Abstract
Mycobacterium bovis Bacillus Calmette-Guerin (BCG) is the primary treatment for non-muscle-invasive bladder cancer (NMIBC), known to stimulate inflammatory cytokines, notably interferon (IFN)-γ. We observed that prolonged IFN-γ exposure fosters adaptive resistance in recurrent tumors, aiding immune evasion and tumor proliferation. We identify HLA-E and NKG2A, part of a novel NK and T cell checkpoint pathway, as key mediators of resistance in BCG-unresponsive NMIBC. IFN-γ enhances HLA-E and PD-L1 expression in recurrent tumors, with an enrichment of intra-tumoral NKG2A-expressing NK and CD8 T cells. CXCL9+ macrophages and dendritic cells and CXCL12-expressing stromal cells likely recruit CXCR3/CXCR4-expressing NK and T cells and CXCR7+ HLA-EHIGH tumor cells. NK and CD8 T cells remain functional within BCG-unresponsive tumors but are inhibited by HLA-E and PD-L1, providing a framework for combined NKG2A and PD-L1 blockade strategy for bladder-sparing treatment of BCG-unresponsive NMIBC.
Keywords: Non-muscle-invasive bladder cancer, BCG-unresponsive, NK cells, Immunotherapy
Introduction
Intravesical Mycobacterium bovis Bacillus Calmette-Guerin (BCG) is the only FDA-approved, first-line treatment for high risk non-muscle-invasive bladder cancer (NMIBC), and is hypothesized to act in part via the release of inflammatory cytokines, notably interferon (IFN)-γ 1.Unfortunately, tumor recurrence following BCG treatment is common, with local recurrence rates between 32.6 to 42.1% and progression rates between 9.5 to 13.4% 2. Radical cystectomy is the only definitive treatment option for BCG-unresponsive disease, leaving patients at high risk for complications and the need for urinary diversion which can lead to diminished quality of life 3, 4. Currently, there is substantial interest and numerous ongoing clinical trials to identify novel therapeutic options for BCG-unresponsive NMIBC.
Despite the use of BCG as the first-line immunotherapy for NMIBC for over 40 years, the mechanisms underlying resistance to BCG’s therapeutic benefits are poorly understood. This lack of understanding has further hindered progress in developing novel approaches to treat tumors resistant to BCG. The six-dose weekly induction regimen was devised anecdotally based on rudimentary urine studies performed in the 1970’s 5. Only a single dosing study has ever been performed to assess the immunologic response with this dosing regimen, which concluded that six doses stimulated an immune response, but did not investigate the ramifications of the magnitude nor the duration of that immune response 6. Contributing to the difficulty in predicting BCG response is the variety of immune lineages implicated in both its success and failure, including neutrophils, monocytes, macrophages, dendritic cells (DCs), T cells, and natural killer (NK) cells 7.
Adaptive immune resistance, in which cancers are recognized by immune cells, but accommodative changes prevent effective clearance, is an important mechanism of therapeutic resistance 8. IFN-γ has been shown to have both pro- and anti-tumorigenic functions across an array of tumor types9. The equilibrium established by IFN-γ signaling may be critical in preventing therapeutic resistance. In melanoma and lung cancer, dysfunctional and overactive IFN-γ signaling is known to drive T cell inhibition via upregulation of PD-L1 10. In muscle-invasive and metastatic urothelial carcinoma, IFN-γ associates with improved as well as poor responses to PD-1/PD-L1 blockade and exists as part of a balance (coined 2IR score) between pro- and anti-tumorigenic pathways expressed in the TME 11. The impact of long-term intravesical BCG, which drives chronic immune stimulation via IFN-γ production, and its role in immune dysregulation and exhaustion in BCG unresponsive NMIBC is emerging but still unclear 12, 13.
Previous work comparing BCG-unresponsive to treatment-naïve tumors showed a significant increase in PD-L1 expression, but only in a minority of unresponsive samples 14. Immunotherapeutic strategies, especially those engaging the PD-1/PD-L1 axis to unleash exhausted immune cells, have modestly improved NMIBC treatment. Unfortunately, response rates to PD-1 blockade monotherapies in NMIBC remain low, at ~19% durable response 15, 16, 17, 18. Further, PD-L1 expression levels alone have failed to accurately predict response to immunotherapy in NMIBC, suggesting that alternative mechanisms of immune suppression are engaged 19.
The HLA-E/NKG2A axis has been identified as a potent immune checkpoint regulating both CD8 T and NK cells 20, 21, 22, 23, 24. Abrogation of HLA-E interactions with NKG2A on NK cells increased effector activity against tumor cells and potentiated anti-tumor function. Further, blocking NKG2A on CD8 T cells in a murine model increased tumor vaccine response, even in a PD-1 resistant setting 20, 22. Additionally, the combination of anti-NKG2A antibody (monalizumab) and antiPD-L1 antibody (durvalumab) has been studied in a Phase 2 clinical trial in patients with nonresectable, stage III NSCLC. The combination demonstrated an objective response rate of 35% vs 17.9% with durvalumab alone, as well as improved progression-free survival (PFS) with durvalumab and monalizumab (72.7%) compared to durvalumab alone (33.9%) 25.
Most recently, BCG was shown to directly infect a proportion of bladder tumor cells in NMIBC patients driving impairment in autophagy flux resulting in downregulation of HLA class I expression along with a shift towards epithelial-to-mesenchymal transitioning (EMT) by BCG-infected tumor cells 26. In a cohort of BCG-treated NMIBC patients with tumor recurrence, the authors observed that abundance of class I HLA-negative tumor cells associated with disease progression and metastasis and overall survival. This study highlighted two different tumor cell types within BCG-unresponsive settings and suggested that a tumor phenotype defined by high HLA class I expression correlated with improved long-term survival post-cystectomy.
In this study, we confirm the presence of both tumor cell phenotypes but focus on their interactions with stromal and immune cells. We present a novel mechanism of resistance, whereby HLA class I-positive tumor cells are activated in response to NK and CD8 T cell-derived IFN-γ and recruited to CXCL12-expressing stromal-rich regions of the tumor where myeloid cells, regulatory T cells (Tregs), and immune effector cells (NK, CD8 T cells) are enriched. Consequently, class I HLA-negative tumor cells can subvert immune cell trafficking. We pose a novel hypothesis that BCG more broadly induces changes in the bladder tumor microenvironment (TME), promoting expression of NKG2A and PD-1 on CD8+ T cells, NKG2A on NK cells, and HLA-E, PD-L1, and CXCR7 on recurring, cytokine-activated tumor cells, and these phenotypic changes are mechanistically linked to treatment resistance and immune evasion by recurring tumors. This study investigates HLA-E and NKG2A as a novel axis of resistance induced by chronic inflammation and provides a mechanistic framework for a bladder-sparing combination immunotherapy strategy of NKG2A and PD-L1 blockade for overcoming resistance to BCG immunotherapy for treating high-grade NMIBC.
Results
Clinical cohort description.
Cohort descriptions of patient samples used for the experiments below are described in Table 1. Patient samples were collected and processed at The Mount Sinai Hospital, and the validation cohort was collected and processed at Aarhus University Hospital, Denmark. Median patient age ranged from 64 to 70.7 years old; median months to recurrence ranged from 1.7 to 2.9 months; median months to progression ranged from 4.43 to 23.28 (Table 1).
Table 1.
| Clinical Cohort Characteristics | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| HTG | OLINK Urine (Sinai) | Single Cell | Spatial | Cytof | ||
|
|
||||||
| n samples, patients | 36, 27 | 42, 26 | 7, 6 | 20, 9 | 16, 16 | |
| Female | 4 | 2 | 0 | 0 | 1 | |
| BCG Naïve | 19 | 26 | 2 | 4 | 0 | |
| BCG Unresponsive | 17 | - | 4 | 5 | 16 | |
| Median Age | 64 | 69.5 | 66.9 | 70.6 | 68.5 | |
| Median Months to Recurrence | 2.9 | 2.1 | 2.3 | 1.7 | 2.73 | |
| Median Months to Progression | 6.7 | - | 14.3 | 23.28 | 4.43 | |
| Stage at Collection | ||||||
| CIS | 4 | - | - | - | - | |
| Tis/Ta | 5 | 13 | - | 2 | 8 | |
| T1 | 10 | 14 | 4 | 6 | 5 | |
| T2 | - | - | - | - | 2 | |
| T3 | - | - | 2 | - | 0 | |
| T4 | - | - | - | - | 1 | |
IFN-γ initiates adaptive resistance including increased tumor expression of HLA-E and PD-L1.
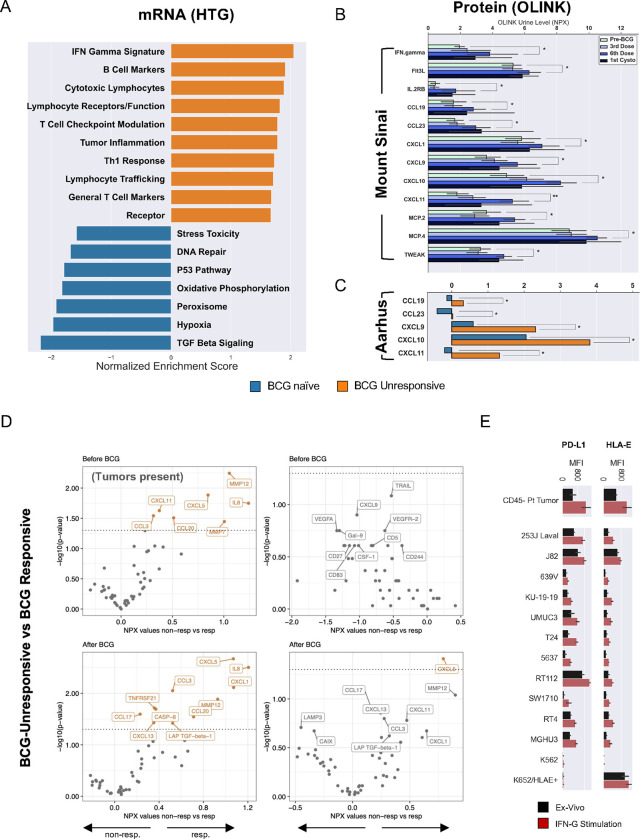
To investigate potential mechanisms underlying BCG resistance in NMIBC, we used tumor specimens from BCG-naïve patients, prior to initiating induction intravesical BCG therapy and BCG-unresponsive tumors, with a recurrence after at least an induction course of intravesical BCG, collected from a prospective cohort of NMIBC patients. Targeted in situ hybridization and mRNA sequencing and gene-set enrichment analysis (GSEA) comparing BCG-unresponsive tumors to pre-treatment timepoints were performed (Fig. 1A) 27. Significant gene sets are shown ranked by their normalized enrichment score (NES) (Fig. 1A, Supplemental Table S1). The top enriched gene-sets comparing BCG-unresponsive to BCG-naïve tumors (n=36) included, but were not limited to, IFN-γ response, cytotoxic lymphocytes, T cell checkpoint modulation, tumor inflammation, TH1 response, and lymphocyte trafficking (Supplemental Table S1). Of note, the “Lymphocyte trafficking” pathway was largely driven by a combination of genes encoding activating cytokines and receptors as well as inhibitory immune checkpoint genes (PDCD1 (PD-1), CD274 (PD-L1), PDCD1LG2 (PD-L2), CTLA4, TIGIT, KLRC1 (NKG2A), HLA-E) indicating a balance between antitumor and pro-tumorigenic features in the TME. Gene sets downregulated in the BCG-unresponsive state and upregulated in the BCG naïve state included TGF beta signaling, hypoxia, peroxisome, oxidative phosphorylation, P53 pathway, DNA repair, and stress toxicity (Fig. 1A).
Fig. 1: Chronic activation within the tumor microenvironment of BCG-unresponsive tumors promotes adaptive resistance.
A, Targeted mRNA gene-set enrichment analysis showing statistically significant differences between BCG-naïve (N=20) and BCG-unresponsive cases (N=20). All gene sets are significant at p<0.05 by Kruskal-Wallis or independent two-sided t-test.
B, Longitudinal protein analysis of urine supernatants across four timepoints in BCG-naïve patients with NMIBC (N=27) with comparisons between BCG-naïve and 6th induction dose timepoints (* p<0.05, ** p<0.001).
C, An additional validation cohort of patients with NMIBC receiving BCG therapy at Aarhus University (N=66) showing BCG-naïve vs BCG-exposed timepoints in urine samples (p-values in both cohorts assessed via independent two-sided t-test or Kruskal-Wallis with Benjamini-Hochberg correction for multiple comparisons).
D, Differential protein expression analysis of urine before (top row) and after (bottom row) BCG exposure and sampled before (left-side) and after (right-side) transurethral resection of bladder tumors. Number of patients included in analyses: (Top-left) BCG-naïve when tumor present, non-responders, N= 47; responders, N= 60. (Top-right) BCG-naïve after tumor resection, non-responders, N= 5; responders, N= 6. (Bottom-left) BCG-exposed when tumor present, non-responders, N= 49; responders, N= 34. (Bottom-right) BCG-exposed after tumor-resection, non-responders, N= 19; responders, N= 26. P-values assessed with two-sided Wilcoxon rank sum. Proteins listed in orange text indicate unadjusted p-values <0.05. BCG-unresponsive NMIBC was defined as high-grade recurrence within two years after receiving BCG therapy.
E, Median fluorescence intensity (MFI) for PD-L1 and HLA-E staining on primary CD45- cells from patients with NMIBC and immortalized bladder tumor lines ex vivo or after 24 hours of incubation with recombinant human (rh) IFN-γ stimulation (all comparisons, p<0.05). All stimulation experiments were performed in triplicate.
To better understand the inflammatory state observed in BCG-unresponsive tumors, we used cell-free urine supernatants from a prospective cohort of BCG-treated NMIBC patients and performed an analysis using the OLINK Proteomics™ “inflammation” panel, a 92-analyte protein proximity extension assay, to profile urine cytokines reflective of the urothelial microenvironment 28, 29. Independent of clinical response to BCG therapy, all patients experienced elevated cytokine levels in their urine. Significantly upregulated cytokines of interest at the sixth dose compared with the BCG-naïve state included IFN-γ, Flt3L, IL2RB, CCL8, CCL13, CCL19, CCL23, CXCL1, CXCL9, CXCL10, CXCL11, and TWEAK (Fig. 1B; all p<0.05). Importantly, urine cytokine levels kept increasing with each consecutive dose of BCG, where even after a six-week treatment interruption (time of first cystoscopy,) urine cytokine levels never equilibrated back to baseline, although the differences were not significant. The inflammatory signature was validated in an independent cohort, “Aarhus” (Aarhus University Hospital, Denmark), consisting of BCG-treated patients with long term clinical follow-up. Urine samples from 66 patients were collected at the BCG-naïve time point and compared to patient-matched urine samples collected within four months after completion of BCG induction course. CXCL9, CXCL10, CXCL11, CCL19, and CCL23 had lasting upregulation at the post-BCG timepoint (p < 0.05) (Fig. 1C). Additionally, urinary protein profiles at these timepoints were compared between responsive and unresponsive patients considering whether visible bladder tumors were present at the time of urine sampling (Fig. 1D). With exception of CXCL5, when assessing urine from BCG-unresponsive patients (before the start of BCG induction therapy) we identified significantly elevated levels of cytokines and chemokines only when tumors were present at time of sampling, where the presence of CCL3, CCL20, CXCL11, IL-8, MMP7 and MMP12 were attributable to bladder tumors rather than in response to BCG. Patients unresponsive to BCG showed a distinct urinary protein profile after BCG treatment with a greater magnitude of inflammatory cytokine and chemokine production as well as Caspase-8 (a marker of apoptosis) and TGF-β1 in its pre-processed inactive state interacting with its latency associated peptide (LAP) (Fig. 1D).
