examined particles assembled in vitro from recombinant Gag polyproteins and nucleic acids to determine the role of the membrane in organizing the internal Gag domains. We have recently demonstrated that HIV-1 GagΔMAΔp6 protein (Fig. 1) forms spherical particles under in vitro assembly conditions (24). The in vitro-assembled particles were concentrated by brief centrifugation and studied by cEM. A large number of spherical particles were present in the sample, reflecting efficient assembly of recombinant Gag polyprotein. The resultant particles are shown in Fig. 5 and display a well-defined diameter of 111.7 ± 7.3 nm (n = 67). Although nonenveloped, the in vitro-assembled particles shared features with the enveloped immature particles produced by the expression of Gag polyproteins in insect cells (Fig. 2A). In particular, in vitro-assembled particles displayed two layers that were similar to the two internal layers of Gag particles that we had attributed to CA and NC. The outer layer was formed by rod-like subunits indistinguishable in size and shape from the CA rods of wild-type VLPs. This layer was separated from an internal layer by a 15-Å-thin low-density gap corresponding to that between the CA and NC protein layers in the VLPs. The average radial density profile for the in vitro-assembled particles (Fig. 5B) matches that of the HIV-1 Gag wt particles with the exception of the outer layers, which represent the membrane; the protein layers are arranged in a very similar manner.
FIG. 5.
cEM of in vitro-assembled HIV GagΔMAΔp6. (A) cEM of in vitro-assembled particles. Recombinant Gag polyprotein lacking the entire p6 domain and most of the MA domain (including the N-terminal myristic acid) can be assembled into spherical particles. The two major layers of density are marked by arrows (right). Note the rod-like shape of the outer protein layer (left, arrowheads), reminiscent of the CA protein layer in Gag particles. A second protein layer is located further inside and is similar in appearance to the innermost protein layer of Gag particles released from cells. Bar, 50 nm. (B) Radial density profile of HIV-1 GagΔMAΔp6 particles. The two main peaks of density corresponding to the CA and NC protein layers can easily be aligned with the radial density profiles of enveloped HIV-1 Gag particles. The plot shows density (y axis) as a function of distance from the particle center in angstroms (x axis).
Immature HIV-1 particles: are VLPs like the virus?
The results obtained with the wild-type and mutant Gag VLPs lead to a better understanding of the multilayered immature state of HIV-1, allowing the assignment of Gag domains to the protein layers. Several lines of evidence suggest that the VLPs produced in insect cells are a good model for the study of immature HIV-1. Nevertheless, the HIV-1 Gag particle lacks several potentially important structural components, including the Gag-Pol precursor, the virus glycoproteins, and the authentic viral RNA. Their presence might modify the arrangement of the Gag polyprotein and result in a changed organization of the immature virion.
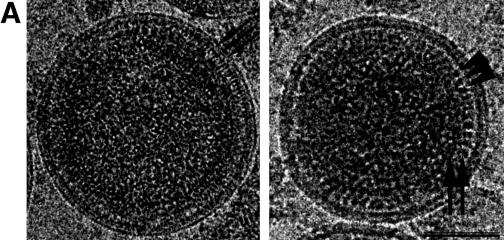
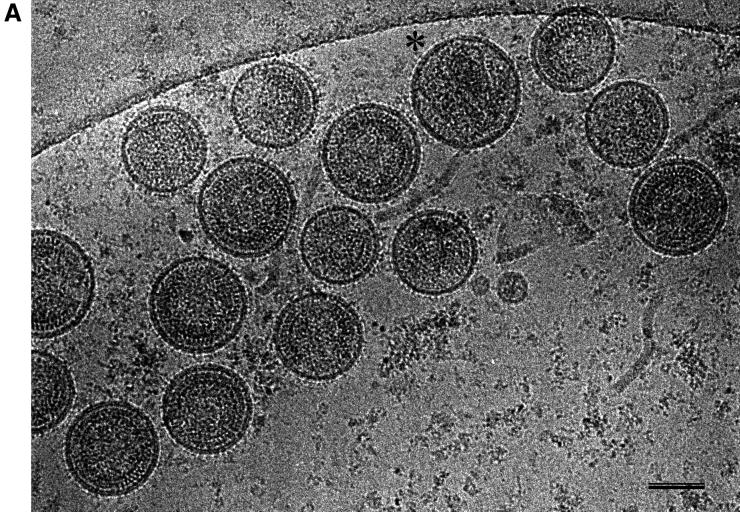
We explored the role of additional virus proteins on the organization of immature HIV-1 by analyzing virions harvested from infected T cells after treatment of cells with an inhibitor of the viral protease (Fig. 6A). The majority of the particles had the radial arrangement typical of immature virions. Less than 1% of particles showed the internal cone produced by the maturation cleavage.
