Abstract
We have studied coxsackievirus A9 (CAV9) mutants that each have a single amino acid substitution in the conserved 29-PALTAVETGHT-39 motif of VP1 and a reduced capacity to produce infectious progeny virus. After uncoating, all steps in the infection cycle occurred according to the same kinetics as and similar efficiency to the wild-type virus. However, the particle/infectious unit ratio in the progeny was significantly increased. The differences were apparently due to altered stability of the capsid: there were mutant viruses with enhanced or hindered uncoating, and both of these characteristics were found to reduce fitness under standard passaging conditions. At 32°C the instable mutants had an advantage, while the wild-type and the most stable mutant grew poorly. When comparing the newly published CAV9 structure and the other enterovirus structures, we found that the PALTAVETGHT motif is always in exactly the same position, in a cavity formed by the 3 other capsid proteins, with the C terminus of VP4 between this motif and the RNA. In the 7 enterovirus structures determined to date, the most conserved residues of the studied motif have identical contacts to neighboring residues of VP2, VP3, and VP4. We conclude that (i) the mutations affect the uncoating step necessary for infection, resulting in an untimely or hindered externalization of the VP1 N terminus together with the VP4, and (ii) the reason for the studied motif being evolutionarily conserved is its role in maintaining an optimal balance between the protective stability and the functional flexibility of the capsid.
Coxsackievirus A9 (CAV9) is a member of the Enterovirus genus in the family Picornaviridae. Enteroviruses have icosahedral capsids that consist of 60 copies of each of the four capsid proteins VP1 to VP4. Their genomes are single molecules of positive-stranded RNA, approximately 7,500 nucleotides in length.
In order to initiate a new infection cycle, enteroviruses have to attach to the host cell, uncoat to reveal the RNA, and at least the RNA must cross a membrane to enter the cell cytoplasm. For CAV9 there are at least two alternative receptors, the integrin αvβ3 (25, 27), and an unidentified cell surface receptor (17, 28). Several other specific receptors are known for various enteroviruses, but the mechanism and location of crossing the membrane are still unclear. It is known that the A particles, altered virus particles lacking the VP4 and having the N terminus of VP1 externalized, of poliovirus type 1 (PV1) are capable of binding to liposomes through the N terminus of VP1 (12). Furthermore, A particles as well as native polioviruses induce channel formation in an artificial membrane (30). The N terminus of VP1 might thus form a pore structure in one of the cell membranes, possibly together with VP4, allowing the RNA to be externalized from the capsid and enter the cytoplasm (12). The pseudoatomic models of the poliovirus 135S and 80S particles were recently determined by cryo-electron microscopy and image reconstruction (5). Based on these structures, the model was revised to include a possible “float valve” in the form of the VP3 plug that was found to be at the fivefold axis in the 135S particle as well as in the 160S particle.
The N terminus of VP1 has been shown to be reversibly externalized from the capsid, being recognized in the native poliovirus by peptide antibodies (26). It has also been reported that unidentified parts of VP4 are similarly recognized by antibodies, but only when coincubating the poliovirus and antisera at 37°C (20). This dynamic nature of the capsid also explains the temperature-dependent formation of channels upon incubating native poliovirus with artificial lipid bilayers (30). The mechanisms of these transient modifications are still not known. Likewise, the location of externalization of the VP4 and the N terminus of VP1 still remains open. Two recent studies give support to two different models, assuming externalization to occur either near the base of the canyon, judging by the structures of the poliovirus 135S and 80S particles (5), or through the fivefold axis, as judged on the basis of the CAV9 structure (14).
In order to gain more understanding about the uncoating process of enteroviruses, we decided to study the effects of mutations in a highly conserved motif of approximately 11 amino acids that is in the externalized N terminus of the capsid protein VP1 in all enteroviruses (16). This region contains several amino acids that were found to be completely conserved in an alignment of 79 different enterovirus sequences (2). Due to the strong conservation of the studied motif among enteroviruses, CAV9 can be considered to serve here as a model virus: we presume that the CAV9 characteristics found in this study should reflect the properties of other enteroviruses as well.
MATERIALS AND METHODS
Virus strains and cell cultures.
We have used CAV9 mutants that were generated earlier (1, 2), originally derived from an infectious cDNA clone obtained from Glyn Stanway (University of Essex, Colchester, United Kingdom) (6, 17). The viruses were propagated in the monkey kidney cell line LLC Mk2, which was maintained in the minimal essential medium of Eagle, supplemented with 10% fetal calf serum.
Buffers.
The following buffers, with compositions as indicated, were used: phosphate-buffered saline (PBS), pH 7.3; DPBS, PBS containing 1 mM MgCl2 and 1 mM CaCl2; blocking buffer, PBS containing 2% bovine serum albumin (BSA), 30 mM MgCl2, and 1 mM CaCl2; lysis buffer, 0.5 % Triton X-100 in DPBS containing 0.1% BSA; RSB, 10 mM Tris, 10 mM NaCl, 1.5 mM MgCl2 (pH 7.4); washing buffer 1, 1% Triton X-100 in Tris-HCl (pH 7.4), 5 mM EDTA, 500 mM NaCl, 5 mM sucrose; and washing buffer 2, Tris-HCl (pH 6.8), 1 mM EDTA.
RNA purification, RT-PCR, and sequencing.
The whole capsid coding region was sequenced from all viruses. The viral RNA was purified using the Qiagen RNeasy kit (Qiagen Ltd., Crawley, West Sussex, United Kingdom). Reverse transcriptase PCR (RT-PCR) was performed by using either the RobusT RT-PCR kit (Finnzymes, Espoo, Finland) or a combination of avian myeloblastosis virus reverse transcriptase (Promega) and Dynazyme (Finnzymes). The amplicons were sequenced with an ABI sequencer, using the PCR primers. The primers are shown in Table 1.
TABLE 1.
