Abstract
Here we present the first complete genomic sequence of Marek's disease virus serotype 3 (MDV3), also known as turkey herpesvirus (HVT). The 159,160-bp genome encodes an estimated 99 putative proteins and resembles alphaherpesviruses in genomic organization and gene content. HVT is very similar to MDV1 and MDV2 within the unique long (UL) and unique short (US) genomic regions, where homologous genes share a high degree of colinearity and their proteins share a high level of amino acid identity. Within the UL region, HVT contains 57 genes with homologues found in herpes simplex virus type 1 (HSV-1), six genes with homologues found only in MDV, and two genes (HVT068 and HVT070 genes) which are unique to HVT. The HVT US region is 2.2 kb shorter than that of MDV1 (Md5 strain) due to the absence of an MDV093 (SORF4) homologue and to differences at the UL/short repeat (RS) boundary. HVT lacks a homologue of MDV087, a protein encoded at the UL/RS boundary of MDV1 (Md5), and it contains two homologues of MDV096 (glycoprotein E) in the RS. HVT RS are 1,039 bp longer than those in MDV1, and with the exception of an ICP4 gene homologue, the gene content is different from that of MDV1. Six unique genes, including a homologue of the antiapoptotic gene Bcl-2, are found in the RS. This is the first reported Bcl-2 homologue in an alphaherpesvirus. HVT long repeats (RL) are 7,407 bp shorter than those in MDV1 and do not contain homologues of MDV1 genes with functions involving virulence, oncogenicity, and immune evasion. HVT lacks homologues of MDV1 oncoprotein MEQ, CxC chemokine, oncogenicity-associated phosphoprotein pp24, and conserved domains of phosphoprotein pp38. These significant genomic differences in and adjacent to RS and RL regions likely account for the differences in host range, virulence, and oncogenicity between nonpathogenic HVT and highly pathogenic MDV1.
Turkey herpesvirus (HVT) is a ubiquitous, nonpathogenic virus of domestic turkeys (15, 78–80), and it is classified as the third serotype within the Marek's disease virus (MDV) group of antigenically and genetically related lymphotropic avian herpesviruses (15). This group also includes MDV serotype 1 (MDV1), which is the etiologic agent of the globally and economically significant Marek's disease in chickens (15). MDV1 is pathogenic in chickens, causing cytolytic infection in B cells, latent infection in T cells, and induction of T-cell lymphoma. MDV2 is nonpathogenic or of low pathogenicity in chickens (15).
HVT is nonpathogenic in chickens, but it does induce a viremia which is associated with induction of protective immune responses against MDV1 (15). Chickens infected with HVT become persistently infected and maintain long-lasting immunity (15, 29, 44, 53, 80). HVT appears to replicate less efficiently in skin than does MDV1, a phenotype that may be involved in the relatively infrequent transmission of HVT among chickens compared to attenuated strains of MDV1 and MDV2 (15–17).
HVT and combinations of HVT and nonpathogenic strains of MDV1 and MDV2 have been used extensively and effectively as vaccines against virulent MDV1 since the early 1970s (15, 44, 53, 80). Other benefits of HVT vaccines include the ability to genetically modify the virus for expression of protective heterologous antigens and the potential for in ovo vaccination. HVT vectors expressing genes from Newcastle disease virus, infectious bursal disease virus, and infectious bronchitis virus confer protective systemic immune responses against these pathogens (19, 54, 58, 86). In ovo vaccination of late-stage (16- to 18-day) embryos with HVT leaves newly hatched chickens immune to challenge with pathogenic MDV1 (62, 63).
Although current vaccination strategies will effectively protect against most MDV1 isolates, they are not completely effective against newly emerging MDV1 strains of greater virulence (77). These strains which pose a constant threat to the poultry industry, are characterized by higher cytolytic activity, unusual tissue tropism, increased atrophy of lymphoid organs, immunosuppression, enhanced capacity to transform T cells, and earlier host death (77). The genetic basis underlying differences in viral virulence, oncogenicity, and host range among MDV strains is poorly understood.
Comparative genomics has proven useful in identifying genes with functions involving virulence and host range. The genomes of two MDV1 strains (GA and Md5) have been sequenced (45, 74), and the unique long and unique short (UL and US, respectively) regions of MDV2 are available (37). However, less than 30% of the HVT genome has been sequenced (5, 41, 43, 48, 68, 83, 85). Here we present the complete sequence of HVT strain FC-126 (Burmenester), with analysis and comparison to MDV1 and MDV2.
MATERIALS AND METHODS
DNA isolation, cloning, and sequencing.
HVT strain FC-126 (Burmenester), originally isolated from the blood of an asymptomatic turkey (79), was obtained from the American Type Culture Collection (Manassas, Va.). The virus was propagated in primary chicken embryo fibroblast cell culture, and viral DNA was extracted from the cytoplasm of infected cells as previously described (76). Random DNA fragments were obtained by incomplete enzymatic digestion with endonucleases TaqI and AciI (New England Biolabs, Beverly, Mass.). DNA fragments of 1.5 to 3 kbp were isolated after separation on agarose gels, cloned into the dephosphorylated AccI site of pUC19 plasmids, and grown in Escherichia coli DH10B cells (Gibco BRL, Gaithersburg, Md.). Plasmids were purified by alkaline lysis as instructed by the manufacturer (Eppendorf 5 Prime, Boulder, Colo.). DNA templates were sequenced from both ends with M13 forward and reverse primers, using dideoxy-chain terminator sequencing chemistries (60) and an Applied Biosystem PRISM 377 automated DNA sequencer (PE Biosystems, Foster City, Calif.). ABI sequencing analysis software (version 3.3) was used for lane tracking and trace extraction. Bases were called from chromatogram traces with Phred (27), which also produced a quality file containing a predicted probability of error at each base position.
