Abstract
Exploration of the diversity among primate lentiviruses is necessary to elucidate the origins and evolution of immunodeficiency viruses. During a serological survey in Cameroon, we screened 25 wild-born guereza colobus monkeys (Colobus guereza) and identified 7 with HIV/SIV cross-reactive antibodies. In this study, we describe a novel lentivirus, named SIVcol, prevalent in guereza colobus monkeys. Genetic analysis revealed that SIVcol was very distinct from all other known SIV/HIV isolates, with average amino acid identities of 40% for Gag, 50% for Pol, 28% for Env, and around 25% for proteins encoded by five other genes. Phylogenetic analyses confirmed that SIVcol is genetically distinct from other previously characterized primate lentiviruses and clusters independently, forming a novel lineage, the sixth in the current classification. Cercopithecidae monkeys (Old World monkeys) are subdivided into two subfamilies, the Colobinae and the Cercopithecinae, and, so far, all Cercopithecidae monkeys from which lentiviruses have been isolated belong to the Cercopithecinae subfamily. Therefore, SIVcol from guereza colobus monkeys (C. guereza) is the first primate lentivirus identified in the Colobinae subfamily and the divergence of SIVcol may reflect divergence of the host lineage.
Simian immunodeficiency viruses (SIVs) and the closely related human immunodeficiency virus type 1 (HIV-1) and HIV-2 belong to the lentivirus subfamily of retroviruses. So far, the primate lentiviruses for which full-length genome sequences are available fall into five major lineages, on the basis of comparisons of their sequences and the functional similarity of their genes (21). These five lineages are approximately equidistant and are represented by (i) SIVcpz from chimpanzees (Pan troglodytes) together with HIV-1 (9, 16, 26, 27, 43, 52), (ii) SIVsm from sooty mangabeys (Cercocebus atys) together with HIV-2 (6, 15, 24), (iii) SIVagm from four species of African green monkeys (members of the Chlorocebus aethiops superspecies) (2, 10, 13, 14, 23, 28, 30, 36), (iv) SIVsyk from Sykes' monkeys (Cercopithecus mitis albogularis) (22) and (v) SIVmnd from a mandrill (Mandrillus sphinx) (50) together with SIVlhoest from l'Hoest monkeys (Cercopithecus lhoesti) (20) and SIVsun from Sun-tailed monkeys (Cercopithecus solatus) (3). Recently, SIVs from red-capped mangabeys (Cercocebus torquatus torquatus) (18), drills (Mandrillus leucophaeus) (8), and talapoin (Miopithecus talapoin) (40) monkeys have been partially characterized. They may represent additional distinct lineages, but analysis of the complete genome will be necessary to establish the exact phylogenetic relationship between these SIVs and other nonhuman primate (NHP) lentiviruses.
The phylogeny of the NHP lentiviruses indicates that some of the viruses have coevolved with their natural host species, as is the case for African green monkeys (2, 28, 36) and within the C. lhoesti superspecies (3). On the other hand, differences between the phylogenetic relationships of some viruses and primate phylogeny indicate also that there have been multiple cross-species transmissions (review in reference 48).
Although these NHP viruses are called immunodeficiency viruses, the most striking feature of these natural host models is the lack of AIDS-like disease, despite continuous high-level replication of the virus in some natural hosts, suggesting that these viruses have been associated with their hosts for an extended period of time (37, 45). However, if cross-species transmission occurs, the virus may be pathogenic for the new host. For example, SIV isolated from sooty mangabey monkeys (SIVsm) causes AIDS when transmitted to a new host such as rhesus or pig-tailed macaques (Macaca mulatta or Macaca nemestrina), which are not infected by SIV in their natural habitat (11). Both groups of viruses giving rise to AIDS in humans appear to have resulted from several independent transmissions from NHP species. The close phylogenetic relationships between HIV-2 and SIVsm (80% identity), the geographic coincidence, and the fact that only these two lentiviruses share the accessory gene vpx provide strong evidence that sooty mangabeys are the natural reservoir for the original pandemic in West Africa (7, 17). In a similar way, it seems highly probable that HIV-1 arose as a consequence of SIVcpz transmissions from chimpanzees (P. troglodytes) to humans (16). Other transmissions have occurred in the wild (to a yellow baboon [Papio hamadryas cynocephalus] in Tanzania [29], a chacma baboon [Papio ursinus] in South Africa [51], and a patas monkey [Erythrocebus patas] in Senegal (5), all of which were infected by viruses derived from the local sympatric species of African green monkeys).
The viruses identified to date probably represent only a small number of the lentiviruses present in African NHP. Indeed, serological surveys have indicated that numerous species may harbor lentiviruses, including Allen's swamp monkey (Allenopithecus nigrovidis), Diana monkey (Cercopithecus diana), greater white-nosed monkey (Cercopithecus nictitans), moustached monkey (Cercopithecus cephus), Hamlyn's monkey (Cercopithecus hamlyni), and Wolf's mona monkey (Cercopithecus wolfi) (19, 34, 39). Given that viruses from chimpanzees and sooty mangabeys have both crossed the species to humans on multiple occasions, the possibility of ongoing zoonotic transfers has to be considered. The characterization of new SIV strains is important to better understand the origins of HIV-1 and HIV-2 but is even more important to assess new lentiviruses which could potentially infect the human population, since zoonotic transfer from other primates cannot be excluded. This paper describes the genetic characterization of a new SIV, designated SIVcol, according to its species of origin, the guereza colobus monkey (Colobus guereza) from Cameroon, which represents the sixth lineage among the NHP lentiviruses.
MATERIALS AND METHODS
Animals and serologic testing.
