Abstract
Protein-encoding nucleotide sequences of the N, P, M, F, H, and L genes were determined for a low-passage isolate of the Edmonston wild-type (wt) measles virus and five Edmonston-derived vaccine virus strains, including AIK-C, Moraten, Schwarz, Rubeovax, and Zagreb. Comparative analysis demonstrated a high degree of nucleotide sequence homology; vaccine viruses differed at most by 0.3% from the Edmonston wt strain. Deduced amino acid sequences predicted substitutions in all viral polypetides. Eight amino acid coding changes were common to all vaccine viruses; an additional two were conserved in all vaccine strains except Zagreb. Comparisons made between vaccine strains indicated that commercial vaccine lots of Moraten and Schwarz had identical coding regions and were closely related to Rubeovax, while AIK-C and Zagreb diverged from the Edmonston wt along slightly different paths. These comparisons also revealed amino acid coding substitutions in Moraten and Schwarz that were absent from the closely related reactogenic Rubeovax strain. All of the vaccine viruses contained amino acid coding changes in the core components of the virus-encoded transcription and replication apparatus. This observation, combined with identification of noncoding region nucleotide changes in potential cis-acting sequences of the vaccine strains (C. L. Parks, R. A. Lerch, P. Walpita, H.-P. Wang, M. S. Sidhu, and S. A. Udem, J. Virol. 75:921–933, 2001), suggest that modulation of transcription and replication plays an important role in attenuation.
Measles is a highly contagious disease that most commonly strikes children. The causative agent, measles virus (MV), is generally transmitted by aerosolized secretions deposited on upper-respiratory-tract mucosal surfaces. Exposure leads to local respiratory tract replication; infection of regional lymphoid tissues then occurs followed by viremia and systemic dissemination as revealed by the characteristic skin rash. Most children recover uneventfully from the illness, but serious complications can occur, including pneumonia and involvement of the central nervous system (17, 27, 28). Despite the highly contagious nature of the disease, MV can be controlled effectively by immunization with live attenuated vaccines. The effectiveness of MV vaccines is well illustrated by the epidemiology of the disease in the United States. Prior to 1963, before use of the earliest vaccines, there were over 500,000 reported cases per year. Twenty years later, MV incidence was less than 2,000 cases per year (11, 28). The availability of these effective vaccines has not eliminated the threat from MV, and measles still causes significant levels of morbidity and mortality in developing countries largely because of inadequate and unsustained vaccination efforts (17).
Several effective MV vaccines were derived from a single clinical viral isolate called the Edmonston strain (28, 66). Enders et al. (20) developed the first MV vaccine by the classical approach (1) of propagating the pathogen in heterologous cells and tissues. Specifically, MV was serially propagated in semi-permissive chicken embryos and chick fibroblast cells. Variations of the Enders approach have led to the development of a number of independently derived but effective Edmonston-based vaccines (28, 66).
MV is a member of the genus Morbillivirus in the Paramyxoviridae family and, like other members of this family, it is an enveloped RNA virus that contains a single-strand, negative-sense, nonsegmented genome (28, 47). The 16-kb MV genome encodes eight known proteins from six nonoverlapping cistrons arranged 3′-N-P-M-F-H-L-5′. The major structural polypeptide is encoded by the N (nucleocapsid) gene. The N protein is essential for packaging the genome into a ribonucleoprotein complex that serves as template for transcription, replication, and packaging into progeny virions. The P cistron specifies three polypeptides: P, C, and V. The P (phosphoprotein) polypeptide is a subunit of the viral RNA polymerase. P protein also acts as a chaperone that interacts with and regulates the cellular localization of N protein and probably assists in nucleocapsid assembly (28, 33, 70). The C and V polypeptides are nonstructural proteins that are translated from P mRNAs through the use of alternative reading frames; C protein is synthesized from a downstream translation start signal, whereas V protein is translated from an edited mRNA that contains an extra G residue (28, 33, 70). The M gene encodes the matrix protein that lines the inner surface of the viral envelope and participates in virion maturation (28, 83). The F (fusion) and H (hemagglutinin) genes encode envelope glycoproteins that mediate cell surface recognition, membrane fusion, and virus entry (28, 83). Finally, the L (large) gene encodes the multifunctional catalytic subunit of the RNA-dependent RNA polymerase (28, 33, 70).
How changes in individual MV proteins may influence vaccine virus attenuation is not well understood. Partial sequence data for MV vaccine virus genomes clearly indicates that multiple mutations have accumulated in more than one protein coding region, but these analyses have so far failed to point to an underlying mechanism of MV attenuation (28, 66). To facilitate further analysis of the molecular basis of MV attenuation, we have determined the nucleotide sequence of all protein coding regions from an early-passage laboratory isolate of the original Edmonston virus (see Fig. 1, Edmonston wild type and reference (66) and five Edmonston vaccine strains. Comparison of deduced amino acid sequences has revealed amino acid coding substitutions common to all of the vaccines, as well as changes found only in subsets of the vaccine viruses. The results of these comparisons identified a number of mutations that appear to be strong candidates for attenuation determinants. In addition, we also suggest a model that correlates modulation of gene expression with the attenuated phenotype.
FIG. 1.
Edmonston vaccine lineage. Passage history of the Edmonston vaccines as described by Rota et al. (66) and adapted with permission from Elsevier Science. The protein coding region nucleotide sequence was determined for each virus highlighted in the gray boxes.
MATERIALS AND METHODS
Cells and virus.
Vero cells were maintained in Dulbecco modified Eagle medium (Life Technologies) supplemented with 5% fetal bovine serum. Stocks of MV were prepared by infection of Vero cell monolayers at a multiplicity of infection of approximately 0.1 PFU per cell. Infected cells were harvested by scraping the monolayer when the cytopathic effect was detectable in 70 to 80% of the cell monolayer. Harvested cells were collected by centrifugation and resuspended in serum-free OPTIMEM (Life Technologies) and lysed by one freeze-thaw cycle. The Edmonston wt isolate (21, 28) was kindly provided by Judy Beeler (Center for Biologics Evaluation and Research). Edmonston B (Rubeovax) (20, 28, 45), AIK-C (52), Schwarz (28, 45, 69), and Zagreb (28, 38) were generously provided by William Bellini and Paul Rota (Centers for Disease Control and Prevention) (66). Moraten (Attenuvax, Merck & Co.) (28, 31) and a second sample of Schwarz (Rimevax, SmithKline Beecham) (28, 45, 69) were obtained from commercially available vaccine preparations. The Schwarz virus strain from each source was analyzed independently.
Viral genome sequencing.
