Abstract
The noncoding sequence of five Edmonston vaccine viruses (AIK-C, Moraten, Rubeovax, Schwarz, and Zagreb) and those of a low-passage Edmonston wild-type (wt) measles virus have been determined and compared. Twenty-one nucleotide positions were identified at which Edmonston wt and one or more vaccine strains differed. The location of some of these nucleotide substitutions suggests that they may influence the efficiency of mRNA synthesis, processing, and translation, as well as genome replication and encapsidation. Five nucleotide substitutions were conserved in all of the vaccine strains. Two of these were in the genomic 3′-terminal transcriptional control region and could affect RNA synthesis or encapsidation. Three were found within the 5′-untranslated region of the F mRNA, potentially altering translation control sequences. The remaining vaccine virus base changes were found in one to four vaccine strains. Their genomic localization suggests that some may modify cis-acting regulatory domains, including the Kozak consensus element of the P and M genes, the F gene-end signal, and the F mRNA 5′-untranslated sequence.
Measles virus (MV) is a highly contagious human pathogen best known as the cause of one of the classical “rash” illnesses of children. Few escaped this acute infection and disease prior to the development of the currently used live attenuated virus vaccines. Severe, even fatal complications, particularly involving the respiratory and central nervous systems, were not uncommon even in industrialized countries. Unfortunately, in many parts of the less-well-developed world, measles continues to be the major cause of preventable childhood mortality (9, 15, 16).
MV is an enveloped RNA virus of the genus Morbillivirus, family Paramyxoviridae (32). Like other members of this family, its genome is nonsegmented and of negative sense. The 16-kb linear RNA contains six nonoverlapping cistrons (3′-N-P-M-F-H-L-5′) that encode eight known polypeptides (16, 27). In addition to the protein coding regions, nearly 11% of the 16-kb MV genome is composed of noncoding RNA. Presumably, this relatively small viral genome has maintained a significant noncoding nucleotide sequence content for its functionally important cis-acting elements.
cis-acting regulatory sequences found in all viral genomes are essential for the orderly progression of the viral life cycle. These elements serve to specify, organize, and control gene expression and genome replication, often exerting these effects through interaction with virus-encoded proteins or host cell factors. The important role of cis-acting sequence elements is well illustrated by the complex transcriptional regulatory schemes employed by large DNA viruses such as herpes simplex virus or adenovirus (38, 42). It is equally clear that cis-acting regulatory elements are exploited by the smaller genomes of negative-strand RNA viruses to control their replicative strategy (27). In MV, like other paramyxoviruses, cis-acting sequences have been identified that have roles in genome replication, genome packaging, translation, and mRNA synthesis, processing, and editing (2, 16, 19, 27, 41).
The known or proposed cis-acting signals in the MV genome are summarized in Fig. 1 (2, 16, 19, 27, 41). The noncoding 107 nucleotides at the 3′ end of the negative-strand genome (called the leader transcriptional control region [TCR]) include promoter sequences that initiate two distinct RNA synthesis pathways: (i) production of an end-to-end copy of the genome to generate the positive-strand replication intermediate and (ii) an elongation-termination-reinitiation transcription pathway that produces mRNAs corresponding to the six cistrons. Transcription termination and reinitiation during mRNA synthesis is mediated by conserved sequence elements located in each intergenic region: a gene-end (GE) plus a gene-start (GS) signal separated by the characteristic GAA nucleotide triplet forms the GE/GS signal. The highly conserved GAA triplet is found between all intergenic GE and GS signals except between the H and L genes, where a GCA triplet is found. In addition to guiding transcription termination and reinitiation, the GE/GS signal also directs mRNA polyadenylation.
FIG. 1.
MV genome map. The location of protein coding regions (white boxes: N, P, V, C, M, F, H, and L) and noncoding regions including the leader and trailer (black terminal boxes) and intergenic regions (shaded blue) are shown, along with specialized sequence motifs (2, 16, 19, 27, 41, 52). The TCRs at the 3′ end of the genomic RNA (leader TCR, nucleotides 1 to 107) and the 3′ end of the antigenomic RNA (trailer TCR, nucleotides 15785 to 15894) are enlarged below the genome map. Genomic RNA synthesis initiation is shown as open arrowheads in the enlargement of the leader and trailer TCR regions. Sequence motifs shown in the expanded TCR maps include the leader, trailer, GE and GS signals, the 16-base conserved terminal sequence (shown 3′ to 5′), the repeated B- and B'-box motif, the G(N)5 motif, the ATG codon for the N gene adjacent to the leader, and the L protein stop codon outside of the 5′ end of the trailer. The positions of GE/GS signals are designated with a combination of solid arrowhead (GS) and a black box (GE). The leader TCR GE/GS symbol lacks a GE box, and the trailer TCR GE/GS lacks a GS arrowhead to signify that these sequences may not function identically to GE/GS elements in the intergenic regions. The mRNA editing site is indicated below the V protein coding region (6). Part of the F gene mRNA 5′-untranslated nucleotide sequence is shown to illustrate the three in-frame AUG initiator codons (5). At the bottom of the figure, several symbols found in the TCR maps are defined.
cis-acting sequences similar to those in the leader TCR are also located in the 3′-terminal 109 nucleotides of the positive-stranded antigenome (the trailer TCR; Fig. 1). The trailer TCR exclusively directs initiation of negative-strand genome synthesis. Important sequence motifs found in the leader and trailer TCRs (Fig. 1) include the terminal 16 nucleotides that are thought to be part of the primary site of RNA polymerase recognition (20, 28), the G(N)5 (52) and B-box motifs (2) that have been implicated as regulators of MV transcription and replication, and a potential GS signal in the leader TCR prior to the N gene and a GE signal in the trailer TCR that terminates L gene mRNA synthesis (16, 19, 27, 41).
Two additional functional cis-acting sequences have been identified in the MV genome. One is the RNA editing site in the P cistron that permits addition of a G residue in some P mRNAs, resulting in the translational frameshift that directs synthesis of V protein (6). The second cis-acting sequence is found within the rather long noncoding intergenic region between the M and F genes. Part of this region specifies the nontranslated leader sequence of the F gene mRNA which has been shown to be an important determinant of translational efficiency and AUG codon selection (5, 13). Deletion of this translational control element from recombinant viruses also appears to compromise replication in vivo (55). Identification of yet other cis-acting elements is likely to occur as more of the MV noncoding sequences are scrutinized with transient assay systems and recombinant viruses.
cis-acting elements can be an important determinant of virus attenuation. For example, mutations in the leader sequence of human parainfluenza virus type 3 (PIV3) vaccine candidates have been shown to specify the temperature sensitivity, cold adaptation, and attenuation phenotypes (46). Some attenuated respiratory syncytial virus A2 (RSV A2) strains contain a nucleotide change in the GS signal of the M2 gene that contributes to their temperature-sensitive and attenuated phenotypes (56). Also, a GE signal mutation in the M gene of an attenuated RSV B strain has been found to compromise expression of the downstream SH gene (D. A. Buonagurio, personal communication). Similarly, a cis-acting sequence mutation has been noted within the GE signal of the mumps vaccine virus F gene that appears to also disrupt normal expression of the downstream SH gene (51). Taken together, these studies indicate that specific alterations of noncoding cis-acting sequence elements of negative-strand RNA viruses can modulate virus virulence and attenuation.