Blood samples were obtained from 25 guereza colobus monkeys (C. guereza). All the animals had been caught in the wild in Cameroon and were sampled on their arrival at the bushmeat market in Yaounde, Cameroon, or in the villages around the city, except for one animal, which was kept as a pet. In order to avoid an increase of the trade in bushmeat by our studies, the monkeys were given back to their owners after sampling. Blood was obtained by intracardiac puncture or by venipuncture. Whole blood or plasma was tested for the presence of HIV/SIV antibodies by commercially available HIV antibody tests: Innolia HIV-1/HIV-2 (Innogenetics, Ghent, Belgium), HIV-1/2 Western blots (HIV Blot 2.2; Genelabs Diagnostics, Singapore), or HIV-2-specific Western blots (LAV blot 2; Diagnostics Pasteur, Marnes La Coquette, France).
PCR amplification and cloning.
DNA was isolated from whole blood or from primary peripheral blood lymphocytes using the QIAamp blood kit (Qiagen), according to the manufacturer's instructions. PCR amplification was performed with an automated DNA thermal cycler (GeneAmp PCR system 2400). For amplification of the pol region, degenerated primers designed to amplify a small region of all primate lentivirus pol sequences were used, as follows: DR1 (5′ TRCAYACAGGRGCWGAYGA 3′) and DR2 (5′ AIADRTCATCCATRTAYTG 3′) for the first round and DR4 (5′ GGIATWCCICAYCCDGCAGG 3′) and DR5 (5′ GGIGAYCCYTTCCAYCCYTGHGG 3′) for the second round of amplification. PCR conditions were as previously reported (8). In addition, a second region of pol was amplified with PolOR (5′ ACBACYGCNCCTTCHCCTTTC 3′; positions 5233 to 5253 in SIV(MM251) [M19499]) in combination with DR1 for the first round and Polis4 [5′ CCAGCNCACAAAGGNATAGGAGG 3′; positions 4433 to 4455 in SIV(MM251)] and UNIPOL2 (35) for the second round. Amplification conditions were as follows for the first round: 95°C for 2 min and then 10 cycles of 15 s at 94°C, 30 s at 45°C, and 3 min at 72°C, followed by 25 cycles with extension at 72°C for 3 min with an increment of 5 s per cycle. The second PCR round was as follows: 35 cycles of 15 s at 95°C, 30 s at 50°C, and 45 s at 72°C. The resulting fragment, DR4/DR5, DR1/DR2, or Polis4/UNIPOL2, was cloned into pGEM-T vector (Promega) and sequenced.
To obtain the full-length sequence of SIVcolCGU1 (from the animal designated CGU1), two sets of primers were designed based on the DR1/DR2 sequence, as follows: CGA (5′ CGGATCCAAGGGAATTGAGAAATAGG) and CGB (5′ TCCCATCAGTGAACAATTTGGCACCAG) for the first round and CGF (5′ TAGAACCGTTCAGGAAGAGAGG) and CGG (5′ GCTGTCCAAGCGCCTGTTAATTG) for the second round of amplification. These primers were used to amplify the complete genome of SIVcol by targeting unintegrated circular SIV DNA. PCRs were performed using a Long Expand High Fidelity PCR kit (Roche Molecular Biochemicals) and included a hot start (94°C for 3 min) with the following cycle conditions: 10 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 68°C for 10 min, followed by 20 cycles with extension at 68°C for 10 min with an increment of 20 s per cycle. Amplification was completed by a final extension at 68°C for 12 min. Then, a fraction (1/20) of the first PCR product was used as template in a nested amplification with the same cycling conditions except for the annealing temperature (55°C). The PCR amplification products were then purified and cleaved with EcoR1, and the three resulting fragments were subcloned into pBluescript KS(+) cleaved by EcoRI or by EcoRI and SmaI. Double-stranded recombinant plasmids DNA were sequenced using cycle sequencing and dye terminator methodologies (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq FS DNA polymerase [PE Biosystems, Warrington, England]) on an automated sequencer (ABI 373, Stretch model; Applied Biosystems) using a primer-walking approach.
Sequence comparisons.
To reconstitute the full-length genome sequence, overlapping sequences were joined using contig assembly program (25). The protein sequences predicted to be encoded by SIVcol were compared with other published SIV and HIV sequences representing each of the five known NHP lentivirus lineages, as follows: HIV-1 group M (U455 and M62320), HIV-1 group O (MVP5180 [accession no. L20571]), SIVcpzUS (accession no. AF103818), SIVsyk (173 [accession no. L06042]), SIVsm (PBj [accession no. M31325]), SIVlhoest (accession no. AF075269), SIVmnd (GB1 [accession no. M27470]), SIVagm from grivet (gri677 [accession no. M58410]).
Nucleotide and protein sequences were aligned by Clustal W software program (49) with subsequent manual adjustments. Sites containing a gap in any sequence were excluded from the analyses.
The extent of sequence differences, along the genome, between SIVcol and other primate lentiviruses was examined in a diversity plot in which protein (Gag, Pol, Vif, Env, and Nef) sequences were concatenated. Segments encoded by overlapping genomic regions (between Gag and Pol, Pol and Vif, and Env and Nef) were represented only once. The fractional amino acid sequence difference was calculated for a window of 200 residues, moved in steps of 20 residues. Diversity plots were performed using the online program DIVERT (http://igs-server.cnrs-mrs.fr/anrs/phylogenetics).
The phylogenetic relationship of SIVcol to other primate lentivirus sequences was estimated for the full-length genome sequence, for each of the genes, and from subregions to investigate whether SIVcol might have a mosaic genome. Relationships were estimated using the neighbor-joining and maximum-likelihood methods. The neighbor-joining method was applied to matrix distances with Kimura's correction (32) and 1,000 bootstrap replicates and was implemented using the Clustal W package (49). The maximum-likelihood method was implemented with PROTML using the JTT model (31) using the MOLPHY package (1). TREEVIEW was used to draw trees for illustrations (41). The following viruses were used in our analyses (GenBank accession numbers are in parentheses): HIV-1 group N (AJ006022), SIVcpzGAB (X52154), SIVcpzANT (U42720), HIV-2-EHOA (U27200), HIV-2-ROD (M15390), SIVsm251 (M19499), SIVagmVER155 (M29975), SIVagmTAN (U58991), and SIVsun (AF131870).