DNA sequence analysis.
DNA sequences were assembled with Phrap (26), using the quality files and default settings to produce a consensus sequence which was manually edited with Consed (33). An identical sequence was assembled using the TIGR assembler with quality files and clone length constraints (72). The final DNA consensus sequence represented an average eightfold redundancy at each base position. Gap closure was achieved by primer walking of gap-spanning clones and sequencing of PCR products. A total of 5,074 usable traces were assembled into a 140,525-bp contig by bidirectional sequencing of random clones and 28 PCR products. The assembled contig had an estimated error rate of <0.03% and no evidence of polymorphism using Polyphred analysis (26). Because the terminal and internal UL repeats (TRL and IRL, respectively) are identical and the internal and terminal US repeats (IRS and TRS, respectively) are identical, the coverage (redundancy at each base position) of the internal long and short repeats was approximately doubled. The assembled contig contained 663 bp of the TRS, all of the UL, IRL, IRS, and US, and 395 bp of the TRS (i.e., the full genome length minus the length of the TRL and IRL). TRL and TRS sequences were assembled separately with clones containing the TRL/UL and US/TRS junctions and overlapping clones, using clone length constraints and position as provided by the computer assembly programs. The TRL and IRL contigs were then joined to the main contig at the overlapping region in the TRL/UL and US/TRS boundaries, thus providing the complete genome. The predicted restriction map for HVT matched previously published data (13). For descriptive purposes, we have presented HVT in a linearized fashion as described by Dolan et al. (24). Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed as previously described (2). Open reading frames (ORFs) encoding proteins of ≥60 amino acids with a methionine start codon (70, 71) were evaluated for coding potential using the Hexamer (ftp.sanger.ac.uk/pub/rd), Framefinder (www.hgmp.mrc.ac.uk /∼gslater/ESTate/), and Glimmer (59) computer programs. Other criteria included similarity to other herpesvirus or cellular proteins and compact gene arrangement with little gene overlap (21, 73). Homology searches were conducted using BLAST (3), PSIBLAST (4), FASTA (52), BLIMPS (75), and HMMER (69) programs with the Prosite, Pfam, Prodom, Sbase, Blocks, Domo, and GenBank databases (14). GCG (22), MEMSAT (40), and SAPS (10) programs were used for gene analysis.
Nucleotide sequence accession number.
The HVT genome sequence has been deposited in GenBank under accession no. AF291866.
RESULTS AND DISCUSSION
Genome organization.
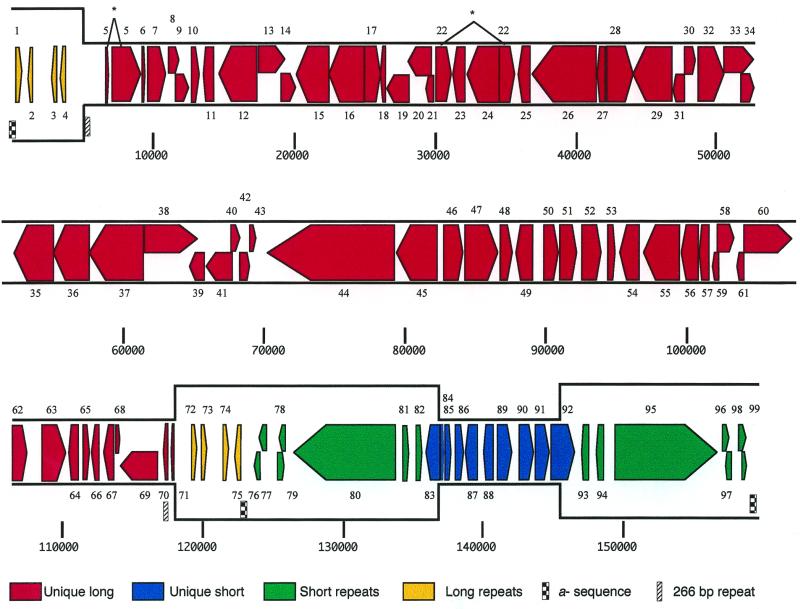
The HVT genome is 159,160 bp long and contains a 47.5% G+C base composition. HVT is organized in the same overall manner as are other alphaherpesviruses (55) (Fig. 1). UL and US regions are 111,868 and 8,617 bp in length, respectively. Each unique region is bounded by identical inverted repeats. The TRL and IRL are 5,658 bp, and the IRS and TRS are 13,303 bp. As with other herpesviruses, the G+C content in the repeat regions is higher than in unique regions (55% in repeats versus 45% in unique regions) (21, 32, 73). HVT contains no retrovirus long terminal repeat sequences as have been described for cell culture-adapted MDV1 strains (36, 38, 39, 45). The assembled HVT sequence contains a 251-bp sequence that is identical at each terminus and at the IRL/IRS junction. This repeat contains sequences similar to alphaherpesvirus α-type sequences (42) and includes 17 copies of the GGGTTA motif found in MDV1 (64 copies), human herpesvirus 6, and eukaryotic telomers (42, 74). In addition, a 266-bp identical inverted repeat is located adjacent to the TRL at positions 5910 to 6175 and near the IRL at positions 116856 to 117121 (Fig. 1). A 656-bp unique sequence (117122 to 117177) separates the second 266-bp repeat from the IRL (Fig. 1).
FIG. 1.