Sequencing was performed on DNA fragments generated by reverse transcription and PCR amplification (RT-PCR). Total RNA was extracted from infected Vero cells by the guanidinium-phenol extraction procedure (12) using Trizol reagent (Gibco-BRL). RT-PCR required first RT of approximately 1 μg of total infected-cell RNA with avian myeloblastosis virus (AMV) reverse transcriptase (Pharmacia) and random hexamer primers (Pharmacia), followed by Taq DNA polymerase-mediated PCR amplification (Amplitaq; Perkin-Elmer) with sequence-specific primers. Some single-tube RT-PCR reactions were performed by RT in the presence of gene-specific primers and AMV reverse transcriptase, followed by amplification with the high-fidelity enzyme mixture in the Titan RT-PCR kit (Roche Molecular Biology). Terminal fragments were either amplified by RT-PCR performed across the junction formed after circularization of the RNA genome with RNA ligase (71) or amplified by using the RACE (rapid amplification of cDNA ends) procedure (24). Amplified DNA fragments were purified for sequencing by electrophoresis in low-melting-temperature agarose (FMC). DNA was recovered from gel slices with the Wizard DNA Clean-Up System (Promega) or by digestion of gel slices with β-agarase (New England Biolabs). Cycle sequencing (29, 44) was executed with dye-labeled terminators and Taq DNA polymerase (Applied Biosystems), followed by analysis on an ABI Prism 377 automated sequence apparatus (Applied Biosystems). Primers for PCR amplification and sequencing of both cDNA strands were designed based on published MV sequences (GenBank accession numbers K01711 and S58435). Sequence data were analyzed using the MacVector (Oxford Molecular Group) and Lasergene (DNAstar, Inc.) software packages. A rooted phylogram was prepared from a CLUSTAL W alignment by the neighbor-joining method and plotted by using NJplot (61, 79).
RESULTS AND DISCUSSION
Nucleotide sequence analysis.
MV coding region sequences were analyzed to reveal genetic modifications in vaccine strains and thereby identify candidate vaccine-specific coding changes for future studies of MV attenuation. The virus strains sequenced are shown on the Edmonston vaccine lineage in Fig. 1 (28, 66). Figure 1 also includes the passage history as described by Rota et al. (66). The virus, referred to as Edmonston wt (Fig. 1), was the lowest-passage stock available of the original Edmonston clinical isolate. It had been passaged 13 times (66) prior to our analysis. At several points in the passage history of this isolate, the virus has been shown to retain pathogenicity (2, 3). The virus samples obtained for sequence analysis were passaged minimally (one to three times) in Vero cells to generate virus stocks, and these stocks were then used to infect cells for the isolation of infected-cell RNA. Purified RNA was used to generate RT-PCR products from all protein coding regions. These PCR fragments were sequenced on both strands (see Materials and Methods) to provide a “consensus” sequence representing the population of viruses replicating within the infected cells. Direct sequencing of these PCR products helped alleviate concerns associated with sequencing cloned RT-PCR products that could represent a selected subpopulation of viral genomes or include nucleotide substitutions introduced by PCR amplification. Minor virus populations accumulated during limited Vero cell passage should not notably affect consensus sequence determinations since the RT-PCR products should reflect the majority sequence in the viral RNA pool.
The compiled sequence data, summarized in Fig. 2 and 3, demonstrated that the coding region nucleotide sequence of the vaccine viruses differed from Edmonston wt by at most 0.3% (Fig. 2A). Comparison of predicted amino acid sequences revealed substitutions in the five vaccine strains that differentiated these viruses from the low-passage Edmonston wt isolate (Fig. 2B). Eight amino acid coding substitutions were shared by all of the Edmonston vaccine strains, and two substitutions were found in all vaccine strains except Zagreb (Fig. 3). Less-well-conserved amino acid coding substitutions were also found that affected one to three of the vaccine strains. Generation of a phylogram (Fig. 2C) indicated that the Moraten and Schwarz vaccine viruses contained identical coding sequences and were closely related to Rubeovax. Zagreb and AIK-C were obviously distinguishable from this group of vaccine viruses.
FIG. 2.
Comparison of MV protein coding region sequences. (A) The total number of vaccine virus protein coding region nucleotide substitutions is shown, and values are categorized further into coding and silent substitutions. GenBank accession numbers are provided. (B) Number of predicted amino acid changes in vaccine virus protein coding regions. The size of each protein is given below the gene designation. (C) The phylogram was generated using Edmonston wt as the out group. The sequences were aligned using CLUSTAL W (79), and the phylogram was generated by the neighbor-joining method using NJplot (61). The scale represents the number of substitutions per nucleotide.
FIG. 3.
Comparison of Edmonston wt and vaccine virus genomes. Nucleotide changes are shown for each coding region. Whether the nucleotide substitution results in an amino acid change or is silent is illustrated in the grid. Amino acid changes from wt are presented using one-letter amino acid symbols. Yellow shading in the grid highlights an amino acid substitution. Red shading of the amino acid position indicates a residue that is substituted in four or five of the vaccine strains. Blue shading in the grid without an amino acid symbol denotes strains that contain a silent base change; a line through a blue box indicates that the change is not silent in an overlapping reading frame. The F protein amino acid numbers are given relative to the predominant AUG codon at genomic nucleotide position 5458 (9). Abbreviations in the green header: NUC POS, MV genomic nucleotide position; BASE SUB, nucleotide substitution; Wt-Vac, wt and vaccine virus nucleotides; AA POS, amino acid position; W, Edmonston wt; A, AIK-C; M, Moraten; R, Rubeovax; S, Schwarz; and Z, Zagreb.
The most highly conserved vaccine virus amino acid coding changes shown in Fig. 3 (shaded in red) represent a “vaccine strain signature” that should prove useful for comparison with viruses isolated during MV outbreaks or isolated from suspected cases of vaccine-related illness. Although these coding changes are characteristic of the vaccine viruses, some of them may not be entirely unique to Edmonston vaccine strains. Several have been detected in circulating wt virus strains and in subacute sclerosing panencephalitis brain tissue RNA (amino acid positions 73 in C, 61 in M, 46 and 481 in H, and 1717 in L [data not shown and references 4, 16, 43, and 67]). It is possible that some degree of cell culture adaptation during viral propagation contributes to this observation.
The presence of amino acid substitutions shared by nearly all five vaccine strains also implies a strong correlation between at least some of these genome changes and the attenuated phenotype. These conserved nucleotide and amino acid changes represent important targets for future studies aimed at understanding the molecular basis of attenuation. Additionally, it will likely be important to consider some of the less-well-conserved amino acid substitutions in these studies. These changes could in fact play important roles in determining the attenuation level of individual strains.
