Abstract
Extrachromosomal DNA (ecDNA) is a hallmark of aggressive cancer, contributing to both oncogene amplification and tumor heterogeneity. Here, we used Hi-C, super-resolution imaging, and long-read sequencing to explore the nuclear architecture of MYC-amplified ecDNA in colorectal cancer cells. Intriguingly, we observed frequent spatial proximity between ecDNA and 68 repetitive elements which we called ecDNA-interacting elements or EIEs. To characterize a potential regulatory role of EIEs, we focused on a fragment of the L1M4a1#LINE/L1 which we found to be co-amplified with MYC on ecDNA, gaining enhancer-associated chromatin marks in contrast to its normally silenced state. This EIE, in particular, existed as a naturally occurring structural variant upstream of MYC, gaining oncogenic potential in the transcriptionally permissive ecDNA environment. This EIE sequence is sufficient to enhance MYC expression and is required for cancer cell fitness. These findings suggest that silent repetitive genomic elements can be reactivated on ecDNA, leading to functional cooption and amplification. Repeat element activation on ecDNA represents a mechanism of accelerated evolution and tumor heterogeneity and may have diagnostic and therapeutic potential.
Introduction
Extrachromosomal DNA (ecDNA) is a prevalent form of oncogene amplification found across cancers, present in approximately 15% of cancers at diagnosis1-5. EcDNAs are megabase-scale, circular DNA elements lacking centromeric and telomeric sequences and found as distinct foci apart from chromosomal DNA. Recent work has underscored the importance of ecDNA in tumor initiation and various aspects of tumor progression, such as accelerating intratumoral heterogeneity, genomic dysregulation, and therapeutic resistance6-10. The biogenesis of ecDNA is complex and tied to mechanisms that induce genomic instability, such as chromothripsis and breakage-fusion-bridge cycles, which are prevalent in tumor cells11-17.
In addition to amplifying oncogenes, a key aspect of ecDNA is their ability to hijack regulatory elements that increase oncogene expression beyond the constraints imposed by standard chromosomal architecture18-23. Given that ecDNA has been shown to hijack enhancers and regulate expression of both endogenous chromosomal sequences and oncogenes present on separate ecDNA21-23, the nuclear organization of these molecules is tightly tied to their ability to amplify gene expression. As such, we sought to better characterize the nuclear architecture of ecDNA in COLO320DM cells which is a colorectal adenocarcinoma cancer cell line with MYC oncogene amplification24,25 through Hi-C, long-read sequencing and high-resolution imaging approaches26-28.
Results
Pervasive ecDNA contacts at confined chromosomal locations
To interrogate the conformational state of circular extrachromosomal DNA (ecDNA), we performed Hi-C on COLO320DM colorectal cancer cells (Fig. 1A). Previous investigation of COLO320DM utilizing DNA fluorescent in situ hybridization (FISH) and whole-genome sequencing (WGS) had identified a highly-rearranged (up to 4.3 MB) ecDNA amplification containing several genes including the oncogene MYC and the long non-coding RNA PVT118,22. As a large fraction of the ecDNA in COLO320DM is derived from chromosome 8, with smaller contributions from chromosomes 6,16, and 13, we elected to focus on the chromosome 8 amplified locus containing MYC and PVT122.
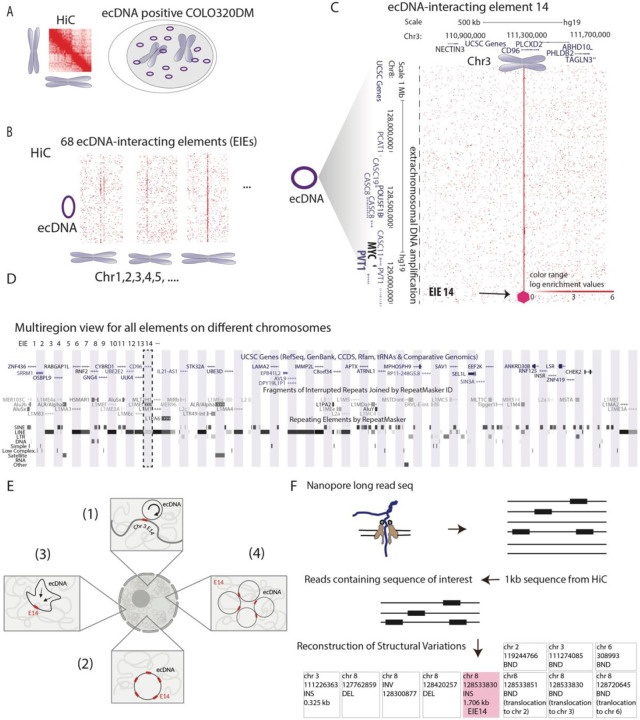
Figure 1: Identification of ecDNA interacting elements (EIEs).
A. Hi-C analysis in COLO320DM cell line: method schematics
B. Identification of 68 ecDNA-interacting elements (EIEs). The visualization represents the ecDNA from Chromosome 8 with lines indicating the interactions with ecDNA-interacting elements (EIEs) localized on other chromosomes.
C. An example of a specific interaction, EIE 14 on Chromosome 3, is enlarged and associated genes are shown for both loci. Arrow and purple hexagon indicate EIE.
D. UCSC Genome Browser multiregion view snapshot showing the genomic context of the EIEs. Each element on different chromosomes is indicated by a vertical bar, with EIE 14 highlighted. The browser displays the annotations for genes and repetitive elements such as Alu, LINE, and LTR elements (RepeatMasker). Detailed mapping of each EIE is in Supplementary Table T2, T3
E. Hypothetical models for interaction between ecDNA and elements: (1) the ecDNA is interacting with the endogenous EIE making frequent contacts with the endogenous sequence (2) multiple copies of the EIE integrate at multiple locations on the ecDNA becoming extra-chromosomally amplified; (3) a single copy of the element integrates in the ecDNA at a specific site, but the ecDNA is folded in a way that allows extensive contact within the ecDNA molecule; or (4) whether a copy of the EIE inserts into the ecDNA, and clustering of multiple ecDNA copies enables the extensive contact.
F. Schematic representation of the Nanopore long-read sequencing methodology used to identify structural variations associated with the 1 kb EIEs. Reads containing the sequence of interest were extracted, aligned, and analyzed for structural variation. Reconstruction of Structural Variations is shown for EIE 14.
Upon inspection of the Hi-C maps, we observed 68 putative interactions between ecDNA and other chromosomes (Fig. 1B; Supplementary Table T1). The structure of these interactions was atypical: we found that upon binning the data at 1kb resolution, linear elements appeared to contact the entirety of the megabase-scale ecDNA amplification in a distinctive stripe (Fig. 1C). These contacts were widely spread across all chromosomes in the genome (Supplementary Table T1). This atypical pattern of interaction suggested a complex structural relationship between the ecDNA and the endogenous chromosome regions (hereafter referred to as ecDNA-interacting elements or EIEs), specific to ecDNA amplifications. (Fig. 1B-C). Further inspection revealed these EIEs contained Alu sequences, LINEs, LTRs, and various other retrotransposons (Fig. 1D; Supplementary Table T2 and T3). As these retrotransposons can acquire the ability to regulate transcription when active, the spatial relationship with oncogenes like MYC may be important for enhanced expression in COLO320DM cells29,30.
To clarify the nature of such distinct contact interactions, we considered various hypotheses that may explain the observed stripes where the ecDNA is either contacting chromosomal EIEs or the EIEs have been amplified as part of the ecDNA itself. These hypotheses were as follows: (1) the ecDNA is interacting with chromosomal EIEs (2) multiple copies of the EIE integrate at multiple locations on the ecDNA becoming extra-chromosomally amplified; (3) a single copy of the element is integrated in the ecDNA at a specific site (or indeed may have pre-existed in the haplotype that gave rise to the ecDNA), but the ecDNA is folded in a way that allows extensive contact within the ecDNA molecule; or (4) a single copy of the EIE is integrated in the ecDNA and clustering of multiple ecDNA copies enables the extensive contact (Fig. 1E). Each of these possibilities has implications for the function of these EIEs in regard to ecDNA segregation and oncogene regulation. For example, in scenario (1), proximity to an EIE may aid ecDNA in their segregation during mitosis by facilitating contact with endogenous chromosomes. In contrast, scenarios (3-4) provide a possible mechanism to amplify MYC transcription if these EIEs can act as enhancers.