Genomic modifications that attenuate MV have only recently begun to be defined. For example, several studies have shown that viruses defective for V or C protein expression display attenuated growth characteristics in some model systems (12, 31, 34, 53, 55). Additionally, mutations in genes encoding C, V, P, and L proteins have been associated with reduced viral replication in the B95 lymphocyte cell line (50). Although these studies have started to define roles for various proteins in MV attenuation, the potential role of cis-acting sequences has so far received less attention. In one case, deletion of most of the F gene mRNA 5′-untranslated region from a recombinant MV strain did result in less-efficient replication in human thymus-liver implants engrafted into SCID mice (55), demonstrating that modification of this cis-acting sequence can modulate attenuation.
To learn more about the potential role of noncoding cis-acting sequences in MV attenuation, a comparative sequence analysis was performed on viruses in the Edmonston vaccine lineage. This study was based on posing two relatively simple questions. (i) Do the noncoding sequences from several optimally attenuated vaccine viruses differ from the Edmonston wt progenitor strain or an underattenuated Edmonston vaccine strain? (ii) If differences exist, do they affect noncoding sequences that may function as cis-acting sequences controlling gene expression or replication? The Edmonston virus lineage (39) is attractive for this type of comparative study for a variety of reasons. Six viruses from the vaccine lineage are available for comparison, including five independently generated vaccine strains and a low-passage laboratory isolate of Edmonston wt (10, 11, 16, 18, 21, 26, 30, 39). This provides a unique opportunity to examine the molecular consequences of similar but independent vaccine derivation schemes (39). Comparison of Edmonston wt and five different vaccine strains also provides an opportunity to examine the diversity of molecular mechanisms by which the attenuated phenotype is produced by different genotypes. Furthermore, the vaccine strains differ in the level of attenuation. Four of the five vaccines are adequately attenuated, while Rubeovax proved to be reactogenic (26), and this is an important point for comparison that should provide insight into what genome changes influence the degree of attenuation. Finally, transient expression systems and cDNA rescue technologies provide experimental systems to further analyze the genetic changes identified by sequence analysis (35).
To identify potential attenuation determinants within MV cis-acting regulatory elements we have sequenced the noncoding regions of a low-passage isolate of Edmonston wt and five vaccine derivatives. Base changes were identified at 21 noncoding positions when the vaccine genomes were compared to Edmonston wt. Five nucleotide substitutions were common to all vaccine strains, while the remaining substitutions were present in one to four vaccine viruses. The sequence comparison also revealed that Moraten and Schwarz contained identical noncoding region sequences and differed at five nucleotide positions from closely related Rubeovax.
MATERIALS AND METHODS
Cells, virus, and genome sequencing.
Cell culture and MV propagation was performed as described in an accompanying article (33). The Edmonston wt isolate (11, 16) was a gift from Judy Beeler (Center for Biologics Evaluation and Research). Edmonston B (Rubeovax) (10, 26), AIK-C (30), Schwarz (Rimevax; SmithKline Beecham) (26, 40), and Zagreb (21) were generously provided by William Bellini and Paul Rota (Centers for Disease Control) (39). Moraten (Attenuvax; Merck & Co) (18) and a second preparation of Schwarz (Rimevax, SmithKline Beecham) (26, 40) were obtained from commercially available vaccine preparations. The two separate sources of Schwarz were analyzed independently.
Viral genome sequence was determined directly from DNA fragments generated by reverse transcription-PCR (RT-PCR) of RNA extracted from infected Vero cells (33). Cycle sequencing (17, 25) was performed using dye-labeled terminators and Taq DNA polymerase (Applied Biosystems), followed by analysis on an ABI Prism automated sequence apparatus. Primers used for PCR amplification and sequencing on both cDNA strands were designed based on published MV sequences (GenBank accession numbers K01711 and S58435). Data analysis was performed using the MacVector (Oxford Molecular Group) and Lasergene (DNAstar, Inc.) software packages. The MV sequences have been deposited in GenBank (Edmonston wt, AF266288; AIK-C, AF266286; Moraten AF266287; Rubeovax, AF266289; Schwarz, AF266291; Zagreb, AF266290).
RESULTS
Comparative sequence analysis.
Evidence that the base changes within the cis-acting elements of the MV genome may contribute to viral attenuation was sought through comparative analysis of the noncoding region nucleotide sequence of five Edmonston vaccine viruses and a low-passage Edmonston wt isolate. Potentially attenuating cis-acting nucleotide substitutions were indeed located, providing the framework for design of future genetic studies aimed at dissecting the molecular basis of attenuation using the MV minireplicon and recombinant MV systems.
Interpreting such comparative sequence analyses requires caution given the lack of the true Edmonston wt progenitor virus. Fortunately, a low-passage derivative of the Edmonston clinical isolate was available for these studies. This virus was passaged 13 times (39) prior to the analysis presented here, so it is possible that some degree of tissue culture adaptation may be reflected in the Edmonston wt sequence. Nevertheless, it is the best approximation of the original clinical isolate available, and the passage number has been kept to a minimum in hopes of obtaining a meaningful comparison with the vaccine viruses.
The issue of additional nucleotide changes arising during cell culture passage also applies to sequence analysis of vaccine virus genomes. Propagation of vaccine virus strains in cell culture to produce RNA for analysis can lead to genome changes that reflect adaptation to culture conditions and host cell type used for infection. This concern has been addressed in two ways. First, the number of cell culture passages was limited to three or less while generating viral stocks and RNA. Although this may result in some degree of cell culture adaptation, the additional minimal passage number is unlikely to affect greatly the majority of genomes in the infected cell population. Second, the sequence determination used RT-PCR products rather than cloned cDNA fragments. Sequence obtained from RT-PCR products should represent the majority sequence in a population of viral genomes and help alleviate the influence from minor viral populations that began to evolve during passage in Vero cells.
Leader and trailer transcriptional control regions.
The 107 3′-terminal noncoding nucleotides include the MV genomic promoter (43, 44) and are referred to here as the leader TCR (Fig. 1). Within this TCR lie several discrete sequence elements that are thought to act in cis to modulate MV transcription, replication, and gene expression. Two of these are highly conserved, well-accepted cis-acting regulatory elements of the TCR. One is the terminal 16 bases of the leader (Fig. 1). The second is the GS signal that directs N gene mRNA synthesis (2, 16, 19, 23, 27, 41). Additional cis-acting elements in the leader TCR [B-box and G(N)5 sequences] have been proposed based on genomic sequence comparisons (2, 8) and analyses of defective interfering RNAs (44), and the existence of these proposed elements has been substantiated by studies with Sendai virus (52). Nucleotide substitution in any of these elements or yet-undefined cis-acting sequences could have a significant effect on virus-cell interaction through changes in replication or gene expression efficiency.
