Abstract
Diffusion tensor imaging (DTI) is a magnetic resonance imaging (MRI) technique that can be used to assess microstructural features of skeletal muscle that are related to tissue function. Although widely used, direct correlations between DTI derived metrics such as fractional anisotropy and spatially matched tissue microstructure assessed with histology have not been performed. This study investigated the relationship between scalar-based DTI measurements and histologically derived muscle microstructural measurements in rat rotator cuff muscles. Despite meticulous co-localization of MRI and histology data, negligible correlations were found between DTI metrics and histological measurements including muscle fiber diameter, cross-sectional area, and surface-to-volume ratio. These findings highlight the challenges in validating DTI with histology due to requirements in anatomical co-localization, necessity of high-quality histology, and consideration of diffusion measurement scales. Our findings underscore the need for further research with optimized imaging parameters to enhance our knowledge regarding the sensitivity of DTI to important features of muscle microstructure.
Keywords: diffusion tensor imaging, rotator cuff, skeletal muscle, histological correlation
Introduction
Diffusion magnetic resonance imaging (dMRI) is an imaging technique that exploits the anisotropic motion of water molecules to generate image contrast, enabling quantification of the restricted diffusion profile of a tissue1. It is well-established that dMRI provides unique clues to the microstructure and architecture of tissues that have biological and clinical relevance2. Although most widely utilized in neuroimaging, dMRI has been increasingly used for musculoskeletal applications, including skeletal muscle imaging3.
dMRI data can be mathematically modeled to fully characterize how diffusion in space varies according to direction, a technique referred to as diffusion tensor imaging (DTI)2. One of the goals of DTI applications in skeletal muscle is to assess key features of muscle microstructure related to function including muscle fiber size. Muscle fiber diameter and cross-sectional area are directly related to the isometric force-generating capacity of a muscle fiber4 and thus, a noninvasive tool that can measure changes in fiber size associated with injury, pathology, or treatment holds great clinical value. DTI in skeletal muscle has been applied to study clinically relevant applications including assessment of severity of muscular dystrophy5–8, inflammatory myositis9–11, and denervation12,13, as well as age-related muscle changes14,15, monitoring recovery after injury16, and prediction of return to sport17.
Despite these successes, key questions on a more fundamental level remain. For instance, the ability of DTI to resolve specific features of muscle microstructure is unknown. Validation with histology is required for an answer to this question, and the studies to date using histological tissue have provided conflicting results. A few studies have utilized the random permeable barrier model (RPBM), which leverages diffusion time (Δ) dependent stimulated echo DTI data into a mathematical framework used to relate diffusion measurements in tissue to discrete microstructural features of muscle such as fiber size18. Some have found no significant associations between these DTI-based features of muscle microstructure and muscle biopsy data, including fiber diameters15,19. This has been hypothesized to be due to insufficient anatomic co-localization between modalities; it is highly difficult to precisely colocalize biopsy measurements (< 1 mL in volume)with whole muscle DTI based measurements, especially in large human muscles (hundreds to thousands of mLs in volume). In contrast, systematic simulations of DTI data and histology informed models of skeletal muscle injury indicate that significant relationships should exist, with the strength of the correlation dependent on the specific pulse sequence parameters and mathematical model used for the data20–22.
Another important parameter is spatial resolution of the imaging sequence, which is one of the most important practical considerations of any MRI protocol. dMRI/DTI is sensitive to molecular motion on a micrometer scale, but imaging protocols are often on a millimeter scale. Since a muscle fiber is approximately 30–70 μm in diameter, hundreds to tens of thousands of muscle fibers can pass through a single voxel. Thus, the resulting diffusion signal is sensitive to macroscopic anisotropy of the underlying muscle tissue and is not sensitive to heterogeneity within the voxel. It has been suggested that diffusion measurements remain stable among different voxel sizes in phantoms and rat brain tissue23, but the effects of imaging resolution on correlations between DTI measures and histologically-derived measurements remains unknown.
The purpose of this exploratory study was to quantify the relationship between scalar-based measures of DTI and histologically-derived microstructural measurements in precisely co-localized rat rotator cuff muscle tissue and to compare the results when imaged at 0.25 mm and 0.5 mm isotropic resolutions.
Methods and Materials
Ethical Approval
All methods were performed in accordance with the relevant guidelines and regulations. All animal research was approved by the VA San Diego Healthcare System Institutional Animal Care and Use Committee, with protocol number: A20–005. The reporting in this manuscript follows the recommendations in the ARRIVE guidelines (PLoS Bio 8(6), e1000412, 2010).
