Abstract
Doublet microtubules (DMTs) are flagellar components required for the protist Trichomonas vaginalis (Tv) to swim through the human genitourinary tract to cause trichomoniasis, the most common non-viral sexually transmitted disease. Lack of DMT structures has prevented structure-guided drug design to manage Tv infection. Here, we determined the cryo-EM structure of native Tv-DMTs, identifying 29 unique proteins, including 18 microtubule inner proteins and 9 microtubule outer proteins. While the A-tubule is simplistic compared to DMTs of other organisms, the B-tubule features specialized, parasite-specific proteins, such as TvFAP40 and TvFAP35 that form filaments near the inner and outer junctions, respectively, to stabilize DMTs and enable Tv locomotion. Notably, a small molecule, assigned as IP6, is coordinated within a pocket of TvFAP40 and has characteristics of a drug molecule. This first atomic model of the Tv-DMT highlights the diversity of eukaryotic motility machinery and provides a structural framework to inform rational design of therapeutics.
Keywords: cryo-EM, doublet microtubule, Trichomonas vaginalis, trichomoniasis, antiparasitic
Introduction
Trichomonas vaginalis (Tv) is a flagellated, extracellular parasite of the human genitourinary tract and causative agent of trichomoniasis, the most common non-viral sexually transmitted infection (STI), with 250 million infections per annum and global prevalence over 3%1–3. Tv infection is linked to increased rates of preterm delivery and mortality, genitourinary cancers, and HIV transmission, with disproportionate impact on women in developing countries1–5. Though the antibiotic metronidazole can be curative, its carcinogenicity concern, increasing metronidazole resistance in Tv, and frequency of reinfection underscore the need for alternative precision therapies1,6–8. Tv relies on its four anterior and one membrane-bound, recurrent flagellum to propel itself through the genitourinary tract and attach to the mucosa of its human hosts, making the mechanisms driving locomotion potential therapeutic targets9. Unfortunately, no high-resolution structures related to Tv flagella are currently available, and even tubulin remains uncharacterized in Tv despite a putative role in antimicrobial resistance10–12.
As observed in low-resolution, thin-section transmission electron microscopy (TEM) studies13, the locomotive flagella originate from cytosolic basal bodies, and extend into the flagellar membrane with decorations along the microtubule filaments that stabilize the tubules and facilitate intraflagellar transport. The flagellar core, or axoneme, conforms to the canonical “9+2” axonemal arrangement wherein a central pair of singlet microtubules (MTs) is connected via radial spokes to nine surrounding doublet-microtubules (DMTs) which transduce force through the flagella (Fig. 1)13,14. Studies in other organisms revealed DMTs are coated with different combinations of microtubule inner and outer proteins (MIPs and MOPs) that facilitate assembly, stability, and function (Fig. 2a)15–20.
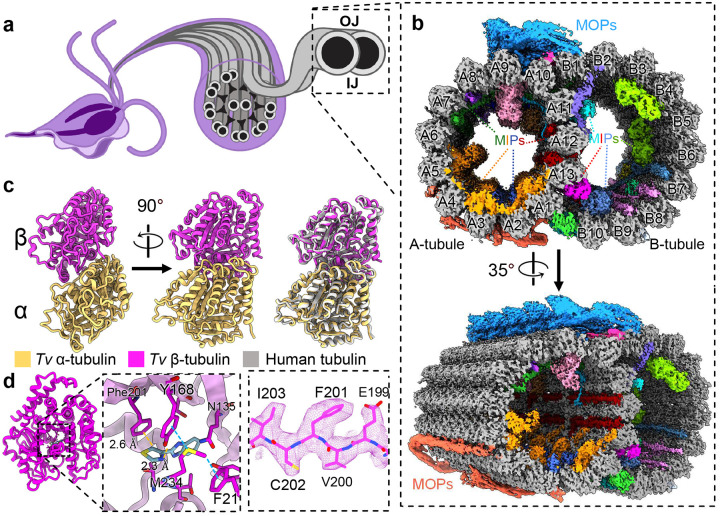
Figure 1. Cryo-EM reconstruction of the doublet microtubules from Tv.
(a) Diagram of axoneme from the flagella of T. vaginalis. (b) Cross-section of Tv-DMTs with microtubule inner proteins (MIPs) and microtubule outer proteins (MOPs) indicated with various colors. A- and B-tubules, as well as protofilaments, are labeled. (c) Atomic models of α and β tubulin, superimposed with human tubulin (right). (d) Alternate view of Tv β tubulin (left) and docked thiabendazole molecule (blue) fit into putative binding site with adjacent residues shown (right) with cryo-EM map density. IJ: inner junction; OJ: outer junction.
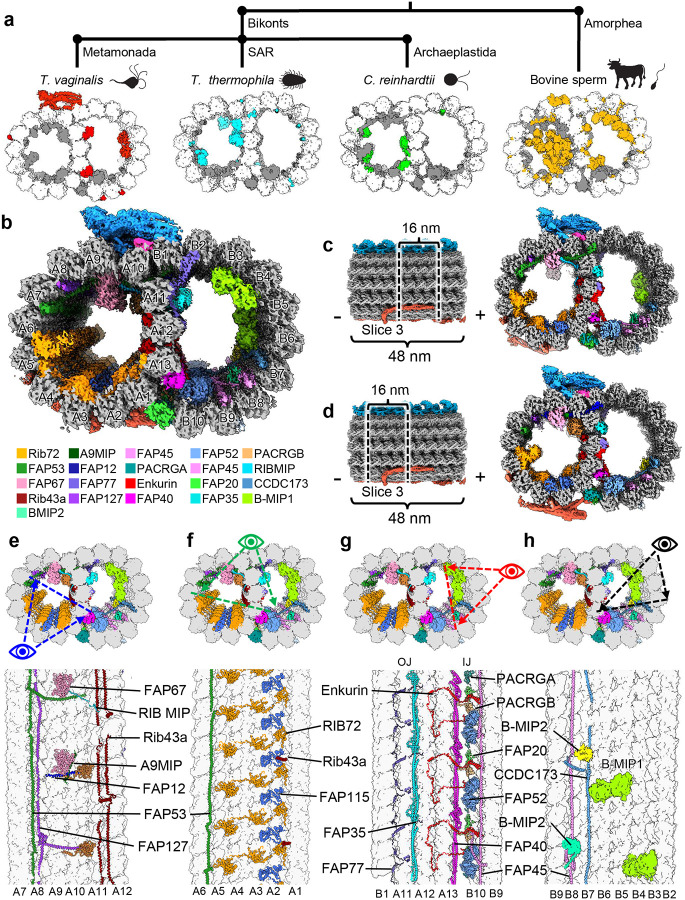
Figure 2. Tv-DMTs reveal conserved and novel MIPs.
(a) Phylogeny tree illustrating proposed divergence between Bikonts and Amorphea (top), with example organisms from these branches and accompanying DMTs (bottom) with tubulin (white), conserved flagella associated proteins (FAPs) (grey), and species-specific FAPs (colored) (b) Cross-sectional view of cryo-EM reconstruction of 48 nm repeat with MIP protein densities colored to demonstrate arrangement. (c and d) Cross-sectional view of DMTs from the 48 nm repeat map, shown as different 16 nm long sections through out the DMT. (e-h) Cross-sectional views of Tv-DMTs from different perspectives to illustrate MIP arrangement and periodicity.
Dozens of MIPs and MOPs have been identified across numerous studies of eukaryotic flagella, of which about half are conserved15–19. DMTs from multicellular eukaryotes incorporate more complex MIP arrangements, particularly along the highly variable ribbon protofilaments (PFs) that compose the inner and outer junctions (IJ and OJ) where the A- and B-tubules meet (Fig. 1a)15–19. In sperm flagella, filamentous tektin bundles near the ribbon PFs are thought to reinforce the long flagella as they swim through the viscous milieu of the genitourinary tract21,22. Though the Tv genome lacks tektin genes, the parasite swims through the same environment as sperm, coordinating its much shorter flagella into a distinct beating pattern23. Despite these apparent differences, it is unclear how the parasite propagates motion under these conditions and suggests a species-specific adaptation which may be exploited for therapeutic development.
Here, we leveraged mass spectrometry, cryogenic electron microscopy (cryo-EM), and artificial intelligence to analyze the DMTs derived from Tv parasites and elucidate the structures of the proteins that compose them. Our structure contains 29 atomic models, including the α- and β-tubulin, 18 MIPs and 9 MOPs. Among these, we identified three Tv-specific proteins, including one bound to a ligand not observed in the DMTs of other organisms. This first structure from the Tv flagella highlights remarkable simplicity in the species’ DMT architecture compared to more complex organisms such as mammals, as well as other protists like Tetrahymena thermophila. Despite this simplicity, Tv can still traverse the same viscous environment as the more complex mammalian sperm, suggesting a key to parasite locomotion lies in the short list of Tv-DMT proteins.
