Abstract
Our group and others have recently demonstrated the ability of recombinant baculoviruses to transduce mammalian cells at high frequency. To further characterize the use of baculovirus as a mammalian gene delivery system, we examined the status of transduced DNA stably maintained in Chinese hamster ovary (CHO) cells. Four independent clones carrying two introduced markers, the genes for neomycin resistance (Neo) and green fluorescent protein (GFP), were selected. PCR analysis, Southern blotting, and DNA sequencing showed that discrete portions of the 148-kb baculovirus DNA were present as single-copy fragments ranging in size from 5 to 18 kb. Integration into the CHO cell genome was confirmed by fluorescent in situ hybridization (FISH) analysis. For one clone, the left and right viral/chromosomal junctions were determined by DNA sequencing of inverse PCR products. Similarly, for a different clone, the left viral/chromosomal junction was determined; however, the right junction sequence revealed the joining to another viral fragment by a short homology (microhomology), a hallmark of illegitimate recombination. The random viral breakpoints and the lack of homology between the virus and flanking chromosomal sequences are also suggestive of an illegitimate integration mechanism. To examine the long-term stability of reporter gene expression, all four clones were grown continuously for 36 passages in either the presence or absence of selection for Neo. Periodic assays over a 5-month period showed no loss of GFP expression for at least two of the clones. This report represents the first detailed analysis of baculovirus integrants within mammalian cells. The potential advantages of the baculovirus system for the stable integration of genetic material into mammalian genomes are discussed.
A number of procedures are available for stably introducing foreign DNA into mammalian cell genomes. The predominant method is transfection by one of several means, including electroporation, calcium phosphate precipitation, lipofection, and microinjection (1). Although transfection procedures vary in their effects on cell viability, frequency of integration, and concatenation of the vector, all transfection protocols result in stable integration generally via illegitimate recombination. Illegitimate integration is frequently associated with rearrangements in both the introduced DNA and the chromosomal target (4, 28).
Several mammalian viruses, such as retrovirus and adeno-associated virus, are commonly used as gene delivery vehicles. In choosing a virus, one must carefully consider its particular features, including the ease of producing recombinant virus, possible size limitations for cloned inserts, and the ability to generate sufficiently high titers. One must also consider its mode of integration. Retrovirus, for example, encodes a viral integrase which directs nearly full-length, single-copy integration events (9); adeno-associated virus, on the other hand, has the propensity to form concatemers (23) and to integrate preferentially into human chromosome 19 (16).
A recent advance is the use of recombinant baculovirus derived from the insect virus Autographa californica nucleopolyhedrovirus (AcMNPV) as a vector for gene transfer into mammalian cells. Baculovirus has traditionally been used to overexpress proteins for purification from insect-derived host cells (14, 15, 19, 27). Indeed, much has been learned about the life cycle of baculovirus in insect cells. There is no evidence that baculovirus DNA integration occurs within a permissive host during productive infection; rather, the virus buds from the cell to allow for reinfection (25). Recently, our laboratory and other groups have demonstrated the ability of recombinant baculoviruses carrying reporter genes to transiently and stably transduce a wide variety of mammalian cell types at high frequencies (3, 5, 30, 31). The mechanism by which baculovirus DNA is introduced into a mammalian cell remains unclear, although several studies have provided insights (2, 7, 10, 13). Likewise, little is known concerning the state of the viral genome once inside the cell's nucleus. One formal possibility is that all or part of the AcMNPV genome is maintained episomally following transduction. Tjia et al. (33) used Southern blotting to examine populations of mammalian cells treated with baculovirus, demonstrating that viral DNA persisted in the nuclei for 24 to 48 h but apparently not beyond that time. However, having no selection for individual clones, they did not exclude the possibility that portions of the baculovirus genome integrated into mammalian chromosomes in a percentage of the cells.
In this study, we wished to determine if transduced baculovirus integrates into a mammalian genome and to examine the state of the viral DNA in terms of its integrity and copy number. In addition, we wished to study the stability of the virally introduced reporter gene over extended periods. We addressed these issues by mapping baculovirus breakpoints, confirming integration by fluorescent in situ hybridization (FISH) analysis, cloning chromosomal junctions, and by long-term analysis of reporter gene expression.
MATERIALS AND METHODS
Generation of recombinant baculovirus.