Linear map of the HVT genome. Genes (colored arrows) are numbered from left to right based on positions of methionine initiation codons and transcribed in the direction indicated. Genes included in different genomic regions are defined by the color key. Nucleotide positions are indicated below the map. Spliced genes are indicated by asterisks.
Gene characterization.
HVT contains 397 ORFs of 60 to 2,164 codons, of which 99 are estimated to be functional genes (Fig. 2). Seventy-five genes are present as single copies and initiate within unique regions. Two genes initiate within the US region but are partially located within repeat regions (HVT083 and HVT092). Twenty-two genes initiate and are completely located within repeat regions. HVT gene products are most similar to homologues from MDV1 (36 to 82% amino acid identity) and MDV2 (34 to 81% amino acid identity). Among nonavian herpesviruses, ORFs of equine herpesviruses 1 and 4 are most similar to those of HVT (27 to 62% amino acid identity).
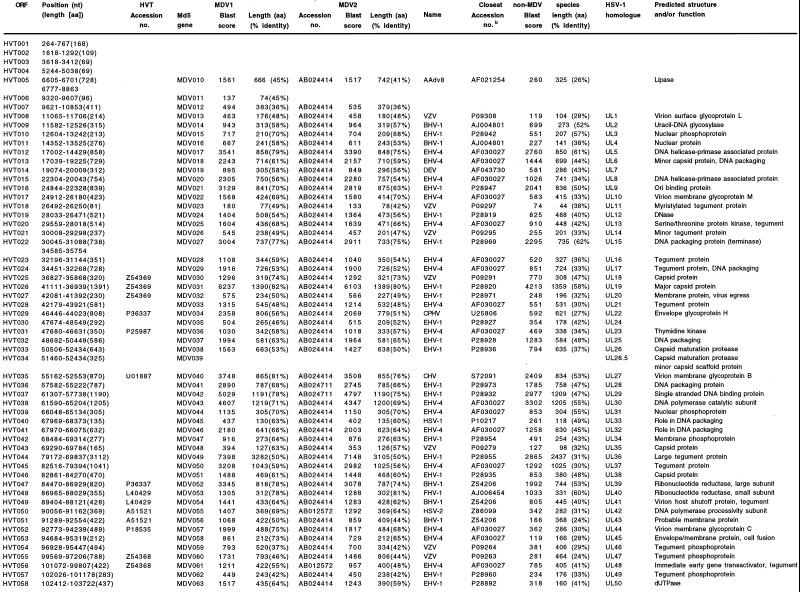
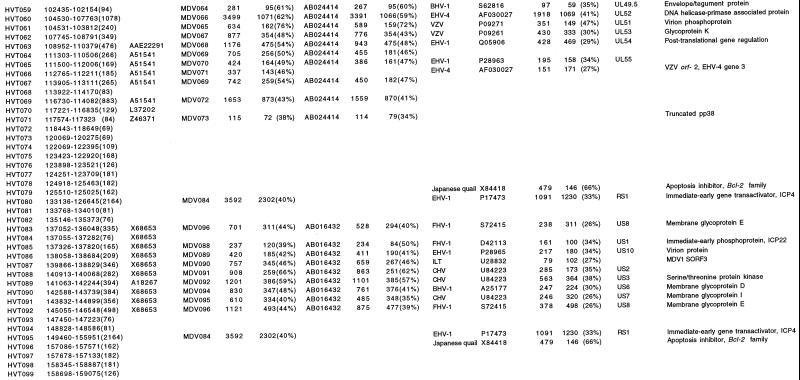
FIG. 2.
Accession numbers are from GenBank, SwissProt, and PIR databases; the accession number for Md5 genes is AF243438. Names for viruses: bovine herpesvirus (BHV), canine herpesvirus (CHV), duck enteritis virus (DEV), equine herpesviruses 1 and 4 (EHV-1 and -4), feline herpesvirus (FHV), herpes simplex virus (HSV), herpesvirus of turkeys (HVT), infectious laryngotracheitis virus (ILT), Marek's disease virus serotypes 1 and 2 (MDV1 and -2), varicella-zoster virus (VZV), cercopithecine herpesvirus 9 (CPHV), and avian adenovirus type 8 (Aadv8). Function was deduced either from the degree of amino acid (aa) similarity to known genes or by the presence of Prosite signatures.
HVT and MDV1 share 76 conserved gene homologues, 92% of which are within the UL and US regions. Only a single MDV1 gene homologue is present in the repeats. HVT contains 13 genes for which there are no MDV1 homologues, and it lacks homologues of 16 genes present in MDV1.
UL region.
The UL region, extending from nucleotide positions 5910 to 117777, contains 67 probable genes (Fig. 1 and 2). HVT008 to HVT063 represent 62% of the HVT genome and are colinear with genes UL1 to UL54 of herpes simplex virus type 1 (HSV-1) and MDV013 to MDV068 of MDV1 (74). Predicted proteins are 37 to 82% identical to MDV1 homologues. Those with the greatest (>70%) amino acid identity to MDV1 proteins correspond to capsid proteins (HVT025 and HVT026), viral replication proteins (HVT012, HVT016, HVT022, HVT037, HVT038, HVT039, HVT047, and HVT048), membrane proteins (HVT035, HVT052, and HVT053), and phosphoproteins (HVT010 and HVT061). These proteins are also the most conserved in comparison with MDV2 and other nonavian herpesviruses. HVT tegument proteins are the least conserved within the UL, with seven (HVT018, HVT021, HVT028, HVT044, HVT054, HVT055, and HVT057) sharing less than 51% amino acid identity with MDV1 homologues. Other, less conserved UL proteins include homologues of HSV virion surface glycoprotein L (HVT008), glycoprotein K (HVT062), and proteins of unknown function (HVT030, HVT051, HVT064, HVT065, and HVT066). HVT044 (large tegument protein) is 230 amino acids shorter than its MDV1 homologue due to the lack of a highly repetitive central proline-rich domain.