Results
T. vaginalis DMTs feature both familiar and novel MIPs
We optimized a protocol to isolate DMTs from T. vaginalis and limit perturbations to the internal structures, then subjected them to single-particle analysis using cryo-EM. The resultant cryo-EM maps of the 48 nm repeat DMT had a global resolution of 4.2 Å and focused refinement improved local resolution to between 3.2 Å and 3.8 Å (Fig. 1b, Table S1). Reconstructions of the 16 nm and 96 nm repeat structures were resolved to 3.8 Å and 4.3 Å respectively. We also collected mass spectrometry data for our cryo-EM sample to produce a library of potential Tv-DMT proteins and utilized cryoID to identify most likely candidates for certain map densities24. AlphaFold predicted structures served as initial models for atomic modeling of both conserved and species-specific cryo-EM map densities25,26. From our structures we built 29 unique atomic models, including 18 MIPs, 9 MOPs and the α/β tubulin of Tv (Movie S1, Table S2). Of these proteins, 15 MIPs and all 9 MOPs are conserved between Tv and previous DMT structures, whereas 3 MIPs are novel. There are also 5 unassigned MIP and 3 MOP densities that appear to play an important role in DMT function, but for which we lacked sufficient resolution to model.
Consistent with their ~80% sequence identities, the atomic models of Tv’s α- and β-tubulin are nearly identical to those of their human homologs (Fig. 1c), including the region of β-tubulin where many antiparasitic, benzimidazole-derived drugs (BZs) bind (Fig. 1d). Previous studies in Tv suggest mutations aromatic residues at codons 168 and 201 in β-tubulin confer BZ resistance12,27,28. Indeed, like human β-tubulin’s Phe169 and Tyr202, Tv orients Tyr168 and Phe201 into the BZ binding pocket where they are stabilized by Aro-Met-Aro interactions with adjacent Met234 and Phe21 residues and sterically occlude BZ drugs like thiabendazole (TBZ) (Fig. 1d). To corroborate this, we performed docking experiments using AutoDock Vina and found TBZ docked β-tubulin produced large positive binding free energy values (ΔG) (Fig. S2). By contrast a virtual β-tubulin Y168A, P201A mutant exhibited a negative binding free energy when TBZ was docked (Fig. S2). Interestingly, we observe the swapped positions of phenylalanine and tyrosine residues between human and Tv β-tubulin, which may help to explain species-specific sensitivity to different BZs.
Like other organisms, the α/β tubulin heterodimers polymerize and assemble into rings of 13 and 10 PFs that compose the A- and B-tubules respectively (Fig. 2b). Within the A-tubule, molecular rulers FAP53, FAP127, and Rib43a impose a 48 nm MIP periodicity and facilitate the organization of other MIPs like FAP67 and RIB72 (Fig. 2e–h). Consistent with studies in T. thermophila18, FAP115 repeats every 32 nm and creates a mismatch with the 48 nm periodicity of the ruler proteins, leading to 96 nm periodicity (Fig. 2f). Interestingly, FAP141 from other organisms is replaced by the smaller TvFAP12 which lashes FAP67 to the A-tubule lumen like the N-terminal helices of FAP53 and FAP127 (Fig. 2e)15. Along with the N-terminal helices of FAP53 and FAP127, TvFAP12 passes into the B-tubule to maintain 16 nm a repeating crosslink between the A- and B-tubules as observed in FAP141 expressing organisms15. Unlike other species, the Tv ribbon PFs (A11-A13) that divide A- and B-tubules are sparsely decorated with A-tubule MIPs suggesting alternative strategies of ribbon arc stabilization.
In the B-tubule lumen, we found assembly-related MIPs FAP45, CCDC173, enkurin, FAP77, FAP52, FAP20, and PACRGA/B that are conserved amongst other organisms. Interestingly, along the B-ubule side of the ribbon arc, we identified the filamentous MIPs TvFAP35 and TvFAP40, which run lengthwise along the A11 and A13 PFs respectively and may compensate for the dearth of MIPs along the ribbon arc in the A-lumen (Fig. 2g). Further, we observed globular MIPs that span PFs B3-B4 and B5-B6 and exhibit 96 nm periodicity (Fig. 2h). While the map resolution was insufficient to model these proteins, their interactions with the neighboring ruler proteins like CCDC173, indicate an enforced periodicity of 96 nm which is the first of this length from any DMT MIP to date. Though we observed several novel proteins, the Tv-DMTs have the simplest MIP organization in the A-tubule with just 10 MIPs (eight identified and two unidentified) compared to the next simplest species of record, C. reinhardtii, with 22 A-tubule MIPs15. The comparatively simple MIP organization observed in Tv suggests the few novel MIPs may play a substantial role in flagellar function.
T. vaginalis microtubules reinforce the inner junction with species-specific protein
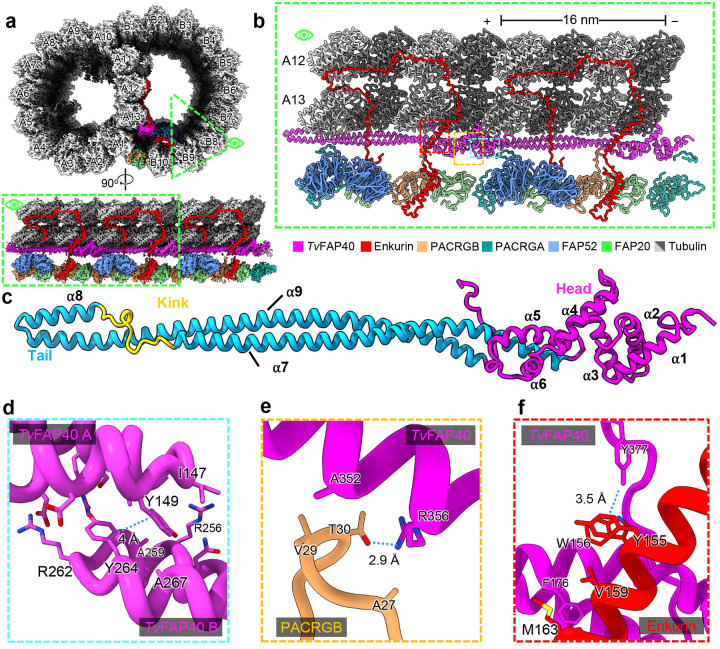
The DMT IJs of other organisms are typically composed of FAP52, enkurin/FAP106, PACRG isoforms (PACRGA and PACRGB), and FAP20, while Tetrahymena and mammalian DMTs include globular proteins atop FAP52 that mediate interactions with PF A1318,22,29. Interestingly, the Tv-DMT cryo-EM map revealed the long, filamentous protein TvFAP40, running atop PF A13 at the IJ which alters the topography of this important protofilament. TvFAP40 monomers repeat every 16 nm and are arranged head-to-tail, where head-tail polarity corresponds to the − and +-ends of the DMT respectively (Fig, 3B–C). Each TvFAP40 monomer consists of a globular N-terminal ‘head’-domain (residues 1–145) connected to a coiled-coil ‘tail’ (residues 149–361). The tail consists of 3 coiled-helices (α7–9) where a proline-rich kink connects α7 and α8 while a 180° turn at the linker between α8 and α9 forms the ‘tip’ of the tail. The tip includes α8 and neighboring residues of α9 (residues 246–282), with both a polar face oriented towards the MT and a hydrophobic face oriented towards a neighboring TvFAP40 monomer (Fig. 3c–d). As the kink reaches into the cleft between tubulin heterodimers, α8 is brought into close contact with tubulin, and establishes electrostatic interactions. The kink also offsets α7 from α8, creating an overhang to bind the head of a neighboring TvFAP40 monomer which may help stabilize the interaction (Fig. 3d).
Figure 3. TvFAP40 alters the inner junction arrangement in parasite DMTs.
(a) Cross-sectional view of cryo-EM reconstruction of 16nnm repeat with protofilaments labeled and proteins near inner junction colored (top) and cutaway view of region of interest (bottom). (b) View of atomic models built from map in a. (c) atomic model of TvFAP40 colored by domain. (d) Zoomed-in view of dimerization domain between two TvFAP40 monomers (labeled TvFAP40 A and B). (e and f) Close-up view of interaction between PACRGB (tan) and TvFAP40 and Enkurin (red), with residues shown to highlight interactions.