How these genome modifications produce an attenuated phenotype is unknown, but their origin must reflect selection that favors growth in the cells of animals other than the natural host (Fig. 1). An indication that the particular tissue culture passage scheme determines the composition of genome modifications can be seen in the sequence data when it is examined in light of the details of the MV lineage (Fig. 1) (66). The phylogram (Fig. 2C) showed that Moraten, Schwarz, and Rubeovax were the most closely related vaccine viruses, while AIK-C and Zagreb have distinguishing genetic characteristics. In the lineage (Fig. 1) we can see that all of the vaccines were derived from the Edmonston-Enders strain; thereafter, however, their passage histories differ. The Moraten-Schwarz-Rubeovax group was propagated under very similar conditions, most notably the exclusive use of chicken cells. In contrast, AIK-C and Zagreb were both propagated in nonavian cell types that may account for their divergence from the Moraten-Schwarz-Rubeovax group. These distinctions in the lineage correlate well with the phylogram displayed in Fig. 2C.
It was unexpected to find that Moraten and Schwarz contained identical coding region nucleotide sequences given the fact that they were passaged independently. It was a further surprise to find that the two viruses also contained identical noncoding sequences (59). It is possible that quasispecies subpopulation differences in the vaccines exist and were below the detection levels of consensus sequencing; but even if this was the case, the results still indicated that the two viruses were remarkably similar. Consensus sequence analysis of a second Schwarz vaccine specimen was performed, confirming the initial observation. The basis for the perplexing sequence identity of these two independently derived MV vaccine strains is unknown. Possibly, the convergence of sequence in the Moraten and Schwarz viruses reflects a highly selective and delimited spectrum of nucleotide changes imposed by extensive passage in chicken embryo fibroblasts at reduced temperatures (Fig. 1).
The process of adapting MV to semipermissive cell types obviously generates strong selective pressure that favors the evolution of viral polypeptides that function more effectively in the new host cell environment. Although the following is speculative, it seems likely that early in the adaptation process the interaction between some viral proteins and proteins in the semipermissive cell are inefficient. This gives rise to slow viral growth and selective pressure that favors mutations leading to alterations in viral polypeptides that enhance functional interaction with host cell proteins. A prime target for some of the earliest mutations would be the genes encoding the transcription and replication apparatus since these proteins must adjust the initial stages of viral replication (mRNA synthesis and positive-strand synthesis) to the semipermissive cell environment. A potential disadvantage associated with these earliest mutations is that they may create viral proteins that enhance interaction with the host cell at the expense of other segments of the viral replication cycle. For example, a mutation that improves P protein interaction with a semipermissive host cell factor could have a negative effect on another function such as the P-N protein-protein interaction or recognition of template sequences by the P-L polymerase complex. This will generate additional selective pressure for “second-site repressor” mutations that help compensate for the effect of the primary mutations. We could imagine that these second-site repressor mutations would evolve in viral protein coding regions as well as cis-acting sequences as the virus attempts to fine-tune the replicative capacity to the semipermissive cell environment during the course of a prolonged passage scheme. In theory, these genetic adjustments give rise to the best-fit virus for growth in the semi permissive cells but ultimately render the virus less effective at interaction with the cellular proteins of the natural host. Thus, when this vaccine virus infects permissive human host cells, its accrued mutations lead to less-effective virus-host cell protein interaction and thereby reduce virus replication efficiency. Adequate time for the vaccine virus to revert some of the genetic change after vaccination is not available before an immunocompetent host clears the viral infection. We present below the data analysis for each gene region and describe how some of the genetic changes may relate to virus attenuation.
N gene.
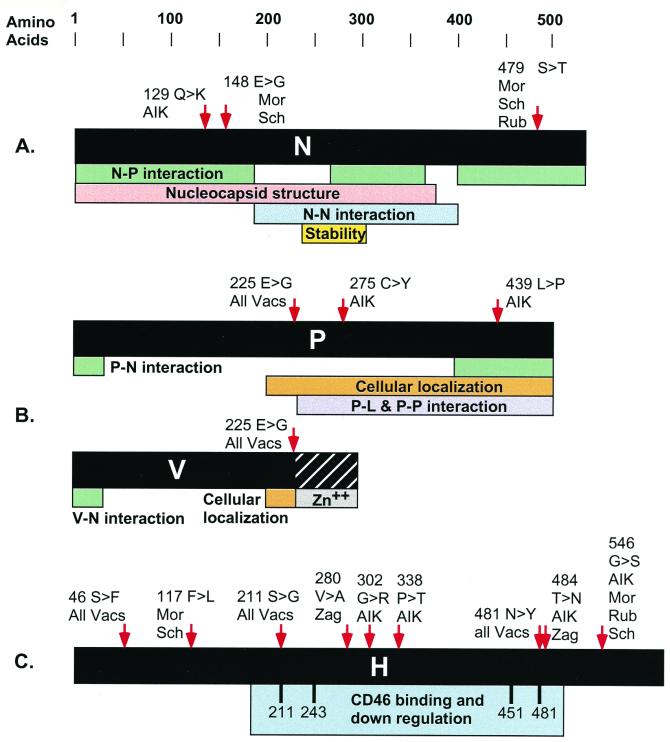
Analysis of the N gene identified three predicted amino acid substitutions (Fig. 2B and 3); none of these changes were conserved by all of the vaccine strains, and no single vaccine strain contained all three substitutions. Although the N protein substitutions were not conserved by all vaccine strains, they still represent attractive candidates for attenuating modifications. N protein plays a key role in the virus life cycle during genome packaging, genome replication, and gene expression (28). The role of the N protein in these diverse activities makes it seem probable that amino acid substitutions would have some impact on the virus. The nonconservative amino acid substitution found in the amino terminus of the Moraten and Schwarz N protein (position 148, glutamic acid to glycine) is a obvious candidate to alter N protein function. It occurred in a region of N protein that likely plays a role in several functions, including RNA binding (47), the formation of the nucleocapsid structure, and interaction with P protein (Fig. 4A). Computer predictions also indicate that this amino acid substitution would disturb an alpha-helical region of the protein. Interactions with P protein may also be influenced by the amino acid substitutions in N protein at position 479 in Moraten, Rubeovax, and Schwarz, as well as the position 129 substitution in AIK-C (Fig. 4A) (5, 50). No coding region changes were detected in the Zagreb N gene. This indicates that successful attenuation can occur without substitutions in N, but that the accumulated strain-specific changes in N may be important contributors to the degree of attenuation in viruses such as Moraten, Schwarz, and AIK-C.
FIG. 4.