Six predicted HVT genes located at the ends of the UL region are MDV specific. The absence of HVT006 (MDV011), HVT007 (MDV012), HVT064 and HVT067 (MDV069), HVT069 (MDV072), and HVT071 (MDV073) (Fig. 2) in non-MDV alphaherpesviruses suggests an avian host range function, perhaps involving some aspects of lymphotropism. On the left end, HVT lacks homologues of MDV008 (pp24) and MDV009, a gene of unknown function (74). On the right end, HVT contains two novel genes (HVT068 and HVT070) which are absent in MDV1 and encodes two homologues (HVT064 and HVT067) of MDV069.
HVT005 is similar to eukaryotic and viral lipases (GenBank accession no. AF007578 and L43561) and shares 41 to 45% identity with MDV homologues (6, 74). HVT005 contains the serine active site at amino acid position 287 within the lipase signature motif (IxxIGHSxS, Prosite PS00120) and conserved cysteines involved in disulfide bond formation (amino acid positions 387 and 397). The region between amino acids 120 and 265 is similar to corresponding regions in eukaryotic lipases such as phospholipase A1 (26% over 154 amino acids). Similarity to predicted proteins from MDV1, MDV2, and fowl adenovirus extends beyond this region, suggesting the presence of additional virus-specific domains. The presence of a signal peptide in the amino-terminal domain and a transmembrane domain at amino acid positions 479 to 501 suggests that HVT005 may be membrane localized. HVT005 may perform host range functions involving alteration of host lipid metabolism and/or modification of second-messenger signaling pathways (1, 23, 31, 35, 50, 61, 66). Altered lipid metabolism has been observed both in vivo and in cell cultures during MDV infection (28, 34).
US region.
The US region, extending from positions 136990 to 145606, has been previously sequenced (83). This region encodes 10 likely genes (HVT083 to HVT092), of which 7 are homologues of the HSV-1 genes US1, US2, US3, US6, US7, US8, and US10. Proteins encoded in the HVT US region are 39 to 66% identical to their homologues in MDV1 and overall are less conserved (48%) than those in the UL region (61%). The arrangement of genes in the HVT US region is similar to that in MDV1 and MDV2 (37, 74), including the inversion-translocation of the UL10 homologue, compared to HSV-1 (11, 21, 49, 56, 73, 87). HVT087, a homologue of MDV090, is found only in MDV. In addition, an HVT-specific gene of unknown function (HVT084) is found near the IRS/US boundary. HVT lacks a homologue of MDV093 (SORF4), a gene found in MDV1.
The US short repeat (RS) junction region is variable among MDV1, MDV2, and HVT (11, 37, 74, 83). Unlike MDV1 and MDV2, the carboxyl terminus of the HVT US8 gene homologue (glycoprotein E [gE] gene) is duplicated and inverted to the other end of the US. The presence of two copies of the gE ORF does not imply that the virus expresses two forms of gE. Thus, the HVT US/RS boundaries are located within the US8 homologues (HVT083 and HVT092 genes) and reduce the size of the US compared to MDV1 and MDV2. Duplication of US genes and resulting changes in US/RS boundary position has been noted between strains of MDV1 and with other herpesviruses (11, 20, 74). Like MDV2, HVT lacks a homologue of the MDV087 (SORF2) gene. SORF2 is nonessential for MDV1 replication in cell culture, but it is duplicated in the very virulent MDV1 strain Md5 (11, 51, 74). Although the function of this gene is unknown, its presence in fowlpox virus and fowl adenovirus suggests a significant avian host range function (2, 65).
RL.
The RL are 5,658 bp in length and are located at nucleotide positions 252 to 5909 and 117778 to 123435. HVT RL are 7,407 bp shorter than MDV1 RL and lack homologues of all MDV1 RL genes. Each RL contains four possible HVT-specific genes, which predict proteins of unknown function (HVT001 to HVT004 and HVT072 to HVT075). Although there are no other readily identifiable genes in this region based on our criteria, the RL does contain 21 ORFs of more than 60 codons. Thus, additional genes may be present.
Most notably, and consistent with its nonpathogenic phenotype, HVT lacks homologues of all MDV1 RL proteins with putative functions involving viral virulence, host range, and oncogenicity. HVT contains no homologues of the Marek's EcoRI Q protein (MEQ) (MDV005 and MDV076), which is associated with cellular transformation and oncogenicity (45, 74). In addition, HVT lacks pp24 (MDV008) and conserved domains of pp38 (MDV073), phosphoproteins associated with cellular transformation (18, 81, 88). The functional homology of HVT071 to pp38 is unclear. HVT071 lacks the amino terminus (75 amino acids) and a centrally located (22-amino-acid) conserved region containing a DLLVEAE motif found in pp38 and pp24 (74). Moreover, recent experiments have shown that deletion of this putative gene from the HVT genome has no effect on viral growth or virulence (12). HVT lacks a homologue of the MDV1 CxC chemokine (MDV003 and MDV078) (74) and the 132-bp repeat region whose expansion has been associated with altered viral transcription and loss of oncogenicity (7, 9, 30, 47, 57, 67).
RS.