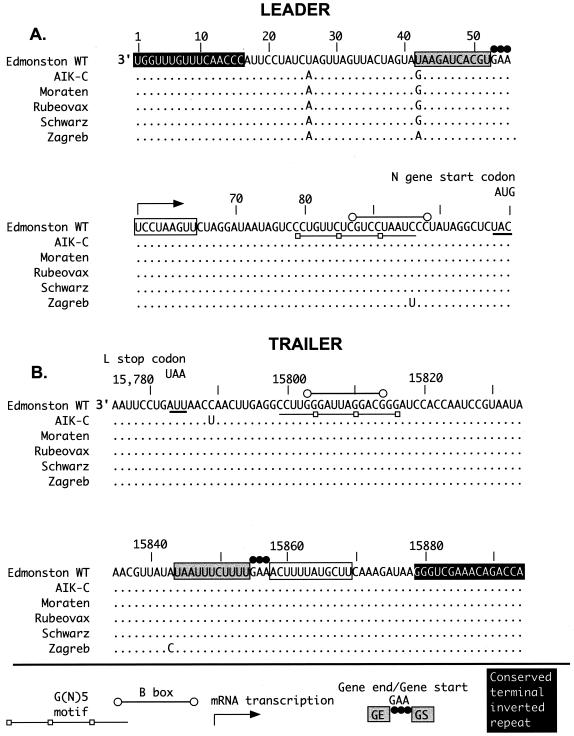
Comparison of leader TCR sequences of Edmonston wt and the five vaccine strains revealed nucleotide differences at three positions (Fig. 2A). Nucleotide transversions were detected in all vaccines at positions 26 and 42. The wt U residue in the negative-sense genome strand was changed to an A residue at position 26. The wt U residue at position 42 was substituted with a G in AIK-C, Moraten, Schwarz, and Rubeovax, and an A residue in Zagreb. The location of these base substitutions is shown in Fig. 2A relative to previously described sequence motifs (Fig. 1). The position 26 and 42 base substitutions were located between the terminal leader TCR domain and the GS sequence (Fig. 2A; see also Fig. 7A). The third leader TCR substitution was detected only in Zagreb at position 96. This C-to-U transition occurred within the B box sequence and at the 5′ boundary of the G(N)5 motif.
FIG. 2.
Sequence comparison of Edmonston wt and vaccine leader and trailer TCRs. Terminal 110 bases of genomic sequence (3′ to 5′) containing the leader TCR (nucleotides 1 to 107; part A) and trailer TCR (nucleotides 15785 to 15894; part B) of Edmonston wt are shown and labeled with genome nucleotide positions. Sequence motifs are illustrated on the sequence according to the key at the bottom of the figure. Relevant protein start or stop codons also are included. The sequence corresponding to the GE region in the leader TCR and the GS region of the trailer TCR are not shaded in gray to indicate that these sequence motifs may not function identically to their intergenic GE/GS counterparts. Illustrated below the wt sequence is the sequence comparison with vaccine strains. Nucleotide identity is given as a dot, and a typed nucleotide indicates disagreement with wt.
FIG. 7.
Vaccine nucleotide substitutions in cis-acting sequence motifs. (A) Alignment of MV TCRs containing the GE/GS signal. Similarity between the signals and a consensus sequence is outlined by the box (yellow). The leader TCR GE and trailer TCR GS regions are presented in lower case to indicate that these sequences may not function like the corresponding sequences of the intergenic GE/GS elements and that these sequences were not considered in evaluating the consensus. Below the consensus are the two GE/GS sequences that contained vaccine virus base substitutions. The nucleotide substitution is highlighted by a blue box. (B) Summary of the base changes found in the leader and trailer TCRs. The description of the leader and trailer TCRs can be found in Fig. 1. Nucleotide changes are illustrated below the leader and trailer TCRs. Base changes are highlighted in blue. (C) Comparison of the Kozak element (24) found in wt MV mRNAs. Variation in vaccine virus P and M gene Kozak sequences is shown below the wt sequences.
Little variability was detected in the trailer TCR and no base substitution was common to all of the vaccine viruses. Transitions were identified at position 15,789 (C to U) in AIK-C and at position 15,843 (U to C) in Zagreb. Neither mutation was located in a previously described sequence motif, although the Zagreb mutation was adjacent to the 3′ boundary of the GE signal that terminates transcription following the L gene (Fig. 1 and 2B). Both mutations changed bases in the 3′ noncoding region of the L mRNA.
Intergenic regions.
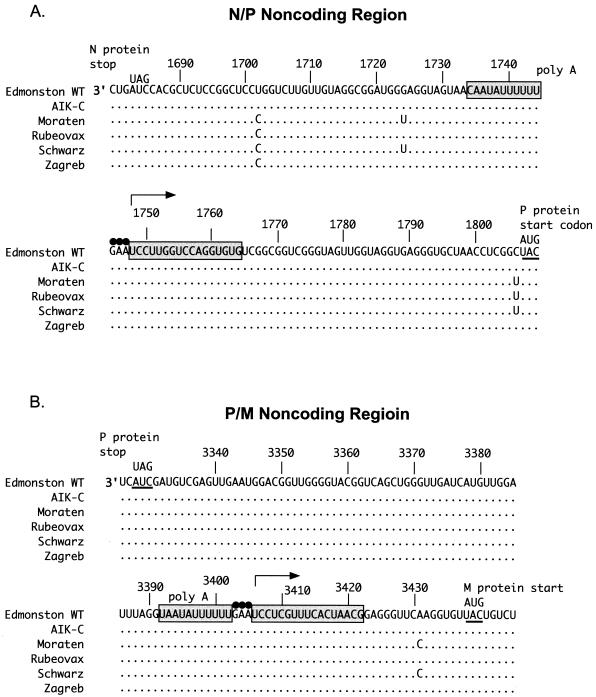
Three variable nucleotide positions (Fig. 3A) were found in the intergenic region between the N and P genes (N/P). None of these nucleotide substitutions were common to all vaccines. Two were in the N cistron affecting the region that specifies the 3′-untranslated region of the N mRNA. These substitutions included a U-to-C substitution at position 1702 shared by all vaccine viruses except AIK-C. In the same region, Moraten and Schwarz contained a G-to-U transversion at position 1724. Within the P cistron, a C-to-U transition was identified at position 1806. This nucleotide substitution was found in Moraten, Schwarz, and Rubeovax, and it changed the base immediately upstream of the P protein translation start codon.
FIG. 3.
Comparison of N/P and P/M intergenic regions of Edmonston wt and vaccine viruses. Description of this figure is as described for Fig. 2.
Only one nucleotide change was identified in the P/M intergenic region (Fig. 3B). This occurred in the M cistron at nucleotide 3431, where a wt A nucleotide was changed to a C residue in Moraten and Schwarz. This nucleotide substitution alters the sequence of the 5′ noncoding region of the M mRNA at nucleotide position −7 relative to the M gene translation initiation codon.
Nine different nucleotide positions differed from wt in the long M/F intergenic region (Fig. 4). None of these mutations were found within the GE/GS signal. Four of the base changes were detected in the 3′-untranslated region of the M gene mRNA. These were at genomic positions 4536 (C to A in AIK-C), 4574 (C to U in AIK-C), 4608 (A to G in Rubeovax), and 4611 (G to A in AIK-C and Zagreb). The five remaining M/F intergenic region changes occurred within sequences encoding the 5′-untranslated region of the F gene mRNA. Two of these base substitutions were found only in one vaccine strain, at genomic positions 5030 in AIK-C (G to A) and 5308 in Rubeovax (A to G). The other three were conserved in all vaccine strains. These changes were two A-to-G transitions at 4978 and 5349 and a G-to-C transversion at 5073.