TvFAP40’s unique location along PF A13 has not been seen in other MIPs and alters conserved MIP interactions at the inner junction. In other organisms, the PACRGB N-terminus binds the groove between A11 and A13, but in our structure, TvFAP40 blocks this groove and replaces A13 as the binding partner. Additionally, the TvFAP40 C-terminus hooks around α2 of enkurin, where the C-terminal tyrosine (Tyr377) participates in hydrophobic interactions with adjacent aromatic residues from enkurin (Tyr155 and Trp156) (Fig. 3f). This C-terminal hook acts in concert with the TvFAP40 head that binds the other side of enkurin α2 and restricts it such that the bottom end of the helix is 1 nm closer to A13 than in other structures.
Tv-specific FAP40 head domain binds a stabilizing ligand
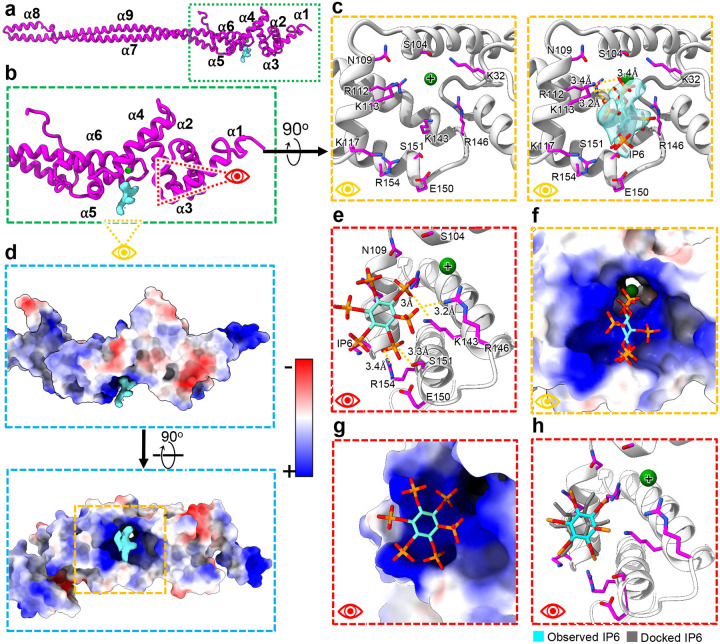
In addition to binding neighboring monomers, TvFAP40 incorporates a unique ligand binding pocket. Our cryo-EM maps indicate the TvFAP40 head-domain binds a six-pointed, star-shaped ligand, and our atomic model indicates this pocket is positively charged (Fig. 4a–c, Movie S2). Indeed, the putative binding site features seven positively charged side chains oriented towards the points of the star, and density from a likely metal cation coordinated by additional arginine residues (Fig. 4c), which suggests negatively charged functional groups (Fig. 4c–g). Sequence and structural homology searches within UniProt or the RCSB protein database could not identify similar binding sites30,31, but the high local resolution of our map in this pocket revealed the stereochemistry of the functional groups at the points of the star, consistent with bonding to a non-planar six-membered ring. Together these features suggested an inositol polyphosphate, in this case inositol hexakisphosphate (IP6) which was a good fit for the map density (Fig. 4c).
Figure 4. TvFAP40 binds IP6 in a positively charged pocket.
(a) Atomic model of TvFAP40 with (b) zoomed-in view of the head domain and (c) perspectives of the putative IP6 binding site with (right) and without (left) IP6 fit into the cryo-EM map. (d) Coulombic potential map of head domain from B (top) and rotated (bottom) views with blue and red indicating positive and negative coulombic potentials respectively. (e) Side-view of IP6 in binding pocket with adjacent residues shown. (f-g) Views from C and E shown with electrostatic potential maps of TvFAP40. (h) Comparison of observed IP6 binding site and docked IP6.
IP6 is an abundant cellular polyanion known to stabilize positive interfaces such as the pore of HIV nucleocapsids32, a trait which may be useful to DMT reinforcing proteins. To confirm whether IP6 was a reasonable ligand assignment, we carried out in silico molecular docking using Swissdb’s AutoDock Vina webserver33–36. Restricting the docked ligand to the observed binding pocket resulted in docked arrangements consistent with the observed ligand density, and binding energies (ΔG) of −3.5 kcal/mol or less (Fig. 4h). The docking experiments suggested interactions with the same arginine and lysine residues as the real ligand structure in the binding pocket. These results support the notion that IP6 acts as a ligand within TvFAP40 and may stabilize the head to reinforce its interactions with enkurin and the tail of its neighboring monomer and microtubule PF. Interestingly, in zebrafish embryos the IP6 producing enzyme (Ipk1) was found to localize to basal bodies of cells, and Ipk1 knockdown disrupted cilia growth and beating37,38. Combined with our structures, these studies point to an uncharacterized role for IP6 in flagellar function and stability.
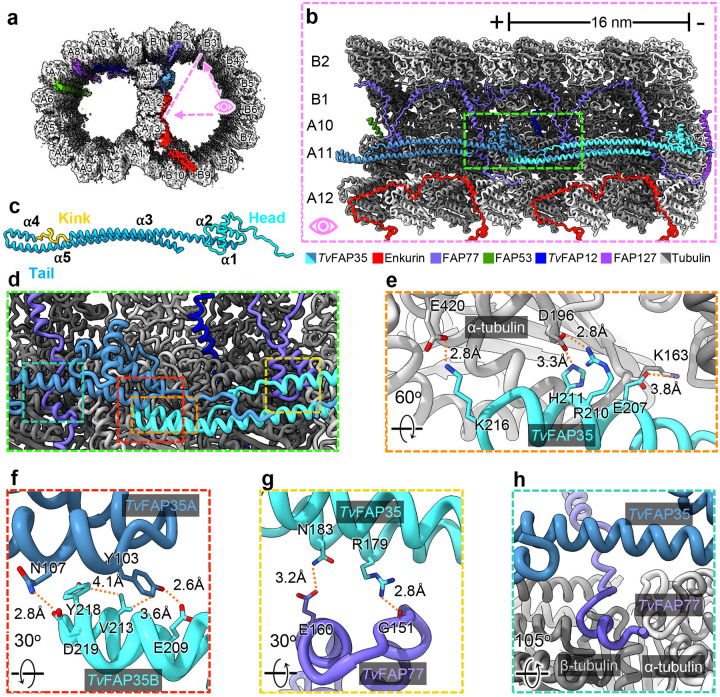
TvFAP35 secures FAP77 and buttresses the outer junction of Tv-DMTs
Directly above TvFAP40 in the B-tubule we identified a novel, filamentous density along the ribbon arc PF A11 as TvFAP35, another Tv-specific protein (Fig. 5a–c). In the B-tubule, TvFAP35 repeats every 16 nm in a head-to-tail fashion with the heads and tails oriented to the − and +-ends respectively (Fig. 5b). The TvFAP35 tail domain has the same ‘kinked-coiled-coil’ fold as TvFAP40 including the tips that mediate MT binding and dimerization (Fig. 5c–f). The head-domain of TvFAP35 differs from TvFAP40, as it includes only a flexible N-terminus and helix-turn-helix (Fig. 5c), as opposed to the six helices found in the TvFAP40 head (Fig. 3c).
Figure 5. TvFAP35 stabilizes ribbon PF A11 and outer junction proteins.
(a) Cross-sectional view of the Tv-DMT cryo-EM map with enkurin and outer junction proteins colored. (b) 32 nm section of protofilaments A10, A11, A12, B1, and B2, along with their associated MIPs, shown with atomic models. (c) TvFAP35 monomer labeled with head (cyan), tail (blue), and kink (yellow), with helix numbers. (d) Zoomed-in view including important interactions of TvFAP35. (e) Electrostatic interactions at the MT-binding motif of TvFAP35. (f) Mixed residue interactions at the dimerization interface between TvFAP35 monomers. (g) Interactions between TvFAP35 and the helix-turn-helix (residues 140–164) of TvFAP77. (h) Residues 238–246 of TvFAP77 pass near the TvFAP35 coiled-coil. Residues 255 and after of TvFAP77, which stretch further down, are omitted for clarity.