Green fluorescent protein (GFP)-expressing recombinant baculovirus BacMam1 GFP (hereafter referred to as vBMGFP) was generated using the Bac-to-Bac system (Life Technologies) with the shuttle plasmid pFastBacMam1 GFP as described previously (5). Briefly, recombinant virus is generated by site-specific transposition of shuttle plasmid sequences flanked by the left and right ends of Tn7 into its target site on the baculovirus vector, which is derived from AcMNPV (20). Virus was propagated in Spodoptera frugiperda (Sf9) cells grown in Grace's supplemented insect media containing 10% fetal bovine serum, 0.1% pluronic F-68, and 25 μg of gentamicin/ml. Virus titer was determined by plaque assay on Sf9 cells according to the standard protocol (25).
Cell culture.
Chinese hamster ovary (CHO) cells were grown in a 1-to-1 mixture of Dulbecco's modified Eagle's medium and F-12 medium, supplemented with 5% fetal bovine serum and 25 μg of gentamicin/ml. Stably transduced clones were grown in the same medium supplemented with 500 μg of active Geneticin (G418)/ml. Cells were grown at 37°C in a humidified incubator with 5% CO2. Fetal bovine serum was purchased from HyClone; all other tissue culture reagents were from Life Technologies.
Stable transduction of CHO cells.
Clones V2, V4, V6, and V8 were generated from separate transduction experiments. CHO cells were seeded at 2 × 105 cells per well of a six-well culture dish. The following day, culture medium was replaced with virus at a multiplicity of 200 PFU per cell, and incubation was continued at 37°C for 1 h. Following removal of virus inoculum, fresh medium was added. To select for stable transductants, cells were grown in the presence of G418 (500 μg/ml). Cells expressing GFP were single-cell cloned by sorting bulk selected pools via flow cytometry.
Flow cytometry.
To isolate clones V2, V4, V6, and V8, G418-resistant populations of cells from separate transductions were washed and resuspended in phosphate-buffered saline with 1% fetal bovine serum. Cells expressing GFP were sorted by flow cytometry into wells of a 96-well plate with a Becton Dickinson FACStar Plus flow cytometer. Clones V2, V4, V6, and V8 were subcultured for 5 months by diluting cells 1:50 every 5 to 7 days both in the presence and in the absence of G418. Every 30 days, cultures were assayed by flow cytometry for GFP expression. To allow comparison between different assays and calculation of fluorescence intensity, Quantum GFP particles (Flow Cytometry Standards, San Juan, Puerto Rico) were analyzed to generate a standard curve of molecules of equivalent soluble fluorochrome of GFP.
PCR.
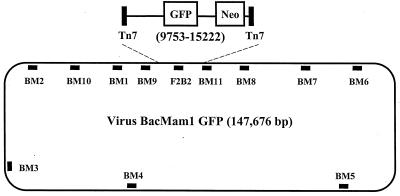
A series of primers (20 to 22 nucleotides) were designed to amplify 1-kb segments of the 148-kb recombinant baculovirus DNA. The product names and corresponding baculovirus map positions amplified in PCR reactions were as follows: BM1 (3579 to 5000), BM9 (7311 to 8381), F2B2 (11597 to 12707), BM11 (15025 to 16226), BM8 (21032 to 22065), BM7 (29967 to 31079), BM6 (38096 to 39356), BM5 (68001 to 69252), BM4 (98981 to 100200), BM3 (119201 to 120170), BM2 (139931 to 141054), and BM10 (146120 to 147233). Sequences are available upon request. Each PCR mixture contained 500 ng of genomic DNA or 1 ng of virus DNA. Reactions were performed under standard conditions with Perkin-Elmer Taq DNA polymerase in an MJ Research PT-200 thermal cycler with the following conditions: an initial denaturation of 94°C for 5 min, then 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, followed by a final cycle of 72°C for 7 min.
Southern blot analysis.
Purified recombinant baculovirus DNA and cellular genomic DNA were digested with restriction endonucleases and subjected to electrophoresis on 0.8% agarose gels. Purification of DNA, digestion of DNA, and Southern blotting were performed according to standard procedures (29). Gels were blotted onto Biodyne B membranes (Pall). Shuttle plasmid pFastBacMam1 GFP, linearized with EcoRI, was radioactively labeled with the Life Technologies Random Primers DNA labeling system for use as a probe.
FISH analysis.