FIG. 4.
Comparison of M/F intergenic regions from Edmonston wt and vaccine viruses. The description of this figure is similar to that for Fig. 2. One line of dots indicating homology to all vaccines is shown under the wt sequence if there were no base changes to report. The sequence shown in this figure extends beyond the noncoding intergenic region to show the position of the three F gene AUG codons. The second AUG codon is the predominately used initiation codon (5). Vaccine virus names are abbreviated as follows: AIK-C (AIK), Moraten and Schwarz (Mor/Sch) Rubeovax (Rubeo) and Zagreb (Zag).
Few nucleotide changes were identified in the F/H and H/L intergenic regions (Fig. 5). One nucleotide substitution was present in the F/H intergenic region at nucleotide position 7243 (Fig. 5A). This A-to-G transition was present within the boundaries of the F gene GE signal of Moraten and Schwarz (Fig. 5A). In the H/L intergenic region, two base substitutions were located in the 3′-untranslated region of the H gene mRNA (Fig. 5B). At position 9139 a G-to-A substitution was present in AIK-C, and a U-to-A transversion at position 9144 was detected in both Moraten and Schwarz.
FIG. 5.
Comparison of F/H and H/L intergenic regions from Edmonston wt and vaccine viruses. The description of this figure parallels that for Fig. 2.
DISCUSSION
These analyses, though limited by the considerations described earlier, revealed distinctive base substitutions within multiple noncoding region sequences that perform potential cis-acting functions (Fig. 2 to 5). Analyses of human PIV3 and RSV A and B vaccine candidates also have identified base substitutions in cis-acting regions (7, 14, 18; D. A. Buonagurio, personal communication), indicating that this characteristic is shared by negative-strand vaccine virus strains. None of the five Edmonston vaccine viruses accumulated base changes resulting in gross alteration of a noncoding region, suggesting that these sequences do contain important components of the viral regulatory apparatus and that alteration is not well tolerated. It may also suggest that only subtle adjustments to cis-acting sequences controlling gene expression and replication were required to facilitate growth in the semipermissive cells used for vaccine virus selection.
Why MV noncoding region mutations accumulate in potential cis-acting sequences is currently speculative, but it seems reasonable to suggest that at least some of these mutations confer a selective advantage for viral replication under conditions used for vaccine derivation. Whether any of these mutations contribute to the attenuated phenotype remains to be established. Answering this question will require further studies using reverse genetics and recombinant viruses in studies like those ongoing with RSV and PIV3 vaccine candidates (7, 45, 46, 56–58).
Nucleotide substitutions in MV noncoding cis-acting sequences may be a favored response to changes in viral polypeptides induced by passage in heterologous cell types. In one simple model describing the biological selection process (33), it has been proposed that selective pressure is initially driven by a requirement for interaction between semipermissive host cell factors and viral proteins involved in the earliest stages of viral infection, particularly transcription and replication. It follows that the early stages of infection in semipermissive cells would be inefficient because viral proteins are not well equipped to deal with the semipermissive host cell environment. As the passage process progresses, mutations in viral protein coding regions are favored if they enhance the ability of viral proteins to functionally interact with proteins found in the semipermissive host cell. Although these protein modifications may enhance the level of interaction between viral and cellular proteins, they may be costly to other aspects of the virus life cycle. To help compensate for any adverse effects of amino acid substitutions, the virus evolves second-site compensatory mutations that subtly change gene expression and replication through base changes in cis-acting regulatory sequences and additional amino acid substitutions. As implied by this model, many of the noncoding region base changes identified in the Edmonston vaccines seem to have the potential to modify cis-acting regulatory components of the virus genome.
The noncoding region base substitutions can be grouped into three categories based on their potential to modify cis-acting regulatory functions: (i) base substitutions that may affect protein translation through changes in mRNA stability or modification of translational regulatory elements in message, (ii) base substitutions that could affect mRNA synthesis by altering promoter activity in the leader TCR or GE/GS signal function, and (iii) base substitutions that could affect the activity of replication promoters in the leader or trailer TCRs. Below, the identified MV vaccine noncoding base substitutions and their attenuation potential are discussed in the context of these categories. Figures 2 to 5 provide the sequence data for each gene region, while Fig. 6 summarizes these results schematically.
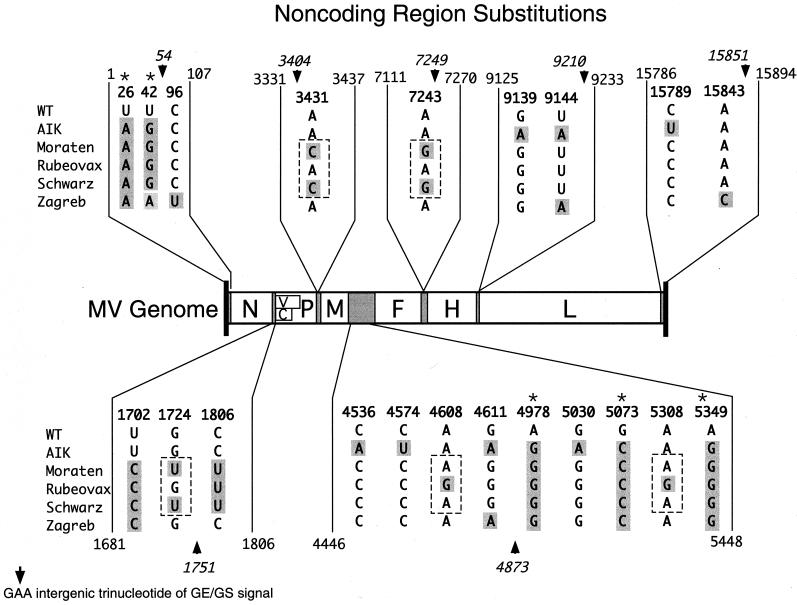
FIG. 6.
Summary of noncoding region nucleotide substitutions. A genome map similar to Fig. 1 is shown along with the nucleotide positions that varied in vaccine strains. The relative position of the GE/GS signals is shown by an arrow head plus the nucleotide position corresponding to the central A residue in the conserved GAA motif. Boundaries of the noncoding regions are indicated at the ends of the brackets containing the nucleotide substitutions. Nucleotides highlighted in gray depict changes from the Edmonston wt sequence. An asterisk above a column of nucleotides indicates that a base change occurred in all vaccine strains. The dashed box outlining groups of three nucleotides shows positions where Rubeovax differs from Moraten and Schwarz.
Base substitutions and cis-acting regulation of translation.
Mutations within the 5′- or 3′-untranslated region of mRNAs can alter protein expression by affecting mRNA stability or translation efficiency (22, 29). The combination of noncoding sequence and poly(A) tail at the 3′ end of mRNAs is known to play an important role in determining mRNA stability (22). Destabilizing sequences are generally AU-rich and commonly contain the pentamer AUUUA. It is plausible that mutations in the 3′-untranslated region of MV mRNAs could affect a destabilizing sequence by interrupting an AU-rich motif or altering the secondary structure near a destabilizing motif. Base substitutions in 3′ noncoding regions were found in multiple locations, including the N, M, H, and L genes. None of the mutations affecting the 3′-untranslated regions of MV mRNAs, except possibly the base substitution at position 15843 in Zagreb (Fig. 2B), obviously affects an AU-rich element (Fig. 2 to 6), but the potential of any of these base changes to alter mRNA stability can only be determined experimentally.