Primers used in PCR and sequencinga
| Primer | Position | Primer sequence (5′ to 3′) |
|---|---|---|
| 91856 | 641–676 | CGGTGAGCAACAGAGCTATTGTGTATCTGTTTACTG |
| 91857 | 1347–1323 | CCATCTCAGCCTCGGGCACACACAC |
| 92066 | 1230–1263 | CAGAACATGCAATACCACTACCTGGGTCGTGCAG |
| 92067 | 1892–1860 | CACGACCGAGTCCACTTCTGCTATTTCCATTAG |
| 91860 | 1801–1829 | GGCCTTTGCCACAATTTGATGTGACGCCT |
| 91861 | 2481–2451 | CAACTGCCCTCTCAATGGCTTCTTCCACAT |
| 7625 | 2379–2410 | GCGTGCAACGACTTCTCAGTAAGAATGTTGAG |
| 9354 | 3043–3012 | CACCCGTCATAAAAATTGCTGTATGCATTTCC |
| 91862 | 2903–2946 | GATTCCAGCAAAAGTTGATGATTATGCCTGGCAG |
| 91863 | 3436–3408 | CAATTTTGCCAATCCGTGTGTGTTG |
Numbering of the nucleotide positions is according to Chang and coworkers (6).
Infection kinetics.
The kinetics of the production of infectious virus was measured in 24-well plates of LLC Mk2 cells. The cells were infected with equal multiplicities of infection (MOIs) of the various mutant viruses and the wild-type virus. The inoculum was removed after incubating 1 h at room temperature. The cells were washed twice and covered with 1 ml of medium containing 1% fetal calf serum. The infection was allowed to proceed at 37°C, and the cells were scraped from two wells of each virus at 2-h intervals. The cells were freeze-thawed three times together with the medium, and cell debris was removed by centrifuging 10 min in an Eppendorf centrifuge. Viral titers were measured from the supernatants by the end-point dilution method.
Attachment to cells.
LLC Mk2 cells on 24-well plates were washed with DPBS and then incubated for 2 h under 50 μl of blocking buffer. Purified 35S-labeled virus was added in 40 μl of blocking buffer, in four parallel wells for each virus. The plates were incubated for 12 h at 4°C and then washed twice with DPBS. The cells were lysed with 100 μl of 0.3 N NaOH and agitated for 30 min at 37°C. Radioactivity was measured from these samples in a MicroBeta counter (Wallac, Espoo, Finland), using 50-μl samples and 200 μl of Optiphase Supermix SC/9235/21 scintillation liquid (Wallac).
Uncoating kinetics.
LLC Mk2 cells on six-well plates were washed with DPBS. About 106 cpm of labeled virus (MOI, approximately 1) was incubated on the cells for 1 h at 18°C, after which the inoculum was removed, the cells were washed twice with DPBS, and 2 ml of prewarmed (37°C) medium was added. The infection was allowed to proceed for 0 to 3 h at 37°C, and the plates were moved onto ice. The medium was collected, and the cells were washed twice with cold DPBS. The cells were lysed by adding 200 μl of lysis buffer, and cell debris was removed by centrifuging for 2 min in an Eppendorf centrifuge. Sodium dodecyl sulfate (SDS) was added in the supernatant to a final concentration of 0.2%, and sedimentation of the virus particles was analyzed by centrifuging on a 15 to 30% sucrose gradient at 38,000 rpm for 1 h 40 min (Beckman SW40 rotor; 18°C). Fractions of 200 μl were collected from the top, and the radioactivity was measured as described above.
Spontaneous disintegration of viruses.
Purified 35S-labeled virus was diluted in minimal essential medium containing 1% fetal calf serum to a final volume of 270 μl, and the samples were incubated for either 0, 1, 3, 6, 24, or 72 h at 37°C. From the samples, 20 μl was used in measuring the viral titers, and 250 μl was used to assess the different particle forms by centrifuging on a 15 to 30% sucrose gradient at 38,000 rpm for 1 h 40 min (Beckman SW40 rotor; 4°C). Fractions of 270 μl were collected from the top, and the radioactivity was measured as described above.
Protein synthesis.
LLC Mk2 cells in 24-well plates were infected with standard titers of wild-type and mutant viruses. The cells were incubated for 30 min at room temperature and 30 min at 37°C, after which the virus was replaced by 1.5 ml of medium without methionine. At various time points (3, 4, 5, 6, and 7 h postinfection [hpi]), 5 μl (71.5 μCi total radioactivity) of 35S-Pro-Mix (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) was added in the medium. Infection was allowed to proceed for 30 min, after which the medium was discarded and the cells were lysed by adding 50 μl of 0.5% NP-40 in PBS. Cell debris was removed by centrifuging the samples for 5 min in an Eppendorf centrifuge. SDS was added to a final concentration of 1%. Protein production was assayed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by blotting on a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) and immunostaining with the polyclonal antibody 861 against CAV9 capsid proteins. Relative protein quantities were estimated by measuring the integrated optical densities of the bands and comparing the integrated optical density values to those from a standard curve. The reliability of immunostaining as a semiquantitative measure of protein concentrations was confirmed by comparing the autoradiography band densities with the immunostained bands.
Assembly intermediates.
LLC Mk2 cells in six-well plates were infected with wild-type and mutant viruses as described above. At 3.5 hpi, 5 μl (71.5 μCi) of 35S-Pro-Mix (Amersham) was added in the medium. At 4.5 hpi, the radioactively labeled medium was replaced by regular medium. The cells were lysed at 4.5, 5.5, or 6.5 hpi with 400 μl of cold RSB containing 0.5% NP-40. Cell debris was removed by centrifuging 5 min in an Eppendorf centrifuge. Assembly intermediates were analyzed by centrifuging the supernatants at 38,000 rpm (Beckman SW40 rotor; 4°C) either for 16 h on a 5 to 20% sucrose gradient in RSB or for 2.5 h on a 15 to 30% sucrose gradient in RSB. Fractions of 270 μl were collected from the top, and the radioactivity was measured. The protein composition of the peak fractions was analyzed by SDS-PAGE. After electrophoresis, the proteins were blotted on a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) and immunostained using the polyclonal antibody 861 against CAV9 capsid proteins.
Survival in competition.