Analysis of clone V6 by FISH was performed by SeeDNA Biotech Inc. Cells were harvested, and slides were prepared using standard procedures including hypotonic treatment and methanol/acetic acid fixation. Shuttle plasmid pFastBacMam1 GFP was biotinylated with dATP using a BioNick labeling kit (Life Technologies) for use as a probe. The detection procedure was performed as described previously (11, 12). Briefly, slides were treated with RNase A, denatured in 70% formamide in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 70°C for 2 min, and dehydrated with ethanol. The probe was denatured at 75°C for 5 min in a hybridization mix consisting of 50% formamide and 10% dextran sulfate. Following overnight hybridization, FISH signals and DAPI (4′,6-diamidino-2-phenylindole) banding patterns were recorded separately by photography.
Inverse PCR.
To clone baculovirus integration junctions, we used inverse PCR essentially as described previously (18, 24). PCR primers were designed to baculovirus sequences based on mapping data for clones V2 and V6 (see Results). Sequences of primers are available upon request. To prepare template DNA for inverse PCR, genomic DNA was digested with the appropriate restriction enzyme, diluted, and then circularized with T4 DNA ligase (New England Biolabs). Ethanol-precipitated template DNA was subjected to 40 rounds of PCR using Platinum PCR Supermix (Life Technologies). A 3-μl aliquot was reamplified with nested primers.
DNA sequencing.
Inverse PCR products were subcloned into either the pCR vector (Invitrogen) or pGEM-T (Promega) and subjected to DNA sequencing of both strands. Sequencing was performed in the Glaxo Wellcome DNA Sequencing Facility using PE Biosystems ABI Prism 377 and 3700 sequencers with Taq FS dye termination chemistry.
RESULTS
PCR analysis of clones stably transduced with baculovirus.
The recombinant baculovirus vBMGFP, which possesses the two mammalian expression cassettes Neo (neomycin resistance) and GFP, was used to transduce CHO cells. Four independent clones (V2, V4, V6, and V8) from separate transduction experiments were isolated by first selecting for neomycin resistance and then sorting of single cells expressing GFP. Genomic DNA was extracted from each clone and characterized by PCR with a series of baculovirus primer pairs designed to generate ∼1-kb products (Fig. 1). Results of this analysis are presented in Table 1. As expected, all four clones were positive for F2B2, a PCR product representing a portion of the GFP gene. However, analysis with other primer pairs suggested that only a fragment of the 148-kb baculovirus DNA was present in each clone. Clones V2 and V6 were negative for all other PCR products, V4 was positive for one additional PCR product, and V8 was positive for two additional PCR products.
FIG. 1.
vBMGFP and relative positions of 12 regions amplified in PCRs. To generate vBMGFP, insert sequences carried on shuttle plasmid pFastBacMam1 GFP (5) were transposed to the Tn7 target site on a baculovirus vector (20) via the Life Technologies Bac-to-Bac system. The expanded view of the insert (map positions 9753 to 15222) shown. This sequence includes left and right Tn7 sequences and the genes for GFP and neomycin resistance (Neo). The only PCR product located entirely in this region is F2B2.
TABLE 1.
PCR analysis of vBMGFP-transduced CHO cells
| PCR product | Expected size (kb) | DNA template
|
|||||
|---|---|---|---|---|---|---|---|
| vBMGFP | CHO | V2 | V4 | V6 | V8 | ||
| BM5 | 1.2 | + | − | − | − | − | − |
| BM4 | 1.2 | + | − | − | − | − | − |
| BM3 | 1.0 | + | − | − | − | − | − |
| BM2 | 1.1 | + | − | − | − | − | − |
| BM10 | 1.1 | + | − | − | − | − | − |
| BM1 | 1.4 | + | − | − | − | − | + |
| BM9 | 1.1 | + | − | − | − | − | + |
| F2B2 | 1.1 | + | − | + | + | + | + |
| BM11 | 1.2 | + | − | − | + | − | − |
| BM8 | 1.0 | + | − | − | − | − | − |
| BM7 | 1.1 | + | − | − | − | − | − |
| BM6 | 1.2 | + | − | − | − | − | − |
Mapping of viral breakpoints by Southern blotting.
With the PCR data serving as a reference, genomic DNA from clones V2, V4, V6, and V8 was subjected to Southern blotting to more accurately map the baculovirus breakpoints. Initially, XbaI-digested DNA was probed with the shuttle plasmid (pFastBacMam1 GFP), which carries GFP and Neo flanked by Tn7 sites (5). Results are presented in Fig. 2A. Since XbaI cleaves once in the probe region between the Neo and GFP genes, the two-band pattern observed for each clone was consistent with the previous PCR analysis. Two of the clones (V4 and V8) showed a common 2.6-kb band, suggesting the presence of viral sequence containing an XbaI site immediately outside the probe region (Fig. 2B). The remaining two clones (V2 and V6) lacked this 2.6-kb band, suggesting breakpoints within the probe region near the Neo gene.
