Abstract
Live-attenuated human immunodeficiency virus type 1 (HIV-1) variants have shown great promise as AIDS vaccines, but continued replication can lead to the selection of faster-replicating variants that are pathogenic. We therefore designed HIV-1 genomes that replicate exclusively upon addition of the nontoxic effector doxycycline (dox). This was achieved by replacement of the viral TAR-Tat system for transcriptional activation by the Escherichia coli-derived Tet system for inducible gene expression. These designer “HIV-rtTA” viruses replicate in a strictly dox-dependent manner both in a T-cell line and in primary blood cells, and the rate of replication can be fine-tuned by simple variation of the dox concentration. These HIV-rtTA viruses provide a tool to perform genetics, e.g., selection and optimization experiments, with the E. coli-derived Tet reagents in a eukaryotic background. Furthermore, such viruses may represent improved vaccine candidates because their replication can be turned on and off at will.
Live-attenuated virus vaccines (such as vaccinia, polio, and measles viruses) have been enormously successful and have made a dramatic and historic impact on public health. Replicating virus vaccines also demonstrated superior performance in AIDS vaccine trials (1, 14, 32, 40, 45, 46, 49, 54). However, safety concerns remain for the human immunodeficiency virus type 1 (HIV-1) about either the reversion of attenuated vaccine strains to virulent phenotypes or the induction of fulminant infection in (immunocompromised) individuals. Testifying to the genetic instability of such strains is the recent demonstration that the HIV-1 Δ3 vaccine candidate, which contains three deletions in nonessential parts of the genome, is able to regain full replication capacity within 4 months of replication in tissue culture (11). It has also been reported that the viral load of attenuated simian immunodeficiency virus (SIV) variants increased after several years in some infected monkeys, concomitant with the onset of AIDS (2). Furthermore, although there is some evidence that attenuated HIV-1 variants lacking the nef gene result in a benign course of infection in humans (17), a decline in CD4+ T-cell numbers has been reported recently for some of these individuals, which is an early sign that these persons could develop AIDS (20, 25).
To improve the safety of potential HIV-1 vaccine strains, we designed an HIV-1 variant for which replication depends on the addition of the nontoxic, selective effector doxycycline (dox). This was done by incorporation of the Tet system for inducible gene expression (5, 23, 24) into the viral genome. This system is based on two elements from the Escherichia coli tet operon, the tetracycline-inducible repressor protein (TetR) that has been converted into a eukaryotic transcriptional activator (tTA or rtTA), and the tetO operator DNA sequence. The Tet system has found wide application and allows strict and graded regulation of gene expression in many experimental setups, for example, in the breeding of transgenic animals and in gene therapy approaches. The Tet system has been successfully used to regulate the expression of transgenes in a variety of viral vector systems, including lentiviral vectors. We now report a completely novel strategy to impose control over HIV-1 replication by replacement of the viral trans-activator protein Tat and its binding site TAR by the two components of the Tet system, such that an exogenous agent (dox) can be used to turn virus replication on in a reversible manner.
MATERIALS AND METHODS
Construction of the HIV-rtTA molecular clones.
The following primers were used in mutagenesis and cloning: BglIINef (5′-AGCTGTAGATCTTAGCCAC-3′, sense), deltaU3 (5′-GACAAGATATCCTTGATCTG-deletion-GAAGTGTTAGAGTGGAGGT-3′, sense), Sp6BspEI (5′-ATTTAGGTGACACTATAGGTACTCCGGATGCAGCTCTCG-3′, antisense), TARB123L13 (5′-CCAGAGA GCTCCAATGCTCCTTTCTGGTCTAACCAGAGAGACC-3′, antisense), XcmIdeltaNef (5′-GCTTGGAAAGGATTTTGCTATAACCATGTCTAGACTGG-deletion-CCAGTCACACCTCAGGTACC-3′, sense), BamHIEnv (5′-GAACTAGTGGATCCTTAGCACTTATC-3′, sense), SmaIanti-Nef (5′-TCCCCCGGGGTGGCTAAGATCTACAGCTGC-3′, antisense), Anti-U3att (5′-GGAGTGAATTAGCCCTTCCA-3′, antisense), SalISp1 (5′-GACATCGAGCTTGCTACAA-deletion-GTCGACAGGGAGGCGTGGCCTG-3′, sense), κBSalISp1 (5′-TCCGCTGGGGACTTTCCGTCGACAGGGAGGCGTGGCCTG-3′, sense), Sp6anti-luc (5′-ATTTAGGTGACACTATAGGCAGTTGCTCTCCAGCGGTTCC-3′, antisense), KB12− (5′-CTGGAAAGTCCCCAGCGGAAAGTCCCTT-3′, antisense), M13reverse (5′-AGGAAACAGCTATGACCAT-3′, sense), and ExtN4Not (5′-NNCGCCGGCGACTCAAGGCAAGCTTTATTGAGGCTTAAG-3′, antisense). Changes compared with the sequence of LAI are marked as follows: insertions are underlined, substitutions are in boldface, deletions are denoted as such, and non-template 5′ extensions are shown in italics.
