Abstract
Paddy fields include two interconnected ecosystems—soil and floodwater. Microbes and viruses are an integral component of these ecosystems, yet the viral communities have not been extensively studied. We present an analysis of the viromes of paddy floodwater collected during the two cropping seasons in India, the kharif and rabi seasons respectively. The overall taxonomic and functional characteristics appeared to be similar in both seasons, suggesting stability of the viral community. Taxonomically, the families of tailed bacteriophages dominated. The predominance of functional roles related to lytic phages further confirmed this. We reconstructed two complete and several partial viral genomes from the assembled data. The genomes did not align with any known sequences, thus representing novel viruses of the floodwater ecosystem.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-024-01292-9.
Keywords: Virome, Aquatic viruses, Paddy floodwater, Rice field
Rice (Oryza sativa L.) is the most widely cultivated cereal crop worldwide. In India, rice is grown in two seasons, the kharif season (June–November), coinciding with the southwest monsoon and the rabi season (December–May), a dry season necessitating irrigation from non-rainwater sources [1]. Rice paddies are generally submerged during the vegetative phase. The shallow water layer above the soil—the floodwater—is considered part of the paddy soil ecosystem, due to the proximity and constant exchanges occurring between the two media [2]. Paddy soil and floodwater are rich in micro- and macro-biodiversity. The microbial component maintains soil fertility and promotes plant growth and health [3]. Concurrent with a thriving microbial community is the viral community, majorly bacteriophages that propagate in bacterial hosts. Viruses are ecological drivers of aquatic and soil ecosystems – they regulate microbial community composition, alter nutrient cycling patterns and influence evolution through horizontal gene transfer [4]. These activities assume greater significance in light of the very high abundances of viruses in natural environments, often an order of magnitude higher than bacteria [5]. The existing research on paddy soil and floodwater microbiomes is tilted in favour of bacterial community studies [3, 6, 7]. Viral community studies have largely been limited to enumeration [8] and marker-gene based studies [9]. We report for the first time, virome-level analysis of paddy floodwater during the kharif and rabi cultivation seasons.
Surface water samples (25 cm depth) from paddy floodwaters, were collected in sterile polypropylene containers, 3 weeks after transplantation of rice seedlings, in the respective seasons. Locations of sampling sites and physicochemical parameters of water samples are detailed in Table S1. The kharif sample is hereafter referred to as RW1, and the rabi sample as RW2. Following 0.22 µm-filtration of water samples, viral metagenomes were prepared by a previously described protocol [10]. Shotgun metagenomic sequencing was carried out at Medgenome Labs, India.
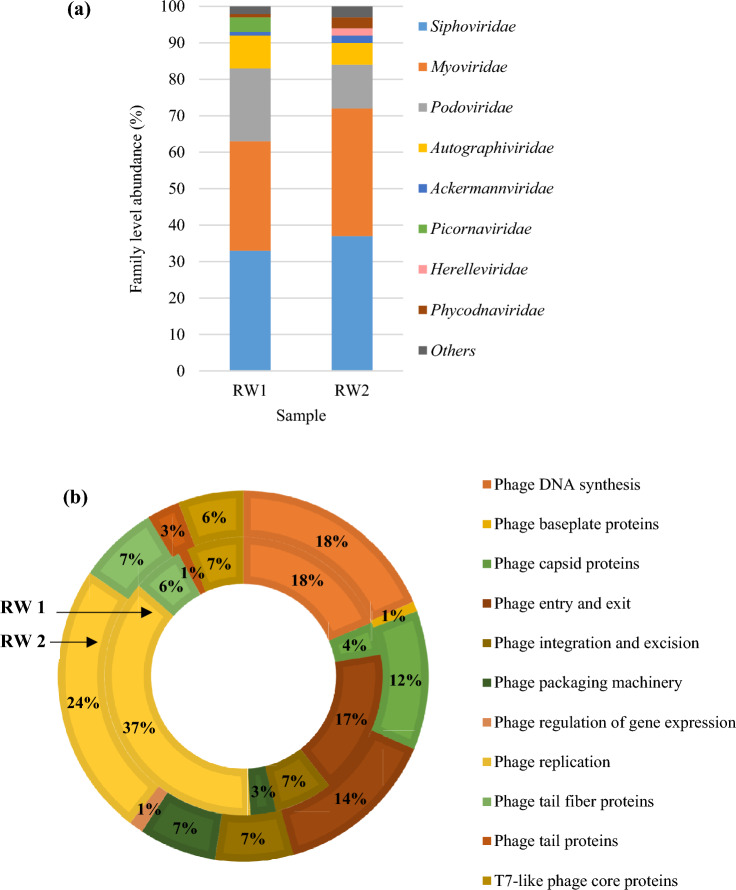
