Abstract
SeqCode is a nomenclatural code for naming prokaryotes based on genetic information. With the majority of prokaryotes being inaccessible as pure cultures, they are not eligible for naming under the International Code of Nomenclature of Prokaryotes. To address this challenge, a new concept that is SeqCode, which assign names to prokaryotes on the basis of genome sequence, has been announced in 2022. The valid publication of names for prokaryotes based on isolated genome, metagenome-assembled genomes, or single-amplified genome sequences. It operates through a registration portal, SeqCode Registry, where metadata is linked to names and nomenclatural types. This code provides a framework for reproducible nomenclature for all prokaryotes, either culturable or not and facilitates communication across all microbiological disciplines. Additionally, the SeqCode includes provisions for updating and revising names as new data becomes available. By providing a standardized system for naming and classifying these microorganisms based on their genetic information, the SeqCode will facilitate the discovery, understanding and comparison of these microorganisms, helping us to understand their role in the environment and how they contribute to the functioning of the Earth.
Keywords: SeqCode, Taxonomy, Uncultured prokaryotes, ICNP
Introduction
Why Do We Need Seqcode?
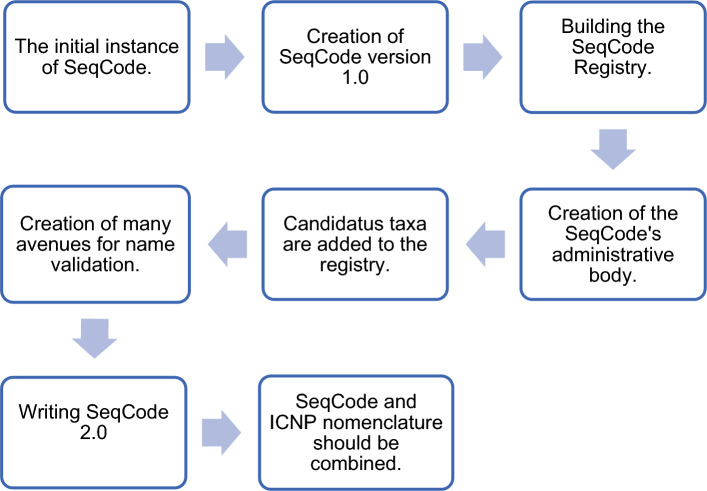
It is widely acknowledged that a significant proportion of prokaryotes, over 99%, cannot be cultivated and isolated in pure form under laboratory conditions and as such these uncultured prokaryotes cannot be taxonomically characterized by conventional methods as recommended in ICNP.1 A new taxonomy is required for these unculturable prokaryotes [1]. They represent more than 85% phylogenetic diversity in prokaryotes [2]. This creates a gap in our understanding of “Tree of life.” And as such hinders us from identifying new species and understanding how they fit into the larger picture of evolution of life on earth. To overcome this problem, a new concept for nomenclature has been discovered by Brian et al., 2022, in the form of SeqCode. Figure 1, shows the need of the SeqCode. The typification of both cultivated and uncultivated microorganisms is based on sequence of genome and MAGs2 and the priority of validly published names is recognised by the rules similar to those of the ICNP [3]. Goal of SeqCode is to validly publish names before using in primary literature.
Fig. 1.
Showing the need of the SeqCode. As INCP cannot name 99% of prokaryotes, a new code of nomenclature is required which is internationally accepted
Seqcode Has 3 Main Bodies
Seqcode Legislative Commission and its Objectives
The SeqCode legislative commission has been created as the legislative arm of the SeqCode Committee in compliance with its regulations (Fig. 2). This commission is the sole entity authorized to modify the SeqCode.
Fig. 2.