The short repeat regions (RS) are 13,303 bp and are located at nucleotide positions 123687 to 136989 and 145607 to 158909. Each RS encode seven possible genes, of which five lack homology to other known genes (Fig. 1 and 2). HVT080 and HVT095 encode homologues of the HSV-1 major immediate-early trans-activating protein ICP4. These genes, which encode proteins of 2,164 amino acids, comprise over 48% of the RS. Interestingly, HVT080 and HVT095 contain an 810-amino-acid amino-terminal extension which is absent in ICP4 homologues of other nonavian alphaherpesviruses. A similarly sized ICP4 homologue has been previously found in MDV1 and predicted to exist in HVT (74, 84). HVT lacks identifiable homologues of immediate-early proteins ICP0, which is nonessential for replication of HSV-1 in cell culture (55), and ICP47, a host range protein which blocks HSV-1 antigen presentation on infected cells (82). HVT also lacks identifiable homologues of MDV085, a protein of unknown function, and MDV086, a gene present in a region transcribed during viral latency (74).
HVT079 and HVT096 encode identical 162-amino-acid proteins with significant similarity (66% amino acid identity) to the quail Nr13 protein, a homologue of Bcl-2 (Fig. 3) (46). Amino acid identity occurs over the entire length of the protein and includes Bcl-2 BH1, BH2, and BH3 domains and a novel form of the BH4 domain (46). HVT079 and HVT096 lack the 15-amino-acid carboxyl-terminal transmembrane domain present in Nr13 and in other mammalian Bcl-2 homologues. Although all sequenced gammaherpesvirus genomes encode Bcl-2 homologues (8), HVT079 and HVT096 represent the first Bcl-2 homologues found in an alphaherpesvirus.
FIG. 3.
Multiple amino acid sequence alignment of HVT096 with Bcl-2 family members. Asterisks mark anti- and proapoptotic domains, and shaded residues indicate identity to HVT096. Gaps generated during the alignment with Pileup are indicated with dashes. Amino acid positions are indicated on the right. Rat, Rattus norvegicus, accession no. U34963; Chicken, Gallus gallus, accession no. Q07816; Quail, Coturnix japonica, accession no. X84418; BHV-4, bovine herpesvirus 4, accession no. AF129421.
HVT079 and HVT096 likely function in promoting infected cell survival and may account for differences in cell and tissue tropism (15, 62, 64). Nr13, a developmentally regulated gene which is maximally expressed at 15 to 21 days in the chicken bursa of Fabricius, may play a role in maintaining bursal stem cell populations (46). Additionally, in vitro studies have shown the Nr13 can protect bursal lymphoma cells from low-serum-induced apoptosis (46). Recent experimental evidence suggests that HVT, but not MDV1, contains genes capable of preventing cell death (25). Induction of apoptosis by serum deprivation in transformed chicken B-cell lines was prevented by infection with HVT but not by MDV1 (25).
Conclusions.
HVT resembles other alphaherpesviruses in genome organization and gene content. A close relationship among MDV serotypes is evident by the high degree of amino acid identity of protein homologues and by the presence of MDV-specific proteins. Significant genomic differences occur between HVT and MDV1 in and adjacent to RL and RS, and these may account for differences in tissue tropism, host range, and the nonpathogenic phenotype of HVT (15). HVT lacks putative MDV1 virulence and host range genes, including MEQ, pp24, pp38, CxC chemokine, MDV087 (SORF2), and MDV093 (SORF4). There are 13 HVT-specific genes, which include a Bcl-2 homologue. Thus, comparative analysis of nonpathogenic HVT and MDV2 and virulent MDV1 genomes enhances our overall understanding of MDV virulence and host range. This information will permit the engineering of novel MDV vaccine viruses and avian expression vectors with enhanced efficacy and greater versatility.
ACKNOWLEDGMENTS
We thank A. Ciupryk and G. Smoliga for excellent technical assistance and W. H. Martinez, F. P. Horn, and R. G. Breeze for their interest and encouragement.
REFERENCES
- 1.AbuBakar S, Boldogh I, Albrecht T. Human cytomegalovirus stimulates arachidonic acid metabolism through pathways that are affected by inhibitors of phospholipase A2 and protein kinase C. Biochem Biophys Res Commun. 1990;166:953–959. doi: 10.1016/0006-291x(90)90903-z. [DOI] [PubMed] [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandyopadhyay P K. Characterization of a highly transcribed DNA region of herpesvirus of turkeys. Gene. 1989;79:361–367. doi: 10.1016/0378-1119(89)90218-7. [DOI] [PubMed] [Google Scholar]
- 6.Becker Y, Asher Y, Tabor E, Davidson I, Malkinson M. Open reading frames in a 4556 nucleotide sequence within MDV-1 BamHI-D DNA fragment: evidence for splicing of mRNA from a new viral glycoprotein gene. Virus Genes. 1994;8:55–69. doi: 10.1007/BF01703602. [DOI] [PubMed] [Google Scholar]
- 7.Becker Y, Tabor E, Asher Y, Davidson I, Malkinson M, Witter R L. PCR detection of amplified 132 bp repeats in Marek's disease virus type 1 (MDV-1) DNA can serve as an indicator for critical genomic rearrangement leading to the attenuation of virus virulence. Virus Genes. 1993;7:277–287. doi: 10.1007/BF01702588. [DOI] [PubMed] [Google Scholar]