FIG. 6.
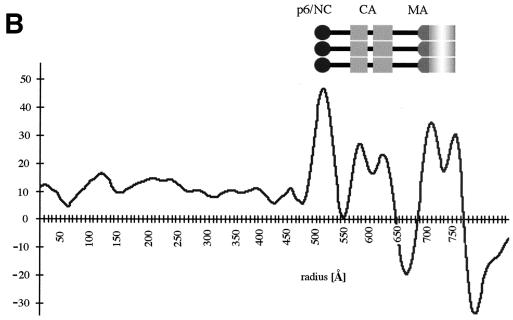
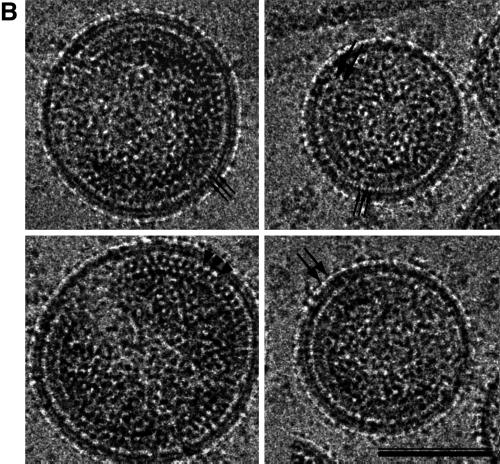
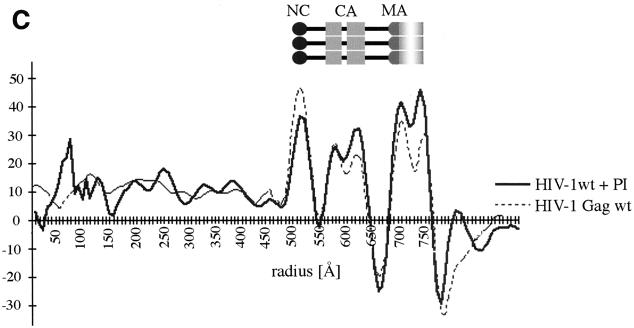
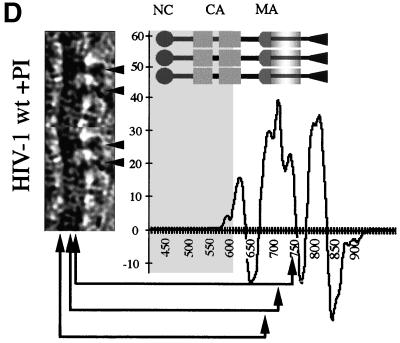
cEM of immature HIV-1. (A) Overview of immature HIV-1 released from infected T cells. The image shows a typical low-magnification view of particles that have been released from T cells after treatment of the cells with an inhibitor of the viral protease (bar = 100 nm). Note the presence of spikes on the surface of the particles and the heterogeneity of diameters within the population. The asterisk marks a rare particle in the preparation (<1%) which shows a mature phenotype. (B) The details of the structure of the immature HIV-1 particle are seen in the views of individual particles. The two major layers of density are marked by arrows (upper right). Note the rod-like shape of the outer protein layer (arrowheads, lower left), reminiscent of the CA protein layer in Gag particles (bar = 100 nm). Small arrows mark the tethers in the upper-right panel and the glycoproteins in the lower-right panel. (C) The comparison of the radial density profiles of the immature virion and the wild-type VLP shows that the internal features are conserved. The plot shows density (y axis) as a function of distance from the particle center in angstroms (x axis). The two particles differ in the depth of the features in the membrane region and the presence of a weak average external density corresponding to that of the envelope proteins. (D) The CTF-corrected image of a portion of an immature particle reveals the extra density in the membrane region. The correspondence between the CTF-corrected image and the average radial density profile from 50 particles is indicated by the arrows matching their corresponding features with a schematic of the arrangement of the proteins. The internal density is masked (grey region).
