Abstract
Most of our understanding of the fundamental processes of mutation and recombination stems from a handful of disparate model organisms and pedigree studies of mammals, with little known about other vertebrates. To gain a broader comparative perspective, we focused on the zebra finch (Taeniopygia castanotis), which, like other birds, differs from mammals in its karyotype (which includes many micro-chromosomes), in the mechanism by which recombination is directed to the genome, and in aspects of ontogenesis. We collected genome sequences from three generation pedigrees that provide information about 80 meioses, inferring 202 single-point de novo mutations, 1,174 crossovers, and 275 non-crossovers. On that basis, we estimated a sex-averaged mutation rate of 5.0 × 10−9 per base pair per generation, on par with mammals that have a similar generation time. Also as in mammals, we found a paternal germline mutation bias at later stages of gametogenesis (of 1.7 to 1) but no discernible difference between sexes in early development. We also examined recombination patterns, and found that the sex-averaged crossover rate on macro-chromosomes (1.05 cM/Mb) is again similar to values observed in mammals, as is the spatial distribution of crossovers, with a pronounced enrichment near telomeres. In contrast, non-crossover rates are more uniformly distributed. On micro-chromosomes, sex-averaged crossover rates are substantially higher (4.21 cM/Mb), as expected from crossover homeostasis, and both crossover and non-crossover events are more uniformly distributed. At a finer scale, recombination events overlap CpG islands more often than expected by chance, as expected in the absence of PRDM9. Despite differences in the mechanism by which recombination events are specified and the presence of many micro-chromosomes, estimates of the degree of GC-biased gene conversion (59%), the mean non-crossover conversion tract length (~23 bp), and the non-crossover to crossover ratio (6.7:1) are all comparable to those reported in primates and mice. The conservation of mutation and recombination properties from zebra finch to mammals suggest that these processes have evolved under stabilizing selection.
Introduction
Germline mutation and meiotic recombination are fundamental biological processes and the sources of heritable variation. The rates at which they occur are key parameters in evolutionary models and enable phylogenetic dating. Yet their properties have been studied in depth in only a handful of model organisms–primarily yeast species, mice, Arabidopsis thaliana, and a couple of Drosophila species (e.g., Lang & Murray, 2008; Mancera et al., 2008; Comeron et al., 2012; Drouaud et al., 2013; Keightley et al., 2014; Li et al., 2019). More recently, such studies have been complemented by sequencing pedigrees, primarily of mammals (e.g., Williams et al., 2015; Li et al., 2019; Wall et al., 2022; Versoza et al., 2024; Porubsky et al., 2024), as well as by high-fidelity (HiFi) long-read sequencing of male germ cells of humans and other primates (Charmouh et al., 2024; Porsborg et al., 2024; Schweiger et al., 2024).
To date, few similar studies have been conducted in non-mammalian vertebrates. For example, among ~11,000 species of birds, recombination events have only been called from genome sequencing of pedigrees in the collared flycatcher (Smeds et al., 2016a) and the great weed warbler (Zhang et al., 2024). Pedigree studies of germline mutation have been conducted in a somewhat larger number of species, including the collared flycatcher (Smeds et al., 2016b), the great reed warbler (Zhang et al., 2023), as well as 18 other species (Bergeron et al., 2023), but inferences were limited by the small number of trios considered per species (from one to eight, with a median of one trio).
Intriguingly, comparisons among mammalian species indicate that mutation and recombination parameters are relatively stable over time: for instance, the mutation rate per base pair (bp) per generation in mice (0.5×10−8; Uchimura et al., 2015; Milholland et al., 2017; Lindsay et al., 2019) is only around two times lower than that in humans (1.2 × 10−8, e.g., Kong et al., 2012; Rahbari et al., 2015) despite their generation time being around 50 times shorter (Milholland et al., 2017), and the sex-averaged recombination rate in dogs (0.8 cM/Mb) is intermediate between the rates in humans (1.2 cM/Mb) and mice (0.5 cM/Mb), despite dogs having nearly twice as many chromosomes and a shorter genome than that of humans (Dumont & Payseur, 2008; Williams et al., 2015; Campbell et al., 2016). In the soma, in turn, the mutation burden in colonic crypts appears relatively constant across 16 mammalian species at their typical lifespan (i.e., on average, a mammalian epithelial cell carries around 3,000 de novo mutations at the time of death) (Cagan et al., 2022). Such findings suggest that aspects of mutation and recombination are evolving under stabilizing selection across mammals. It remains an open question, however, whether this conservation extends to broader phylogenetic distances.
Birds offer an interesting comparison in this regard, as they diverged from mammals 320 million years ago (Kumar et al., 2022) and differ in many potentially salient features. For one, most passerines are seasonal breeders with spermatogenesis occurring during the breeding season, unlike many mammals (e.g., mice and humans), which typically produce sperm continuously (Bentley et al., 2000; Wikelski et al., 2003). Moreover, sex differences may appear earlier in bird ontogenesis, given that the avian sexual phenotype is directly determined by the sex chromosome content of individual cells (Zhao et al., 2010; Ioannidis et al., 2021), consistent with reported sex differences in primordial germ cell phenotypes before gonadal development (Soler et al., 2021). These features may help explain the weaker paternal bias in germline mutation in birds compared to mammals (de Manuel et al., 2022). More generally, it is unclear if the mutational processes dominant in the mammalian germline, notably those accounted for by COSMIC mutational signatures SBS1 and SBS5 (Rahbari et al., 2015; Sasani et al., 2024; Spisak et al., 2024), are also active in birds.
Recombination dynamics may also differ between birds and mammals. Notably, avian genomes often harbor a large number of micro-chromosomes, which have higher average crossover rates, replicate earlier and have a higher repeat content (Srikulnath et al., 2021; Waters et al., 2021). Further, birds lack the gene PRDM9, which encodes the protein directing recombination in several mammals (e.g., Baudat et al., 2010; Myers et al., 2010; Parvanov et al., 2010) and other vertebrates (Schield et al., 2020; Hoge et al., 2024; Raynaud et al., 2024), but was lost in archosaurs (Baker et al., 2017). In rodent knockouts for PRDM9 and dogs that carry a pseudogene for PRDM9, recombination preferentially occurs at promoter-like features, in particular CpG islands (Brick et al., 2012; Auton et al., 2013; Mihola et al., 2021). Analyses of patterns of linkage disequilibrium (LD) suggest that in birds as well, population recombination rates are elevated near CpG islands (Smeds et al 2016a, Singhal et al., 2015; Kawakami et al., 2017). However, LD patterns mostly reflect the effects of crossovers, and carry limited information about non-crossover events (Hellenthal & Stephens, 2006). To our knowledge, there is only one study of non-crossover events in birds, in collared flycatcher (Smeds et al., 2016a). On that basis, it remains unclear if non-crossovers are also concentrated at promoter-like features. At a broad scale, the number of double-strand breaks per meiosis and the ratio of crossover to non-crossover in birds is also unknown.
In addition to crossover and non-crossover rates, other key parameters, such as the non-crossover mean conversion tract length, appear to be conserved between mice and primates (<100 bp; Li et al., 2019; Wall et al., 2022; Charmouh et al., 2024; Porsborg et al., 2024). Yet this length is substantially shorter than that in Drosophila melanogaster or in yeast (Saccharomyces cerevisiae), estimated to be ~500 bps and 1.8 kb, respectively (Mancera et al., 2008; Comeron et al., 2012). Therefore, it is unclear how far the similarity observed in mammals extends phylogenetically. In turn, estimates of magnitude of the GC-biased gene conversion in mammals vary between 57% and 68% (Williams et al., 2015; Li et al., 2019; Wall et al., 2022; Versoza et al., 2024). The extent of bias may depend on nucleotide diversity levels, as a recent study reported that only non-crossovers with a single mismatch in the gene conversion tract experience a transmission bias in mice and potentially humans (Li et al., 2019). In collared flycatcher, in which heterozygosity is ~0.4% (Dutoit et al., 2017), the authors reported a point estimate of 59% based on 229 non-crossover events (Smeds et al., 2016a). Whether the same is true in a species with higher diversity remains to be tested.
To explore whether fundamental mutation and recombination parameters are similar in birds, we focused on the zebra finch, a well-studied passerine with a contiguous and high-quality genome assembly (Rhie et al., 2021) and high levels of nucleotide diversity (~1%) (Balakrishnan & Edwards,2009). To this end, we sequenced genomes of extended pedigrees to identify de novo point mutations (DNMs), as well as infer crossovers and non-crossovers.
Results
Germline de novo mutation
Identification of sex-specific de novo mutations from pedigrees
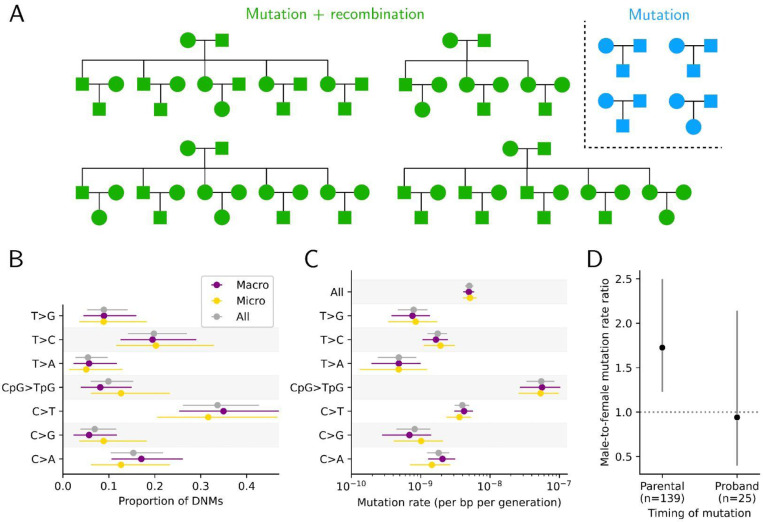
We generated whole genome sequences from 74 zebra finch individuals in four three-generation pedigrees and four trios, comprising 40 trios, on average at 25-fold coverage (Fig 1A and S1 Table). After mapping the reads of each individual to the zebra finch reference genome (RefSeq ID: GCF_003957565.2), we identified autosomal regions in the assembly to which short-read sequencing data could be reliably mapped and where the three individuals in the focal trio had sufficient but not unusually high depth of coverage (see Methods). This approach led us to retain an average of 502.8 Mb per trio (min=475.3, max=511.1), or 52% of the autosomal genome. We then identified genomic positions where the parents are homozygous for the same reference allele and the offspring heterozygous, a Mendelian violation consistent with a DNM. We filtered these candidate DNMs using current best practices, notably by checking that the non-reference allele is not present on the sequencing reads in either parent (see Methods). In the end, we identified 202 putative DNMs in the 40 probands (Fig 1).
Fig 1. Pedigree structure and mutation rate estimates in zebra finch.
(A) Familial relationships among the 74 sequenced zebra finch individuals. We identified DNMs in all families (blue and green), and studied recombination events in the nuclear families with multiple siblings (green) (see Methods). (B) Proportion of DNMs for the seven mutation types, in all autosomal sequences (gray), macro-chromosomes (purple), and micro-chromosomes (gold). (C) Mutation rate estimates for the seven mutation types and all mutations (top). Horizontal lines show the 95% CIs, assuming mutation counts are Poisson-distributed. (D) The ratio of male-to-female mutations for events occurring in the parental germlines or in early development of the proband. Horizontal lines show the binomial 95% CIs.
To determine the parental chromosome on which the DNMs occurred, we employed two different strategies. For the 88 DNMs identified in the 18 probands with sequenced partners and offspring, we phased 91% of mutations based on their pattern of inheritance to the next generation (see Methods). For the 114 DNMs identified in the 22 probands without sequenced offspring, we phased by read tracing, i.e., by linking DNMs to phase informative heterozygous alleles found in the same sequenced DNA fragment (see Methods). Given the high genetic diversity in zebra finches (π ≈1%), we were able to phase 81% of the 114 DNMs by this strategy. Applying both approaches to probands with sequenced offspring, 94.3% of the DNMs are assigned to the same parental chromosome as inferred by transmission, confirming the reliability of the phasing.