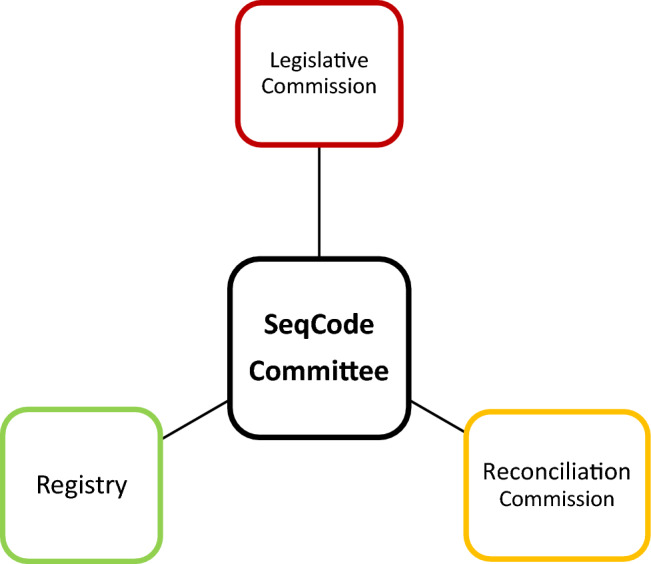
Showing the decentralized nature of SeqCode Committee and its various bodies
Seqcode Reconciliation Commission and its Objectives
The establishment of the SeqCode Reconciliation Commission as the judicial arm of the SeqCode Committee aims to address matters concerning the implementation of the SeqCode in line with its statutes. In this regard, the SeqCode Reconciliation Commission holds exclusive authority in rendering decisions pertaining to the application of the SeqCode.
Seqcode Registry and its Objectives
For efficient working of SeqCode a registration portal was created through which registers names and nomenclatural types, validate them and then link to metadata. The objectives of SeqCode Registry involve: (1)—To register and evaluate names proposed according to SeqCode. (2)—To Automatically identify Candidatus names which are being used in literature to standardise them through validation under SeqCode. (3)—To keep a uniform and publicly accessible list of names certified by the SeqCode. After completion of registration process, a user-friendly interface is provided access to its resources (Fig. 3) [4].
Fig. 3.
Blueprint of development of SeqCode Registry
Pathways of Naming Under Seqcode
There are two mechanisms or pathways on which SeqCode operates for validation of names.
In the first path as described in Fig. 4, the users will have to submit the name of prokaryote along with metadata for draft registration into the SeqCode Registry portal (draft version https://seqco.de/). At same time users can prepare and submit manuscript for publications. Within the Registry, the curators check the data quality and synonymous names, which leads to proposal’s provisional acceptance only if they comply with rules of SeqCode. By this process data quality is ensured and errors before or after publications are avoided. The DOI3 of the publication must also be submitted into the SeqCode Registry in order to determine the date and time of priority, Since the oldest taxon given name should be used, and only the priority date may determine the precedence of name validation.
Fig. 4.

Paths of naming under SeqCode. Path 1 involves naming of one newly published name. Path 2 involves correction of already published names including Candidatus names
The second path (Fig. 4) is meant for those names which are published already such as those under Genus Candidatus. In such cases, the name and supporting metadata are uploaded via the Registration portal [5]. After automated checks and review by curators of SeqCode, the proposed name is accepted and registration along with time and date of priority.
The third mechanism has been proposed for the future and will be developed in partnership with one or more scientific journals. It would involve an integrated path to the validation of proposed names by simultaneous review by peer and curator.
Major goal of SeqCode is to provide an alternative to restrictions put by ICNP of strains being culturable and accessible for nomenclature [6]. The SeqCode solves the problem by providing an effective and straightforward to use resource to the scientific community. Features of SeqCode like findability, accessibility, interoperability and reusability (FAIR) will definitely be appreciated by researchers globally. SeqCode also facilitates communication across microbiological disciplines by providing meaningful and consistent names for prokaryotic diversity.
Principles and Rules of Nomenclature Under Seqcode
Following are the principles and rules for SeqCode, one must follow to submit the data on SeqCode registry. There are currently 10 main principles and 50 rules in SeqCode version 1.0.3.
Mentioned below are some of the most important rules.
Principles
| Provide a standardized, resilient, and stable naming system that is compatible with taxonomic classification flexibility |
| Botanical, zoological, and viral nomenclature are all connected with prokaryotic nomenclature |
| Latinization of names |
| The objective of giving a taxon a name is to make it easier to remember; names should aid memorability |
| Taxon names are linked to their nomenclatural types, which serve as a reference point for clear taxon identification |
| Accepted publication, validity, systematic location and publication priority all contribute to a taxon's right name |
| A name has no status in nomenclature unless it is officially published according to SeqCode standards |
| A taxon can only have one correct name. A taxon's position or rank indicates its link to a parent taxon |
| There should be no mistakes, confusion, or misunderstandings |
| Names should not be changed or updated unless there are compelling reasons or a requirement to correct a name that violates SeqCode guidelines. (Hedlund et al., 2022) |
Rules
1. General
SeqCode became operational on January 1, 2022. The SeqCode Legislative Commission, a part of the SeqCode committee, is the only entity with the authority to alter the SeqCode. The SeqCode registry was created to store and maintain names as well as their valid publications.