The immature virions were clearly not uniform in diameter; however, they appeared significantly less heterogeneous than wild-type VLPs. The size distribution of authentic immature virions peaked around their average diameter (133 ± 17 nm; n = 118), with less than 30% of the particles deviating by more than 10 nm from this value (Fig. 1E).
cEM of authentic immature HIV at high magnification revealed an overall organization reminiscent of that of the VLPs (Fig. 6B). In particular, internal radial arrangements in the immature virion and in the VLP were very similar. The internal protein layers were separated from the membrane by a prominent gap, as in the VLPs. Similar thin tethers were observed crossing the gap and connecting the internal densities to the virus membrane (Fig. 6B). One clear difference between immature virions and the VLP structure was seen at the position of the virus membrane, which contained regularly spaced viral spike proteins (Fig. 6). These protruded ∼120 Å from the center of the membrane and were visible on all of the particles in the preparation, forming an additional layer of density on the outside of the immature virion (Fig. 6A and B).
The radial density measurement of immature HIV-1 particles confirmed the visual impression of the internal organization of the virion (Fig. 6C). It shows a radial density distribution which superimposes well with that of the wild-type VLPs, thereby demonstrating that the layers of density observed in the authentic immature virion are exclusively created by the Gag polyprotein. The virus glycoproteins which are easily discernible in the images (Fig. 6A and B) show up only weakly in the radial density profile because they are a less consistent feature than the radially arranged internal structural proteins.
We corrected the images for the effect of the CTF (Fig. 6D) to obtain a more accurate view of the membrane organization. CTF-corrected images show that the membrane of authentic immature HIV-1 is composed of three layers of density. Careful examination of a large number of images revealed several interesting features. The leaflets of the bilayer retained the same spacing as in the VLPs: however, the separation of layers was less obvious because the space between them appeared filled. Frequently, density appeared to connect the MA-derived layer with the lipid bilayer and the inner to the outer leaflet of the membrane (Fig. 6D). This contrasts with the VLP images, which showed the membrane layers of Gag particles clearly separated.
We cannot completely interpret the more complex organization of the membrane of the immature virion, however; the virus glycoprotein adds a considerable amount of mass to the virus membrane and could easily account for the observed features. On the outside of the particles, regularly spaced protrusions were observed, reflecting the presence of the surface portion of the HIV-1 glycoprotein. Measurements show that the glycoprotein protrudes ∼120 Å from the center of the membrane. The radial density measurement (Fig. 6D) confirms these observations. The three peaks of density superimpose well with the radial density distribution in the Gag particle, indicating the same spacing of the bilayer leaflets and the MA domain-derived layer. The shallower density trough between these layers in immature HIV-1 indicates the presence of additional mass in this region (Fig. 6D). The additional density found outside the membrane reflects the presence of glycoprotein on the surface. The HIV-1 TM protein contains a cytoplasmic subdomain of ∼17 kDa. Several lines of evidence suggest that this portion of the TM protein is located near the viral membrane; however, the radial density profile did not resolve it as a separate feature.
DISCUSSION
Gag determines the radial organization of immature HIV.
The radial organization of the immature HIV particle is revealed by cEM. Our previous work showed that VLPs contain a radially arranged Gag polyprotein with the N terminus (MA domain) located at the membrane and the C terminus (NC and p6 domains) toward the center of the particle (47, 48). The domains of the Gag polyprotein fold separately and create individual protein layers in the particle (13). Here, we use an analysis of Gag deletion mutants to confirm this arrangement in the VLP.