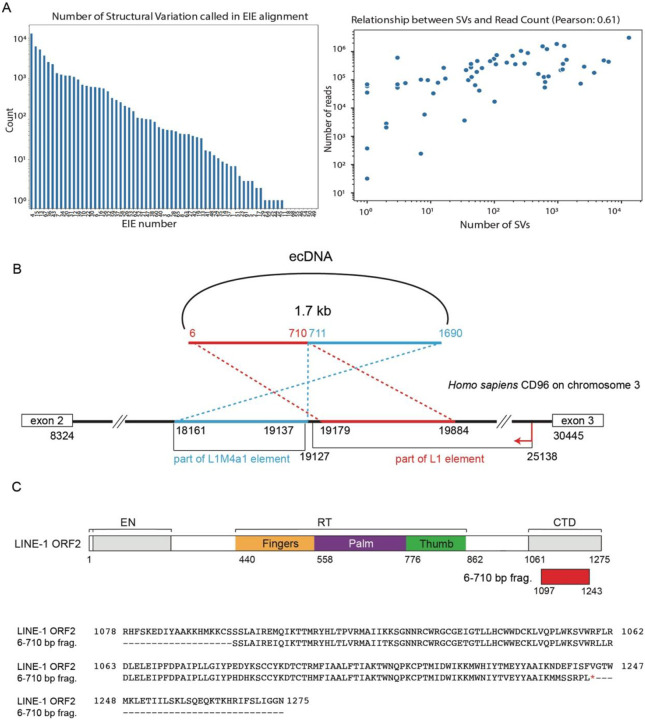
To begin investigating these hypotheses, we performed Nanopore long-read sequencing to distinguish purely topological interactions of the MYC containing ecDNA with these EIEs versus integration events onto the ecDNA. We chose ultra-long-read sequencing to also capture potential heterogeneity in insertion sites in the case of single or multiple integrations. (Fig. 1F; Methods). We generated median read lengths of 67,000 bp with the longest read spanning 684,457 bases. Across the 68 EIEs identified, we determined that each participated in a broad spectrum of structural variation - some involved with hundreds or thousands of different rearrangement events (Extended Data Fig. 1A). We found a strong correlation between the number of reads mapping to an element and the number of structural variants associated with said element (Extended Data Fig. 1A).
EIE 14 is a “passenger” on MYC ecDNA
To extensively characterize the insertion site, spatial organization, and potential function of an EIE we chose to focus on EIE 14, which contains an approximately 1 kb segment similar to L1M4a1, an ancient element distantly related to human LINE-1. The percent identity of the L1M4a1 segment to the consensus sequence is consistent with the L1M4a1’s Kimura divergence value of 34%. This degree of divergence suggests that the L1M4a1 portion of EIE 14 lacks the ability to jump autonomously and was moved en bloc with adjacent sequences by an unknown mechanism. Despite the age of this segment, a second component of EIE 14 contains a fragment of a LINE-1 PA2 element and encodes a segment of an ORF-2 like protein. EIE 14 also contains a portion of the adjacent intron 2 of the CD96 gene on chromosome 3 part of which is annotated as L1M4a1 (Extended Data Fig. 1B). The region encoding the segment of ORF2 lacks the full sequence to encode a functional ORFp2 (Extended Data Fig.C)31,32. Since a portion of EIE 14 mapped to a single genomic position in the reference genome we were able to specifically target and interrogate its function.
Because repetitive elements are typically discarded during ecDNA reconstruction by sequencing methods33, we turned to CRISPR-CATCH - a method for isolating and sequencing specific ecDNA - to elucidate the structure of ecDNAs containing EIE 14 (Fig. 2A)21. Targeting EIE 14, we successfully isolated ecDNA fragments from the COLO320DM cell line for sequencing (Fig. 2B). Sequence analysis of these bands confirmed the presence of EIE 14 pulled down in CRISPR-CATCH, and the flanking sequence identified from these bands indicates an integration between the CASC8 and CASC11 genes approximately 200 kilobases away from MYC, in agreement with long-read sequencing (Fig. 1F, Fig.2C, Extended Data Fig.2B & D and Supplementary Table T4-T6). Multiple bands and different sizes on the gel indicate the genomic position of EIE 14 is consistent across ecDNA species (Fig. 2B-C). Beyond EIE 14, the CRISPR-CATCH approach allowed us to capture and sequence a subset of EIEs initially identified through Hi-C analysis (Fig. 2D). The identification of the additional EIEs observed in the Hi-C data suggest that the “striping” between the ecDNA and endogenous chromosomes is an artifact of these sequences’ presence on ecDNAs, rather than true trans contacts, at least for this identified subset. Though the recent T2T genome build34 annotates EIE 14 to chromosome 3 (Extended Data Fig. 2C), we found evidence that the structural variant described here between EIE 14 and the MYC-containing amplicon region is identified as a translocation event between Chr8:128,533,830 and Chr3:111,274,086 in approximately 46% (minor allele frequency of 0.467646) of non-disease individuals35 (Supplementary Table T4 (row 7)). This suggests that this structural variant was pre-existing prior to cancer formation but was amplified as a passenger on ecDNA.
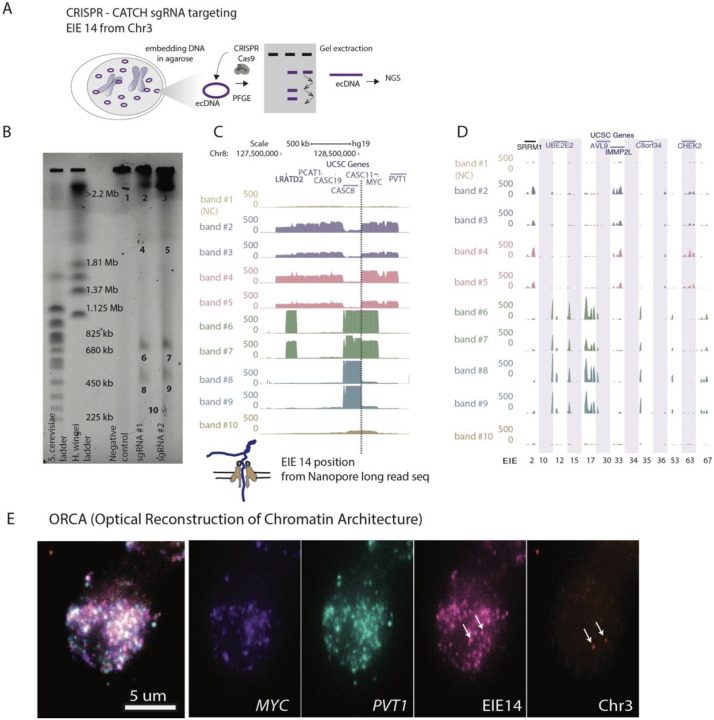
Figure 2: CRISPR-CATCH Elucidates ecDNA Composition and EIE Insertions.
A. Schematic diagram illustrating the CRISPR-CATCH experiment designed to isolate and characterize ecDNA components. The process involves the use of guide RNA targeting the EIE 14 from Chromosome 3. DNA is embedded in agarose, followed by pulse-field gel electrophoresis (PFGE), allowing for the band extraction and subsequent next-generation sequencing (NGS) of ecDNA fragments.
B. PFGE gel image displays the separation of DNA fragment, lines from left ladder, ladder, empty lane, Negative control, sgRNA #1, sgRNA #2 and band numbers for NGS. The EIE 14 targeted by the guide RNAs leads to cutting of the ecDNA's chromosome 8 sequences to form multiple discrete bands. sgRNA #1 ATATAGGACAGTATCAAGTA; sgRNA #2 TATATTATTAGTCTGCTGAA; Full EIE 14 sequences from long-read sequencing is in Supplementary Table T6.
C. Visualization of the sequencing results confirms the presence of EIE 14, originally annotated on Chromosome 3, within the ecDNA, between the CASC8 and CASC11 genes, approximately 200 kilobases upstream from MYC. The dotted line indicates the position of this insertion.
D. Additional EIEs identified in the initial Hi-C screen, captured, and sequenced in the CRISPR-CATCH, each EIE is one one vertical shaded box with coordinates.
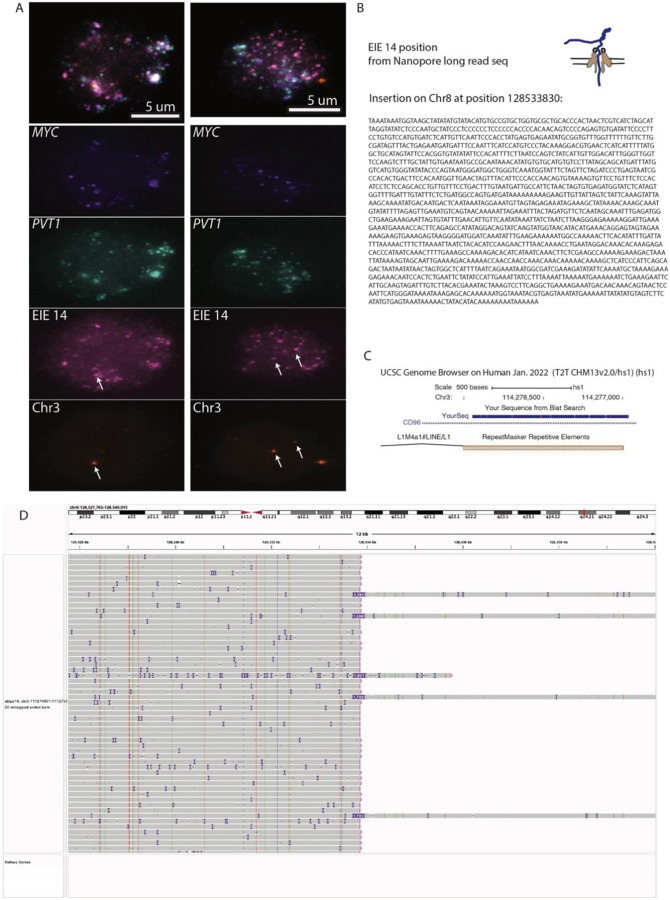
E. ORCA (Optical Reconstruction of Chromatin Architecture) visualization of the COLO320DM cell nucleus. The images show the spatial arrangement of the MYC oncogene, EIE 14 and the PVT1 locus, labeled in different colors. The scale bar represents 5 micrometers. Chr3 probe maps to the breakpoints of the EIE 14 origin inside CD96 intron.
