The rise of full transcriptome acquisition technologies has fueled the rapid proliferation of molecular-level biological data. These large datasets require interpretation beyond the single-gene level to connect them to meaningful biology and clinical impacts. In 2003, we pioneered the gene set enrichment analysis (GSEA) approach1 to enable the identification of the coordinate activation or repression of sets of genes that share common biology, thereby distinguishing even subtle differences between phenotypes or cellular states. We first released our GSEA software and a companion resource of gene sets, the Molecular Signatures Database (MSigDB), in 2005 (ref. 2).
Historically, the GSEA/MSigDB resource (https://gsea-msigdb.org) focused on the analysis of human-specific datasets. The gene sets in MSigDB were offered exclusively in the human gene space, and analysis of mouse data was minimally supported through basic ortholog mapping to human genes. In recognition of the importance of model organisms, particularly mice, for research into the mechanisms of human disease, we recently expanded the MSigDB to address this by introducing a database of mouse-native gene sets and by substantially improving MSigDB’s ortholog mapping.
Mouse-native gene sets
While ortholog mapping is useful, some degree of uncertainty exists when discriminating between orthologs and paralogs, and even orthologous genes in human and mouse can have different functions. The inclusion of all genes — that is, not excluding species-specific genes — may be key to gaining mechanistic insights and understanding species-specific biology. Therefore, we invested substantial effort to support analysis of mouse data through the release of Mouse MSigDB, which consists of gene sets curated directly from mouse-centric databases and datasets and specified in native mouse gene identifiers. These sets can be used without the need for ortholog mapping. The new Mouse MSigDB leverages the organizational paradigm of Human MSigDB and includes the following collections of sets (see Supplementary Note 1):
M1 – A ‘positional’ collection of the genes in each cytogenetic band
M2 – Molecular pathways mined from public mouse databases, and a subcollection of mouse signatures of chemical and genetic perturbations curated from peer-reviewed literature
M3 – Regulatory targets, including micro-RNA (miRNA) targets and transcription factor targets for mouse
M5 – Gene sets derived from ontology hierarchies
M8 – Curated cell type markers from single-cell data
MH – An ortholog-converted version of the popular MSigDB Hallmarks collection3; the only collection not derived from mouse native data
Although Mouse MSigDB is a new resource, it benefits from the maturity of the existing Human MSigDB and accompanying website (Fig. 1). The gene sets were prepared following the procedure that we have honed over the years for Human MSigDB (see Supplementary Note 2).
Fig. 1 |. Home page of the MSigDB resource.
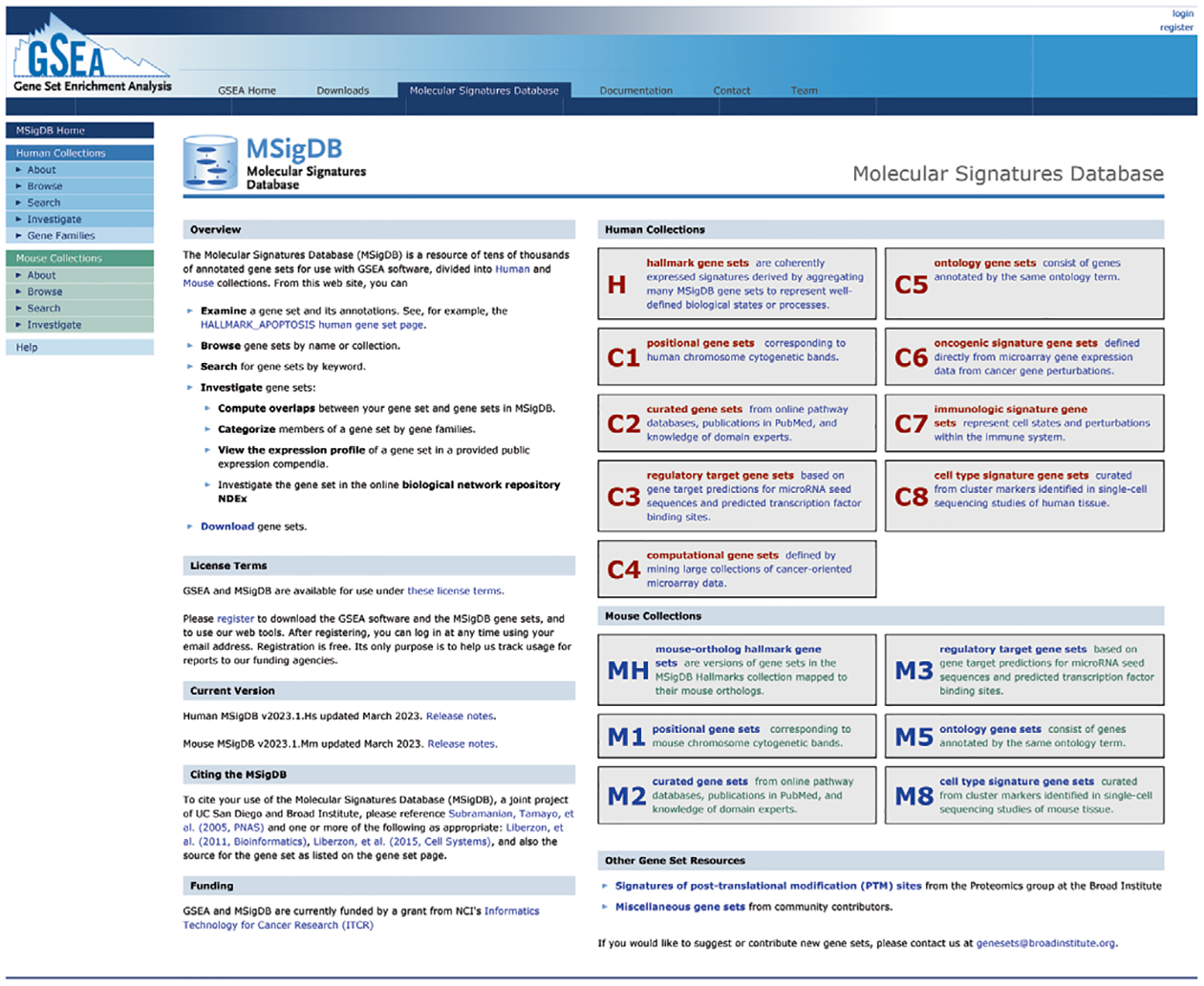
The updated MSigDB home page at https://msigdb.org provides access to both the mouse gene set collections and the human collections. Separate color schemes are used for the two different collections throughout the website to allow the user to easily distinguish between the two. A green-and-blue color scheme indicates information about the mouse collections, blue-and-red about the human collections.
Improved ortholog mapping