To better understand the effects of IFN-γ on recurring bladder tumors, primary tumor cells from NMIBC patients (n=10) and urothelial tumor lines were cultured for 24 hours in the presence or absence of recombinant human IFN-γ (Fig. 1E) and HLA-E and PD-L1 expression were measured via fluorescence flow cytometry. Tumor cells expressed significantly higher levels of PD-L1 and HLA-E following stimulation with IFN-γ, which was also observed across the collection of bladder tumor lines (n=11) (Fig. 1E). We saw no effects of IFN-γ on wild type (WT) K562 tumors (lack expression of HLA class I, including HLA-E). HLA-E-stably–transduced K562 cells were used as a positive control for maximal HLA-E expression. Overall, these results suggest adaptive resistance by recurring tumor cells after BCG therapy that respond to IFN-γ by increasing HLA-E and PD-L1 expression and in a microenvironment replete with immune effector cells.
Selective upregulation of tumor HLA-E in regions near NKG2A-expressing NK and CD8 T cells
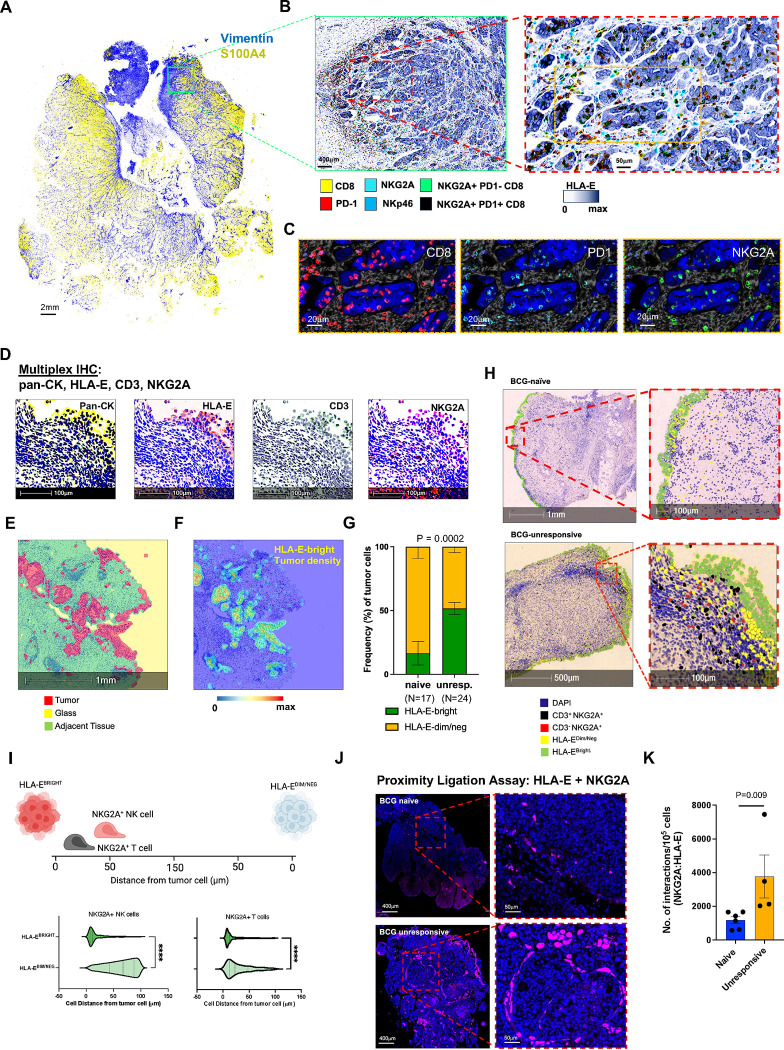
To validate in situ hybridization mRNA analyses and to better understand the spatial relationship between tumor cells and tumor infiltrating lymphocytes (TILs), we performed multiplex imaging analyses using both immunofluorescence (IF) as well as immunohistochemistry (IHC) with whole slide scanning for targeted high-resolution analyses on a cohort BCG naïve (N=17) and BCG unresponsive (N=24) NMIBC patients (Fig. 2). We first performed a subset analysis of NMIBC sections (N=4) using a PhenoCycler™ (formerly CODEX) imaging system (Akoya Biosciences). Representative imaging for one BCG-unresponsive NMIBC section highlights S100A4+ tumor cells intermixed with stroma (Vimentin) (Fig. 2A). We observed a large abundance of NKp46+ NK cells and CD8 T cells with variegated expression of NKG2A and/or PD-1 (Figs. 2B and 2C). While the analysis was underpowered for robust analyses, the data demonstrated substantially greater abundance of NKG2A-expressing NK and CD8 T cells in BCG-unresponsive sections and in regions of tumor tissue with high HLA-E expression by tumor cells and stroma.
Fig: 2: HLA-E tumor expression is highest when near tumor-infiltrating NKG2A+ NK and CD8 T cells.
A, Representative multiplexed immunofluorescence (IF) analysis by PhenoCycler™ (also known as CODEX) of bladder tumor sections from one patient with BCG-unresponsive NMIBC. Representative staining of the entire section is shown for presence of stroma (vimentin, blue) and tumor (S100A4, yellow). Scale bar indicates 2mm.
B, Magnified inset from Panel A, highlighting tumor HLA-E expression of and use of digital pathology software to identify NKp46+ NK and CD8 T cells with and without co-expression of NKG2A +/− PD-1. Left image scale bar indicates 400μm and right image scale bar indicates 50μm.
C, Magnified inset from Panel B, right image, highlighting individual cells expressing CD8, PD-1, and NKG2A. Scale bars indicate 20μm.
D, Representative multiplexed immunohistochemistry (IHC) of bladder tumor from one patient with BCG-unresponsive NMIBC measuring expression of pan-cytokeratin (pan-CK), HLA-E, CD3, and NKG2A. Scale bar indicates 100μm.
E, Representative digital pathology analyses identifying tumor (red) and adjacent/non-tumor (green) tissue along with exposed areas of glass (yellow) to be excluded from subsequent analyses. Scale bar indicates 1mm.
F, Density map highlighting regions of high HLA-E tumor expression. Blue-red color scale indicates HLA-E expression intensity. Scale bar indicates 1mm.
G, Summary analysis of frequency of tumor cells that are HLA-E-bright (green) and HLA-E-dim/negative (dim/neg, yellow) in BCG-naïve (N=17) and BCG-unresponsive (unresp., N=24) NMIBC tumors.
H, Representative digital pathology analysis on one BCG-naïve (top row) and one BCG-unresponsive (bottom row) NMIBC tumor section highlighting nuclear expression of DAPI (blue) and identification of CD3+NKG2A+ CD8 T cells (black), CD3- NKG2A+ NK cells (red), and tumor cells with bright (green) or dim/negative (yellow) expression of HLA-E.
I, Proximity analysis (N=41) measuring cell distance (0–150μm) from HLA-E-bright and HLA-E-dim/regative tumors by NKG2A+ NK cells (left side) and NKG2A+ T cells (right side). ****, p<0.00001. P-values were assessed via independent two-sided t-test.
J, Representative proximity ligation assay (PLA) using immunofluorescence to profile interactions between HLA-E and NKG2A in one BCG-naïve (top row) and one BCG-unresponsive (bottom row) patient with NMIBC.
K, Summary analysis of PLA measuring interactions between HLA-E and NKG2A. P-values were assessed via independent two-sided t-test.
To validate the observation, we stained the cohort of 41 NMIBC sections by multiplexed IHC for Pan-cytokeratin (pan-CK), HLA-E, CD3, and NKG2A (Fig. 2D). Among tumors from the cohort, we analyzed 840,348 tumor cells in total (277,315 for BCG-naïve and 563,033 for BCG-unresponsive). We used Halo digital pathology software (Indica Labs, Albuquerque, NM) to annotate cell lineages, and to measure tumor HLA-E expression within the for density and spatial relationships with stromal and T/NK cell infiltration and proximity (Fig. 2E–2I). Here, we confirmed that HLA-EBRIGHT tumor cells were significantly more abundant in BCG-unresponsive tumors (p= 0.0002) and represented 43.6%% of total tumor cells compared to 10.3% in BCG-naïve tumors (Fig. 2G). Further, we identified NKG2A+ CD3− NK cells and NKG2A+ CD3+ T cells (Fig. 2H) and determined they were in significantly closer proximity to HLA-EBRIGHT tumor cells (p<0.00001)(Fig. 2I). Finally, we performed a proximity ligation assay by immunofluorescence using antibodies targeting HLA-E and NKG2A to quantify the numbers of interactions between NKG2A+ NK/CD8 T cells and HLA-E+ tumor cells and found a significantly greater number of interactions in BCG-unresponsive tumors (p = 0.009) (Fig. 2J and 2K).
Chronic activation of tumor-infiltrating NK and CD8 T cells in BCG-unresponsive tumors
To better understand the composition of the tumor microenvironment and to characterize the potential drivers of adaptive resistance, single-cell RNA sequencing (scRNA-seq) of nine tumors (BCG-naïve (n=3) and unresponsive (n=6)), was performed. In total, 65,324 tumor-derived cells containing 26,520 gene features passed quality control and were used for subsequent analyses, with a median of 958 genes detected per cell.
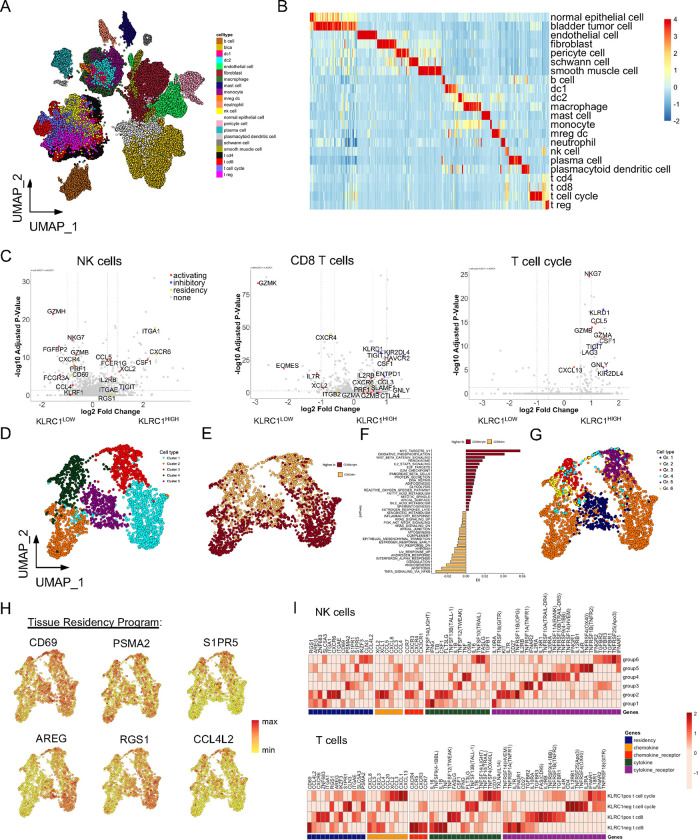
Twenty-two identifiable cell populations, including CD4 and CD8A T cells, regulatory T cells (Tregs), NK cells, dendritic cells (DCs), monocytes and macrophages, granulocytes, B cells, plasma cells, and nonhematopoietic stromal, and tumor cells were identified after graph-based clustering (Fig. 3A and 3B, Supplementary Table S3 and S4). To identify the cell origin(s) of IFN-γ and related pro-inflammatory gene signatures, we examined the expression of IFNG in each of the cell populations. NK cells, CD8 T cells, and a subset comprised of both CD4 and CD8 T cells defined by cell cycle genes (T cell-cycle) demonstrated the highest levels of IFNG expression among all cell clusters and had distinct IFN-γhigh cell populations (Extended data Fig. S1A). Additionally, we profiled the median gene expression across all major canonical checkpoint receptors and observed variegated expression on T cell subsets. However, tumor-derived NK cells mostly expressed KLRC1, and to a lesser extent HAVCR2 (Tim3) and TIGIT (Extended data Fig. S1B).
Fig. 3: Single-cell RNA sequencing analysis of NKG2A+ NK and CD8 T cells from bladder tumors reveals strong antitumor potential but with co-expression of inhibitory receptors.
A, UMAP visualization of cell lineages from single-cell RNA sequencing analysis of urothelial tumor samples (N=9, 65,324 cells).
B, Heatmap summary of genes most differentially expressed on each cell lineage identified.
C, Differentially expressed gene (DEG) analysis of bladder tumor-derived NK cells (left), CD8 T cells (center), and proliferating T cell cycle cells (right) when stratified by high versus dim/negative KLRC1 expression.
D and E, UMAP visualizations of bladder tumor-derived NK cells from unsupervised clustering revealing (D) five clusters or (E) expression of NCAM1/CD56 for annotation of CD56BRIGHT and CD56DIM NK cells (N=2,580 cells).
F, Pathway analysis of Hallmark gene networks that are significantly differentially expressed (at least p<0.05) on bladder tumor-derived CD56BRIGHT and CD56DIM NK cells. P-values were determined using a two-sided t-test.
G and H, UMAP visualization of bladder tumor-derived NK cells (G) clustered according to Groups 1–6 defined by Netskar et al. 30 and (H) highlighting expression and distribution of representative tissue residency genes.
I, Heatmap summary showing average expression for genes associated with tissue residency, chemokines, cytokines, and their receptors on bladder tumor-derived Group 1–6 NK cells (top heatmap) and CD8 T cells and T cell cycle cells when stratified according to KLRC1 expression (bottom heatmap).
To better understand the functional significance associated with NKG2A expression, we stratified NK cells, CD8 T and T cell-cycle cells by low and high KLRC1 expression and performed a differential expression of genes (DEG) analysis (Fig. 3C). Our analysis revealed a balance between cytokine and cytolytic activity and expression of inhibitory checkpoint receptors. KLRC1high NK cells expressed significantly higher CCL5, CSF1, XCL2, IL2RB, and FCER1G, but additionally expressed higher levels of TIGIT. Tumor-infiltrating NK cells also expressed significantly higher levels of CD2, ITGA1 (CD49a), ITGAE (CD103) and RGS1 suggesting they are tissue-resident. Additionally, we observed that KLRC1high CD8 T cells expressed elevated levels of CCL3, CSF1, GNLY, GZMA, GZMB, and PRF1 (perforin), but they additionally expressed higher levels of CTLA4, ENTPD1 (CD39), HAVCR2, LAG3, KIR2DL4 and TIGIT. KLRC1high T cell-cycle cells showed significant upregulation in CCL5, CSF1, CXCL13, NKG7, GNLY, GZMA, GZMB, but additionally expressed LAG3, KIR2DL4, and TIGIT. Collectively, the data suggest that BCG therapy drives strong immune activation and that many repeated doses of treatment sustain activation over prolonged period driving upregulation of inhibitory checkpoint pathways.