2. Ranks of Taxa
The following categories are covered: phylum, class, order, family, genus, species, and subspecies. It is best to use taxonomic classifications of subspecies. SeqCode does not handle intermediate ranks, which are not stated above.
3. Naming of taxa
Latinization is required for scientific names. Species names are binary, consisting of a genus name and a species epithet. A name can only refer to one kind at each taxonomic rank. The genus name, the species epithet abbreviation "subsp," and the subspecies epithet, which begins with a lowercase letter, are combined to form the name of a subspecies. Table 1 shows the suffixes that are used for naming.
Table 1.
Name of Taxa above Genus with their commonly used Suffix
| Rank | Suffix |
|---|---|
| Phylum | -ota |
| Class | -ia |
| Order | -ales |
| Family | -aceae |
Table 1 lists the appropriate Suffixes for naming Phylum, Class, Order and Family.
4. The Nomenclature types and their designation
After taxa have been named, they must be assigned a Nomenclatural type based on their Taxonomic Category, as shown in Table 2.
Table 2.
Taxonomic rank nomenclature types
| Classification of taxa | Designated nomenclatural type |
|---|---|
| Subspecies | DNA sequence |
| Species | DNA sequence |
| Genus | species |
| Family | genus |
| Order | genus |
| Class | genus |
| Phylum | genus |
Table 2 delineates the various Nomenclatural Types for their respected Taxonomic Categories in ascending order from Subspecies to Phylum.
5. Valid and Priority Publication Names
A taxon can only have one right name if it has a definite circumscription, location, and hierarchy. The date and timing of a name's publication define its legitimacy. Only legally registered names are considered for priority purposes. Under ICNP, authentic names are published. remain valid in SeqCode. The taxon's name and proof must have been published in a peer-reviewed journal or book for effective publishing under SeqCode.
6. Authors' and Names' Citation
The name of a previously proposed taxon should be cited in the optimal publication. The publication date, justification, and circumscription of the taxon may all be found with the proper citation of the name. Valid names and the date of publication for SeqCode names should be obtained from the SeqCode Registry. When a circumscription is changed, the words "emend." (emendavit) are added, then the author who made the change, followed by their name.
7. Changes in Taxa Name Due to Transfer, Union, or Rank Change
If a taxon's type is rejected, an alternative form that bears a new name must be created for the remaining members of the taxon. If a genus is divided into two or more genera, only the genus with the type species must keep its name. The species epithet must be kept when a species is moved to another genus without any rank modifications, unless it is already in use in the new genus. In that instance, a new species epithet must be assigned to the transferred species. This rule prevents the emergence of a subsequent homonym. The genus was created when two or more species—including type species—from various genera came together to establish a single genus.
8. Illegitimate Names and Epithets
Replacement, Rejection, and Preservation
A name that disobeys a regulation is invalid and ought not to be used. On the other hand, a taxon name that is illegitimate in one taxonomic position may not always be unlawful in another. A species or subspecies epithet for the same genus that is identical to one that has already been published but whose name is based on a different type is considered invalid. A name that conflicts with the broad ideas or considerations of the code may be rejected by the SeqCode Reconciliation Commission.
9. Orthography
All names are formed using the ISO basic Latin alphabet's 26 letters. Using judgmental signs is not advised. Any name or epithet should be written using the same spelling as the word it is derived from and following the grammar norms of Latin. This prohibition does not apply to typographic and orthographic variations. Unintentional spelling, grammar, or orthographic mistakes that the author corrects after they are reported are accepted in their rectified form without changing the status or publication date of the original work. Another author could correct it, who might or might not notice the spelling correction. The word "corrig" can be abbreviated [4].
Recommendations to Follow for SeqCode Registry
1. Genome Quality
Completion rate more than 90%.
Percentage of Contamination 5%.
There is more than 80% tRNA present.
The presence of 16 s rRNA is greater than 75%
2. Assembly Quality
N50 > 25 Kb (defined as the length of the shortest contig in the collection of biggest contigs that together account for at least half of the entire assembly size).