The ability to compare the radial density profiles of the immature virion and the in vitro-assembled particle with the VLP allowed us to look for a role of the specific RNA and of the membrane in the organization of the particle. Our analysis of the immature virion showed that the radial profile of its internal structure matches that of the VLP. Hence, the additional components present in the authentic particle do not have a perceptible effect on its internal organization. Interestingly, the inner portions of this region are also unchanged in the in vitro-assembled particle without a membrane, showing that the membrane plays a limited role in the organization. HIV-1 resembles D-type retroviruses (45) in this respect. Hence, the Gag polyprotein itself plays the dominant role in defining the internal organization of the immature particle.
The bipartite organization of MA is retained during maturation.
We previously suggested that the matrix domain is tightly associated with the inner face of the viral membrane and is separated from the underlying capsid derived protein layer by a space of approximately 40-Å thickness (13). Here, we visualize a discrete protein layer at the inside of the lipid bilayer. Further, thin tethers are seen crossing the underlying 40-Å low-density space. We tested the assignment of these feature to the MA domain by comparing wild-type Gag particles to Gag particles lacking residues 41 to 143. Deleting most of the MA domain and 11 N-terminal residues of CA removed the protein layer next to the membrane and reduced the space underneath. This result confirms that both of these distinct structural elements are formed by the MA domain of the Gag polyprotein.
A bipartite organization of the MA domain is consistent with other structural data. The precise sequence that spans the 40-Å space must be defined by smaller deletions; however, the appearance of extended thin tethers is consistent with the presence of α-helices. X-ray crystallographic studies have shown that the C terminus of the mature MA domain forms a 25-amino-acid-long α-helix that is distinct from the rest of the molecule (25). This helix (helix 5) points away from the globular head of the MA domain and hence should extend toward the center of the virion. Its 37-Å length would span most of the 40-Å space observed. Although evidence suggests that the structure of the MA domain in the uncleaved (i.e., immature) state of Gag may differ from the cleaved (mature) state, particularly in the neighborhood of the CA domain (24), a helix in this position would certainly be consistent with our observations. This arrangement of helix 5 would position the globular head of the MA molecule next to the inner leaflet of the lipid bilayer. The detection of a distinct MA domain-derived protein layer just below the inner leaflet of the immature particle lends strong support to the idea that the bipartite organization seen in crystals of the cleaved protein also corresponds to the immature state and is retained after maturation.
A model for the arrangement of the MA protein layer in mature lentiviruses has been proposed from its arrangement in the simian immunodeficiency virus (38) and HIV (25) MA domain crystal structures. The observed ∼25-Å width of the membrane proximal matrix layer is consistent with the arrangement of the globular head structure in these models. The arrangement of the MA domain as a network of six membered rings linked by two MA trimers (38) results in a center-to-center spacing of neighboring MA trimers (∼66 Å) and would yield a ∼33-Å spacing in projection along the membrane. The tethers which cross the low-density region in the VLPs display a 33 ± 5 Å separation (Fig. 2B). This spacing would be consistent with the hexamer network (25, 38), if the tethers represent bundles of MA domain carboxy-terminal extensions. The analysis of the repeating unit in the VLP (13) supported a similar local arrangement.
The data presented here reveals the striking arrangement of the MA domain in immature HIV-1. It reveals a thin MA domain protein layer next to the membrane, resting on thin stalks which connect it to CA. The role of this construction is unclear. The MA layer may support the viral membrane from the inside. The fact that a deletion in MA significantly affects particle stability provides some evidence for this suggestion. We think it is likely that some space in proximity to the domain boundaries is helpful during viral maturation because it will significantly increase the accessibility of the protease cleavage sites. This speculation is supported by the presence of a similar, albeit thinner, space between the CA and NC protein layers. This stretch of amino acids is bordered by two viral protease cleavage sites and has been identified as the spacer peptide, sp1 (36). sp1 has been shown to be an important conformational determinant with profound influence on the symmetry of assembly of Gag particles in vitro (1, 16, 24). Although no direct structural information is available, molecular modeling suggests that the residues between CA and NC form a continuous α-helix (1).
TM contributes density to the bilayer region.