The 5′ end of several vaccine virus mRNA sequences included base substitutions located in positions that may affect translation initiation (Fig. 6 and 7C). In both the P (Fig. 3A and 6) and M (Fig. 3B and 6) genes, base substitutions were identified near the AUG translation initiation codon in the Kozak element (Fig. 7C). The Kozak consensus sequence influences AUG codon selection and efficiency of translation initiation (24). In the P gene of Moraten, Schwarz, and Rubeovax, a G-to-A transition (mRNA sense) was found at position −1 relative to the AUG codon for the P open reading frame (Fig. 7C). In the Moraten and Schwarz M gene, a U-to-G transversion (mRNA sense) occurred at position −7 relative to the AUG codon (Fig. 7C).
The initiator codon context of both the M and P mRNAs deviate from the Kozak sequence at positions considered most important for AUG strength (24). The M mRNA deviates at the highly conserved +4 position, resulting in a less-favorable context for translation initiation. This potentially increases the influence of other nucleotides in the vicinity of the AUG such as the U residue at position −7 that was changed in the Moraten and Schwarz vaccines. Similarly, the P mRNA lacks a purine at position −3, which is the most highly conserved base in the Kozak element (24). Absence of a purine at −3 will likely increase the importance of other bases in the P mRNA Kozak element, such as the base at position −1 that was changed in Moraten, Schwarz, and Rubeovax. If these changes in initiation codon context alter translation efficiency, the effect may be quite subtle since the codon context is changed only by a single base substitution. This does not mean that these base substitutions are not important since it is possible that attenuation is the result of the cumulative effect of numerous relatively small adjustments in the virus life cycle.
The poor agreement between the P gene AUG context and the Kozak element consensus may have evolved to permit a certain percentage of ribosomes to scan through the P AUG codon and engage the C protein mRNA AUG codon located downstream. This also suggests that base changes in the P Kozak element may influence the ratios of P, V, and C protein synthesis in the infected cell. The possibility that the P AUG codon context may influence translation of C protein has been examined previously. Alkhatib et al. (1) placed the P gene into a recombinant adenovirus and analyzed the levels of P and C protein synthesis in infected cells. Deletion of the P AUG codon had little effect on the synthesis of C protein in this system. This implies that C protein mRNA AUG codon strength is independent of the presence of the upstream P gene AUG codon and further implies that altering the P mRNA Kozak element would have little if any effect on C protein synthesis. This conclusion, however, may be worth reexamining in an alternative system since the P gene cDNA was fused to the adenovirus major late transcription unit tripartite leader sequence. These adenovirus sequences function as a cis-acting translational control element in adenovirus-infected cells (42) and may negate some of the intrinsic cis-acting elements found in the measles virus P mRNA. Clarification of possible interplay between the P and C mRNA AUG codons may best emerge from study of appropriately designed recombinant MVs.
Further evidence linking translation and attenuation may be found by examining the F gene. MV has maintained the 1-kb intergenic region between the M and F genes that in part specifies a long untranslated 5′ end for the F mRNA (Fig. 1 and 4). This untranslated sequence is dispensable for recombinant virus growth in a Vero cell culture (36). However, it has been found to modulate translation and play a role in the selection of a predominant initiation codon from several closely positioned AUG sequences that are in frame with the F coding region (Fig. 1 and 4 and reference 5). In comparing Edmonston strains, five base changes were detected in the vaccine virus F gene mRNA untranslated 5′ end (Fig. 4). Given that the F mRNA 5′-untranslated sequence functions as a cis-acting element and that three base substitutions in this sequence were common to all vaccine strains, it seems likely that modification of this sequence may have been favored for MV growth in semipermissive cell types. Whether the F mRNA untranslated region plays a role in attenuation remains uncertain, but studies showing that recombinant virus lacking most of this sequence replicates less efficiently in human thymus-liver tissue transplanted in SCID mice (55) suggests that the untranslated region may play a role in pathogenicity.
Base substitutions and the control of mRNA synthesis and processing.
The second category encompasses mutations with potential to influence viral mRNA synthesis, specifically mutations in GE/GS signals and the leader TCR. One such base substitution occurred within the F/H intergenic GE/GS signal (Fig. 7A). This mutation was found in Moraten and Schwarz, where an A-to-G transition occurred in the GE signal (nucleotide position 7243; Fig. 7A). The effect of this base substitution is not known as yet, but it may perturb normal transcription termination and reinitiation at the F-H gene boundary, leading to altered expression of the downstream H gene. This phenomenon has been observed in some strains of RSV, PIV, and simian virus 5, where less-efficient GE signal variants cause reduced transcription termination with overproduction of readthrough bicistronic transcripts at the expense of normal monocistronic mRNA synthesis of the downstream gene (3, 37, 47, 54) (D. A. Buonagurio, personal communication). Reduction in correctly initiated mRNA synthesis from the downstream gene, coupled with the likelihood that the fused bicistronic transcripts are inadequate templates for translation of the gene distal to the 5′ mRNA cap (24, 60), will restrict protein synthesis encoded by the downstream gene. This effect could result in diminished H protein expression if Moraten and Schwarz produce elevated levels of a F/H bicistronic message.
It also may be relevant to mention the mutation at position 42 in this context even though it is possible that the GE signal located in the leader TCR may not function analogously to GE signals found in intergenic regions. The position 42 mutation was present at the 3′ boundary of the GE/GS signal consensus drawn in Fig. 7A, raising the possibility that it may modify GE/GS signal function. This could be significant if transcription initiating at the genomic 3′ end must terminate and reinitiate to effectively transcribe an N mRNA and the position 42 mutation alters termination or reinitiation function. Whether a termination and reinitiation mechanism applies to mRNA synthesis initiated from the MV leader TCR is uncertain given the failure to detect abundant small leader RNA products in infected cells that would be indicative of termination prior to the N gene GS signal (4, 8). However, it is worth noting that attempts to detect the small leader RNA in infected cells have relied on infection with laboratory-adapted Edmonston strains that have vaccine leader mutations.
If the base 42 substitution negatively affects termination efficiency, greater accumulation of leader-N mRNA fusions will result. These readthrough transcripts introduce an AUG sequence upstream of the authentic N gene initiation codon, potentially reducing N protein expression. In addition, leader sequences attached to N mRNA should serve as substrates for interaction with N protein. Encapsidation of leader-N gene fusion mRNAs should render them unavailable for translation. Given these considerations, the position 42 base substitution could reduce the level of N protein synthesis in all vaccine viruses by altering GE function.
mRNA levels also may be influenced by promoter strength of the leader TCR. As noted above, three base changes were observed in this region of the vaccine viruses. One was a C-to-U transition that was unique to Zagreb at nucleotide position 96 (Fig. 2B). The other two mutations were at positions 26 and 42. The position 26 and 42 pyrimidine-to-purine transversions were conserved in all vaccine strains. Alignment of leader TCR mutations to previously described sequence motifs (Fig. 1, 2, and 7B) showed that the position 26 mutation was excluded from any currently described motifs while, as described earlier, the base change at 42 lay within the boundaries of the GE/GS consensus sequence (Fig. 7A). The unique Zagreb mutation at position 96 (Fig. 7B) was present within the boundaries of the B box (2, 8) and the overlapping G(N)5 motif (52).