The position of TvFAP35 along A11 is similar to that of tektin-like protein 1 (TEKTL1) which is thought to reinforce OJ stability during flagellar beating in sperm DMTs22. Further, the coiled-coil structure of TvFAP35 resembles the 3-helix bundle architecture of TEKTL1. Unlike TEKTL1, the coiled coils of TvFAP35 include a proline-rich kink that occupies the cleft between tubulin heterodimers. As PFs bend, gaps form at the interface between tubulin heterodimers39, and the TvFAP35 kink may create stress relief points along A11 by acting as a flexible linker which accommodates bending. Thus, like TEKTL1, the coiled-coils of TvFAP35 may provide structural stability to the DMT while the kink allows bending and greater flexibility. TvFAP35 also interacts with FAP77, a MIP that aids in B-tubule formation and tethers complete A- and B-tubules together at the OJ (Fig. 5g & h)18. The FAP77 helix-turn-helix motif (residues 140–164) is braced to PF A11 via electrostatic interactions with the coiled-coil of TvFAP35 (Fig. 5b, c, and f). Further, the tail-domain of TvFAP35 passes over residues 238–246 of TvFAP77 which run between a cleft of the A11 PF and reinforces TvFAP77’s association to A11 (Fig. 5g). These observations suggest that, like TvFAP40, TvFAP35 plays an integral role in the stabilization of the ribbon PFs and their associated MIPs. Additionally, because FAP77 is implicated in B-tubule assembly18, the interactions of TvFAP35 with FAP77 and A11 suggest that TvFAP35 may also contribute to DMT assembly.
Novel Tv proteins share an ancient MT binding motif
Upon comparison, we noticed both TvFAP35 and TvFAP40 have kinked-coiled-coils composed of three helices (ɑ1–3, Fig. S2) with similar lengths, dimerization domains, and MT binding motifs (Fig. 3–4). This similarity prompted us to search for homologous proteins via amino acid sequence alignment, but this returned few candidates. Interestingly, the coiled-coils of TvFAP35 and TvFAP40 share just 23% identity despite similar folds. We next turned to structural alignment using FoldSeek40, and identified numerous structural homologs. After curating homolog candidates by removing those without kinked-coiled-coil domains or with TM-scores below 0.4, 31 homolog candidates were selected for further comparison. Initial analysis revealed all kinked-coiled-coil containing homologs belonged to the group Bikonta, which includes many protists, and excludes animals, fungi and amoebozoans.
Six kinked-coiled-coil homologs from protists, representing different clades were selected for multiple sequence alignment with TvFAP35 and TvFAP40 (Fig. S3a) which revealed several conserved residues from the dimerization and MT binding domains. Based on the TvFAP35 sequence, the dimerization domains include hydrophobic residues at Val213 and aromatic residues at Tyr103 and Tyr218, which form hydrophobic interfaces between neighboring monomers (Figs. S2g). Further, the MT binding motif on α2 has a high proportion of charged residues, which are likely important in tubulin binding (Fig. S3f). Outside of the dimerization and MT binding motifs, the kinked-coiled-coils exhibit an average sequence conservation of ~20% which may be necessary to accommodate different MIPs, as observed in our novel proteins (Fig. 3 and 5).
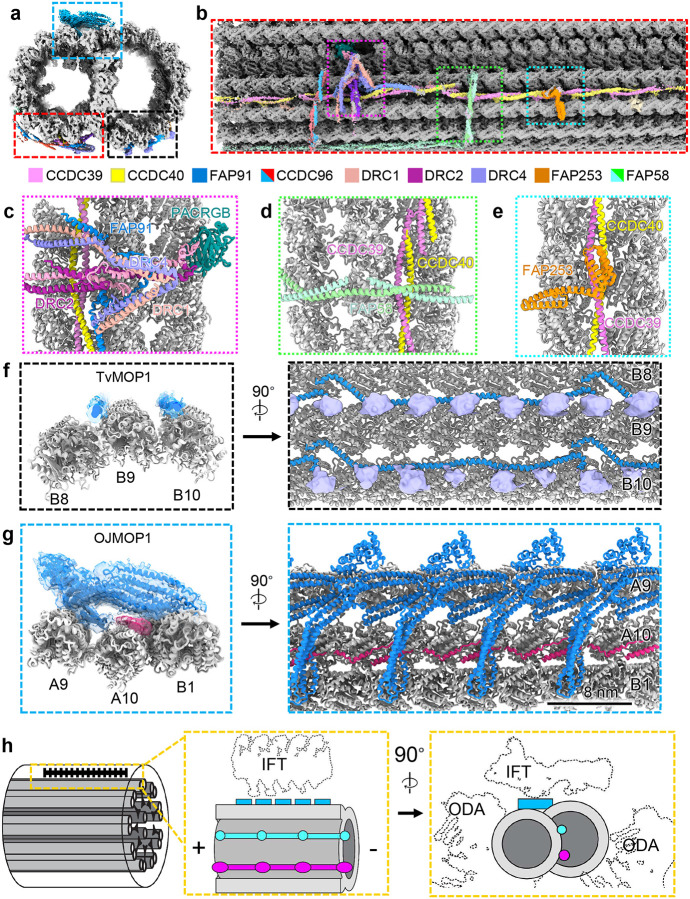
T. vaginalis microtubule outer proteins exhibit 8 nm periodicities
Considering the marked simplicity of the Tv-DMT MIP arrangement, we expected to find comparably simple MOP organization. Along the A-tubule we observed both the canonical N-DRC and radial spoke complexes that mediate inter-axoneme connections and flagellar bending (Fig. 6a–c). We see that, like other DMT structures, the axoneme-related proteins exhibit 96 nm periodicity enforced by the molecular ruler proteins CCDC39 and CCDC40, which coil their way between PFs A3 and A2 (Fig. 6b–e). Besides N-DRC and radial spoke proteins, a diverse arrangement of filamentous MOPs occupies the clefts and the surface of several PFs. Previous DMT structures from other species found the shortest MOP periodicity to be 24 nm18,22. TvMOP1 is 24 nm repeating MOP that arranges head-to-tail in the furrows between PFs A3, A4, B8, B9, & B10 and contacts the flexible C-terminal tails of α and β tubulin in the B-tubule (Fig. 6f). Interestingly, though the exterior of the outer junction is sparsely decorated in DMTs structures from other organisms15,22, we found this area to contain a large filamentous protein that repeats every 8 nm and a smaller filament that runs in a zig-zag beneath it and between A10 and B1 (Fig. 6g). The large protein density fashions an ankyrin-like domain seated atop a large coiled-coil domain, which spans the gap between PF A9 and B1 (Fig. 6g).
Figure 6. Microtubule organization reveals novel 8nm periodicity.
(a) Cross-sectional view of 96 nm repeat map, colored by MOP. (b) external view of Tv-DMT and zoomed in views of MOPs (c-e). (f) TvOJMOP1 demonstrating 24 nm periodicity as cross-section (left) and external view (right). (g) TvMOP1 demonstrating 8nm periodicity with cross-sectional (left) and external views (right). (h) Schematic view of Tv-DMT organization with dotted lines to indicate positions of IFT and inner and outer dynein arm attachment (IDA and ODA).
Due to limited local resolution, we were unable to confidently assign the identities of these proteins and instead dubbed them Tv outer junction microtubule outer protein 1 and 2 (TvOJMOP1 and TvOJMOP2) for the large and zig-zag MOPs respectively. TvOJMOP1 exhibits an 8 nm periodicity like that of tubulin heterodimers, an unusual repeat length amongst DMT MOPs that crosslinks PF B1 to A9 and A10 (Fig. 6g). TvOJMOP1 was observed in only 1/5 of particles, suggesting that some may have been lost during DMT isolation or that TvOJMOP1 localizes to certain regions of the axoneme. Exhaustive search through AlphaFold predicted structures from our proteomic data using a strategy similar to that of DomainFit41 yielded the following 5 candidate proteins which contain both ankyrin and coiled-coil domains: TVAGG3_0305310, TVAGG3_0421180, TVAGG3_0431750, TVAGG3_0596110, and TVAGG3_0415080. However, none of these candidates could fully account for the observed density, and so it remains unclear if TvOJMOP1 is composed of one or more of these proteins. Recent work in C. reinhardtii has demonstrated that anterograde intraflagellar transport (IFT) brings IFT-B complexes directly over this area (Fig. 6h)42. However, as components are often lost during DMT isolation they are unlikely candidates. As TvOJMOP1 features an ankyrin domain oriented towards the would-be IFT-B cargo (Fig. 6g–h), it may interact with TPR-rich proteins of IFT-B to stabilize the cargo. Additionally, others have documented the tendency for cytoplasmic dynein motors to jump between PFs43. TvOJMOP1 may therefore create tracks to keep the dynein motors on their preferred A-tubule PFs.