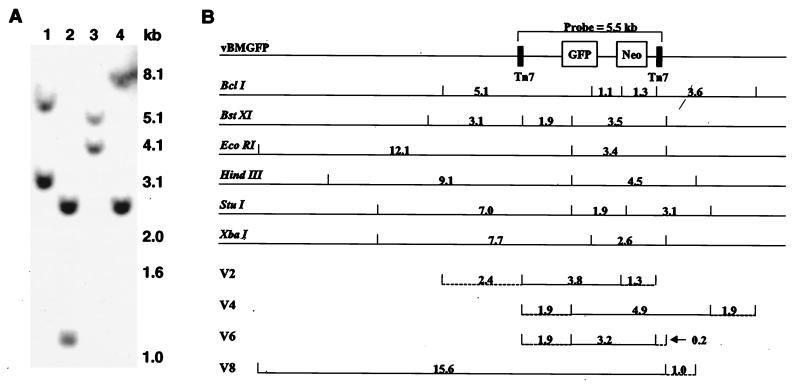
FIG. 2.
Representative Southern blot and schematic map of viral integrants. (A) Southern blot analysis of XbaI-digested genomic DNA from clones V2, V4, V6, and V8 (lanes 1 to 4, respectively), probed with shuttle plasmid pFastBacMam1 GFP (containing a 5.5-kb insert). (B) Band sizes for intact vBMGFP digested with given restriction endonucleases (top) and composite map of viral breakpoints in four clones (bottom). A portion of the recombinant baculovirus genome, including the 5.5-kb probe region, is illustrated. Positions of the restriction sites in relation to the baculovirus genome are marked by short vertical lines, with intervening distances given in kilobases. A composite map of each clone was derived from Southern blot data. Horizontal solid lines represent sequences known to be integrated; horizontal dashed lines represent breakpoint regions. For example, for clone V2, the left breakpoint fell somewhere within the 2.4-kb region between BclI and BstXI sites (drawing not exactly to scale).
A similar analysis was performed with five additional restriction endonucleases (Fig. 2B, top) to generate a composite map for each of the four clones (Fig. 2B, bottom). Thus, based solely on restriction mapping, clone V2 showed a baculovirus fragment of minimally 3.8 kb, but with a maximum possible size of 7.5 kb given the defined left and right breakpoint regions. Clone V4 demonstrated a fragment of 4.9 to 8.7 kb, and V6 showed a fragment of 3.2 to 5.3 kb. Clone V8 carries the largest baculovirus fragment, with a size of at least 15.6 kb. Combining the Southern blotting data with the PCR results indicates that the V8 fragment has a maximum size of 17.9 kb. To confirm these results, we used several PCR products (generated using purified virus as template) as probes with stripped filters, giving the predicted positive and negative signals for all four clones (data not shown).
FISH analysis of V6 clone.
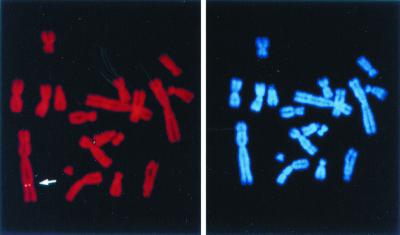
To confirm integration of baculovirus DNA into the CHO genome, clone V6 was subjected to FISH. Probe pFastBacMam1 GFP was detected on one specific chromosome (Fig. 3). Other loci were detected on different chromosomes at a very low frequency (<5% of chromosome spreads examined) and were regarded as background. Thus, the FISH results for clone V6 were consistent with our previous mapping data, demonstrating a single integration event within the CHO genome.
FIG. 3.
Example of FISH analysis of the V6 clone. Left, FISH signal on a specific chromosome (marked by an arrow) using shuttle plasmid pFastBacMam1 GFP as the probe; right, the same mitotic figure stained with DAPI.
Isolation and sequencing of viral/chromosomal junctions.