100 contigs; largest contig > 100 Kb.
3. Naming Conventions
Using SeqCode validation, automatically identifying the Candidatus names currently used in literature in order to normalize and standardize.
4. Description Requirements
Prediction of phenotype or metabolic state based on DNA sequence.
Environmental and biogeographic considerations.
Extra metadata such as protein-coding genes and GC content.
5. MAGs and SAGs4
The Genomic Standards Consortium (GSC) has established guidelines for reporting the quality of bacterial and archaeal genome sequences, encompassing both the genome itself and the associated assembly. These guidelines are embodied in two specific standards: Minimum Information about a Metagenome-Assembled Genome (MIMAG7) for genomes reconstructed from metagenomic data, and Minimum Information about a Single Amplified Genome (MISAG8) for genomes obtained through single-cell amplification [8]. Following are the list of modern Bioinformatics tool which are used meet the specific standards set by MIMAG and MISAG in Table 3.
Table 3.
Commonly Used Bioinformatics Tools
| S. No | Tool | Function |
|---|---|---|
| 1 | Checkm | Completeness check, 16S recovery & Decontamination |
| 2 | anvi’o | Completeness check, Decontamination |
| 3 | rnammer | 16S recovery software |
| 4 | metaxa2 | 16S recovery software |
| 5 | trnascan-se | tRNA extraction software |
| 6 | aragorn | tRNA extraction software |
| 7 | prodege | Decontamination software |
| 8 | metabat | Binning software |
| 9 | maxbin | Binning software |
| 10 | concoct | Binning software |
| 11 | Metawatt | Binning software |
| 12 | Bwa | MAG Coverage software |
| 13 | Bbmap | MAG Coverage software |
| 14 | bowtie | MAG Coverage software |
Table 3 briefly mentions about some of the most widely used bioinformatics tools used for checking and filtering the MAGs and SAGs data according to SeqCode guidelines [8].
Data Submission and Curation Process
SeqCode is still a relatively new system, and the specific details of the review process used by curators haven't been fully established but we have mentioned the Standard Operating Procedure (SOP9) that is being currently used in Fig. 5.
Fig. 5.
Showing the Standard Operating Procedure used when a manuscript is submitted to SeqCode Registry [9]
Advantages of SeqCode Over ICNP
SeqCode allows for the valid publication of names for all prokaryotes, independent of cultivability.
A reproducible and impartial framework for naming based on genomic data is provided by SeqCode.
By giving meaningful and consistent labels for bacterial diversity, SeqCode improves communication across microbiological fields.
SeqCode strives to maximize efficiency and scalability while avoiding human mistakes through automation.
Prevents confusion caused by name changes after publication by executing pre-checks throughout the pre-registration process.
Concerns Related to SeqCode Implementation and Future
Although SeqCode implementation will finally solve a long-lasted grievance of many scientists and researchers through out the worldwide. It does not mean it is readily accepted everywhere. There are many pockets of scientists who raise different types of concerns and effects of SeqCode implementation. Mentioned Below are some most pressing concerns which need to be addressed and resolved quickly by SeqCode Committee.
Will it affect the availability of cultures? Although Implementation of SeqCode does not discourage culturing of prokaryotes. But depending upon which field a microbiologist belongs to—he/she will have a different opinion regarding SeqCode implementation, like for example—an environmental microbiologists will be benefitted from SeqCode, whereas it’s not that practically useful for clinical microbiologists.
Will the studies be reproducible? As the algorithms for metagenome assembly are constantly getting updated and changing, it is difficult to get same results from the same sample using different NGS technologies and assembly algorithms.
Issues related to Candidatus names priority and correction of typographic and orthographic errors [10].
How will the names validly published under SeqCode be treated by IJSEM as it follows the rules of ICNP nomenclature [11, 12]?
Conclusion
Although SeqCode uses genome and metagenome metadata for nomenclature and taxonomy, it is not designed to hinder prokaryote production. Pure or mixed culture cultivation will provide circumstantial and direct proof for Genomic analysis findings. Researchers are also strongly encouraged to add strains to global culture collections.
Additionally, SeqCode does not provide recommendations or criteria for taxonomic demarcation, and applications for previously unrecognized species must be settled through peer review using the nomenclature systems currently in use.