Paired-end sequencing reads from RW1 and RW2 were submitted to the NCBI sequence read archive under the BioProject accession number PRJNA958449. The reads were quality-filtered with FastQC v. 0.11.9 [11] and submitted to the Kaiju web server, v. 1.9.0 [12] for taxonomic annotation against the NCBI-nr database. A large majority of reads in both viromes, i.e. 94% in RW1 and 92% in RW2, were unclassified. Among classified sequences, 11% were identified as viral in each of RW1 and RW2. The distribution of classified viral sequences at key taxonomic hierarchical levels was analysed. Duplodnaviria was the dominant realm of both viromes, constituting 94% of sequences in RW1 and 95% in RW2 (Fig. S1). The most abundant viral families in both RW1 and RW2 were Siphoviridae, Myoviridae and Podoviridae, in that order (Fig. 1a). Aquatic viromes ranging from marine [13] to lake [14], estuarine [15], creek [16], and wastewater [17] consistently contain a majority of sequences mapped to these families. This reflects both the true abundance of tailed bacteriophages in aquatic and soil environments and the predominance of their genomic sequences in the reference databases, due to the ease of their isolation and cultivation. Families Autographiviridae, Ackermannviridae and Phycodnaviridae (both RW1 and RW2), Picornaviridae (RW1) and Herelleviridae (RW2) were minor constituents of respective viromes. The similarity in viral community structure between RW1 and RW2 viromes suggests stability of the paddy floodwater viral community under different cropping seasons and irrigation regimes. This finding is in consonance with previous large-scale studies, where the viromes of similar ecosystems have been found similar, even across geographical distances [18–20].
Fig. 1.
Taxonomic and functional annotation of the RW1 and RW2 viromes. a Taxonomic annotation of classified viral reads at the family level; b Functional annotation of the viromes through MG-RAST—the figure depicts functional descriptions at Level 3 specificity; The RefSeq filter “Viruses” was applied to limit results to viral functions; Non-specific annotations were omitted
Further, the species composition of the viromes was analysed (Table S2). In RW1, Lentibacter virus vB_LenP_ICBM2 (5% of classified virus sequences) and Enterovirus C (4%) were the most abundant viral species. Enteroviruses (Family: Picornaviridae) are indicators of sewage pollution in water and cause polio, viral meningitis and encephalitis in humans. The possibility of sewage runoff into paddy fields with rainwater in the kharif season is a public health risk, and necessitates better biological monitoring of floodwater. No single viral species constituted more than 2% of the RW2 virome.
Functional annotation of raw reads was carried out in MG-RAST v. 4.0.3 [21] against the Subsystems database. The dominant viral functions identified in both the viromes were phage replication and DNA synthesis (Fig. 1b). This finding suggests that the large abundance of viruses in paddy floodwater [22] constitutes an actively replicating viral community. The functions indicative of active lytic infections support the significant ecological role of viruses in the paddy floodwater ecosystem [23, 24]. Phage integration and excision functions were at an intermediate level in both viromes, suggesting a relatively minor presence of lysogenic phages, as with other nutrient-rich freshwater environments [4].
Quality-filtered sequencing reads were further trimmed and assembled into contigs. Contigs representing complete viral genome sequences were annotated with PHASTEST [25]. The workflow for bioinformatics analysis is provided in Table S3 and details of complete genomes in Table S4.