The possibility that these leader TCR mutations affect mRNA synthesis is speculative but intriguing to consider further. It is noteworthy that the leader TCR changes at 26 and 42 were located within a region that is not highly homologous to the trailer TCR (Fig. 8). Considering the functional similarity between the leader and trailer TCRs, it seems reasonable to expect significant sequence homology in regions that perform largely identical functions. Alignment of both TCRs shows, as described before (2, 8), that they share two regions of strong homology. One includes the terminal 16 bases, and the second encompasses the B and B′ boxes. The fact that only the Zagreb mutation (position 96; Fig. 2 and 6 to 8) fell within one of these homologous sequences (the B-box region) may indicate that these sequence elements are relatively intolerant of nucleotide substitutions. Alternatively, it may imply that virus passage in semipermissive cells favored changes in sequences that were unique to leader TCR. This may further imply that these leader TCR changes were favored because they modulated a leader TCR-specific function such as the initiation of mRNA synthesis.
FIG. 8.
Alignment of the Edmonston wt leader (minus strand) and trailer (plus strand). The alignment illustrates the homology between the terminal 16 nucleotides at the 3′ end of the leader and trailer, as well as the B-box regions. The motifs identified in the figure are described in the legend to Fig. 1. Vaccine virus leader TCR base changes are shown above the wt sequence, and vaccine trailer TCR changes are shown below.
That the position 26 and 42 base changes may primarily affect mRNA transcription function of the leader TCR is consistent with the base 42 mutation localization to the leader TCR GE consensus sequence boundary (Fig. 1A and 7). Moreover, it is possible that the region between nucleotides 17 and 42 performs a function unique to mRNA transcription initiation. Perhaps it acts as a polymerase pause site where the decision is made between genome synthesis and mRNA synthesis. It could also serve as a contact site that has specificity for a modified polymerase complex that is used for mRNA synthesis. Both of these possibilities imply that replacing two pyrimidine residues at 26 and 42 with purines may affect the efficiency of mRNA transcription initiation by interrupting important polymerase contacts in the leader. These contacts could be perturbed if the substitutions replace important contact nucleotides or induce a subtle change in phasing caused by substitution of pyrimidines with bulkier purines.
Leader and trailer TCR base substitutions and the control of genome replication.
Base substitutions in the leader or trailer TCR could influence replication by altering the efficiency of genome-length RNA synthesis. As described above, three base substitutions were found in the leader TCR. While it was suggested that these mutations might act at the level of mRNA transcription, they may additionally or alternatively have an impact on the initiation of antigenome synthesis or modulate the pathway that controls the preferential selection of antigenome or mRNA synthesis. In that context, the unique Zagreb mutation at position 96 present in the B box is interesting because its location in a motif that is conserved in both the leader and trailer TCRs suggests that this homologous sequence domain regulates the replication pathway.
Only two base changes were found in the trailer TCR, and these were specific to individual vaccine strains. Neither the AIK-C mutation (position 15789) nor the Zagreb mutation (position 15843) lie within the 16-base conserved terminal promoter sequences or the overlapping B box-G(N)5 domains. Although localized to the trailer TCR, these base substitutions also fall into the category of mutations that modify untranslated regions of mRNAs. Both mutations reside in the 3′ noncoding region of the L mRNA. Were they to affect the phenotype of these vaccine strains, it would likely be through the changes in the L mRNA stability. Additionally, the Zagreb trailer TCR mutation is positioned at the GE/GS consensus region (Fig. 8). Although it does not function as a GE/GS signal in the positive genome strand, this relative positioning could indicate that this region makes contact with the polymerase complex bound at the trailer. It remains to be determined if this Zagreb mutation affects the replicative function of the trailer.
Beyond their putative capacity to influence transcription and replication, the mutations in leader and trailer TCRs can theoretically affect encapsidation efficiency. During genome synthesis, the first sequences that become accessible for encapsidation are the leader and trailer, suggesting that nucleation sites mediate encapsidation within these regions. Thus, base substitutions in the leader or trailer could affect the interaction of N protein with nascent genomes. This seems a less likely scenario given studies with vesicular stomatitis virus delimiting the encapsidation signal to the terminal 14 bases (41), a region that was unaffected by base substitutions amongst the Edmonston MV strains.
Contribution to attenuation.
Not all of the vaccine virus noncoding region base changes are expected to have equivalent impact on replication or gene expression and the levels of attenuation. The base changes found in the P and M mRNAs, within the Kozak consensus sequence, might affect protein expression either positively or negatively. The alteration in the M mRNA AUG context could influence virion maturation and possibly gene expression (16, 49, 59) if the levels of M protein synthesis are altered. The case of the P initiation codon is particularly interesting because changes in the AUG codon strength could affect the translation of P (and V) as well as the translation of C protein from the downstream AUG. Regulation of P, V, and C mRNA translation as a means of achieving attenuation is particularly attractive because each of the P gene-encoded proteins, and their balance, likely play a central role in controlling genome replication and mRNA transcription (16, 19, 41). Given these multiple and vital functions, even small changes in the relative concentrations of P, V, and C proteins in the infected cells could significantly influence viral replication in the infected host (12, 31, 34, 53, 55).
Base changes in the untranslated leader of the F gene also may contribute to the attenuated phenotype. Expression of F protein is essential for cell fusion and effective cell-to-cell spread of MV (16, 59), and it has been demonstrated that the F gene 5′-untranslated region plays a role in determining translation efficiency and AUG codon selection (5). The fact that three mutations were common to all vaccine F gene mRNAs implies that passage in semipermissive cells selected for alteration in this cis-acting element. The most obvious advantage accrued by base substitutions in this region is their ability to alter the levels of F protein expression levels, perhaps through changes in the higher-order structure of the mRNA 5′ end. Perturbation of this function may also be responsible for the reduced viral titers observed in the human thymus-liver tissue-SCID mouse model system (55) when the thymic implants are infected with a recombinant Edmonston B strains that lacks much of the sequence for the F mRNA 5′-untranslated region.
Base changes in the leader TCR also are likely attenuation candidates. It was proposed above that the position 26 and 42 pyrimidine-to-purine transversions may influence gene expression by changing the interaction between the promoter and the viral polymerase. In this scenario, base substitutions disturbing normal levels of mRNA transcription certainly might contribute to an attenuated phenotype. It is also noteworthy that amino acid coding changes were identified in vaccine virus genes for the polymerase complex proteins (P and L) and accessory proteins (C and V) (33). Possibly, the vaccine virus base changes in the leader TCR and the amino acid changes in protein components of the gene expression apparatus have evolved together to optimize expression in semipermissive cells such as chicken fibroblasts. A by-product of these changes is that upon infection of the human host this adapted form of the transcriptional apparatus is less efficient and leads to an attenuated phenotype.