Functionally distinct states of bladder tumor-derived NK and CD8 T cells in BCG-unresponsive NMIBC
NK cells are very heterogenous and remain poorly described in solid tumors with virtually nothing known in human bladder cancer. We performed unbiased clustering on all NK cells to derive five major clusters (Fig. 3D), which were divided across conventional definitions of CD56bright and CD56dim NK cells (Fig. 3E). A pathway analysis revealed significant enrichment in CD56bright NK cells for genes involved in metabolism, IL-2 signaling, and immune ‘stress’ (ROS and DNA repair) (Fig. 3F). Conversely, CD56dim NK cell enriched pathways included response to TNFα and IFNα and apoptosis, MAPK activity but also reflect immune ‘stress’ (hypoxia, complement regulation). Building on two recent seminal resources for transcriptional reference mapping of human NK cells in solid tissues30, 31, we integrated the newly defined gene signatures to annotate ‘functional states’ to bladder tumor-derived NK cells (Fig. 3G, Extended data Fig. S2). Our analysis confirmed that clusters 1 and 2 are very similar to Group 2 typical CD56bright NK cells, and cluster 5 strongly correlates with Group 1 “stressed” CD56bright NK cells, described by Netskar et al.30. Collectively clusters 1,2, and 5 reflecting Groups 1 and 2 NK cells represent ~50% of the total tumor-infiltrating NK cells. Additionally, the remaining fraction of CD56dim NK cells were distributed among groups 3–6 NK cells, defined by effector functions (Gr. 3), adaptive NKG2C+ NK cells (Gr. 4), activated (Gr. 5), and typical (Gr. 6) NK cells.
We similarly applied gene signatures for NK1, NK2, and NK3 subsets as recently demonstrated by Rebuffet et al31, 32, and these definitions more closely reflect a developmental or ontogeny trajectory of NK cells across human tissues, NK1 and NK-intermediate cells represent immature CD56bright NK cells and NK3 cells include terminally mature and adaptive NK cells (Extended data Fig. S2). Our comparative analysis revealed an enrichment of NK-intermediate cells representing the original clusters 1 and 2, and NK1B cells representing the original clusters 3 and 4. Both NK1B and NK-intermediate signatures were shared among the original cluster 5 suggesting transitional properties of this NK cell subset. Finally, we profiled tumor-infiltrating NK cells according to the six functional states (Gr. 1–6) (and CD8 T cells and T cell-cycle cells according to KLRC1+/−) for expression of tissue residency genes as well as cytokines/chemokines and cytokine/chemokine receptors to understand how NK cells and T cells are trafficking to the bladder, and we confirmed they are functionally competent (Fig. 3H and 3I). We also observed clear evidence suggesting functional specialization within the NK cells and within the T cells. For instance, KLRC1+ CD8 T cells express high levels of IL2RB, TNFRFS9 (4–1BB), CSF1 and FASLG compared to their KLRC1− counterparts. However, KLRC1− CD8 T cells expressed higher levels of IFNG, and FLT3LG (Fig. 3I).
BCG-unresponsive tumors are characterized by a segregated microenvironment, where activated HLA-EHIGH tumor cells are recruited to stromal regions replete with immune cells.
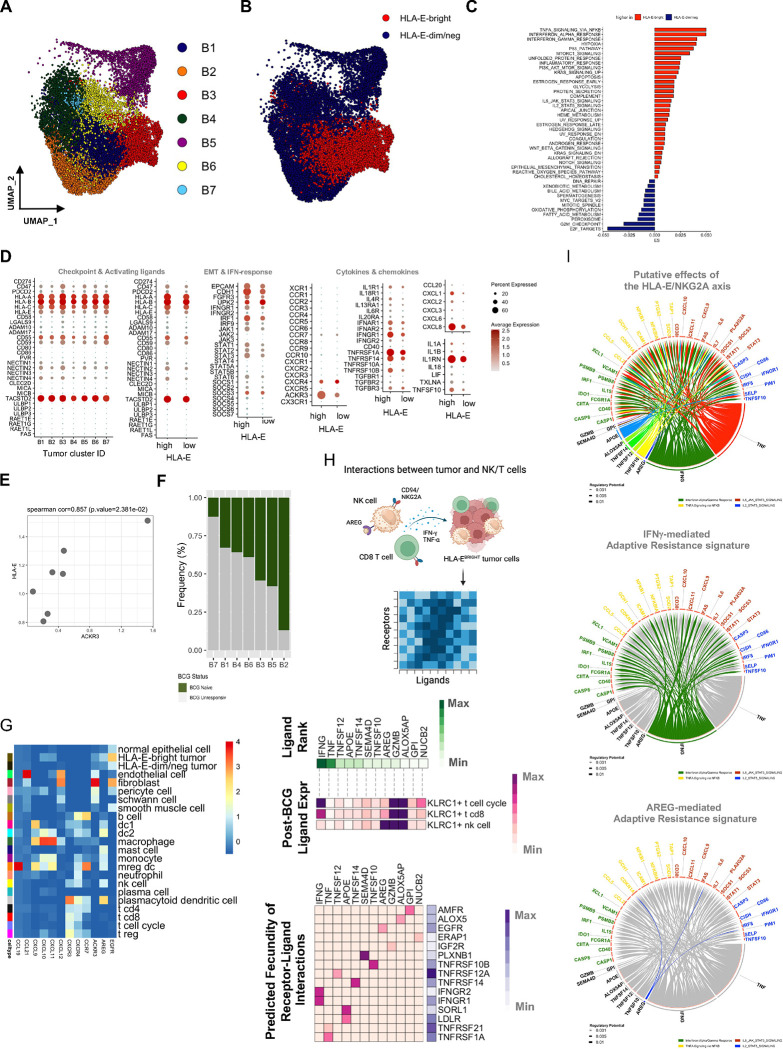
We next sought to understand the composition of tumor cells in tumors from both treatment-naïve (N=14,852) and BCG-unresponsive (N=29,858) NMIBC patients. UMAP clustering of scRNAseq analysis revealed seven major subsets of tumor cells (Fig. 4A), where clusters B1 and B3 had significantly higher HLA-E expression (Fig. 4B) and were significantly more responsive to cytokines and inflammatory signals (Fig. 4C). We profiled tumor clusters for expression of known inhibitory and activating ligands as well as for cytokines/chemokines and cytokine/chemokine receptors (Figs. 4D and 4E). Importantly, of all chemokine receptors, we observed that HLA-EHIGH bladder tumor cells had significantly higher ACKR3 (CXCR7) expression, and pseudo-bulking the scRNAseq data revealed that HLA-E and CXCR7 tumor expression were strongly correlated (Fig. 4F). Additionally, we profiled the abundance of each tumor cluster according to BCG treatment status and observed cluster B2 was uniquely enriched in BCG-naïve tumors (Fig. 4G). The other tumor clusters were either evenly distributed between naïve and unresponsive tumors or they were disproportionately skewed towards unresponsive.
Fig. 4: Single-cell RNA sequencing analysis of bladder tumor cells reveals functional differences that segregate the tumor microenvironment.
A, UMAP visualization of bladder tumor cells from unsupervised clustering revealing seven clusters (B1-B7).
B, UMAP visualization of HLA-E expression defined by tertiles, highlighting the top tertile (red dots) as HLA-E-bright (N=5,461) and bottom tertile (blue dots) as HLA-E-dim/negative (N=9,256) tumor cells.
C, Pathway analysis of Hallmark gene networks that are significantly differentially expressed (at least p<0.05) on HLA-E-bright and HLA-E-dim/negative bladder tumor cells. P-values were determined using a two-sided t-test.
D, Bubble plot showing targeted gene expression across bladder tumor clusters B1-B7 and stratified by HLA-EHIGH or HLA-ELOW tumor clusters. Size of bubble indicates percent of cluster or group and color indicates average gene expression.
E, Scatterplot highlighting pseudobulking of bladder tumor cells and expression of HLA-E and ACKR3 (CXCR7). P-value was determined using a two-sided correlation.
F, Stacked bar plot showing the frequency of bladder tumor clusters, B1-B7 that are identified in BCG-naïve (green) and BCG-unresponsive (grey) NMIBC tumors.
G, Heatmap summary of average gene expression of select chemokines, chemokine receptors, amphiregulin (AREG) and EGFR across all major clusters identified by scRNAseq of bladder tumors (N=9, 65,324 cells).
H, Ligand-receptor interaction analysis of KLRC1HIGH cells (NK cells, CD8 T cells, T cell cycle) and HLA-E-bright tumor cells (schematic diagram at top) showing the top 12 ranked ligands by KLRC1-expressing cells and cognate receptors expressed by HLA-EHIGH tumor cells. Ligand-receptor pairs were selected by rank-ordering and using cut-off weight of 0.2.
I, Circos plot showing top ranked ligands by KLRC1-expressing cells at the bottom and significant genes expressed by HLA-EHIGH tumor cells as a result of ligand-receptor interactions highlighting putative effects of the HLA-E/NKG2A axis (top) or the IFNG-mediated (middle) or AREG-mediated (bottom) resistance signatures.
Collectively, our analyses of NK cells, T cells and tumor cells suggest immune crosstalk within the TME. Further, the data are supported by our observations that NKG2A+ NK and CD8 T cells are significantly closer in proximity to HLA-EHIGH tumor cells (Fig. 2). NK cells and T cells expressed a variegated array of CXCR3, CXCR4, and CCR7 (Fig. 3I) and HLA-EHIGH tumor cells uniquely express ACKR3 (CXCR7) (and to lesser extent CXCR4) (Fig. 4E), indicating an important axis with CXCL12. Additionally, KLRC1-expressing NK and CD8 T cells expressed significantly higher AREG (amphiregulin) which signals through EGFR. To test the hypothesis that chemokine signaling explains why NKG2A+ NK and CD8 T cells are biased towards HLA-EHIGH tumor cells, we measured across all major clusters expression of chemokines and their receptors: a) CCL19, CCL21 → CCR7; b) CXCL9, CXCL10, CXCL11 → CXCR3; c) CXCL12 → CXCR4 and CXCR7; and d) amphiregulin → EGFR (Fig. 4H). We observed HLA-EHIGH tumor cells expressed higher levels of EGFR compared to HLA-EDIM/NEG tumor cells, whereas AREG was expressed by NK cells as well as dendritic cells (DCs), monocytes, mast cells, and even HLA-EHIGH tumor cells at low levels. CXCL12 was mostly expressed by stromal cells, whereas CXCR4 was expressed by NK cells, B and T cells (including Tregs), and myeloid cells. CXCR3 was mostly relegated to NK and T cells and DC1 cells, and CXCL9, CXCL10, and CXCL11 were mostly expressed by monocytes, macrophages and DCs. Finally, CCL19 was very specific for mregDCs and CCL21 by endothelial cells suggesting important interactions with CCR7-expressing B and T cells.
In a recent study of BCG-unresponsive NMIBC by Rouanne et al26, the authors demonstrated that BCG can directly infect bladder tumor lines driving epithelial to mesenchymal transitioning (EMT), autophagy flux impairment, a loss of class I HLA expression, and increased cell cycling. The study focused on BCG-unresponsive NMIBC and found that patients with a dominant tumor phenotype showing these hallmarks experienced significantly poorer metastasis-free survival. In our study, we next sought to stratify tumor cells according to their HLA-E expression and measure average gene expression across the hallmark pathways for EMT, autophagy, and cell cycle (Extended data Fig. S3). We observed a remarkably similar phenotype of HLA-E-negative tumors as in vitro BCG-infected tumor lines showing significantly higher EMT phenotype along with reduced autophagy flux genes and increased cell cycle. The data might suggest that a proportion of recurring tumor cells in the face of BCG therapy, undergo in vivo infection rendering them less sensitive to cytokine stimulation and chemokine attraction and promoting this stratification where HLA-EHIGH tumor cells are in significantly closer proximity to CXCL12+ stromal cells and immune cells.
To study the potential interactions between NK and CD8 T cells and HLA-EHIGH tumor cells, we modeled the interactions using NicheNet, an algorithm that uses a combination of transcriptomic data, known ligand-receptor interactions, and gene-gene signaling and regulatory relationships to predict potential interactions between cells of interest (Fig. 4I) 33. The top twelve ranked ligands from KLRC1+ NK, KLRC1+ CD8 T cells, and KLRC1+ T cell-cycle cells predicted to interact with HLA-EBRIGHT tumor cells were IFNG, TNF, TNFSF12 (TWEAK), APOE, TNFSF14 (LIGHT), SEMA4D, TNFSF10 (TRAIL), AREG, GZMB, ALOX5AP, GPI, NUCB2 (Fig. 4J). KLRC1-expressing T cell-cycle and CD8 T cells expressed the highest levels of IFNG, while AREG expression was highest in NK cells. Putative receptors for these ligands, including IFNGR1, IFNGR2, TNFRSF1A/10B/12A/14/21, SORL1, LDLR, AMFR, ALOX5, EGFR, IGF2R, and PLXNB1 were most highly expressed in the BCG-unresponsive samples, suggesting the presence of corresponding signaling activity by recurring tumors (Fig. 4J). To further examine genes upregulated in the tumor as a consequence of signals from NK and T cells, a circos plot revealed the ligand-target weights of the top third of predicted relationships between ‘sender’ or effector ligands and ‘receiver’ or target genes, defining the ‘putative effects of the HLA-E/NKG2A axis” in bladder tumors as well a targeted ‘IFNγ-mediated’ and ‘AREG-mediated’ adaptive resistance signatures (Fig. 4K).
Organization of the TME through immune crosstalk with stroma and HLA-Ebright tumors
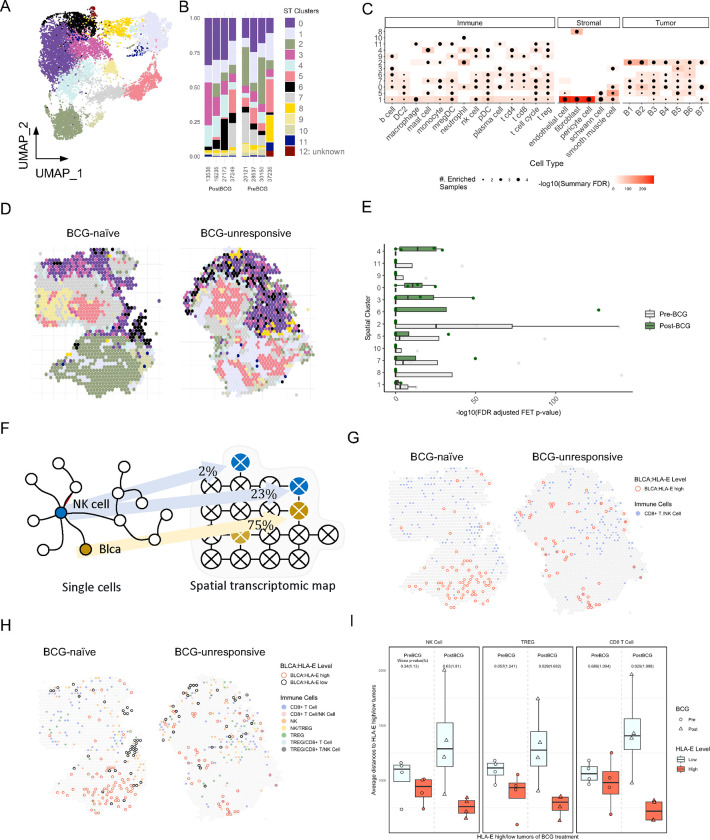
To spatially resolve the relationships between tumor cells, stroma and immune infiltrates, we performed spatial transcriptomics (ST-seq) analysis of bladder tumor sections from four treatment-naïve and four BCG-unresponsive tumors (Fig. 5). Clustering analysis revealed 12 unique clusters, where cluster 12 remained unclassified and was removed from subsequent analyses (Fig. 5A–5C). Clusters were annotated by their proportional composition by immune, stromal, and/or tumor cells defined using gene signatures derived from scRNAseq analysis (Figs. 3 and 4). Clusters were defined both by the gene content defining immune, stromal, and/or tumor cells but additionally integrating regional features of the tumor tissue (Fig. 5C and 5D, Extended Figure S4). We profiled the relative abundance of each spatial cluster between BCG-naïve (pre-BCG) and BCG-unresponsive (post-BCG) tumors (Fig. 5E). We observed clear differences, where spatial clusters #0, 3, 4, and 6 were substantially enriched in BCG-unresponsive tumors. Conversely, there was a trend for enrichment of clusters # 1, 2, 5, 7, 8, and 11 in BCG-naïve tumors. Only spatial cluster # 8 lacked the presence of immune cells, and cluster 10 was defined by neutrophils exclusively, where they were near regions enriched with DCs, Tregs, T cell-cycle and tumor cells.