Further evidence of EIE 14’s amplification on ecDNA can be visualized by utilizing Optical Reconstruction of Chromatin Architecture (ORCA)27,36. Barcoded probes were designed targeting the unique portion of EIE 14 (1kb), MYC exon 2 (3.1kb), PVT1 exon 1 (2.5kb), and the endogenous chromosome 3 region flanking of EIE 14 (3kb) (Supplementary Table T7) to determine the spatial organization of EIE 14 relative to the ecDNA. These specific exons were chosen to account for the fact that amplicon reconstruction of ecDNA in the COLO320DM cell line demonstrated an occasional rearrangement of MYC exon 2 replacement by PVT1 exon 1 22. EIE 14 visually colocalized with the ecDNA and amplified to a similar copy number per cell (Fig. 2E, Extended Data Fig. 2A). This definitively ruled out a model where the observed stripe in the Hi-C data was a result of ecDNA solely interacting with the endogenous chromosomal EIE 14. Between the extensive structural variation detected in the long-read sequencing and the amplification of EIE 14 visualized by ORCA (Extended Data Fig. 2), it was suggestive of a model where the element resides in the sequence amplified on ecDNA and participates in cis and/or trans-contacts with other ecDNA molecules (supporting hypotheses 3 and 4, Fig. 1E).
Three-dimensional conformation of EIE 14 in MYC ecDNA hubs
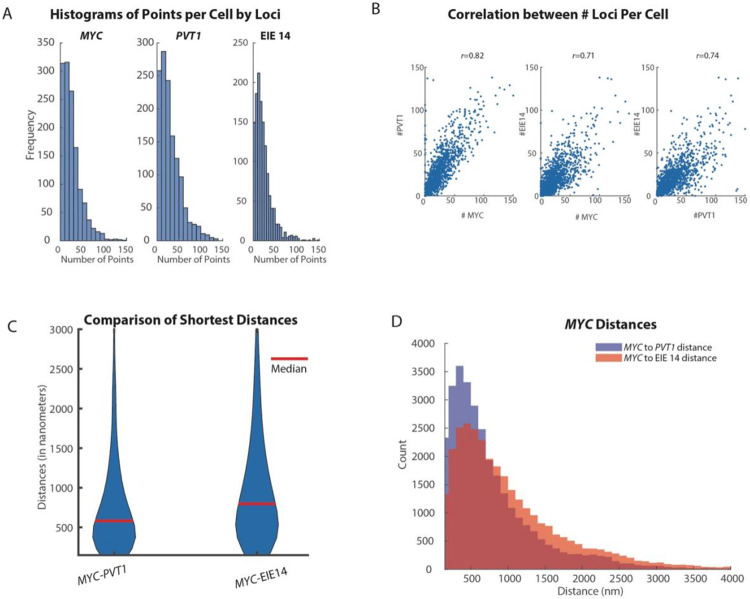
It has been proposed that amplified loci within ecDNA are able to regulate oncogene expression through cis-interactions on the same ecDNA molecule as well as trans-interactions between ecDNA via “hub” formation22. As such it is important to understand not only the structural variations of ecDNA, but also how they are arranged in the nucleus for a comprehensive understanding of potential regulatory function. We quantified the spatial distributions of MYC exon 2, PVT1 exon 1, and EIE 14; the imaged loci were fitted in 3-dimensions with a gaussian fitting algorithm to extract x,y,z coordinates (Fig. 3A-B, Methods). The copy number of identified loci varied from zero detected points to 150 per cell. On average, MYC had 29, PVT1 had 31 and EIE 14 had 22 copies per cell (Extended Data Fig. 3A). Similar distributions of points-per-cell, as well as strong correlation (r>0.7) between number of points per loci per cell (Extended Data Fig. 3B) makes it unlikely that the EIE is inserting into multiple sites on a single ecDNA.
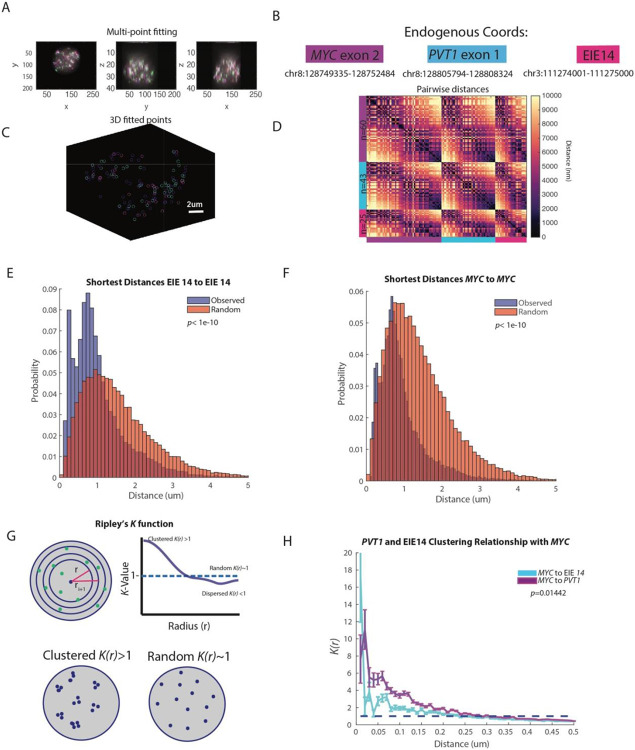
Figure 3: EIE 14 spatially clusters with MYC.
A. X, Y, Z projections of MYC exon (purple), PVT1 (blue), and EIE 14 (pink)
B. Endogenous coordinates of all three measured genomic regions.
C. Single cell projection of the 3D fitted points from (A).
D. Pairwise distances between MYC (purple), PVT1 (blue), and EIE 14 (pink) of a single cell. Number of fitted points per genomic region n=60, n=43, and n=25 respectively.
E. Histogram of distribution of distances of the observed shortest pairwise EIE 14 to EIE 14 distances and the expected shortest pairwise distances of points randomly simulated in a sphere (two-tailed Wilcoxon ranksum p<1e-10) of n=1329 analyzed cells.
F. As in (E) but for MYC to MYC shortest pairwise distances (two-tailed Wilcoxon ranksum p<1e-10).
G. Schematic of Ripley’s K function to describe clustering behaviors over different nucleus volumes. Top shows the nucleus divided into different shell intervals and how the K value is plotted for increasing radius (r). Bottom shows an example of what clustered K(r)>1 vs. random K(r)~1 points could look like. H. The average K(r) value across distance intervals of 0.01 to 0.5um in 0.02um step sizes to describe the clustering relationship of PVT1 and EIE 14 relative to MYC across different distance intervals (um). Error bars denote SEM. (Two-tailed Wilcoxon ranksum p=0.01442).
Once the centroids of each point per cell were identified (Fig. 3C) we calculated the all-to-all pairwise distance relationship (Fig. 3B). The off-diagonal pattern of distances between EIE 14, MYC, and PVT1 suggested a tendency for these loci to cluster at genomic distances <1000nm. We further quantified the spatial relationships across all 1329 imaged cells by calculating the shortest pairwise distances of MYC to EIE 14 and PVT1, as well as MYC to MYC and EIE 14 to EIE 14 distances. To determine if these ecDNA molecules were spatially clustering in cells we leveraged the fact that each ecDNA molecule appeared to have a single copy of MYC and EIE 14. Thus, distances between MYC and other MYC loci should be closer than random if the ecDNA were spatially clustered. Random distances were simulated in a sphere with the identical number of points per a given cell. The distribution of shortest pairwise distances between MYC and MYC and between EIE 14 and EIE 14 were left-shifted compared to the randomly simulated points, suggestive of a nonrandom organization (Fig. 3E-F, p<1e-10). The median observed versus expected distances between each EIE 14 loci were 748 nm and 927 nm respectively and the median observed versus expected distances between each MYC loci were 707nm and 814nm respectively.