Inferences of developmental timing of germline mutations
Mutations that occurred in the early development of the probands can be mistaken for mutations that occurred in the parental germlines. To distinguish among these two possibilities, we looked for DNMs that showed “incomplete linkage” with neighboring heterozygous alleles, i.e., regions in which the (diploid) proband carries three distinct haplotypes (see Methods and S1 Fig for a visual example) (Harland et al., 2017). Among the 174 DNMs with informative variants nearby (~150 bps, the length of our sequencing reads), 25 showed evidence of being post-zygotic, i.e., as having arisen after fertilization of the proband (see Methods). The fraction of post-zygotic mutations in zebra finches (~15%) is larger than that reported in humans (~5%) (p < 3 × 10−5 by a one-sided binomial test), consistent with previous studies showing larger proportions in shorter lived organisms like mice (Lindsay et al., 2019) and cattle (Harland et al., 2017). In addition, these post-zygotic mutations are at lower than 50% frequency among the sequencing reads in the proband (p < 10−7 by one-tailed Wilcoxon signed-rank test; S2 Fig), and as expected for mosaic mutations, have a transmission rate to the next generation significantly lower than 50% (21%, 95% CI: 7% − 47%). In contrast, the transmission rate for the rest of DNMs is 45% (95% CI: 33% − 55%), not significantly different from the 50% expected for a constitutive mutation. Moreover, the 25 putative post-zygotic mutations occur at a similar rate on both parental chromosomes: the ratio of paternally to maternally phased mutations is 1.14 (95% CI: 0.33 − 2.34), again as expected if they occurred after fertilization of the proband. Collectively, these lines of evidence—under-transmission, lower allele frequency compared to constitutive heterozygous variants, and equal occurrence in both parental chromosomes—indicate this set of mutations indeed arose post-zygotically and are mosaic in the proband.
If a DNM occurs during or shortly after parental primordial germ cell specification, it may not be present in parental somatic cells but could be carried by a significant proportion of their gametes, increasing the likelihood of inheritance by multiple descendants. By analyzing the four multi-sibling pedigrees, we found only one instance of DNMs shared between siblings (S2 Table). While there are only a small number of siblings, finding only one such DNM suggests that the mutation rate during this early developmental stage is relatively low in zebra finches.
Estimation of mutation rates
Given the total length of genome sequence that we analyzed across the 40 pedigrees (~40 Gbs) and the number of DNMs identified (202) (see Methods), the point mutation rate is 5.0 × 10−9 (95% CI: 4.3 × 10−9 − 5.7 × 10−9; Fig 1C) per bp per generation. Dividing by the average parental age for both sexes in our pedigrees (2.5 years; S1 Table) yields an estimated mutation rate per year of 2.0 × 10−9 per bp. The rate per generation is similar to what was previously inferred by pedigree sequencing in zebra finch (5.8 × 10−9, the average of rates in two trios; (Bergeron et al., 2023)), as well as in collared flycatcher (4.4 × 10−9, based on seven trios; (Smeds et al., 2016b)), great reed warbler (7.1 × 10−9, based on eight trios;(Zhang et al., 2023a)), and 14 trios from four passerine species (ranging from 5.1 × 10−9 to 6.9 × 10−9 (Bergeron et al., 2023)). Thus, the per generation mutation rate appears to be quite stable among these passerines.
An exact estimate of the mutation rate per generation would be based on a comparison of zygotes to germ cells. Instead, existing estimates, including ours, are based on somatic tissue samples from parents and offspring. This approach leads to the incorrect inclusion of DNMs that arose during early development of the probands (Moorjani et al., 2016). In that respect, we note that if we only consider the DNMs inferred to have occurred in the parental germlines, our estimate reduces to 4.4 × 10−9 per bp per generation. This rate, in turn, may be a slight underestimate of the mutation rate from zygote to germ cell, given that it does not account for early parental developmental mutations that are present at detectable allele frequencies in the parental somatic tissues and thus were excluded by our filtering steps (Moorjani et al., 2016) (see Methods).
The mutation rate is known to be influenced by the immediate sequence context, notably when a CpG site is methylated. To determine if DNMs in zebra finch reflect these and other influences, we classified all DNMs into seven single mutation types (Fig 1B-C). We found a transition-to-transversion ratio of 1.73 (a ratio not significantly lower than the value in humans of ~2 (Durbin et al., 2010), p = 0.33 by a two-sided binomial test), and observed that the CpG>TpG mutation rate is more than an order of magnitude higher than the rest of substitution types (Fig 1C), consistent with findings in vertebrates and beyond (Bird, 1980). In addition, the proportions across the seven mutation types are highly similar to those observed in 19 million low-frequency polymorphisms (at most three copies, see Methods) segregating in a sample of 27 “unrelated” zebra finches (S3 Fig; p= 0.11 by a Chi-square goodness-of-fit test with 6 degrees of freedom). Since rare (young) variants are expected to reflect the mutational process, this close correspondence lends further support for the reliability of the DNM calls.
To explore if the mutation processes active in the mammalian germline also play a role in zebra finches, we assigned mutations to COSMIC single-base mutational signatures (SBS), originally inferred from patterns of genetic variation in human tumor samples (Nik-Zainal et al., 2012; Alexandrov et al., 2013). In the mammalian germline, the so-called “clock-like” mutational signatures, SBS1 and SBS5, have been shown to account for most mutations (Rahbari et al., 2015; Sasani et al., 2024; Spisak et al., 2024). Specifically, most mutations are assigned to SBS5, of unknown etiology, and a smaller proportion to SBS1, which is thought to occur due to spontaneous deamination of methylated CpG sites (Alexandrov et al., 2013). In birds, both of these signatures are also present. In the 19 million relatively low frequency alleles, we found contributions from SBS5 (84% of mutations), SBS84 (14%), and SBS1 (2%) (S3 Table). When analyzing the smaller sample of DNMs instead (202 mutations), we observed contributions from SBS5 (59%), SBS19 (19%), SBS4 (15%), and SBS1 (6%) (S3 Table). These results suggest that SBS5, to a lesser extent SBS1, and possibly other mutation processes, play a role in the zebra finch germline, as reported for SBS5 based on polymorphism data in other taxa (Gelova et al., 2022).
In contrast to most mammalian karyotypes, the genome of zebra finches and other birds contains a large number of micro-chromosomes (here defined as autosomes shorter than 40 Mb), which differ in GC content, replication timing, and other genomic features that could affect mutation rate (Srikulnath et al., 2021; Waters et al., 2021). Nonetheless, as far as we can tell, mutation rates (both the total and seven types) appear to be remarkably similar between both types of autosomes (Fig 1B-C).
Next, we compared the mutation rate between the sexes. For the subset of DNMs that occurred in the parental germline after primordial germ cell specification, we inferred a male-to-female mutation rate ratio of 1.72 (95%CI: 1.22 – 2.43) (Fig 1D). Notably, the parental ages in both sexes are similar in our pedigrees (mothers are only ~1 month older on average, S1 Table). This mutation rate ratio is in good agreement with previous reports of male-biased germline mutation in birds based on putatively neutral substitution rates in sex chromosomes versus autosomes (de Manuel et al., 2022), as well as with direct estimates by pedigree sequencing that pooled a small number of trios from four passerine species (Bergeron et al., 2023). For the post-zygotic DNMs, we used the sex of the proband—instead of the sex of the parental haplotype—to compare mutation rates between the sexes. After accounting for the different number of individuals for each sex among probands, the male-to-female ratio is 0.94 (95% CI: 0.43 – 2.02) (Fig 1D), consistent with there being no sex differences in the number of post-zygotic mutations, as also reported in humans (Sasani et al., 2019), mice (Lindsay et al., 2019), and cattle (Harland et al., 2017). Considering all mutations jointly, regardless of when they arose in development, the estimated paternal bias is 1.6:1 (95% CI: 1.19 −2.27).
Meiotic recombination
Detection of crossover and non-crossover events
To detect autosomal crossover and non-crossover events, we used an approach based on the patterns of inheritance of informative sites along the multi-sibling, three-generation pedigrees (Fig 1A). In brief, we relied on the configuration of informative sites (i.e. sites heterozygous in one parent but not in the other) to track changes of phase in parental haplotypes (following (Coop et al., 2008)). Crossovers were detected by an odd number of changes of phase within a short genomic distance; events involving more than one change of phase were classified as “complex” (see Methods). In turn, non-crossovers were detected by two changes of phase within a short genomic distance (see Methods).
In the 54 zebra finch meioses (28 maternal and 26 paternal) in which recombination events can be called (Fig 1A), we identified 1,174 autosomal crossovers. Their high genetic diversity allows us to delimit recombination events to a median interval of 647 bps (mean = 52 kb) (S4 Fig). Of these events, 604 and 570 occurred in maternal and paternal meioses, respectively, an average of 21.6 and 21.9 crossovers per meiosis. This difference is not statistically significant (p=0.81 by a two tailed binomial test) (S4 Table), in agreement with previous studies reporting an absence of heterochiasmy in zebra finch (Stapley et al., 2008; Backström et al., 2010). Combining events in the two sexes, ~10% of crossovers are complex events involving more than a single change of phase, in agreement with reports in humans (Williams et al., 2015; Halldorsson et al., 2019), mice (Li et al., 2019), and baboons (Wall et al., 2022). There is no discernable difference in the rate of complex events between sexes in our data (p = 0.65 by a Chi-square test).
Considering the number of crossovers per chromosome per meiosis, the estimated crossover rate is consistent with an obligatory crossover per tetrad (Page & Hawley, 2003; Massy, 2013) for 30 of the 39 chromosomes (p<0.05; S5A Fig). Given that, in the absence of a back up mechanism for achiasmatic tetrads, we would only expect a couple of chromosomes to fall below this p-value by chance, this observation suggests that we are missing a few events, particularly on short and highly repetitive chromosomes (S5B-C Fig). Indeed, a study of MLH1 foci in zebra finch reported an average of 46 autosomal foci, which would correspond to 23 transmitted crossovers (Calderón & Pigozzi, 2006).
This caveat notwithstanding, the sex-averaged genetic map length is estimated to be 2,174 cM (2,157 cM and 2,192 cM for females and males, respectively), corresponding to a mean recombination rate of 2.28 cM/Mb. Despite the fact that this map length is likely to be a slight under-estimate, it is substantially higher than the two previous reports for zebra finch based on pedigrees: 1,068 cM (1.06 cM/MB) (Stapley et al., 2008) and 1,341 cM (1.50 cM/Mb) (Backström et al., 2010). Part of the explanation likely lies in which chromosomes were included, as most of the micro-chromosomes, which experience high recombination rates, were not included in these studies. Indeed, if we restrict our estimate to the same set of autosomal chromosomes as Backström et al., 2010, our estimate (1.78 cM/Mb) is in better agreement with theirs. Moreover, previous estimates were based on few genetic markers (~900 and ~2,000, respectively), so may have missed a number of crossover events, even on larger chromosomes. The average rate for the six macro-chromosomes is 1.05 cM/MB–much more similar to several estimates in placental mammals, who have only macro-chromosomes (Dumont & Payseur, 2008). In contrast, the average rate for the 33 micro-chromosomes is 4.21 cM/Mb.