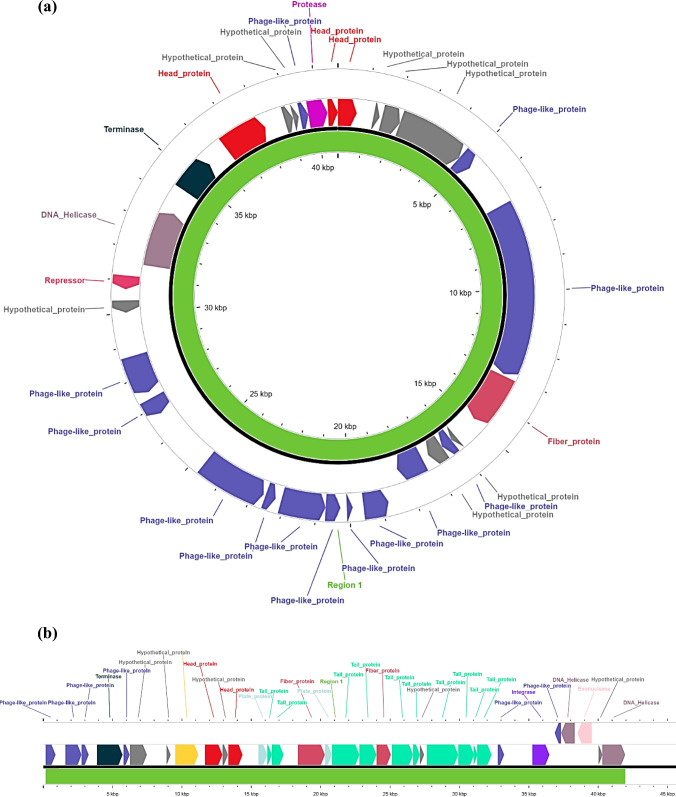
Two intact viral (phage) genomes were identified in the RW1 dataset and designated Φ RW1.1 and Φ RW1.2 respectively. Φ RW1.1 appeared to have a circular genome (Fig. 2a) of size 40.7 kb, with 29 phage genes identified, the majority of which, closely matched with Bordetella phage BPP-1. Specific protein identifications included phage head proteins, terminase, protease, DNA helicase, repressor and fiber protein. The remaining regions were non-specifically annotated. Thus, it could not be concluded whether Φ RW1.1 is a tailed phage or not. Φ RW1.2 appeared to have a linear genome (Fig. 2b) of size 41.7 kb, with 33 phage genes identified, most of which closely matched with Enterobacter phage Arya. Specific protein annotations included several tail proteins, in addition to head proteins, terminase, DNA helicase, endonuclease and integrase. Thus, Φ RW1.2 is highly likely to be a tailed phage. Additionally, three genome sequences were partially reconstructed (Table S4). A BLAST search of the respective sequences revealed no significant alignment to any sequence in the NCBI nr database. Thus, these are novel phages of the floodwater viral community, and merit further exploration through metagenomic as well as laboratory cultivation-based approaches.
Fig. 2.
Protein annotation of phage genomes identified as intact, a Φ RW1.1, a circular genome and b Φ RW1.2, a linear genome. Functional terms such as “Head protein” or “Tail protein” are provided by the software; “Hypothetical protein” and “Phage-like protein” indicate sequences with phage-gene-like characteristics, but which could not be assigned specific functions; “Region” indicates the contiguous sequence identified as a complete phage genome
Studies on paddy soil microbiomes have reported a relatively constant bacterial community under different fertilization practices [26] and cropping patterns [27]. The similar taxonomic and functional composition of the RW1 and RW2 viromes is in line with previous findings, as the viral community structure generally parallels that of the microbial hosts. Moreover, paddy floodwater is closely connected with the soil. Thus, the stability of soil communities may be reflected in the floodwater communities. This is particularly significant given the fact that the kharif crop is rainfed, while the rabi crop is irrigated. Several studies have also indicated that microbial community composition is highly dependent on the growth phase of the rice plant [3, 28]. Samples in the present study were collected during the vegetative phase in both seasons. In conclusion, the viral community of paddy floodwater was dominated by actively replicating tailed bacteriophages, during both kharif and rabi cropping seasons. Further research in this area could include the analysis of floodwater viromes during the reproductive phase of rice plants and the exploration of floodwater bacterial communities in parallel with the viral communities. The novel phage genomes ΦRW1.1 and ΦRW1.2 represent our contribution to nucleotide databases, which would help to annotate viral sequence data in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Mr. Venkatesh Prabhu for his help with running the bioinformatics software.