Discussion
T. vaginalis pathogenesis relies on the parasites’ locomotive flagella to establish infection and spread between human hosts23,44. This study reports the first high-resolution structure of Tv flagellar doublet microtubules, elucidating their molecular composition, architectural arrangement, atomic structures, and small molecule ligands. In addition to the first atomic structures of the Tv tubulin subunits comprising the DMTs, we have identified 20 MIPs and 13 MOPs distributed across the A- and B-tubules. These MIPs and MOPs mediate Tv-DMT function in the flagella with several novel proteins. As the first near-atomic structure of flagellar microtubules in the major human parasite Trichomonas vaginalis, our results provide a structural framework to understand the parasite’s distinct locomotion, offer insights into antibiotic drug resistance, and identify new targets for precision medicine.
With a relatively short list of both conserved and Tv-specific MIPs, the Tv-DMT is perhaps the simplest among known DMT structures. Notably, the Tv A-tubule fashions the fewest MIPs of any characterized organism (Fig. 2). Among them, the ruler proteins Rib43a, FAP53, and FAP127 are conserved, but lack many of the interacting partners of their homologs in other species, such as mammalian sperm, that traverse the same environment. The sparsity of A-tubule ribbon proteins in Tv suggests these proteins are less essential for locomotion in the human genitourinary tract, which contrasts with the complex MIP arrangement of sperm-specific proteins and tektin bundles seen in the A-tubule of mammalian sperm22,29. While Tv and sperm exhibit distinct flagellar beating patterns, the sinusoidal beating pattern of the recurrent flagella in Tv suggests the additional MIP complexity observed in sperm is not essential to this style of beating. However, human sperm swim five times faster than Tv and must propagate beating over longer flagella, so sperms’ complex A-tubule MIP arrangement may facilitate rapid propulsion through their viscous environment9,45.
Remarkable specialization is observed in the B-tubule, where several novel proteins reinforce the ribbon arc in a manner similar to tektin bundles from other organisms15. Like tektin, the TvFAP35 and TvFAP40 proteins exhibit 16 nm periodicity and similarly interact with other MIPs along their respective protofilaments (Fig. 3 and 5). However, unlike tektin, TvFAP35 and TvFAP40 have variable head domains which seemingly confer different functionalities. To this end, the positively charged pocket of TvFAP40 putatively binds an IP6 pocket factor (Fig. 4). In HIV, IP6 acts as a pocket factor to stabilize the nucleocapsid lattice32. Considering TvFAP40 likely plays a stabilizing role along the ribbon arc, IP6 binding may augment that stabilization by reinforcing the interactions between the monomers at the head-tail interface. Binding abundant biomolecules is a common strategy amongst pathogens, particularly viruses46–48, but this is the first instance such pocket factors have been documented in DMTs.
The Tv-specific MIPs and MOPs are particularly significant in light of their role in propagating the pathogenesis of the most widespread non-viral STI3. Specialization differentiates the parasite’s DMTs from those of other organisms, including their human hosts, thus drugs targeting these specialized components would have minimal toxicity. For instance, the unique cofactor binding pocket found in the Tv-specific TvFAP40 protein has a structure with no known homologs and appears to specifically bind IP6 (Fig. 4). This pocket could be targeted by antimicrobial compounds to destabilize parasite DMTs with limited off-target effects. Notably, the only homologous proteins to TvFAP40 belonged to other Bikonts, and include other human-borne parasites like T. brucei and Leishmania donovani, that may incorporate similar species-specific proteins (Fig. S3). While this study represents the first of its kind on the DMT from a human-borne parasite, it has demonstrated the power of in situ cryo-EM over other structural or in silico methods, to open new avenues for rational drug design. Together, our findings provide a basis to explore the contribution of microtubule-associated proteins to the unique aspects that allow T. vaginalis to swim through the human genitourinary tract, and the diversity of eukaryotic motility in general. Moreover, the atomic details revealed in species-specific proteins and bound small molecules can inform the rational design of therapeutics.
Methods and Data Availability
Cell culture
T. vaginalis strain G3 was cultured in Diamond’s modified trypticase-yeast extract-maltose (TYM) medium supplemented with 10% horse serum (Sigma-Aldrich), 10 U/mL penicillin, 10 μg/ml streptomycin (Gibco), 180 μM ferrous ammonium sulfate, and 28 μM sulfosalicylic acid49. 2L of parasites, grown at 37 °C and passaged daily, were harvested by centrifugation, and washed twice with phosphate-buffered saline and pelleted at low speed. Cells were resuspended in 50 mL lysis buffer (2% IGEPAL CA-630, 2% Triton X-100, 10% glycerol, 10 mM Tris, 2 mM EDTA, 150 mM KCl, 2 mM MgSO4, 1 mM dithiothreitol [DTT], 1× Halt protease inhibitors [pH 7.4]) and lysed in a Stansted cell disrupter, operated at 30 lb/in2 front pressure and 12 lb/in2 back pressure.
Cytoskeletal elements were harvested similar to what has been previously described50. Lysates were recovered and maintained at 4 °C for all subsequent steps. Nuclei were removed via low-speed centrifugation (1000 × g) for 10 mins to generate pellet 1 (P1) and lysate 1 (L1). L1 was centrifuged (10,000 × g for 40 mins) to pellet cytoskeletal components into P2 and L2. Cytoskeleton pellets (P2) were resuspended in 1 mL low salt (LS) buffer (150 mM NaCl, 50 mM Tris, 2 mM MgCl2, 1 mM DTT, 1× complete protease inhibitor (Sigma-Aldrich)) and centrifuged at low speed (1000 × g, 10 mins) to pellet cellular debris into P3. The resulting lysate (L3) was placed a sucrose cushion (30% w/v sucrose in LS buffer) and centrifuged at low speed (1,800 × g, 10 mins). The supernatant atop the cushion was collected and resuspended in 1 mL LS buffer prior to centrifugation (5,000 × g 15 mins) to pellet larger cytoskeletal components (P4). The lysate was finally centrifuged at high speed (16,600 × g, 40 minutes) to pellet axoneme related cytoskeletal elements (P5). The P5 was then resuspended in minimal volume of LS buffer supplemented with 5 mM ATP and left at RT for 1 hour.
In-solution digestion, Mass Spectrometry Data Acquisition and Analysis
T. vaginalis cytoskeleton pellets P4 and P5 resuspended in low salt (LS) buffer were mixed with 4× volume of ice-cold acetone and kept at −20°C for 2 h. The mixtures were centrifuged at 4°C with 14,000 rpm for 15 min and supernatants discarded. The air-dried pellets were fully dissolved in 8 M Urea in 100 mM Tris-HCl (pH 8) at 56 °C and the proteins reduced with 10 mM Tris(2-carboxyethyl) Phosphine for 1 h at 56 °C. The reduced proteins were then alkylated with 40 mM iodoacetamide for 30 min in dark at room temperature and the reaction was quenched with Dithiothreitol at a final concentration of 10 mM. The alkylated samples were subsequently diluted with 7× volume of 100 mM Tris-HCl pH 8, to 1M Urea concentration. To generate peptides, Pierce Trypsin Protease (Thermo Fisher Scientific) was added to the samples and the ratio of trypsin:protein was 1:20 (w/w). The digestion reaction was incubated at 37 °C overnight, and the residue detergents in the protein samples were removed using a HiPPR Detergent Removal Spin Column Kit (Thermo Fisher Scientific) on the next day. Prior to the mass spectrometry assay, the samples were desalted with Pierce C18 Spin Columns (Thermo Fisher Scientific) and lyophilized.
Three biological replicates were prepared and trypsin-digested following the steps above for fractions P4 and P5, respectively. The lyophilized protein pellets were dissolved in sample buffer (3% Acetonitrile with 0.1% formic acid) and ~1.0 μg protein from each sample was injected to an ultimate 3000 nano LC, which was equipped with a 75μm × 2 cm trap column packed with C18 3μm bulk resins (Acclaim PepMap 100, Thermo Fisher Scientific) and a 75μm × 15 cm analytical column with C18 2μm resins (Acclaim PepMap RSLC, Thermo Fisher Scientific). The nanoLC gradient was 3−35% solvent B (A = H2O with 0.1% formic acid; B = acetonitrile with 0.1% formic acid) over 40 min and from 35% to 85% solvent B in 5 min at flow rate 300 nL/min. The nannoLC was coupled with a Q Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA), operated with Data Dependent Acquisition mode (DDA) with inclusion list for the target peptides. The ESI voltage was set at 1.9 kV, and the capillary temperature was set at 275 °C. Full spectra (m/z 350 – 2000) were acquired in profile mode with resolution 70,000 at m/z 200 with an automated gain control (AGC) target of 3 × 106. The most abundance 15 ions were subjected to fragmentation by higher-energy collisional dissociation (HCD) with normalized collisional energy of 25. MS/MS spectra were acquired in centroid mode with resolution 17,500 at m/z 200. The AGC target for fragment ions is set at 2 × 104 with maximum injection time of 50 ms. Charge states 1, 7, 8, and unassigned were excluded from tandem MS experiments. Dynamic exclusion was set at 45.0 s.