The following tetO motif was used: 5′-CTCGAGTTTACCACTCCCTATCAGTGATAGAGAAAAGTGAAAGTCGAC-3′ and 3′-GAGCTCAAATGGTGAGGGATAGTCACTATCTCTTTTCACTTTCAGCTG-5′. The sequence of a single tetO double-stranded DNA site is shown (underlined), flanked by XhoI and SalI restriction sites. The dimeric, tetrameric, and hexameric tetO linkers were generated by XhoI/SalI digestion and subsequent ligation of the compatible sticky ends. Because ligation of the XhoI/SalI sticky ends does not reform these restriction enzyme sites, the multimerized head-to-tail tetO elements could be cloned as XhoI/SalI fragments in pBluescript II SK(+) (R. Loew, personal communication).
We used standard DNA cloning and amplification methods (41), and all mutations were verified by dideoxy sequencing using ET Dyeprimer technology (Amersham) on an Applied Biosystems automated DNA sequencer. The stepwise cloning procedure was as follows.
Cloning. (i) Introduction of a SalI site and deletion of NF-κB elements in the HIV-1 LTR promoter.
To facilitate the introduction of tetO operator elements in the long terminal repeat (LTR) promoter, a SalI restriction site was introduced immediately upstream of the Sp1 sites. A mutagenesis PCR (42) was done on pBlue3′LTR-luc (31) with one of the mutagenic primers (κBSalISp1 or SalISp1 [primer M]) and the general primers M13reverse (primer 1), ExtN4Not (primer 2), and Sp6anti-luc (primer 3). After we performed separate PCR reactions with primer sets 1+2 and M+3, the PCR products were purified from a gel and used as templates in a final PCR with primer set 1+3. The PCR products from the final reactions were digested with XhoI and HindIII and used to replace the corresponding fragments in the pBlue3′LTR-luc construct, thereby generating plasmids pBlue3′LTR-luc-κBSalISp1 (K promoter) and pBlue3′LTR-luc-SalISp1 (S promoter; NF-κB sequences deleted).
(ii) Introduction of tetO elements in the HIV-1 LTR promoter.
The pBlue3′LTR-luc-κBSalISp1 and pBlue3′LTR-luc-SalISp1 constructs were linearized by SalI digestion. The 2-mer, 4-mer, and 6-mer tetO repeats were isolated from the pBluescript backbone through digestion with XhoI and SalI and then ligated into the linearized luciferase reporter construct. Clones were selected in which the operator sequences are present in the sense orientation. We thus generated LTR-luc constructs containing 2, 4, and 6 tetO element repeats placed between the NF-κB and Sp1 binding sites (κB-2xtetO-Sp1/κB-4xtetO-Sp1/κB-6xtetO-Sp1) and a similar set of reporter constructs in which both NF-κB elements are deleted (2xtetO-Sp1/4xtetO-Sp1/6xtetO-Sp1). Fortuitously, we isolated a clone in which two tetrameric tetO repeats were ligated in tandem, yielding a promoter with eight tetO elements between the NF-κB and Sp1 sites (κB-8xtetO-Sp1).
(iii) Inactivation of the TAR RNA element.