In parallel, we inferred a genetic map from patterns of LD by combining data from six of the founders of the pedigrees and sequencing data that was previously generated for 19 unrelated zebra finches (Singhal et al., 2015) (see Methods). LD-based genetic maps provide estimates of the population recombination , where is the effective population size and is the recombination rate per generation. Estimates for micro-chromosomes are likely less reliable, given the high background recombination rate (S6 Fig) (Singhal et al., 2015). For macro-chromosomes alone, the mean in our LD-based map is estimated to be 0.082 per bp. Given the observed diversity level in our sample (Watterson estimator θw = 0.0159) and our estimate of the mutation rate, an estimate of the effective population size of zebra finch is 792,000. If we use this estimated value to estimate from the population recombination rate, then the mean recombination rate is 2.8 cM/Mb on macro-chromosomes (S6 Fig). This estimate is of the same order as what we obtain directly from pedigrees but two- to three-fold higher; this discrepancy is not surprising given the numerous assumptions that come into play in estimating from population data.
Next, we inferred non-crossover events by considering two phase changes among the parental haplotypes (see Methods). Given the mean tract lengths estimated in other vertebrates and the diversity levels in zebra finches, we expected such events to typically involve only a single informative variant, and thus for their identification to be highly sensitive to sequencing and genotyping errors (Williams et al., 2015; Li et al., 2019; Wall et al., 2022). In order to minimize the number of spurious non-crossover calls, we excluded the nine shortest micro-chromosomes, which together account for 1.6% of the assembled autosomal genome, and which may be less reliably mapped (S7 Fig). We focused on the second generation of three generation pedigrees, because there are several siblings, allowing us to detect changes of phase. This subset of the data represents a total of 36 meioses (18 paternal and 18 maternal). The F1 individuals from these crosses all have sequenced offspring, in which we can verify transmission of any putative non-crossover event.
To make non-crossover calls, we tried three variant callers (GATK, freebayes and bcftools) and stringently filtered the results (see Methods). To estimate the reliability of the non-crossover calls, we quantified their transmission rate to the next generation (S5 Table), with the expectation that true events should be transmitted ~50% of the time (assuming complete power to detect transmission). On that basis, we determined that freebayes provides the most reliable set of non-crossover calls, with an observed transmission rate of 0.47 (95% CI: [0.38 – 0.57]). Using this caller, we identified a total of 275 non-crossovers in the 36 meiosis. As expected, most (235) of the non-crossover events involve a single informative site in the conversion tract (S8 Fig). As for crossovers, there is no detectable sex difference:141 were maternal and 134 paternal (p = 0.72 by a two tailed binomial test) (S4 Table).
Distribution of recombination events along the genome
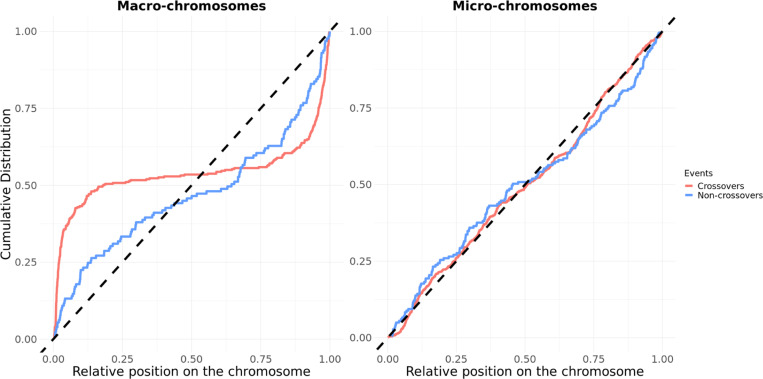
In many vertebrates, the crossover rates increase near telomeric regions, and this elevation is typically higher in males than in females (Haenel et al., 2018; Sardell & Kirkpatrick, 2020). The distribution of non-crossovers, however, remains poorly described, especially in non-mammalian vertebrates and species with micro-chromosomes. Considering the distribution of crossover resolutions in macro-chromosomes and micro-chromosomes separately, we found that rates are differentially distributed (Kolmogorov-Smirnov test p< 2.2 × 10−16 ; Fig 2). Whereas in macro-chromosomes, crossover rates are elevated towards the telomeres, in micro-chromosomes they are much more uniformly distributed (Fig 2, S6 Table for all p-values). Similarly, LD-based recombination rates, which primarily reflect crossovers, show an elevation near telomeres on macro-chromosomes, but not micro-chromosomes (S9 Fig). A caveat, however, is that inferring recombination rates through LD-based approaches is challenging when background recombination rates are high (Singhal et al., 2015; Raynaud et al., 2023), as may be the case for micro-chromosomes. In principle, the difference between macro- and micro-chromosomes could arise simply from a proportionally shorter effect of telomeres on large chromosomes.
Fig 2. Cumulative distribution of recombination events along both types of chromosomes.
The cumulative distribution of crossovers (in red) and non-crossovers (in blue) on the autosomes, for macro-chromosomes (left) and micro-chromosomes (right). Macro-chromosomes are defined as autosomes longer than 40 Mb. The position of the events is normalized by the length of the chromosome (see Method). The black dashed lines represent the uniform distribution. S10 Fig shows similar results, separating micro-chromosomes above and below 20 Mb in length. S6 Table reports p-values from Kolmogorov-Smirnov tests for various comparisons (crossover versus non-crossover, macro versus microchromosomes).
In contrast to crossovers, the spatial distribution of non-crossovers does not differ between the two sets of chromosomes (p=0.16, Fig 2, S6 Table). Accordingly, the distribution of crossovers along the chromosomes is significantly different from that of non-crossovers on macro-chromosomes (p=4.8 × 10−6), with less of a skew towards telomeres among non-crossovers; the same is not seen in micro-chromosomes (p=0.36) (Fig 2 S6 Table). None of these patterns differ significantly between sexes (S11 Fig).
Overlap with genomic features
Zebra finch, like other birds, lacks a functional copy of PRDM9 (Oliver et al., 2009; Baker et al., 2017). Consistent with findings in mammals without PRDM9, meiotic recombination is enriched in promoter-like features (Singhal et al., 2015; Campbell et al., 2016; Kawakami et al., 2017). Specifically, analyses of LD suggest that increase in recombination rates appears to be mainly attributable to CpG islands, with no further increase explained by TSSs (Singhal et al., 2015). To test this hypothesis more directly, we first improved the annotation of CpG islands in zebra finch. To this end, we relied on the fact that CpG islands are usually hypomethylated in vertebrates (Deaton & Bird, 2011) and quantified DNA methylation levels in the testes of two males using bisulfite sequencing (see Methods). We then considered both DNA methylation levels and local sequence composition in order to identify 46,205 CpG islands, on par with numbers reported for other vertebrate species (Antequera & Bird, 1993; Hoge et al., 2024).
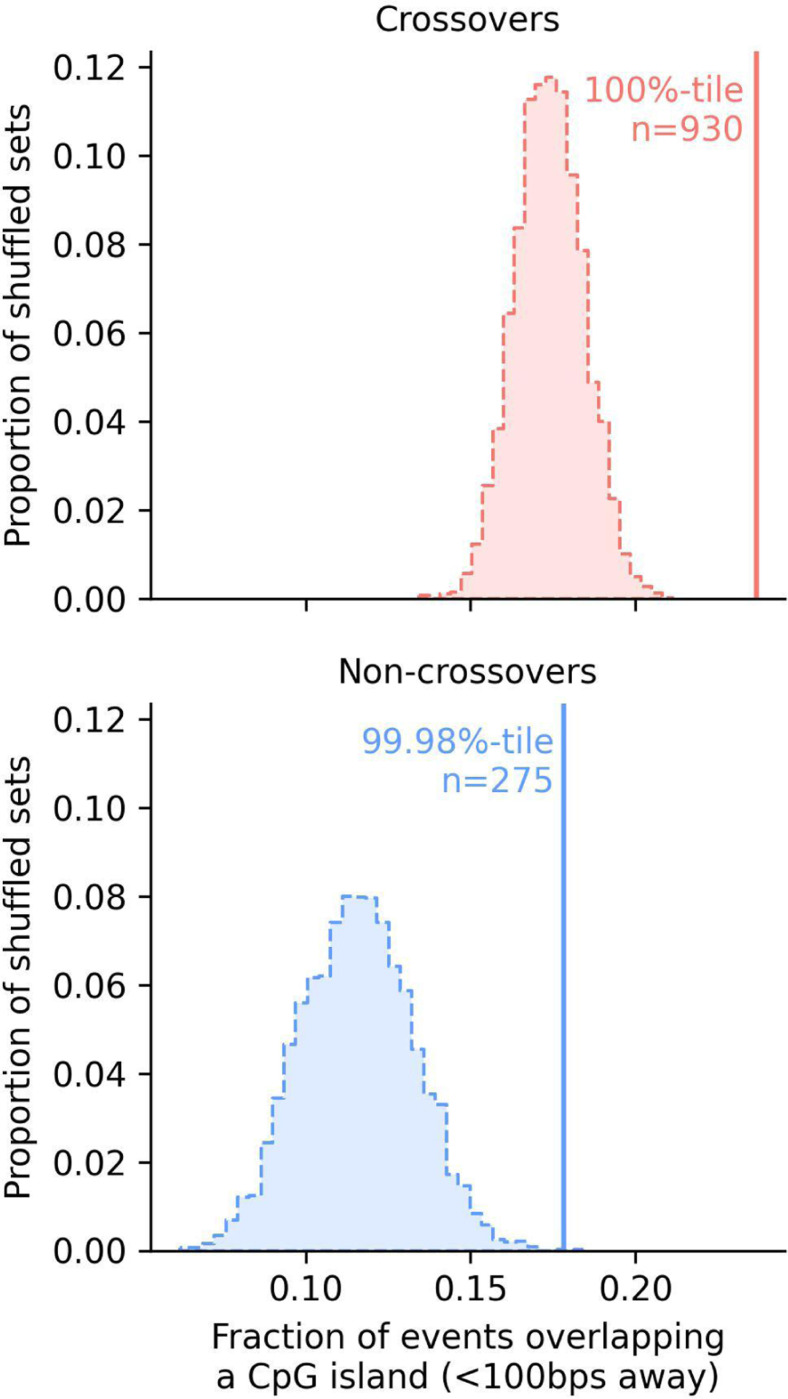
Using this annotation, we asked whether the recombination events identified in pedigrees are enriched at CpG islands. As expected, significantly more recombination events occur close to a CpG island than expected by chance: 23.6% of crossovers occur within 100 bps of a CpG island when only 17.3% are expected to do so by chance on average (a 1.36-fold enrichment), and 17.8% of non-crossovers when 11.5% are expected to do so by chance (a 1.54-fold enrichment) (Fig 3). The fact that we expect (and observe) more overlap between crossovers and CpG islands than we do for non-crossovers likely reflects a difference in the genomic distribution of the two types of recombination events: crossovers are relatively more likely to occur in in telomeric regions and micro-chromosomes (Fig 2), which have increased CpG island densities (S12 Fig) (Han et al., 2008).
Fig 3. Overlap of both types of recombination events with CpG islands.
Fractions of crossovers (top) and non-crossovers (bottom) detected within 100 bps of a CpG island. The vertical lines show the observed overlap. The distribution for the overlap expected by chance are shown as a histogram, obtained by randomly shuffling all the events within a 2.5 Mb window on each side of their original location, matching for the GC content and ensuring a similar mappability (see Methods for details).
As previously reported on the basis of LD patterns, recombination events were not enriched at TSSs conditional on the presence of a CpG island nearby (whereas they were enriched at CpG islands irrespective of the presence of a TSS) (S13 and S14 Figs). The analogous analysis had not been conducted in the collared flycatcher. To do so, we used the 443 recombination events called in (Smeds et al., 2016a) and identified CpG islands in collared flycatcher genome with cpgplot (default parameters, (Madeira et al., 2022)). As shown in S15 Fig, the enrichment of recombination events at CpG islands, and not at TSSs, is very similar to what we obtained in zebra finch. Therefore, in birds as in rodents lacking PRDM9 (Brick et al., 2012; Mihola et al., 2021), proximity to a CpG island is predictive of both crossover and non-crossover events. A rough calculation suggests that the degree of overlap is consistent with all recombination hotspots occurring at CpG islands: if each CpG island is assigned the mean heat inferred for hotspots in our LD-based map (9.93), then given that CpG islands (+/− 100 bps) cover 3.48% of the autosomes, we would expect 26.4% of crossovers to overlap CpG islands.