Fig. 5: Spatial analysis of NMIBC tumors reveals an organization within the TME.
A and B, UMAP visualization of spatial transcriptomics sequencing (ST-seq) analysis of BCG-naïve (N=4) and BCG-unresponsive (N=4) NMIBC tumors identifying 12 ST-clusters and their distributions across NMIBC specimens.
C, Bubble plot showing ST-clusters 1–11 (rows) and their relative composition across immune, stromal, and tumor subtypes (columns) as defined by scRNAseq analysis. Size of bubble indicates its relative enrichment and shading of surrounding boxes indicate significance by FDR (corrected p values).
D, Representative ST-seq images showing the distribution of ST-clusters 1–11 in one BCG-naïve (left) and one BCG-unresponsive (right) NMIBC tumor specimens.
E, Summary histogram comparing relative enrichment of each ST-cluster in BCG-naïve (grey, N=4) and BCG-unresponsive (green, N=4) NMIBC tumor specimens.
F, Schematic diagram illustrating deconvolution of NK cells and tumor cells across 10x Visium spots. Cutoff of 20% of spot’s composition was used for defining a true signal.
G and H, Representative ST-seq images from one BCG-naïve (left) and one BCG-unresponsive (right) NMIBC tumor specimens showing proximity analyses of HLA-ELOW (black circles) and HLA-EHIGH (red circles) tumor cells to (G) NK/CD8 T cells (blue spots) or (H) NK cells, CD8 T cells, and Tregs alone or in combination (colored spots).
I, Summary comparisons of proximity of HLA-EHIGH (red bars) and HLA-ELOW (grey bars) tumor cells to NK cells (left), Tregs (middle) and CD8 T cells (right). P-values were determined using two-sided Wilcoxon test.
ST-seq Visium™ spots are 55μm in diameter and require deconvolution to understand which cell lineages are represented as well as their relative abundances (Fig. 5F). We performed neighborhood analyses to measure proximity of HLA-EHIGH and HLA-EDIM/NEG tumor Visium spots to CD8 T cells and NK cells (Fig. 5G) as well as Tregs (Fig. 5H). A summary of the average distances to HLA-EHIGH and HLA-EDIM/NEG tumor Visium spots from BCG-naïve and BCG-unresponsive tumors revealed significantly closer proximity of HLA-EHIGH tumor spots to NK cells, Tregs, and CD8 T cells. This observation was restricted to BCG-unresponsive tumors (Fig. 5I).
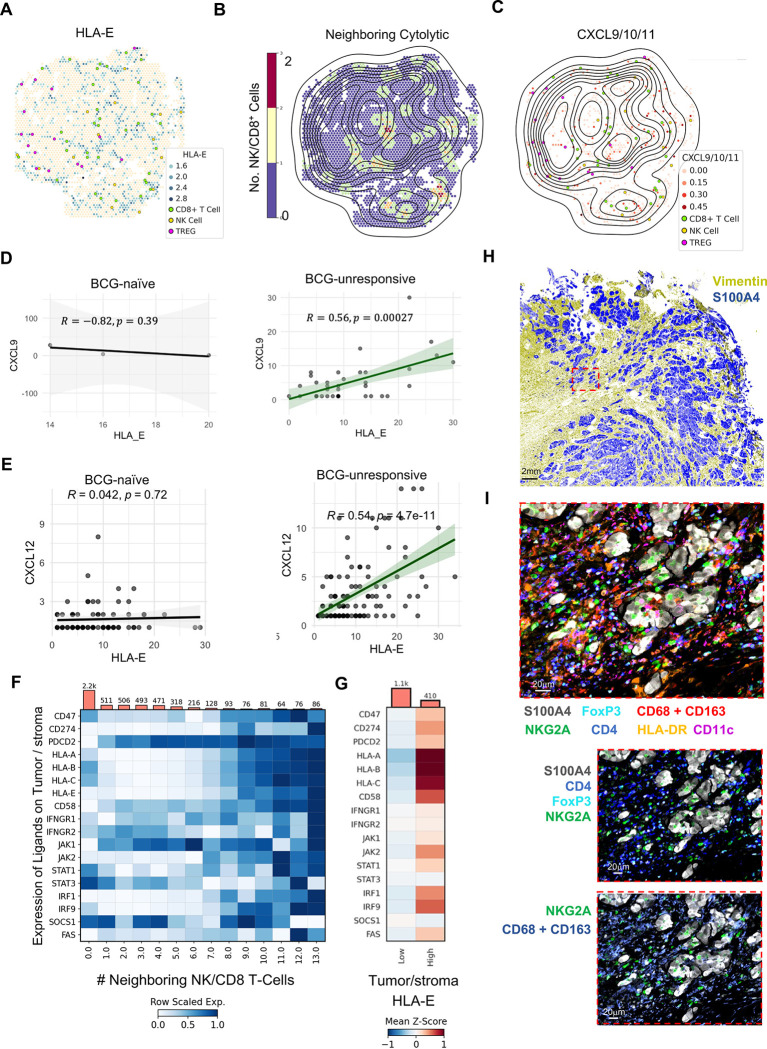
To better understand the mechanisms underlying preferential interactions of immune cells with HLA-EHIGH tumor cells, we tested the hypothesis that stroma-produced CXCL12 mediates recruitment of CXCR7+ HLA-EHIGH tumor cells to regions where macrophages and DCs produce CXCL9, CXCL10, and/or CXCL11 that mediate recruitment of NK cells and T cells (Fig. 4H). Spatial relationships were investigated on the ‘nano-environment’ level, defined as a Visium spot with a radius of 2.5 adjacent Visium spots, or ~138 microns. A representative BCG unresponsive tumor section with brightfield image of underlying tissue is shown with HLA-EHIGH tumor/stroma Visium spots shown in blue, and HLA-EDIM/NEG Visium spots shown in tan represented in Fig. 6A. Additional profiling of NK and CD8 T cells and Tregs revealed their distribution being close in proximity to HLA-EHIGH tumor/stroma Visium spots. BCG-unresponsive tumors revealed significantly higher levels of tumor/stroma spot infiltration with cytotoxic NK and CD8 T cells (7.05 times closer) and regulatory T cells (Tregs, 2.66 times closer) when compared to the BCG-naïve state (p < 0.0001 for both). Using the same representative BCG-unresponsive tumor section, a topographical map of HLA-E expressing tumor/stroma spot density is shown overlaid on top of a neighborhood analysis highlighting the intensity of neighboring NK and CD8 T cell infiltration (Fig. 6B).
Fig. 6: Recruitment of HLA-EHIGH tumor cells to stromal regions replete with immune infiltration correlates with BCG-unresponsive NMIBC.
A-C, Representative ST-seq images from one BCG-unresponsive NMIBC tumor highlighting (A) proximity of NK cells, CD8 T cells, and Tregs to tumor and stromal cells according to HLA-E expression, (B) overlaid topographical map of HLA-E tumor/stromal cell expression with color indicating abundance of NK and cytolytic CD8 T cells, and (C) proximity of NK cells, CD8 T cells, and Tregs to myeloid cells expressing any combination of CXCL9, CXCL10, and/or CXCL11.
D, Correlation analysis of Visium spots, defined as tumor cell with or without stromal cells, for co-expression of CXCL9 and HLA-E expression in BCG-naïve (N=4, left) and BCG-unresponsive (N=4, right) NMIBC tumor specimens.
E, Correlation analysis of Visium spots, defined as tumor cell with or without stromal cells, for co-expression of CXCL12 and HLA-E expression in BCG-naïve (N=4, left) and BCG-unresponsive (N=4, right) NMIBC tumor specimens.
F, Meta-analysis of all tumor/stroma-labelled Visium spots showing row-scaled expression of pertinent genes stratified by number of neighboring infiltrating NK cells and/or CD8 T-cells.
G, Z-scored heatmap showing HLA-ELOW, low-cytotoxic infiltrating tumor spots (N = 1,200 Visium spots) vs HLA-EHIGH and high-NK/CD8 T cell-infiltrated tumor spots (N = 242 Visium spots).
H, Representative multiplexed IF analysis by PhenoCycler™ of bladder tumor sections from one patient with BCG-unresponsive NMIBC highlighting the presence of stroma (vimentin, blue) and tumor (S100A4, yellow). Scale bar indicates 2mm.
I, Magnified inset from Panel H, highlighting tumor S100A4 expression of and use of digital pathology software to identify NKG2A+ NK and CD8 T cells along FoxP3+ CD4+ Tregs, macrophages (CD68 and CD163) and dendritic cells (CD11c and/or HLA-DR). Image scale bar indicates 20μm.
To investigate the biased distribution of NK and T cells near HLA-EHIGH tumor-stroma spots, we profiled bladder tumor-stroma spots for expression of cytokines and chemokines (Fig. 4H). Among the chemokines with detectable gene expression by ST-seq analysis, we observed a strong correlation between CXCL9 (Fig. 6C–6E, Extended Figure S5) and HLA-E expression as well as between CXCL12 and HLA-E expression (Fig. 6F and 6G). We next applied our neighborhood analysis to profile tumor expression of known checkpoint ligands, HLA class I expression as well as the IFN-response pathway as a function of proximity to neighboring NK cells and CD8 T cells (Fig. 6H). Analysis of 5,319 tumor-stroma spots (defined by >95% tumor- and stroma-specific genes revealed a clear program by tumor/stroma regions increasing expression of inhibitory ligands CD47, CD274 (PD-L1), and HLA-E as a greater number of NK cells and CD8 T cells were in contact with tumor-stroma spots (Fig. 4G). Additionally, we observed increased tumor-stroma spot expression of the apoptotic genes PDCD2 and FAS and the adhesion molecule CD58 (LFA-3), known to interact with CD2-expressing NK and T cells. Finally, tumor-stroma spot expression of IFNGR1 and IFNGR2 along with JAK1/JAK2, STAT1, IRF1 and IRF9 were increased as a greater number of NK and CD8 T cells were in contact. STAT3 expression did not change as a function of neighboring NK and CD8 T cells, and SOCS1 expression was reduced with neighboring NK and CD8 T cells, suggesting that tumor-stroma regions are actively responding to IFN signals. Tumor-stroma expression of these genes was further stratified according to high vs dim/negative expression of HLA-E (Fig. 6I).
To validate the ST-seq analyses, we used immunofluoresecence and the PhenoCycler/CODEX platform to profile 59 antibodies (Supplemental Table S2) in order to better define the interactions between tumor cells and multiple cell lineages. Antibodies targeted markers distinguishing tumor cells (pan-cytokeratin (CK), GATA-3, S100A4) from stromal endothelial cells and fibroblasts (Collagen Type IV, podoplanin, vimentin, PECAM1/CD31), and immune cells (CD45), including monocytes, macrophages, dendritic cells (CD14, CD16, CD68, CD11b, CD11c, HLA-DR), neutrophils (CD66b, CD16), B cells (CD20, CD138), CD4 and CD8 T cells (CD3, CD4, CD8), Tregs (CD3, CD4, FoxP3), and NK cells (NKp46, Gzm.B, CD16) (Fig. 6J and 6K).
Tumor-derived NK and CD8 T cells retain functional competence at time of tumor recurrence.
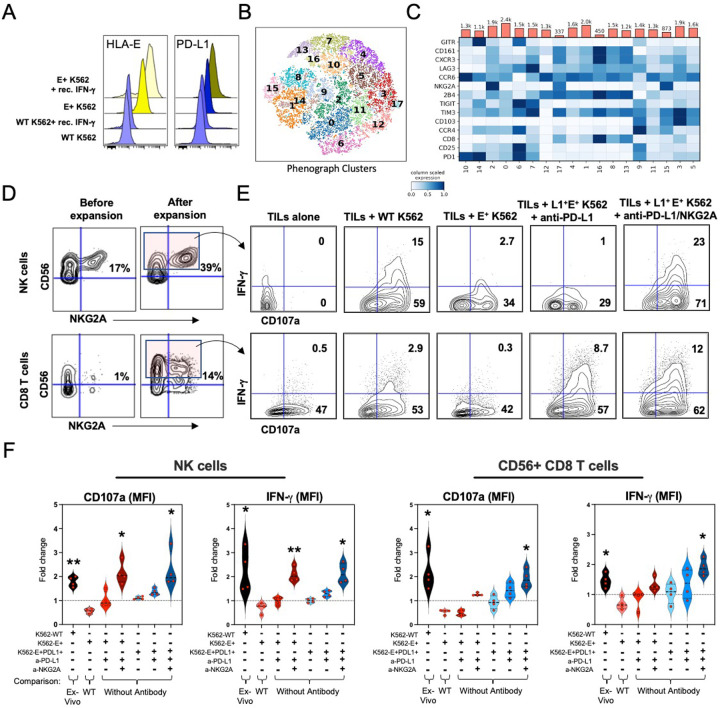
To better understand if an elevated expression of cytolytic genes in tumor-derived NKG2A-expressing NK and CD8 T cell populations correlated with improved functional sensitivity, we used mass cytometry (CyTOF) to profile the in vitro responsiveness of tumor-derived cells from BCG-unresponsive NMIBC patients at time of tumor recurrence. We tested their ability to respond to HLA-E+ PD-L1+ tumors in the presence/absence of durvalumab (anti-PD-L1 antibody) and/or monalizumab (anti-NKG2A antibody), using an artificial K562 acute myeloid leukemia tumor model that allows for uniform HLA-E and PD-L1 expression via pre-stimulation of tumor lines with exogenous recombinant IFN-γ. Briefly, after 24 hours incubation with recombinant IFN-γ [400ng/ml], stably transduced HLA-E+ K562 cells saw 2.7-fold and 2.1-fold-increase in expression of HLA-E and PD-L1, respectively, when compared with baseline expression levels (Fig. 7A). Separately, tumor-infiltrating lymphocytes from four BCG-unresponsive tumors were isolated and then expanded up to 13 days in the presence of low dose IL-2, IL-7, and IL-15 as well as tetramers targeting CD3 and CD28. Finally, TGF-β, was included in the stimulation cocktail for an additional three days to promote NKG2A expression by CD8 T cells 24. Phenograph clustering analysis 34 of expanded TILs revealed NK cells uniformly expressing NKG2A as well as 10 unique clusters of CD8 T cells with variegated expression of inhibitory checkpoint receptors (CD161, GITR, LAG3, PD-1, TIGIT, Tim-3, NKG2A) as well as markers of activation, trafficking, and tissue residence (CD25, CCR4, CCR6, 2B4, CD103, CXCR3) (Fig. 7B and 7C). Four clusters (ID#: 2, 9, 15, 17) expressed elevated levels of NKG2A, representing ~32.5% of the total CD8 TILs and reflecting the fraction of TILs predicted to react to NKG2A-blockade. Additionally, ~9% (range, 3.91% - 14.04%) of all expanded TILs were comprised of NK cells (CD14− CD19− CD3− CD56+) and NK and CD56+ CD8 T cells were strongly enriched for co-expression of NKG2A. CD56 was then used as a surrogate for NKG2A as use of monalizumab precluded our ability for detecting NKG2A with antibodies by CyTOF. Further, NKG2A and PD-1 expression by CD8 T cells was variegated highlighting the dynamic and non-redundant mechanisms for regulating CD8 T cell activation 24.