Nucleotide sequence accession numbers.
The complete sequence of SIVcolCGU1 has been submitted to GenBank under accession number AF301156, and the partial sequences have been submitted to GenBank under accession no. AF301154 and AF301155 (for SIVcol163 and SIVcol216, respectively).
RESULTS
Animals and serology.
A total of 25 wild-born guereza colobus monkeys (C. guereza) were tested for HIV/SIV-specific antibodies. Among the 25 animals, 1 was an infant, 3 were juveniles, and 21 were adults. Overall, 14 of the monkeys tested were males and 11 were females.
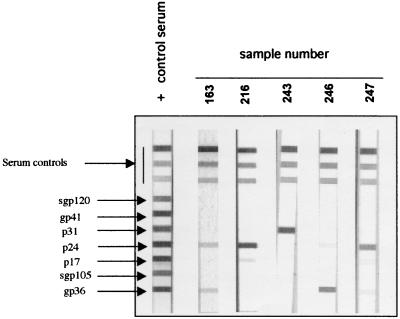
In the Innolia HIV-1/HIV-2 test, the following HIV-1 and HIV-2 recombinant proteins and synthetic peptides were coated as discrete bands on a nylon strip: gp120 (HIV-1 synthetic peptide); gp41 (HIV-1 recombinant protein); p31, p24, and p17 (recombinant proteins of HIV-1 which also cross-react with HIV-2 antibodies); gp36 (HIV-2 recombinant protein), and gp105 (HIV-2 synthetic peptide). Table 1 summarizes the antibody profiles obtained with the Innolia HIV-1/HIV-2 and Western blot assays, and Fig. 1 illustrates some antibody profiles observed with the Innolia HIV-1/HIV-2 tests. We slightly adapted the already-developed interpretation criteria to human sera as follows: samples reactive with any HIV protein were considered positive if the intensity of the bands observed was equal to or higher than either that at the immunoglobulin G cutoff point for the Innolia HIV-1/HIV-2 test (Fig. 1) or that observed with the weakly positive control serum in the Western blot assay; sera were considered indeterminate when the intensity of the bands on the strips was lower than the benchmarks described above; and the absence of reactivity with any of the bands was considered to be a negative result. We further categorized the antibody profiles of positive samples into reaction to Gag, Pol, HIV-1 envelope, or HIV-2 envelope peptides. Interestingly, seven sera had a strong antibody reaction with one or more peptides in the Innolia HIV-1/HIV-2 test, including five that had a reaction with the HIV-2 envelope protein (four strongly positive and one with a weak reaction). Of these seven sera, six also had a clear reaction with Gag or Pol peptides on a commercial HIV-1 Western blot, but none was reactive with the HIV-2-specific peptide, gp36, added on the strips. One particular serum sample (from animal 246), with a weak p24 reaction and a strong gp36 reaction in the Innolia HIV-1/HIV-2 test, was only weakly reactive with p24 on a Western blot (reaction even weaker than that of the weakly positive control serum) and was therefore considered indeterminate on a Western blot. Moreover, the serum sample from animal CGU1, showing a strong reaction with gp36 in the Innolia HIV-1/HIV-2 test, was even completely negative on a HIV-2-specific Western blot (LAV Blot 2; Diagnostics Pasteur) (data not shown).
TABLE 1.
Antibody profiles of sera from guereza colobus monkeys, obtained with commercial HIV antibody confirmatory assays described in Materials and Methods
| Animal | Agea | Sexb | Result ofc:
|
Interpretation of antibody profilesd | PCR result for pol | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Innolia HIV-1/HIV-2
|
Western blot HIV-1/2
|
|||||||||||
| gag | pol | env1 | env2 | gag | pol | env1 | env2 | |||||
| 10 | J | M | − | − | − | − | − | − | − | − | −/− | − |
| 11 | A | M | wk | − | − | − | wk | − | − | − | Ind/Ind | + |
| 12 | A | F | − | − | − | − | − | − | − | − | −/− | − |
| 39 | A | F | − | − | wk | − | − | − | − | − | Ind/− | − |
| 43 | A | F | + | + | − | + | + | + | − | − | +/+ | + |
| 77 | A | M | − | − | − | − | − | − | − | − | −/− | − |
| 92 | I | M | − | − | − | − | − | − | − | − | −/− | nae |
| 147 | A | M | − | − | − | − | wk | − | − | − | −/Ind | − |
| 151 | J | F | − | − | − | − | − | − | − | − | −/− | − |
| 153 | A | M | − | − | − | − | − | − | − | − | −/− | − |
| 161 | A | F | − | − | − | − | wk | − | − | − | −/Ind | − |
| 162 | J | F | − | − | − | − | − | − | − | − | −/− | − |
| 163 | A | M | + | − | − | + | + | − | − | − | +/+ | + |
| 193 | A | M | wk | − | wk | − | wk | − | − | − | Ind/Ind | − |
| 216 | A | F | + | wk | − | − | + | wk | − | − | +/+ | + |
| 229 | A | F | wk | − | − | − | wk | wk | − | − | Ind/Ind | − |
| 234 | A | M | − | − | − | − | − | − | − | − | −/− | − |
| 235 | A | M | − | − | − | − | − | − | − | − | −/− | − |
| 238 | A | M | wk | − | − | − | − | − | − | − | Ind/− | − |
| 243 | A | F | − | + | − | − | + | + | − | − | +/+ | + |
| 246 | A | M | wk | − | − | + | wk | − | − | − | +/Ind | na |
| 247 | A | F | + | − | − | wk | + | wk | − | − | +/+ | + |
| 248 | A | M | wk | − | − | − | − | − | − | − | Ind/− | − |
| 252 | A | F | − | − | wk | − | − | − | − | − | Ind/− | − |
| CGU1 | A | M | wk | − | − | + | + | − | wk | − | +/+ | + |
I, infant; J, juvenile; A, adult.