Large-scale analyses in humans have reported that meiotic recombination is mutagenic, with 1/200 mutations arising from a double strand break (Halldorsson et al., 2019; Hinch et al., 2023). Given the relatively small number of de novo mutations identified in our study, we should have very limited power to detect an effect in birds of a similar magnitude. Accordingly, we did not find evidence for the co-occurrence of de novo mutations and crossover events in zebra finch (S16 Fig).
Estimation of fundamental parameters of gene conversion
To the best of our knowledge, GC-biased gene conversion (hereafter gBGC) has only been examined directly in one bird species, the collared flycatcher (Smeds et al., 2016a), for which the authors estimated that 59% of events that include a AT/GC polymorphism are resolved towards GC rather than AT (CI: [52 – 65]; Smeds et al., 2016a). This point estimate is on par with that has been reported for mice and primates (57%−68%; Williams et al., 2015; Halldorsson et al., 2016; Li et al., 2019; Versoza et al., 2024; Porsborg et al., 2024; Schweiger et al., 2024).
A challenge in estimating gBGC is that even a small fraction of false positive calls can bias the estimate downwards (i.e., towards 50%). To minimize the problem, we only used the events detected in the three pedigrees with five siblings, which appear to be the most reliable (S7 Tables). This approach led to the identification of 191 non-crossovers. We also considered the subset of non-crossover events that occur less than 100 bps from a CpG island, reasoning that such events are more likely to be true positives (Singhal et al., 2015) (Fig 3).
Considering the 177 cases of AT/GC variants within all the non-crossover events, the gBGC bias in zebra finch is estimated to be 59% (95% CIs: [52 – 66]), the same point estimate as reported in flycatchers. Moreover, we can reject a null model of no GC-biased gene conversion for zebra finch (p=0.008, by a one-tailed binomial test). When focusing on the 18 cases when the non-crossover event is close to a CpG island, the point estimate is higher but with large uncertainty (72%; 95% CIs: [47 – 90]).
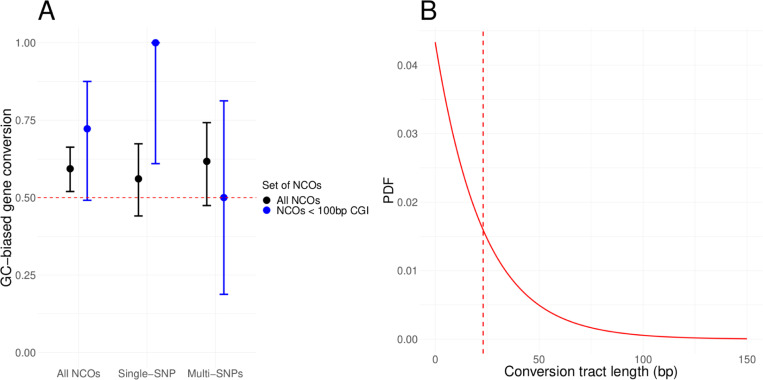
A recent study reported that the gBGC depends on the number of heterozygous sites present in the conversion tract: specifically, a bias was only seen for conversion events with a single such site both in mice and, more tentatively, in humans (Li et al., 2019). Two recent studies, one focusing on humans and the other on humans, chimpanzee and gorilla found no evidence for this claim (Porsborg et al., 2024; Schweiger et al., 2024). To examine this question in zebra finch, we separated the non-crossovers into those with one heterozygous site versus more than one (see Methods section Phasing NCOs for detail). We also find no evidence supporting the hypothesis that gBGC depends on the number of heterozygous sites in the conversion tract, but the data are insufficient to reach a firm conclusion (Fig 4A).
Fig 4. GC-biased gene conversion and conversion tract length distribution estimates.
(A) Point estimates of the GC-biased gene conversion (gBGC) for different sets of non-crossovers. The 95% CI were obtained from an exact binomial test (see Methods). The black dots are the point estimates for the set of all non-crossovers and the blue dots are the estimates for non-crossovers within 100 bps of a CpG island. For each set of non-crossovers, we looked either at (i) all the events, (ii) the event with only one heterozygous site in the conversion tract, (iii) the events with more than one heterozygous site. (B) Estimated distribution of the conversion tract lengths. The mean is 23 bps, indicated with a vertical red dashed line.
Next, we estimated the mean conversion tract length, following Li et al., 2019. Specifically, we relied on the distances between co-converted and non co-converted informative sites and assumed a single exponential distribution of tract lengths (see Methods). While the assumption of an exponential distribution is not valid, previous results in mammals suggest that it is a reasonable approximation for the vast majority of non-crossover events and it allows us to compare our findings to those previously reported (Li et al., 2019; Wall et al., 2022; Charmouh et al., 2024; Schweiger et al., 2024). In baboons and humans, it appears that a tiny fraction of events (<2%) are many kilobases in length and likely arise from a distinct process (Wall et al., 2022; Schweiger et al., 2024). While the small number of non-crossover events prevent us from exploring that possibility in depth, there are a couple of cases where the minimum tract length is likely kilobases in length: for example, among the tracts that include more than one informative site, the distance between sites is 642 bp in one case, and 4169 in another. We excluded the event that is minimally 4 kb in length in what follows. By this approach, we estimated the mean conversion tract length of non-crossovers in zebra finch to be 23 bps on average (central 95%-tile: 15 – 35 bps; see Methods). If we add back the one longer event, the estimate of the mean shifts to 35 bps. These results are similar to mammalian species, for which estimates range from 24 to 50 bps (Li et al., 2019; Wall et al., 2022; Charmouh et al., 2024; Porsborg et al., 2024; Schweiger et al., 2024).
Total number of non-crossovers per meiosis and non-crossover to crossover ratio
Given the mean conversion tract length and the density of informative sites, we can infer the expected number of non-crossovers per meiosis. This inference relies on our power to detect a non-crossover, i.e., on the probability that a gene conversion event overlaps an informative site, which in turn depends on the mean conversion tract length and the density of informative sites (see Methods). We inferred 191 non-crossovers in 30 meiosis or 6.37 events per meiosis (considering only families of five siblings; S7 Table). For a mean gene conversion tract length of 23 bps, we should detect an estimated 5.4% of non-crossover events in the genome, averaged over all founders. In other words, the total number of non-crossover events per genome (i.e., chromatid) is ~18.5-fold higher than observed. Taking into account differences in power among founders, our prediction is that there are 123.1 non-crossover events per genome per meiosis on average; given the uncertainty in the conversion tract length estimate, between 85.2 and 180.0 non-crossover events (Table 1).
Table 1.
Estimates of number of non-crossovers per chromatid
| Tract length estimate | Observed number of crossovers | Predicted number of non-crossovers | NCO:CO per chromatid |
|---|---|---|---|
| 15 | 18.5 | 180.0 | 9.7 |
| 23 | 18.5 | 123.1 | 6.7 |
| 35 | 18.5 | 85.2 | 4.6 |
Predicted number of non-crossovers and estimated non-crossover to crossover ratio for different estimates of the mean conversion tract length. We report the ratio per chromatid (i.e., the number of predicted non-crossovers divided by the number of observed crossovers).
In turn, our estimates of the number of non-crossovers suggest that the non-crossover:crossover ratio per genome is 6.7 (between 4.6 and 9.7, given the tract length uncertainty). We note that this ratio is provided per chromatid and not per tetrad, as sometimes done (Cole et al., 2012). Our estimate falls between values in mice and humans, for which reported ratios are 5 and 10, respectively (Cole et al., 2012; Halldorsson et al., 2019; Hinch et al., 2023).
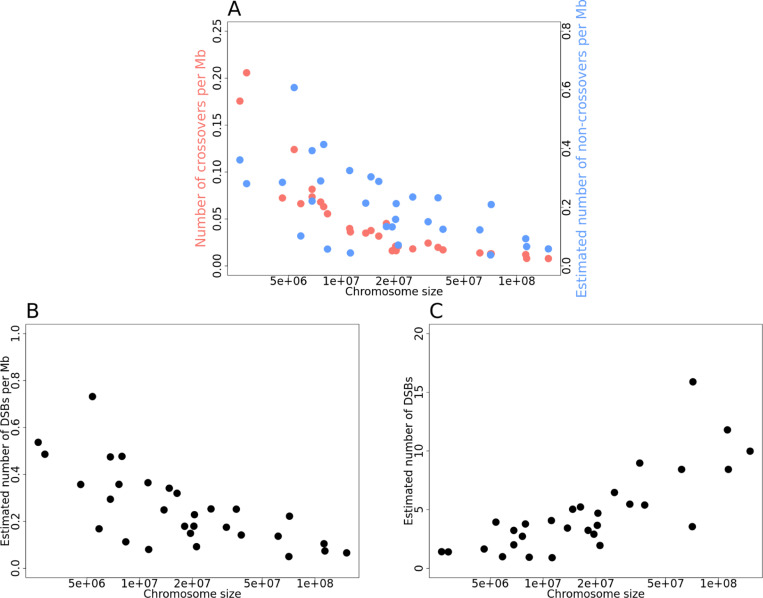
The densities of crossovers and non-crossovers both decrease with chromosome length (Fig 5A). If we ignore the subset of events repaired off of the sister chromatid, which leave no discernable mark, or through mechanisms other than homologous recombination (Massy, 2013), and consider the number of DSBs to be given by the sum of crossovers and non-crossovers, it follows that the same is true of DSBs (Fig 5B). Thus, either larger chromosomes have a lower density of DSBs or a larger fraction of DSBs are resolved non-canonically (i.e., as neither a crossover nor a non-crossover). While the density of DSBs appears to decrease with chromosome length, the estimated number of DSBs increases (Fig 5C).
Fig 5.
(A) Density of crossovers (blue) and non-crossovers (red) as a function of chromosome size for the 30 autosomes that passed mapping quality and coverage thresholds (see Methods). We note that the x-axis is on a log scale. (B) The estimated density of double-strand breaks (DSBs) as a function of the chromosome size, for the same set of chromosomes. (C) The estimated number of DSBs as a function of the chromosome size.
Discussion
To date, meiotic recombination events and DNMs have only been directly identified in a handful of non-mammalian vertebrate species, based on a small number of trios. There is therefore little comparative information about properties of mutation and recombination, beyond average rates in the genome per generation. In this study, we collected whole genome sequences from 74 zebra finches. The genomic data provided information about 80 meioses, and allowed us to infer key mutation and recombination parameters.
As we had previously reported on comparisons of substitution rates on the Z chromosome and autosomes for passerines, we found that the paternal bias in germline mutation is lower in zebra finches (1.6:1, S2 Table) than in most mammals (about 3:1, de Manuel et al., 2022). This difference could arise from lower rates of endogenous DNA damage in avian males or from a greater proportion of DNMs occurring during early developmental stages, when both sexes have similar mutation rates (Fig 1D). We note that our estimate is substantially lower than the ratio of ~7.5:1 (95% CI: ~4.5:1 − ~13.5:1) reported for passerines in a recent study, which pooled phased germline mutations across four different species (including zebra finch) (Bergeron et al., 2023). Other studies in passerines based on pedigree sequencing have reported lower paternal biases, in better agreement with our estimate: ~1.5:1 in collared flycatcher (Smeds et al., 2016b) and ~3:1 in the great reed warbler (Zhang et al., 2023). In principle, variation among estimates could reflect, at least in part, differences in the parental ages of the sequenced pedigrees. Regardless, sex-averaged per year and per generation mutation rates in zebra finch and other passerines are similar to what is seen in mammals with the same generation time (Bergeron et al., 2023) (S17 Fig).