We carried out a competition study of six mutants and the wild-type virus by performing five blind passages of a mixture of the viruses on six-well plates of LLC Mk2 cells. In addition to the mutants described in detail in this article, the mixture contained the mutant A30N, initially characterized as having similar properties to those of V34A. The experiment was begun by infecting the cells simultaneously with an MOI of 0.1 for each of the viruses. Infection was allowed to proceed in four parallel wells at 37 or 32°C until full cytopathogenic effect was seen after 2 or 3 days. The medium was collected and centrifuged in an Eppendorf centrifuge to remove cells and cell debris, and a 1:100 dilution of the supernatant was used to infect the next set of cells. After five passages, plaque titration was performed, and well-isolated plaques were collected in 2 μl of medium. The sequences in the mutated region were determined as described above, using primers 7625 and 9354.
Immunoprecipitation.
Radioactively labeled virus samples (10 μl; 10 000 cpm; [35S]methionine) were mixed with 10 μl of peptide antiserum 910 (26), either undiluted or in 10−1 or 10−2 dilutions. The mixture was agitated for 2 h at room temperature, after which 1 ml of Pansorbin (Calbiochem, San Diego, Calif.) (10−1 dilution in DPBS containing 0.1% BSA) was added, and agitation was continued for 30 min at room temperature. The precipitate was removed by 1 min of centrifugation in an Eppendorf centrifuge. The unprecipitated radioactivity was measured from 50 μl of supernatant as described above. The precipitate was washed twice with washing buffer 1 and once with washing buffer 2, and it was dissolved in 70 μl of 2% SDS–2% β-mercaptoethanol and heated for 5 min at 100°C. It was centrifuged for 1 min in an Eppendorf centrifuge, and radioactivity was measured from the supernatant as described above.
Molecular graphics.
In visualizing the molecular structures, we used the RasMol 2.6 Molecular Visualisation Program by Roger Sayle, Glaxo Wellcome Research and Development, Stevenage, Hertfordshire, United Kingdom.
RESULTS
From a larger set of mutants, we selected five mutants that covered as wide a range of properties as possible, judging from the preliminary characterization. These mutants were then studied in more detail in comparison with the wild-type CAV9. Each of the selected mutants had one of the following amino acid substitutions in VP1: A30G, A30S, T32S, V34A, and V34I. The mutant viruses are hereafter referred to by using these codes only.
Production of infectious virus.
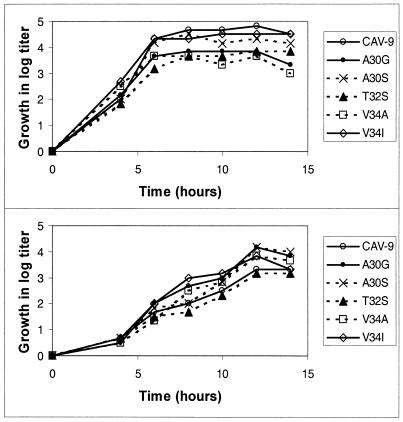
We studied the kinetics of producing infectious virus at 32 and 37°C. The results indicate that the kinetics appears identical for all the viruses (Fig. 1), reaching the plateau phase after 7 to 8 h at 37°C and after 12 h at 32°C. At 37°C, the wild-type virus produces highest amounts of virus, with the mutants A30S and V34I being almost as efficient. At 32°C, all mutants except for T32S produce more progeny virus than the wild type.
FIG. 1.
Kinetics of production of infectious progeny virus at 37°C (top) and 32°C (bottom). To clarify the picture, the growth curves are shown as increase in titers after the attachment and washing steps.
Efficiency of attachment to cells.
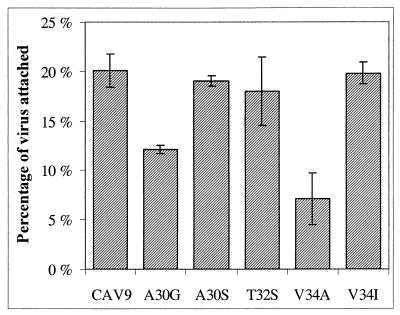
The wild-type CAV9 was found to be most efficient in attaching to LLC cells, but A30S, T32S, and V34I were almost as effective (Fig. 2). These differences are within the error margin. In each of these cases, 18 to 20 % of the radioactive label bound to cells. In contrast to these viruses, only 12% of A30G and 7% of V34A attached to the cells.
FIG. 2.
Efficiency of attachment of CAV9 mutants to LLC Mk2 cells. The results shown are averages from four parallel experiments, and error bars indicate standard deviations.
Kinetics of formation of subviral particles.
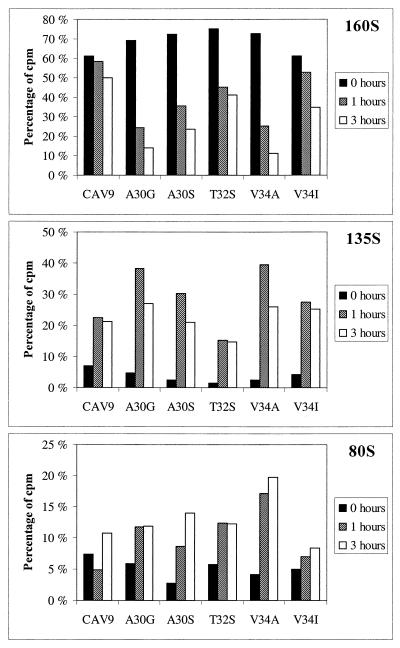
The irreversible shift from the native virion sedimenting at 160S to the altered virus particle sedimenting at 135S is a measure that is frequently used in assaying the uncoating of enteroviruses, although the role of the 135S particle as an intermediate in infection has been questioned (9). We assayed the relative amounts of 160S, 135S, and 80S particles after incubating cell-bound virus with cells at 37°C for 0, 1, or 3 h. Formation of subviral particles was clearly faster for A30G and V34A, compared to the other mutants or the wild-type, and the alteration to 135S particles was almost quantitative already after 1 h (Fig. 3). Alteration of A30S and V34I was somewhat more efficient than that of the wild-type, while T32S showed a reduced rate of alteration. In none of the cases was faster uncoating found to accelerate the subsequent steps in infection.
FIG. 3.
Sedimentation analysis of CAV9 uncoating after incubating cell-bound virus for 0, 1, or 3 h at 37°C. Prior to this incubation, 35S-labeled viruses (about 2.5 × 106 cpm) were allowed to adsorb to cells for 2 h at 18°C. Percentages of total label forming each peak (160S, 135S, and 80S) are shown.
Spontaneous disintegration of virions.