M, male; F, female.
See Materials and Methods; −, negative, +, positive; wk, weak antibody reaction (below the cutoff).
Interpretation of Innolia HIV-1/HIV-2 test results/interpretation of Western blotting results. Ind, indeterminate.
na, not available.
FIG. 1.
Antibody profiles of guereza colobus monkeys (Colobus guereza) on a commercial line-immunoassay, Innolia HIV-1/HIV-2 (Innogenetics). The correspondence of the different bands observed are indicated on the left of the positive control strip. The numbers 163, 216, 243, 246, and 247 were assigned to five different colobus monkeys, from whom whole-blood samples were collected.
Six additional sera showed only a weak reaction with Gag and/or Env HIV peptides in the Innolia HIV-1/HIV-2 test, and four of them were also weakly reactive with Gag proteins in a Western blot. Two additional sera were also only weakly reactive with Gag proteins on a Western blot. All these sera were considered indeterminate for HIV/SIV antibodies.
PCR amplification of viral sequences was performed using primers which are highly conserved among the NHP lentiviruses in the pol region on available samples (23 out of 25). Only six samples out of the seven classified as positive (with strong serological reactions with one or more peptides in Western blot or Innolia HIV-1/HIV-2 assays) were available, and all six were PCR positive. Interestingly, one serum sample (from animal 11) out of seven considered indeterminate by Innolia HIV-1/HIV-2 test, was also PCR positive (Table 1). Obviously, there is no clear specific serological pattern indicative of SIV infection in guereza colobus monkeys, but given our PCR results, a strong reactivity against at least one antigen in an Innolia HIV-1/HIV-2 test seems to correlate with the presence of virus. Among the sera considered indeterminate by our criteria, some are from SIV-infected animals and some correspond to nonspecific reactions.
Genomic organization of SIVcol.
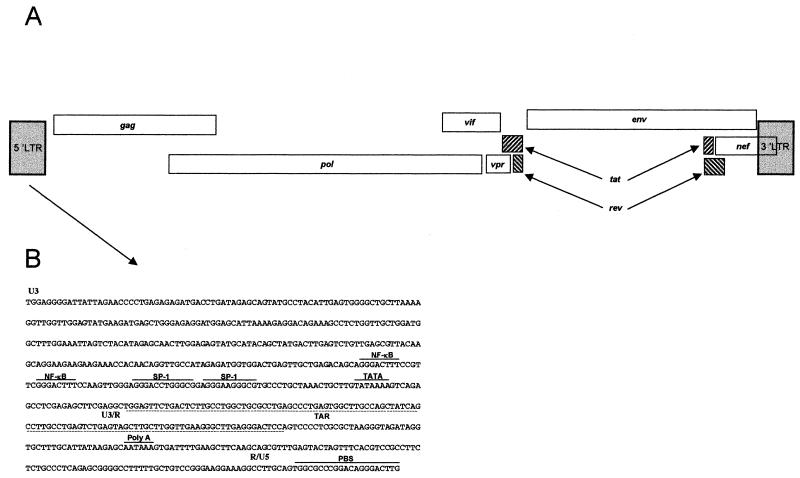
In order to genetically characterize the lentivirus from the seropositive guereza colobus monkeys, the full-length genome of one SIVcol virus was sequenced. Degenerated primers were used to amplify a 787-bp fragment of the pol gene from DNA extracted from a whole-blood sample from animal CGU1. This fragment was cloned and sequenced, and comparison with sequences from the available SIV sequences confirmed that the fragment was related to NHP lentivirus pol sequences. The complete genome of SIVcolCGU1 was amplified by targeting unintegrated circular DNA. Based on the partial pol sequence, spanning the region between the DR1 and DR2 primers, two reverse primers at the 5′ end and two forward primers at the 3′ end were designed. A nested long PCR was done to amplify the near-full-length genome. The long PCR products were cleaved, and the different fragments were cloned. The SIVcol genome was sequenced in its entirety (8,728 bp) by the primer-walking approach, and the full-length genome sequence was compared to those of other primate lentiviruses. The SIVcol genome displayed the genomic organization typical of most primate lentiviruses, lacking vpu (group SIVcpz/HIV-1 lineage) and vpx (group SIVsmm/HIV-2 lineage) (Fig. 2A). The long terminal repeat (LTR) of SIVcol contained all the characteristic features of other primate lentivirus. The U3 region of SIVcol contained two NF-κB sites and two potential SP-1 binding sites (Fig. 2B). Secondary structure analysis of its LTR sequences revealed a duplicated TAR stem-loop structurally similar to the one found in SIVmnd (4).
FIG. 2.
(A) Genomic organization of SIVcol from guereza colobus monkeys; (B) sequence of the SIVcol LTR. The U3, R, and U5 regions are indicated. The NF-κB and SP-1 sites, the TATA box, the poly(A) signal, and the primer binding site (PBS) are noted with a line above the sequence. The TAR region is shown by a dashed line below the sequence.
SIVcol is a member of a sixth lineage.