The raw data was searched against total T. vaginalis annotated proteins (version 63) downloaded from TrichDB, using ProteomeDiscoverer 2.5. Following parameters were set: precursor mass tolerance ±10 ppm, fragment mass tolerance ±0.02 Th for HCD, up to two miscleavages by semi trypsin, methionine oxidation as variable modification, and cysteine carbamidomethylation as static modification. Protein abundance was quantified using Top 3 approach, i.e., the sum of the three most intense peptides coming from the same protein. Only proteins that were detected in all three replicates of P4 or P5 were included for further analyses, which resulted in a total of 386 and 311 proteins identified from P4 and P5, respectively. Among these common proteins, contaminants that are obviously not cytoskeletal proteins were identified from the datasets based on the GO terms and function annotations. For instance, proteins annotated as histone, kinase or DNA binding proteins or proteins located in subcellular compartments e.g., translational apparatus, nucleus, plasma membrane, were removed from the datasets. As a consequence, the numbers of putative cytoskeletal proteins identified in P4 and P5 were reduced to 303 and 239, respectively. The union of dataset P4 and P5, which consists of 371 distinct proteins, represent the entire cytoskeletal proteome of T. vaginalis identified by this study. DeepCoil 2.0 program was employed to predict coiled-coil domains (ccds) from the 371 putative cytoskeletal proteins based on protein sequence51. Three indices, i.e., number of ccds within each protein, the average length of ccds in each protein and percentage of total protein length occupied by ccds were calculated based on the output of DeepCoil 2.0. In addition to the cytoskeletal proteome in this study, the presence of ccds was also investigated for the hydrogenosome proteome of T. vaginalis and a randomly picked T. vaginalis protein dataset52.
Cryo-EM sample preparation and image aquisition
To prepare DMTs for single particle analysis, 2.5 μL of DMT lysate was applied to glow discharged carbon holey grids (R2/1) (Ted Pella) and incubated on the grid for 1 minute prior to blotting and plunge freezing into a 50:50 mixture of liquid ethane and propane using a Vitrobot Mark IV (Thermo-Fisher). Flash frozen grids were stored under liquid nitrogen until cryo-EM imaging.
Dose fractionated cryo-EM movies were recorded on a K3 direct electron detector (Gatan) equipped Titan Krios electron microscope (FEI/Thermo-Fisher) fitted with a Gatan Imaging Filter (GIF) and operated at 300 keV. Movies were recorded at a nominal magnification of 81,000 x and calibrated pixel size of 0.55 Å at the specimen level, operated in super resolution mode. Using SerialEM53, 30,834 movies were recorded with a cumulative electron dose of ~ 45 e−/A2.
Cryo-EM image processing and 3-dimensional reconstruction
Movie frame alignment and motion correction were performed in CryoSPARC54, to generate cryo-EM micrographs from each movie. Patch-aligned and dose weighted micrographs were binned 2X to improve processing speeds and transferred for processing in Relion 4.0 and Topaz automated particle picking, using the filament option “-f” incorporated by Scheres and colleagues55–57. Picked particles coordinates were extracted in Relion using the particle extract job with helical option enabled to extract particles every 8.2 nm along the picked filaments. The extracted particles were transferred back to CryoSPARC for further analysis and 3D reconstruction. 942,986 DMT particles were initially screened for quality using 2D classification job type, and those classes with good features were chosen for further data processing leaving 868,683 particles. Initial 3D reconstructions were made using 2X binned particles to expedite data processing. CryoSPARC’s Helix refine job type was used to refine the DMT particles and prevent particles from the same filament from being placed in different half-sets during refinment. With half sets determined, the particles were then subjected to non-uniform refinement to yield an initial DMT reconstruction based on the 8.2 nm repeating tubulin heterodimer organization.
We next carried out focused classification and refinements as described previously15, using CryoSPARC. Briefly, cylindrical masks over MIPS or MOPs with known periodicities were used to relax the 16, 48, and 96 nm periodicity from the initial 8.2 nm repeating DMT structure in stepwise fashion. To improve local resolutions, we performed focused local refinements wherein cylindrical masks were placed over specific protofilaments so that CryoSPARC could be used to align those protofilaments and their MIP and MOP features. This resulted in 8, 16, 48, and 96 nm reconstructions with 3.8, 3.8, 4.2, and 4.3 Å global resolutions, respectively. Local resolutions were improved using the local refinement job types in CryoSPARC, with maps over the regions of interest.
Atomic Modeling and Docking
The tubulin models were built using AlphaFold predicted models of α- and β-tubulin and using molecular dynamics flexible fitting software in UCSF ChimeraX58,59. To model MIPs, homologs from other organisms with existing structures roughly fit into our DMT maps before using NCBI’s basic local alignment search tool (BLAST) to identify homologs in Tv and confirmed their identity using our mass spectrometry data60. For densities lacking homologous proteins, initial models were built using DeepTracer61, followed by refinement in Coot62.
The identities of unknown densities were confirmed using automated building in ModelAngelo and standard Protein BLAST of the predicted amino acid sequences against TrichDB database60,63,64. Alternatively, or often in combination with ModelAngelo predicted models, cryoID was used to identify the most likely candidates for cryo-EM densities24. Further attempts to fit proteins in low resolution regions were made using a strategy similar to that of the DomainFit software package41. Briefly, visual inspection of AlphaFold predicted structures also aided in matching of candidates with map density shapes to assess potential matches. Models were fit using Coot and ISOLDE as described previously58,65 and refined using Phenix Real Space Refinement66.
Docking of thiabendazole (SMILES: C1=CC=C2C(=C1)NC(=N2)C3=CSC=N3) into Tv β-tubulin was performed using SwissDock tools and AutoDock Vina version 1.2.034,35. Tyr168 and Phe201 were mutated to alanine residues using the “swap amino acid” function in UCSF ChimeraX59. The box center was placed at 361 – 472 – 277 for each run with dimensions 10 – 10 – 15 and sampling exhaustivity set to the default value of 4.
We used the same software as above for docking IP6 (SMILES: C1(C(C(C(C(C1OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O) O) into TvFAP40, except that the box center was placed at 393 – 277 – 260 and box size was left at default of 20 – 20 – 20 with sampling exhaustivity of 4. Grid box size was chosen to constrain ligands to putative binding sites from previous studies (thiabedazole) or observed localization (IP6)27. Structure visualization and figure preparation were done with UCSF ChimeraX59 and Adobe illustrator, respectively.
Supplementary Material
Figure S1. Fitted models in cryo-EM densities. Examples of cryo-EM maps with fitted atomic models of MIP and MOP proteins.
Figure S2. Docking experiments of β-tubulin and TvFAP40. (a) Atomic model of β-tubulin with putative BZ drug binding site boxed. (b) WT Tv β-tubulin with docked thiabendazole (TBZ), fit into putative binding site. (c) Tv β-tubulin Y168A, P201A mutant with docked TBZ in putative binding site. (d) Atomic model of TvFAP40 with putative IP6 binding site boxed. (e) TvFAP40 binding pocket with docked IP6. (f) TvFAP40 binding pocket with observed IP6.
Figure S3. Analysis of TvFAP40 and TvFAP35 and structural homologs. (a) AlphaFold-predicted models of TvFAP35 and TvFAP40 (top) colored by AlphaFold confidence interval (blue more confident, red less confident) and their atomic models (bottom) colored in cyan and magenta respectively. (b) AlphaFold-predicted structures for structural homologs from selected species, colored by AlphaFold confidence interval. (c) Sequence alignment of dimerization and MT binding domain regions from proteins in a and b aligned to TvFAP35, with conserved residues highlighted and those at the active site indicated with arrows. (d and e) α-carbon backbone aligned models from the MT-binding and dimerization domains of the kinked-coiled-coil domains. (f) Conserved proteins from c shown at their locations at the MT-binding interface on 40 TvFAP35. (g-h) Same as f but based on both faces of the dimerization domain. (i) Phylogeny tree including organisms in which FoldSeek identified similar protein structures.
Movie S1. overview of Tv-MIPs. Cross sectional view down the Tv-DMT with MIP and MOP densities colored. Model view of all modeled MIPs rotated to show detail and models of TvFAP35 and TvFAP40 in cyan and magenta respectively.