Recombination patterns also conform to expectations based on mammals: as seen in dogs as well as in mouse and rat knockouts for PRDM9, recombination events are enriched near CpG islands (Brick et al., 2012; Auton et al., 2013; Mihola et al., 2021). At a broader scale, the constraint of an obligate crossover per chromosome leads to higher rates of recombination on micro-chromosomes than typically seen in mammals (outside of the pseudo-autosomal region), but rates on macro-chromosomes are highly comparable (S6 Fig). Moreover, other recombination parameters–including the mean gene conversion tract length, the degree of GC-biased gene conversion, and the ratio of crossovers to non-crossovers–are all very similar to what has been reported in mice and humans (Fig 4, Table 1). The conservation of all these parameters from mammals to zebra finch suggests that a number of mutation and recombination properties evolve under stabilizing selection in vertebrates.
The increasing availability of genome sequences should make it possible to test this hypothesis more systematically, and to identify which aspects of mutagenesis and meiotic recombination are most strongly conserved. As this study demonstrates, many of these parameters can now be estimated in non-model species by sequencing a relatively modest number of large three-generation pedigrees. Recent advancements in long-read sequencing offer further possibilities: in particular, high-fidelity (HiFi) long-read sequencing of germ cells will enable the detection of a large number of mutation and recombination events (Liu et al., 2024; Charmouh et al., 2024; Porsborg et al., 2024; Schweiger et al., 2024; Porubsky et al., 2024). Application of such approaches across the tree of life will help address long-standing questions about the selective pressures that shape the evolution of mutation and recombination (Otto & Lenormand, 2002; Coop & Przeworski, 2007; Lynch, 2010; Dapper & Payseur,2019; Johnston, 2024).
Materials and Methods
The command lines used for each of the following subsections can be found in the Supplementary Section “Command lines”.
Pedigree breeding and sequencing
The ancestors of the zebra finch individuals sequenced here were taken from the wild at Fowlers Gap (New South Wales in southeastern Australia), under NSW Scientific license SL100378. Wild-derived zebra finches were held and bred at Macquarie University (Sydney, Australia), under Animal Ethics Approval 2018/027. Zebra finch pairs were bred in one of 20 outdoor aviaries (4.1×1.85×2.24 m) with a single pair per aviary, with access to ad libitum water, finch seed mix (Avigrain Finch Blue, Berkeley Vale, Australia), and a ‘greens’ mix (blended spinach and frozen vegetables, as well as micro- and macronutrient supplements from Naturally For Birds, Pullenvale, Australia). Each aviary had three nest boxes and nesting material (‘November’ grass and emu feathers). Blood was collected from F0, F1 and some F2 individuals via venipuncture of the brachial vein and stored in 99% ethanol. Tissue samples for the remaining F2 individuals were collected from terminated embryos between day 5–10 of incubation (euthanised via decapitation). Legs and wings of the embryos were stored in 99% ethanol before DNA extraction.
Whole blood (n = 53) and whole tissue (n = 21) samples were shipped to Genewiz (Azenta Life Sciences, South Plainfield, NJ, USA) for DNA extraction, library preparation, and whole genome sequencing. To reduce the possibility of false positives due to amplification errors, the 150 bp paired-end DNA libraries were prepared using a PCR-free protocol. The libraries were sequenced on an Illumina Hiseq X with a target of ~350 million reads per lane. Genotype calling errors in the parental generation can greatly increase the false positive rate during mutation calling, as true heterozygous sites that have been incorrectly genotyped as homozygous will appear to be a DNM. To guard against this possibility, we targeted the parental generation for higher coverage sequencing relative to the other individuals in the pedigree.
Whole genome sequencing alignment
We combined the whole genome sequences that we generated from the pedigrees (S1 Table) with previously generated genome sequences for 19 “unrelated” individuals sampled from an Australian population located in the same area as the eight founders of the pedigrees (Singhal et al., 2015). After removing adapters from all sequencing reads using cutadapt, we mapped them to the zebra finch reference genome sequence bTaeGut1.4 (NCBI RefSeq ID: GCF_003957565.2) using the Nextflow pipeline sarek (v 2.7.1) with default parameters (Rhie et al., 2021; Garcia et al., 2024). The familiar relationship among individuals, along with the mapping coverage for each sample, are presented in S1 Table. Given that the sex chromosomes have a lower depth of coverage in the heterogametic sex, making it harder to reliably detect DNMs and recombination events, we focused our analyses on autosomal sequences, as had been done in previous studies (e.g., Smeds et al., 2016a; Besenbacher et al., 2019; Bergeron et al., 2023).
Variant calling
We called variants using GATK (McKenna et al., 2010). First, HaplotypeCaller was used to generate per sample GVCF files for each assembled chromosome (McKenna et al., 2010), including non-variant sites and setting the heterozygosity parameter to 0.01 (version 4.2.6.1; McKenna et al., 2010). We then combined GVCFs from each sample using GenomicsDBImport and called a final set of genotypes using GenotypeVCFs, setting again the heterozygosity to 0.01. With this approach, we identified a total of 101 million single-nucleotide polymorphisms (SNPs) and 17 million indels.
Mappability
To restrict our analyses to locations where short read sequences can be confidently mapped, we built a mappability mask for the zebra finch genome assembly following the SNPable pipeline, with a read length of 150 bps and stringency parameter of 1 (version 0.2.4;https://lh3lh3.users.sourceforge.net/snpable.shtml). We then used the script makeMappabilityMask.py from msmc-tools to generate a final BED file of mappable genomic regions (Schiffels & Durbin, 2014). The mappability mask excludes 33.4% of the genome assembly. We removed all single nucleotide variants closer than 5 bps of an indel. In mappable regions, we called a total of 45.6 million SNPs of which we retained the 43.5 million that are biallelic.
Detection of single-point de novo mutations
To detect DNMs, we analyzed the genomes of the 40 parents-offspring trios in the seven pedigrees (Fig 1A and S1 Table), looking for mutations in the proband and not found in their parents. For each trio, we examined which positions fulfilled all the following requirements using bedtools intersect (Quinlan & Hall, 2010a): (i) all individuals in the trio had a depth of coverage of at least eight reads and at most two times their autosomal average coverage, (ii) the position was at least 5 bps away from an indel called in any individual in the trio, (iii) it was not found within repetitive regions annotated in the reference assembly, and (iv) it was within the mappable section of the genome (see Mappability section). By this approach, we retained an average of 502.8 autosomal Mb per trio (min=475.3, max=511.1), which represents 52% of the autosomal genome.
For each trio, we then identified positions with a putative DNM as those that were heterozygous in the proband, homozygous for the reference allele in both the parents, and not polymorphic in all genotyped individuals outside that nuclear family. We only considered biallelic sites and required the genotype quality to be at least 30 in all individuals in the trio. A potential source of false positives are inherited variants that were erroneously called as homozygous in the parents. To minimize this problem, we discarded any candidate mutation if GATK reported the presence of a read-based assembled haplotype supporting the non-reference allele in either parent, regardless of their called genotype. We noticed that the local reassembly of reads performed by GATK to improve variant calling sometimes “erases” the presence of rare reads carrying the non-reference allele; we therefore verified the absence of reads carrying the DNM in the parents using bcftools mpileup (Danecek et al., 2021).
Another potential source of error is when a homozygous reference allele is miscalled as heterozygous in the proband. To mitigate this issue, we required at least two reads carrying the non-reference allele in the proband, and discarded sets of putative DNMs within 10 bps of each other. While a tiny fraction of germline mutations in humans have been shown to occur in close proximity (Harris & Nielsen, 2014; Francioli et al., 2015), this filter allows to discard false positives resulting from alignment to collapsed paralogous regions in the reference assembly (Wu et al., 2020). Following this pipeline, we detected 226 candidate DNMs (S2 Table).
Assignment of parent of origin of de novo mutations
To determine the parent of origin for each DNM detected in the 18 probands with sequenced partners and descendants, we phased alleles by transmission. Specifically, we searched for informative sites within 200 kb of each DNM. An informative site was defined as position where an allele is carried by only one of the two parents (either as in a heterozygous or homozygous state); present in the proband (who is therefore heterozygous); not carried by the partner; and carried by the descendant. We then assigned a parental origin to the DNM when at least 75% of all informative alleles came from a single parent: if the DNM was transmitted to the descendant, it was assigned to that parent, and if not, it was assigned to the other parent.
Additionally, we phased DNMs detected in the 22 probands without sequenced descendants using ‘read tracing’ (Goldmann et al., 2016; Jónsson et al., 2017). Briefly, for each DNM, we first searched for informative sites nearby (± 300 bps), i.e., positions where an allele inherited by the proband is carried by only one of the parents. If the DNM was carried on the same read pair as the inherited allele, we assigned the DNM as having occurred in the germline of the parent carrying the informative allele. In turn, if the DNM was not on the same read as the informative allele, we inferred that the DNM occurred in the other parental germline.
Distinction of mutations by their developmental timing
Mutations that occurred in the early development of the offspring can be mistakenly identified as mutations that occurred in the cell lineage leading to the parental germlines. To distinguish these events, we looked for candidate DNMs that showed “incomplete linkage” with informative heterozygous alleles nearby. In particular, we identified all heterozygous sites within ± 300 bps from a DNM in an offspring. In 37 of the mutation events, there were no neighboring heterozygous positions within this distance. For the rest of mutations, we searched for indications of three distinct haplotypes in the offspring by assessing the linkage of the mutation and nearby heterozygous alleles (S18 Fig). By this approach, we discovered that 25 of the apparent parental germline DNMs exhibited evidence of being post-zygotic mutations that arose after the fertilization of the proband (see main text).
We also detected 32 DNMs in which reads assembled into more than two haplotypes in the parents (S18 Fig), violating the expectation for diploid individuals, and suggesting genotyping errors. We discarded such events.
Estimation of mutation rate per generation
We calculated an autosomal point mutation rate per site per generation by dividing the number of DNMs by two times the length of the autosomal genome considered (see “Detection of single-point de novo mutations”). To calculate mutation rates for specific mutation types (e.g., C>T), we divided by two times the number of mutational opportunities of that type in the subset of the genome considered (e.g., C). The 95% confidence intervals for the mutation rates were calculated assuming a Poisson distribution, based on the number of observed mutations and the total sequence length.
Mutational spectrum in low frequency polymorphisms
To analyze mutation types in polymorphism data, we considered low frequency SNPs in a sample of 27 non-closely related zebra finch individuals (i.e., polymorphic sites where the alternative allele is found at most in three chromosomes), using the --max-maf parameter in VCFtools. We then classified these mutations in seven distinct substitution types (see Fig 1), assuming the ancestral allele is the major allele, and using the flanking nucleotides in the reference assembly to distinguish C>T mutations that occurred in CpG context.
Inference of mutational signature activity
We inferred COSMIC mutational signatures (Alexandrov et al., 2013) from low frequency polymorphisms and DNMs identified by pedigree sequencing. To this end, we classified mutations into 96 substitution types depending on their trinucleotide context in the zebra finch reference assembly, and generated mutation matrices with the number of events for each type. To infer signature activity, we used the cosmic_fit function in SigProfilerAssignment (Díaz-Gay et al., 2023), which assumes the genome-wide frequencies of the 32 trinucleotide contexts are consistent with those observed in humans. To account for the differences between human and zebra finch genomes, we adjusted the mutation counts by scaling them according to the ratio of genome-wide trinucleotide frequencies. Specifically, for each mutation type, we multiplied the observed count by the ratio of the trinucleotide frequency in zebra finches to that in humans.