Author Contribution Statement
Sarvesh R. Halankar: Investigation, Formal Analysis, Writing—original draft; Judith M. Noronha: Conceptualization, Investigation, Formal Analysis, Supervision, Writing—original draft, Writing—review and editing.
Funding
This research was partly supported by the Seed Money Grant No. GU/D-RDRM/Seed money/JN/Microbiology/2021/12 from Goa University.
Data Availability
The raw sequencing reads of RW1 and RW2 were submitted to the NCBI sequence read archive under the BioProject Accession Number PRJNA958449.
Declaration
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Frolking S, Yeluripati JB, Douglas E (2006) New district-level maps of rice cropping in India: a foundation for scientific input into policy assessment. F Crop Res 98:164–177. 10.1016/j.fcr.2006.01.004 10.1016/j.fcr.2006.01.004 [DOI] [Google Scholar]
- 2.Watanabe I, Furusaka C (1980) Microbial ecology of flooded rice soils. Adv Microbial Ecol, pp 125–168. 10.1007/978-1-4615-8291-5_4
- 3.Pittol M, Scully E, Miller D et al (2018) Bacterial community of the rice floodwater using cultivation-independent approaches. Int J Microbiol. 10.1155/2018/6280484 [DOI] [PMC free article] [PubMed]
- 4.Sime-Ngando T (2014) Environmental bacteriophages: viruses of microbes in aquatic ecosystems. Front Microbiol 5:1–14. 10.3389/fmicb.2014.00355 10.3389/fmicb.2014.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mushegian AR (2020) Are there 1031 virus particles on Earth, or more, or less? J Bacteriol, pp 1–14. 10.1128/JB.00052-20 [DOI] [PMC free article] [PubMed]
- 6.Ezeokoli OT, Nuaila VNA, Obieze CC, et al (2021) Assessing the impact of rice cultivation and off-season period on dynamics of soil enzyme activities and bacterial communities in two agro-ecological regions of mozambique. Agronomy 11. 10.3390/agronomy11040694
- 7.Imchen M, Kumavath R, Vaz ABM et al (2019) 16S rRNA Gene amplicon based metagenomic signatures of Rhizobiome community in rice field during various growth stages. Front Microbiol 10:1–15. 10.3389/fmicb.2019.02103 10.3389/fmicb.2019.02103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakayama N, Okumura M, Inoue K et al (2007) Abundance of bacteriophages of common heterotrophic bacteria in the floodwater of a Japanese paddy field. Soil Sci Plant Nutr 53:595–605. 10.1111/j.1747-0765.2007.00189.x 10.1111/j.1747-0765.2007.00189.x [DOI] [Google Scholar]
- 9.Wang G, Liu J, Yu Z et al (2014) Unique distribution of cyanobacterial podoviruses and their potential hosts in a paddy field of northeast China. FEMS Microbiol Ecol 90:331–334. 10.1111/1574-6941.12401 10.1111/1574-6941.12401 [DOI] [PubMed] [Google Scholar]
- 10.Thurber RV, Haynes M, Breitbart M et al (2009) Laboratory procedures to generate viral metagenomes. Nat Protoc 4:470–483. 10.1038/nprot.2009.10 10.1038/nprot.2009.10 [DOI] [PubMed] [Google Scholar]
- 11.Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. In: Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 15 Jan 2023
- 12.Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7. 10.1038/ncomms11257 [DOI] [PMC free article] [PubMed]
- 13.Gao C, Liang Y, Jiang Y, et al (2022) Virioplankton assemblages from challenger deep, the deepest place in the oceans. iScience 25:104680. 10.1016/j.isci.2022.104680 [DOI] [PMC free article] [PubMed]