To clone the left and right junctions of baculovirus and chromosomal DNA, we used inverse PCR, a technique that allows amplification of sequences that fall outside known sequences. Using this procedure, left and right junctions were obtained for clones V2 and V6 (Fig. 4). Isolation of each junction required a separate inverse PCR strategy. In some cases, our previous mapping by Southern blotting proved helpful in deciding which restriction endonucleases to use in the procedure. For instance, given the sizes of the two bands seen in our Southern blotting with XbaI-digested V2 genomic DNA (Fig. 2A, lane 1), we knew the two possible locations of XbaI sites in both the left and right unknown sequences. Genomic DNA extracted from V2 was digested with XbaI, diluted, and circularized with ligase. Nested primer pairs to left and right known sequences were used in attempts to amplify junctional sequences. This procedure resulted in a 1.4-kb PCR product representing the right junction. DNA sequencing using primers to known sequence revealed a baculovirus breakpoint at position 14907 (between the Neo cassette and the right Tn7 site) and 895 nucleotides of unknown chromosomal junction sequence (Fig. 4A). The linkage of this sequence to the integrated baculovirus DNA fragment of clone V2 was confirmed by Southern blotting, using the 895-nucleotide chromosomal sequence as a probe. This 895-nucleotide sequence did not demonstrate homology to the flanking baculovirus sequence, nor did it show significant homology to sequences in the GenBank database.
FIG. 4.
Sequences at viral integration junctions. The three viral/chromosomal junctions (A and B) and two viral/viral junctions (B and C) were obtained by inverse PCR and DNA sequencing as described in the text. The observed junction sequences are underlined. Sequences not underlined are deleted viral sequences beyond the breakpoint, shown for the sake of comparison. Twenty nucleotides of each cloned chromosomal junction sequence (bold lowercase) are given (A and B). Microhomologies (bold uppercase) are illustrated for viral/viral junctions (B and C). Above or below the breakpoint nucleotide is the first numeral of the vBMGFP map position.
In a similar manner, the left junction for clone V2 was isolated. Starting with EcoRI-digested genomic DNA, we generated an inverse PCR product containing the baculovirus breakpoint at position 9947 (within the left Tn7 site) and 51 nucleotides of chromosomal junction sequence (Fig. 4A). Taken together, these results revealed a 5-kb baculovirus fragment in the V2 clone. To confirm linkage, PCR primers were designed to the left and right chromosomal junction sequences. Using V2 genomic DNA as template, long-distance PCR generated the expected 5-kb product, diagnostic for left and right chromosomal sequences linked to the integrated baculovirus fragment (data not shown).
To obtain the left junction for clone V6, genomic DNA was digested with EcoRI. Inverse PCR and DNA sequencing revealed a baculovirus breakpoint at position 9979 and provided 1198 nucleotides of chromosome sequence at the junction (Fig. 4B). Using the 1,198-nucleotide chromosomal sequence as a probe, the linkage of this sequence to the integrated baculovirus DNA fragment was confirmed by Southern blotting of V6 genomic DNA. This 1,198-nucleotide sequence did not demonstrate significant homology to flanking baculovirus sequences or to sequences in the GenBank database.
The right junction for clone V6 was isolated by an inverse PCR strategy using genomic DNA digested with restriction enzymes (SacII and SstI) not used in our previous mapping experiments. Starting with SacII-digested genomic DNA, we generated an inverse PCR product that, when sequenced, revealed a baculovirus breakpoint at position 15082 (Fig. 4B), indicating a baculovirus fragment of 5.1 kb. In this case, the right end is joined not to unknown sequence but rather to a second baculovirus fragment normally located over 37 kb away in inverse orientation on intact baculovirus DNA. This junctional fragment is of undetermined length but is at least 285 nucleotides based on DNA sequence analysis. Since we had not designed PCR primers to this particular region of the baculovirus genome, this second fragment went undetected in our previous mapping experiments. At the junction between these two baculovirus fragments is a three-nucleotide homology.
Starting with SstI-digested V6 genomic DNA, another inverse PCR product was obtained for the right junction. Again, sequence analysis revealed the joining of the 5.1-kb fragment to a second baculovirus fragment normally found over 37 kb away. However, unlike the previously described case where only two joined fragments were detected, here a third baculovirus fragment (>290 nucleotides) was observed (Fig. 4C). In this case, this third fragment has disrupted the second fragment, leaving it with only 42 of its original >285 nucleotides. At the junction between the second and third fragments is a four-nucleotide homology. We believe this particular junction to represent a very minor rearrangement product in V6 cells since it was not detected in our previous mapping by Southern blotting, even though it introduces a diagnostic XbaI site and is homologous to the probe. Most likely, this third fragment was derived from the 5.1-kb fragment (positions 9979 to 15082) through an illegitimate recombination event in late-passage cells.
Long-term stability of reporter gene expression.