Animals and Rotator Cuff Tear Model
Experiments were performed on Four Lewis rats, which were originally purchased from Charles River Laboratories (Portage, MI, USA). A rat model of chronic massive rotator cuff tearing was adapted for the purpose of increasing the range of myofiber sizes24. Specifically, smaller sizes are seen post-injury24,25. At 13-weeks of age, supraspinatus and infraspinatus tenotomies were performed on the right shoulders of each animal and the distal 2–3 mm of the tendons were removed. A thin, L-shaped silicone (polydimethylsiloxane [PDMS]) implant was placed over the greater tuberosity and firmly fixed with a size 2–0 nonabsorbable polyester suture to prevent spontaneous tendon healing. After closure, rats were allowed unrestricted cage activity. At 20-weeks post injury, the rats were deeply anesthetized with isoflurane and euthanized via terminal perfusion with 250 ml saline and 300 ml 4% paraformaldehyde (PFA) in phosphate buffer. Each scapula containing all rotator cuff muscles was individually dissected and postfixed in PFA at 4°C for 48 hours.
Magnetic Resonance Imaging
All scapulae were individually imaged using a 30 mm receive-only surface coil and 82 mm volume coil for transmission on a 3T pre-clinical MRI scanner (BioSpec 3T, Bruker, Billerica, MA, USA). Relaxometry was performed on two randomly selected fixed rotator cuff muscle samples using the Image Sequence Analysis tool on ParaVision 360 v3.3 (Bruker) to inform the optimal parameters for the DTI sequences. Using a variable repetition time (TR) rapid acquisition with relaxation enhancement sequence with 5 TRs ranging from 850–4000 ms, mean mono-exponential T1 was approximately 900 ms. For optimal SNR efficiency (~1.3x T126), a TR of 1200 was selected for the DTI sequences. Using a Carr−Purcell−Meiboom−Gill sequence with TR = 7500 ms and 8 echo times (TEs) ranging from 9–72 ms was, mean mono-exponential T2 was 35 ms. A TE of 23–25 ms was selected as for the DTI sequences since approximately 50% of the transverse signal would remain based on the T2 decay.
Two sets of spin echo single-pulsed field gradient (sPFG) DTI sequences with fat-suppressed spin-echo readout were employed, one with 0.25 mm isotropic resolution and the other with 0.5 mm isotropic resolution. The 0.25 mm isotropic sequence included the following imaging parameters: TR/TE = 1200/25 ms, number of excitations (NEX) = 2, bandwidth = 30 kHz, 30 diffusion encoding directions with b = 500 s/mm2 (gradient duration/gradient separation = 3.1/15 ms), five b = 0 s/mm2 images, with a total imaging time of approximately 80 hours. The 0.5 mm isotropic sequences included the following imaging parameters: TR/TE = 1200/23 ms, NEX = 1, bandwidth = 30 kHz, 30 diffusion encoding directions with b = 500 s/mm2 (gradient duration/gradient separation = 3.1/15 ms), five b = 0 s/mm2 images, with a total imaging time of approximately 10 hours.
Magnetic Resonance Imaging Data Analysis
The supraspinatus and infraspinatus muscles were manually segmented from the 0.25mm isotropic dMRI b = 0 s/mm2 images using Horos (v4.0, Purview, Annapolis, MD, USA). Segmented volumes were scaled to the 0.5mm resolution using the analysis of functional neuroimaging (AFNI) command 3dresample27,28. Diffusion weighted images were denoised using a local principal component analysis filter29. The diffusion tensor was calculated using the AFNI command 3dDWItoDT and fractional anisotropy (FA), mean diffusivity (MD), and radial diffusivity (RD) were the parameters that were included in the analysis.
Histological Preparation and Acquisition
Samples were decalcified in 20% ethylenediaminetetraacetic acid (EDTA) for 4 weeks with a medium change every 72 hours that was checked weekly with radiography. Following decalcification, the samples were copiously washed in phosphate buffered saline, treated with 30% sucrose for cryoprotection, snap-frozen in liquid nitrogen-cooled isopentane, and then embedded in optimal cutting temperature (OCT) compound.
Sectioning and staining performed by a subspecialized MSK pathologist (X.C., with 24 years of experience). Twelve μm-thick cryosections were collected using a cryostat microtome in the transverse direction relative to the spine and body of the scapula, corresponding to the “sagittal oblique” plane on human MRI. At least three locations were sectioned: medial scapular, mid-scapular in the central region of the muscle belly, and myotendinous junction near the lateral margin where the scapular spine and body merged. All sections were stained with hematoxylin and eosin (H&E) and Masson trichrome (Epredia Richard-Allan Scientific Masson Trichrome Kit, 22110648, Fisher Scientific, Hampton, NH, USA). Brightfield image digitization was performed using a slide scanner at 0.22 μm/pixel (AxioScan.Z1, ZEISS, Thornwood, NY, USA).
Histologic Image Analysis
Quality control (QC) was performed on each digitized histology slide with the following requirements for inclusion in the study: scapular bone must be entirely preserved to permit co-localization with MRI, and at least 50% of either the supraspinatus or infraspinatus muscle must be preserved without tissue loss, folds, or distortion artifacts.