The GSEA method is species agnostic, but the software requires that the identifiers of the genes in the analyzed data match those of the querying gene sets. However, if there are mismatches, MSigDB provides a critical set of gene-identifier mapping-table files that GSEA uses to convert the identifiers in a dataset to the gene symbols namespace of MSigDB. The files can map between different name spaces and versions of human genes, as well as provide ortholog mapping between mouse genes and human genes, and vice versa. We made substantial improvements to our procedure for creating MSigDB’s ortholog mapping tables, leveraging premiere ortholog databases and considering the particular requirements of GSEA. We adopted a systematic ‘best match ortholog’ approach, using annotations from the Alliance of Genome Resources4, which integrates annotations from multiple independent5 methods via the Drosophila RNAi Screening Center’s Integrative Ortholog Prediction Tool, and supplementing with ortholog data from Ensembl6. The resulting files improve the analysis of mouse datasets when used with Human MSigDB, but also allow the analysis of human datasets with the mouse gene set database. (See Supplementary Notes 3 and 4 for details of the new ortholog mapping method and an assessment of its performance.)
The mouse collections will be updated on the same frequent release schedule as Human MSigDB (see Supplementary Note 2). Our first priority is to add other mouse collections modeled on the Human MSigDB — for example, oncogenic and immunologic sets. For other future work, the infrastructural changes we have invested in developing and releasing the Mouse MSigDB open the possibility of supporting the curation of gene sets from other model organisms (for example, rat) and species of interest.
Few comprehensive resources exist for mouse native gene sets that can be used for GSEA (see Supplementary Note 5). By offering both support for native analysis of mouse data and support for mapping mouse data to use with human gene sets and human data to use with mouse-native sets, the new MSigDB enhancements represent a significant step forward in the potential translational impact of GSEA results and put the enrichment analysis of mouse and human data on equal footing.
Supplementary Material
Acknowledgements
This work was supported by US National Institutes of Health grant U24CA220341 to J.P.M.
Footnotes
Code availability
The GSEA software is open source under a BSD-style license; the source code is available at http://github.com/GSEA-MSigDB. MSigDB is available under a Creative Commons license. The full license terms can be found at http://gsea-msigdb.org.
Competing interests
The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-023-02014-7.
Data availability
The MSigDB resource, including both mouse and human gene sets, is freely available at http://msigdb.org. The full GSEA/MSigDB resource is available at http://gsea-msigdb.org.
References
- 1.Mootha VK et al. Nat. Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Subramanian A et al. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liberzon A et al. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alliance of Genome Resources Consortium. Genetics 213, 1189–1196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y et al. BMC Bioinformatics 12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrero J et al. Database (Oxford) 2016, baw053 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MSigDB resource, including both mouse and human gene sets, is freely available at http://msigdb.org. The full GSEA/MSigDB resource is available at http://gsea-msigdb.org.