Movie S2. TvFAP40 ligand binding pocket. View flying into putative ligand binding site of TvFAP40. Rotations around the ligand binding site with and without the cryo-EM density.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (R01GM071940 to Z.H.Z. and R01AI103182/R33AI119721 to P.J.J.). K.A.M. and A.S. received support from NIH Ruth L. Kirschtein National Research Service Award AI007323. We acknowledge the use of resources at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057, 1S10OD018111) and NSF (DBI-1338135 and DMR-1548924). We acknowledge support from the UCLA AIDS Institute, the James B. Pendleton Charitable Trust, and the McCarthy Family Foundation.
Footnotes
Conflict of interest
The authors declare no competing interests.
Data availability
Cryo-EM maps of the 16, 48, and 96 nm repeats have been submitted to the Electron Microscopy Data Bank and can be found under accession numbers EMD-XXXXX, EMD-XXXXX, and EMD-XXXXX respectively. The coordinates for the complete atomic models were deposited in the Protein Data bank under accession number XXXX.
References:
- 1.Kissinger P. (2015). Trichomonas vaginalis: a review of epidemiologic, clinical and treatment issues. BMC Infectious Diseases 15, 307. 10.1186/s12879-015-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston V.J., and Mabey D.C. (2008). Global epidemiology and control of Trichomonas vaginalis. Current opinion in infectious diseases. [DOI] [PubMed] [Google Scholar]
- 3.WHO (2012). Global incidence and prevalence of selected curable sexually transmitted infections.
- 4.Kissinger P., and Adamski A. (2013). Trichomoniasis and HIV interactions: a review. Sexually Transmitted Infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z., Li D., Li Y., Zhang R., Xie X., Yao Y., Zhao L., Tian X., Yang Z., Wang S., et al. (2023). The correlation between Trichomonas vaginalis infection and reproductive system cancer: a systematic review and meta-analysis. Infectious Agents and Cancer 18, 15–15. 10.1186/s13027-023-00490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kissinger P., Secor W.E., Leichliter J.S., Clark R.A., Schmidt N., Curtin E., and Martin D.H. (2008). Early Repeated Infections with Trichomonas vaginalis among HIV-Positive and HIV-Negative Women. Clinical Infectious Diseases 46, 994–999. 10.1086/529149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirkcaldy R.D., Augostini P., Asbel L.E., Bernstein K.T., Kerani R.P., Mettenbrink C.J., Pathela P., Schwebke J.R., Secor W.E., Workowski K.A., et al. (2012). Trichomonas vaginalis antimicrobial drug resistance in 6 US cities, STD Surveillance Network, 2009–2010. Emerging infectious diseases 18, 939–943. 10.3201/eid1806.111590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Toxicology Program. Report on Carcinogens. Fifteenth Edition. (2021). [Google Scholar]
- 9.Malli S., Loiseau P.M., and Bouchemal K. (2020). Trichomonas vaginalis Motility Is Blocked by Drug-Free Thermosensitive Hydrogel. ACS Infectious Diseases 6, 114–123. 10.1021/acsinfecdis.9b00243. [DOI] [PubMed] [Google Scholar]
- 10.Korosh T., Bujans E., Morada M., Karaalioglu C., Vanden Eynde J.J., Mayence A., Huang T.L., and Yarlett N. (2017). Potential of bisbenzimidazole-analogs toward metronidazole-resistant Trichomonas vaginalis isolates. Chemical Biology & Drug Design 90, 489–495. 10.1111/cbdd.12972. [DOI] [PubMed] [Google Scholar]
- 11.Aguirre G., Boiani M., Cerecetto H., Gerpe A., González M., Sainz Y.F., Denicola A., De Ocáriz C.O., Nogal J.J., Montero D., and Escario J.A. (2004). Novel antiprotozoal products: Imidazole and benzimidazole N-oxide derivatives and related compounds. Archiv der Pharmazie 337, 259–270. 10.1002/ardp.200300840. [DOI] [PubMed] [Google Scholar]
- 12.Katiyar S.K., and Edlind T.D. (1994). β-Tubulin genes of Trichomonas vaginalis. Molecular and Biochemical Parasitology 64, 33–42. 10.1016/0166-6851(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 13.Coceres V.M., Iriarte L.S., Miranda-Magalhães A., Santos de Andrade T.A., de Miguel N., and Pereira-Neves A. (2021). Ultrastructural and Functional Analysis of a Novel Extra-Axonemal Structure in Parasitic Trichomonads. Frontiers in Cellular and Infection Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manton I., and Clarke B. (1950). Electron Microscope Observations on the Spermatozoid of Fucus. Nature 166, 973–974. 10.1038/166973a0. [DOI] [PubMed] [Google Scholar]
- 15.Ma M., Stoyanova M., Rademacher G., Dutcher S.K., Brown A., and Zhang R. (2019). Structure of the Decorated Ciliary Doublet Microtubule. Cell. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walton T., Wu H., and Brown A. (2021). Structure of a microtubule-bound axonemal dynein. Nature Communications 12, 477–477. 10.1038/s41467-020-20735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walton T., Gui M., Velkova S., Fassad M.R., Hirst R.A., Haarman E., O’Callaghan C., Bottier M., Burgoyne T., Mitchison H.M., and Brown A. (2023). Axonemal structures reveal mechanoregulatory and disease mechanisms. Nature 618, 625–633. 10.1038/s41586-023-06140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo S., Black C.S., Joachimiak E., Yang S.K., Legal T., Peri K., Khalifa A.A.Z., Ghanaeian A., McCafferty C.L., Valente-Paterno M., et al. (2023). Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins. Nature Communications 14, 2168–2168. 10.1038/s41467-023-37868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gui M., Farley H., Anujan P., Anderson J.R., Maxwell D.W., Whitchurch J.B., Botsch J.J., Qiu T., Meleppattu S., Singh S.K., et al. (2021). De novo identification of mammalian ciliary motility proteins using cryo-EM. Cell. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imhof S., Zhang J., Wang H., Bui K.H., Nguyen H., Atanasov I., Hui W.H., Yang S.K., Zhou Z.H., and Hill K.L. (2019). Cryo electron tomography with volta phase plate reveals novel structural foundations of the 96-nm axonemal repeat in the pathogen Trypanosoma brucei. eLife 8, 1–30. 10.7554/eLife.52058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z., Shiozaki M., Haas K.M., Skinner W.M., Zhao S., Guo C., Polacco B.J., Yu Z., Krogan N.J., Lishko P.V., et al. (2023). De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking. Cell 186, 5041–5053.e5019. 10.1016/j.cell.2023.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tai L., Yin G., Huang X., Sun F., and Zhu Y. (2023). In-cell structural insight into the stability of sperm microtubule doublet. Cell Discovery 9, 116–116. 10.1038/s41421-023-00606-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenaghan S.C., Nwandu-Vincent S., Reese B.E., and Zhang M. (2014). Unlocking the secrets of multi-flagellated propulsion: drawing insights from Tritrichomonas foetus. Journal of The Royal Society Interface 11, 20131149–20131149. 10.1098/rsif.2013.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho C.-M., Li X., Lai M., Terwilliger T.C., Beck J.R., Wohlschlegel J., Goldberg D.E., Fitzpatrick A.W.P., and Zhou Z.H. (2020). Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nature Methods. Springer US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. Springer US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research 50, D439–D444. 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguayo-Ortiz R., Méndez-Lucio O., Medina-Franco J.L., Castillo R., Yépez-Mulia L., Hernández-Luis F., and Hernández-Campos A. (2013). Towards the identification of the binding site of benzimidazoles to β-tubulin of Trichinella spiralis: Insights from computational and experimental data. Journal of Molecular Graphics and Modelling 41, 12–19. 10.1016/j.jmgm.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Juliano C., Martinotti M.G., and Cappuccinelli P. (1985). “In vitro” effect of microtubule inhibitors on Trichomonas vaginalis. Microbiologica 8, 31–42. [PubMed] [Google Scholar]
- 29.Leung M.R., Zeng J., Wang X., Roelofs M.C., Huang W., Zenezini Chiozzi R., Hevler J.F., Heck A.J.R., Dutcher S.K., Brown A., et al. (2023). Structural specializations of the sperm tail. Cell 186, 2880–2896.e2817. 10.1016/j.cell.2023.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Helge W., Shindyalov I.N., Bourne P.E., Weissig H., Shindyalov I.N., and Bourne P.E. (2000). The Protein Data Bank. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bateman A., Martin M.-J., Orchard S., Magrane M., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bye-A-Jee H., Cukura A., et al. (2023). UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Research 51, D523–D531. 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallery D.L., Márquez C.L., McEwan W.A., Dickson C.F., Jacques D.A., Anandapadamanaban M., Bichel K., Towers G.J., Saiardi A., Böcking T., and James L.C. (2018). IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 7. 10.7554/eLife.35335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grosdidier A., Zoete V., and Michielin O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Research 39, W270–W277. 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eberhardt J., Santos-Martins D., Tillack A.F., and Forli S. (2021). AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. Journal of Chemical Information and Modeling 61, 3891–3898. 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugnon M., Röhrig U.F., Goullieux M., Perez M.A.S., Daina A., Michielin O., and Zoete V. (2024). SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Research, 1–9. 