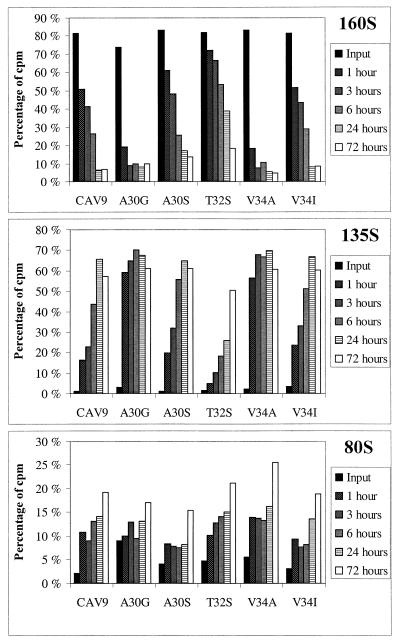
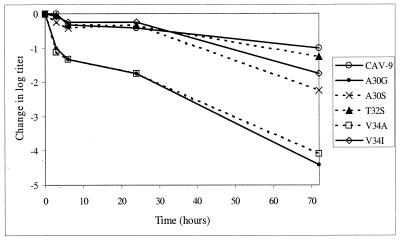
Two of the mutant viruses, A30G and V34A, turned out to be extremely unstable. Already after an incubation of 1 h at 37°C in the regular growth medium, the majority of these virus particles were sedimenting at 135S instead of 160S. Two other mutants, A30S and V34I, were only slightly less stable than the wild-type virus. The 160S particles of these three viruses had half-lives of approximately 3 to 4 h. In contrast, T32S turned out to be significantly more stable, having a half-life of almost 24 h. These results are shown in Fig. 4. The disintegration of the virus particles was also seen as a decrease in viral titers, as is shown in Fig. 5. Differences seen between A30G and V34A are negligible, as are also differences between the rest of the viruses before the final time point at 72 h. It appears that for all the viruses, the decrease in infectious units occurs in three separate phases. The first phase is fast, occurring during the first 3 or 6 h. After this point, the decrease is significantly slower until at least the time point of 24 h, being accelerated again before the time point of 72 h. It was surprising that the wild-type virus was found to disintegrate spontaneously already in a few hours.
FIG. 4.
Sedimentation analysis of spontaneous formation of subviral particles in medium. Percentages of total label as in the legend to Fig. 3 are shown.
FIG. 5.
Decrease of viral titers after incubation of 3, 6, 24, or 72 h in regular medium at 37°C. The results shown are averages of four parallel dilution series, comprising a total of 24 cell wells per dilution.
Particle/infectious unit ratio.
We used purified viruses, sedimenting at 160S, with known relative protein contents that were adjusted on the standard curve (Materials and Methods) for maximal accuracy of concentration determination. Aliquots of the samples were used in determining the amounts of VP1, as described in Materials and Methods, and in measuring the viral titers. Relative ratios of virus particle to infectious unit were calculated from these two parameters. All the mutants were found to have higher particle/infectious unit ratios than the wild-type. A30S, T32S, and V34I were moderately reduced, whereas A30G and V34A had only 2 and 4% of the infectivity of the wild-type (Table 2). The numbers given are unavoidably imprecise due to the inaccuracies in both methods used in quantitation.
TABLE 2.
Relationships between infectious units and the amount of virus particles
| Virus | Relative amount of VP1 | Log titer/50 μl | Infectious units per virion compared to wild-type (%) |
|---|---|---|---|
| CAV9 | 1 | 8.8 | 100 |
| A30G | 2.0 | 7.5 | 2 |
| A30S | 1.7 | 8.7 | 40 |
| T32S | 0.77 | 8.3 | 40 |
| V34A | 1.7 | 7.7 | 4 |
| V34I | 1.2 | 8.7 | 60 |
Survival in competition.
When the viruses had to compete against each others, three different viruses were found to survive at both temperatures. At 37°C, when sequencing 45 plaques isolated after five passages of four parallel passaging chains, the wild-type was found to survive in addition to A30S and V34I. These viruses also had the largest plaque sizes, and the stabilities of the two mutants were similar to that of the wild-type. At 32°C, the wild-type virus was not found among the 45 viruses recovered, but rather only mutants A30G, A30N, and A30S were found. The last one of these is thus the only one surviving at both temperatures, presumably due to properties that are close to wild type, but with the capsid being slightly less stable. These results are completely in accordance with the kinetics of producing infectious virus (Fig. 1).
Plaque phenotypes.
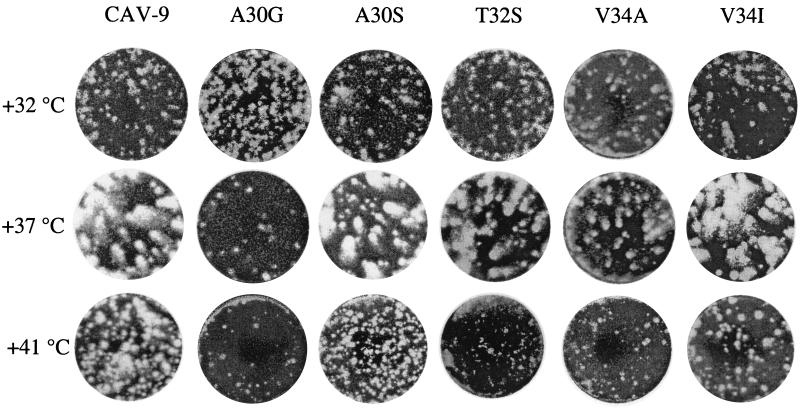
Plaque assays were performed at 32, 37, and 41°C. The plaque phenotypes are shown in Fig. 6. At the two higher temperatures, the wild-type virus had the largest plaque sizes, whereas at 32°C it was among the poorest ones. The two unstable mutants, A30G and V34A, had a clear advantage at the decreased temperature, having very small plaque sizes at 37 and 41°C. The intermediate ones, A30S and V34I, had only slightly reduced plaque sizes at both these temperatures, while at 32°C they had some advantage over the wild-type. The most surprising plaque phenotype was seen in the T32S mutant. It had moderately reduced plaque sizes at 37°C but very small plaques at both 32 and 41°C, indicating a very narrow optimum temperature for infection.