Whole-muscle myofiber boundary segmentation was performed using HALO AI (v3.6.4134, Indica Labs, Albuquerque, NM, USA). Specifically, the pre-trained Membrane Segmentation network feature in the software was fined-tuned using at least 50 manually segmented myofibers per slide by an MSK pathologist (X.C.). Each slide employed its own fine-tuned algorithm and QC was double checked in consensus by a MSK pathologist (X.C.) and MSK radiologist (E.Y.C.). Tendon regions and artifacts were manually excluded.
MATLAB software (R2024a, Mathworks Inc., Natick, MA) was used for size measurements on the histologic slides and processing of MRI data. Myofiber diameter, cross-sectional area, and surface area to volume (S/V) ratio measurements were determined from the whole-muscle segmentations. While myofiber diameter and cross-sectional area are commonly assessed features of muscle microstructure related to function, S/V is a relatively understudied feature of muscle microstructure that may have high correlations with DTI metrics, including RPBM modeling or diffusion tensor subspace imaging analysis22. Thus, while not a standard muscle microstructural measurement, as its relationship to function is still relatively unknown, this feature was additionally included as a biomarker for histologic assessment. Artifactual gaps between myofibers are expected as part of the histological preparation process and this was compensated through down-sampling of the spatial resolution of the histological slides using an averaging filter.
MRI data was reviewed by a MSK radiologist (E.Y.C., with 13 years of experience) and co-localization with the histological section locations was meticulously performed based on scapular bone and muscle dimensions. Signal-to-noise (SNR) was measured for all included samples at each resolution, defined as mean signal over the muscle divided by the standard deviation of the noise. Non-rigid body registration of muscle boundaries between the DTI and histology maps was performed. Further down-sampling was performed on these co-registered histological sections and the 0.25 mm isotropic resolution DTI data (2×2 kernel) to match the dimensions of the 0.5 mm isotropic resolution DTI data. Histograms were reviewed during each down-sampling step to ensure that the measurements and distributions remained consistent.
Statistical analyses
Scatter plots were generated on a pixel-by-pixel basis from co-localized DTI and histology data. Pearson’s correlations were performed between each scalar-based DTI metric (MD, RD, and FA) and each histologic metric (diameter, area, S/V) for all included samples and interpreted as: 0–0.29, negligible; 0.3–0.49, low; 0.5–0.69, moderate; 0.7–0.89, high; 0.9–1.0, very high30. Due to the exploratory nature of this study utilizing a very small sample size, adjustments for internal dependence and multiple comparisons were not made. Thus, all results are reported only informally and will need to be confirmed with independent studies. Statistical analysis was performed in MATLAB.
Results
Out of 48 possible muscle locations (4 rats, 2 shoulders per rat, each containing both supraspinatus and infraspinatus muscles, with 3 locations per muscle), 26 did not pass histologic QC. After QC exclusions, data from 2 rats remained (4 shoulders with 8 rotator cuff muscles) in the form of 22 distinct muscle locations, which were included in this study (Supplemental Figure 1).
Mean SNR of the rotator cuff muscles was: 92 on the b = 0 s/mm2 0.25 mm isotropic images, 46 on the b = 500 s/mm2 0.25 mm isotropic images, 156 on the b = 0 s/mm2 0.5 mm isotropic images, and 76 on the b = 500 s/mm2 0.5 mm isotropic images.
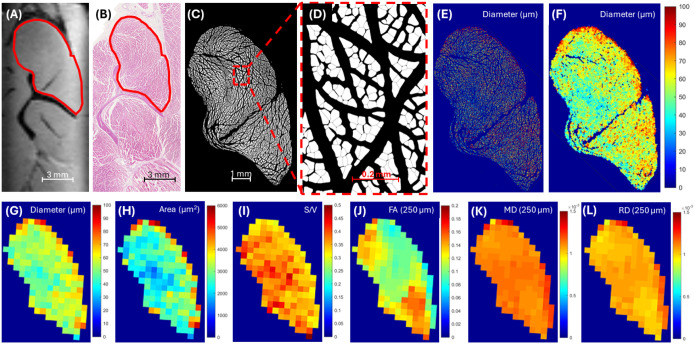
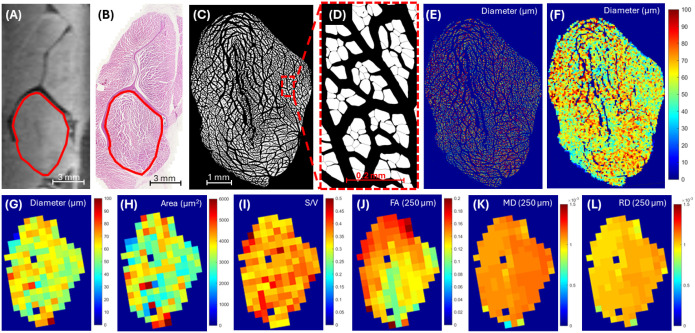
Figures 1 and 2 show representative examples for two muscle locations, including one in the supraspinatus and one in the infraspinatus, respectively. Guided by the scapular bone landmarks and muscle contours, co-registration between the DTI data (Figures 1A and 2A) and corresponding histological sections (Figures 1B and 2B) for all included samples was successful. Myofiber segmentation (Figures 1C/D and 2C/D) was also successful. Gaps introduced by the histological preparation process were successfully minimized through down-sampling with an averaging filter (Figures 1E/F and 2E/F). The resultant pixel maps of histological measures (Figures 1G–I and 2G–I) after down-sampling were readily correlated with DTI metric pixel maps (Figures 1J–L and 2J–L).