Mutation of the TAR bulge and loop sequences was performed in a PCR on the pBlue3′LTR-luc plasmid (31) with the primers κBSalISp1 and TARB123L13. The PCR product was cut with SalI and SacI and exchanged with the homologous fragment from the κB-8xtetO-Sp1 and 6xtetO-Sp1 LTR-luc constructs. This procedure generated plasmids pBlue3′LTR-luc-κB-8xtetO-Sp1TAR* (pBlue3′LTR-lucK8TAR*) and pBlue3′LTR-luc-6xtetO-Sp1TAR* (pBlue3′LTR-lucS6TAR*).
(iv) Construction of a Nef-3′LTR shuttle vector.
To facilitate the modification of the 3′ end of the HIV-1 genome, we constructed a shuttle vector to exchange Nef-3′LTR sequences with the full-length HIV-1 infectious clone pLAI (43). To this end, pLAI was cut with BamHI and BglI enzymes, and the resulting 2.3-kb fragment was cloned into the corresponding sites of pBluescript KS(+), generating plasmid pBlue3′LTRext.
(v) Deletion of U3 sequences.
Mutagenesis PCRs (42) were performed on pBlue3′LTRext with mutagenic primer deltaU3 (primer M) and general primers BglIINef (primer 1), Sp6BspEI (primer 2), and KB12− (primer 3). Separate PCRs were performed with primer sets 1+2 and M+3, after which the products were purified from a gel and used as templates in a final PCR with primer set 1+3. The resulting PCR product was digested with BglII and BspEI and used to replace the corresponding fragment of plasmid pBlue3′LTRext, generating pBlue3′LTRext-deltaU3.
(vi) Deletion of nef sequences and introduction of an XcmI site.
A mutagenesis PCR (42) was performed on pBlue3′LTRext-deltaU3 with mutagenic primer XcmIdeltaNef (primer M) and general primers BamHIEnv (primer 1), SmaIanti-Nef (primer 2), and Anti-U3att (primer 3). Separate PCRs were performed with primer sets 1+2 and M+3, after which the PCR products were purified from a gel and used as templates in a final PCR with primer set 1+3. The resulting PCR product was used to replace wild-type nef sequences in the pBlue3′LTRext-deltaU3 construct through the SpeI (vector-encoded) and BglII sites, generating pBlue3′LTRext-deltaU3XcmI.
(vii) Insertion of the rtTA gene in the nef locus.
The plasmid pUHD 52-1, containing the novel rtTA-S2 gene (48), was cut with XcmI and BamHI, and the rtTA gene fragment was ligated into the nef locus via XcmI and BglII restriction sites in pBlue3′LTRext-deltaU3XcmI. The resulting construct was named pBlue3′LTRext-deltaU3rtTA.
(viii) Introduction of tetO elements and TAR mutations in the 5′ and 3′ LTR shuttle vectors.
pBlue3′LTR-lucK8TAR* and pBlue3′LTR-lucS6TAR* were cut with BspEI and HindIII and used to replace the homologous wild-type fragments of pBlue5′LTR (36) and pBlue3′LTRext-deltaU3rtTA. Constructs were named pBlue5′LTRK8TAR*/pBlue5′LTRS6TAR* and pBlue3′LTRext-deltaU3rtTA-K8TAR*/pBlue3′LTRext-deltaU3rtTA-S6TAR*. At this step all the desired modifications have been introduced in the 5′ and 3′ LTR shuttle vectors, and these modified LTRs can now be introduced in the full-length HIV-1 molecular clone pLAI.
(ix) Introduction of the Tet system elements in the 5′ and 3′ LTRs of pLAI.