- 14.Butina TV, Bukin YS, Petrushin IS et al (2021) Extended evaluation of viral diversity in lake baikal through metagenomics. Microorganisms 9:1–31. 10.3390/microorganisms9040760 10.3390/microorganisms9040760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai L, Zhang R, He Y et al (2016) Metagenomic analysis of Virioplankton of the subtropical Jiulong river estuary, China. Viruses 8:1–13. 10.3390/v8020035 10.3390/v8020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chopyk J, Nasko DJ, Allard S et al (2020) Metagenomic analysis of bacterial and viral assemblages from a freshwater creek and irrigated field reveals temporal and spatial dynamics. Sci Total Environ 706:135395. 10.1016/j.scitotenv.2019.135395 10.1016/j.scitotenv.2019.135395 [DOI] [PubMed] [Google Scholar]
- 17.Salih H, Karaynir A, Yalcin M et al (2022) Metagenomic analysis of wastewater phageome from a University Hospital in Turkey. Arch Microbiol 204:1–15. 10.1007/s00203-022-02962-2 10.1007/s00203-022-02962-2 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Brito B, Li LL, Wegley L et al (2010) Viral and microbial community dynamics in four aquatic environments. ISME J 4:739–751. 10.1038/ismej.2010.1 10.1038/ismej.2010.1 [DOI] [PubMed] [Google Scholar]
- 19.Dávila-Ramos S, Castelán-Sánchez HG, Martínez-ávila L et al (2019) A review on viral metagenomics in extreme environments. Front Microbiol 10:1–19. 10.3389/fmicb.2019.02403 10.3389/fmicb.2019.02403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux S, Enault F, Robin A et al (2012) Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7. 10.1371/journal.pone.0033641 [DOI] [PMC free article] [PubMed]
- 21.Keegan KP, Glass EM, Meyer F (2016) MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol Biol 1399:207–233. 10.1007/978-1-4939-3369-3_13 10.1007/978-1-4939-3369-3_13 [DOI] [PubMed] [Google Scholar]
- 22.Noronha JM, Mulla AB, Gauns MU, Ghadi SC (2018) Enumeration of total virioplankton and isolation of specific cyanophages from selected aquatic ecosystems in Goa, India. Curr Sci 115:2147–2150. 10.18520/cs/v115/i11/2147-2150
- 23.Trubl G, Hyman P, Roux S, Abedon ST (2020) Coming-of-age characterization of soil viruses: a user’s guide to virus isolation, detection within metagenomes, and viromics. Soil Syst 4:1–34. 10.3390/soilsystems402002333629038 10.3390/soilsystems4020023 [DOI] [Google Scholar]
- 24.Li Y, Sun H, Yang W et al (2019) Dynamics of bacterial and viral communities in paddy soil with irrigation and urea application. Viruses 11. 10.3390/v11040347 [DOI] [PMC free article] [PubMed]
- 25.Arndt D, Marcu A, Liang Y, Wishart DS (2019) PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief Bioinform 20:1560–1567. 10.1093/bib/bbx121 10.1093/bib/bbx121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorfer M, Borruso L, Deltedesco E et al (2022) The effect of environmental parameters and fertilization practices on yield and soil microbial diversity in a Kenyan paddy rice field. Appl Soil Ecol 176. 10.1016/j.apsoil.2022.104495
- 27.Maguire VG, Bordenave CD, Nieva AS et al (2020) Soil bacterial and fungal community structure of a rice monoculture and rice-pasture rotation systems. Appl Soil Ecol 151:103535. 10.1016/j.apsoil.2020.103535 10.1016/j.apsoil.2020.103535 [DOI] [Google Scholar]
- 28.Fernández-Baca CP, Rivers AR, Kim W et al (2021) Changes in rhizosphere soil microbial communities across plant developmental stages of high and low methane emitting rice genotypes. Soil Biol Biochem 156:108233. 10.1016/j.soilbio.2021.108233 10.1016/j.soilbio.2021.108233 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing reads of RW1 and RW2 were submitted to the NCBI sequence read archive under the BioProject Accession Number PRJNA958449.