Figure 1.
Representative example of the supraspinatus muscle. 3D spin-echo fat-saturated DTI image at 0.25 mm isotropic resolution (A) and the corresponding histological section after H&E staining (B) with the supraspinatus muscle outlined in red. Automated whole-muscle myofiber boundary segmented images at low (C) and high (D) magnification. Automated fiber size (diameter) color map before (E) and after (F) 100x down-sampling of spatial resolution shows that the majority of the gaps introduced from histological preparation are omitted. After manual co-registration to the DTI images and further down-sampling to match the resolutions, diameter, area, and surface-to-volume (S/V) ratio pixel maps are generated (G-I). Corresponding fractional anisotropy (FA), mean diffusivity (MD), and radial diffusivity (RD) pixel maps at 0.25 mm isotropic resolution are shown (J-L).
Figure 2.
Representative example of the infraspinatus muscle. 3D spin-echo fat-saturated DTI image at 0.25 mm isotropic resolution (A) and the corresponding histological section after H&E staining (B) with the supraspinatus muscle outlined in red. Automated whole-muscle myofiber boundary segmented images at low (C) and high (D) magnification. Automated fiber size (diameter) color map before (E) and after (F) 100x down-sampling of spatial resolution shows that the majority of the gaps introduced from histological preparation are omitted. After manual co-registration to the DTI images and further down-sampling to match the resolutions, diameter, area, and surface-to-volume (S/V) ratio pixel maps are generated (G-I). Corresponding fractional anisotropy (FA), mean diffusivity (MD), and radial diffusivity (RD) pixel maps at 0.25 mm isotropic resolution are shown (J-L).
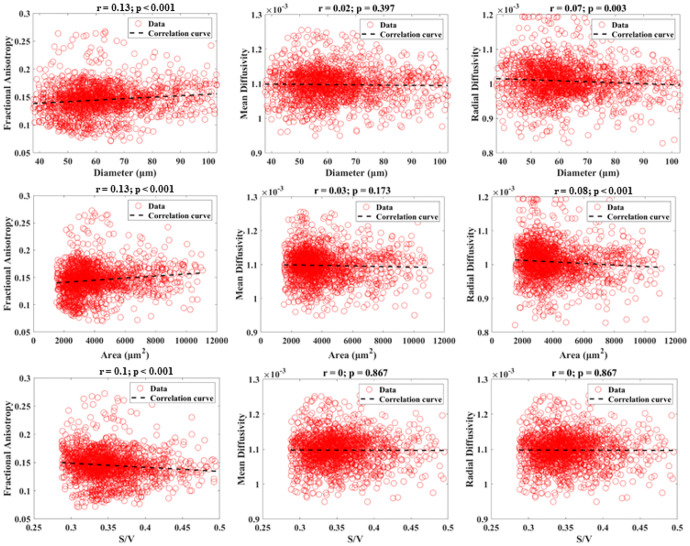
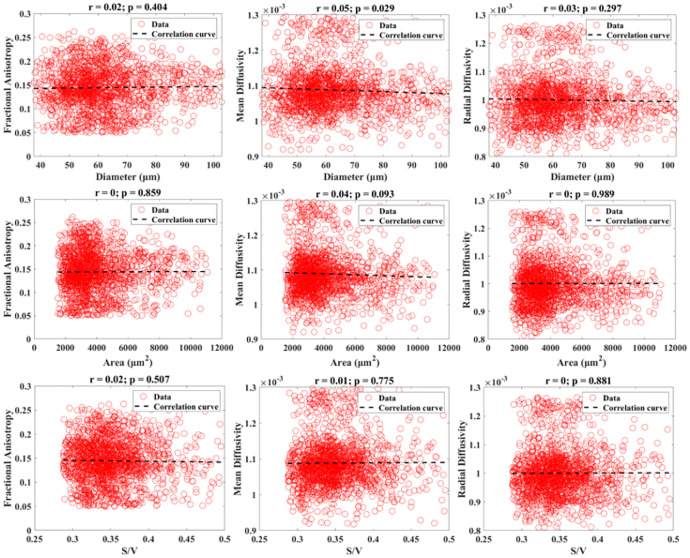
Figures 3 and 4 show the scatter plots and Pearson correlations for the 0.25 mm isotropic resolution and 0.5 mm isotropic resolution DTI data with their corresponding histologic measurements of all included muscle locations. All DTI measures yielded negligible correlations (r=0–0.13) with histologically derived microstructural measurements. No consistent trend was found between the 0.25 mm and 0.5 mm isotropic resolution correlations.