- 8.Bellows D S, Chau B N, Lee P, Lazebnik Y, Burns W H, Hardrick J M. Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J Virol. 2000;74:5024–5031. doi: 10.1128/jvi.74.11.5024-5031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley G, Lancz G, Tanaka A, Nonoyama M. Loss of Marek's disease virus tumorigenicity is associated with truncation of RNAs transcribed with BamHI-H. J Virol. 1989;63:4129–4135. doi: 10.1128/jvi.63.10.4129-4135.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunovskis P, Velicer L F. The Marek's disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 12.Bublot M, Laplace E, Audonnet J-C. Non-essential loci in the BamHI-1 and -F fragments of the HVT FC126 genome. Acta Virol. 1999;43:181–185. [PubMed] [Google Scholar]
- 13.Buckmaster A E, Scott S D, Sanderson M J, Boursnell M E G, Ross N L J, Binns M M. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J Gen Virol. 1988;69:2033–2042. doi: 10.1099/0022-1317-69-8-2033. [DOI] [PubMed] [Google Scholar]
- 14.Burks C. Molecular biology database list. Nucleic Acids Res. 1999;27:1–9. doi: 10.1093/nar/27.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calnek B W, Witter R L, editors. Marek's disease. 9th ed. Ames, Iowa: Iowa State University Press; 1991. [Google Scholar]
- 16.Cho B R. Horizontal transmission of turkey herpesvirus to chickens. IV. Viral maturation in the feather follicle epithelium. Avian Dis. 1974;19:136–141. [PubMed] [Google Scholar]
- 17.Cho B R, Kenzy S G. Horizontal transmission of turkey herpesvirus to chickens. III. Transmission in three different lines of chickens. Poult Sci. 1975;54:109–115. doi: 10.3382/ps.0540109. [DOI] [PubMed] [Google Scholar]
- 18.Cui Z, Lee L F, Liu J-L, Kung H-J. Structural analysis and transcriptional mapping of the Marek's disease virus gene encoding pp38, an antigen associated with transformed cells. J Virol. 1991;65:6509–6515. doi: 10.1128/jvi.65.12.6509-6515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darteil R, Bublot M, Laplace E, Bouquet J-F, Audonnet J-C, Riviere M. Herpesvirus of turkey recombinant viruses expressing infectious bursal disease virus (IBDV) VP2 immunogen induce protection against an IBDV virulent challenge in chickens. Virology. 1995;211:481–490. doi: 10.1006/viro.1995.1430. [DOI] [PubMed] [Google Scholar]
- 20.Davidson A J, McGeoch D J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986;67:597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- 21.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 22.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitrov T, Krajcsi P, Hermiston T W, Tollefson A E, Hannink M, Wold W S M. Adenovirus E3–10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J Virol. 1997;71:2830–2837. doi: 10.1128/jvi.71.4.2830-2837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewert D, Duhadaway J. Inhibition of apoptosis by Marek's disease viruses. Acta Virol. 1999;43:133–135. [PubMed] [Google Scholar]
- 26.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 27.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 28.Fabricant C G, Hajjar D P, Minick C R, Fabricant J. Herpesvirus infection enhances cholesterol and cholesteryl ester accumulation in cultured arterial smooth muscle cells. Am J Pathol. 1981;105:176–184. [PMC free article] [PubMed] [Google Scholar]
- 29.Fabricant J, Calnek B W, Schat K A. The early pathogenesis of turkey herpesvirus infection in chickens and turkeys. Avian Dis. 1982;26:257–264. [PubMed] [Google Scholar]
- 30.Fukuchi K, Tanaka A, Schierman L W, Witter R L, Nonoyama M. The structure of Marek's disease virus DNA: the presence of unique expansion in nonpathogenic viral DNA. Proc Natl Acad Sci USA. 1985;82:751–754. doi: 10.1073/pnas.82.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geras-Raaka E, Arvanitakis L, Bais C, Cesarman E, Mesri E A, Gershengorn M C. Inhibition of constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J Exp Med. 1998;187:801–806. doi: 10.1084/jem.187.5.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 33.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:192–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 34.Hajjar D P, Fabricant C G, Minick C R, Fabricant J. Virus-induced atherosclerosis. Herpesvirus infection alters aortic cholesterol metabolism and accumulation. Am J Pathol. 1986;122:62–70. [PMC free article] [PubMed] [Google Scholar]
- 35.Higgs H N, Glomset J A. Purification and properties of a phosphatidic acid-preferring phospholipase A1 from bovine testis. J Biol Chem. 1996;271:10874–10883. doi: 10.1074/jbc.271.18.10874. [DOI] [PubMed] [Google Scholar]
- 36.Isfort R J, Qian Z, Jones D, Silva R F, Witter R, Kung H-J. Integration of multiple chicken retroviruses into multiple chicken herpesviruses: herpesviral gD as a common target of integration. Virology. 1994;203:125–133. doi: 10.1006/viro.1994.1462. [DOI] [PubMed] [Google Scholar]
- 37.Jang H-K, Ono M, Kim T-J, Izumiya Y, Damiani A M, Matsumura T, Niikura M, Kai C, Mikami T. The genetic organization and transcriptional analysis of the short unique region in the genome of nononcogenic Marek's disease virus serotype 2. Virus Res. 1998;58:137–147. doi: 10.1016/s0168-1702(98)00110-5. [DOI] [PubMed] [Google Scholar]