FIG. 6.
Plaque phenotypes at various temperatures.
Immunoprecipitation.
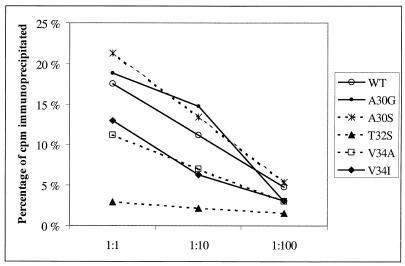
To study whether the mobility of the N-terminal extension was changed due to the mutations, we performed an immunoprecipitation experiment using antiserum 910, which is a peptide antiserum raised against the peptide KEVPALTAVETGATC (26). This antiserum has been shown to recognize peptides corresponding to the CAV9 VP1 sequence ASVPALTAVETGHT and single amino acid modifications of this peptide, corresponding to mutations A30G, A30S, T32S, and V34A (Taina Härkönen, unpublished results). The peptide corresponding to V34I has not been studied. Both of the Ala30 mutants were precipitated by the antiserum 910 similarly or even better than the wild-type CAV9 (Fig. 7). Both of the Val34 mutants were also precipitated by this antiserum, but to a lower degree than the wild type. In contrast, precipitation of the T32S mutant was nearly abolished.
FIG. 7.
Immunoprecipitation of CAV9 and mutants using three dilutions of peptide antiserum 910.
Unaltered properties.
We found no differences in the time scale of infection, protein synthesis, accumulation of assembly intermediates, or release from cells. In all cases, measurable concentrations of protein were synthesized within 3 to 4 h pi, and the peak concentrations were reached after 7 h. There were no differences in the accumulation patterns of particles sedimenting at approximately 15S, 80S, or 160S (presumably pentamers, empty capsids, and mature virions, respectively), and PAGE analysis showed that maturation cleavage occurs normally. Release of virus from cells was unaltered, judging by viral titers inside or outside cells at various time points. Maximum titers were reached at approximately 7 hpi (in cells) or 9 hpi (in media).
DISCUSSION
We have shown that mutations in the conserved PALTAVETGHT region of the capsid protein VP1 of CAV9 result in defects in producing infectious progeny virus. The differences between the viruses were found in six different parameters: (i) attachment to cells, (ii) uncoating, (iii) stability of the virions, (iv) particle/infectious unit ratio, (v) production of infectious progeny virus, and (vi) immunoprecipitation. All these phenomena can be explained by the observed variation in capsid stability.
Stability and uncoating.
The particle/infectious unit ratios and the capability of the viruses to produce infectious progeny were in complete accordance with the differences observed in stability, allowing the conclusion that nonspecific, and thereby nonproductive, conversion to the 135S form of the mutant viruses resulted in accumulation of noninfectious particles (most notably, A30G and V34A) or hindered initiation of infection (T32S). The two opposing effects, the uncoating that is triggered either too easily or ineffectively, reflect the two requirements that the virus capsid must meet: protection of the RNA and the capacity to uncoat upon binding to a susceptible cell.
The stability of T32S appears to reflect a decrease in mobility of the N-terminal extension of VP1. Unlike the other mutants, the amino acid sequence of this mutant was recognized only as a peptide but not in a virus sample, which suggests, but does not prove, that the studied motif is less dynamic in T32S than it is in the wild-type virion. An alternative explanation is that the epitope is recognized as a peptide, but not in the conformation seen in the virus. The other studied mutant sequences were recognized both as peptides and in virus samples. It is noteworthy that the enhanced uncoating did not give any measurable advantage to any of the mutant viruses, even though the 160S forms could be purified and three of the five mutants attached to cells to a similar degree with the wild-type virus. It therefore appears that although the mutants are capable of correct uncoating, they perform it in a hasty and nonspecific manner, many of the virions being irreversibly converted into 135S particles before getting the correct signal to initiate uncoating, thereby becoming noninfectious. The fact that a considerable proportion of virions are still found in the native form after 3 h of incubation at 37°C after binding to cell demonstrates the high particle/infectious unit ratio that is common to enteroviruses and suggests that a significant proportion of the bound viruses may have nonspecific interactions with the cell. This would be consistent with the finding that the proportion of the unaltered 160S form becomes clearly smaller for the labile mutants both in the presence and in the absence of cells (Fig. 3 and 4). It appears that uncoating is efficiently triggered upon meeting the correct conditions, which is not merely binding to cells, and easing the uncoating step per se will not necessarily give any advantage to the virus.
Differences in the attachment step are unlikely to explain the altered properties of the mutants. Attachment to cells was very close to equal for all viruses except for the two highly unstable mutants, A30G and V34A. On the other hand, it is known that 135S particles of poliovirus are incapable of or extremely inefficient at (8) binding to cells and initiating infection. Moreover, the differences in attachment efficiencies were merely threefold at most, whereas differences in infectivity were more than 1 log. It therefore seems safe to assume that differences in attachment to cells did not have a major role in the defects found in the mutants.
Role and topology of the VP1 N terminal extension.