Figure 3.
Combined scatter plots showing the relationships between diffusion measurements (FA, MD, and RD) measured from DTI at 0.25 mm isotropic resolution versus histological measurements (diameter, area, and S/V). Pearson correlation results are displayed.
Figure 4.
Combined scatter plots showing the relationships between diffusion measurements (FA, MD, and RD) measured from DTI at 0.5 mm isotropic resolution versus histological measurements (diameter, area, and S/V). Pearson correlation results are displayed.
Discussion
In this study we investigated the relationship between scalar-based measures of DTI, including MD, RD, and FA, and histologically derived microstructural measurements, including myofiber diameter, area, and S/V, in precisely co-localized rat rotator cuff muscle tissue. Unfortunately, we failed to find a substantial relationship between any of the comparisons, for either the 0.25 mm or 0.5 mm isotropic datasets.
To potentially explain our results requires a discussion of some of the variables with DTI, particularly the conditions used in this study, which can be viewed in broad categories relating to the methods of measurement and the conditions of the tissue. We utilized a diffusion time (Δ) of 15 ms, which is in the intermediate range31, and typical of that which can be acquired with the spin echo sPFG method for diffusion preparation to study muscle microstructure3. Although the most commonly used method for diffusion sensitization, the main drawback to the spin echo SPFG method is that Δ is constrained by the TE (23–25 ms in our protocol). The TE of the sequence must be sufficiently short as muscle is a short T2 tissue (measured at 35 ms in our case), and SNR must be a minimum of 25 for accuracies of the DTI metrics to be within 5% or 50 to be within 1%32,33. Δ may also be substantially shorter, but either very high-performance pre-clinical gradients are required or an oscillating gradient spin echo (OGSE) diffusion encoding strategy must be employed. Earlier nuclear magnetic resonance studies found that short Δ values of a few seconds or less were able to probe S/V ratios in porous media34 and erythrocytes35. OGSE diffusion encoding has also shown promising results for S/V ratio measurements in anisotropic fiber phantoms36 and was more recently employed to study human muscle37, but the sensitivity of short Δ values to microstructural measurements in muscle remain to be determined.
Longer Δ can be achieved using stimulated echo diffusion preparations. Although SNR may be up to 2.9x lower compared lower compared with spin echo sPFG38, Δ values of greater than 1 second can be obtained39. Using Monte-Carlo simulations, Berry et al. previously demonstrated that Δ should be ~170 ms to maximally discriminate fiber size if only a single diffusion experiment can be performed, though ideally multiple diffusion times should be sampled21. This was independently supported by McDowell et al., where Δ = 190 ms yielded the largest effect size in discriminating between muscles from healthy adult volunteers, boys with Duchenne muscular dystrophy, and age-matched controls7. The studies to date using long Δ and RPBM analysis have provided conflicting results regarding the sensitivity of diffusion measures to muscle fiber geometries/sizes. Winters et al. used Δ values ranging from 20–700 ms on mice and found significant correlations between histological perimeter-to-area (P/A) and DTI-RPBM S/V ratios, for both ex vivo (r=0.71) and in vivo (r=0.56) conditions40. However, Cameron et al. used Δ values of 100 and 300 ms on muscle biopsy samples from 5 participants19 and Malis et al. used 10 Δ values ranging from 15–590 on biopsy samples from 10 participants15, with neither being able to demonstrate a statistically significant association between DTI-RPBM measures and fiber sizes. Notably, the authors of these prior studies acknowledge that there were challenges with localizing the same anatomic area between histology and MRI. Thus, this underscores the importance of studies such as ours where great care is taken for anatomic co-localization.
The muscle used in this study was post-mortem, post-fixed, and at room temperature during imaging, each resulting in reductions in the apparent diffusion coefficient compared with the in vivo condition. Sapienza et al. found that the apparent diffusion coefficient of muscle progressively decreased with longer post-mortem intervals41. Rats were perfused in our study, so only minimal reductions would be expected. The fixation, however, reduces diffusivity. Although the magnitude is unknown in muscle, decreases from 30–80% are seen in neurological tissue42,43. Scanning at room temperature results in a further decrease in apparent diffusion coefficient of approximately 30% compared with live tissue (~2%/°C)44. A rough estimate of diffusion length (ℓD) is provided by the square of the product of λ1 and Δ45. λ1 is the measurement of diffusion along the length of a muscle fiber and thus, is the closest measurement of unrestricted molecular diffusion that can be performed given this application. λ1 values of ~1.6 mm2/s have been reported in normal rat muscles in vivo46. Assuming a decrease of 30% due to temperature differences, and a further decrease of 30–80% due to fixation, an expected ℓD of 2–3 μm would be expected with the intermediate Δ of 15 ms used in our study. This is at least an order of magnitude below the expected muscle fiber diameter and appears to be insufficient to characterize the interactions with and restriction by the sarcolemma.