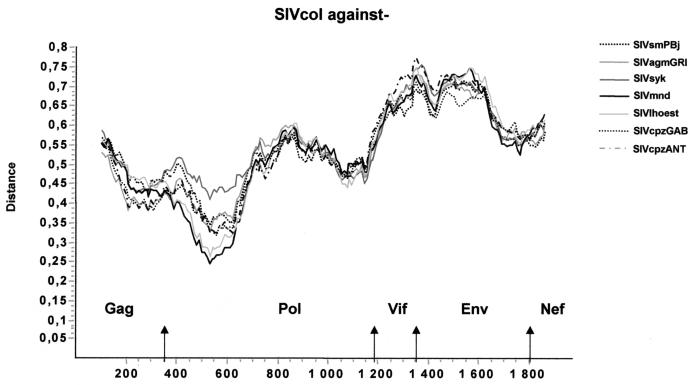
Comparisons of the protein sequences predicted to be encoded by the eight genes common among other primate lentiviruses revealed that SIVcol was quite distant from all other SIVs and HIVs (Table 2). Globally, SIVcol is approximately equidistant to the HIV/SIV representatives of all five primate lineages, with average amino acid identities of 40% for Gag, 50% for Pol, and 23, 27, 26, 22, 28, and 25% for Vif, Vpr, Tat, Rev, Env, and Nef, respectively. Comparisons of the predicted protein sequences and the conserved Gag and Pol protein sequences of other SIVs reveal several well-conserved regions. Amino acid sequences corresponding to the surface unit of the envelope of SIVcol show a similar structure to those of previously described SIVs. Seventeen out of 18 cysteine residues are conserved, but only 2 out of 17 potential N-linked glycosylation sites of SIVcol were at positions conserved among NHP lentiviruses, with 1 in the V3 loop analog. Nevertheless, the highest similarities were seen in the V3 analog region and also in the CD4 binding domain, where out of 28 amino acids, 8 are conserved and 8 are strongly similar between SIVs (Fig. 3). Diversity plots were constructed to further investigate the extent of sequence difference from the other primate lentiviruses across the genome. Constructing such diversity plots is a useful technique to detect discordant sequence relationships suggestive of recombination. A multiple alignment of concatenated predicted gene products was generated. Sites that could not be aligned unambiguously, as well as sites containing a gap in any sequence, were removed from the alignment in order to ensure that all comparisons were made across the same sites. The percent sequence diversity between sequence pairs was then calculated for a window of 200 amino acids moved in steps of 20 amino acids. The resulting diversity plots confirmed that SIVcol is equally divergent from all SIVs throughout the entire genome (Fig. 4). This could indicate that the times since SIVcol shared a common ancestor with each lineage and the constraints on sequence evolution in SIVcol versus each lineage are largely similar. However, one region (120 amino acids) of smaller divergence was found in the pol gene: only around 25% divergence was observed between SIVmnd/SIVlhoest and SIVcol.
TABLE 2.
Percent amino acid identities between SIVcol and other HIV/SIV group strainsa
| SIV/HIV | % Identity with SIVcol and other SIV/HIV group strains
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpr | Tat | Rev | Env | Nef | |
| HIV-1U455 | 42.8 | 48.7 | 24.9 | 26.4 | 33.3 | 23.1 | 27.9 | 24.3 |
| HIV-1MVP180 | 43.7 | 51 | 23.1 | 24.5 | 22.6 | 23.1 | 27 | 23.4 |
| SIVcpzUS | 39.8 | 48.8 | 24.4 | 26.4 | 31.1 | 16.1 | 28.8 | 22 |
| SIVsmPBj | 37.4 | 48.2 | 22.9 | 30.6 | 30.2 | 25.8 | 28.8 | 25.5 |
| SIVmnd | 38.2 | 51.2 | 23.6 | 31.8 | 31.3 | 26.2 | 28.9 | 26.4 |
| SIVlhoest | 38.9 | 50.1 | 19.4 | 29.6 | 27.6 | 18.2 | 26.5 | 25.9 |
| SIVagmGRI | 41.9 | 47.3 | 22.9 | 27.4 | 17.5 | 25.7 | 29.1 | 26.5 |
| SIVsyk | 39.2 | 47.8 | 22.1 | 22.7 | 18.7 | 20 | 29.2 | 26.4 |
The strains used correspond to those described in Materials and Methods.
FIG. 3.
Alignment of gp120 of SIVcol and an SIV representative of each of the five other lineages. The amino acid sequence of SIVcol is shown on the top line, with variable regions (V1 to V5) analogous to those observed in other SIVs indicated. The CD4 binding domain is shown. An asterisk represents a conserved cysteine residue. Potential N-linked glycosylation sites are noted with a line above. Dots indicate amino acid identity at a residue, and dashes indicate gaps introduced to optimize alignment.
FIG. 4.
Diversity plot comparing SIVcol with representatives of the five major lineages of primate lentiviruses, i.e., SIVcpz, SIVmnd, SIVsyk, SIVsm, and SIVagm. Protein sequence difference is plotted for windows of 200 amino acids, moved in steps of 20.
In addition to diversity plots, many phylogenetic trees derived from alignments of individual or partial gene products were generated to examine the evolutionary relationship of SIVcol to the other primate lentiviruses (data not shown). Whatever the gene in consideration, the results were similar: SIVcol clusters independently from the other HIV/SIV group viruses. However, in trees derived from the alignment spanning the pol region highlighted in the diversity plot, SIVcol clusters with the SIVmnd/SIVlhoest group with high bootstrap values (82%). This exceptional region was further investigated by maximum-likelihood phylogenetic analysis, and similar results were obtained. So, throughout the genome, SIVcol is a highly divergent virus compared to other HIV/SIV group viruses and is not the result of a recombination event between other known isolates.