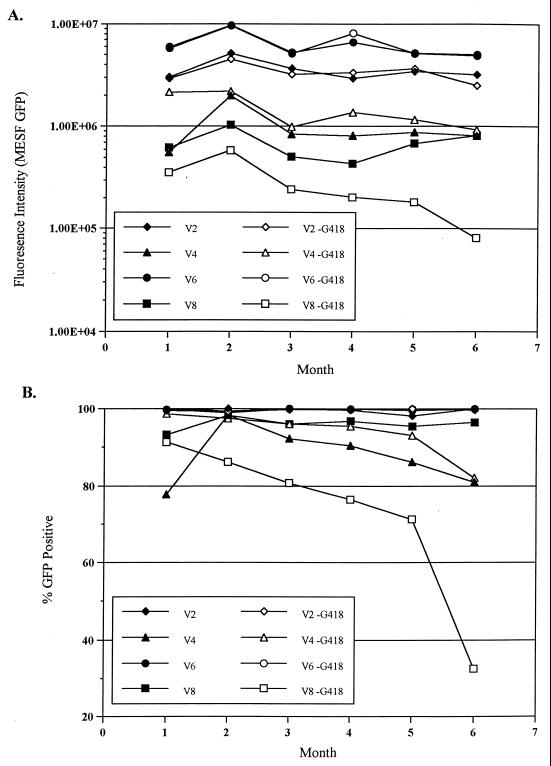
Knowing that baculovirus DNA is stably integrated into the CHO genome, we wished to further characterize baculovirus as a gene delivery system by determining the abilities of clones V2, V4, V6, and V8 to maintain GFP expression over extended growth periods. The four clones were passaged continuously in medium with or without G418, which selects for maintenance of the Neo gene. Cells were monitored periodically for GFP expression in two ways: (i) by assaying for total GFP expression, as measured by fluorescence intensity (Fig. 5A), and (ii) by assaying for the percentage of GFP-positive cells in the population (Fig. 5B). Cells were grown for a total of 36 passages. During that period, flow cytometry assays for GFP were performed once a month for 5 months. Two clones, V2 and V6, maintained starting levels of fluorescence, with or without selection for Neo. Similarly, both clones demonstrated nearly 100% GFP-positive cells during the course of these experiments. Clone V8 cells grown under G418 selection maintained starting levels of both total GFP expression and the percentage of GFP-positive cells. In contrast, V8 cells without selective pressure showed a large decrease in GFP by both assays. Clone V4 showed an intermediate response, demonstrating some decrease in total fluorescence and percent GFP-positive cells over time, with and without selection for Neo.
FIG. 5.
Stability of GFP expression in vBMGFP-transduced clones. V2, V4, V6, and V8 were grown in the presence (closed symbols) and absence (open symbols) of G418, which selects for the Neo gene adjacent to the GFP gene. Flow cytometry assays were performed once a month for 5 months to determine the total fluorescence intensity (A) and percent GFP-positive cells (B) for each clone. MESF, molecules of equivalent soluble fluorochrome.
DISCUSSION
Previous studies in our laboratory described the use of a recombinant baculovirus to direct the transient and stable expression of the GFP reporter gene in a wide variety of mammalian cells (5). In these previous studies, we observed one stable cell clone for approximately every 50 to 100 transiently transduced CHO cells. In this report, we explored the state of the transduced viral sequences that are stably maintained in CHO cells. The baculovirus system is largely unlike any other method currently used for DNA delivery into mammalian cells. Indeed, insect cells, not mammalian cells, are the natural host of baculovirus derived from AcMNPV. Given this, we were uncertain what findings to expect a priori. By mapping breakpoints, examining transduced cells by FISH analysis, and cloning viral/chromosomal junctions, we demonstrated that recombinant baculovirus DNA integrated into the CHO genome as small, discrete fragments.
The breakpoint positions and sequences at integration junctions suggest that the baculovirus DNA integrates into the CHO genome via illegitimate recombination. We found that only a fraction of the 148-kb baculovirus genome was present in the four examined clones. These fragments, which ranged in size from 5 kb to about 18 kb, had randomly distributed breakpoints outside of the selected region, suggestive of an illegitimate mode of integration. We cannot rule out the possibility that regions of the baculovirus genome other than those immediately surrounding the selected markers might be found in cells early in the process of selecting for the clones. However, in our limited PCR analysis of the CHO clones presented here (Table 1), we detected only regions closely linked to the selected markers.