This study highlights the challenges in directly validating dMRI measurements in real tissue. While these experiments are highly controlled and precise, some concessions in the form of imaging live animals and imaging within a clinically relevant scan time had to be sacrificed. This study supports the use of simulation-based experiments to develop theoretical frameworks between the dMRI signal and underlying microstructure, as it allows the user to precisely control microstructure, SNR, pulse sequence parameters, and diffusion characteristics20–22,32,33,47–50. Translating the findings between simulation-based platforms to actual DTI experiments is still a challenge, as in a tissue-based experiment, the ground truth (i.e. microstructural features) is unknown until the tissue is processed and precisely co localized, as was shown in this study. Diffusion phantoms with muscle informed microstructure51 provide an excellent balance between these two platforms, as they provide user defined features of microstructure that can be directly measured and validated against a ground truth.
In addition to the aforementioned limitations in tissue quality, a limitation of this study includes low sample size. However, this is an exploratory study and the methodology and findings should inform more optimal parameters and experimental conditions in future studies with greater numbers. Additionally, the myofiber dimensions in our study are smaller than the in vivo condition due to the histological processing. Specifically, the sucrose concentration was hypertonic, resulting in cell shrinkage. However, this should not have affected correlation analysis.
In conclusion, we were unable to find a substantial relationship between scalar-based measures of DTI and histology-derived microstructural measures in muscle using a diffusion time of 15 ms for either 0.25 mm or 0.5 mm isotropic resolution datasets. Our study confirms that the experimental conditions greatly influence the sensitivity to muscle fiber size and intermediate diffusion times used with spin echo sPFG DTI sequences can produce negligible sensitivity to direct measures of muscle tissue microstructure.
Acknowledgements
The authors acknowledge Tanya Wolfson for her guidance with statistical considerations. The authors also acknowledge funding from the VA Research and Development Services (I01CX001388, I01BX005952, and I01CX002211) and the National Institutes of Health (R01AR079484, R01AR075825, R21AR083046, and RF1AG075717).
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Supplementary Files
Contributor Information
James Lo, University of California, San Diego.
David B. Berry, University of California, San Diego
Qingbo Tang, University of California, San Diego.
Xin Cheng, University of California, San Diego.
Marco Toto-Brocchi, University of California, San Diego.
Jiang Du, University of California, San Diego.
Samuel R. Ward, University of California, San Diego
Yajun Ma, University of California, San Diego.
Eric Y. Chang, VA San Diego Healthcare System
Data availability statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files] and available from the corresponding author at reasonable request.
References
- 1.Basser P. J., Mattiello J. & LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B 103, 247–254, doi: 10.1006/jmrb.1994.1037 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci 4, 469–480, doi: 10.1038/nrn1119 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Berry D. B., Gordon III J. A., Adair V., Frank L. R. & Ward S. R. From Voxels to Physiology: A Review of Diffusion Magnetic Resonance Imaging Applications in Skeletal Muscle. Journal of Magnetic Resonance Imaging n/a, doi: 10.1002/jmri.29489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minamoto V. B. et al. Increased efficacy and decreased systemic-effects of botulinum toxin A injection after active or passive muscle manipulation. Dev Med Child Neurol 49, 907–914, doi: 10.1111/j.1469-8749.2007.00907.x (2007). [DOI] [PubMed] [Google Scholar]
- 5.Ponrartana S. et al. Effectiveness of diffusion tensor imaging in assessing disease severity in Duchenne muscular dystrophy: preliminary study. Pediatr Radiol 45, 582–589, doi: 10.1007/s00247-014-3187-6 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Maggi L. et al. Quantitative Muscle MRI Protocol as Possible Biomarker in Becker Muscular Dystrophy. Clin Neuroradiol 31, 257–266, doi: 10.1007/s00062-019-00875-0 (2021). [DOI] [PubMed] [Google Scholar]