Co-localization between recombination and mutation events
We calculated the distance between each phased DNM and the closest crossover or non-crossover recombination event, discarding five mutations that were phased both by read-tracing and transmission but were assigned to a different parent (see Methods). We took this approach for events that occurred in the germline of the same parent and were detected in the same proband (“within”), as well as for events that occurred in different parents and probands (“between”). For cases where no recombination event occurs on the same chromosome as a DNM, we set the distance to 200 Mb, i.e., longer than the largest chromosome in the zebra finch assembly. To determine whether mutation and recombination events co-localize more than expected by chance, we compared the distribution of “within” and “between” distances by a Kolmogorov-Smirnov test.
Individuals used to estimate diversity levels and infer an LD-based genetic map
Using the biallelic SNPs in mappable regions, we obtained the Weir and Cockerham’s Fst estimator between the individuals sequenced by (Singhal et al., 2015) and the founders in our pedigrees with VCFtools (Weir & Cockerham, 1984; Danecek et al., 2011). We calculated Fst in 10 kb windows; the average Fst was 0.009. Given the low levels of genetic differentiation, we combined the two sets of samples for analysis, for a total of 27 zebra finches.
We estimated the population mutation rate , where is the effective population size and the mutation rate per generation, using Watterson’s estimator for the 27 finches (Watterson, 1975). We used bedtools coverage to count the number of biallelic segregating sites in regions within autosomal mappable regions, then divided this count by the total length of these regions (v2.30.0; Quinlan & Hall, 2010b). With this approach, we estimated ; given a mutation rate estimate of 5.02 × 10−9 (see main text), this yields an estimated of ~792,000.
Because LDhelmet can take as input a maximum of 50 haplotypes, we based the inference of LD-based recombination rates on a subset of 25 individuals. To remove two from the 27, we used the relatedness2 option in VCFtools on the same set of biallelic SNPs as in the Fst analysis (see S19 Fig for distribution of pairwise relatedness). We choose the two (both founders of the pedigree) so as to decrease the highest pairwise relatedness between the individuals used in the LD-based recombination rate; after excluding them, the maximal value of the kinship coefficient decreased from 0.09 to 0.032.
Ancestral alleles and mutation transition matrix
To infer the ancestral allele state at biallelic sites, we tested if the frequency of the major allele among the 27 non-closely related individuals significantly exceeded 0.5 (exact binomial test, one-sided). In cases where the major allele frequency was significantly greater than 0.5 (96.5% of the SNPs), we assigned to the major allele a probability of 0.91 of being the ancestral allele, and a probability of 0.03 to each of the three remaining alleles. For SNPs where the major allele frequency did not significantly exceed 0.5, we assigned a probability of 0.47 to the two observed alleles, and a probability of 0.03 to two other alleles. These polarized SNPs were then used to compute the mutation transition matrix following the method described in Chan et al, (2012) (Chan et al., 2012).
Variant phasing
We phased variants using information from sequencing reads that span multiple heterozygous sites (called “phase informative variants”, PIRs) and the patterns of haplotype inheritance across the pedigrees. The PIRs were extracted with the SHAPEIT suite and a file with the familial relationships in the pedigrees was formatted using plink2. Variants were then phased using the assemble tool in SHAPEIT, and the outputs were converted into the LDhelmet VCF format (v2.r904; Delaneau et al.,2012).
Implementation of LDhelmet
We used the estimate of θw and of the effective population size to run LDhelmet (version 1.9; Chan et al., 2012). Apart from the set of p values (see code for p values), we used the recommended parameters for all the steps of LDhelmet. For the Markov chain Monte Carlo analysis, we used three block penalties (10, 20 and 50), a burn in of 100,000 steps and a total of one million steps.
To identify LD-hotspots, we computed the mean recombination rate in windows of 1 kb, and assessed the relative recombination rate of each window compared to the mean recombination rate of the 20 kb each side, with a buffer a of 2 kb between the focal window and the surrounding region (Hoge et al., 2024). Windows with a relative recombination rate greater than five were kept, and merged if adjacent. We identified 5,241 hotspots, on par with previous results in finches (Singhal et al., 2015). These LD-based hotspots are highly enriched at CpG islands (S20 fig), as expected from previous LD-based maps and from our findings for crossovers and non-crossovers (Fig 3) (Auton et al., 2013; Singhal et al., 2015).
Previous simulations indicate that there is limited power to detect hotspots in zebra finch where the population recombination rate is high (Singhal et al., 2015). One implication is that we expect to detect many fewer hotspots in telomeric regions of macro-chromosomes, as well as on micro-chromosomes. Perhaps for that reason, there is only a weak overlap of crossovers and non-crossover events with LD-based hotspots (S21 Fig). As this observation makes clear, although LD-based inferences have been transformative to our understanding of recombination (e.g., Johnston, 2024), it remains useful to complement them with the direct identification of recombination events.
Identification of CpG islands
We quantified the methylation status of CpG sites with bisulfite sequencing. To this end, we used testis samples from two male zebra finches. Samples were kindly provided by Sarah London (University of Chicago). All procedures were conducted in accordance with the NIH Guidelines for the Care and Use of Animals and were approved by the University of Chicago Institutional Animal Care and Use Committee (ACUP #72220). Birds were bred in the London laboratory flight aviaries, and housed on a 14:10 h light:dark cycle. Food and water were provided ad libitum. Testis samples were dissected and flash-frozen in liquid nitrogen. Sequencing was performed by Azenta Life Sciences (Indianapolis) on a Illumina NovaSeq 6000 (PE150 technology) to generate ~50 Gb per sample. We processed and analyzed the bisulfite sequencing with the Nextflow pipeline methylseq (v 11.0.13) with default parameters (Ewels et al., 2020).
CpG islands are regions in the genome that contain a large number of hypomethylated CpG dinucleotide repeats and often serve as sites of transcription initiation (Deaton & Bird, 2011). We initially used the cpgplot (Madeira et al., 2022) tool in EMBOSS:6.6.0.0 to detect regions where, over an average of ten windows of 100 bases and a minimum of 250 consecutive bases, the GC content is more than 50% and the calculated observed-to-expected ratio of the number of CpG sites is >0.6. To expand on the set identified by cpgplot, we combined this information with the DNA methylation data from bisulfite sequencing. To this end, we defined hypomethylated CpG sites as positions where <50% of the reads support a methylated cytosine. We then implemented a hidden Markov model with six emission probabilities, corresponding to the four ‘standard’ nucleotides, as well as a hypomethylated C and a hypomethylated G—cytosine in the complementary strand—and eight hidden states (corresponding to an ‘island’ and ‘non-island’ state for each ‘standard’ nucleotide). Since we do not have a gold standard set of CpG islands in zebra finch, to set the transition and emission probabilities, we based ourselves on the CpG islands identified by cpgplot. We then used the function MultinomialHMM in the Python package hmmlearn to identify positions in the zebra finch genome supporting the ‘island’ states (Rabiner, 1989), merging those <50 bps away from each other, and keeping merged stretches longer than 150 bps and shorter than 2 kb. By this approach, the CpG islands identified contain the vast majority of those identified by cpgplot plus some islands that were presumably too short or missed for other reasons.
Detection of crossover events
To identify autosomal crossover events in the zebra finch pedigrees, we analyzed a total of 54 meiosis across the four multi-sibling pedigrees (Fig 1A), and implemented a previously described algorithm to call crossovers (Coop et al., 2008). The algorithm uses the transmission of “informative sites” (i.e., positions that are heterozygous in one parent but not in the other) to phase the haplotypes inherited by the offspring and identify crossover events in the germline of the parents. We focused on polymorphic positions where all individuals in the focal pedigree had a depth of coverage of at least eight reads, none of the individuals had an indel within 10 bps, and masked heterozygous genotypes with evidence of allelic imbalance (based on the p-value of a two-sided exact binomial test > 0.01).
Briefly, for each F0 parent with multiple sequenced offspring, we considered each offspring in turn as the “template” individual. The alleles in the non-template offspring are re-coded to “1” if they match the parental allele transmitted to the template, “2” if they match the untransmitted allele, or “0” if they have a missing genotype. After this re-coding, positions at which a non-template offspring changes from copying one allele to the other are defined as a “switch” (e.g., sequences such as “..1 1 2 2..”). We called putative crossovers in the template individual as having occurred within intervals in which the majority (in practice, usually all) of the non-template individuals with genotype calls showed a switch. In turn, for F1 individuals with sequenced offspring, we re-coded informative sites as “1” and “2” states in the offspring depending on the F0 individual they were inherited from. We then called putative crossovers as transitions from one state to another.
This procedure is susceptible to genotyping errors, e.g., missed heterozygotes in the focal individual, which generate changes of phase in close physical succession. To address this issue, we grouped putative crossovers within ten informative sites of each other in each of the template individuals, and kept only those clusters with an odd number of changes of phase, classifying events with more than a single phase change as “complex”. In addition, we removed crossovers within ten informative markers of the edges of the chromosomes.
Finally, to enhance the resolution of putative crossover locations, we relaxed the filtering criteria for polymorphic positions. Specifically, we identified previously masked informative sites within each putative crossover interval by reducing the minimum depth of coverage to 5X and the distance to indels to 5 bps. We redefined the crossover interval if a single phase switch occurred in the same template individual as the original crossover location. This approach improved the resolution of 439 crossovers, reducing the median interval length from 849 to 647 bps.
Detection of non-crossover events
While crossover events manifest as a single change of phase among the parental haplotypes, non-crossovers are indicated by two changes of phase within a relatively short genomic distance. In most of the cases, non-crossovers involve a single SNP, making it challenging to distinguish genuine non-crossover events from sequencing and genotyping errors. To keep the number of false positives at a minimum, we compared non-crossover calls obtained from GATK with those based on two extra variant callers (bcftools and freebayes).
We called variants with freebayes, using a minimum mapping quality of 30, a minimum base quality of 30, and limited the analysis to the best four alternative alleles per site (v1.3.6; Garrison & Marth, 2012). To standardize the output format of variants occurring in phase and close to each other, which are represented as haplotypes in freebayes, we used vcfallelicprimitives from vcflib suite (version 1.0.10; https://github.com/vcflib/vcflib).
We used bcftools mpileup to generate a pileup format of sequencing reads with a minimum base quality of 30 and a minimum mapping quality of 30, restricting the depth to a maximum of 150 reads (version 1.9; Li, 2011). We then used bcftools call to identify variants, and bcftools filter to exclude low-quality variants with a quality score below 20.
To minimize false positives, we implemented extra stringent filtering criteria. Specifically, we restricted the detection of non-crossover events to the 30 autosomes with an average mapping coverage above 20 and an average mapping quality above 40 across all individuals (S7 Fig). Moreover, we only considered genotypes meeting all of these criteria: (i) the site(s) in the putative conversion tract are consistent with the rules of Mendelian segregation, (ii) the depth of coverage is above half and below twice the average for that chromosome in that individual, and (iii) there is no evidence of allelic imbalance in heterozygous calls of the parents or any of the siblings (based on the p-value of a two-sided exact binomial test > 0.05).
To identify non-crossovers, we followed a similar approach as for crossovers, but focused on genomic segments that included two changes of phase in the same meiosis. To minimize assembly and mapping problems, and given the small number of meiosis considered, we further required no change of phase within the same region in another meiosis. To further avoid the challenge of calling events at repetitive loci that are collapsed in the reference assembly, we only included pairs of phase changes surrounded by at least 10 congruent informative sites on each side (i.e., informative sites that do not change phase in any individual). With these criteria, we identified 158, 398 and 384 instances with two changes of phase (i.e., putative non-crossovers) with GATK, Bcftools and freebayes, respectively.
We removed non-crossover events located within 1 kb from any other. To estimate the impact of repetitive regions that are challenging to analyze with short-read sequencing, we analyzed transmission rates to the next generation as a function of the mappability around putative non-crossover events. Specifically, for each non-crossover event, we computed the fraction of mappable bps (see Mappability section) found within the two closest non-converted informative sites. Based on this analysis, we kept events in which at least 50% of bps are mappable (S22 Fig). This approach led us to detect 121, 288 and 278 non-crossovers with GATK, Bcftools and freebayes, respectively.