Fig. 7: In vitro combination NKG2A and PD-L1 blockade restores tumor-derived NK and CD8 T cell-mediated antitumor activity.
A, Median fluorescence intensity (MFI) of HLA-E and PD-L1 expression by wild-type (WT) or HLA-E+ K562 tumors cultured overnight in the presence or absence of recombinant human IFN-γ.
B and C, Phenograph clustering meta-analysis of tumor-derived and expanded CD56+ CD8 T cells by CyTOF before co-culture with K562 tumors showing B, tSNE analysis of identified CD8 T cell clusters, and C, expression of individual inhibitory and activating receptors by CD8 T cells and their distribution across clusters.
D, Representative CyTOF analysis of CD56 and NKG2A expression on tumor-derived NK (top row) and CD8 T cells (bottom row) profiled before (left column) and after (right column) expansion with low dose recombinant human IL-2, IL-7, IL-15 and CD3/CD28 tetramers.
E, Representative fluorescence flow cytometric analysis of IFN-γ and CD107a expression by CD56+ NK (top row) and CD8 T cells (bottom row) after 6-hour culture alone or in presence of K562 tumor lines with or without pre-treatment with anti-PD-L1 antibodies or with anti-PD-L1 + anti-NKG2A antibodies.
F, Fold-change differences between the frequencies of NK and CD56+ CD8 T cells when comparing 1271 between experimental conditions (defined by “+”). Individual comparisons are indicated at the bottom. *, p< 0.05; **, p< 0.001. P values determined by paired Wilcoxon matched rank test.
Following expansion and characterization, we performed 6-hour co-cultures of TILs with K562 cells with and without HLA-E and/or PD-L1 expression (Fig. 7D–7F). Additionally, we measured the effects of PD-L1-blockade or combined PD-L1- and NKG2A-blockade on NK and CD8 T cell degranulation (CD107a) and expression of IFN-γ. WT K562 cells were used as positive control condition for measuring NK and NKG2A-expressing CD8 T cell activation. We observed that NK cells and CD56+ CD8 T cells were sensitive to HLA class I−/− K562 cells compared to ex vivo activity, and their response was diminished when K562 cells stably expressed HLA-E or HLA-E and PD-L1. NK cell activation was restored in response to both HLA-E+ and HLA-E+ PD-L1+ K562 cells when pre-treated with anti-NKG2A Ab alone or in combination with anti-PD-L1 Ab (CD107a: 2.1-fold increase, p=0.02; IFN-γ: 1.6-fold increase, p=0.05).
Unlike NK cells, CD56+ CD8 T cells were more dynamic in their co-expression profiles for NKG2A and PD-1 staining. Similar to NK cells, untreated CD56+ CD8 TILs were not reactive to HLA-E+ K562 cells with or without co-expression of PD-L1. Activation of CD56+ CD8 TILs was only significant in the presence of both anti-NKG2A and anti-PD-L1 Abs (CD107a: 1.75-fold increase, p=0.02; IFN-γ: 1.8-fold increase, p=0.007) (Fig. 7F). Collectively, the data strongly suggest sensitivity of NK and CD8 T cells to combination immunotherapy strategies following tumor recurrence after failing BCG therapy.
Discussion:
Adaptive resistance can be driven by tumor-intrinsic and/or extrinsic mechanisms in overcoming immune pressures 11, 12, 13, 35. Evidence from mouse and human studies demonstrates that interferons, while stimulating a robust anti-tumor response, also upregulate immune-suppressive factors in the setting of prolonged activation. In melanoma, IFN-γ from CD8 T cells was shown to upregulate tumor PD-L1 expression and mediate infiltration of FOXP3+ regulatory T cell pathways within the TME 36. In a broader meta-analysis across 18 tumor indications, including bladder cancer, inflammatory mediators including IFN-γ were associated with inhibitory immune checkpoints, including PD-L1/L2 37, 38. Despite emerging evidence positioning pro-tumorigenic roles for IFN-γ, there are well-established anti-tumor functions mediated through IFN-γ that are critical for anti-PD-L1 immunotherapy 39. Anti-tumor inflammation, therefore, exists along a continuum where an equilibrium is necessary for appropriate immunotherapeutic efficacy40.
In a recent investigation conducted by Rouanne et al.26, distinct patterns of disease progression after BCG therapy and cystectomy were identified in the context of NMIBC. The study revealed that BCG-unresponsive NMIBC patients characterized by tumors bearing a ‘BCG infection’ phenotype correlated with poorer survival. Conversely, BCG-unresponsive patients exhibiting HLA class I proficiency in tumor cells after BCG therapy (before cystectomy) demonstrated greater abundance of CD8 T cell tumor infiltrates, upregulation of inflammatory cytokines, and the presence of inhibitory checkpoint molecules. This latter group experienced significantly improved metastasis-free long-term survival post-cystectomy.
In our own investigation, we delved into the primary resistance mechanisms to BCG therapy and explored the impact of BCG-induced inflammation on the dynamic interplay between tumor and immune cell populations. Utilizing both prospective and retrospective NMIBC specimens collected before, during, and after BCG treatment, our study uncovered a pervasive inflammatory response in both BCG responders and non-responders alike. Our findings implicated IFN-γ in directly contributing to the upregulation of HLA-E by approximately 43% of recurring NMIBC tumor cells, and uniquely evident in all BCG-unresponsive patients. Spatial analyses of tumor cells, stratified by their proximity to infiltrated NK and T cells, unveiled a program of tumor activation (and potential dysregulation) and selective recruitment to stromal-rich regions replete with immune effector and regulatory cells that are responding to the same collection of chemotactic signals.
Distinctive to BCG-unresponsive tumor cells, localized areas or ‘nests’ exhibiting elevated expression of HLA-E were enriched for expression of CXCL9 and CXCL12. This activation correlated with the infiltration of NK cells, CD8 T cells, and Tregs. Notably, even within confined regions of tumor tissue featuring a mix of HLA-EDIM/NEG and HLA-EHIGH tumor cells, a significant preference was observed for the proximity of NK and CD8 T cells to HLA-EHIGH tumor cells. These findings hint at an evasion mechanism wherein HLA-EHIGH tumor cells (responding to stroma-derived CXCL12) appear to play a role in safeguarding HLA class I negative tumor cells. The expression of the NKG2A receptor also emerged as a critical factor in categorizing the anti-tumor cytolytic functions of NK and CD8 T cells. However, our observations revealed that NKG2A+ NK and CD8 T cells also expressed several well-established secondary checkpoint receptors, emphasizing the necessity for a comprehensive understanding of the inhibitory landscape within patients’ tumors to inform effective second-line immunotherapy strategies.
Upon removing NK and CD8 T cells from their suppressive microenvironment, these cells demonstrated retained functional capacity. Both NK and CD8 T cells, enriched for NKG2A expression, exhibited highly enhanced cytolytic reactivity to HLA-E+ PD-L1+ K562 tumors in the presence of combination NKG2A/PD-L1 blockade. Collectively, these findings portray a landscape of immune dysregulation prominently characterized by elevated tumor expression of HLA-E and PD-L1 in the context of BCG-induced chronic activation and inflammation. Importantly, HLA-EHIGH tumor cells similarly expressed higher levels of CD47, Nectin-2 (CD112), Nectin-4, PVR and at higher levels compared to PD-L1 expression. Further, HLA-EHIGH tumor cells expressed higher levels of decay accelerating factor (DAF, CD55) and CD59, which block C3 convertase and formation of the membrane attack complex, two critical steps in driving complement-mediated inflammation and phagocytosis, and apoptosis, respectively. Finally, the trophoblast-2 (Trop2) antigen was highly expressed on all recurring NMIBC tumor cells. These data suggest future combination immunotherapy strategies may benefit from additional targets that are strongly engaged in BCG-unresponsive NMIBC patients. Emphasizing this point, antibody-drug conjugates targeting Trop2 (sacituzumab govitecan)41 and Nectin4 (enfortumab vedotin)42 have demonstrated promising improvements in patient outcomes for metastatic triple-negative breast cancers and muscle-invasive bladder cancer, respectively; the latter which, when combined with pembrolizumab (PD-1 blockade), has recently been approved FDA clearance in an all comer population for 1st line treatment of metastatic urothelial carcinoma 43.
Intravesical administration of BCG is initiated as adjuvant immunotherapy, as complete tumor resection is the initial diagnostic and therapeutic step. Thus, initiation of standard of care (SOC) BCG immunotherapy presupposes inflammation without an active tumor to clear 44. Treatment improvements for NMIBC and BCG-resistant disease have lagged. This may be, in part, because few studies have attempted to understand the relationship between the timing of tumor recurrence, reasoning for the recurrence, and the state of the immune system at the time of recurrence. Poor dosing study designs and lack of understanding of the mechanisms underlying a therapeutic response to intravesical BCG have led to a significant gap in knowledge and benefit for patients with NMIBC compared to muscle-invasive or metastatic disease.
Our findings suggest that all NMIBC patients at the time of tumor recurrence show signs of a hallmark anti-tumor immune response dominantly driven by IFN-γ. BCG-unresponsive tumors see uniform increases in chemotactic cytokines and inflammatory pathways that should otherwise function to suppress tumor growth. Further, increased expression of inhibitory ligands on BCG-unresponsive tumor cells was observed, suggesting that inflammatory stimuli had been prolonged and triggered feedback mechanisms responsible for immune evasion. These results are seen on multiple levels, including bulk and single-cell RNA sequencing, spatial sequencing, multiplex imaging, and mass cytometry. This implies that when tumors recur, for reasons beyond the scope of this study, they are met by an overactive status of the immune system ill-equipped to combat them.
Previous analyses profiling urine analytes between the BCG naïve and third dose timepoints demonstrated that three doses of BCG induced an inflammatory response hundreds of times above baseline levels of cytokines, including but not limited to IP-10, MIP-1β, IL-8, IL-6, and TNFα 45. Importantly, the third dose-response dwarfed the magnitude of the first dose-response, suggesting that repeated exposure increases the magnitude of inflammation 45. While these data did not profile out to the time of tumor recurrence, they lend credence to the theory that all patients experience a ubiquitous and increasingly powerful immune response to repeated doses of BCG. Importantly, our findings confirm these previous observations. Further, they demonstrate that inflammation is sustained even throughout the six-week treatment interruption at time of first cystoscopy and do not equilibrate to baseline levels.
Novel to our study is the spatially-resolved insights into potential consequences of IFN-γ dysregulation and tumor cell and immune homing patterns. Preferential trafficking of HLA-EHIGH tumor cells to immune-rich regions was seen in areas of high expression of CCL19, CCL21, CXCL9/10/11, and CXCL12, and chemokines were detected in the urine early following start of therapy and were sustained throughout therapy. Further, BCG unresponsive tumor-stroma proximity to cytolytic cells and Tregs was increased by a median of 7-fold and 2.6-fold, respectively, when comparing CXCL9/10/11HIGH to CXCL9/10/11DIM/NEG tumor nests.
Clinical efficacy in checkpoint blockade is dependent on reinvigorating effector cells expressing high levels of the targeted checkpoint(s). Congruent with analogous studies, we found that NKG2A+ NK and CD8 T cells were enriched in BCG-unresponsive tumors and tumor-derived NK and CD8 T cell anti-tumor functions were intact at the time of tumor recurrence. Additionally, co-culture experiments with patient TILs and K562s with and without HLA-E and PD-L1 confirmed the ability of combination anti-NKG2A plus anti-PD-L1 blockade to enhance the cytolytic function of stimulated TILs from patients at time of tumor recurrence. This reinforces both the use of NKG2A to reinvigorate cytolytic effector functions by NK and CD8 T cells, as well as the potential to exploit a critical mechanism of tumor resistance, whereby chemokines lure in effector cells to take on exhausted phenotypes.
Recent clinical evidence emerging from interim results of the randomly controlled Phase II COAST trial demonstrated significantly prolonged survival in non-small cell lung cancer (NSCLC) patients treated with monalizumab combined with durvalumab compared to standard of care durvalumab alone for treatment of unresectable stage 3 NSCLC 25. This combination is being further evaluated in a larger planned Phase III trial (PACIFIC-9, NCT05221840) following these promising early results. Collectively, our data demonstrate that elevated tumor expression of HLA-E and PD-L1 and chronic activation of NKG2A-expressing NK and CD8 T cells are hallmark features of resistance to BCG immunotherapy in NMI bladder tumors and that integrating NKG2A-blockade into combination immunotherapy strategies may prove particularly effective in settings of BCG-unresponsive, high-grade NMIBC. Based on the data generated from this study, we recently designed a Phase 2 trial of durvalumab (MEDI4736) and monalizumab in NMIBC. The Phase 2 ENHANCE (Elevated NKG2A and HLA-E Amplify NK and CD8 T-cell Engagers) (clinicaltrials.gov ID: NCT06503614) will investigate the effects of monalizumab (anti-NKG2A Ab) plus durvalumab (anti-PD-L1 Ab) for combination checkpoint blockade in high-grade BCG-unresponsive NMIBC as a bladder-sparing immunotherapy strategy. In this trial, an exploratory objective is to assess the relationship between baseline tumor HLA-E expression and clinical outcomes to facilitate development of HLA-E as a potential biomarker for patient selection in future clinical trials intending to target NKG2A.
Our study has notable limitations. Our sample sizes, while being the largest NMIBC spatial sequencing and single-cell RNA sequencing cohorts available, are small and present a potentially limited view of the disease. Additionally, as BCG is administered as an adjuvant therapy, we are only afforded access to bladder tissues after treatment if tumor recurs. Thus, it was not feasible to profile bladder tissues in the absence of tumor following exposure to BCG. Future studies might benefit from a neoadjuvant approach, where tumor tissue can be sampled and profiled for potential therapeutic responses to BCG.
Collectively, our analyses suggest that the current guidelines on immunotherapy for BCG-unresponsive NMIBC could be improved via a multi-cell-targeting immunotherapy approach focused on NK cells alongside T cells. Randomized trials from bladder and other tumor indications have shown that PD-1/PD-L1 stratification fails to predict response to immunotherapy with anti-PD-1/PD-L1 antibodies: IMvigor210, JAVELIN bladder 100, and CheckMate-275 (NCT02108652, NCT01772004, NCT02387996) all saw that PD-1/PD-L1 biomarker stratification alone did not effectively stratify response rates. In comparison, recent results using KLRC1 (NKG2A) expression in the pre-treatment tumor significantly improved predicted anti-PD-L1 response rates in the IMvigor210 cohort, but only in the CD8HIGH PDCD1 (PD-1)HIGH group 24. In fact, where IHC stratification of PD-L1 expression has failed to predict immunotherapeutic responses, KLRC1 stratification in IMvigor210 showed protective effects were restricted to the PD-L1 IC high group. In conclusion, while our analysis does not exclude the presence of alternative checkpoints, it lends evidence to the hypothesis that combination NK and T cell immunotherapeutic approaches may hold the key to improved outcomes for NMIBC patients.