Finally, these comparative analyses included comparison of highly related vaccine viruses that differ in attenuation level. Optimally attenuated Moraten and Schwarz were found to contain identical noncoding region sequences, and these sequences differed from the underattenuated Rubeovax strain by only five nucleotides (Fig. 6, nucleotides highlighted with a dashed box). Two of these differences were substitutions unique to Rubeovax, and these were both T-to-C transitions (mRNA sense) in the M/F intergenic region. One of these base substitutions was present in the M gene mRNA 3′-untranslated sequence, and the other was present in the F gene mRNA 5′-untranslated region. These mutations could affect expression by changing mRNA stability or efficiency of translation. The third base that distinguished Rubeovax from Moraten and Schwarz was found in the 3′-untranslated region of the N mRNA. In this case, Rubeovax contained a wt nucleotide compared to the base change contained in the other two vaccines. Again, if this base contributes to a difference in attenuation levels it would probably be the result of altered mRNA stability. The final two bases that distinguish Rubeovax from Moraten and Schwarz have somewhat more compelling links to attenuation. These two substitutions in Moraten and Schwarz remained wt in Rubeovax. They altered a base in the M mRNA Kozak consensus sequence at position 3431 and a base in the F gene GE signal at position 7243. As mentioned earlier, the base change in the M gene could influence translation of M protein resulting in altered virion maturation (59) and possibly transcription regulation (49). The F GE signal mutation may similarly downregulate expression of the downstream H gene if it compromises transcription termination or subsequent reinitiation. One or more of these five features may be responsible for subtle differences between Moraten and Schwarz and the underattenuated Rubeovax strain that helps account for the difference in attenuation levels.
ACKNOWLEDGMENTS
C.L.P. and R.A.L. contributed equally to this work.
We thank William Bellini and Paul Rota (Centers for Disease Control) for providing MV vaccine strains. We also thank Judy Beeler (Center for Biologics Evaluation and Research, FDA) for providing a low-passage wt virus. We also appreciate Martin Billeter's careful review of the manuscript.
The initial stages of these studies were supported by NIH grant AI35286 to S.A.U.
REFERENCES
- 1.Alkhatib G, Massie B, Briedis D J. Expression of bicistronic measles virus P/C mRNA by using hybrid adenoviruses: levels of C protein synthesized in vivo are unaffected by the presence or absence of the upstream P initiator codon. J Virol. 1988;62:4059–4069. doi: 10.1128/jvi.62.11.4059-4069.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumberg B M, Chan J, Udem S A. Function of paramyxovirus 3′ and 5′ end sequences. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 235–247. [Google Scholar]
- 3.Bousse T, Takimoto T, Murti K G, Portner A. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology. 1997;232:44–52. doi: 10.1006/viro.1997.8524. [DOI] [PubMed] [Google Scholar]
- 4.Castaneda S J, Wong T C. Measles virus synthesizes both leaderless and leader containing polyadenylated RNAs in vivo. J Virol. 1989;63:2977–2986. doi: 10.1128/jvi.63.7.2977-2986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cathomen T, Buchholz C J, Spielhofer P, Cattaneo R. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology. 1995;214:628–632. doi: 10.1006/viro.1995.0075. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo R, Kaelin K, Baczko K, Billeter M A. Measles virus editing provides an additional cysteine-rich protein. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- 7.Collins P L, Whitehead S S, Bukreyev A, Fearns R, Teng M N, Juhasz K, Chanock R M, Murphy B R. Rational design of live-attenuated recombinant vaccine virus for human respiratory syncytial virus by reverse genetics. Adv Virus Res. 1999;54:423–451. doi: 10.1016/s0065-3527(08)60374-7. [DOI] [PubMed] [Google Scholar]
- 8.Crowley J C, Dowling P C, Menonna J, Silverman J I, Schuback D, Cook S D, Blumberg B M. Sequence variability and function of measles virus 3′ and 5′ ends and intercistronic regions. Virology. 1988;164:498–506. doi: 10.1016/0042-6822(88)90564-8. [DOI] [PubMed] [Google Scholar]
- 9.Cutts F T, Markowitz L E. Successes and failures in measles control. J Infect Dis. 1994;170:S32–S41. doi: 10.1093/infdis/170.supplement_1.s32. [DOI] [PubMed] [Google Scholar]
- 10.Enders J F, Katz S L, Milovanovic M L, Holloway A. Studies of an attenuated measles virus vaccine. N Engl J Med. 1960;263:153–159. doi: 10.1056/NEJM196007282630401. [DOI] [PubMed] [Google Scholar]
- 11.Enders J F, Peebles T C. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med. 1954;86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 12.Escoffier C, Mainie S, Vincent S, Muller C P, Billeter M A, Gerlier D. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J Virol. 1999;73:1695–1698. doi: 10.1128/jvi.73.2.1695-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans S A, Belsham G J, Barrett T. The role of the 5′ nontranslated regions of the fusion protein mRNAs of canine distemper virus and rinderpest virus. Virology. 1990;177:317–323. doi: 10.1016/0042-6822(90)90486-b. [DOI] [PubMed] [Google Scholar]
- 14.Firestone C Y, Whitehead S S, Collins P L, Murphy B R, Crowe J E., Jr Nucleotide sequence analysis of the respiratory syncytial virus subgroup A cold-passaged (cp) temperature sensitive (ts) cpts-248/404 live attenuated virus vaccine candidate. Virology. 1996;225:419–422. doi: 10.1006/viro.1996.0618. [DOI] [PubMed] [Google Scholar]
- 15.Gershon A A. Measles virus. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. Vol. 2. New York, N.Y: Churchill Livingstone; 1995. pp. 1519–1526. [Google Scholar]
- 16.Griffin D E, Bellini W J. Measles virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1267–1312. [Google Scholar]
- 17.Griffin H G, Griffin A M. DNA sequencing: recent innovations and future trends. Appl Biochem Biotech. 1993;38:147–159. doi: 10.1007/BF02916418. [DOI] [PubMed] [Google Scholar]
- 18.Hilleman M R, Buynak E B, Weibel R E, Stokes J, Jr, Whitman J E, Leagus M B. Development and evaluation of the Moraten measles virus vaccine. JAMA. 1968;206:587–590. [PubMed] [Google Scholar]
- 19.Horikami S M, Moyer S A. Structure, transcription, and replication of measles virus. Curr Top Microbiol Immunol. 1995;191:35–50. doi: 10.1007/978-3-642-78621-1_3. [DOI] [PubMed] [Google Scholar]
- 20.Horikami S M, Moyer S A. Synthesis of leader RNA and editing of the P mRNA during transcription by purified measles virus. J Virol. 1991;65:5342–5347. doi: 10.1128/jvi.65.10.5342-5347.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikic D, Juzbasic M, Beck M, Hrabar A, Cimbur-Schreiber T. Attenuation and characterization of Edmonston-Zagreb measles virus. Ann Immunol Hung. 1972;16:175–181. [PubMed] [Google Scholar]
- 22.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 23.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. The scanning model of translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretz K, Callen W, Hedden V. Cycle sequencing. Genome Res. 1994;3:S101–S112. doi: 10.1101/gr.3.5.s107. [DOI] [PubMed] [Google Scholar]