- 7.McDowell A. R., Feiweier T., Muntoni F., Hall M. G. & Clark C. A. Clinically feasible diffusion MRI in muscle: Time dependence and initial findings in Duchenne muscular dystrophy. Magn Reson Med 86, 3192–3200, doi: 10.1002/mrm.28945 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Nava S. et al. Diffusion tensor imaging reveals subclinical alterations in muscles of patients with Becker muscular dystrophy. Br J Radiol 97, 947–953, doi: 10.1093/bjr/tqae070 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ai T. et al. Diffusion tensor imaging in evaluation of thigh muscles in patients with polymyositis and dermatomyositis. Br J Radiol 87, 20140261, doi: 10.1259/bjr.20140261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigmund E. E. et al. MRI assessment of the thigh musculature in dermatomyositis and healthy subjects using diffusion tensor imaging, intravoxel incoherent motion and dynamic DTI. Eur Radiol 28, 5304–5315, doi: 10.1007/s00330-018-5458-3 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Wang F. et al. Simultaneous Multislice Accelerated Diffusion Tensor Imaging of Thigh Muscles in Myositis. AJR Am J Roentgenol 211, 861–866, doi: 10.2214/AJR.17.19318 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Tan E. T. et al. Quantitative MRI Differentiates Electromyography Severity Grades of Denervated Muscle in Neuropathy of the Brachial Plexus. J Magn Reson Imaging 56, 1104–1115, doi: 10.1002/jmri.28125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morena J. et al. MR Neurography and Quantitative Muscle MRI of Parsonage Turner Syndrome Involving the Long Thoracic Nerve. J Magn Reson Imaging 59, 2180–2189, doi: 10.1002/jmri.29007 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cameron D. et al. Age-related changes in human skeletal muscle microstructure and architecture assessed by diffusion-tensor magnetic resonance imaging and their association with muscle strength. Aging Cell 22, e13851, doi: 10.1111/acel.13851 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malis V. et al. Time-dependent diffusion tensor imaging and diffusion modeling of age-related differences in the medial gastrocnemius and feasibility study of correlations to histopathology. NMR Biomed 36, e4996, doi: 10.1002/nbm.4996 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monte J. R. et al. Diffusion tensor imaging and quantitative T2 mapping to monitor muscle recovery following hamstring injury. NMR Biomed 36, e4902, doi: 10.1002/nbm.4902 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Torres-Velazquez M. et al. MRI radiomics for hamstring strain injury identification and return to sport classification: a pilot study. Skeletal Radiol 53, 637–648, doi: 10.1007/s00256-023-04449-7 (2024). [DOI] [PubMed] [Google Scholar]
- 18.Novikov D. S., Fieremans E., Jensen J. H. & Helpern J. A. Random walk with barriers. Nat Phys 7, 508–514, doi: 10.1038/nphys1936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cameron D. et al. Diffusion-tensor magnetic resonance imaging captures increased skeletal muscle fibre diameters in Becker muscular dystrophy. J Cachexia Sarcopenia Muscle 14, 1546–1557, doi: 10.1002/jcsm.13242 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry D. B., Regner B., Galinsky V., Ward S. R. & Frank L. R. Relationships between tissue microstructure and the diffusion tensor in simulated skeletal muscle. Magn Reson Med 80, 317–329, doi: 10.1002/mrm.26993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry D. B., Englund E. K., Galinsky V., Frank L. R. & Ward S. R. Varying diffusion time to discriminate between simulated skeletal muscle injury models using stimulated echo diffusion tensor imaging. Magn Reson Med 85, 2524–2536, doi: 10.1002/mrm.28598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry D. B. et al. Double pulsed field gradient diffusion MRI to assess skeletal muscle microstructure. Magn Reson Med 90, 1582–1593, doi: 10.1002/mrm.29751 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y. L. et al. The effect of spatial resolution on the reproducibility of diffusion imaging when controlled signal to noise ratio. Biomed J 42, 268–276, doi: 10.1016/j.bj.2019.03.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto E. et al. Macroscopic and histologic evaluation of a rat model of chronic rotator cuff tear. J Shoulder Elbow Surg 25, 2025–2033, doi: 10.1016/j.jse.2016.04.024 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gumucio J. P. et al. Rotator cuff tear reduces muscle fiber specific force production and induces macrophage accumulation and autophagy. J Orthop Res 30, 1963–1970, doi: 10.1002/jor.22168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilling K. G.