Methods
Patients and samples
Patients at Mount Sinai Hospital (MSH) were enrolled in the study following Institutional Review Board (IRB) approval (protocol 10–1180). 10–1180 covers the use of patient tissues in a biorepository and allows for prospective collection of blood, urine, and tissue samples from enrolled patients. Formalin-fixed paraffin-embedded (FFPE) blocks from BCG patients were obtained retrospectively from the biorepository and prospectively for patients receiving treatment. For prospective patients, samples were collected on the day of surgery and throughout BCG immunotherapy. Due to IRB limits on the collection, blood and urine samples were taken at the first dose, third dose, and sixth dose of the induction cycle; at every follow-up cystoscopy; and at the third maintenance cycle dose. Tumor samples were taken at every possible timepoint. BCG naïve was defined as any patient who had yet to receive BCG, regardless of past treatment with other chemotherapies. BCG-unresponsive was defined as any patient with recurrent tumors following at least five of six induction doses of BCG at time of first evaluation.
Sample processing
Blood and urine samples from bladder cancer patients were processed to collect PBMCs, serum, and cell-free urine. Blood was spun down (4C, 2000rpm, 10 min) to isolate serum, and PBMCs were collected using Ficoll-Paque isolation. PBMCs were frozen down in a Mr. Frosty at −80 C for 24 hours and stored at −160 C in 10% DMSO and 90% fetal bovine serum; PBMCs and cell-free urine were stored at −80 C.
Tumor tissues obtained from transurethral resections of bladder tumor (TURBT) were placed into RPMI medium immediately after removal and transferred to the laboratory for additional processing. Bladder and lymph nodes obtained from radical cystectomies were sent from the operating room directly to the pathology suite after completion of the lymph node dissection. The bladder was bivalved; samples of visible tumor were extracted and placed in RPMI medium, and the tumor was transferred to the laboratory for additional processing.
Fresh tumor samples underwent a variety of different processing techniques based on the planned experiment. Tumor used for spatial sequencing was placed in a 10 mm × 10 mm cryomold with optimal cutting temperature (OCT) media and frozen down on a thermal block immersed in liquid nitrogen. Tumor tissues were digested using tumor dissociation enzymes (Miltenyi, 130–095-929) and a GentleMACS machine (program 37C_h_TDK_3) at 37C. Mechanically and enzymatically separated tissues were filtered through a 70μM cell strainer and assessed on Countess II (ThermoFisher) for viability and cell numbers.
In situ hybridization and targeted RNA sequencing
FFPE sections from 40 retrospective NMIBC cases were obtained from the institutional biorepository and used for targeted RNA sequencing. RNA was extracted from five and ten μm sections. HTG EdgeSeq lysis buffer was added to lyse and permeabilize the samples. Nuclease protection probes (NPPs) were added to the lysed samples and hybridized to the target mRNA. A nuclease was added to digest non-hybridized mRNA and excess NPPs. The nuclease digestion reaction was finalized with a termination solution followed by heat-mediated denaturation of the enzyme.
Each sample was used as a template for PCR reactions with specially designed primers. Each primer contains a unique barcode that is used for sample identification and multiplexing. Samples were analyzed simultaneously on an Illumina sequencing platform to prepare the library. All samples and controls were quantified in triplicates. No template control (NTC) reactions were made for each master mix used during the qPCR process to test the absence of a probe or qPCR contamination. Molecular-grade water was used in place of a test sample in the NTC reactions using the same volume as the template.
Sufficient concentration of sample for library pooling, appropriate dilution for the library pool, and volume of denaturation reagents to add to the library were determined by HTG library calculator. 2N NaOH and heat (98C, 4 minutes) were used for library denaturation. The denatured library was loaded into the well of the NextSeq sequencing cartridge. Sequencing was performed using an Illumina NextSeq sequencer.
The sequencing data on mRNA expression of target genes were imported into HTG EdgeSeq parser software. HTG biostatistics department performed quality control analyses and normalized the data. Data were returned from the sequencer as demultiplexed FASTQ files with four files per assay well. The HTG EdgeSeq parser software aligned the FASTQ files to the probe list and collated the data.
Prior to any analyses, clinical data on all patients was gathered. The following items were collected for all samples processed by HTG Molecular: patient date of birth; gender; age at BCG induction; BCG status (unresponsive vs. naïve vs. exposed); date of sample collection; stage at collection; grade at collection; recurrence date; time to recurrence; time to progression; BCG start date; BCG last exposure; BCG induction cycle last dose; prior chemotherapy; cystectomy; date of cystectomy; stage at cystectomy.
Following clinical data collection, a gene set enrichment analysis (GSEA) was performed on the targeted RNA sequencing data. Specifically, we used paired patient samples before and after BCG exposure in the BCG recurrent patient population only. We used custom gene sets, as well as all Hallmark gene sets from the Broad Institute’s MSigDB, as inputs for the enrichment analysis 27. Statistical significance was set at p < 0.05, and gene sets found to be significant are listed in Supplementary Table S1. All gene sets found to be statistically significant were evaluated for leading-edge genes, defined as the genes that contribute most to the enrichment score and associated p-value.
The leading-edge genes from statistically significant gene sets in the GSEA were collated and used to assess for group differences between the paired HTG patient samples. We performed these analyses specifically on the BCG-recurrent cohort. Prior to any analyses, a Shapiro-Wilk test, chosen for suitability in small sample sizes, was used to assess for normality 46. All samples with p < 0.05 were considered not normally distributed, and a Kruskal-Wallis test was performed to assess for group differences 47. All other samples were assessed using an independent T-test. Genes with statistically significant differences between the BCG naïve and the BCG-unresponsive populations were then visualized on radar plots.
Protein concentration measurement
Cell-free urine supernatant and serum samples were randomized in a 96-well plate. Diluted samples and positive and negative controls were incubated overnight with an incubation mix (incubation solution, incubation stabilizer, A-probes, and B-probes) at 4°C. Samples were then incubated with an extension mix (High purity water, PEA solution, PEA enzyme, PCR polymerase) for 5 min and placed on a thermal cycler. Following the thermal cycler, samples were incubated with a detection mix (detection solution, High purity water, detection enzyme, PCR polymerase) and transferred to a chip. Primers were loaded onto the chip, and the chip was profiled using the Fluidigm IFC controller HX with the Fluidigm Biomark Reader. Data were normalized using extension and interplate controls and a correction factor. The resulting data were reported in normalized protein expression (NPX) units on a log2 scale.
IFN-γ co-culture
Cell lines and CD45- isolated primary tumor cells were incubated in media optimized for high viability for 72 hours (RPMI-1640 supplemented with 20% fetal bovine serum). Tumor cells were expanded until they were confluent in 2 T175 flasks. Following expansion, cells were culture in 100 ng / mL of IFN-γ for a total of 24 hours in a 24 well plate. Following co-culture, cell lines were trypsinized (immortalized), and primary tumors were gently removed from the solid phase by a cell scraper. Samples with and without IFN-γ stimulation were measured for HLA-E and PD-L1 protein expression levels.
HLA-E and PD-L1 levels were assessed via FlowCytometry. Cells were stained in 4C FACS buffer (phosphate-buffered saline (PBS) with 2% heat-inactivated FBS and EDTA 2 mM) for 30 minutes. Subsequently, cells were washed in PBS, incubated for 20 minutes in a viability dye, washed again with PBS, and resuspended in 2% paraformaldehyde. The experiment was performed in triplicate, with three readouts per cell line per experimental condition. FlowCytometry acquisition was performed using an LRS-Fortessa (BD Biosciences), and data were analyzed using the CytoBank software. When staining for HLA-E, cells were first stained 20 minutes with HLA-E prior to staining with additional PD-L1. In CytoBank, several gates were applied to generate the final dataset. A live/dead gate was applied, followed by a gate to remove doublets and isolate singlets. Lastly, the data was arcsinh transformed prior to analysis.
Once the final dataset had been generated, statistical significance between unstimulated and stimulated cell lines was assessed. An independent t-test was run with a significance cutoff of p < 0.05.
Plasma and cell-free urine supernatant protein concentration analysis
We used the OLINK Proteomics®- inflammation panel to profile cell-free urine supernatant. Protein levels were visualized at BCG naïve, third dose, sixth dose, and first-cystoscopy timepoint, comparing NPX values for all patients between the BCG naïve timepoint and the sixth dose timepoint. Additionally, urine supernatants from a validation cohort of NMIBC patients treated with BCG were used from the Aarhus University as previously described 13. In order to determine the suitable statistical test, a Shapiro-Wilk’s test was used to assess for normality, and a Kruskal-Wallis test was used in every instance in which one or both samples were not normally distributed. An independent T-test was used in the event both samples were normally distributed. All statistically significant p values were then used to assess adjusted p values via the Benjamini-Hochberg correction, with an alpha of 0.05. All statistically significant genes between the BCG naïve and sixth induction dose time points are shown.
Multiplex Immunohistochemistry:
Sections of tumor for immunohistochemical (IHC) staining were taken at a thickness of 3-mm from formalin fixed paraffin-embedded (FFPE) blocks. H&E-stained sections were performed every 5 – 10 slices. The Ventana Discovery Ultra (Roche Diagnostics) machine was used to automatically bake, deparaffinize, and condition the slices. The RUO Discovery Universal (v21.00.0019) was used to perform chromagen IHC on sequential slices. Primary antibodies included (CD3, HLA-E, NKp46, and NKG2A) and were utilized for staining on NMIBC tumors. All slices followed the same protocol, which included a 60 minute incubation at 37°C; secondary antibodies using OmniMap HRP or NP DISCOVERY (Roche Diagnostics); signal detection using Discovery OmniMap. Nuclear counterstaining with Mayer’s hematoxylin; and conversion to high-resolution images via the NanoZoomer S10 Digital slide scanner (Hamamatsu).
Tumor and CD3+ T cell expression and proximity analyses
Prior to analysis, all slides were reviewed and regions of interest were annotated by a board-certified pathologist (R.B.). Tissue artifacts, including torn, folded, and damaged tissue, were excluded from any analyses. The HALO™ (Indica Labs, Inc.) digital image analysis platform, a semi-automated platform using machine learning to segment and label stained sections, was utilized for quantitative analyses. Halo AI™ and train-by-example classification, segmentation, and random forest classification was used to separate chromogenic stains and generate tabular data for downstream analysis. Slide features of each tumor, including cell lineages (tumor, stroma, and immune) and slide features (such as glass) were characterized. Glass was excluded from all downstream analyese. Classified cell classes were tabulated, and positive staining cells were stratified into expression tertiles (dim, moderate, and bright). Calibration for intensity expression was performed using tonsil tissues from healthy human tonsil. In addition to cell counts, total surface area (mm2) was recorded to facilitate density calculations. Statistical analyses were performed using Python 3.8.1.
Proximity Ligation Assay
The NaveniFlex™ Proximity Ligation Assay (PLA) was performed according to the manufacturer’s instructions using NaveniFlex Tissue MR ATTO647N (Navinci, Sweden). PLA was performed on sections of tumor taken at a thickness of 3-mm from formalin fixed paraffin-embedded (FFPE) blocks. H&E-stained reviewed with pathologist. Briefly, after deparaffinization, rehydration, and antigen retrieval, slides were blocked with Block NT blocking solution (Navinci, NT.1.100.01) for 60 min at 37 °C in a preheated humidity chamber and then incubated with mouse anti-HLA-E (clone: MEM-E/02, Abcam, 1:200) and rabbit anti-NKG2A (clone: EPR23737–127, Abcam, 1:2000) diluted in Diluent 1 NT solution (Navinci, NB.1.100.02) overnight at 4°C. As negative controls, two (tonsillectomy) slides were incubated in antibody diluent with only one primary antibody each. After washing, the slides were incubated with the PLA probes corresponding to the primary antibodies using anti-mouse Navenibody M1 NT (Navinci, NB.1.100.06) and anti-rabbit Navenibody R2 NT (NB.1.100.07) in Diluent 2 NT solution (Navinci, Navinci, NF.1.100.03) for 60 min at 37 °C. Slides were then processed for ligation using reaction 1 reagent containing Buffer 1 NT (Navinci, NB.2.100.17) and Enzyme 1 NT (Navinci, NF.2.100.11) and subsequently reaction 2 reagent containing Buffer 2 NT (NT.2.100.01) and Enzyme 2 NT (Navinci, NF.2.100.15) and incubated for 30 min at 37 °C and 90 min at 37 °C, respectively. The slides were washed and incubated with post-block NT reagent (Navinci, NF.1.100.01) in post-block supplement NT (Navinci, NT.2.100.04) for 30 min at 37 °C, then processed for detection, counter-stained with DAPI, and mounted with coverslips using Prolong Gold Antifade reagent (Invitrogen, P36930).
Images were captured at the Microscopy and Advanced Bioimaging Core of the Icahn School of Medicine at Mount Sinai. A Leica DMi8 (Leica Microsystems, Germany) was equipped with a HC PL APO CS 10x/0.4 (Part Number 506285; Leica Microsystems, Germany) objective lens. A SpectraX fluorescence illuminator (Lumencor, Oregon, USA) with multiple narrow-band light emitting diodes provided illumination (LEDs used: 395/25nm for DAPI, 470/24nm for autofluorescence channel, and 640/30nm for Navinci signal). The microscope and light source were controlled by LAS X software, version 3.7.5.24914 (Leica Microsystems, Germany). For fluorescence excitation, the following illumination settings were used: a 395nm LED set to 50% (147mW at the SpectraX output port) for DAPI signal, captured at 20 milliseconds of exposure; a 470nm LED set to 44% (86mW), captured at 70 milliseconds for an autofluorescence channel; and a 640nm LED set to 100% (231mW), captured at 150 milliseconds for the target signal. A multi-band pass filter set (Part Number 11525366; Leica Microsystems, Germany) was used to separate fluorophore emission (Dichroic 415/490/570/660nm; Emission bands: 430/35, 515/40, 595/40, 720/100nm). Images were captured using a Leica DFC9000GT monochrome camera set to 12-bit depth, 2×2 (4-pixel) binning and “Low Noise” Gain mode. Images were captured in montage at 10% overlap, merged (“Smooth” blending option) and then saved in the proprietary LIF (“Leica Image File”) format before being converted to IMS (Imaris) format for analysis.
Image analysis was performed using Imaris software 10.1.1 (Oxford Instruments, Concord MA). A surface for the green background channel was created using the surface creation wizard with the following parameters – Enable Region Of Interest = false, Enable Region Growing = false, Enable Tracking = false, Enable Classify = false, Enable Shortest Distance = false, Enable Smooth = true, Surface Grain Size = 2.00 μm, Enable Eliminate Background = false, Active Threshold = true, Enable Automatic Threshold = false, Manual Threshold Value = 1900, Active Threshold B = false. Masked channels were created by subtracting the intensities within the green surface from the blue and far-red channels: the mask intensity was set to 0 for inside the green surface while the outside was set to the original channel’s value. A new surface was created using the surface creation wizard for the masked far-red channel using the following parameters – Enable Region Of Interest = false, Enable Region Growing = false, Enable Tracking = false, Enable Classify = false, Enable Shortest Distance = false, Enable Smooth = true, Surface Grain Size = 2.00 μm, Enable Eliminate Background = false, Active Threshold = true, Enable Automatic Threshold = false, Manual Threshold Value = 1600, Active Threshold B = false. An area filter was applied to this far-red surface to remove surfaces whose area was larger than 50um2. A DAPI surface was created using surface creation wizard with the parameters – Enable Region Of Interest = false, Enable Region Growing = true, Enable Tracking = false, Enable Classify = false, Enable Shortest Distance = true, Enable Smooth = true, Surface Grain Size = 2.00 μm, Enable Eliminate Background = true, Diameter Of Largest Sphere = 7.50 μm, Active Threshold = true, Enable Automatic Threshold = false, Manual Threshold Value = 10, Active Threshold B = false, Region Growing Estimated Diameter = 6.00 μm, Region Growing Morphological Split = false, Filter Seed Points = “Quality” above 60.0, Filter Surfaces = “Number of Voxels Img=1” between 10.0 and 500, to obtain individual nulcei within the field of views. Finally, a fourth surface was created by applying the filter – Overlapped Area to Surfaces (Minimum = 0.050 um2, Maximum = false) to obtain the DAPI surfaces that were in contact with the far-red channel. The counts of total number of nuclei and nuclei overlapping with the masked far-red surfaces were extracted from the statistics tab of Imaris.