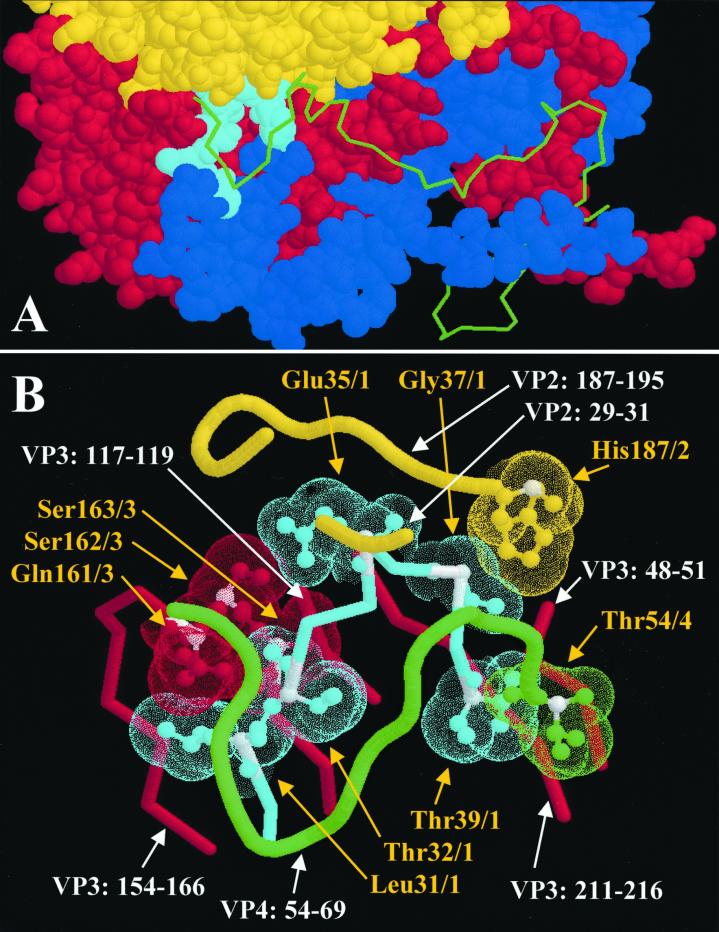
In all enteroviruses, the whole VP1 N terminal extension is an integral part of the stabilizing network of intraprotomer, intrapentamer, and interpentamer interactions that occur on the inner surface of the capsid. In the poliovirus empty capsid (VP0, VP1, VP3), the N-terminal 67 amino acids of VP1 are disordered (4). The stabilizing network, formed as a consequence of the maturation cleavage and/or the presence of RNA, then serves by locking the capsid into the stable form. The three-dimensional structure of CAV9 was recently published (14), including a detailed model of the VP1 N terminus. The N-terminal 12 amino acids form a finger pointing towards the base of the fivefold axis. At position His13, the chain comes into close contact with Ser58 of the same chain. Together the residues 13 to 58 form a loop structure that extends towards the threefold axis facing the interior of the virion, with residues Pro29 to Thr47 forming a hook-like structure, diving into a cavity formed by the three other capsid proteins. The base of the cavity is formed by the BIDG β-sheet of the VP3, the tip of the cavity is formed by the VP2, and it is loosely covered by the C-terminal 16 amino acids of the VP4. This structure repeats in the seven known enterovirus structures and the four rhinovirus structures, with the exceptions that in rhinoviruses 1A and 16, the parts of VP2 and VP4 that could cover the hook are not seen in the structure. The exceptions may reflect the conformational flexibility of the capsid that is required in externalization of the VP4 and the N terminus of VP1. In the hook region, 7 of 24 amino acids (Pro29, Leu31, Ala33, Glu35, Gly37, Thr50, and Arg51) showed no variation in an alignment of 79 different enterovirus sequences, and when trying to mutate Leu31, we could recover no viable mutant viruses, except for the ones with different leucine codons (2). Five of the completely conserved amino acids are in the PALTAVETGHT motif, which is the region covered by VP4 at the tip of the hook.
Conserved intraprotomer contact points.
We mapped the close contacts that the PALTAVETGHT motif has with the other capsid proteins (connected Van der Waals surfaces) and found that there are several contact points that are identical in all known enterovirus structures (10, 11, 14, 15, 19, 24, 29), and these are mostly conserved in rhinoviruses as well (3, 13, 18, 31). In CAV9, these contact points are at the following positions: VP1/Leu31-VP3/Gln161, VP1/Leu31-VP3/Ser163, VP1/Thr32-VP3/Ser163, VP1/Glu35-VP3/Ser162, VP1/Gly37-VP2/His187, and VP1/Thr39-VP4/Thr54 (Fig. 8). All capsid proteins are involved in these interactions.
FIG. 8.
(A) Location of the VP1 hook in the three-dimensional structure of one protomer of CAV9, seen from the inside of the capsid. The tip of the hook dives in a cavity, the base of which is formed by the capsid proteins VP2 and VP3. The N termini of VP1, VP3, and VP4 are extended toward the fivefold axis at the bottom right. VP1, VP2, and VP3 are shown as space-filling models, while VP4 is shown as backbone only. The PALTAVETGHT motif is shown in cyan, while the rest of VP1 is blue. VP2, VP3, and VP4 are shown in yellow, red, and green, respectively. (B) A close view of the PALTAVETGHT motif and its surroundings in CAV9. Intraprotomer contact points that are completely conserved among PV1, PV2, PV3, CAV9, CBV5, echovirus 1, and BEV are shown as ball-and-stick models with dotted Van der Waals surfaces. The intertwined connection between the conserved VP3 amino acids 161 to 163 and VP1 amino acids 31 to 35 may have an important stabilizing role. Yellow text and arrows indicate the conserved amino acids, and white text and arrows indicate the amino acid chains included in this figure. Alpha carbons of conserved residues are colored white. CAV9 coordinates were from the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics (available at http://www.rcsb.org/pdb/).
In a study by Mosser and coworkers (23), the uncoating inhibitor WIN 51711 was used to select drug-resistant mutants of PV3/Sabin. Out of the 13 drug-resistant mutants, 7 were drug dependent, and these were all found to have single amino acid substitutions near Thr53 of the VP4. One of the seven mutations was alanine in place of Thr53 of the VP4, and three of them were mutations of the three amino acids surrounding, but not including, Thr50 of VP1 (Table 3).. Thr53 of VP4 and Thr50 of VP1 form one of the completely conserved contacts in enterovirus and rhinovirus structures. All the drug-dependent mutants were reported to be hyperlabile without the presence of WIN 51711, displaying half-lives of less than 1 min in the absence of drug. This resembles the two labile mutants described here, the mutants A30G and A34V, and suggests that some of the mutations in the site saturation mutagenesis study, from which our collection of mutants originate, might also have been hyperlabile and, as such, were excluded from the viable ones.
TABLE 3.