To further explore the mechanism of integration, we isolated and characterized three viral/chromosomal junctions by inverse PCR and DNA sequencing. A hallmark of illegitimate recombination is the presence of little to no homology between recombining DNA molecules (22). Comparisons of the chromosomal sequences found at the junctions, in fact, revealed no significant homology to the flanking baculoviral sequences. In addition to the three viral/chromosomal junctions, a fourth junction was isolated and found to involve the joining of a 5.1-kb baculovirus fragment to another fragment derived from a distant site in the baculovirus genome. In this fourth case, flanking sequences in both directions, including deleted sequences, are known. Thus, it was possible to examine the flanking sequences for the presence of microhomologies, short homologies (one to seven nucleotides) commonly found at illegitimate recombination junctions, including integration junctions for viruses (32, 35) and transfected DNA (21). Indeed, a three-nucleotide homology was present at the junction between these two fragments, which may have joined via illegitimate recombination prior to integration. Alternatively, insertion of a larger piece of the baculovirus genome may have been followed by rearrangement accompanied by deletion of intervening sequences.
The four clones examined were found to have single-copy integrants of the selected baculovirus region; in contrast, transfection methods frequently result in concatenated integrants. Tandemly arrayed concatemers of transfected DNA form predominantly by homologous recombination, followed by illegitimate integration into the mammalian genome (6, 8). Such a process may be precluded by the size of the baculovirus genome; concatemers of the baculovirus genome may be too large to undergo stable integration events. Alternatively, the lack of concatemers could be related to the mechanism of DNA delivery by baculovirus. Whatever the reason, these results suggest that the baculovirus system may be useful for producing mammalian cell clones containing single copies of an introduced gene, without a loss in the overall frequency of stable clones generated.
To further examine the use of baculovirus as a gene delivery system, we characterized the four clones for the ability to maintain reporter gene expression over an extended period of time. Two of the clones maintained starting levels of GFP expression over a 5-month period, whether or not the clones were maintained under selection for the Neo gene (which is adjacent to the GFP gene). That the remaining two clones exhibited at least some loss of GFP expression is not surprising. The gradual extinction of reporter gene expression is commonly observed in mammalian cells and is largely attributed to epigenetic modifications (17, 26), as well as point mutations and other genetic rearrangements, including deletions (34). Southern blot analysis of our four late-passage clones demonstrated the persistence of the selected region as well as viral sequences flanking the selected region (data not shown), consistent with the quenching of GFP reporter gene expression by epigenetic modifications and/or minor genetic alterations. Thus, the baculovirus system is comparable to other modes of gene delivery in terms of generating stable expression of a reporter gene in mammalian cells. Our studies further underscore the importance of considering long-term transgene stability, especially for studies designed to correct genetic defects in vivo.
The baculovirus system has several advantages over other methods for gene delivery. Our previous studies demonstrated that baculovirus is able to transduce several cell lines that are traditionally difficult to transfect (5). Mammalian viruses used as gene delivery vehicles likewise often have specific limitations, such as low titers, strict size restraints for inserts, or high toxicity in mammalian cells. In contrast, the generation of recombinant baculoviruses with large inserts is straightforward, high titers are readily produced, and toxicity in mammalian cells is low. In addition, baculovirus is unable to replicate in mammalian cells and is therefore nonpathogenic. These studies demonstrate the potential use of baculovirus to deliver single copies of stably integrated genes into mammalian genomes. Since the illegitimate mode of integration and long-term stability of reporter gene expression are similar to that seen for transfected DNA and other viruses, recombinant baculoviruses should provide a viable alternative for DNA transfer into mammalian cells. Indeed, in recent studies with several different cell lines, we have obtained stable clones from any cell line that is successfully transduced transiently, regardless of the transduction efficiency.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. pp. 901–995. [Google Scholar]
- 2.Barsoum J, Brown R, McKee M, Boyce F M. Efficient transduction of mammalian cells by a recombinant baculovirus having the vesicular stomatitis virus G glycoprotein. Hum Gene Ther. 1997;8:2011–2018. doi: 10.1089/hum.1997.8.17-2011. [DOI] [PubMed] [Google Scholar]