The internal regions of the immature virion and the VLP were indistinguishable; however, subtle differences were seen in the region of the membrane. The membrane organization of authentic immature HIV-1 appeared more complex than that in Gag particles. The layers of the membrane were clearly separated in the VLPs by spaces which were filled with density in the virion. This difference may explain the fact that VLP membranes are sensitive to nonionic detergents under conditions which leave the membrane of authentic immature virions intact (T. Wilk, unpublished observation, and reference 48). We believe that the additional density is contributed by the viral glycoprotein complex of HIV-1 which was observed to cover the entire surface of the immature virions. The abundance of surface projections was surprising, since the SU portion is known to be bound weakly to the virion (45). Further work is needed to show whether this projecting density reflects the entire glycoprotein (SU-TM) complex or only the stable, membrane-anchored TM protein. The projections are indeed shorter than those seen in the distantly related spumavirus retrovirus, in which the SU domain is retained (46).
All lentivirus TM proteins contain long cytoplasmic tails (45). The HIV-1 tail contributes a mass of ∼17 kDa, equivalent to the mass of the MA domain. It has been suggested (13) that the 40-Å space below the membrane could harbor the large C-terminal tail of TM. Our analysis of cEM images and radial density measurements provides no support for this suggestion, since no additional density was detected at this position in immature virions or further within the particle. Interestingly, MA mutations that prevent glycoprotein incorporation largely map to the membrane-exposed surface of the MA trimer model (38). This suggests a placement of the C-terminal domain between the inner leaflet of the membrane and the MA domain-derived protein layer. Indeed, multiple palmitoylation events (50) and the presence of amphipathic helices (44) suggest a rather intimate association of the TM C terminus with the lipid bilayer, as proposed previously (50). The observation of additional density within the membrane of immature HIV-1 supports such an arrangement; however, only the analysis of a virus mutant lacking the cytoplasmic domain would address this issue directly.
The CA and NC domains, but not p6, form regular inner-density layers.
The capsid domain of the Gag polyprotein is visible as a pair of peaks spaced ∼45 Å apart, reflecting the division of the CA protein into separately folding N- and C-terminal subdomains. This spacing matches that seen in tubes formed by the CA protein in vitro (22) and the outer two rings seen in tubes assembled from recombinant CA-NC protein and nucleic acid (T. Wilk, unpublished data), confirming that these regions represent CA.
The NC domain contributes the innermost radial density to the immature particle. We originally assigned this layer in immature particles to the NC-p6 domain (13); however, the contribution of the C-terminal p6 domain was unclear. The deletion of the entire p6 domain did not affect the width or the substructure of the innermost protein layer in mutant HIV-1 GagΔp6 VLPs or in vitro-assembled particles. Hence, p6 does not contribute to this density. Since p6 is roughly the same size as the NC domain, its deletion should have a marked effect on the density unless it is disordered or unstructured. This latter possibility is consistent with structural studies of the isolated protein (42).
The function of p6 during or after assembly and budding remains unclear (reviewed in reference 12). When Gag is expressed alone, the p6 domain can be deleted without causing a defect in assembly (39). In contrast, p6 deletions from the full-length viral genome result in the arrest of budding particles at the plasma membrane (21), a phenotype which has been shown to depend on the virus protease (27). It has been suggested recently that p6 is a major determinant of particle size and that p6 deletion results in the generation of large particles (14, 15). Nevertheless, analysis of p6 mutants (16, 21, 26, 39) by conventional electron microscopy revealed no effect on the particle diameter. cEM of wild-type and mutant subviral Gag particles allows visualization of unstained particles and reveals the entire particle in projection, providing a more reliable measure. The cEM results show that deletion of p6 results in the generation of particles with a diameter of 140 ± 23 nm. The similar diameter of authentic immature virions (133 ± 17 nm) and p6-deleted VLPs demonstrates that the control of Gag assembly is not affected by the absence of a functional p6 domain. In fact, the absence of the p6 domain was advantageous in the expression system, since the mutant particles were more regular in size and shape than wild-type VLPs.
Conclusions.
The analysis presented here complements the ability of cEM to provide quantitative density distributions with mutational analysis. Our analysis validates the radial model of domain arrangement (13, 47, 48) and extends it to reveal the details of the interaction between MA and the membrane in the uncleaved Gag protein. It also defines differences between the VLPs and the immature virion in the organization of the membrane region while showing that the membrane is not necessary for the organization of the internal Gag domains.