To determine whether or not EIE 14 and MYC could be within regulatory distance of one another we calculated the shortest pairwise distances between the loci and which percentage were within 300 nm which is a proposed distance at which enhancers can exert transcriptional regulation on promoters via accumulation of activating factors37-40. The median distance between MYC and EIE 14 was 797nm and 12% of MYC loci had an associated EIE 14 locus within 300 nm (Extended Data Fig. 3C-D). In comparison, the median distances between MYC and PVT1 were 585 nm and 20% of measured MYC loci had a corresponding PVT1 within 300 nm. To further describe the spatial relationship between EIE 14 and MYC, controlling for density of loci, we turned to Ripley’s K analysis which is a spatial point pattern analysis technique commonly used to describe the degree of spatial clustering or dispersion within a given distance interval (See Methods). K-values greater than one indicate clustering behavior relative to a random distribution over that given distance interval (r), K values ~ one denote random distribution, while K values less than one indicate dispersion behavior (Fig. 3G). MYC exhibits the strongest clustering with EIE 14 at distances less than 200 nm and this behavior approaches a random distribution at greater distances (Fig. 3E). While, on average, distances between MYC and EIE 14 are further than MYC and PVT1 (Extended Data Fig. 3B-C, at distances <20nm, EIE 14 displays stronger clustering behavior with MYC (Fig. 3H). This contact may suggest that EIE 14 is acting in a proximity-dependent regulatory role of MYC similar to enhancer-promoter interactions41. Due to the spatial clustering behavior of this ecDNA species measured here and previously22, the propensity for MYC to engage in “enhancer hijacking”42, and the ability for reactivated repetitive elements to engage in long-range gene activation29 it is possible that any genomically linear separation of MYC and EIE 14 is overcome in both cis- (interaction with MYC on the same ecDNA molecule) and trans (ecDNA-ecDNA interactions) (hypotheses 3-4, Fig. 1E).
EIE 14 is critical for cancer cell fitness and can act as enhancer
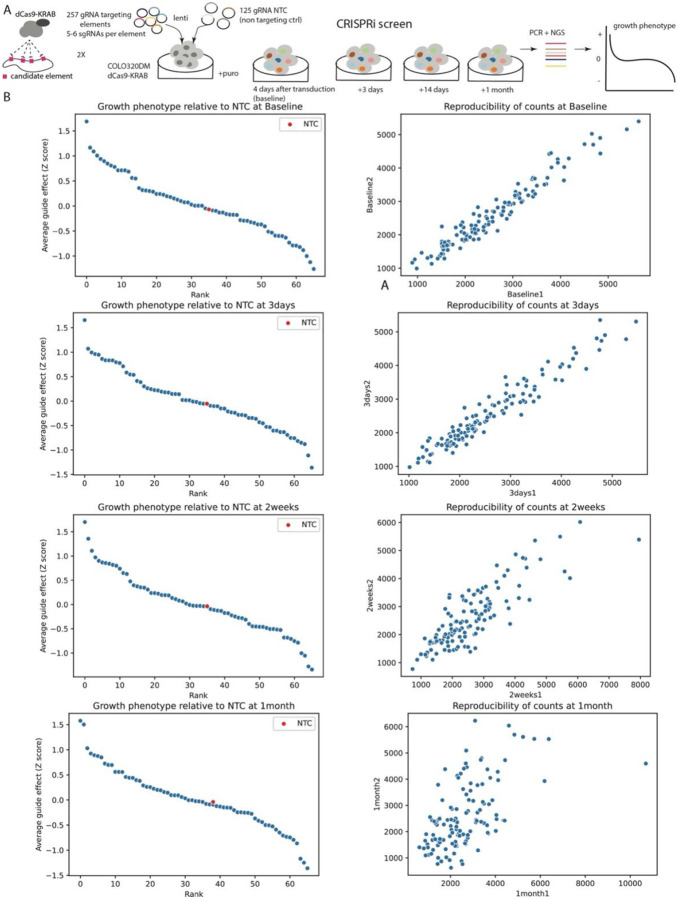
To test whether the identified transposable elements acted as transcriptional enhancers of ecDNA-amplified genes, we first assessed the fitness effects of each transposable element using pooled CRISPR interference (CRISPRi) (Fig. 4A-B). We engineered the COLO320DM cell line such that it contained the necessary dCas9-KRAB CRISPRi components43. For each of the 68 EIEs, we designed between five and six sgRNAs and included an additional 125 non-targeting controls (NTC) that were introduced into cells via lentiviral transduction (Supplementary Table T10). Post-transduction, we monitored cell proliferation at multiple time points: 4 days (baseline), 3 days after baseline, 14 days, and 1 month, followed by deep sequencing to enumerate sgRNA frequencies (Fig. 4A). Our data showed that the growth phenotype curve for three EIEs at various time points indicated a Z-score of less than −1, which suggested a significant negative impact on cell viability (Fig. 4C, Extended Data Fig. 4, Supplementary Tables T8 and T9). This finding points to a functional role of a subset of the identified EIEs in the regulation of cell growth and fitness, possibly through their interaction with the MYC oncogene as knockdown of MYC leads to growth defects and increased apoptosis in COLO320DM cells44,45.
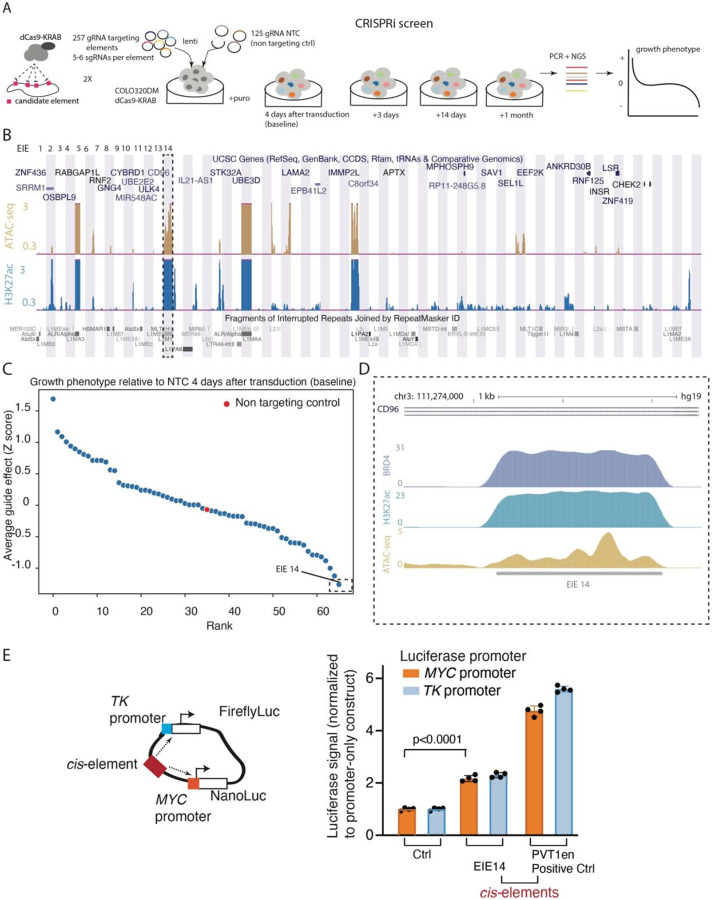
Figure 4: CRISPRi Screen Reveals EIE-Dependent Growth Phenotypes in COLO320DM Cells.
A. Schematic of the CRISPRi screening strategy used to evaluate the regulatory potential of 257 genomic EIEs near the MYC oncogene in COLO320DM cells. For each EIE, 5-6 sgRNAs were designed and 125 non-targeting control sgRNAs. The screen involved the transduction of cells with a lentivirus expressing dCas9-KRAB and the sgRNAs, followed by calculation of cell growth phenotype over a series of time points (4 days, 3 days, 14 days, and 1 month).
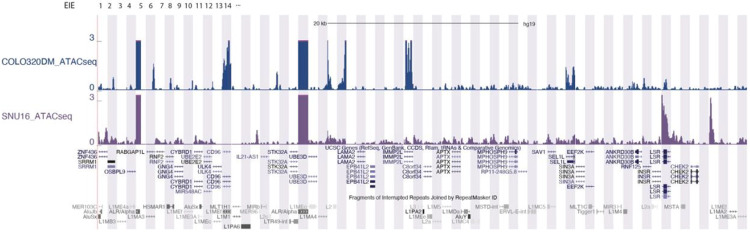
B. UCSC Genome Browser multi-region view showing the locations of the EIEs within the genome. Each EIE is indicated by a vertical bar. The browser displays the annotations for genes and repetitive elements such as Alu, LINE, and LTR elements (RepeatMasker), ATAC-seq and H3K27ac signal.