Finally, we calculated the transmission rate of non-crossovers identified by each caller, expecting that for true calls it should be approximately 50% (assuming complete power to detect transmission). Based on this criterion, we decided to base ourselves on the non-crossovers called from freebayes for the rest of the analyses (S5 Table).
Of the 275 events identified using freebayes, three are very poorly resolved: the distance between the single polymorphic site in the putative conversion tract and the closest non-converted informative site is >10 kb and in two cases close to 1 Mb. In all three cases, heterozygosity is extremely low in these regions. We therefore excluded them from consideration and focused our analyses on the other 280 events.
Overlap between recombination events and CpG islands
To compare the two types of recombination events, we restricted our attention to crossovers that occurred on the 30 chromosomes used to call non-crossovers. Crossovers that could not be resolved in an interval of <10 kb were filtered out, leaving 933 crossovers. We used bedtools closest to measure the distance between a recombination event and a genomic feature, and counted the number of crossovers or non-crossovers within 100 bps from a CpG island.
To estimate the overlap expected by chance, for each recombination event defined within an interval, we generated all the locations to which the same length interval could be randomly assigned. Specifically, we kept all the windows fulfilling the following criteria: (i) within 2.5 Mb from the edge of the original interval (ii) a GC content within 2.5% the GC content of the 100 kb surrounding the original event (iii) a fraction of mappable bps within 20% of the original interval and (v) no overlap with the the original interval. We randomly sampled 5,000 such windows for each recombination event. For each of the 2000 sets of shuffled distribution, we counted the number of shuffled windows within 100 bps from a CpG island.
GC-biased gene conversion (gBGC)
To call non-crossovers, we relied on informative sites and applied stringent filters, potentially leading us to miss additional SNPs in conversion tracts. To examine the number of SNPs in the conversion tract, we reconstructed the haplotypes of each individual in the pedigree. Specifically, for each non-crossover, we extracted all SNPs present between the two flanking non-converted informative sites from the VCF file that had not passed our filtering (see section ‘Detection of non-crossover events’). In order to identify which of the additional SNPs were co-converted with the informative site(s) and thus likely in the conversion tract, we phased the biallelic SNPs using whatshap (v2.1) (Martin et al., 2016). Out of the 275 non-crossovers, we successfully reconstructed the haplotypes of all individuals across the pedigree for 103 events.
Given that the non-crossovers identified in the pedigree with three siblings show suspiciously many pairs of changes of phase per meiosis (S7 Table), suggesting that inferences of non-crossovers in small families is less reliable, we excluded that family from this analysis.
Among the 103 non-crossovers for which we reconstructed the haplotypes, 75 did not include any other SNPs in the conversion tract. To these cases, we added those for which the phasing did not succeed but where there was no other SNP than the informative site in the unfiltered VCF (n = 13), totaling 88 events with only 1 SNP. Among the events where more SNPs were added to a conversion tract, some of those might be false positives; we therefore kept only those SNPS that passed the same stringent filters applied to identify informative sites. Together with the unphased events with more than one informative site, we identified 39 multi-SNPs events, including a total of 101 SNPs. We lacked support to determine if there is a single or multiple SNPs for 146 events, which remain unclassified.
The degree of gBGC was then computed for three sets of non-crossovers: (i) all the non-crossovers, (ii) the non-crossovers for which the phasing analysis confirmed the presence of a single heterozygous position in the conversion tract, and (iii) the non-crossovers with more than one heterozygous position in the conversion tract. For each set, we analyzed the informative heterozygous sites used for the detection of phase changes, which pass all filtering criteria (see “VCF extra filtering step” subsection). To estimate the magnitude of gBGC, we selected all the positions carrying a Strong (G or C) and a Weak (A or T) allele, and quantified the proportion of times the Strong allele was transmitted over the Weak one:
Conversion tract length estimation
To estimate the mean length of the gene conversion tracts, we followed the approach in Li et al, 2019 (Li et al., 2019). This method assumes that the conversion tract length follows an exponential distribution, such that the probability of co-conversion of two informative sites sites apart is given by (An informative site here is defined as in the section “Detection of crossover events”). For each informative site inferred to be in a conversion event, we recorded its distance to the other informative sites in a window of 5000 bps, each side, as well as whether the sites were co-converted with the focal informative site. We estimated the mean conversion tract length (i.e., ) by maximum likelihood over a grid of 1 bp ranging between 1 to 1000. In this approach, each converted site is treated as independent, even when they may not be; the resulting likelihood is therefore a composite likelihood. To estimate uncertainty, we bootstrapped the set of informative sites 1000 times; the central 950 values were used to estimate the 95% confidence interval. We repeated this procedure using a window of 2000 bps instead of 5000, and the estimated mean tract length was identical and the confidence interval very similar.
Estimating the number of non-crossovers in a meiosis
The total number of non-crossovers can be estimated from the number of observed events, given an estimate of the power to detect non-crossovers. The power depends on the probability of having at least one informative site within the conversion tract. This probability hinges on the density of informative sites and the mean conversion tract length.
To estimate our power, we considered each chromosome in each focal individual (a founder of the pedigree) separately. As for the gBGC estimates, we excluded the family with three siblings for this analysis. We took 1M draws from an exponential distribution with parameter , where is the mean conversion length, and placed the intervals along the chromosome at random, recording the fraction that overlapped at least one informative site. We used three values of : our estimate of the mean conversion tract length (23 bps) and the lower and upper bounds on the 95% confidence intervals for L (15 and 35 bps). We considered the fraction of intervals that overlapped an informative site as our estimate of the probability of detecting a non-crossover event on that chromosome in that founder.
We then estimated the total number of non-crossovers on a chromosome by dividing the observed number of events by the estimate of the power. We computed the average number across founders to produce estimates per chromosome and considered the sum for the estimates per chromatid (Fig 5).
Supplementary Material
Acknowledgments
We thank Sarah London (University of Chicago) for providing two zebra finch testis samples and Sarah London as well as members of the Andolfatto, Przeworski, and Sella labs for helpful discussions.
Fundings
The zebra finches were established in captivity and maintained with support from the Australian Research Council to SCG. This work was supported by R35 GM153355 and R01 GM83098 to MP, as well as a Human Frontiers Science Program Fellowship to MM (LT000257/2021-L).
Data Accessibility
The whole genome sequencing data and the bisulfite sequencing data are available in the GenBank Sequence Read Archive under the accession numbers PRJNA1152924 and PRJNA1152957, respectively. The code is available at https://doi.org/10.5281/zenodo.13696268
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al.2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Bird A. 1993. Number of CpG islands and genes in human and mouse.Proceedings of the National Academy of Sciences 90: 11995–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Li YR, Kidd J, Oliveira K, Nadel J, Holloway JK, Hayward JJ, Cohen PE, Greally JM, Wang J, et al.2013. Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs. PLOS Genetics 9: e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Öst T, Schneider M, Kempenaers B, et al.2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Research 20: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Z, Schumer M, Haba Y, Bashkirova L, Holland C, Rosenthal GG, Przeworski M. 2017. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates (de Massy B, Ed.). eLife 6: e24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan CN, Edwards SV. 2009. Nucleotide Variation, Linkage Disequilibrium and Founder-Facilitated Speciation in Wild Populations of the Zebra Finch (Taeniopygia guttata). Genetics 181: 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B.2010. PRDM9 Is a Major Determinant of Meiotic Recombination Hotspots in Humans and Mice.Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron LA, Besenbacher S, Zheng J, Li P, Bertelsen MF, Quintard B, Hoffman JI, Li Z, St. Leger J, Shao C, et al.2023. Evolution of the germline mutation rate across vertebrates.Nature 615: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besenbacher S, Hvilsom C, Marques-Bonet T, Mailund T, Schierup MH. 2019. Direct estimation of mutations in great apes reconciles phylogenetic dating. Nature Ecology & Evolution 3: 286–292. [DOI] [PubMed] [Google Scholar]
- Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Research 8: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan A, Baez-Ortega A, Brzozowska N, Abascal F, Coorens THH, Sanders MA, Lawson ARJ, Harvey LMR, Bhosle S, Jones D, et al.2022. Somatic mutation rates scale with lifespan across mammals. Nature. [DOI] [PMC free article] [PubMed]
- Calderón PL, Pigozzi MI. 2006. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosome Research 14: 605–612. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Bhérer C, Morrow BE, Boyko AR, Auton A. 2016. A Pedigree-Based Map of Recombination in the Domestic Dog Genome. G3 Genes|Genomes|Genetics 6: 3517–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH, Jenkins PA, Song YS. 2012. Genome-Wide Fine-Scale Recombination Rate Variation in Drosophila melanogaster. PLOS Genetics 8: e1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmouh AP, Sørud PP, Bataillon T, Hobolth A, Hansen LT, Besenbacher S, Winge SB, Almstrup K, Schierup MH. 2024. Estimating gene conversion tract length and rate from PacBio HiFi data. : 2024.07.05.601865.
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nature Cell Biology 14:424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S. 2012. The Many Landscapes of Recombination in Drosophila melanogaster. PLOS Genetics 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. 2007. An evolutionary view of human recombination. Nature Reviews Genetics 8: 23–34. [DOI] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. 2008. High-Resolution Mapping of Crossovers Reveals Extensive Variation in Fine-Scale Recombination Patterns Among Humans.Science 319: 1395–1398. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al.2011. The variant call format and VCFtools.Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al.2021. Twelve years of SAMtools and BCFtools. Gigascience 10: giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2019. Molecular evolution of the meiotic recombination pathway in mammals. Evolution 73: 2368–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes & Development 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury J-F. 2012. A linear complexity phasing method for thousands of genomes. Nature Methods 9: 179–181. [DOI] [PubMed] [Google Scholar]
- Díaz-Gay M, Vangara R, Barnes M, Wang X, Islam SMA, Vermes I, Duke S, Narasimman NB, Yang T, Jiang Z, et al.2023. Assigning mutational signatures to individual samples and individual somatic mutations with SigProfilerAssignment. Bioinformatics 39: btad756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Khademian H, Giraut L, Zanni V, Bellalou S, Henderson IR, Falque M, Mézard C. 2013. Contrasted Patterns of Crossover and Non-crossover at Arabidopsis thaliana Meiotic Recombination Hotspots. PLOS Genetics 9: e1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2008. EVOLUTION OF THE GENOMIC RATE OF RECOMBINATION IN MAMMALS. Evolution 62: 276–294. [DOI] [PubMed] [Google Scholar]
- Durbin RM, Altshuler D, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Collins FS, De La Vega FM, Donnelly P, et al.2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutoit L, Burri R, Nater A, Mugal CF, Ellegren H. 2017. Genomic distribution and estimation of nucleotide diversity in natural populations: perspectives from the collared flycatcher (Ficedula albicollis) genome. Molecular Ecology Resources 17: 586–597. [DOI] [PubMed] [Google Scholar]
- Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, Genome of the Netherlands Consortium, van Duijn CM, Swertz M, Wijmenga C, et al.2015. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 47: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MU, Hanssen F, Pedersen AS, Gabernet G, Wacker O, SusiJo, Talbot A, James C, bot nf-core, Ergüner B, et al.2024. nf-core/sarek: Sarek 3.4.2 - Sájtáristjåhkkå.
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing.