Protein domains and vaccine virus amino acid substitutions. Domain maps for N, P, V, and H proteins are shown along with arrowheads marking the position of predicted amino acid substitutions. Vaccine virus names are abbreviated as follows: AIK-C (AIK), Moraten (Mor), Rubeovax (Rub), Schwarz (Sch), and Zagreb (Zag). (A) The domain boundaries illustrated below the linear map of N protein are derived from Bankamp et al. (5) and Liston et al. (50). These include domains involved in N-P complex formation and nucleocapsid formation and a region that affects protein stability. (B) The linear maps of P protein and V protein drawn in black illustrate sequences shared by these proteins. The unique sequence in the carboxy terminus of V is designated with a cross-hatched box. Domain boundaries include regions that promote interaction with N protein, a region that affects the cellular localization of N-P complexes, a region of P protein that promotes interaction with L protein and the formation of P-P multimers, and the carboxy-terminal domain of V that binds zinc (30, 34, 37, 51, 80). (C) Illustrated below the map of H protein are amino acids that have been implicated in mediating binding and downregulation of CD46 (7, 35, 49).
P cistron.
Amino acid substitutions detected in the P gene included a change shared by all of the vaccines at position 225 (Fig. 2B and 3). The amino-terminal portion of the P gene open reading frame (ORF) also encodes two-thirds of the V polypeptide; therefore, all of the vaccine virus V proteins contain the same amino acid substitution. The C protein coding region embedded within the P gene contained three substitutions (Fig. 2B and 3) which were silent with respect to the overlapping P and V ORFs.
P protein is a multifunctional polypeptide that is a component of the polymerase complex. It also plays a role in viral RNA encapsidation and regulates the cellular localization of N protein (28, 33, 37, 47, 70). Given its pleiotropic activities, substitutions in P protein are likely to significantly impact viral replication efficiency. The position 275 and 439 substitutions in AIK-C P (Fig. 3) protein may be significant in the context of this strain. They map within P domains (Fig. 4B) that mediate interaction with N protein as well as promote its own multimerization (30, 37, 80). The position 225 substitution (Fig. 3), found in all of the vaccines, replaced an Edmonston wt glutamic acid with a glycine. Substitution of this glutamic acid with a nonpolar residue occurred in a region of P protein that may be involved in a chaperone function of P protein that regulates the cellular localization of N protein (37, 80). The fact that the N-P protein-protein interaction is essential for several functional activities suggests that any mutations that alter P protein and influence the N-P interaction could affect gene expression, replication and ultimately the degree of attenuation.
The 225 substitution also affects V protein (Fig. 3 and 4B). As mentioned above, this substitution occurred within one of the P protein domains (Fig. 4B) that plays some role in the interaction with N protein (30, 37). This region appears to play a similar role in V protein (80). Thus, it is possible that the position 225 substitution could affect the interaction between the N and P proteins as well as between the N and V proteins. Perturbing these interactions could lead to changes in the relative ratios of N-P and N-V complexes and possibly alter the effective availability of N protein for encapsidation.
V protein is dispensable for growth in cultured cells (68), but several studies indicate that V protein may be a virulence factor. Sendai virus defective for V protein synthesis is less pathogenic in mice (18, 36, 40, 41). Furthermore, a recombinant lab strain of Edmonston B that is defective for V protein expression replicates less efficiently in some experimental model systems (56, 60, 80, 81). The connection between pathogenicity and V proteins suggests that mutations in the V ORF should be considered potential attenuation determinants.
Finally, the variability in the P cistron also affected the C protein ORF (Fig. 3). Like V protein, alterations that affect C protein are intriguing because C protein is dispensable for MV growth in Vero cell culture (63) but may be an important factor influencing pathogenicity. In the SCID mouse model system, transplanted human thymic tissue supports less viral replication if infection is performed with a recombinant Edmonston B strain that cannot express C protein (81). In addition, the C protein defect appears to hinder replication in cultured human peripheral blood mononuclear cells (23). How C protein regulates growth in these model systems is not understood, but it can be inferred from studies with Sendai virus (8, 15, 32) that MV C protein may modulate viral RNA synthesis. Also, in Sendai virus, C protein seems to counter innate immune responses to infection induced by interferon (26), raising the possibility that MV C protein may perform a similar function. Care must be taken when drawing these comparisons between MV and Sendai virus C proteins given that Sendai virus encodes multiple C protein species (48), while MV encodes only one known C protein. Yet it is interesting to note that Sendai virus C protein is dispensable for growth in cell culture like MV C protein and that Sendai virus defective for C protein expression is less virulent in mice (25, 46, 57).
M gene.
Ten coding changes were identified in the vaccine virus M genes (Fig. 2B and 3). Only two of these were common to all vaccine strains (positions 61 and 89); these were two nonconservative changes that replaced a wt nonpolar glycine for an aspartic acid at position 61 and a wt glutamic acid for lysine at position 89. The substitution at position 61 has been detected also in some circulating wt strains (data not shown and references 16 and 67).
Changes in M protein could influence the level of attenuation by perturbing M protein function during virion maturation (83) or transcriptional repression (77). In addition, the accumulation of M gene mutations is one characteristic of latent genomes indicating that changes in the M gene can contribute to an atypical virus life cycle (10).
F and H glycoproteins.
Since the glycoproteins are important determinants of MV host range and cell tropism (39, 76) it was expected that some mutations would accumulate in these genes after serial passage in cells of nonprimate origin. In addition, the role played by the viral glycoproteins in cell entry, cell fusion, and virus maturation (28, 83) raises the possibility that some of the changes in F and H may influence the cell-to-cell spread of the virus and contribute to the attenuated phenotype. The F protein coding region contained a number of codon changes, but none of these were conserved in all vaccine strains (Fig. 3). The specificity of nine codons varied in the H ORF. Three H amino acid substitutions were conserved among all of the vaccine viruses, and a fourth differed in all vaccines strains except Zagreb (Fig. 2B and 3). None of the amino acid substitutions should affect the glycosylation pattern of H (28).
The H gene actually accumulated the highest number of substitutions that were shared by all vaccine viruses (Fig. 3). This may reflect the fact that H protein is the receptor component of the virus envelope and significant changes were required to adapt H protein for effective infection of the heterologous cell types used during vaccine passage (22). These changes in H protein may also contribute to attenuation if they render the virus less efficient at infecting human cells. In fact, several of the amino acid residues that were changed in the vaccine viruses have received considerable attention recently because studies indicate that they play an important role in binding to one of the cellular receptors for MV (Fig. 4C). The amino acids at positions 211 and 481 are both important for interaction with one of the cellular receptors (CD46) for MV (7, 35, 49). Virus isolates with a wt asparagine residue at position 481 do not readily infect monkey kidney cell lines but efficiently infect a transformed marmoset lymphoid cell line (derivatives of B95-8 cells) (42, 54). In contrast, tyrosine at position 481 enhances the ability to infect monkey kidney cell lines. Interaction between H protein and CD46 also promotes clearance of CD46 from the infected-cell surface. This phenomenon also depends on amino acids at positions 211 and 481. H proteins containing the vaccine virus amino acids at these positions displayed an enhanced ability to remove CD46 from the cell surface (7, 49). This has led to speculation that more potent clearance activity of vaccine H protein may contribute to attenuation. Recombinant MV used in primate studies probably will be necessary to definitively determine the role of H protein in attenuation.