The constructs pBlue5′LTR-K8TAR* and pBlue5′LTR-S6TAR* were amplified in the methylation-deficient E. coli strain 3902 and cut with XbaI and ClaI restriction enzymes. Unmethylated wild-type pLAI and Tat mutant pLAI Y26A (51) DNA was cut with these two enzymes, and the U3/R region of the 5′ LTR of these constructs was replaced by the K8TAR* and S6TAR* fragments, yielding constructs KW, KY, SW, and SY (“K” and “S” denote the promoter configuration κB-8xtetO-Sp1-TARB123L13 [K] or 6xtetO-Sp1-TARB123L13 [S]; “W” and “Y” indicate the status of the Tat gene [W, wild type; Y, inactive Y26A Tat mutant]). The rtTA gene and tetO elements were introduced at the 3′ end of the infectious clones by cutting the KW, KY, SW, and SY constructs with BamHI and BglI and replacing this 3′ LTR fragment by the BamHI/BglI fragments of the pBlue3′LTRext-deltaU3rtTA-K8TAR*/pBlue3′LTRext-deltaU3rtTA-S6TAR* shuttle vectors, such that identical promoter sequences are present in both the 5′ and the 3′ LTRs (constructs KWK, KYK, SWS, and SYS).
Cells and viruses.
The SupT1 T-cell line was grown and transfected by electroporation as described previously (3). Infection of SupT1 cells with the SWS virus was performed by incubating 6 × 106 cells with 2,200 ng of CA-p24 of a SupT1-produced virus stock for 4 h at 37°C. Subsequently, the cells were washed twice to remove residual dox that was present in the virus stock and split into six different culture wells, after which each culture received the appropriate additives. In some cultures, dox was added at a final concentration of 1,000 ng/ml; zidovudine (AZT) and saquinavir (SQV) were used at 1,000 and 200 nM, respectively. The culturing and transfection by CaPO4 precipitation of the adherent C33A cervical carcinoma cell line was as described previously (16). Peripheral blood mononuclear cells (PBMCs) were isolated, cultured, and transfected by electroporation as described earlier (3). At 7 days after transfection, half of the culture medium was replaced by medium containing 2 × 106 freshly stimulated PBMCs. Virus production in the culture medium was quantitated in a CA-p24 antigen enzyme-linked immunosorbent assay (ELISA).
RESULTS
Construction of HIV-rtTA viruses.
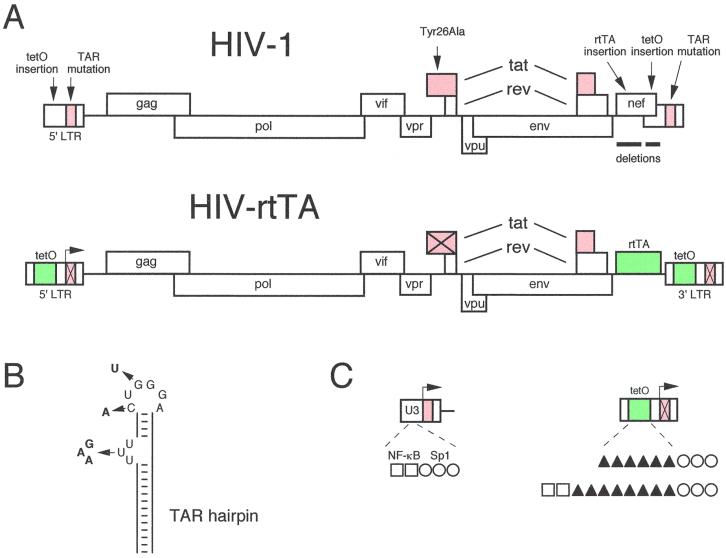
The full-length, infectious HIV-1 molecular clone pLAI was used for construction of an HIV-rtTA virus genome, the transcription of which can be controlled by dox. The viral transcriptional elements TAR and Tat (marked red in Fig. 1A) were replaced by the prokaryotic tetO-rtTA elements (green). In total, nine cloning steps were required to construct these putative dox-dependent viral genomes, and all details of the construction are provided in Materials and Methods. In general, we took a conservative approach with regard to the type of mutations that were introduced in the HIV-1 genome in order to minimize the chance of inactivating important sequences that constitute replication signals.
FIG. 1.