C. The growth phenotype of COLO320DM cells 4 days post-transduction, relative to non-targeting control (NTC). Each point represents the average guide effect (Z-score) for sgRNAs targeting a specific EIE, ranked by their impact on cell growth. EIE 14 is indicated by dashed rectangle with negative Z-score < −1 (significant negative impact on cell viability). See Extended Data for additional timepoints.
D. Zoom-in of EIE 14’s histone marks: enrichment of H3K27 acetylation, BRD4 binding, and ATAC-seq peaks. (H3K9me3 ChIP-seq is in Extended Data Fig. 5)
E. Luciferase enhancer assay schematics and fold change in luciferase signal driven by either MYC or TK promoter normalized to promoter-only construct. 4 biological replicates. EIE 14 compared to positive control (PVT1 positive control from22)
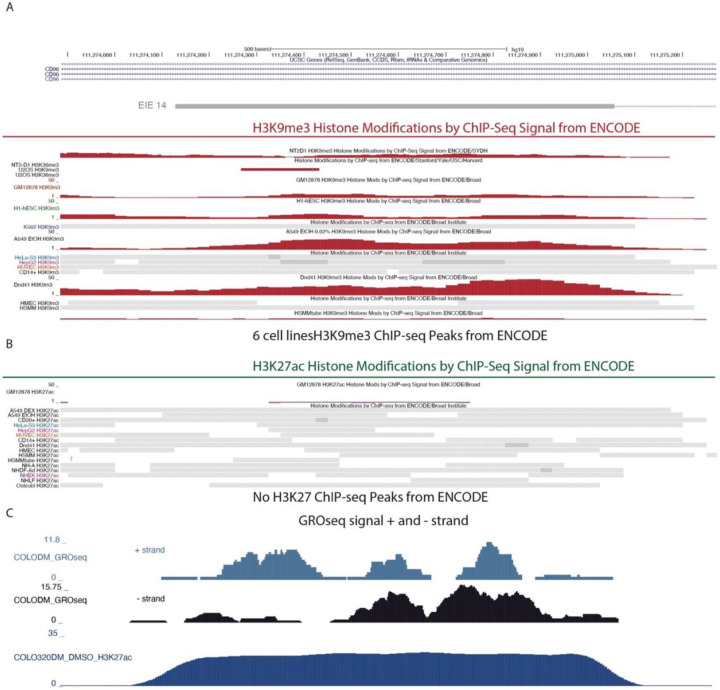
The strongest growth defect was observed in cells that silenced EIE 14 (Fig. 4C), which when combined with evidence that EIE 14 co-localized with ecDNA-amplified MYC, is suggestive of a potential enhancer-like regulatory role (Fig. 3H). To further analyze this locus, and other identified EIEs, we integrated existing genomic data measuring histone H3 lysine 27 acetylation (H3K27ac), a histone modification associated with active enhancers, and BRD4 occupancy, a key transcriptional regulator often present at active enhancers to examine potential enhancer signatures22,46-48 (Fig 4B) . We also utilized ATAC-seq to assay accessibility as enhancers tend to be highly accessible genomic regions49,50. Notably, not only were EIE 14 and others amplified in COLO320DM, they are also characterized by accessible chromatin and significant enrichment of H3K27ac and BRD4 occupancy (Fig. 4B, D). While the increased copy number of ecDNA can make it challenging to quantitatively interpret enrichment, these signatures of activation contrast the normally silenced H3 lysine 9 trimethylation (H3k9me3) state of EIE 14 across annotated human cell lines (Extended Data Fig. 5)51,52. Altogether, the accessibility and proximal clustering, combined with BRD4 and H3K27ac enrichment, points towards the active regulatory potential gained in COLO320DM cells once EIEs are ecDNA amplified (Fig 4D)49,50. Indeed, EIE 14 displays clear evidence of bi-directional transcription on both strands as shown by GRO-seq, a hallmark of active regulatory regions53,54 (Extended Data Fig. 5)
To directly test the ability for the EIE 14 sequence to act as an enhancer of MYC expression, we performed a luciferase assay measuring transcription activation of a TK promoter and MYC promoter22,55 (Fig. 4E). EIE 14 significantly increased MYC promoter-mediated reporter gene expression relative to the promoter only control, signifying bonafide enhancer activity (Fig. 4E), albeit to a lower extent than the positive control PVT1 enhancer sequence. Altogether, the enhancer-associated features and regulatory activity of the luciferase assay suggested that EIE 14, and possibly other EIEs, have been co-opted as regulatory sequences when found on ecDNA, influencing the expression of ecDNA-borne oncogenes.
Discussion
This study elucidates a novel mechanism by which transposable elements traditionally silenced by heterochromatin may gain oncogenic potential when amplified on ecDNA56-58. Somatic retrotransposons such as LINEs and SINEs are abundant in the human genome and are a major source of genetic variation59. Analysis of retrotransposon insertion across cancer types suggests a pervasive role in structural variation, implicated in various genomic rearrangements, copy number alterations, and mutations, including in colorectal cancer 60-67. The transposition of these elements in cancer can lead to genomic instability and potentially drive the gain of malignant traits. For example, when inserted into the APC tumor suppressor gene associated with colorectal tumors, reactivated LINE-1 disrupts the tumor suppressor and provides a fitness advantage to the cell68. In other cases, they are able to amplify oncogenic gene expression and directly promote oncogenesis by acting as bonafide transcriptional enhancers69. Here we describe enhancer-like activity of EIE 14 without an active retrotransposition, but rather the element presumably becomes active as a side effect of hitching a ride on ecDNA from a naturally occurring structural variation upstream of MYC. Because ecDNAs are randomly segregated in every cell division as a potent selection7, the co-amplification of the transposable elements identified in this study on extant ecDNAs indicates that they likely promote ecDNA fitness and function.
We demonstrated that retrotransposons such as EIE 14 escape the repressive chromatin state typically imposed on them in their native chromosomal context. This escape is facilitated by their location on ecDNA, an ectopic genomic compartment that is transcriptionally active and less subject to epigenetic silencing18. The presence of these EIEs on ecDNA may allow them to escape heterochromatin-mediated silencing and influence the expression of adjacent oncogenes such as MYC. As LINEs have demonstrated enhancer-like behavior when reactivated29,30,70, the spatial clustering of ecDNA molecules observed with ORCA may potentiate both cis- and trans-regulatory interactions of EIE 14 with oncogenic targets. Our findings highlight the need for caution when interpreting trans- interactions between chromosomal DNA and ecDNA as these may, in fact, represent incorporations of chromosomal elements into ecDNA. Therefore, additional assays like the long-read sequencing and imaging performed in this study should accompany Hi-C to confirm true trans- contacts.
This recontextualization highlights a dynamic interplay where normally silenced genomic loci can be 'reactivated' when found on ecDNA and acquire new functional roles driving oncogene expression. As EIE 14 is fragmented, containing a segment of L1M4a1 and LINE1 PA2, it would normally be unable to jump autonomously, however ecDNA (and the many structural variations associated with it), may allow for even the most divergent sequences to find the “right time and right place” for reactivation. Thus, revealing a new mechanism for inherited genetic variation to contribute to cancer development and progression. Previous studies of single nucleotide polymorphisms associated with familial cancer risk indicated that these variants impact the biochemical activity of noncoding enhancer elements linked to oncogenes that become activated in cancer71,72. Our results suggest that inherited variation of ancient TE insertion near oncogenes, such as EIE 14 near MYC, creates a latent enhancer that becomes activated if the oncogene locus becomes liberated as an ecDNA.
Perturbation of EIE 14 resulted in an impaired cell growth phenotype, suggesting that this particular reactivation may play a driving role in the colorectal cancer cell phenotype. While there is a strong growth defect upon CRISPRi inhibition of EIE 14 in COLO320DM cells, future work and analysis in in vivo patient samples will be necessary to determine if the presence of transposable elements on ecDNA is sufficient to drive a survival advantage or result in poor patient prognosis. However, the observation of recurrent LINE-1 on ecDNA in primary esophageal cancer provides in vivo evidence that this phenomenon is potentially clinically relevant73. Finally, amplification of retrotransposable elements onto ecDNA provides a mechanism for increased structural variation of ecDNA via the 40% of normally silenced repetitive regions of the genome. In fact, retrotranspositions are the second-most frequent type of structural variants identified in colorectal adenocarcinomas74. Transposons are classically recognized as a major driving force of plasmid evolution, via cycles of insertions and recombination in bacteria75. Our results suggest a convergent evolutionary tale in human oncogenic ecDNAs. The transcriptionally permissive state of ecDNA, beyond the normal confines of endogenous chromosomes, provides a landscape of activation where these elements can further enhance the activation and selection of oncogenes, making them both prognostic and therapeutic targets.
Methods
Cell culture
Cell lines were obtained from ATCC. COLO320-DM cells were maintained in RPMI; Life Technologies, Cat# 11875-119 supplemented with 10% fetal bovine serum (FBS; Hyclone, Cat# SH30396.03) and 1% penicillin-streptomycin (pen-strep; Thermo Fisher, Cat# 15140-122).