10.1093/nar/gkae300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott O., and Olson A.J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarmah B., Winfrey V.P., Olson G.E., Appel B., and Wente S.R. (2007). A role for the inositol kinase Ipk1 in ciliary beating and length maintenance. Proceedings of the National Academy of Sciences 104, 19843–19848. 10.1073/pnas.0706934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarmah B., Latimer A.J., Appel B., and Wente S.R. (2005). Inositol Polyphosphates Regulate Zebrafish Left-Right Asymmetry. Developmental Cell 9, 133–145. 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Fedorov V.A., Orekhov P.S., Kholina E.G., Zhmurov A.A., Ataullakhanov F.I., Kovalenko I.B., and Gudimchuk N.B. (2019). Mechanical properties of tubulin intra- and inter-dimer interfaces and their implications for microtubule dynamic instability. PLOS Computational Biology 15, e1007327–e1007327. 10.1371/journal.pcbi.1007327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Kempen M., Kim S.S., Tumescheit C., Mirdita M., Lee J., Gilchrist C.L.M., Söding J., and Steinegger M. (2024). Fast and accurate protein structure search with Foldseek. Nature Biotechnology 42, 243–246. 10.1038/s41587-023-01773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J., Tong M., Lee C., Gaertig J., Legal T., and Bui K.H. (2024). DomainFit: Identification of protein domains in cryo-EM maps at intermediate resolution using AlphaFold2-predicted models. Structure, 1–12. 10.1016/j.str.2024.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lacey S.E., Foster H.E., and Pigino G. (2023). The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains. Nature Structural & Molecular Biology 30, 584–593. 10.1038/s41594-022-00905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reck-Peterson S.L., Yildiz A., Carter A.P., Gennerich A., Zhang N., and Vale R.D. (2006). Single-Molecule Analysis of Dynein Processivity and Stepping Behavior. Cell 126, 335–348. 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Miguel N., Riestra A., and Johnson P.J. (2012). Reversible association of tetraspanin with Trichomonas vaginalis flagella upon adherence to host cells. Cellular Microbiology 14, 1797–1807. 10.1111/cmi.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beauchamp P.J., Galle P.C., and Blasco L. (1984). Human Sperm Velocity and Postinsemination Cervical Mucus Test in the Evaluation of the Infertile Couple. Archives of Andrology 13, 107–112. 10.3109/01485018408987508. [DOI] [PubMed] [Google Scholar]
- 46.Smyth M., Pettitt T., Symonds A., and Martin J. (2003). Identification of the pocket factors in a picornavirus. Archives of Virology 148, 1225–1233. 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 47.Hardy J.M., Newton N.D., Modhiran N., Scott C.A.P., Venugopal H., Vet L.J., Young P.R., Hall R.A., Hobson-Peters J., Coulibaly F., and Watterson D. (2021). A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses. Nature Communications 12, 3266–3266. 10.1038/s41467-021-22773-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flatt J.W., Domanska A., Seppälä A.L., and Butcher S.J. (2021). Identification of a conserved virion-stabilizing network inside the interprotomer pocket of enteroviruses. Communications Biology 4, 250–250. 10.1038/s42003-021-01779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark C.G., and Diamond L.S. (2002). Methods for Cultivation of Luminal Parasitic Protists of Clinical Importance. Clinical Microbiology Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens A., Muratore K., Cui Y., Johnson P.J., and Zhou Z.H. (2021). Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2. mBio 12, 1–17. 10.1128/mBio.02924-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludwiczak J., Winski A., Szczepaniak K., Alva V., and Dunin-Horkawicz S. (2019). DeepCoil—a fast and accurate prediction of coiled-coil domains in protein sequences. Bioinformatics 35, 2790–2795. 10.1093/bioinformatics/bty1062. [DOI] [PubMed] [Google Scholar]
- 52.Garg S., Stölting J., Zimorski V., Rada P., Tachezy J., Martin W.F., and Gould S.B. (2015). Conservation of Transit Peptide-Independent Protein Import into the Mitochondrial and Hydrogenosomal Matrix. Genome Biology and Evolution 7, 2716–2726. 10.1093/gbe/evv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mastronarde D.N. (2003). SerialEM: A program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microscopy and Microanalysis 9, 1182–1183. 10.1017/s1431927603445911. [DOI] [Google Scholar]
- 54.Punjani A., Rubinstein J.L., Fleet D.J., and Brubaker M.A. (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods. [DOI] [PubMed] [Google Scholar]
- 55.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A.J., and Berger B. (2019). Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nature Methods. Springer US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheres S.H.W. (2013). Single-particle processing in RELION. Manuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lövestam S., and Scheres S.H.W. (2022). High-throughput cryo-EM structure determination of amyloids. Faraday Discussions 240, 243–260. 10.1039/D2FD00034B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Croll T.I. (2018). ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallographica Section D: Structural Biology 74, 519–530. 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., and Ferrin T.E. (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGinnis S., and Madden T.L. (2004). BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research 32, W20–W25. 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfab J., Phan N.M., and Si D. (2021). DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proceedings of the National Academy of Sciences 118. 10.1073/pnas.2017525118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P., Lohkamp B., Scott W.G., and Cowtan K. (2010). Features and development of Coot. Acta Crystallographica Section D Biological Crystallography 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jamali K., Käll L., Zhang R., Brown A., Kimanius D., and Scheres S.H.W. (2024). Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457. 10.1038/s41586-024-07215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alvarez-Jarreta J., Amos B., Aurrecoechea C., Bah S., Barba M., Barreto A., Basenko E.Y., Belnap R., Blevins A., Böhme U., et al. (2024). VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center in 2023. Nucleic Acids Research 52, D808–D816. 10.1093/nar/gkad1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu I., Nguyen L., Avaylon J., Wang K., Lai M., and Zhou Z.H. (2018). Building atomic models based on near atomic resolution cryoEM maps with existing tools. Journal of Structural Biology. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., and Adams P.D. (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallographica Section D: Structural Biology 74, 531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Fitted models in cryo-EM densities. Examples of cryo-EM maps with fitted atomic models of MIP and MOP proteins.
Figure S2. Docking experiments of β-tubulin and TvFAP40. (a) Atomic model of β-tubulin with putative BZ drug binding site boxed. (b) WT Tv β-tubulin with docked thiabendazole (TBZ), fit into putative binding site. (c) Tv β-tubulin Y168A, P201A mutant with docked TBZ in putative binding site. (d) Atomic model of TvFAP40 with putative IP6 binding site boxed. (e) TvFAP40 binding pocket with docked IP6. (f) TvFAP40 binding pocket with observed IP6.
Figure S3. Analysis of TvFAP40 and TvFAP35 and structural homologs. (a) AlphaFold-predicted models of TvFAP35 and TvFAP40 (top) colored by AlphaFold confidence interval (blue more confident, red less confident) and their atomic models (bottom) colored in cyan and magenta respectively. (b) AlphaFold-predicted structures for structural homologs from selected species, colored by AlphaFold confidence interval. (c) Sequence alignment of dimerization and MT binding domain regions from proteins in a and b aligned to TvFAP35, with conserved residues highlighted and those at the active site indicated with arrows. (d and e) α-carbon backbone aligned models from the MT-binding and dimerization domains of the kinked-coiled-coil domains. (f) Conserved proteins from c shown at their locations at the MT-binding interface on 40 TvFAP35. (g-h) Same as f but based on both faces of the dimerization domain. (i) Phylogeny tree including organisms in which FoldSeek identified similar protein structures.
Movie S1. overview of Tv-MIPs. Cross sectional view down the Tv-DMT with MIP and MOP densities colored. Model view of all modeled MIPs rotated to show detail and models of TvFAP35 and TvFAP40 in cyan and magenta respectively.
Movie S2. TvFAP40 ligand binding pocket. View flying into putative ligand binding site of TvFAP40. Rotations around the ligand binding site with and without the cryo-EM density.
Data Availability Statement
Cryo-EM maps of the 16, 48, and 96 nm repeats have been submitted to the Electron Microscopy Data Bank and can be found under accession numbers EMD-XXXXX, EMD-XXXXX, and EMD-XXXXX respectively. The coordinates for the complete atomic models were deposited in the Protein Data bank under accession number XXXX.