- 26.Krugman S, Giles J P, Jacobs A M, Friedman H. Studies with a further attenuated live measles-virus vaccine. Pediatrics. 1963;31:919–928. [PubMed] [Google Scholar]
- 27.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 28.Leppert M, Rittenhouse L, Perrault J, Summers D F, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 29.Lewin B. Genes VI. New York, N.Y: Oxford University Press; 1997. [Google Scholar]
- 30.Makino S. Development and characteristics of live AIK-C measles virus vaccine: a brief report. Rev Infect Dis. 1983;5:504–505. doi: 10.1093/clinids/5.3.504. [DOI] [PubMed] [Google Scholar]
- 31.Mrkic B, Odermatt B, Klein M A, Billeter M A, Pavolic J, Cattaneo R. Lymphatic dissemination and comparative pathology of recombinant measles viruses in genetically modified mice. J Virol. 2000;74:1364–1372. doi: 10.1128/jvi.74.3.1364-1372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy F A. Virus taxonomy. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 15–57. [Google Scholar]
- 33.Parks C L, Lerch R A, Walpita P, Wang H-P, Sidhu M S, Udem S A. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J Virol. 2000;75:910–920. doi: 10.1128/JVI.75.2.910-920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson J B, Thomas D, Lewicki H, Billeter M A, Oldstone M B A. V and C proteins of measles virus function as virulence factors in vivo. Virology. 2000;267:80–89. doi: 10.1006/viro.1999.0118. [DOI] [PubMed] [Google Scholar]
- 35.Radecke F, Billeter M A. Reverse genetics meets the nonsegmented negative-strand RNA viruses. Rev Med Virol. 1997;7:49–63. doi: 10.1002/(sici)1099-1654(199704)7:1<49::aid-rmv181>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 36.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rassa J C, Parks G D. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of paramyxovirus SV5. Virology. 1998;247:274–286. doi: 10.1006/viro.1998.9266. [DOI] [PubMed] [Google Scholar]
- 38.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2296. [Google Scholar]
- 39.Rota J S, Wang Z-D, Rota P A, Bellini W J. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 1994;31:317–330. doi: 10.1016/0168-1702(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz A J F. Preliminary tests of a highly attenuated measles vaccine. Am J Dis Child. 1962;103:216–219. doi: 10.1001/archpedi.1962.02080020398042. [DOI] [PubMed] [Google Scholar]
- 41.Sedlmeier R, Neubert W J. The replicative complex of paramyxoviruses: structure and function. Adv Virus Res. 1998;50:101–139. doi: 10.1016/s0065-3527(08)60807-6. [DOI] [PubMed] [Google Scholar]
- 42.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T O, Melnick J L, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2111–2148. [Google Scholar]
- 43.Sidhu M S, Chan J, Kaelin K, Spielhofer P, Radecke F, Schneider H, Masurekar M, Dowling P C, Billeter M A, Udem S A. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology. 1995;208:800–807. doi: 10.1006/viro.1995.1215. [DOI] [PubMed] [Google Scholar]
- 44.Sidhu M S, Crowley J, Lowenthal A, Karcher D, Menonna J, Cook S, Udem S, Dowling P. Defective measles virus in human subacute sclerosing panencephalitis brain. Virology. 1994;202:631–641. doi: 10.1006/viro.1994.1384. [DOI] [PubMed] [Google Scholar]
- 45.Skiadopoulos M H, Durbin A P, Tatem J M, Wu S-L, Paschalis M, Tao T, Collins P L, Murphy B R. Three amino acid substitutions in the L protein of the human parainfluenza virus type 3 cp45 live attenuated vaccine candidate contribute to its temperature-sensitive and attenuation phenotype. J Virol. 1998;72:1762–1768. doi: 10.1128/jvi.72.3.1762-1768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skiadopoulos M H, Surman S, Tatem J M, Paschalis M, Wu S-L, Udem S A, Durbin A P, Collins P L, Murphy B R. Identification of mutations contributing to the temperature-sensitive, cold-adapted, and attenuated phenotypes of live-attenuated cold-passage 45 (cp45) human parainfluenza virus 3 candidate vaccine. J Virol. 1999;73:1374–1381. doi: 10.1128/jvi.73.2.1374-1381.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spriggs M K, Collins P L. Human parainfluenza virus type 3: messenger RNAs, polypeptide coding assignments, intergenic sequences and genetic map. J Virol. 1986;59:646–654. doi: 10.1128/jvi.59.3.646-654.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stokes A, Tierney E L, Sarris C M, Murphy B R, Hall S L. The complete nucleotide sequence of two cold-adapted, temperature-sensitive attenuated mutant vaccine viruses (cp12 and cp45) derived from the JS strain of human parainfluenza virus type 3 (PIV3) Virus Res. 1993;30:43–52. doi: 10.1016/0168-1702(93)90014-e. [DOI] [PubMed] [Google Scholar]
- 49.Suryanarayana K, Baczko K, ter Meulen V, Wagner R R. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J Virol. 1994;68:1532–1543. doi: 10.1128/jvi.68.3.1532-1543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda M, Kato A, Kobune F, Sakata H, Li Y, Shioda T, Sakai Y, Asakawa M, Nagai Y. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J Virol. 1998;72:8690–8696. doi: 10.1128/jvi.72.11.8690-8696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi K, Tanabayashi K, Hishiyama M, Yamada A. The mumps virus SH protein is a membrane protein not essential for virus growth. Virology. 1996;225:156–162. doi: 10.1006/viro.1996.0583. [DOI] [PubMed] [Google Scholar]
- 52.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tober C, Seufert M, Schneider H, Billeter M A, Johnston I C D, Niewiesk S, ter Meulen V, Schneider-Schaulies S. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J Virol. 1998;72:8124–8132. doi: 10.1128/jvi.72.10.8124-8132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsurudome M, Bando H, Kawano M, Matsumura H, Komada H, Nishio M, Ito Y. Transcripts of simian virus 41 (SV41) matrix gene are exclusively discistronic with the fusion gene which is also transcribed as a monocistron. Virology. 1991;184:93–100. doi: 10.1016/0042-6822(91)90825-v. [DOI] [PubMed] [Google Scholar]
- 55.Valsamakis A, Schneider H, Auwaerter P G, Kaneshima H, Billeter M A, Griffin D E. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J Virol. 1998;72:7754–7761. doi: 10.1128/jvi.72.10.7754-7761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitehead S S, Firestone C-Y, Collins P L, Murphy B R. A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of temperature-sensitive and attenuation phenotypes. Virology. 1998;247:232–239. doi: 10.1006/viro.1998.9248. [DOI] [PubMed] [Google Scholar]
- 57.Whitehead S S, Firestone C Y, Karron R A, Crowe J E, Elkins W R, Collins P L, Murphy B R. Addition of a missense mutation present in the L gene of respiratory syncytial virus (RSV) cpts530/1030 to RSV vaccine candidate cpts248/404 increases its attenuation and temperature sensitivity. J Virol. 1999;73:871–877. doi: 10.1128/jvi.73.2.871-877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehead S S, Juhasz K, Firestone C Y, Collins P L, Murphy B R. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wild T F, Buckland R. Functional aspects of envelope-associated measles virus proteins. Curr Top Microbiol Immunol. 1995;191:51–64. doi: 10.1007/978-3-642-78621-1_4. [DOI] [PubMed] [Google Scholar]
- 60.Wong T C, Hirano A. Structure and function of a bicistronic RNA encoding phosphoprotein and matrix protein of measles virus. J Virol. 1987;61:584–589. doi: 10.1128/jvi.61.2.584-589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]