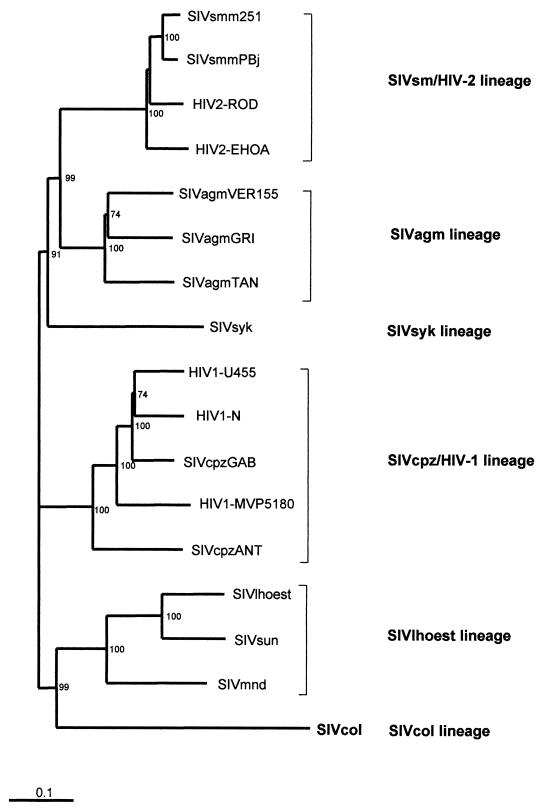
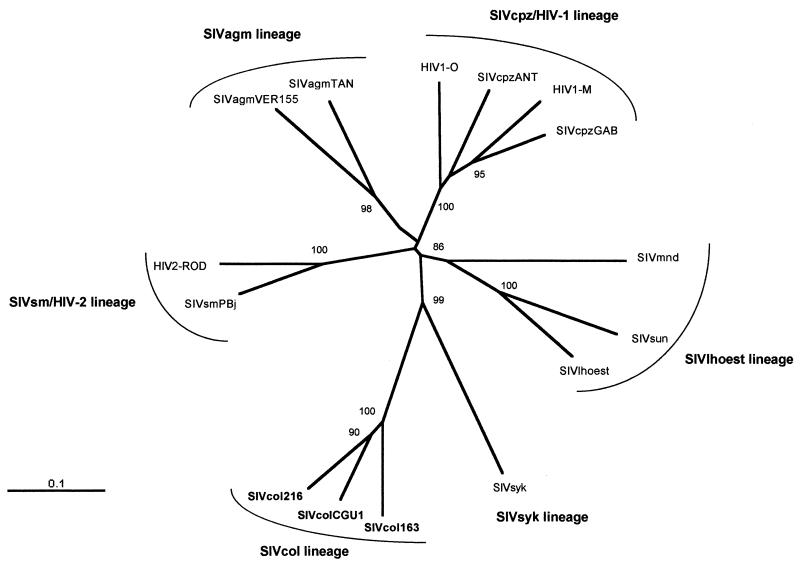
An overview of the phylogenetic relationships between SIVcol and other fully characterized HIV/SIV group viruses representative of the five major lineages is shown in Fig. 5. This tree was obtained from an analysis of an alignment of concatenated Gag-Pol-Vif-Env-Nef proteins using the neighbor-joining method. The results confirmed that SIVcol, the first isolate characterized in the Colobinae subfamily of Cercopithecidae family, clustered separately from all other HIV and SIV strains, forming thus a novel independent lineage, the sixth in the NHP lentivirus classification.
FIG. 5.
Phylogenetic relationship of SIVcol to other primate lentiviruses. This tree was derived by neighbor-joining analysis from a concatenated Gag-Pol-Vif-Env-Nef protein alignment. The reliability was estimated by 1,000 bootstrap replicates; only values above 75% are shown. Branch lengths are drawn to scale. Bar, 0.1 amino acid replacement per site.
Natural infection with SIVcol in the wild.
In our study, the seroprevalence of SIVcol in wild-born guereza colobus monkeys was at least 28% (7 of 25 animals). To investigate if these monkeys are a natural reservoir for SIVcol, we genetically characterized partial SIV sequences from two other animals (no. 163 and 216) to study the extent and nature of genetic diversity among SIVs from this species. A 650-bp fragment of the integrase gene was amplified by PCR from DNA extracted from whole blood and then cloned and sequenced. These two new SIVcol sequences were closely related to each other, with 86% amino acid sequence identity and were also closely related to SIVcolCGU1, with 80% amino acid sequence identity. The extent of sequence similarity among SIVcol isolates is similar to that observed between African green monkeys and l'Hoest monkeys in the corresponding region. The phylogenetic relationship of SIVcol163 and SIVcol216 sequences to SIVcolCGU1 and other primate lentiviruses was investigated by the neighbor-joining and the maximum-likelihood methods. Since both methods generated similar tree topologies, only the neighbor-joining results are shown (Fig. 6). SIVcolCGU1, SIVcol163, and SIVcol216 pol sequences cluster together. Thus, SIVcol appears to be a natural infection in the wild, present in guereza colobus monkeys (C. guereza).
FIG. 6.
Phylogenetic tree showing the relationships of the 3 SIVcol pol sequences (650 bp) to equivalent sequences from selected HIV/SIV group viruses representing the five different lineages. Sites at which there was a gap in any of the aligned sequences were not used to calculate distances. Phylogenetic relationships were computed from these distances by the neighbor-joining method. The significance of the branching order was estimated by the bootstrap method (1,000 resampling).
DISCUSSION
This study presents evidence that guereza colobus monkeys (C. guereza) are infected with a new SIV, SIVcol, representing a sixth lineage in the NHP lentivirus family, and that they are the natural reservoir for this virus. The Cercopithecidae, or Old World monkeys, are subdivided into two distinct subfamilies: Colobinae and Cercopithecinae (12). With the exception of SIVcpz, all the SIVs identified to date originate from NHP belonging to the Cercopithecinae subfamily. This new SIV represents the first primate lentivirus identified in the Colobinae subfamily of the Cercopithecidae.
Phylogenetic analysis of primate lentiviruses shows that all lineages radiate from a single point; this is consistent with evolution and divergence from a common ancestral primate lentivirus. Phylogenetic analyses have shown also that certain SIV lineages appear to have coevolved with their hosts and thus provide further evidence of the ancient nature of the primate lentivirus family. Examples of such host-dependent evolution are SIVagm and SIVlhoest. There are four main species of African green monkeys, and all are naturally infected at high prevalence with SIVagm (2). Viruses from each of the four species form four distinct monophyletic clusters in phylogenetic tree analysis, and all cluster together in the SIVagm lineage (2, 28, 36). A second example of host-dependent evolution is SIVs from l'Hoest and Sun-tailed monkeys, two species classified into the same superspecies (3). It has been suggested that the primate lentiviruses originated and coevolved within monkeys of the Cercopithecus genus (20). However, molecular clock extrapolation analyses of the extent of divergence among primate lentiviruses have estimated maximal divergence times around 200 years ago (33, 44, 47), whereas these examples of host-dependent evolution of SIV suggest divergence times within the primate lentivirus phylogeny on the order of thousands or even millions of years (minimal time of infection). The fact that SIVcol is very divergent from all known SIVs suggest that SIVcol in guereza colobus monkeys is not the result of a recent cross-species transmission. The presence of this virus in this species could be very ancient (although we do not know the specific rate of evolution for this virus), and the divergence of SIVcol may reflect divergence of the host lineage. Colobids split off from other Old World monkeys at least 11 million years ago (42), so the screening of other colobus species, including Asian colobus monkeys, will help to clarify (i) whether the common ancestor of SIV was already present in the common ancestor of the Cercopithecidae family or (ii) whether a cross-species transmission occurred between Cercopithecinae and Colobinae or originated from a yet-unidentified species. Colobus monkeys share habitats with Cercopithecus species and with mangabeys; therefore, an exchange of ancestral SIVs between these species could have been possible in the past.