L gene.
Nine amino acid coding changes were detected in the L gene. The only one shared by all vaccines was at position 1717, where a wt glutamic acid was substituted with an alanine. The L gene had one of the lowest frequencies of amino acid substitution (Fig. 3), presumably reflecting the intolerance of functional domains of this enzyme to amino acid substitution.
To better assess the changes in L protein, they were displayed on a domain map (Fig. 5). Domains of homology exist among various RNA-dependent RNA polymerases, including the paramyxovirus L proteins (6, 53, 62, 72). It has been proposed that these homologous regions (RNA-dependent RNA polymerase domains I to VI) (6, 53, 62, 72) are functional domains. Further comparison of morbillivirus L proteins has led to a somewhat different model of three large domains (morbillivirus domains 1 to 3) and two hinge regions (53). Curiously, only one vaccine virus substitution was located within the RNA-dependent RNA polymerase domains I to VI. This relatively conservative isoleucine-to-threonine substitution at position 331 was found in Moraten and Schwarz. This region of L protein is within a domain that interacts with P protein (34), suggesting that this amino acid substitution may affect L-P protein-protein interaction. This suggestion needs to be examined experimentally since a recent report indicates that a valine substitution found at this site in some circulating virus strains does not seem to affect the interaction between the L and P proteins (4). The remaining amino acid coding changes affecting L protein occurred outside the boundaries of RNA-dependent RNA polymerase domains I to VI. This may indicate that domains I to VI indeed contain regions essential for enzymatic function and that these regions are less tolerant of amino acid substitutions. Furthermore, it is attractive to speculate that the vaccine virus L protein amino acid substitutions occurred in regions of the protein that may modulate enzymatic activity rather than directly affect a region that contains an active site.
FIG. 5.
Position of amino acid substitutions relative to the domain map of the L protein. The domain organization deduced from analysis of RNA-dependent RNA polymerase sequences is labeled with the roman numerals I to VI (6, 53, 62, 72), and the domain and hinge structures predicted for morbillivirus L proteins are labeled 1 to 3 (53). The location of amino acid substitutions is marked with an arrowhead, and circles identify changes from the Edmonston wt sequence.
Silent mutations.
Silent nucleotide substitutions were identified at 17 positions (Fig. 3). Three of these base substitutions were common to all of the vaccine strains (one each in the M, F, and L coding regions). An additional silent substitution in P and V is conserved in all vaccines, but it is not truly silent; three of the four silent base changes in the P and V coding regions result in amino acid substitutions in the overlapping C ORF. Identification of several silent mutations that were conserved in multiple vaccine strains could support the idea that some of these base changes provide a cis-acting advantage such as increased mRNA stability or more favorable secondary structure for translation or may reflect selection due to codon bias. More likely it simply reveals that some silent base substitutions were incorporated early during vaccine virus passage. These are tolerated after incorporation and can be maintained in the genome if they do not affect the fitness of the virus.
It was somewhat surprising to find that so few silent changes have accumulated in light of the fact that the negative-strand RNA virus polymerases have a relatively high error rate (19) and the vaccine viruses had been passaged extensively (Fig. 1). Although this is difficult to explain, it is not unique to the Edmonston vaccines. Biologically derived respiratory syncytial virus vaccine candidiates also have been noted to accumulate relatively few nucleotide substitutions after extensive passage in culture (13).
Comparison of AIK-C sequences.
The complete genomic sequence of AIK-C was carefully analyzed previously in 1993 by Mori et al. (55). Comparison of the coding and noncoding (59) sequences generated by this laboratory (GenBank accession number AF266286) to the earlier AIK-C sequence (GenBank accession number S58435) revealed 21 nucleotide differences. Ten of these predict amino acid substitutions, six were silent, and five were located in noncoding regions. A likely explanation for many of these differences lies in the types of tools and methods used to analyze the virus genomes. Since 1993, sequencing technology, including the enzymatic steps and gel systems, has advanced dramatically and now allows for increased resolution of sequences containing secondary structure. Inspection of regions containing the nucleotide differences indicated that about 10 of the 21 discrepancies lie in areas of locally high G+C content or contain sequences that may form intramolecular duplexes. These regions would likely complicate sequence determination and explain some of the differences between the presented sequence and that of Mori et al. (55). A second source of variation may reflect the fact that Mori et al. analyzed cloned cDNA fragments, whereas we employed direct sequencing of RT-PCR fragments. Finally, a third difference lay in the passage history of the sequenced AIK-C viruses. Here, the virus was passaged a limited number of times in Vero cells, whereas Mori et al. used chicken embryo cells. Taken together, these technical variations probably account for the majority of nucleotide differences in the two sequence determinations.
Only two of the nucleotide differences between our sequence and that of Mori et al. (55) affected amino acids that distinguish between Edmonston wt and the currently proposed AIK-C sequence. Amino acid 362 in F protein was a serine in Edmonston wt and a tyrosine in AIK-C, while the sequence of Mori et al. predicted no amino acid change. Similarly, in F protein, we found that all of the viruses except AIK-C encoded an H residue at position 419, while AIK-C encoded an N. The sequence of Mori et al. again predicted no change from the wt.
Edmonston vaccine attenuation.
Comparison of a low-passage isolate of the Edmonston wt strain with five vaccine virus derivatives revealed distinguishing genetic changes in all coding (Fig. 3) and noncoding regions (59). Finding genetic change in multiple genes and noncoding regions may indicate that viral replication in semipermissive cells requires modification of several components of the virus replication cycle, including cell entry, gene expression, genome replication, and virus maturation. Although a constellation of genomic changes may be essential for adaptation to semipermissive growth conditions, it remains to be determined whether all of these changes are indeed necessary for effective attenuation. Clues from other negative-strand RNA vaccine viruses and vaccine virus candidates suggest that only a subset of these genome changes may be essential for attenuation and that modification of the gene expression and replication apparatus is a particularly critical target. For example, mutations in the L polymerase gene of RSV have been found to effectively modulate the degree to which live virus vaccine strains are attenuated (82). Similarly, cold-adapted parainfluenza virus type 3 vaccine candidate strains contain polymerase gene mutations and base substitutions in the 3′ transcription promoter region that are attenuating (73, 74). Live influenza virus vaccines generated by replacing the genes for the hemagglutinin and neuraminidase glycoproteins retain an attenuated phenotype specified by mutations within the components of the replication apparatus (75). Taken together, these studies imply that polymerase mutations, as well as cis-acting signal mutations, are important contributors to the attenuated phenotype.