Hi-C
Ten million cells were fixed in 1% formaldehyde in aliquots of one million cells each for 10 minutes at room temperature and combined after fixation. We performed the Hi-C assay following a standard protocol to investigate chromatin interactions within colorectal cancer cells76. HiC libraries were sequenced on an Illumina HiSeq 4000 with paired-end 75 bp read lengths. Paired-end HiC reads were aligned to hg19 genome with the HiC- Pro pipeline77. Pipeline was set to default and set to assign reads to DpnII restriction fragments and filter for valid pairs. The data was then binned to generate raw contact maps which then underwent ICE normalization to remove biases. HiCCUPS function in Juicer78 was then used to call high confidence loops. Visualization was done using Juicebox https://aidenlab.org/juicebox/
Whole Genome Sequencing (WGS) with Oxford Nanopore
High-molecular weight (HMW) genomic DNA was extracted from approximately 6 million COLO320-DM cells using the Monarch HMW DNA Extraction Kit for Tissue (NEB #T3060L) following the Oxford Nanopore Ultra-Long DNA Sequencing Kit V14 protocol. After extracting HMW gDNA, we constructed Nanopore libraries using the Oxford Nanopore Ultra-Long DNA Sequencing Kit V14 (SQK-ULK114) kit according to manufacturer’s instructions. We sequenced libraries on an Oxford Nanopore PromethION using a 10.4.1. Flow Cell (FLO-PRO114M) according to manufacturer’s instructions. Basecalls from raw POD5 files were computed using Dorado (v.0.2.4).
Identification of element-specific structural variants from Nanopore data
We first identified Nanopore reads containing a single element by aligning reads with minimap279 and filtered out reads that were not mapped by the algorithm (denoted by “*” in the RNAME column of the BAM entry). Then, taking these reads we performed genomic alignment once again using minimap2 against hg19. From these new alignments of only the reads found to contain the element under consideration, we performed structural variant detection using Sniffles280. We repeated this procedure for each element individually.
Stable CRISPR cell line generation
The pHR-SFFV-dCas9-BFP-KRAB (Addgene, Cat# 46911) plasmid was modified to dCas9-BFP-KRAB-2A-Blast as previously described81. Lentivirus was produced using this vector plasmid. Cells were transduced with lentivirus, incubated for 2 days, selected with 1ug/ml blasticidin for 10–14 days, and BFP expression was analyzed by flow cytometry.
CRISPR interference
sgRNAs targeting elements were designed using the Broad Institute sgRNA designer online tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). The oligo pool encoding guides (Supplementary table T10) were synthesized by Twist Bio and inserted into addgene Plasmid #52963 lentiGuide-Puro digested with Esp3I enzyme (NEB). To evaluate the effects of CRISPR interference cells were transduced with sgRNA lentiviruses, incubated for 2 days and selected with 0.5ug/ml puromycin for 4 days. Cells were harvested after 4 days (baseline), 3 days, one week and one month. gRNA was amplified from the genome using two-step PCR and sequenced.
CRISPRi fitness screen analysis
To compute the effect of each guide on cell fitness, we first quantified guide counts from sequencing libraries. To normalize counts across libraries, we converted raw guide counts to counts-per-million (CPM) and retained guides that had CPM values of at least 20 across all days tested. After confirming that normalized guide abundances were robust across replicates, we proceeded with our analysis using the average of guide replicates at each time point. We next scored the relative fitness of each guide against the non-targeting controls (NTC) by computing the ratio of CPM values between a guide and the NTC at the particular time point. Finally, we transformed this distribution to z-scores and reported this as the relative fitness effect of each guide.
CRISPR-CATCH
CRISPR-CATCH was performed according to standard procedure21 using following sgRNA and marker: sgRNA #1 ATATAGGACAGTATCAAGTA; sgRNA #2 TATATTATTAGTCTGCTGAA; S. cerevisiae ladder, H. wingei ladder.
Probe Design
Probes were designed against human genome assembly hg19, tiling the regions in supplemental table T7 using the probe designing software described previously27,36. We restricted choice of the 40mer targeting region of the probes to a GC range of 20-80%, a melting temperature of 65-90 degrees centigrade, and excluded sequences with non-unique homology (cut off of 17mer homology to any other sequence in the genome) or with homology to common repetitive elements in the human genome listed in repbase (cut off of 14mer). Targeting probes were then appended with a 20mer barcode per target region. Probe design software is available at https://github.com/BoettigerLab/ORCA-public. Finalized probe libraries were ordered as an oligo-pool from Genscript.
ORCA imaging
ORCA hybridization was performed as previously described27,36. Briefly, 40mm Bioptechs coverslips were prepared with EMD Millipore™ Poly-D-Lysine Solution (1 mg/mL, 20mL, dilute 1:10)(Sigma, cat. No. A003E) for 40 minutes. Coverslips were then rinsed 3x in 1x PBS. Cells were passaged onto the coverslips and allowed to adhere overnight. The next day, the coverslip with cells were rinsed 3 times in 1x PBS and then fixed for 10 minutes in 4% PFA. Cells were then permeabilized in 0.5% Triton-x 1x PBS for 10 minutes followed by 5 minutes of denaturing in 0.1M HCL. A 35-minute incubation in hybridization buffer prepared samples for primary probe. Primary probes were added (1ug) directly to the sample in hybridization solution and then the sample was heated to 90 degrees celsius for 3 minutes. An overnight 42-degree incubation (or at least 8 hour incubation) was performed followed by post-fixation in 8% PFA + 2% glutaraldehyde in 1× PBS before being stored in 2x SSC or used immediately for imaging.
Samples were imaged on one of two different homebuilt setups designed for ORCA, “scope-1”, “scope-3”, depending on instrument availability. Microscope design parameters were deposited in the Micro-Meta App82. The design and assembly of the “scope-1” system is described in detail in our prior protocol paper36. Both systems use a similar auto-focus system, fluidics system, and sCMOS camera (Hamamatsu FLASH 4.0), though scope-3 had a larger field of view (2048x2048 108 nm pixels) compared to scope-1 (1024x1024 154 nm pixels).
Automated fluidics handling is described in detail in our prior protocol paper36. Briefly, fluid exchange between each imaging step was performed by a homebuilt robotic setup. The system used a 3-axis CNC router engraver, buffer reservoirs and hybridization wells (96-well plate) on the 3-axis stage, ETFE tubing, imaging chamber (FCS2, Bioptechs), a needle, and peristaltic pump (Gilson F155006). The needle was moved between buffers or hybridization wells and was flown across the samples through tubing using the peristaltic pump. Open-source software for the control of the fluidics system is described in the “Software Availability” section below.
Sequential imaging of ORCA probes was conducted alternating between hybridization of fluorescent readout probes, imaging, and stripping of probes, as described previously27,36. Briefly, a z-stack was acquired over 10um at 250nm step size where each step alternated lasers between data channel and fiducial. Readout probes were labeled with Alexa-750 fluorophores. Fiducial probe was labeled in cy3 and added only in the initial round.
Image processing
Image processing was performed with custom MATLAB functions available: https://github.com/BoettigerLab/ORCA-public. Briefly, cells were max projected and pixel-scale alignment was computed across all fields of view off of the fiducial signal. This alignment was then applied in 3D across all 250 nm z steps. Cellpose83 was then used to segment individual cells. A cell-by-cell fine scale (subpixel) alignment was then computed and aligned individual cells were then ready for 3D-spot calling. The individual ecDNA spots and their 3D positions computed to sub-pixel accuracy using the corresponding raw 3D image stacks and the 3D DaoSTORM function in storm-analysis toolbox [DOI: 10.5281/zenodo.3528330] an open source software for single-molecule localization, adapted for dense and overlapping emitters following the DaoSTORM algorithm84. DaoSTORM was run in the 2d-fixed mode, as the 3D fitting modes are for estimating axial position from astigmatism in the xy plane, rather than computing it directly from a z-stack. The fixed-width PSF of the microscope is pre-computed using 100 nm (sub-diffraction) fluorescent beads. A minimum detection threshold of 30 sigma was used for the fit. The z-position of the localizations was computed using Gaussian fit to the vertically stacked localizations, with an axial Gaussian width also pre-computed from z-stack images with 100 nm fluorescent beads. Additional information can be found in the read-the-docs for storm-analysis: https://storm-analysis.readthedocs.io/en/latest/.
Minimum pairwise distance quantification
All pairwise distances between genomic regions were calculated on a per-cell basis. The shortest distances were saved for each MYC centroid and EIE 14 and PVT1 such that each MYC centroid has one corresponding shortest distance per EIE 14 and PVT1. For each cell, a sphere radius r=4um (the average radius of cells calculated with Cellpose mask) with randomly simulated points corresponding to the number of MYC, EIE 14, and PVT1 centroids. The same minimum pairwise distance quantification was calculated on the randomly simulated points.