- Gelova SP, Doherty KN, Alasmar S, Chan K. 2022. Intrinsic base substitution patterns in diverse species reveal links to cancer and metabolism. Genetics 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann JM, Wong WSW, Pinelli M, Farrah T, Bodian D, Stittrich AB, Glusman G,Vissers LELM, Hoischen, Roach JC, et al.2016. Parent-of-origin-specific signatures of de novo mutations. Nature Genetics 48: 935–939. [DOI] [PubMed] [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, Berner D. 2018. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Molecular Ecology 27: 2477–2497. [DOI] [PubMed] [Google Scholar]
- Halldorsson BV, Hardarson MT, Kehr B, Styrkarsdottir U, Gylfason A, Thorleifsson G, Zink F, Jonasdottir A, Jonasdottir A, Sulem P, et al.2016. The rate of meiotic gene conversion varies by sex and age. Nature Genetics 48: 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldorsson BV, Palsson G, Stefansson OA, Jonsson H, Hardarson MT, Eggertsson HP, Gunnarsson B, Oddsson A, Halldorsson GH, Zink F, et al.2019. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363: eaau1043. [DOI] [PubMed] [Google Scholar]
- Han L, Su B, Li W-H, Zhao Z. 2008. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biology 9: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland C, Charlier C, Karim L, Cambisano N, Deckers M, Mni M, Mullaart E, Coppieters W, Georges M. 2017. Frequency of mosaicism points towards mutation-prone early cleavage cell divisions in cattle. bioRxiv.
- Harris K, Nielsen R. 2014. Error-prone polymerase activity causes multinucleotide mutations in humans. Genome Research: gr.170696.113. [DOI] [PMC free article] [PubMed]
- Hellenthal G, Stephens M. 2006. Insights into recombination from population genetic variation.Current Opinion in Genetics & Development 16: 565–572. [DOI] [PubMed] [Google Scholar]
- Hinch R, Donnelly P, Hinch AG. 2023. Meiotic DNA breaks drive multifaceted mutagenesis in the human germ line. Science 382: eadh2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoge C, de Manuel M, Mahgoub M, Okami N, Fuller Z, Banerjee S, Baker Z, McNulty M, Andolfatto P, Macfarlan TS, et al.2024. Patterns of recombination in snakes reveal a tug-of-war between PRDM9 and promoter-like features. Science 383: eadj7026. [DOI] [PubMed] [Google Scholar]
- Ioannidis J, Taylor G, Zhao D, Liu L, Idoko-Akoh A, Gong D, Lovell-Badge R, Guioli S, McGrew MJ, Clinton M. 2021. Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine adult secondary sex characteristics. Proceedings of the National Academy of Sciences 118: e2020909118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE. 2024. Understanding the Genetic Basis of Variation in Meiotic Recombination: Past, Present, and Future. Molecular Biology and Evolution 41: msae112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, Hardarson MT, Hjorleifsson KE, Eggertsson HP, Gudjonsson SA, et al.2017. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549: 519–522. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Mugal CF, Suh A, Nater A, Burri R, Smeds L, Ellegren H. 2017. Whole-genome patterns of linkage disequilibrium across flycatcher populations clarify the causes and consequences of fine-scale recombination rate variation in birds. Molecular Ecology 26: 4158–4172. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Ness RW, Halligan DL, Haddrill PR. 2014. Estimation of the Spontaneous Mutation Rate per Nucleotide Site in a Drosophila melanogaster Full-Sib Family. Genetics 196: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, et al.2012. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, Stecher G, Hedges SB. 2022. TimeTree 5: An Expanded Resource for Species Divergence Times. Molecular Biology and Evolution 39: msac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW. 2008. Estimating the Per-Base-Pair Mutation Rate in the Yeast Saccharomyces cerevisiae. Genetics 178: 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Bitoun E, Altemose N, Davies RW, Davies B, Myers SR. 2019. A high-resolution map of non-crossover events reveals impacts of genetic diversity on mammalian meiotic recombination. Nature Communications 10: 3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay SJ, Rahbari R, Kaplanis J, Keane T, Hurles ME. 2019. Similarities and differences in patterns of germline mutation between mice and humans. Nat. Commun. 10: 4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MH, Costa BM, Bianchini EC, Choi U, Bandler RC, Lassen E, Grońska-Pęski M, Schwing A, Murphy ZR, Rosenkjær D, et al.2024. DNA mismatch and damage patterns revealed by single-molecule sequencing. Nature 630: 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2010. Evolution of the mutation rate. Trends in Genetics 26: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R. 2022. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50: W276–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, Wu FL, Przeworski M. 2022. A paternal bias in germline mutation is widespread in amniotes and can arise independently of cell division numbers. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Patterson M, Garg S, Fischer SO, Pisanti N, Klau GW, Schöenhuth A, Marschall T. 2016. WhatsHap: fast and accurate read-based phasing. : 085050.
- Massy B de. 2013. Initiation of Meiotic Recombination: How and Where? Conservation and Specificities Among Eukaryotes. Annual Review of Genetics 47: 563–599. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al.2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Landa V, Pratto F, Brick K, Kobets T, Kusari F, Gasic S, Smagulova F, Grey C, Flachs P, et al.2021. Rat PRDM9 shapes recombination landscapes, duration of meiosis, gametogenesis, and age of fertility. BMC Biology 19: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milholland B, Dong X, Zhang L, Hao X, Suh Y, Vijg J. 2017. Differences between germline and somatic mutation rates in humans and mice. Nature Communications 8: 15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Gao Z, Przeworski M. 2016. Human Germline Mutation and the Erratic Evolutionary Clock. PLOS Biology 14: e2000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive Against Hotspot Motifs in Primates Implicates the PRDM9 Gene in Meiotic Recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP. 2009. Accelerated Evolution of the Prdm9 Speciation Gene across Diverse Metazoan Taxa. PLOS Genetics 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. 2002. Resolving the paradox of sex and recombination. Nature Reviews Genetics 3: 252–261. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2003. Chromosome choreography: the meiotic ballet. Science (New York, N.Y.) 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 Controls Activation of Mammalian Recombination Hotspots. Science 327: 835–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsborg PS, Charmouh AP, Singh VK, Winge SB, Hvilsom C, Pelizzola M, Laurentino S, Neuhaus N, Hobolth A, Bataillon T, et al.2024. Insights into gene conversion and crossing-over processes from long-read sequencing of human, chimpanzee and gorilla testes and sperm. : 2024.07.05.601967.
- Porubsky D, Dashnow H, Sasani TA, Logsdon GA, Hallast P, Noyes MD, Kronenberg ZN, Mokveld T, Koundinya N, Nolan C, et al.2024. A familial, telomere-to-telomere reference for human de novo mutation and recombination from a four-generation pedigree.
- Quinlan AR, Hall IM. 2010a. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010b. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner LR. 1989. A tutorial on hidden Markov models and selected applications in speech recognition. Proceedings of the IEEE 77: 257–286. [Google Scholar]
- Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Al Turki S, Dominiczak A, Morris A, Porteous D, Smith B, et al.2015. Timing, rates and spectra of human germline mutation. Nat. Genet. 48: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud M, Gagnaire P-A, Galtier N. 2023. Performance and limitations of linkage-disequilibrium-based methods for inferring the genomic landscape of recombination and detecting hotspots: a simulation study. Peer Community Journal 3. [Google Scholar]
- Raynaud M, Sanna P, Joseph J, Clément J, Imai Y, Lareyre J-J, Laurent A, Galtier N, Baudat F, Duret L, et al.2024. PRDM9 drives the location and rapid evolution of recombination hotspots in salmonids. : 2024.03.06.583651.
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al.2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, Kirkpatrick M. 2020. Sex Differences in the Recombination Landscape. The American Naturalist 195: 361–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasani TA, Pedersen BS, Gao Z, Baird L, Przeworski M, Jorde LB, Quinlan AR. 2019. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. Elife 8: e46922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasani TA, Quinlan AR, Harris K. 2024. Epistasis between mutator alleles contributes to germline mutation spectrum variability in laboratory mice (Gao Z and Weigel D, Eds.). eLife 12: RP89096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield DR, Pasquesi GIM, Perry BW, Adams RH, Nikolakis ZL, Westfall AK, Orton RW, Meik JM, Mackessy SP, Castoe TA. 2020. Snake Recombination Landscapes Are Concentrated in Functional Regions despite PRDM9. Molecular Biology and Evolution 37: 1272–1294. [DOI] [PubMed] [Google Scholar]
- Schiffels S, Durbin R. 2014. Inferring human population size and separation history from multiple genome sequences. Nature Genetics 46: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger R, Lee S, Zhou C, Yang T-P, Smith K, Li S, Sanghvi R, Neville M, Mitchell E, Nessa A, et al.2024. Insights into non-crossover recombination from long-read sperm sequencing. : 2024.07.05.602249.
- Singhal S, Leffler EM, Sannareddy K, Turner I, Venn O, Hooper DM, Strand AI, Li Q, Raney B, Balakrishnan CN, et al.2015. Stable recombination hotspots in birds. Science 350: 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Mugal CF, Qvarnström A, Ellegren H. 2016a. High-Resolution Mapping of Crossover and Non-crossover Recombination Events by Whole-Genome Re-sequencing of an Avian Pedigree. PLOS Genetics 12: e1006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Qvarnström A, Ellegren H. 2016b. Direct estimate of the rate of germline mutation in a bird. Genome Res. 26: 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spisak N, Manuel M de, Milligan W, Sella G, Przeworski M. 2024. The clock-like accumulation of germline and somatic mutations can arise from the interplay of DNA damage and repair. PLOS Biology 22: e3002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K, Ahmad SF, Singchat W, Panthum T. 2021. Why Do Some Vertebrates Have Microchromosomes? Cells 10: 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Birkhead TR, Burke T, Slate J. 2008. A Linkage Map of the Zebra Finch Taeniopygia guttata Provides New Insights Into Avian Genome Evolution. Genetics 179: 651–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versoza CJ, Weiss S, Johal R, La Rosa B, Jensen JD, Pfeifer SP. 2024. Novel Insights into the Landscape of Crossover and Noncrossover Events in Rhesus Macaques (Macaca mulatta). Genome Biology and Evolution 16: evad223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Robinson JA, Cox LA. 2022. High-Resolution Estimates of Crossover and Noncrossover Recombination from a Captive Baboon Colony. Genome Biology and Evolution 14: evac040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Grützner F, et al.2021. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proceedings of the National Academy of Sciences 118: e2112494118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-Statistics for the Analysis of Population Structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Williams AL, Genovese G, Dyer T, Altemose N, Truax K, Jun G, Patterson N, Myers SR, Curran JE, Duggirala R, et al.2015. Non-crossover gene conversions show strong GC bias and unexpected clustering in humans (de Massy B, Ed.). eLife 4: e04637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FL, Strand AI, Cox LA, Ober C, Wall JD, Moorjani P, Przeworski M. 2020. A comparison of humans and baboons suggests germline mutation rates do not track cell divisions. PLoS Biol. 18: e3000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lundberg M, Ponnikas S, Hasselquist D, Hansson B. 2024. Male-biased recombination at chromosome ends in a songbird revealed by precisely mapping crossover positions. G3 Genes|Genomes|Genetics: jkae150. [DOI] [PMC free article] [PubMed]
- Zhang H, Lundberg M, Tarka M, Hasselquist D, Hansson B. 2023a. Evidence of Site-Specific and Male-Biased Germline Mutation Rate in a Wild Songbird. Genome Biology and Evolution 15: evad180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lundberg M, Tarka M, Hasselquist D, Hansson B. 2023b. Evidence of Site-Specific and Male-Biased Germline Mutation Rate in a Wild Songbird. Genome Biology and Evolution 15: evad180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, McBride D, Nandi S, McQueen HA, McGrew MJ, Hocking PM, Lewis PD, Sang HM, Clinton M. 2010. Somatic sex identity is cell autonomous in the chicken. Nature 464: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole genome sequencing data and the bisulfite sequencing data are available in the GenBank Sequence Read Archive under the accession numbers PRJNA1152924 and PRJNA1152957, respectively. The code is available at https://doi.org/10.5281/zenodo.13696268