- 38.Jones D, Brunovskis P, Witter R, Kung H-J. Retroviral insertional activation in a herpesvirus: transcriptional activation of US genes by an integrated long terminal repeat in a Marek's disease virus clone. J Virol. 1996;70:2460–2467. doi: 10.1128/jvi.70.4.2460-2467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones D, Isfort R, Witter R, Kost R, Kung H-J. Retroviral insertions into a herpesvirus are clustered at the junctions of the short repeat and short unique sequences. Proc Natl Acad Sci USA. 1993;90:3855–3859. doi: 10.1073/pnas.90.9.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones D T, Taylor W R, Thornton J M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 41.Kato A, Sato I, Ihara T, Ueda S, Ishihama A, Hirai K. Homologies between herpesvirus of turkey and Marek's disease virus type-1 DNAs within two co-linearly arranged open reading frames, one encoding glycoprotein A. Gene. 1989;84:399–405. doi: 10.1016/0378-1119(89)90514-3. [DOI] [PubMed] [Google Scholar]
- 42.Kishi M, Harada H, Takahashi M, Tanaka A, Hayashi M, Nonoyama M, Josephs S F, Buchbinder A, Schachter F, Ablashi D V, Wong-Staal F, Salahuddin S Z, Gallo R C. A repeat sequence, GGGTTA, is shared by DNA of human herpesvirus 6 and Marek's disease virus. J Virol. 1988;62:4824–4827. doi: 10.1128/jvi.62.12.4824-4827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopacek J, Klucar L, Koptidesova D, Turna J, Pastorek J, Zelnik V. Nucleotide sequence of the gene encoding the major capsid protein of herpesvirus of turkey. Virus Genes. 2000;20:107–115. doi: 10.1023/a:1008158228591. [DOI] [PubMed] [Google Scholar]
- 44.Lee K. Long term effects of Marek's disease vaccination with cell-free herpesvirus of turkey and age at debeaking on performance and mortality of white leghorns. Poult Sci. 1980;59:2002–2007. doi: 10.3382/ps.0592002. [DOI] [PubMed] [Google Scholar]
- 45.Lee L F, Wu P, Sui D, Ren D, Kamil J, Kung H J, Witter R L. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc Natl Acad Sci USA. 2000;97:6091–6096. doi: 10.1073/pnas.97.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee R-M, Gillet G, Burnside J, Thomas S J, Neiman P. Role of Nr13 in regulation of programmed cell death in the bursa of Fabricius. Genes Dev. 1999;13:718–728. doi: 10.1101/gad.13.6.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maotani K, Kanamori A, Ikuta K, Ueda S, Kato S, Hirai K. Amplification of a tandem direct repeat within inverted repeats of Marek's disease virus DNA during serial in vitro passage. J Virol. 1986;58:657–660. doi: 10.1128/jvi.58.2.657-660.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin S L, Aparisio D I, Bandyopadhyay P K. Genetic and biochemical characterization of the thymidine kinase gene from herpesvirus of turkeys. J Virol. 1989;63:2847–2852. doi: 10.1128/jvi.63.6.2847-2852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 50.Nagai Y, Aoki J, Sato T, Amano K, Matsuda Y, Arai H, Inoue K. An alternative splicing form of phosphatidylserine-specific phospholipase A1 that exhibits lysophosphatidylserine-specific lysophospholipase activity in humans. J Biol Chem. 1999;274:11053–11059. doi: 10.1074/jbc.274.16.11053. [DOI] [PubMed] [Google Scholar]
- 51.Parcells M S, Anderson A S, Cantello J L, Morgan R W. Characterization of Marek's disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short-component open reading frames. J Virol. 1994;68:8239–8253. doi: 10.1128/jvi.68.12.8239-8253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 53.Purchase H G, Okazaki W, Burmester B R. Long-term field trials with the herpesvirus of turkeys vaccine against Marek's disease. Avian Dis. 1972;16:57–71. [PubMed] [Google Scholar]
- 54.Reddy S K, Sharma J M, Ahmad J, Reddy D N, McMillen J K, Cook S M, Wild M A, Schwartz R D. Protective efficacy of a recombinant herpesvirus of turkeys as an in ovo vaccine against Newcastle and Marek's diseases in specific-pathogen-free chickens. Vaccine. 1996;14:469–477. doi: 10.1016/0264-410x(95)00242-s. [DOI] [PubMed] [Google Scholar]
- 55.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 56.Ross L J N, Binns M M, Pastorek J. DNA sequence and organization of genes in a 5.5 kbp EcoRI fragment mapping in the short unique segment of Marek's disease virus (strain RB1B) J Gen Virol. 1991;72:949–954. doi: 10.1099/0022-1317-72-4-949. [DOI] [PubMed] [Google Scholar]
- 57.Ross N, Binns M M, Sanderson M, Schat K A. Alterations in DNA sequence and RNA transcription of the BamHI-H fragment accompany attenuation of oncogenic Marek's disease herpesvirus. Virus Genes. 1993;7:33–51. doi: 10.1007/BF01702347. [DOI] [PubMed] [Google Scholar]
- 58.Ross N, O'Sullivan G, Coudert F. Influence of chicken genotype on protection against Marek's disease by a herpesvirus of turkeys recombinant expressing the glycoprotein B (gB) of Marek's disease virus. Vaccine. 1996;14:187–189. doi: 10.1016/0264-410x(95)00215-m. [DOI] [PubMed] [Google Scholar]
- 59.Salzberg S L, Delcher A L, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato T, Aoki J, Nagai Y, Dohmae N, Takio K, Doi T, Arai H, Inoue K. Serine phospholipid-specific phospholipase A that is secreted from activated platelets. J Biol Chem. 1997;272:2192–2198. doi: 10.1074/jbc.272.4.2192. [DOI] [PubMed] [Google Scholar]
- 62.Sharma J M. Delayed replication of Marek's disease virus following in ovo inoculation during late stages of embryonal development. Avian Dis. 1987;31:570–576. [PubMed] [Google Scholar]