Ripley’s K quantification
To calculate the density corrected distance ratios a distance cutoff of 2um and an interval density of 0.01:0.01:2 was used. The spatial relationship between MYC and EIE 14 and MYC and PVT1 were quantified as follows: On a per-cell basis the distance density function was calculated, truncated at the specified cutoff. A uniform distribution was then computed over the same interval and a ratio of these values was taken. This ratio was then corrected by the volume of the interval shell.
Reporter plasmid construction and transfection
All plasmids are made with Gibson assembly (NEB HIFI DNA assembly kit) according to manufacturer’s protocol. We used a plasmid from22 containing the MYC promoter (chr8:128,745,990-128,748,526, hg19) driving NanoLuc luciferase (PVT1p-nLuc) and a constitutive thymidine kinase (TK) promoter driving Firefly luciferase, this plasmid was used as negative control. pGL4-tk-luc2 (Promega) constructing plasmids with a cis-enhancer, an enhancer (chr8:128347148–128348310) was used as positive control22. In the test plasmid, the cis-enhancer was replaced by 1.7 kb sequence of EIE 14: TAAATAAATGGTAAGCTATATATGTATACATGTGCCGTGCTGGTGCGCTGCACCCAC TAACTCGTCATCTAGCATTAGGTATATCTCCCAATGCTATCCCTCCCCCCTCCCCCCA CCCCACAACAGTCCCCAGAGTGTGATATTCCCCTTCCTGTGTCCATGTGATCTCATTG TTCAATTCCCACCTATGAGTGAGAATATGCGGTGTTTGGTTTTTTGTTCTTGCGATAG TTTACTGAGAATGATGATTTCCAATTTCATCCATGTCCCTACAAAGGACGTGAACTC ATCATTTTTATGGCTGCATAGTATTCCACGGTGTATATATTCCACATTTTCTTAATCC AGTCTATCATTGTTGGACATTTGGGTTGGTTCCAAGTCTTTGCTATTGTGAATAATGC CGCAATAAACATATGTGTGCATGTGTCCTTATAGCAGCATGATTTATGGTCATGTGG GTATATACCCAGTAATGGGATGGCTGGGTCAAATGGTATTTCTAGTTCTAGATCCCT GAGTAATCGCCACACTGACTTCCACAATGGTTGAACTAGTTTACATTCCCACCAACA GTGTAAAAGTGTTCCTGTTTCTCCACATCCTCTCCAGCACCTGTTGTTTCCTGACTTT GTAATGATTGCCATTCTAACTAGTGTGAGATGGTATCTCATAGTGGTTTTGATTTGTA TTTCTCTGATGGCCAGTGATGATAAAAAAAAAGAAGTTGTTATTAGTCTATTCAAAG TATTAAAGCAAAATATGACAATGACTCAATAAATAGGAAATGTTAGTAGAGAAATA GAAAGCTATAAAACAAAGCAAATGTATATTTTAGAGTTGAAATGTCAGTAACAAAA ATTAGAAATTTACTAGATGTTCTCAATAGCAAATTTGAGATGGCTGAAGAAAGAATT AGTGTATTTGAACATTGTTCAATATAAATTATCTAATCTTAAGGGAGAAAAAGGATT GAAAGAAATGAAAACCACTTCAGAGCCATATAGGACAGTATCAAGTATGGTAACAT ACATGAAACAGGAGTAGTAGAAAAAGAAGTGAAAGAGTAAGGGGATGGATCAAAT ATTTGAAGAAAAAATGGCCAAAAACTTCACATATTTGATTATTTAAAAACTTTCTTA AAATTAATCTACACATCCAAGAACTTTAACAAAACCTGAATAGGACAAACACAAAG AGACACCCATAATCAAACTTTTGAAAGCCAAAAGACACATCATAATCAAACTTCTCG AAGCCAAAAAGAAAGACTAAATTATAAAAGTAGCAATTGAAAAGACAAAAACCAA CCAACCAAACAAACAAAAACAAAAGCTCATCCCATTCAGCAGACTAATAATATAAC TAGTGGCTCATTTTAATCAGAAATAATGGCGATCGAAAGATATATTCAAAATGCTAA AAGAAAGAGAAACAATCCACTCTGAATTCTATATCCATTGAAATTATCCTTTAAAAT TAAAAATGAAAAAATCTGAAAGAATTCATTGCAAGTAGATTTGTCTTACACGAAAT ACTAAAGTCCTTCAGGCTGAAAAGAAATGACAACAAACAGTAACTCCAATTCATGG GATAAAATAAAGAGCACAAAAAATGGTAAATACGTGAGTAAATATGAAAAATTATA TATGTAGTCTTCATATGTGAGTAAATAAAAACTATACATACAAAAAAAATAAAAAA To assess luciferase reporter expression, COLO320-DM cells were seeded into a 24-well plate with 100,000 cells per well. Reporter plasmids were transfected into cells the next day with lipofectamine 3000 following the manufacturer’s protocol, using 0.25 μg DNA per well. Luciferase levels were quantified using Nano-Glo Dual reporter luciferase assay (Promega).
Extended Data
Extended Data Fig. 1:
A. The graph (left) demonstrates the number of structural variations called in stripe alignments. Relationship between structural variations and read count for each element (right). Pearson correlation is 0.61.
B. Schematics of ecDNA harboring 1.7 kb sequence obtained from long-read analysis of EIE 14. The region spanning 6-710 bp shows alignments with 3’ end of the LINE-1 element, whereas the region from 711-1690 bp is notably unique to intron 2 of the CD96 locus on chromosome 3.
C. Top panel, alignment of predicted protein from 6-710 bp with LINE-1 ORF2. Bottom panel, amino acids alignment of LINE-1 ORF2 and 6-710 bp coding protein by clustalW. Google doc
Extended Data Fig. 2:
A. ORCA (Optical Reconstruction of Chromatin Architecture) visualization of the COLO320DM cell nucleus. The images show the spatial arrangement of the MYC oncogene, Element 14 and the PVT1 locus, labeled in different colors. The scale bar represents 5 micrometers. Chr3 probe maps to the breakpoints of the EIE 14 origin inside CD96 intron.
B. EIE 14 position and structural variant “insertion” on Chr8 between CASC8 and CASC11, full sequence is listed in the methods part for the luciferase enhancer assay. The full SV list is in Supplementary Tables T4 and T5.
C. Sequence alignment of the T2T genome.
D. Screenshot of the IGV viewer with selected long reads depicting insertion sizes in purple.
Extended Data Fig. 3:
A. Quantification of copy number of MYC, PVT1 and EIE 14 across all measured cells (n=1329). Mean copy number of MYC is 29 copies per cell, PVT1 is 31 copies per cell and EIE 14 is 22 copies per cell. Copies for all species ranged from 0 to 150 copies.
B. Correlation plots between the loci per cell. Pearson’s correlation coefficient calculated for PVT1-MYC r=0.82, EIE 14-MYC r=0.71, EIE 14-PVT1 r=0.74.
C. Violin plots of shortest distances of MYC to PVT1 and EIE 14 (median distance denoted by red line). Red line denotes median distance. C. Histogram of shortest distances of MYC to PVT1 (blue) and MYC to EIE 14 (orange) (Wilcoxon two-sided ranksum p=1.23e^-05).
Extended Data Fig. 4:
A. Schematic of the CRISPRi screening strategy used to evaluate the regulatory potential of 257 genomic elements near the MYC oncogene in COLO320DM cells. For each element, 5-6 sgRNAs were designed and 125 non-targeting control sgRNAs. The screen involved the transduction of cells with a lentivirus expressing dCas9-KRAB and the sgRNAs, followed by calculation of cell growth phenotype over a series of time points (4 days (baseline), 3 days, 14 days, and 1 month).
B. The growth phenotype of COLO320DM cells and reproducibility of counts between two biological replicates at different timepoints. Each point represents the average guide effect (Z-score)
Extended Data Fig. 5:
Zoom-in on EIE14 in UCSC genome browser. From top to down:ENCODE Histone modifications, H3K9me3 in red and H3K27ac in green ChIP-Seq Signal and called Peaks. H3K27ac modification is absent in all cell lines in ENCODE51,52, GRO-Seq54 COLO320DM cell line plus strand and minus strand, H3K27ac in COLO320DM.
Extended Data Fig. 6:
Comparison in chromatin accessibility between COLO320DM (top) and SNU16 (bottom) ATAC-seq signal22 displayed in UCSC browser.