Consistent with this notion, all Edmonston vaccine viruses were found to contain mutations in the L and P genes (Fig. 3) as well as within the leader region (59). That polymerase gene mutations in combination with cis-acting signal mutations significantly contribute to attenuation by altering gene expression is a relatively simple and attractive hypothesis. Modulating the abundance of viral gene expression to achieve suitable immunogenicity while limiting virus replication, dissemination, and injury is an essential element of an optimally attenuated virus.
Modulation of MV gene expression as a mechanism of attenuation may involve more than the core polymerase complex and cis-acting signals in the leader. This concept also was proposed by Takeda et al. (78) after they found that the coding differences between a pathogenic MV strain and a Vero cell-adapted derivative resided in the core polymerase genes (L and P), as well as in the V and C coding regions. As described above, the vaccine viruses of the Edmonston lineage also have substitutions outside of the core polymerase genes that appear to have the potential to affect gene expression. These included substitutions within the N gene, the genes encoding accessory proteins (V, C, and M), and additionally in noncoding regions that may have important cis-acting functions (59).
The possibility that regulation of gene expression and replication plays a role in attenuation gains support from our analysis and can be illustrated by sequence comparison between the underattenuated Rubeovax strain and the identical genomes of the desirably attenuated Moraten and Schwarz vaccine viruses (Fig. 6). Presumably, some of the genetic differences between these viruses were responsible for the underattenuated phenotype of Rubeovax. Comparison of these virus genomes revealed differences in the L protein, the N protein (Fig. 3 and 6), and several putative cis-acting sequences. The noncoding region changes (Fig. 6) (59) included nucleotide substitutions in sequences corresponding to the long untranslated region of the F mRNA (nucleotides 4608 and 5308), the M gene translation start codon context (nucleotide 3431), and the gene end signal of the F gene (7234). It is also worth noting that most of the changes that distinguish Rubeovax from Moraten and Schwarz were due to substitutions in Moraten and Schwarz that did not occur in the Rubeovax genome. Thus, it is possible that the slightly more wt genotype in parts of the gene expression apparatus of Rubeovax is responsible for its underattenuated phenotype.
FIG. 6.
Comparison between Moraten, Schwarz, and Rubeovax. Substitutions that distinguish between wt and this group of vaccine viruses are illustrated in the boxes representing each gene region. The amino acid positions are indicated above the columns of amino acids. Noncoding region changes (59) in the intergenic regions that distinguish Moraten and Schwarz from Rubeovax are shown along with the nucleotide position below the boxed gene designations. Black highlighting identifies substitutions that differentiate the identical genomes of Moraten and Schwarz from Rubeovax. Morat/Schw, identical genomes of Moraten and Schwarz.
An attractive feature of the hypothesis that links gene expression to attenuation is the pathway it suggests for rational negative-strand RNA virus vaccine design—that downregulating viral RNA synthesis in human cells will effectively decrease replication and contribute to attenuation. Current molecular technology (14, 58, 64, 65) will facilitate the development of recombinant measles viruses with targeted mutations in different elements of the gene expression apparatus that can be used to test this model. If a recombinant MV can be developed that displays altered gene expression, reduced replicative ability, and safe levels of attenuation, it may be possible to apply these findings to a wider range of paramyxoviruses.
ACKNOWLEDGMENTS
We thank William Bellini and Paul Rota (CDC) for providing vaccine strains, and Judy Beeler (CBER, FDA) for providing the Edmonston wt virus isolate. We also thank Martin Billeter and the anonymous journal reviewers for their thoughtful reviews and suggestions. We are grateful to Shuo Lin for assistance with sequence comparisons.
The early part of this research was supported by NIH grant AI35286 to S.A.U.
REFERENCES
- 1.Ada G L. Vaccines. In: Paul W E, editor. Fundamental immunology. 3rd ed. New York, N.Y: Raven Press Limited; 1993. pp. 1309–1352. [Google Scholar]
- 2.Albrecht P, Lorenz D, Klutch M J. Encephalitogenicity of measles virus in marmosets. Infect Immun. 1981;34:581–587. doi: 10.1128/iai.34.2.581-587.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht P, Lorenz D, Klutch M J, Vickers J H, Ennis F A. Fatal measles infection in marmoset pathogenesis and prophylaxis. Infect Immun. 1980;27:969–978. doi: 10.1128/iai.27.3.969-978.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankamp B, Bellini W J, Rota P A. Comparison of L proteins of vaccine and wild-type measles viruses. J Gen Virol. 1999;80:1617–1625. doi: 10.1099/0022-1317-80-7-1617. [DOI] [PubMed] [Google Scholar]
- 5.Bankamp B, Horikami S M, Thompson P D, Huber M, Billeter M, Moyer S A. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology. 1996;216:272–277. doi: 10.1006/viro.1996.0060. [DOI] [PubMed] [Google Scholar]
- 6.Barik S, Rud E W, Luk D, Banerjee A K, Kang C Y. Nucleotide sequence analysis of the L gene of vesicular stomatitis virus (New Jersey serotype): identification of conserved domains in L proteins of nonsegmented negative-strand RNA viruses. Virology. 1990;175:332–337. doi: 10.1016/0042-6822(90)90218-g. [DOI] [PubMed] [Google Scholar]
- 7.Bartz R, Brinckmann U, Dunster L M, Rima B, Ter Meulen V, Schneider-Schaulies J. Mapping amino acids of the measles virus hemagglutinin responsible for receptor (CD46) downregulation. Virology. 1996;224:334–337. doi: 10.1006/viro.1996.0538. [DOI] [PubMed] [Google Scholar]
- 8.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cathomen T, Buchholz C J, Spielhofer P, Cattaneo R. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology. 1995;214:628–632. doi: 10.1006/viro.1995.0075. [DOI] [PubMed] [Google Scholar]
- 10.Cattaneo R, Billeter M A. Mutations and A/I hypermutations in measles virus persistent infections. Curr Top Microbiol Immunol. 1992;176:63–74. doi: 10.1007/978-3-642-77011-1_5. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control. Measles—United States, 1990. Morbid Mortal Wkly Rep. 1991;40:369–372. [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Collins P L, Whitehead S S, Bukreyev A, Fearns R, Teng M N, Juhasz K, Chanock R M, Murphy B R. Rational design of live-attenauted recombinant vaccine virus for human respiratory syncytial virus by reverse genetics. Adv Virus Res. 1999;54:423–451. doi: 10.1016/s0065-3527(08)60374-7. [DOI] [PubMed] [Google Scholar]