ACKNOWLEDGMENTS
We are pleased to acknowledge the discussions and help of our colleagues at the European Molecular Biology Laboratory and the Heinrich-Pette-Institut who made the work presented both easier and more enjoyable.
This work was supported by a DFG grant (Fu354/1-1) to H.-G.K. and S.D.F., a Wellcome Trust Programme Grant to S.D.F., and a grant from the Agence National de Recherche sur la SIDA (ANRS-1998) to P.B. S.D.F. is a Wellcome Trust Principal Research Fellow.
REFERENCES
- 1.Accola M A, Hoglund S, Göttlinger H G. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T S, Olson N H, Fuller S D. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrière C, Gay B, Chazal N, Morin N, Boulanger P. Sequence requirements for encapsidation of deletion mutants and chimeras of human immunodeficiency virus type 1 Gag precurson into retrovirus-like particles. J Virol. 1995;69:2366–2377. doi: 10.1128/jvi.69.4.2366-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 6.Cullen B R. Human immunodeficiency virus as a prototypic complex retrovirus. J Virol. 1991;65:1053–1056. doi: 10.1128/jvi.65.3.1053-1056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen B R. Mechanism of action of regulatory proteions encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haas F, Paatero A O, Mindich L, Bamford D H, Fuller S D. A symmetry mismatch at the site of RNA packaging by the polymerase complex of dsRNA bacteriophage phi-6. J Mol Biol. 1999;294:357–372. doi: 10.1006/jmbi.1999.3260. [DOI] [PubMed] [Google Scholar]
- 9.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Vedin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson H P, Klug A. Measurement and compensation of defocusing and aberrations by Fourier processing of micrographs. Philos Trans R Soc Lond B Biol Sci. 1971;261:105–118. [Google Scholar]
- 11.Fäcke M, Janetzko A, Shoeman R L, Kräusslich H-G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O. HIV-1 Gag protein: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 13.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains within the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 14.Garnier L, Parent L J, Rovinski B, Cao S X, Wills J W. Identification of retroviral late domains as determinants of particle size. J Virol. 1999;73:2309–2320. doi: 10.1128/jvi.73.3.2309-2320.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier L, Ratner L, Rovinski B, Cao S X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gay B, Tournier J, Chazal N, Carriere C, Boulanger P. Morphopoietic determinants of HIV-1 Gag particles assembled in baculovirus-infected cells. Virology. 1998;247:160–169. doi: 10.1006/viro.1998.9237. [DOI] [PubMed] [Google Scholar]
- 17.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 18.Gelderblom H R, Hausmann E H S, Örzel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 19.Gentile M, Adrian T, Scheidler A, Ewald M, Dianzani F, Pauli G, Gelderblom H R. Determination of the size of HIV using adenovirus type 2 as an internal length marker. J Virol Methods. 1994;48:43–52. doi: 10.1016/0166-0934(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 20.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thinnes D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 21.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grättinger M, Hohenberg H, Thomas D, Wilk T, Muller B, Krausslich H G. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology. 1999;257:247–260. doi: 10.1006/viro.1999.9668. [DOI] [PubMed] [Google Scholar]
- 23.Grief C, Nermut M V, Hockley D J. A morphological and immunolabelling study of freeze-substituted human and simian immunodeficiency viruses. Micron. 1994;25:119–128. doi: 10.1016/0968-4328(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 24.Gross I, Hohenberg H, Wilk T, Wiegers K, Grättinger M, Fuller M B S, Kräusslich H G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hockley D J, Nermut M V, Grief C, Jowett J B M, Jones I M. Comparative morphology of Gag protein structures produces by mutants of the gag gene of human immunodeficiency virus type 1. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 27.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancini E J, Clarke M, Gowen B, Rutten T, Fuller S D. Cryo-electron microscopy reveals the functional anatomy of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 30.Mancini E J, de Haas F, Fuller S D. High resolution icosahedral reconstruction: fulfilling the promise of cryo-electron microscopy. Structure. 1997;5:741–750. doi: 10.1016/s0969-2126(97)00229-3. [DOI] [PubMed] [Google Scholar]