- 63.Sharma J M, Burmester B R. Resistance to Marek's disease at hatching in chickens vaccinated as embryos with the turkey herpesvirus. Avian Dis. 1982;26:134–149. [PubMed] [Google Scholar]
- 64.Sharma J M, Lee L F, Wakenell P S. Comparative viral, immunologic, and pathologic responses of chickens inoculated with herpesvirus of turkeys as embryos or at hatch. Am J Vet Res. 1984;45:1619–1623. [PubMed] [Google Scholar]
- 65.Sheppard M, Werner W, Tsatas E, McCoy R, Prowse S, Johnson M. Fowl adenovirus recombinant expressing VP2 of infectious bursal disease virus induces protective immunity against bursal disease. Arch Virol. 1998;143:915–930. doi: 10.1007/s007050050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shibutani T, Johnson T M, Yu Z-X, Ferrans V J, Moss J, Epstein S E. Pertussis toxin-sensitive G proteins as mediators of the signal transduction pathways activated by cytomegalovirus infection of smooth muscle cells. J Clin Investig. 1997;100:2054–2061. doi: 10.1172/JCI119738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva R F, Witter R L. Genomic expansion of Marek's disease virus DNA is associated with serial in vitro passage. J Virol. 1985;54:690–696. doi: 10.1128/jvi.54.3.690-696.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith G D, Zelnik V, Ross L J N. Gene organization in herpesvirus of turkeys: identification of a novel open reading frame in the long unique region and a truncated homologue of pp38 in the internal repeat. Virology. 1995;207:205–216. doi: 10.1006/viro.1995.1067. [DOI] [PubMed] [Google Scholar]
- 69.Sonnhammer E L L, Eddy S R, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Staden A, McLachlan A D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sutton G G, White O, Adams M D, Kerlavage A R. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- 73.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 74.Tulman E R, Afonso C L, Lu Z, Zsak L, Rock D L, Kutish G F. The genome of a very virulent Marek's disease virus. J Virol. 2000;74:7980–7988. doi: 10.1128/jvi.74.17.7980-7988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallace J C, Henikoff S. PATMAT: a searching and extraction program for sequence, pattern and block queries and databases. CABIOS. 1991;8:249–254. doi: 10.1093/bioinformatics/8.3.249. [DOI] [PubMed] [Google Scholar]
- 76.Wesley R D, Tuthill A E. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev Vet Med. 1984;2:53–62. [Google Scholar]
- 77.Witter R L. Increased virulence of Marek's disease virus field isolates. Avian Dis. 1997;41:149–163. [PubMed] [Google Scholar]
- 78.Witter R L. Turkey herpesvirus: lack of oncogenicity for turkeys. Avian Dis. 1972;16:666–670. [PubMed] [Google Scholar]
- 79.Witter R L, Nazerian K, Purchase H G, Burgoyne G H. Isolation from turkeys of a cell-associated herpesvirus antigenically related to Marek's disease virus. Am J Vet Res. 1970;31:525–538. [PubMed] [Google Scholar]
- 80.Witter R L, Solomon J J. Experimental infection of turkeys and chickens with a herpesvirus of turkeys (HVT) Avian Dis. 1972;16:34–44. [PubMed] [Google Scholar]
- 81.Xie Q, Anderson A S, Morgan R W. Marek's disease virus (MDV) ICP4, pp38, and meq genes are involved in the maintenance of transformation of MDCC-MSB1 MDV-transformed lymphoblastoid cells. J Virol. 1996;70:1125–1131. doi: 10.1128/jvi.70.2.1125-1131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 83.Zelnik V, Darteil R, Audonnet J C, Smith G D, Riviere M, Pastorek J, Ross L J N. The complete sequence and gene organization of the short unique region of herpesvirus of turkeys. J Gen Virol. 1993;74:2151–2162. doi: 10.1099/0022-1317-74-10-2151. [DOI] [PubMed] [Google Scholar]
- 84.Zelnik V, Kopacek J, Rejholcova O, Kabat P, Pastorek J. ICP4 homologues of both Marek's disease virus and herpesvirus of turkeys are larger than their alphaherpesvirus counterparts. In: Silva R F, Cheng H H, Coussens P M, Lee L F, Velicer L F, editors. Current research on Marek's disease. Kennett Square, Pa: American Association of Avian Pathologists; 1996. pp. 164–169. [Google Scholar]
- 85.Zelnik V, Ross N L J, Pastorek J. Characterization of proteins encoded by the short unique region of herpesvirus of turkey's by in vitro expression. J Gen Virol. 1994;75:2747–2753. doi: 10.1099/0022-1317-75-10-2747. [DOI] [PubMed] [Google Scholar]
- 86.Zelnik V, Tyers P, Smith G D, Jiang C-L, Ross N L J. Structure and properties of a herpesvirus of turkeys recombinant in which US1, US10 and SORF3 genes have been replaced by a lacZ expression cassette. J Gen Virol. 1995;76:2903–2907. doi: 10.1099/0022-1317-76-11-2903. [DOI] [PubMed] [Google Scholar]
- 87.Zhang G, Leader D P. The structure of the pseudorabies virus genome at the end of the inverted repeat sequences proximal to the junction with the short unique region. J Gen Virol. 1990;71:2433–2441. doi: 10.1099/0022-1317-71-10-2433. [DOI] [PubMed] [Google Scholar]
- 88.Zhu G-S, Iwata A, Gong M, Ueda S, Hirai K. Marek's disease virus type 1-specific phosphorylated proteins pp38 and pp24 with common amino acid termini are encoded from the opposite junction regions between the long unique and inverted repeat sequences of viral genome. Virology. 1994;200:816–820. doi: 10.1006/viro.1994.1249. [DOI] [PubMed] [Google Scholar]