Single-cell RNA sequencing:
Data preprocessing
Single-cell RNA sequencing (scRNA-seq) analysis was performed using Scanpy package in Python and Seurat package in R. After loading, genes expressed in fewer than three cells were excluded from later analyses. Cells with < 200 or > 8000 unique genes, as well as cells containing >20% mitochondrial gene transcripts, were discarded. Cells then go through scrublet to detect doublet population. Subsequent data normalization was performed by dividing feature counts for each cell by total counts for that cell, scaling by a factor of 10,000, and natural log transformation. Next, scvi model was set up to correct batch-effects. Then we performed scaling and principal component analysis (PCA) on the batch-corrected data. Using the first 50 principal components (PCs), graph-based clustering and UMAP dimensionality reduction was performed to reveal 16 cell clusters. Cluster-specific marker genes were identified using the FindAllMarkers function, and marker genes with a natural-log fold change (FC) > 0.25 and expressed in >= 25% of cells were used to annotate cell cluster identities based on known cell type markers 48, 49, 50: CD8 T cells (CD8A, CD3E), CD8- T cells (CD3E), tumor cells (MUC1, KRT7/8/13/17/18), B cells (MS4A1, CD79A), monocytes/macrophages (CD14, LYZ, FCGR3A, MS4A7), T regulatory cells (CD3E, FOXP3), granulocytes (C1QA/B/C, ITGAM, FCGR3A), NK (NKG7, GNLY, KLRD1, KLRF1), fibroblasts (COL1A1, SPARC), epithelium (EMP1), DC (FCER1A, CST3, ITGAX), plasma cells (MZB1, JCHAIN, IGHG3), and CD8 TRM cells (CD3E, CD8A, KLRD1). Clusters 22, 24, 25, and 26 containing 224, 142, 141, and 101 cells, respectively, were ultimately excluded from further analyses due to unclear cell identities. All scRNAseq analyses were performed using distinct samples without repeated measurements.
Tumor subclustering analysis
We performed subclustering analysis on 18,520 bladder cancer (Blca) cells. Preprocessing steps for the Blca subset included normalization, scaling, and PCA analysis. We then applied Canonical Correlation Analysis (CCA) integration using the IntegrateLayers() function (dimensional reduction for correction = pca) in Seurat v5 to correct batch effects 51. Leiden clustering (resolution = 0.4) was applied to the shared nearest neighbors (SNN) (dims = 1:10). This resulted in the identification of 9 heterogeneous tumor subclusters, distinctly separated on the UMAP plot. Each subcluster was profiled by the expression of tumor marker genes (EPCAM, UPK2) and cytokines (CXCL1, CXCL2, CXCL3, IFNGR1, etc.). Subcluster B1 was removed from further analysis as it was identified as normal bladder cells with low EPCAM (tumor-related marker genes), and subcluster B8 was removed due to high PTPRC expression, indicating a high presence of immune cells (CD45+). Differentially expressed gene (DEG) analysis (MAST, R version 1.2.1) 52 and enrichment pathway analysis 53 further characterized the subclusters, highlighting their distinct biological profiles (Figure 2).
NicheNet Analysis
We used the NicheNet R package to infer potential ligand-receptor interactions between NK and T cells and tumor cells, as well as potential regulatory effects in tumor gene expression as a result of these interactions 33. We added known ligand-receptor interactions between ligands IgG1 and IgG3 and the receptor CD16 (weight = 1) to NicheNet’s default ligand-receptor interactions database, and also integrated the Harmonizome Pathway Commons Protein-Protein Interactions database: (https://maayanlab.cloud/Harmonizome/dataset/Pathway+Commons+Protein-Protein+Interactions) with NicheNet’s default signaling network (weight = 1) 54, 55. We used the modified ligand-receptor interaction and signaling networks for subsequent analyses. We defined the NK and T cell populations as senders and tumor cells as receivers and derived a list of 92 receptors expressed in the receiver population by intersecting the list of receptors from the ligand-receptor interaction network with genes expressed in at least 10% of tumor cells in the RNA assay. Fifty potential ligands expressed in greater than 10% of the sender NK and T cells were then derived based on their interaction with the list of 92 receptors. We then calculated activity scores for each ligand based on its potential to affect the expression of Hallmark interferon gamma response genes 27, 56. Statistical analyses including Wilcoxon rank-sum tests and Pearson and Spearman correlations were performed using R v4.0.3.
Differential gene expression analysis
Spatial transcriptomic data were generated by Visium Spatial Gene Expression which is a spatial transcriptomics solution by 10x Genomics. We obtained eight spatial sections consisting of four preBCG and four postBCG samples. Sequencing data was aligned on GRCh38 and quantified using the Space Ranger Software Suite (version 1.0, 10x Genomics). We processed the ST data to characterize major cell types and their spatial patterns using Giotto workflow 57. Per section, Low quality spots (number of expressed genes > 100), and lowly expressed genes (expressed in < 10 spots) were removed. The raw counts were log2(x+1) transformed, followed by adjusting for batch effects and spot wise number of features using Pearson residuals approach by Lause et al 58. The spatial network of nearest spots by Delaunay triangulation was calculated to identify spatially co-expressed genes in neighboring spots via BinSpect-kmeans algorithm 57, and distinct spatial domains by Hidden Markov Random Field (HMRF) model 57. Then, different sections were combined into a pan-section transcriptome by Harmony to identify pan-section clusters 59. The pan-section clusters will be identified by Leiden clustering 57 to detect transcriptionally similar spots shared across different sections. Subsequently, we identified 12 spatial clusters to profile the spatial transcriptomic landscape. Differentially expressed genes (DEGs) between KLRC1+ versus KLRC1− cells were found using the FindMarkers function from the Seurat R package, where cells with KLRC1 expression greater than 0 in the RNA assay were labeled as KLRC1+.
Identification of HLA-E high/low tumor subsets in spatial transcriptome
As HLA-E expressions are not specific to tumors and HLA-E high/low tumor markers have not been established, we performed a customized cell type presence inference workflow by utilizing Cell Atlas Reconstruction from Spatially mapped Data (CARD) method 60. CARD is a computational approach that reconstructs cell-type-specific expression profiles from spatially mapped transcriptomic data, facilitating the estimation of cell-type proportions within tissue sections. First, we generated reference single-cell transcriptomes of HLA-Ehigh and HLA-Elow tumor subsets by explicitly labeling tumors with HLA-E expressions (high: HLA-E > 0, low: HLA-E=0). These tumor labels were utilized as the inputs to calculate the relative abundances of the tumor subsets on the spatial voxels by CARD.
To further scrutinize the detection of HLA-E high tumors in the spatial transcriptome (ST) data, we applied additional thresholds to require voxels with i) high HLA-E expressions, and ii) highly abundant HLA-E high tumor/stromal cells as inferred by CARD 60. Per section, high HLA-E expression voxels were identified as those with HLA-E expression values greater than zero (SCT normalized assay) and a z-score of inferred HLA-Ehigh tumor/stromal cell abundance > 1.645 (90% confidence). Conversely, low HLA-E expression voxels were identified as those with an absence of HLA-E expression (HLA-E expression equal to zero) and a z-score of inferred HLA-Elow tumor/stromal cell abundance > 1.645 (90% confidence). Similarly, we detected high confidence voxels with NK cells, CD8 T-cells and Treg cells by further applying the abundance z-score > 1.645 for further analyses.
Proximity analysis between different cell types
We evaluated the proximity between different immune subsets (NK, CD8+ T, and regulatory T (Treg) cells) and HLA-Ehigh/low tumor cells to observe if distinct cytotoxic immune micro-environments are present, conditioned on tumoral HLA-E expression status. Upon identifying high confidence voxels with HLA-Ehigh/low tumor/stromal cells and the immune cell types, we calculated the pairwise Euclidean distances between the tumor/stromal cells and each immune cell type. To retrieve actual distance between voxels, the matrix was refined using the computeCellDistance function from the CellChat v2 library in R 61. For each voxel with HLA-Ehigh/low tumor/stromal cell presence, its overall distance to an immune cell type was calculated as the mean distance over the 10 nearest neighboring voxels with the respective immune cell presence. These distances were summarized across each section by taking the average over all HLA-Ehigh/low tumor/stroma voxels, yielding the overall proximity with each immune cell type per section. These overall proximity values were compared between pre-BCG and post-BCG groups by Wilcoxon Rank-sum test (Figs. 5 and 6).
In-depth analysis of cell type enrichment across spatial clusters
We tested if each pan-section cluster shows enriched presence of distinct cell populations and subsets as characterized by the scRNA-seq. To this end, for each section, we leveraged the inferred abundances of different cell types in the spatial voxels by CARD, and tested if a cell type has significantly higher abundances in each pan-section cluster than the other voxels by Wilcoxon Rank-sum test. Then, the overall enrichments of each cell type within pre-BCG or post-BCG group were summarized by combining the Wilcoxon test p-value across the sections via aggregated Cauchy association test (ACAT). The enriched cell types per pan-section cluster in pre-BCG or post-BCG group were called with ACAT summarized FDR < 0.05 and number of sections with significant enrichments (Wilcoxon FDR < 0.05) > 1 (Figure 4.3).
Mass cytometry antibody preparation and staining
PBMCs from BCG-treated bladder cancer patients at the time of tumor recurrence were isolated using Ficoll-Paque and resuspended in cell medium (RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 1% Penicillin, 1% Streptomycin and 1% L-glutamine). PBMCs were conjugated to antibodies purchased from Fluidigm using the Maxpar X8 and MCP9 labeling kits. Platinum barcodes were prepared as previously described. All antibodies were titrated prior to conjugation.
Prior to mass cytometry staining, all cells were incubated for 20 minutes at 37C in RPMI cell medium (described above) and IdU (Fluidigm, t#201127), Rh103 (Fluidigm, #201103A). Following the incubation, cells were centrifuged, washed with PBS and 0.2% bovine serum albumin (BSA), and incubated for 3 minutes on ice with an Fc-blocking reagent. Samples were washed again with PBS and 0.2% BSA; barcoded samples were pooled together and washed; finally, samples were stained with extracellular antibodies for 30 minutes on ice and in PBS and 0.2% BSA.
When staining four samples, cells were single-barcoded using 194Pt, 195Pt, 196Pt or 198Pt. When staining five or six samples, cells were stained with combination of barcodes. Cells were again washed using PBS 0.2% BSA; barcoded samples were pooled together; samples were washed again and stained with extracellular antibodies for 30 minutes on ice in PBS 0.2% BSA. Cells were co-cultured in the presence of either WT or HLA-E-transduced K562 cells for a total of 6 hours.
Samples were washed with PBS 0.2% BSA and resuspended in Fixation/Perm buffer (Invitrogen, #00–5523-00) for 30 minutes on ice. Cells were centrifuged and washed with Maxpar Barcode Perm Buffer (Fluidigm, #201057) and barcoded using the Cell-ID 20-Plex Pd Barcoding kit (Fluidigm, #201060). Barcoded samples were washed with permeabilization buffer (Invitrogen, #00–5523-00) and pooled. Intracellular staining was then performed in permeabilization buffer with Heparin at a concentration of 100U/mL for 30 minutes on ice. Stained cells were washed with permeabilization buffer and resuspended in PBS with PFA 2.4%, saponin 0.08%, Osmium tetroxide 0.075nM and Ir 0.125uM (Fluidigm, #201192A). Finally, samples were washed and resuspended in PBS 0.2% BSA and data were acquired within four days, or frozen in FBS/DMSO 90/10. The antibody panel in Supplemental Table S4 was used to stain bladder cancer patient samples.
Mass cytometry sample acquisition and processing:
Prior to acquisition, samples were washed with cell staining buffer and acquisition solution (Standard Biotools). Following washing, samples were resuspended in acquisition solution (1 million cells / 1 mL) containing a 1:20 dilution of EQ normalization beads. Data were then acquired using the Standard Biotools Helios mass cytometer with a wide bore injector configuration at an acquisition speed of < 400 cells per second. The output files were normalized and concatenated using Standard Biotools’ CyTOF software, and outputted as FCS files.
The Mount Sinai Human Immune Monitoring Core’s (HIMC) pipeline for processing and cleaning was used to clean the resulting FCS files. Aberrant acquisition time-windows and low DNA intensity events were stripped out by the sample preprocessing pipeline. Samples were then demultiplexed via the cosine similarity of the Palladium barcoding channel on a cell-by-cell basis to every possible barcode used in a batch. Once the cell-barcode labeling has been established, the signal-to-noise (SNR) ratio was calculated by taking the difference between the highest and second highest similarity scores. Cells with low SNR ratios were flagged as multiplets and removed. Finally, acquired multiplets are removed based on the Gaussian parameters residual and offset acquired by the Helios mass cytometer.
Data processing and analysis:
Data were uploaded onto Cytobank and processed for downstream analyses. Several gates were applied: a live dead gate and a doublets gate in sequential order. All data were arc-sinh transformed, and no batch corrections were performed given all samples were run in a single batch. NK cells and CD8 T cell subsets were identified via manual gating assignment. Files were downloaded onto Mount Sinai’s supercomputing cluster, Minerva, concatenated into a single object, and clinical data were assigned to each sample. A phenograph analysis was performed to cluster the cellular data. Wilcoxon tests were run for statistical significance and the Benjamini-Hochburg correction was applied.
Tumor Invading Lymphocyte Expansion
TILs were expanded by seeding single cells tumor dissociate at 105 cells/well in 48-well Costar® flat-bottom plates (Corning Inc., NY) in complete TIL medium, consisting of RPMI 1640 with L-glutamine (Corning Inc., NY), 10% human AB serum (MiliporeSigma, MO), 1% nonessential amino acids (Thermo Fisher Scientific, MA), 1% sodium pyruvate (Thermo Fisher Scientific, MA), 1% penicillin/streptomycin (Thermo Fisher Scientific, MA), and 3000 IU/ml interleukin-2 (Proleukin®, kindly provided by Clinigen). Cells were stimulated using 25 μl ImmunoCult™ Human CD3/CD28/CD2 T Cell Activator (STEMCELL Technologies, Vancouver, BC) and seeded on 1.5 X 106 feeder cells, which were derived from healthy control PBMCs (New York Blood Center, NY) and irradiated at 50 Gy. TILs were incubated at 37°C, 5% CO2 and maintained by replacement with fresh complete TIL medium every 2 days. After three weeks, expanded TIL lines were frozen in 10% DMSO (MiliporeSigma, MO), 90% FBS (MiliporeSigma, MO) and stored in LN2.
IFN-γ Stimulation of K562 tumors
Two separate rounds of stimulation were performed on K562 tumors after reaching steady-state expansion. First, K562 tumors were co-cultured at a concentration of 0.5 X 106 viable cells / mL with 200 and 400 ng / mL of IFN-γ in R10 media in order to assess the induction of HLA-E and PD-L1 with high doses of IFN-γ. At every 24-hour interval, cell concentration and viability were assessed using a hemocytometer, and K562 tumors were split to a concentration of 0.5 X 106 live cells / mL. At 72 hours of incubation, cells were spun at 1650 RPM, washed with FACS buffer (PBS, 5% FBS, 0.2% EDTA), resuspended in 5 mL of FACS buffer, and assessed for concentration and viability. 0.5 X 106 live cells were removed, and spun to remove the FACS buffer. K562 tumors were first incubated with zombie near infrared (NIR) at a dilution of 1 in 1000 for 20 minutes at room temperature. Following the zombie stain, cells were washed in 1 mL of FACS buffer, and incubated with PD-L1 (PE; Biolegend cat #124308), and HLA-E (per-CP CY5.5; Biolegend cat #342609) antibodies in a 1:25 dilution cocktail for 30 minutes on ice in the dark. Cells were washed again in FACS buffer and suspended in 400μLof fixative buffer (2% paraformaldehyde in PBS). HLA-E and PD-L1 levels were then assessed using flow cytometry.