Although host-specific evolution of SIVs is often observed, many examples of simian-to-simian cross-species transmission in both captive and free-living animals have been documented (5, 29, 38, 50). HIV infections have also resulted from cross-species transmission events. SIVs from chimpanzees and mangabeys have both crossed the species to humans on multiple occasions, and the possibility of ongoing zoonotic transmissions has to be considered. Similarly, as are chimpanzees and mangabeys, colobus monkeys are frequently hunted for food. The bushmeat trade has increased in the last few decades, due to the expanding logging industry in some central African countries and to the construction of new roads that extend into the deepest reaches of the forest (46). The potential for human exposure to a wide range of different SIVs has increased substantially, along with the conditions that facilitate the dissemination. In addition, the high prevalence of SIVs in their natural hosts is an additional factor that increases the potential of transmission from SIVs to humans.
Therefore, it remains important to continue to characterize new SIVs from as many different primate species as possible and to study their molecular and biological characteristics. One major public health implication is that these SIVs are not recognized by commercial HIV-1/HIV-2 screening assays. As a consequence, human infection with such variants can initially go unrecognized and thus lead to another epidemic. Although in some cases this transfer can lead to a biological dead-end, the capacity of several SIVs to replicate efficiently in human primary peripheral blood lymphocytes in vitro is compatible with the possibility that some of these viruses would have the potential to infect human populations (3, 18, 20).
Evidence of SIV infection has so far been reported for 26 different primate species, and the potential for human exposure has certainly increased, as have the epidemiological conditions known to support the emergence of new infections. Identification of SIVs in wild primates will help to elucidate the origins and evolution of HIV infection in humans, and these viruses can serve as sentinels by signaling which pathogens may be a risk for humans. The antigens from these animal reservoir viruses could be included in diagnostic tests.
ACKNOWLEDGMENTS
We thank Christelle Butel and Florian Liegeois for technical assistance.
This work was supported by IRD.
REFERENCES
- 1.Adachi J, Hasegawa M. MOLPHY (a program package for MOLecular PHYlogenetics) version 2.2. Tokyo, Japan: Institute of Statistical Mathematics; 1994. [Google Scholar]
- 2.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer B E, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J-P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. doi: 10.1128/jvi.73.9.7734-7744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibollet-Ruche F, Galat-Luong A, Cuny G, Sarni-Manchado P, Galat G, Durand J P, Pourrut X, Veas F. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J Gen Virol. 1996;77:773–781. doi: 10.1099/0022-1317-77-4-773. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Telfer P, Reed P, Zhang L, Getti A, Ho D D, Marx P A. Isolation and characterization of the first simian immunodeficiency virus from a feral sooty mangabey (Cercocebus atys) in West Africa. J Med Primatol. 1995;24:108–115. doi: 10.1111/j.1600-0684.1995.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewley J P, Lewis J C, Brown D W, Gadsby E L. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J Virol. 1998;72:10305–10309. doi: 10.1128/jvi.72.12.10305-10309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbet S, Müller-Trutwin M C, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel M D, Li Y, Naidu Y M, Durda P J, Schmidt D K, Troup C D, Silva D P, MacKey J J, Kestler III H W, Sehgal P K, King N W, Dhta Y, Hayami M, Desrosiers R C. Simian immunodeficiency virus from African green monkeys. J Virol. 1988;62:4123–4128. doi: 10.1128/jvi.62.11.4123-4128.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desrosiers R C. The simian immunodeficiency viruses. Annu Rev Immunol. 1990;8:557–578. doi: 10.1146/annurev.iy.08.040190.003013. [DOI] [PubMed] [Google Scholar]
- 12.Disotell T. The phylogeny of Old World Monkeys. Evol Anthropol. 1996;5:18–24. [Google Scholar]
- 13.Fomsgaard A, Hirsch V M, Allan J S, Johnson P R. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991;182:397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- 14.Fukasawa M, Miura T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333:457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 15.Fultz P N, McClure H M, Anderson D C, Swenson R B, Anand R, Srinivasan A. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys) Proc Natl Acad Sci USA. 1986;83:5286–5290. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 17.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 18.Georges-Courbot M C, Lu C Y, Makuwa M, Telfer P, Onanga R, Dubreuil G, Chen Z, Smith S M, Georges A, Gao F, Hahn B H, Marx P A. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J Virol. 1998;72:600–608. doi: 10.1128/jvi.72.1.600-608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn B H, Shaw G M, De Cock K M, Sharp P M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch V M, Campbell B J, Bailes E, Goeken R, Brown C, Elkins W R, Axthelm M, Murphey-Corb M, Sharp P M. Characterization of a novel simian immunodeficiency virus (SIV) from l'hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J Virol. 1999;73:1036–1045. doi: 10.1128/jvi.73.2.1036-1045.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, Dapolito G, Goeken R, Campbell B J. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr Opin Genet Dev. 1995;5:798–806. doi: 10.1016/0959-437x(95)80014-v. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch V M, Dapolito G A, Goldstein S, McClure H, Emau P, Fultz P N, Isahakia M, Lenroot R, Myers G, Johnson P R. A distinct African lentivirus from Sykes' monkeys. J Virol. 1993;67:1517–1528. doi: 10.1128/jvi.67.3.1517-1528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch V M, McGann C, Dapolito G, Goldstein S, Ogen-Odoi A, Biryawaho B, Lakwo T, Johnson P R. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993;197:426–430. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch V M, Olmsted R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 25.Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- 26.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 27.Janssens W, Fransen K, Peeters M, Heyndrickx L, Motte J, Bedjabaga L, Delaporte E, Piot P, van der Groen G. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpz-gab2 from a wild-captured chimpanzee from Gabon. AIDS Res Hum Retrovir. 1994;10:1191–1192. doi: 10.1089/aid.1994.10.1191. [DOI] [PubMed] [Google Scholar]
- 28.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin M J, Rogers J, Phillips-Conroy J E, Allan J S, Desrosiers R C, Shaw G M, Sharp P M, Hahn B H. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J Virol. 1994;68:8454–8460. doi: 10.1128/jvi.68.12.8454-8460.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson P R, Fomsgaard A, Allan J, Gravell M, London W T, Olmsted R A, Hirsch V M. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64:1086–1092. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones D T, Taylor W R, Thornton J M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 32.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 33.Korber B, Theiler J, Wolinsky S. Limitations of a molecular clock applied to considerations of the origin of HIV-1. Science. 1998;280:1868–1871. doi: 10.1126/science.280.5371.1868. [DOI] [PubMed] [Google Scholar]
- 34.Lowenstine L J, Pedersen N C, Higgins J, Pallis K C, Uyeda A, Marx P, Lerche N W, Munn R J, Gardner M B. Seroepidemiologic survey of captive Old-World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys) Int J Cancer. 1986;38:563–574. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- 35.Miura T, Sakuragi J, Kawamura M, Fukasawa M, Moriyama E N, Gojobori T, Ishikawa K, Mingle J A, Nettey V B, Akari H, et al. Establishment of a phylogenetic survey system for AIDS-related lentiviruses and demonstration of a new HIV-2 subgroup. AIDS. 1990;4:1257–1261. doi: 10.1097/00002030-199012000-00012. [DOI] [PubMed] [Google Scholar]
- 36.Müller M C, Saksena N K, Nerrienet E, Chappey C, Hervé V M, Durand J-P, Legal-Campodonico P, Lang M-C, Digoutte J-P, Georges A J, Georges-Courbot M-C, Sonigo P, Barré-Sinousi F. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norley S G. SIVagm infection of its natural African green monkey host. Immunol Lett. 1996;51:53–58. doi: 10.1016/0165-2478(96)02555-2. [DOI] [PubMed] [Google Scholar]
- 38.Novembre F J, Hirsch V M, McClure H M, Fultz P N, Johnson P R. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology. 1992;186:783–787. doi: 10.1016/0042-6822(92)90047-s. [DOI] [PubMed] [Google Scholar]
- 39.Ohta Y, Masuda T, Tsujimoto H, Ishikawa K, Kodama T, Morikawa S, Nakai M, Honjo S, Hayami M. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int J Cancer. 1988;41:115–122. doi: 10.1002/ijc.2910410121. [DOI] [PubMed] [Google Scholar]
- 40.Osterhaus A D, Pedersen N, van Amerongen G, Frankenhuis M T, Marthas M, Reay E, Rose T M, Pamungkas J, Bosch M L. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin) Virology. 1999;260:116–124. doi: 10.1006/viro.1999.9794. [DOI] [PubMed] [Google Scholar]
- 41.Page R D M. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 42.Page S L, Chiu C, Goodman M. Molecular phylogeny of Old World monkeys (Cercopithecidae) as inferred from gamma-globin DNA sequences. Mol Phylogenet Evol. 1999;13:348–359. doi: 10.1006/mpev.1999.0653. [DOI] [PubMed] [Google Scholar]
- 43.Peeters M, Fransen K, Delaporte E, Van den Haesevelde M, Gershy-Damet G M, Kestens L, van der Groen G, Piot P. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS. 1992;6:447–451. doi: 10.1097/00002030-199205000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Querat G, Audoly G, Sonigo P, Vigne R. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology. 1990;175:434–447. doi: 10.1016/0042-6822(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 45.Rey-Cuille M A, Berthier J L, Bomsel-Demontoy M C, Chaduc Y, Montagnier L, Hovanessian A G, Chakrabarti L A. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol. 1998;72:3872–3886. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson J G, Redford K H, Bennett E L. Wildlife harvest in logged tropical forests. Science. 1999;284:595–596. [Google Scholar]
- 47.Sharp P M, Bailes E, Gao F, Beer B E, Hirsch V M, Hahn B H. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem Soc Trans. 2000;28:275–282. doi: 10.1042/bst0280275. [DOI] [PubMed] [Google Scholar]
- 48.Sharp P M, Robertson D L, Hahn B H. Cross-species transmission and recombination of ‘AIDS’ viruses. Philos Trans R Soc Lond Ser B. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 49.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsujimoto H, Hasegawa A, Maki N, Fukasawa M, Miura T, Speidel S, Cooper R W, Moriyama E N, Gojobori T, Hayami M. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature. 1989;341:539–541. doi: 10.1038/341539a0. [DOI] [PubMed] [Google Scholar]
- 51.van Rensburg E J, Engelbrecht S, Mwenda J, Laten J D, Robson B A, Stander T, Chege G K. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J Gen Virol. 1998;79:1809–1814. doi: 10.1099/0022-1317-79-7-1809. [DOI] [PubMed] [Google Scholar]
- 52.Vanden Haesevelde M M, Peeters M, Jannes G, Janssens W, van der Groen G, Sharp P M, Saman E. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology. 1996;221:346–350. doi: 10.1006/viro.1996.0384. [DOI] [PubMed] [Google Scholar]