Ultimately, SeqCode will help in expanding the taxonomic scope by encompassing uncultivated microorganisms, SeqCode will allow for a significantly completer and more representative picture of prokaryotic diversity. SeqCode will also help in increased Data accuracy as Genome sequences provide precise and objective information for taxonomic assignments. This avoids the potential subjectivity and variability associated with phenotypic traits, which were heavily relied upon in ICNP-based classifications. SeqCode by design will reduce potential for name changes as SeqCode emphasizes on pre-checks reduces naming conflicts and potential changes after publication.
Acknowledgements
RL acknowledges Indian National Science Academy for support under the INSA Senior Scientist Programme and Alexander von Humboldt Foundation for the award of Fellowship under its Renewed Research Program. PL acknowledges Department of Zoology, University of Delhi for providing the necessary infrastructure and space to do this work.
Footnotes
ICNP – International Code of Nomenclature of Prokaryotes.
MAGs—Metagenome Assembled Genomes.
DOI—Digital Object Identifier.
SAGs—Single Amplified Genomes.
INSDC—The International Nucleotide Sequence Database Collaboration.
SRA – Sequence Read Archive.
MIMAG—Minimum Information about a Metagenome-Assembled Genome.
MISAG—Minimum Information about a Single Amplified Genome.
SOP – Standard Operating Procedure.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Konstantinidis KT, Rosselló-Móra R, Amann R (2017) Uncultivated microbes in need of their own taxonomy. ISME J 11(11):2399–2406. 10.1038/ismej.2017.113 10.1038/ismej.2017.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F et al (2021) A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 10.1038/s41587-020-0718-6 10.1038/s41587-020-0718-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitman WB, Chuvochina M, Hedlund BP, Hugenholtz P, Konstantinidis KT, Murray AE et al (2022) Development of the SeqCode: A proposed nomenclatural code for uncultivated prokaryotes with DNA sequences as type. Syst Appl Microbiol. 10.1016/j.syapm.2022.126305 10.1016/j.syapm.2022.126305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedlund BP, Chuvochina M, Hugenholtz P, Konstantinidis KT, Murray AE, Palmer M et al (2022) SeqCode: a nomenclatural code for prokaryotes described from sequence data. Nat Microbiol. 10.1038/s41564-022-01214-9 10.1038/s41564-022-01214-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer M, Sutcliffe I, Venter SN, Hedlund BP (2022) It is time for a new type of type to facilitate naming the microbial world. New Microbes New Infect. 10.1016/j.nmni.2022.100991 10.1016/j.nmni.2022.100991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitman WB, Sutcliffe IC, Rossello-Mora R (2019) Proposal for changes in the international code of nomenclature of prokaryotes: Granting priority to Candidatus names. Int J Syst Evol Microbiol. 10.1099/ijsem.0.003419 10.1099/ijsem.0.003419 [DOI] [PubMed] [Google Scholar]
- 7.Murray AE, Freudenstein J, Gribaldo S, Hatzenpichler R, Hugenholtz P, Kämpfer P et al (2020) Roadmap for naming uncultivated Archaea and Bacteria. Nature Microbiol. 10.1038/s41564-020-0733-x 10.1038/s41564-020-0733-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK et al (2017) Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 10.1038/nbt.3893 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuvochina M et al (2023) Guide: How are names internally curated? Seqcode Registry, 1.0.3, N.D.
- 10.Whitman WB, Hedlund BP, Palmer M, Sutcliffe I, Chuvochina M (2023) Request for public discussion and ballot to amend SeqCode rules on priority of Candidatus names and correction of typographic and orthographic errors. ISME Commun 3:1–3. 10.1038/s43705-023-00303-y 10.1038/s43705-023-00303-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Göker M, Moore ERB, Oren A, Trujillo ME (2022) Status of the SeqCode in the international journal of systematic and evolutionary microbiology. Int J Syst Evol Microbiol. 10.1099/IJSEM.0.005754 10.1099/IJSEM.0.005754 [DOI] [PubMed] [Google Scholar]
- 12.Arahal D, Bisgaard M, Christensen H, Clermont D, Dijkshoorn L, Duim B et al (2024) The best of both worlds: a proposal for further integration of candidatus names into the international code of nomenclature of prokaryotes. Int J Syst Evol Microbiol. 10.1099/IJSEM.0.006188 10.1099/IJSEM.0.006188 [DOI] [PubMed] [Google Scholar]