- 3.Boyce F M, Bucher N L. Baculovirus-mediated gene transfer into mammalian cells. Proc Natl Acad Sci USA. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J M. Molecular mechanisms of nucleic acid integration. J Med Virol. 1990;31:43–49. doi: 10.1002/jmv.1890310109. [DOI] [PubMed] [Google Scholar]
- 5.Condreay J P, Witherspoon S M, Clay W C, Kost T A. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc Natl Acad Sci USA. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czernilofsky A P, Stabel P, Jung C. Studies on cellular tandemization of herpes simplex virus thymidine kinase DNA. DNA. 1985;4:309–318. doi: 10.1089/dna.1985.4.309. [DOI] [PubMed] [Google Scholar]
- 7.Duisit G, Saleun S, Douthe S, Barsoum J, Chadeuf G, Moullier P. Baculovirus vector requires electrostatic interactions including heparin sulfate for efficient gene transfer in mammalian cells. J Gene Med. 1999;1:93–102. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<93::AID-JGM19>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 8.Folger K R, Wong E A, Wahl G, Capecchi M R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982;2:1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 10.Groner A, R. G R, Burand J P. Interaction of Autographa californica nuclear polyhedrosis virus with two nonpermissive cell lines. Intervirology. 1984;21:203–209. doi: 10.1159/000149522. [DOI] [PubMed] [Google Scholar]
- 11.Heng H H Q, Squire J, Tsui L-C. High resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc Natl Acad Sci USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heng H H Q, Tsui L-C. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci USA. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones I, Morikawa Y. Baculovirus vectors for expression in insect cells. Curr Opin Biotechnol. 1996;7:512–516. doi: 10.1016/s0958-1669(96)80054-1. [DOI] [PubMed] [Google Scholar]
- 15.Kost T A, Condreay J P. Recombinant baculoviruses as expression vectors for insect and mammalian cells. Curr Opin Biotechnol. 1999;10:428–433. doi: 10.1016/s0958-1669(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 16.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhlmann I, Doerfler W. Loss of viral genomes from hamster tumor cells and nonrandom alterations in patterns of methylation of integrated adenovirus type 12 DNA. J Virol. 1983;47:631–636. doi: 10.1128/jvi.47.3.631-636.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis N, Evelegh C, Graham F L. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology. 1997;233:423–429. doi: 10.1006/viro.1997.8597. [DOI] [PubMed] [Google Scholar]
- 19.Luckow V A. Baculovirus systems for the expression of human gene products. Curr Opin Biotechnol. 1993;4:564–572. doi: 10.1016/0958-1669(93)90078-b. [DOI] [PubMed] [Google Scholar]
- 20.Luckow V A, Lee S C, Barry G F, Olins P O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrihew R V, Marburger K, Pennington S L, Roth D B, Wilson J H. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol Cell Biol. 1996;16:10–18. doi: 10.1128/mcb.16.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meuth M. Illegitimate recombination in mammalian cells. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 833–860. [Google Scholar]
- 23.Nakai H, Iwaki Y, Kay M A, Couto L B. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J Virol. 1999;73:5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman H, Medhora M M, Medhora D, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press, Inc.; 1990. pp. 219–236. [Google Scholar]
- 25.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Oxford University Press, Inc.; 1994. [Google Scholar]
- 26.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Possee R D. Baculoviruses as expression vectors. Curr Opin Biotechnol. 1997;8:569–572. doi: 10.1016/s0958-1669(97)80030-4. [DOI] [PubMed] [Google Scholar]
- 28.Roth D B, Wilson J H. Illegitimate recombination in mammalian cells. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 621–654. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sandig V, Hofmann C, Steinert S, Jennings G, Schlag P, Strauss M. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum Gene Ther. 1996;7:1937–1945. doi: 10.1089/hum.1996.7.16-1937. [DOI] [PubMed] [Google Scholar]
- 31.Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I, Miyamura T, Matsuura Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J Gen Virol. 1997;78:2657–2664. doi: 10.1099/0022-1317-78-10-2657. [DOI] [PubMed] [Google Scholar]
- 32.Takada S, Gotoh Y, Hayashi S, Yoshida M, Koike K. Structural rearrangement of integrated hepatitis B virus DNA as well as cellular flanking DNA is present in chronically infected hepatic tissues. J Virol. 1990;64:822–828. doi: 10.1128/jvi.64.2.822-828.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tjia S T, zu Altenschildesche G M, Doerfler W. Autographa californica nuclear polyhedrosis virus (AcNPV) DNA does not persist in mass cultures of mammalian cells. Virology. 1983;125:107–117. doi: 10.1016/0042-6822(83)90067-3. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, Yee J-K, Wolff J A, Friedmann T. Factors affecting long-term stability of Moloney murine leukemia virus-based vectors. Virology. 1989;171:331–341. doi: 10.1016/0042-6822(89)90600-4. [DOI] [PubMed] [Google Scholar]
- 35.Yang C C, Xiao X, Zhu X, Ansardi C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]