Effects of mutations in the VP1 hook region
| Mutation | Sequence surrounding mutationa | Virus | Reference | Phenotype |
|---|---|---|---|---|
| A30G | SVPALTAVETGHTSQVT | CAV9 | This study | Labile virion |
| A30S | SVPALTAVETGHTSQVT | CAV9 | This study | Somewhat labilized virion |
| T32S | SVPALTAVETGHTSQVT | CAV9 | This study | Stabilized virion |
| V34A | SVPALTAVETGHTSQVT | CAV9 | This study | Labile virion |
| V34I | SVPALTAVETGHTSQVT | CAV9 | This study | Somewhat labilized virion |
| A43V | EIPALTAVETGATNPLV | PV1 | 7 | Neurovirulence determinant (mouse) |
| E40G | ETPALTAVETGATNPLV | PV2 | 22 | Host range determinant (mouse) |
| P54S | ETPALTAVETGATNPLV | PV2 | 22 | Host range determinant (mouse) |
| A49V | EVPALTAVETGATNPLA | PV3 | 23 | Hyperlabile virion (WIN-resistant) |
| N51S | EVPALTAVETGATNPLA | PV3 | 23 | Hyperlabile virion (WIN-resistant) |
| P52S | EVPALTAVETGATNPLA | PV3 | 23 | Hyperlabile virion (WIN-resistant) |
| A54T | EVPALTAVETGATNPLA | PV3 | 21 | Suppressor of ts phenotype |
| A54V | EVPALTAVETGATNPLA | PV3 | 21 | Suppressor of ts phenotype |
Mutated position shown in boldface type.
Molecular basis for labilization or stabilization.
The methyl group of Ala30 of VP1 has hydrophobic contacts with Ile154 and Val165 of VP3. It appears that these contacts contribute to the virion stability since replacing alanine with glycine (A30G) drastically labilizes the virion. On the other hand, the additional hydroxyl group of a substitutive serine is much less detrimental to the virus (A30S). There is plenty of space for this position, and an even larger side chain could easily be fitted in the gap between VP1 and VP3. In fact, even glutamine, asparagine, and leucine have been found in this position of CAV9 (2). In PV1, a mutation at this position (A43V) was found to result in neurovirulence in mice (7). Earlier, mouse neurovirulence had also been associated with mutations that are at positions −2 and +2 (E40G, P54S) relative to the PALTAVETGAT motif in PV2 (22). In both studies, neurovirulence was suggested to be due to facilitated conformational changes during early steps of mouse nerve cell infection. Curiously, two different Ala54 mutations, A54T and A54V (at position +4 relative to PALTAVETGAT), resulted in suppression of the temperature-sensitive (ts) phenotype of PV3 (21) (Table 3).
Both methyl groups of VP1 Val34 have hydrophobic interactions with Trp189 of VP2. In addition, it has weaker contacts with VP3 amino acids Phe213, Ser163, and Thr117, and the carboxyl oxygen makes a hydrogen bond to Gly37 nitrogen. The VP1 Val34 is in a densely packed region, and any additional mass would have to replace some of the elements mentioned above. Isoleucine was the largest substitutive amino acid found at this position, and the highly labile nature of the V34I mutant may thus be due to structural constraints that distort the stabilizing network on the capsid inner surface. The alanine of the mutant V34A lacks some of the hydrophobic contacts that the wild-type valine has, which may result in the less pronounced labilization observed.
Thr32 of VP1 has a conserved contact with Ser163 of VP3. The side chain of VP1 Thr32 is on the inside of the loop forming the hook, directly facing Ser40 of the same chain. The side chain methyl of Thr32 is in contact with the Ser163 side chain, while the hydroxyl group is not close to other amino acid residues. It appears that the stabilizing effect of the T32S mutation, which removes the methyl of the side chain, might result from the hydroxyl group contacting either VP3 Ser163 or, in a more extended conformation, VP1 Ser40. Water molecules may participate in these interactions. A contact that would be parallel to the Ser32-Ser40 contact in the CAV9 mutant, is seen in bovine enterovirus (BEV). In BEV, there is a glutamine (Gln39) at the position corresponding to Thr32 of CAV9, having close interactions with the polar side chains of Ser47 (corresponding to Ser40 of CAV9) and Thr48 of the same chain. It seems plausible that this contact has been selected in its structural context to stabilize the BEV structure. The increased stability and reduced infectivity of T32S show that an optimal sequence here does not have maximal binding to the cavity, but the balance between stability and mobility must be maintained.
Conclusions.
The hook region of VP1 and the part of VP4 covering it are disordered in the poliovirus empty capsid (4). Formation of the interactions on the capsid inner surface stabilizes the capsid into the mature, metastable form and increases the thermal energy required for uncoating. It seems that the PALTAVETGHT mutants are not affected in their capability to assemble into mature virions. Instead, mutations in this motif decrease the potential energy barrier for uncoating, by preventing the capsid from descending into the minimal energy state.
In view of the conserved contacts between the PALTAVETGHT motif and the other capsid proteins, it is intriguing that this region is known to be so highly dynamic in nature. As we have shown earlier, the periodic hydrophobicity profile of the studied motif could not be changed in a site saturation mutagenesis study, as none of the viable mutants had a polar amino acid replaced by a nonpolar one, or vice versa (2). Then, the mutants that were already selected by their viability had deteriorated properties that were connected with either labilized or stabilized phenotypes. The result that a single mutation in the VP4 side of one of the conserved contact points resulted in a hyperlabile phenotype of poliovirus (23) further underlines the connection between the PALTAVETGHT motif and the cavity. We therefore suggest that the VP1 hook, being held in the VP2-VP3 depression by a VP4 cover, has a decisive role in initiating the viral uncoating.
ACKNOWLEDGMENTS
This study was supported by grants from the Academy of Finland, the Sigrid Juselius Foundation, and the Finnish National Technology Agency (TEKES).
We are grateful for the skillful technical assistance of Pekka Mäntyvaara, Anita Hartikainen, Noora Alakulppi, and Mervi Eskelinen and for expert advice from Mick Mulders and Glyn Stanway.