Supplementary Material
Acknowledgements
This project was supported by Cancer Grand Challenges CGCSDF-2021\100007 with support from Cancer Research UK and the National Cancer Institute (H.Y.C., P.S.M.) and NSF grant EF2022182 (A.N.B). M.G.J. is supported by NIH K99CA286968. S.E.M. was supported by a Stanford Bio-X SIGF Fellowship. K.L.H. was supported by a Stanford Graduate Fellowship and an NCI Predoctoral to Postdoctoral Fellow Transition Award (NIH F99CA274692). H.Y.C. is an Investigator of the Howard Hughes Medical Institute. We thank Mervinaz Koska for help with luciferase measurement. We thank Michael Montgomery for feedback on the manuscript.
Footnotes
Competing Interests
H.Y.C. is a co-founder of Accent Therapeutics, Boundless Bio, Cartography Biosciences, Orbital Therapeutics, and an advisor of 10x Genomics, Arsenal Biosciences, Chroma Medicine, Exai Bio and Spring Discovery. The remaining authors declare no competing interests. P.S.M. is a co-founder and advisor of Boundless Bio. J.D.B. is a founder and director of CDI Labs, Inc.; a founder of and consultant to Opentrons LabWorks/Neochromosome, Inc.; and serves or served on the scientific advisory boards of the following: CZ Biohub New York, LLC; Logomix, Inc.; Modern Meadow, Inc.; Rome Therapeutics, Inc.; Sangamo, Inc.; Tessera Therapeutics, Inc.; and the Wyss Institute. The remaining authors declare no competing interests.
Data availability
All sequencing data generated in this study will be available through the Gene Expression Omnibus (GEO), Accession codes will be available before publication. All raw imaging data is available upon request.
References:
- 1.Yi E., Chamorro Gonzalez R., Henssen A.G. & Verhaak R.G.W., Extrachromosomal DNA amplifications in cancer. Nat Rev Genet 23, 760–771 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan X., Mischel P. & Chang H. Extrachromosomal DNA in cancer. Nat Rev Cancer 24, 261–273 (2024). [DOI] [PubMed] [Google Scholar]
- 3.Kim H. et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet 52, 891–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahl G.M. The importance of circular DNA in mammalian gene amplification. Cancer Res 49, 1333–40 (1989). [PubMed] [Google Scholar]
- 5.Benner S.E., Wahl G.M. & Von Hoff D.D. Double minute chromosomes and homogeneously staining regions in tumors taken directly from patients versus in human tumor cell lines. Anticancer Drugs 2, 11–25 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Luebeck J. et al. Extrachromosomal DNA in the cancerous transformation of Barrett's oesophagus. Nature 616, 798–805 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange J.T. et al. The evolutionary dynamics of extrachromosomal DNA in human cancers. Nat Genet 54, 1527–1533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner K.M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.deCarvalho A.C. et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 50, 708–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abeysinghe H.R., Cedrone E., Tyan T., Xu J. & Wang N., Amplification of C-MYC as the origin of the homogeneous staining region in ovarian carcinoma detected by micro-FISH. Cancer Genet Cytogenet 114, 136–43 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Van Roy N. et al. Translocation-excision-deletion-amplification mechanism leading to nonsyntenic coamplification of MYC and ATBF1. Genes Chromosomes Cancer 45, 107–17 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Ly P. et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat Genet 51, 705–715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nones K. et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun 5, 5224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rausch T. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosswog C. et al. Chromothripsis followed by circular recombination drives oncogene amplification in human cancer. Nat Genet 53, 1673–1685 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Shoshani O. et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 591, 137–141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gisselsson D. et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci U S A 97, 5357–62 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y. et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell 39, 694–707 e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helmsauer K. et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun 11, 5823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung K.L. et al. Targeted profiling of human extrachromosomal DNA by CRISPR-CATCH. Nat Genet 54, 1746–1754 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung K.L. et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature 600, 731–736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung K.L. et al. Coordinated inheritance of extrachromosomal DNA species in human cancer cells. bioRxiv (2023). [Google Scholar]
- 24.Quinn L.A., Moore G.E., Morgan R.T. & Woods L.K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res 39, 4914–24 (1979). [PubMed] [Google Scholar]
- 25.Trainer D.L. et al. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer 41, 287–96 (1988). [DOI] [PubMed] [Google Scholar]
- 26.Belton J.M. et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateo L.J. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X. et al. LINE-1 transcription activates long-range gene expression. Nat Genet (2024). [DOI] [PubMed] [Google Scholar]
- 30.Sundaram V. & Wysocka J. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos Trans R Soc Lond B Biol Sci 375, 20190347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldwin E.T. et al. Structures, functions and adaptations of the human LINE-1 ORF2 protein. Nature 626, 194–206 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adney E.M. et al. Comprehensive Scanning Mutagenesis of Human Retrotransposon LINE-1 Identifies Motifs Essential for Function. Genetics 213, 1401–1414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshpande V. et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun 10, 392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altemose N. et al. Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abel H.J. et al. Mapping and characterization of structural variation in 17,795 human genomes. Nature 583, 83–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo L.J., Sinnott-Armstrong N. & Boettiger A.N. Tracing DNA paths and RNA profiles in cultured cells and tissues with ORCA. Nat Protoc 16, 1647–1713 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J. & Pertsinidis A. New insights into promoter-enhancer communication mechanisms revealed by dynamic single-molecule imaging. Biochem Soc Trans 49, 1299–1309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim B. & Levine M.S. Enhancer-promoter communication: hubs or loops? Curr Opin Genet Dev 67, 5–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander J.M. et al. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. Elife 8(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benabdallah N.S. et al. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol Cell 76, 473–484 e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lancho O. & Herranz D. The MYC Enhancer-ome: Long-Range Transcriptional Regulation of MYC in Cancer. Trends Cancer 4, 810–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerman M.W. et al. MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov 8, 320–335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larson M.H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8, 2180–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penttinen R.P. Biosynthesis, secretion and crosslinking of collagen with reference to aging. Scand J Soc Med Suppl 14, 56–68 (1977). [PubMed] [Google Scholar]
- 45.Hongxing Z. et al. Depletion of c-Myc inhibits human colon cancer colo 320 cells' growth. Cancer Biother Radiopharm 23, 229–37 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Barral A. & Dejardin J. The chromatin signatures of enhancers and their dynamic regulation. Nucleus 14, 2160551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Creyghton M.P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B. et al. BRD4-directed super-enhancer organization of transcription repression programs links to chemotherapeutic efficacy in breast cancer. Proc Natl Acad Sci U S A 119(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulco C.P. et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y. & Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res 48, D882–D889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young R.S., Kumar Y., Bickmore W.A. & Taylor M.S. Bidirectional transcription initiation marks accessible chromatin and is not specific to enhancers. Genome Biol 18, 242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jun Tang N.E.W., Guiping Wang. Rampant transcription replication conflict creates therapeutic vulnerability in extrachromosomal DNA containing cancers. (2024). [Google Scholar]
- 55.Long H.K. et al. Loss of Extreme Long-Range Enhancers in Human Neural Crest Drives a Craniofacial Disorder. Cell Stem Cell 27, 765–783 e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robbez-Masson L. et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res 28, 836–845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castro-Diaz N. et al. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28, 1397–409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu N. et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lander E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Helman E. et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 24, 1053–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beck C.R., Garcia-Perez J.L., Badge R.M. & Moran J.V., LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12, 187–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ardeljan D. et al. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat Struct Mol Biol 27, 168–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott E.C. et al. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res 26, 745–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cajuso T. et al. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat Commun 10, 4022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Payer L.M. & Burns K.H. Transposable elements in human genetic disease. Nat Rev Genet 20, 760–772 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Iskow R.C. et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKerrow W. et al. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proc Natl Acad Sci U S A 119(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miki Y. et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52, 643–5 (1992). [PubMed] [Google Scholar]
- 69.Deniz O. et al. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat Commun 11, 3506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuentes D.R., Swigut T. & Wysocka J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corces M.R. et al. The chromatin accessibility landscape of primary human cancers. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taipale J. The chromatin of cancer. Science 362, 401–402 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Ng A.W.T. et al. Disentangling oncogenic amplicons in esophageal adenocarcinoma. Nat Commun 15, 4074 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodriguez-Martin B. et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet 52, 306–319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen S.N. Transposable genetic elements and plasmid evolution. Nature 263, 731–8 (1976). [DOI] [PubMed] [Google Scholar]
- 76.Rao S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Servant N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durand N.C. et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smolka M. et al. Detection of mosaic and population-level structural variants with Sniffles2. Nat Biotechnol (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho S.W. et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 173, 1398–1412 e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rigano A. et al. Micro-Meta App: an interactive tool for collecting microscopy metadata based on community specifications. Nat Methods 18, 1489–1495 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stringer C., Wang T., Michaelos M. & Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Holden S.J., Uphoff S. & Kapanidis A.N. DAOSTORM: an algorithm for high- density super-resolution microscopy. Nat Methods 8, 279–80 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated in this study will be available through the Gene Expression Omnibus (GEO), Accession codes will be available before publication. All raw imaging data is available upon request.