- 31.Nermut M V. Size, shape and surface structure of mouse mammary tumour virus (MTV). Brief report. Arch Gesamte Virusforsch. 1973;41:284–289. doi: 10.1007/BF01252777. [DOI] [PubMed] [Google Scholar]
- 32.Nermut M V, Grief C, Hashmi S, Hockley D J. Further evidence of icosahedral symmetry in human and simian immunodeficiency virus. AIDS Res Hum Retrovir. 1993;9:929–938. doi: 10.1089/aid.1993.9.929. [DOI] [PubMed] [Google Scholar]
- 33.Nermut M V, Hockley D J, Bron P, Thomas D, Zhang W-H, Jones I A. Further evidence for hexagonal organization of HIV gag protein in prebudding assemblies and immature virus-like particles. J Struct Biol. 1998;123:143–149. doi: 10.1006/jsbi.1998.4024. [DOI] [PubMed] [Google Scholar]
- 34.Nermut M V, Hockley D J, Jowett J B, Jones I M, Garreau M, Thomas D. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 35.Nermut M V, Wallengren K, Pager J. Localization of actin in Moloney murine leukemia virus by immunoelectron microscopy. Virology. 1999;260:23–34. doi: 10.1006/viro.1999.9803. [DOI] [PubMed] [Google Scholar]
- 36.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Press W H, Teukolsky S A, Vetterling W T, Flannery B P. Numerical recipes in FORTRAN: the art of scientific computing. Cambridge, England: Cambridge University Press; 1992. [Google Scholar]
- 38.Rao Z, Belyaev A S, Fry E, Jones I M, Stuart D I. Crystal structure of the SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 39.Royer M, Cerutti M, Gray B, Hong S-S, Devauchelle G, Boulanger P. Functional domain of HIV-1 gag polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 40.Royer M, Hong S-S, Gay B, Cerutti M, Boulanger P. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J Virol. 1992;66:3230–3235. doi: 10.1128/jvi.66.5.3230-3235.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shioda T, Shibuta H. Production of human immunodeficiency virus (HIV)-like particles from cells infected with recombinant vaccinia viruses carrying the gag gene of HIV. Virology. 1990;175:139–148. doi: 10.1016/0042-6822(90)90194-v. [DOI] [PubMed] [Google Scholar]
- 42.Stys D, Blaha I, Strop P. Structural and functional studies in vitro on the p6 protein from the HIV-1 gag open reading frame. Biochim Biophys Acta. 1993;1182:157–161. doi: 10.1016/0925-4439(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 43.Swanstrom R, Wills J W. Synthesis, assembly and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 44.Venable R M, Pastor R W, Brooks B R, Carson F W. Theoretically determined three-dimensional structures for amphipathic segments of the HIV-1 gp41 envelope protein. AIDS Res Hum Retrovir. 1989;5:7–22. doi: 10.1089/aid.1989.5.7. [DOI] [PubMed] [Google Scholar]
- 45.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 46.Wilk T, de Haas F, Wagner A, Alke A, Rutten T, Fuller S, Flügel R M, Löchelt M. The intact retroviral Env glycoprotein of human foamy virus is a trimer. J Virol. 2000;74:2885–2887. doi: 10.1128/jvi.74.6.2885-2887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilk T, Fuller S D. Towards the structure of the human immunodeficiency virus: divide and conquer. Curr Opin Struct Biol. 1999;9:231–243. doi: 10.1016/S0959-440X(99)80033-5. [DOI] [PubMed] [Google Scholar]
- 48.Wilk T, Gowen B, Fuller S D. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J Virol. 1999;73:1931–1940. doi: 10.1128/jvi.73.3.1931-1940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology. 1996;218:269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- 50.Yang C, Spies C P, Compans R W. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc Natl Acad Sci USA. 1995;92:9871–9875. doi: 10.1073/pnas.92.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular design of native and immature murine leukemia virus revealed by electron cryo-microscopy. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]