- 14.Conzelmann K K. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genetics. 1998;32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 15.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA sysnthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 16.Curran M D, Rima B K. Nucleotide sequence of the gene encoding the matrix protein of a recent measles virus isolate. J Gen Virol. 1988;69:2407–2411. doi: 10.1099/0022-1317-69-9-2407. [DOI] [PubMed] [Google Scholar]
- 17.Cutts F T, Markowitz L E. Successes and failures in measles control. J Infect Dis. 1994;170:S32–S41. doi: 10.1093/infdis/170.supplement_1.s32. [DOI] [PubMed] [Google Scholar]
- 18.Delenda C, Taylor G, Hausmann S, Garcin D, Kolakofsky D. Sendai viruses with altered P, V and W protein expression. Virology. 1998;242:327–337. doi: 10.1006/viro.1998.9027. [DOI] [PubMed] [Google Scholar]
- 19.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 20.Enders J F, Katz S L, Milovanovic M L, Holloway A. Studies of an attenuated measles virus vaccine. N Engl J Med. 1960;263:153–159. doi: 10.1056/NEJM196007282630401. [DOI] [PubMed] [Google Scholar]
- 21.Enders J F, Peebles T C. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med. 1954;86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 22.Escoffier C, Gerller D. Infection of chicken embryonic fibroblasts by measles virus: adaptation at the virus entry level. J Virol. 1999;73:5220–5224. doi: 10.1128/jvi.73.6.5220-5224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escoffier C, Mainle S, Vincent S, Muller C P, Billeter M A, Gerlier D. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J Virol. 1999;73:1695–1698. doi: 10.1128/jvi.73.2.1695-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frohman M A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 25.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice but not virus growth in cell culture. Virology. 1997;238:424–431. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]
- 26.Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counter the interferon-mediated induction of the antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gershon A A. Measles virus. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. Vol. 2. New York, N.Y: Churchill Livingstone; 1995. pp. 1519–1526. [Google Scholar]
- 28.Griffin D E, Bellini W J. Measles virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1267–1312. [Google Scholar]
- 29.Griffin H G, Griffin A M. DNA sequencing: recent innovations and future trends. Appl Biochem Biotechnol. 1993;38:147–159. doi: 10.1007/BF02916418. [DOI] [PubMed] [Google Scholar]
- 30.Harty R N, Palese P. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J Gen Virol. 1995;76:2863–2867. doi: 10.1099/0022-1317-76-11-2863. [DOI] [PubMed] [Google Scholar]
- 31.Hilleman M R, Buynak E B, Welbel R E, Stokes J, Jr, Whitman J E, Leagus M B. Development and evaluation of the Moraten measles virus vaccine. J Am Med Assoc. 1968;206:587–590. [PubMed] [Google Scholar]
- 32.Horikami S M, Hector R E, Smallwood S, Moyer S A. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 33.Horikami S M, Moyer S A. Structure, transcription, and replication of measles virus. Curr Top Microbiol Immunol. 1995;191:35–50. doi: 10.1007/978-3-642-78621-1_3. [DOI] [PubMed] [Google Scholar]
- 34.Horikami S M, Smallwood S, Bankamp B, Moyer S A. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology. 1994;205:540–545. doi: 10.1006/viro.1994.1676. [DOI] [PubMed] [Google Scholar]
- 35.Hsu E C, Sarangi F, lorio C, Sidhu M S, Udem S A, Dillehay D L, Xu W, Rota P A, Bellini W J, Richardson C D. A single amino acid change in the hemagglutinin protein of measles virus determines its ability to bind CD46 and reveals another receptor on marmoset B cells. J Virol. 1998;72:2905–2916. doi: 10.1128/jvi.72.4.2905-2916.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C, Kiyotani K, Fujii Y, Fukuhara N, Kato A, Nagal Y, Yoshida T, Sakaguchi T. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J Virol. 2000;74:7834–7841. doi: 10.1128/jvi.74.17.7834-7841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber M, Cattaneo R, Spielhofer P, Orvell C, Norby E, Messerli M, Perriard J-C, Billeter M A. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology. 1991;185:299–308. doi: 10.1016/0042-6822(91)90777-9. [DOI] [PubMed] [Google Scholar]
- 38.Ikic D, Jubasic M, Beck M, Hrabar A, Cimbur-Schreiber T. Attenuation and characterization of Edmonston-Zagreb measles virus. Ann Immunol Hung. 1972;16:175–181. [PubMed] [Google Scholar]
- 39.Johnstone I C D, Ter Meulen V, Schneider-Schaulles J, Schneider-Schaulles S. A recombinant measles vaccine virus expressing wild-type glycoproteins: consequences for viral spread and cell tropism. J Virol. 1999;73:6903–6915. doi: 10.1128/jvi.73.8.6903-6915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagal Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagal Y. Importance of the cysteine-rich carboxy-terminal half of the V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobune F, Sakata H, Sugiura A. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J Virol. 1990;64:700–705. doi: 10.1128/jvi.64.2.700-705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komase K, Rima B K, Pardowitz I, Kunz C, Billeter M A, Ter Meullen V, Baczko K. A comparison of nucleotide sequences of measles virus L genes derived from wild-type viruses and SSPE brain tissues. Virology. 1995;208:795–799. doi: 10.1006/viro.1995.1214. [DOI] [PubMed] [Google Scholar]
- 44.Kretz K, Callen W, Hedden V. Cycle sequencing. Genome Res. 1994;3:S101–S112. doi: 10.1101/gr.3.5.s107. [DOI] [PubMed] [Google Scholar]
- 45.Krugman S, Giles J P, Jacobs A M, Friedman H. Studies with a further attenuated live measles-virus vaccine. Pediatrics. 1963;31:919–928. [PubMed] [Google Scholar]
- 46.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 47.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 48.Latorre P, Cadd T, Itoh M, Curran J, Kolakofsky D. The various sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J Virol. 1998;72:5984–5993. doi: 10.1128/jvi.72.7.5984-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lecouturier V, Fayolle J, Caballero M, Carabana J, Celma M L, Fernandez-Munoz R, Wild T F, Buckland R. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J Virol. 1996;70:4200–4204. doi: 10.1128/jvi.70.7.4200-4204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liston P, Batal R, DiFlumeri C, Briedis D J. Protein interaction domains of the measles virus nucleocapsid protein (NP) Arch Virol. 1997;142:305–321. doi: 10.1007/s007050050078. [DOI] [PubMed] [Google Scholar]
- 51.Liston P, Briedis D J. Measles virus V protein binds zinc. Virology. 1994;198:399–404. doi: 10.1006/viro.1994.1050. [DOI] [PubMed] [Google Scholar]