- 27.Cox R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29, 162–173, doi: 10.1006/cbmr.1996.0014 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Cox R. W. & Hyde J. S. Software tools for analysis and visualization of fMRI data. NMR Biomed 10, 171–178, doi: (1997). [DOI] [PubMed] [Google Scholar]
- 29.Manjon J. V. et al. Diffusion weighted image denoising using overcomplete local PCA. PLoS One 8, e73021, doi: 10.1371/journal.pone.0073021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinkle D. E., Wiersma W. & Jurs S. G. Applied statistics for the behavioral sciences. Vol. 663 (Houghton Mifflin Boston, 2003). [Google Scholar]
- 31.Tanner J. E. Self diffusion of water in frog muscle. Biophys J 28, 107–116, doi: 10.1016/S0006-3495(79)85162-0 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damon B. M. Effects of image noise in muscle diffusion tensor (DT)-MRI assessed using numerical simulations. Magn Reson Med 60, 934–944, doi: 10.1002/mrm.21707 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Froeling M., Nederveen A. J., Nicolay K. & Strijkers G. J. DTI of human skeletal muscle: the effects of diffusion encoding parameters, signal-to-noise ratio and T2 on tensor indices and fiber tracts. NMR Biomed 26, 1339–1352, doi: 10.1002/nbm.2959 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Mitra P. P., Sen P. N., Schwartz L. M. & Le Doussal P. Diffusion propagator as a probe of the structure of porous media. Physical Review Letters 68, 3555–3558, doi: 10.1103/PhysRevLett.68.3555 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Latour L. L., Svoboda K., Mitra P. P. & Sotak C. H. Time-dependent diffusion of water in a biological model system. Proc Natl Acad Sci U S A 91, 1229–1233, doi: 10.1073/pnas.91.4.1229 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemberskiy G., Baete S. H., Cloos M. A., Novikov D. S. & Fieremans E. Validation of surface-to-volume ratio measurements derived from oscillating gradient spin echo on a clinical scanner using anisotropic fiber phantoms. NMR Biomed 30, doi: 10.1002/nbm.3708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzoli V., Moulin K., Kogan F., Hargreaves B. A. & Gold G. E. Diffusion Tensor Imaging of Skeletal Muscle Contraction Using Oscillating Gradient Spin Echo. Front Neurol 12, 608549, doi: 10.3389/fneur.2021.608549 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Deuster C., Stoeck C. T., Genet M., Atkinson D. & Kozerke S. Spin echo versus stimulated echo diffusion tensor imaging of the in vivo human heart. Magn Reson Med 76, 862–872, doi: 10.1002/mrm.25998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigmund E. E. et al. Time-dependent diffusion in skeletal muscle with the random permeable barrier model (RPBM): application to normal controls and chronic exertional compartment syndrome patients. NMR Biomed 27, 519–528, doi: 10.1002/nbm.3087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winters K. V., Reynaud O., Novikov D. S., Fieremans E. & Kim S. G. Quantifying myofiber integrity using diffusion MRI and random permeable barrier modeling in skeletal muscle growth and Duchenne muscular dystrophy model in mice. Magn Reson Med 80, 2094–2108, doi: 10.1002/mrm.27188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sapienza D. et al. Feasibility Study of MRI Muscles Molecular Imaging in Evaluation of Early Post-Mortem Interval. Sci Rep 10, 392, doi: 10.1038/s41598-019-57357-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun S. W. et al. Formalin fixation alters water diffusion coefficient magnitude but not anisotropy in infarcted brain. Magn Reson Med 53, 1447–1451, doi: 10.1002/mrm.20488 (2005). [DOI] [PubMed] [Google Scholar]
- 43.D’Arceuil H. E., Westmoreland S. & de Crespigny A. J. An approach to high resolution diffusion tensor imaging in fixed primate brain. Neuroimage 35, 553–565, doi: 10.1016/j.neuroimage.2006.12.028 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Morvan D., Leroy-Willig A., Jehenson P., Cuenod C. A. & Syrota A. Temperature changes induced in human muscle by radio-frequency H-1 decoupling: measurement with an MR imaging diffusion technique. Work in progress. Radiology 185, 871–874, doi: 10.1148/radiology.185.3.1438778 (1992). [DOI] [PubMed] [Google Scholar]
- 45.Grebenkov D. S. in Diffusion NMR of Confined Systems: Fluid Transport in Porous Solids and Heterogeneous Materials (ed Valiullin Rustem) 0 (The Royal Society of Chemistry, 2016). [Google Scholar]
- 46.Gao Y., Lu Z., Lyu X., Liu Q. & Pan S. A Longitudinal Study of T2 Mapping Combined With Diffusion Tensor Imaging to Quantitatively Evaluate Tissue Repair of Rat Skeletal Muscle After Frostbite. Front Physiol 11, 597638, doi: 10.3389/fphys.2020.597638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naughton N. M. & Georgiadis J. G. Comparison of two-compartment exchange and continuum models of dMRI in skeletal muscle. Phys Med Biol 64, 155004, doi: 10.1088/1361-6560/ab2aa6 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Naughton N., Cahoon S., Sutton B. & Georgiadis J. G. Accelerated, physics-inspired inference of skeletal muscle microstructure from diffusion-weighted MRI. IEEE Trans Med Imaging PP, doi: 10.1109/TMI.2024.3397790 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall M. G. & Clark C. A. Diffusion in hierarchical systems: A simulation study in models of healthy and diseased muscle tissue. Magn Reson Med 78, 1187–1198, doi: 10.1002/mrm.26469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan E. T., Zochowski K. C. & Sneag D. B. Diffusion MRI fiber diameter for muscle denervation assessment. Quant Imaging Med Surg 12, 80–94, doi: 10.21037/qims-21-313 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berry D. B. et al. (*) A 3D Tissue-Printing Approach for Validation of Diffusion Tensor Imaging in Skeletal Muscle. Tissue Eng Part A 23, 980–988, doi: 10.1089/ten.tea.2016.0438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files] and available from the corresponding author at reasonable request.