TIL Co-culture with K562s Expressing Checkpoints
For co-culture and surface stain of PD-L1 and HLA-E, three separate K562 tumor lines of K562 cells were generated using 400 ng/ml of IFN-γ for 72 hours, as described by the protocol listed above: WT, E+, and E+/PD-L1+ lines. Tumor-infiltrating lymphocytes (TILs) from four BCG-unresponsive NMIBC patients which had been expanded using the protocol listed above were thawed and seeded at a concentration of 1 X 106 live cells / mL in a stimulatory media consisting of ImmunoCult, CD3/CD28 tetramer (25 uL/mL), IL-2 (10 IU/mL), IL-7 (10 ng/mL), and IL-15 (10 ng/mL). Cells were seeded and plated in a 96 round-bottom plate. At days three, five, seven, and nine, 50% of the media was aspirated and replaced with fresh media at twice the initial concentration of cytokines, without CD3/28 tetramer. At day 10, TGF-β (Immunocult, IL-2 (10IU/mL), IL-7 (10ng/mL), IL-15 (10ng/mL), CD3/CD28 tetramer (25μL/mL), TGF-β (5ng/mL)) was provided to stimulate expression of NKG2A on the TILS.
At day 13, the stimulation was complete, and the TILs were co-cultured with combinations of the K562 cells expressing forms of PD-L1 and HLA-E. In total, 12 experimental co-culture conditions were conducted for each patients’ TIL sample (n=4): two control wells (ex-vivo, and stimulated with a CD3/28 spike); with K562 WT cells, HLA-E+ cells, or HLA-E+ PD-L1+ cells without antibody; TILs in the presence of HLA-E+ or HLA-E+PD-L1+ K562s with durvalumab alone, or both durvalumab plus monalizumab; and lastly, TILs and WT K562s in the presence of monalizumab and durvalumab.
TILs were cultured in TIL medium, consisting of RPMI 1640 with L-glutamine (Corning Inc., NY), 10% human AB serum (MiliporeSigma, MO), 1% nonessential amino acids (Thermo Fisher Scientific, MA), 1% sodium pyruvate (Thermo Fisher Scientific, MA), 1% penicillin, streptomycin (Thermo Fisher Scientific, MA). Cultures were performed in 96-well u-bottom plates (Corning Inc., NY). TILs were added to each well at a concentration of at 3 X105 cells/condition in TIL medium. K562s were resuspended in TIL medium and added at 5 X 104 singe-cell equivalents/condition. Anti-CD28 (BioLegend, CA) was added to each well at 1 μg/ml and anti-CD107a-BV785 (BioLegend, CA) was added to each well at 0.3 μg/ml. Monalizumab was added to the TILs a final concentration of 10 μg/ml, and durvalumab was added to the K562s for a final concentration of 10 μg/ml. TILs and K562s were cultured separately for 20 minutes at 37°C prior to co-culture. The K562s and TILs were then combined, thoroughly resuspended, and gently spun for 5 seconds before being returned to the incubator. Plates were incubated at 37°C and 5% CO2, and after 1 hour of incubation, 0.5X Brefeldin (BioLegend, CA) and 0.5X Monensin (BioLegend, CA) was added to each tube for a final volume of 500 μl, followed by an additional 15 hours of incubation. At hour 16 plates were spun and supernatant was removed; all samples were washed with 200μLof PBS. Samples were resuspended in 50μLof zombie near infrared (NIR) at a 1:3000 dilition, and incubated for 20 minutes in the dark at room temperature. Samples were washed again, spun, supernatant was removed, and incubated in 50μLof TruStain FCX, diluted 1:200, for 20 minutes in the dark at 4°C. 50μLof the surface antibody master mix was added at 20 minutes, and returned to the fridge to incubate in the dark at 4°C for 30 more minutes. Plates were washed twice more in FACS buffer, and were suspended in 0.2% FBS FACS buffer for storage until intracellular staining.
12 hours prior to acquisition by CyTOF, intracellular staining was performed for IFN-γ. 100 μL of intracellular staining permeabilization wash buffer (BioLegend) was added to each well, the plates were spun, and supernatant discarded. This was performed 2 additional times with 200 μL of permeabilization wash buffer. 50 μL of the intracellular staining antibody mix was added to each well, and the plates were incubated for 20 minutes at 4°C. The plates were washed in 0.2% FBS FACS buffer and resuspended for storage in 0.2% FBS FACS buffer.
Supplementary Material
Extended data Table S1: Genes defining gene-set enrichment pathways
Extended data Table S2: Antibody panel for CODEX multiplexed immunofluorescence staining
Extended data Table S3: Annotated clusters with list of significantly defining genes for major clusters defined by scRNAseq
Extended data Table S4: Annotated scRNAseq clusters with numbers of cells
Extended data Figure S1: Cellular sources of IFN-γ in NMIBC by scRNAseq
ACKNOWLEDGMENTS
We thank Mary Anne O’Donnell (Precision Immunology Institute, Icahn School of Medicine at Mount Sinai) for critically reviewing the manuscript; Deepta Bhattacharya ‘(University of Arizona) for kindly providing HLA-E+ K562 tumors. We acknowledge the expertise and assistance of the Dean’s Flow Cytometry Center of Research Excellence at Mount Sinai. The A.H. and J.P. labs were supported by funding from P30CA196521, R01 CA269954–01, R21 CA274148, BCAN No. 961726, and R21AI130760A. The N.B. lab was supported by funding from the Department of Defense Peer Review Cancer Research program Translational team Award No. W81XWH1910269 and from the Parker Institute for Cancer Immunotherapy No.AGR-11611SOW1.
Footnotes
Conflict of Interest Disclosure Statement: The authors declare no potential conflicts of interest.
DECLARATION OF INTERESTS
L.W., R.P.S., and J.Z. are employees of Sema4. A.H. receives research funds from Astra Zeneca and has recently served on the advisory boards of Immunorizon, Purple BiotechUroGen, and Takeda. N.B. is an extramural member of the Parker Institute for Cancer Immunotherapy, receives research funds from Regeneron, Harbor Biomedical, DC Prime, and Dragonfly Therapeutics and is on the advisory boards of Neon Therapeutics, Novartis, Avidea, Boehringer Ingelheim, Rome Therapeutics, Rubius Therapeutics, Roswell Park Comprehensive Cancer Center, BreakBio, Carisma Therapeutics, CureVac, Genotwin, BioNTech, Gilead Therapeutics, Tempest Therapeutics, and the Cancer Research Institute.
Data Availability Statement:
data were generated by the authors and have been uploaded to the Gene Expression Omnibus (GSE276014 and GSE276015) and will be made publicly available upon publication of this manuscript.
References
- 1.Redelman-Sidi G., Glickman M.S. & Bochner B.H. The mechanism of acEon of BCG therapy for bladder cancer--a current perspective. Nat Rev Urol 11, 153–162 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Kamat A.M. et al. Predicting Response to Intravesical Bacillus Calmette-Guerin Immunotherapy: Are We There Yet? A Systematic Review. Eur Urol 73, 738–748 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Müller G. et al. Association between Development of Metabolic Acidosis and Improvement of Urinary Continence after Ileal Neobladder CreaEon. J Urol 203, 585–590 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Hedgepeth R.C., Gilbert S.M., He C., Lee C.T. & Wood D.P. Jr. Body image and bladder cancer specific quality of life in patients with ileal conduit and neobladder urinary diversions. Urology 76, 671–675 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Morales A. BCG: A throwback from the stone age of vaccines opened the path for bladder cancer immunotherapy. Can J Urol 24, 8788–8793 (2017). [PubMed] [Google Scholar]
- 6.ZloTa A.R. et al. What is the optimal regimen for BCG intravesical therapy? Are six weekly instillaEons necessary? Eur Urol 37, 470–477 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Sfakianos J.P. et al. Bacillus Calmette-Guerin (BCG): Its fight against pathogens and cancer. Urol Oncol 39, 121–129 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Sharma P., Hu-Lieskovan S., Wargo J.A. & Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gocher A.M., Workman C.J. & Vignali D.A.A. Interferon-γ: teammate or opponent in the tumour microenvironment? Nat Rev Immunol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benci J.L. et al. Opposing FuncEons of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell 178, 933–948.e914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L. et al. Myeloid Cell-associated Resistance to PD-1/PD-L1 Blockade in Urothelial Cancer Revealed Through Bulk and Single-cell RNA Sequencing. Clin Cancer Res (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strandgaard T. et al. Field Cancerization Is Associated with Tumor Development, T-cell Exhaustion, and Clinical Outcomes in Bladder Cancer. Eur Urol (2023). [DOI] [PubMed] [Google Scholar]
- 13.Strandgaard T. et al. Elevated T-cell Exhaustion and Urinary Tumor DNA Levels Are Associated with Bacillus Calmette-Guérin Failure in Patients with Non–muscle-invasive Bladder Cancer. European Urology (2022). [DOI] [PubMed] [Google Scholar]
- 14.Kates M. et al. Adaptive Immune Resistance to Intravesical BCG in Non-Muscle Invasive Bladder Cancer: Implications for Prospective BCG-Unresponsive Trials. Clin Cancer Res 26, 882–891 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg J.E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powles T. et al. Efficacy and Safety of Durvalumab in Locally Advanced or Metastatic Urothelial Carcinoma: Updated Results From a Phase 1/2 Open-label Study. JAMA Oncol 3, e172411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma P. et al. Nivolumab in metastatic urothelial carcinoma after plaEnum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 18, 312–322 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Patel M.R. et al. Avelumab in metastatic urothelial carcinoma after platinum failure (JAVELIN Solid Tumor): pooled results from two expansion cohorts of an open-label, phase 1 trial. Lancet Oncol 19, 51–64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman J.E., Vasudevan D., Joyce C.E. & Hildago M. Is PD-L1 a consistent biomarker for anE-PD-1 therapy? The model of balstilimab in a virally-driven tumor. Oncogene 40, 1393–1395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamiya T., Seow S.V., Wong D., Robinson M. & Campana D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J Clin Invest 129, 2094–2106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira B.I. et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8(+) T cell inhibition. Nat Commun 10, 2387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Monpoort N. et al. NKG2A Blockade Potentiates CD8 T Cell Immunity Induced by Cancer Vaccines. Cell 175, 1744–1755.e1715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andre P. et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 175, 1731–1743 e1713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomé B. et al. NKG2A and HLA-E define a novel alternative immune checkpoint axis in bladder cancer. Cancer Cell 40, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbst R.S. et al. COAST: An Open-Label, Phase II, Multidrug Platform Study of Durvalumab Alone or in Combination With Oleclumab or Monalizumab in Patients With Unresectable, Stage III Non-Small-Cell Lung Cancer. J Clin Oncol, JCO2200227 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Rouanne M. et al. BCG therapy downregulates HLA-I on malignant cells to subvert antitumor immune responses in bladder cancer. J Clin Invest 132 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y.H. et al. Urine cytokines as biomarkers for diagnosing interstitial cystitis/bladder pain syndrome and mapping its clinical characteristics. Am J Physiol Renal Physiol 318, F1391–f1399 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Tyagi P. et al. Urine cytokines suggest an inflammatory response in the overactive bladder: a pilot study. Int Urol Nephrol 42, 629–635 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Netskar H. et al. Pan-cancer profiling of tumor-infiltrating natural killer cells through transcriptional reference mapping. Nat Immunol (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebuffet L. et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat Immunol (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivier E. et al. Natural killer cell therapies. Nature 626, 727–736 (2024). [DOI] [PubMed] [Google Scholar]
- 33.Browaeys R., Saelens W. & Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Amir el A.D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cristescu R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spranger S. et al. Up-regulaEon of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 5, 200ra116–200ra116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rooney M.S., Shukla S.A., Wu C.J., Getz G. & Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribas A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov 5, 915–919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grasso C.S. et al. Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell 38, 500–515.e503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gocher A.M., Workman C.J. & Vignali D.A.A. Interferon-γ: teammate or opponent in the tumour microenvironment? Nat Rev Immunol 22, 158–172 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardia A. et al. Sacituzumab Govitecan in Metastatic Triple-Negative Breast Cancer. N Engl J Med 384, 1529–1541 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Powles T. et al. Enfortumab Vedotin and Pembrolizumab in Untreated Advanced Urothelial Cancer. N Engl J Med 390, 875–888 (2024). [DOI] [PubMed] [Google Scholar]
- 43.Heath E.I. & Rosenberg J.E. The biology and raEonale of targeting necEn-4 in urothelial carcinoma. Nat Rev Urol 18, 93–103 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Chang S.S. et al. Diagnosis and Treatment of Non-Muscle Invasive Bladder Cancer: AUA/SUO Guideline. J Urol 196, 1021–1029 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Bisiaux A. et al. Molecular analyte profiling of the early events and Essue conditioning following intravesical bacillus calmette-guerin therapy in patients with superficial bladder cancer. J Urol 181, 1571–1580 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Shapiro S.S. & Wilk M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 52, 591–611 (1965). [Google Scholar]
- 47.Kruskal W.H. & Wallis W.A. Use of Ranks in One-Criterion Variance Analysis. Journal of the American Statistical Association 47, 583–621 (1952). [Google Scholar]
- 48.Zhang X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res 47, D721–d728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glass D.R. et al. An Integrated Multi-omic Single-Cell Atlas of Human B Cell Identity. Immunity 53, 217–232.e215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magri G. et al. Human Secretory IgM Emerges from Plasma Cells Clonally Related to Gut Memory B Cells and Targets Highly Diverse Commensals. Immunity 47, 118–134.e118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nature biotechnology 42, 293–304 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finak G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome biology 16, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberzon A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rouillard A.D. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galon J. et al. Affinity of the interaction between Fc gamma receptor type III (Fc gammaRIII) and monomeric human IgG subclasses. Role of Fc gammaRIII glycosylation. Eur J Immunol 27, 1928–1932 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Liberzon A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dries R. et al. GioTo: a toolbox for integraEve analysis and visualization of spatial expression data. Genome biology 22, 1–31 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lause J., Berens P. & Kobak D. Analytic Pearson residuals for normalization of single-cell RNA-seq UMI data. Genome biology 22, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korsunsky I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma Y. & Zhou X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nature biotechnology 40, 1349–1359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin S., Plikus M.V. & Nie Q. CellChat for systematic analysis of cell-cell communication from single-cell and spatially resolved transcriptomics. BioRxiv, 2023.2011. 2005.565674 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended data Table S1: Genes defining gene-set enrichment pathways
Extended data Table S2: Antibody panel for CODEX multiplexed immunofluorescence staining
Extended data Table S3: Annotated clusters with list of significantly defining genes for major clusters defined by scRNAseq
Extended data Table S4: Annotated scRNAseq clusters with numbers of cells
Extended data Figure S1: Cellular sources of IFN-γ in NMIBC by scRNAseq
Data Availability Statement
data were generated by the authors and have been uploaded to the Gene Expression Omnibus (GSE276014 and GSE276015) and will be made publicly available upon publication of this manuscript.