REFERENCES
- 1.Airaksinen A, Hovi T. Modified base compositions at degenerate positions of a mutagenic oligonucleotide enhance randomness in site-saturation mutagenesis. Nucleic Acids Res. 1998;26:576–581. doi: 10.1093/nar/26.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Airaksinen A, Roivainen M, Stanway G, Hovi T. Site-saturation mutagenesis of the PALTAVETG motif in coxsackievirus A9 capsid protein VP1 reveals evidence of conservation of a periodic hydrophobicity profile. J Gen Virol. 1999;80:1919–1927. doi: 10.1099/0022-1317-80-8-1919. [DOI] [PubMed] [Google Scholar]
- 3.Arnold E, Rossmann M G. The use of molecular-replacement phases for the refinement of the human rhinovirus 14 structure. Acta Crystallogr A. 1988;44:270–282. doi: 10.1107/s0108767387011875. [DOI] [PubMed] [Google Scholar]
- 4.Basavappa R, Syed R, Flore O, Icenogle J P, Filman D J, Hogle J M. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 1994;3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belnap D M, Filman D J, Trus B L, Cheng N, Booy F P, Conway J F, Curry S, Hiremath C N, Tsang S K, Steven A C, Hogle J M. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang K H, Auvinen P, Hyypiä T, Stanway G. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J Gen Virol. 1989;70:3269–3280. doi: 10.1099/0022-1317-70-12-3269. [DOI] [PubMed] [Google Scholar]
- 7.Couderc T, Guédo N, Calvez V, Pelletier I, Hogle J, Colbère-Garapin F, Blondel B. Substitutions in the capsids of poliovirus mutants selected in human neuroblastoma cells confer on the Mahoney type 1 strain a phenotype neurovirulent in mice. J Virol. 1994;68:8386–8391. doi: 10.1128/jvi.68.12.8386-8391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curry S, Chow M, Hogle J M. The poliovirus 135S particle is infectious. J Virol. 1996;70:7125–7131. doi: 10.1128/jvi.70.10.7125-7131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dove A W, Racaniello V R. Cold-adapted poliovirus mutants bypass a postentry replication block. J Virol. 1997;71:4728–4735. doi: 10.1128/jvi.71.6.4728-4735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filman D J, Syed R, Chow M, Macadam A J, Minor P D, Hogle J M. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filman D J, Wien M W, Cunningham J A, Bergelson J M, Hogle J M. Structure determination of echovirus 1. Acta Crystallogr D Biol Crystallogr. 1998;54:1261–72. doi: 10.1107/s0907444998002790. [DOI] [PubMed] [Google Scholar]
- 12.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadfield A T, Lee W M, Zhao R, Oliveira M A, Minor I, Rueckert R R, Rossmann M G. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure. 1997;5:427–421. doi: 10.1016/s0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 14.Hendry E, Hatanaka H, Fry E, Smyth M, Tate J, Stanway G, Santti J, Maaronen M, Hyypiä T, Stuart D. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure. 1999;7:1527–1538. doi: 10.1016/s0969-2126(00)88343-4. [DOI] [PubMed] [Google Scholar]
- 15.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 16.Hovi T, Roivainen M. Peptide antisera targeted to a conserved sequence in poliovirus capsid VP1 cross-react widely with members of the genus Enterovirus. J Clin Microbiol. 1993;31:1083–1087. doi: 10.1128/jcm.31.5.1083-1087.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes P J, Horsnell C, Hyypiä T, Stanway G. The coxsackievirus A9 RGD motif is not essential for virus viability. J Virol. 1995;69:8035–8040. doi: 10.1128/jvi.69.12.8035-8040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S S, Smith T J, Chapman M S, Rossmann M G, Pevear D C, Dutko F J, Felock P J, Diana G D, McKinlay M A. Crystal structure of human rhinovirus serotype 1A (HRV1A) J Mol Biol. 1989;210:91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- 19.Lentz K N, Smith A D, Geisler S C, Cox S, Buontempo P, Skelton A, DeMartino J, Rozhon E, Schwartz J, Girijavallabhan V, O'Connell J, Arnold E. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes. Structure. 1997;5:961–978. doi: 10.1016/s0969-2126(97)00249-9. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Gomez Yafal A, Mie-Ha Lee Y, Hogle J, Chow M. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macadam A J, Arnold C, Howlett J, John A, Marsden S, Taffs F, Reeve P, Hamada N, Wareham K, Almond J, Cammack N, Minor P D. Reversion of the attenuated and temperature-sensitive Phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology. 1989;172:408–414. doi: 10.1016/0042-6822(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 22.Moss E G, Racaniello V R. Host range determinants located on the interior of the poliovirus capsid. EMBO J. 1991;10:1067–1074. doi: 10.1002/j.1460-2075.1991.tb08046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosser A G, Sgro J-Y, Rueckert R R. Distribution of drug resistance mutations in type 3 poliovirus identifies three regions involved in uncoating functions. J Virol. 1994;68:8193–8201. doi: 10.1128/jvi.68.12.8193-8201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muckelbauer J K, Kremer M, Minor I, Diana G, Dutko F J, Groarke J, Pevear D C, Rossmann M G. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure. 1995;3:653–667. doi: 10.1016/s0969-2126(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 25.Roivainen M, Hyypiä T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J Virol. 1991;65:4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roivainen M, Piirainen L, Rysä T, Närvänen A, Hovi T. An immunodominant N-terminal region of VP1 protein of poliovirion that is buried in crystal structure can be exposed in solution. Virology. 1993;195:762–765. doi: 10.1006/viro.1993.1427. [DOI] [PubMed] [Google Scholar]
- 27.Roivainen M, Piirainen L, Hovi T, Virtanen I, Riikonen T, Heino J, Hyypiä T. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology. 1994;203:357–365. doi: 10.1006/viro.1994.1494. [DOI] [PubMed] [Google Scholar]
- 28.Roivainen M, Piirainen L, Hovi T. Efficient RGD-independent entry process of coxsackievirus A9. Arch Virol. 1996;141:1909–1919. doi: 10.1007/BF01718203. [DOI] [PubMed] [Google Scholar]
- 29.Smyth M, Tate J, Hoey E, Lyons C, Martin S, Stuart D. Implications for viral uncoating from the structure of bovine enterovirus. Nat Struct Biol. 1995;2:224–31. doi: 10.1038/nsb0395-224. [DOI] [PubMed] [Google Scholar]
- 30.Tosteson M T, Chow M. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol. 1997;71:507–511. doi: 10.1128/jvi.71.1.507-511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao R, Pevear D C, Kremer M J, Giranda V L, Kofron J A, Kuhn R J, Rossmann M G. Human rhinovirus 3 at 3.0 A resolution. Structure. 1996;4:1205–1220. doi: 10.1016/s0969-2126(96)00128-1. [DOI] [PubMed] [Google Scholar]