- 52.Makino S. Development and characteristics of live AIK-C measles virus vaccine: a brief report. Rev Infect Dis. 1983;5:504–505. doi: 10.1093/clinids/5.3.504. [DOI] [PubMed] [Google Scholar]
- 53.McIlhatton M A, Curran M D, Rima B K. Nucleotide sequence analysis of the large (L) genes of phocine distemper virus and canine distemper virus (corrected) J Gen Virol. 1997;78:571–576. doi: 10.1099/0022-1317-78-3-571. [DOI] [PubMed] [Google Scholar]
- 54.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mori T, Sasaki K, Hashimoto H, Makino S. Molecular cloning and complete nucleotide sequence of the genomic RNA of the AIK-C strain of attenuated measles virus. Virus Genes. 1993;7:67–81. doi: 10.1007/BF01702349. [DOI] [PubMed] [Google Scholar]
- 56.Mrkic B, Odermatt B, Klein M A, Billeter M A, Pavolic J, Cattaneo R. Lymphatic dissemination and comparative pathology of recombinant measles viruses in genetically modified mice. J Virol. 2000;74:1364–1372. doi: 10.1128/jvi.74.3.1364-1372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagai Y. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev Med Virol. 1999;9:83–99. doi: 10.1002/(sici)1099-1654(199904/06)9:2<83::aid-rmv244>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 58.Nagai Y, Kato A. Paramyxovirus reverse genetics is coming of age. Microbiol Immunol. 1999;43:613–624. doi: 10.1111/j.1348-0421.1999.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 59.Parks C L, Lerch R A, Walpita P, Wang H-P, Sidhu M S, Udem S A. Analysis of the noncoding regions of measles virus strains in the Edmonston vaccine lineage. J Virol. 2001;75:921–933. doi: 10.1128/JVI.75.2.921-933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patterson J B, Thomas D, Lewicki H, Billeter M A, Oldstone M B A. V and C proteins of measles virus function as virulence factors in vivo. Virology. 2000;267:80–89. doi: 10.1006/viro.1999.0118. [DOI] [PubMed] [Google Scholar]
- 61.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 62.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmonston B strain of measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 64.Radecke F, Billeter M A. Reverse genetics meets the nonsegmented negative-strand RNA viruses. Rev Med Virol. 1997;7:49–63. doi: 10.1002/(sici)1099-1654(199704)7:1<49::aid-rmv181>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 65.Roberts A, Rose J K. Recovery of negative-strand RNA viruses from plasmid DNAs: a positive approach revitalizes a negative field. Virology. 1998;247:1–6. doi: 10.1006/viro.1998.9250. [DOI] [PubMed] [Google Scholar]
- 66.Rota J S, Wang Z-D, Rota P A, Bellini W J. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 1994;31:317–330. doi: 10.1016/0168-1702(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 67.Rota P A, Bloom A E, Vanchiere J A, Bellini W J. Evolution of the nucleoprotein and matrix genes of wild-type strains of measles virus isolated from recent epidemics. Virology. 1994;198:724–730. doi: 10.1006/viro.1994.1086. [DOI] [PubMed] [Google Scholar]
- 68.Schneider H, Kaelin K, Billeter M A. Recombinant measles virus defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 69.Schwarz A J F. Preliminary tests of a highly attenuated measles vaccine. Am J Dis Child. 1962;103:216–219. doi: 10.1001/archpedi.1962.02080020398042. [DOI] [PubMed] [Google Scholar]
- 70.Sedlmeier R, Neubert W J. The replicative complex of paramyxoviruses: structure and function. Adv Virus Res. 1998;50:101–139. doi: 10.1016/s0065-3527(08)60807-6. [DOI] [PubMed] [Google Scholar]
- 71.Sidhu M S, Husar W, Cook S D, Dowling P C, Udem S A. Canine distemper terminal and intergenic non-protein coding nucleotide sequences: completion of the entire CDV genome sequence. Virology. 1993;193:66–72. doi: 10.1006/viro.1993.1103. [DOI] [PubMed] [Google Scholar]
- 72.Sidhu M S, Menonna J P, Cook S D, Dowling P C, Udem S A. Canine distemper virus L gene: sequence and comparison with related viruses. Virology. 1993;193:50–65. doi: 10.1006/viro.1993.1102. [DOI] [PubMed] [Google Scholar]
- 73.Skiadopoulos M H, Durbin A P, Tatem J M, Wu S-L, Paschalis M, Tao T, Collins P L, Murphy B R. Three amino acid substitutions in the L protein of the human parainfluenza virus type 3 cp45 live attenuated vaccine candidate contribute to its temperature-sensitive and attenuation phenotype. J Virol. 1998;72:1762–1768. doi: 10.1128/jvi.72.3.1762-1768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skiadopoulos M H, Surman S, Tatem J M, Paschalis M, Wu S-L, Udem S A, Durbin A P, Collins P L, Murphy B R. Identification of mutations contributing to the temperature-sensitive, cold-adapted, and attenuated phenotypes of live-attenuated cold-passage 45 (cp45) human parainfluenza virus 3 candidate vaccine. J Virol. 1999;73:1374–1381. doi: 10.1128/jvi.73.2.1374-1381.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Snyder M H, Betts R F, BeBorde D, Tierney E L, Clements M L, Herrington D, Sears S D, Dolin R, Maassab H F, Murphy B R. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol. 1988;62:488–495. doi: 10.1128/jvi.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spielhofer P, Bachi T, Fehr T, Christiansen G, Cattaneo R, Kaelin K, Billeter M A, Naim H Y. Chimeric measles viruses with a foreign envelope. J Virol. 1998;72:2150–2159. doi: 10.1128/jvi.72.3.2150-2159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suryanarayana K, Baczko K, ter Meulen V, Wagner R R. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J Virol. 1994;68:1532–1543. doi: 10.1128/jvi.68.3.1532-1543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takeda M, Kato A, Kobune F, Sakata H, Li Y, Shioda T, Sakai Y, Asakawa M, Nagai Y. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J Virol. 1998;72:8690–8696. doi: 10.1128/jvi.72.11.8690-8696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tober C, Seufert M, Schneider H, Billeter M A, Johnston I C D, Niewiesk S, ter Meulen V, Schneider-Schaulies S. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J Virol. 1998;72:8124–8132. doi: 10.1128/jvi.72.10.8124-8132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valsamakis A, Schneider H, Auwaerter P G, Kaneshima H, Billeter M A, Griffin D E. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J Virol. 1998;72:7754–7761. doi: 10.1128/jvi.72.10.7754-7761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whitehead S S, Firestone C Y, Karron R A, Crowe J E, Elkins W R, Collins P L, Murphy B R. Addition of a missense mutation present in the L gene of respiratory syncytial virus (RSV) cpts530/1030 to RSV vaccine candidate cpts248/404 increases its attenuation and temperature sensitivity. J Virol. 1999;73:871–877. doi: 10.1128/jvi.73.2.871-877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wild T F, Buckland R. Functional aspects of envelope-associated measles virus proteins. Curr Top Microbiol Immunol. 1995;191:51–64. doi: 10.1007/978-3-642-78621-1_4. [DOI] [PubMed] [Google Scholar]