Design of dox-dependent HIV-1 variants. (A) HIV-1 genome and modifications that were introduced to construct HIV-rtTA. Details of the nine-step construction are provided in Materials and Methods. In brief, we inactivated the TAR-Tat transcriptional axis (marked in red), which was replaced by the tetracycline-inducible tetO-rtTA system (marked in green). Inactivation of TAR and Tat is indicated by crosses through the motifs. The size of the HIV-rtTA genome is larger than that of HIV-1 (the maps are not drawn to scale). The RNA genome of HIV-1 LAI is 9,229 nt, and HIV-rtTA is either 9,767 nt (the SYS and SWS variants) or 9,875 nt (KYK and KWK variants). (B) TAR hairpin structure and inactivating mutations that were introduced in the bulge (triple-nucleotide substitution) and in the loop (two point mutations). This RNA target binds the Tat-cyclin T1 complex during transcriptional activation of the LTR promoter (53). Panel C provides some details of the tetO insertion in the LTR promoter. The U3 region of the wild-type LTR (left) encodes two NF-κB sites (□) and three Sp1 sites (○). The modified LTR (right) contains either six or eight tetO operators (▴) upstream of the Sp1 sites. The six tetO variant (mutant S) has no NF-κB sites, whereas both NF-κB sites are present upstream of the eight tetO operators (mutant K). The arrow marks the transcription start site at the U3-R border, which also denotes the 5′ border of the TAR RNA hairpin motif.
TAR and Tat inactivation.
The TAR RNA hairpin motif of nascent HIV-1 transcripts is recognized by the viral Tat protein and the cellular cyclin T cofactor (19, 53). Both TAR and Tat are essential for transcription from the viral LTR promoter and virus replication. TAR was inactivated by mutation of multiple nucleotides in the single-stranded bulge and loop domains, the binding sites for Tat and cyclin T, respectively (Fig. 1B). This produces a fully inactive TAR motif because even single point mutations in one of these single-stranded TAR domains have a dramatic effect on LTR transcription and virus replication (8, 9, 21). We did not introduce more gross sequence changes or deletions in TAR because this sequence is also essential for virus replication as a repeat-R region during strand transfer of reverse transcription (10). Although we demonstrated previously that the TAR element of the 5′ LTR is inherited in both LTRs of the viral progeny (38), the inactive TAR motif was inserted in both LTRs to minimize the chance of reversion to the wild-type virus by a recombination event.
Inactivation of the Tat protein was accomplished by introduction of the Tyr26Ala point mutation. This single amino acid change results in a severe loss of Tat transcriptional activity and virus replication (51). The corresponding codon change (UAU to GCC) was designed to restrict the likelihood of simple reversion to the wild-type amino acid, which requires at least two nucleotide substitutions (50). It has been suggested that Tat may play additional roles in the replication cycle besides its transcriptional function (26, 28, 47). Thus, Tat may facilitate HIV-rtTA replication even in the absence of an intact TAR element, and we therefore also constructed viruses that retained the wild-type tat gene. These constructs are referred to here as Y (tyrosine mutant) and W (wild-type).
rtTA and tetO insertion.
Two deletions were introduced in the 3′-terminal nef gene of the HIV-1 genome to create space for insertion of the components of the Tet system (Fig. 1A). We removed a 250-nucleotide (nt) upstream nef fragment and a 200-nt downstream fragment overlapping the U3 region of the 3′ LTR. This U3 deletion will be inherited by the viral progeny in both LTRs. The exact borders of the nef and U3 deletions were carefully chosen such that important cis-acting sequences for virus replication were not affected. In particular, we maintained approximately 80 nt around the 5′ end of the 3′ LTR (Fig. 1A). This region encodes multiple sequence elements that are critical for reverse transcription (30) and integration (12). In fact, we tried to mimic spontaneous deletions that have been observed in the nef-U3 region of several HIV and SIV variants in a variety of replication studies (22, 29, 33, 34). To prepare for the insertion of the exogenous rtTA gene into the position of the nef gene, a short synthetic sequence was inserted that provides a translational start codon (underlined) in an optimized sequence context (CCAUGU [39]) and convenient restriction enzyme recognition sites. We used the novel rtTA-S2 variant with improved properties for insertion in frame with the optimized start codon (48). Thus, rtTA translation should occur from the subgenomic mRNA that was originally meant for expression of the Nef protein.
To identify the optimal configuration of an LTR promoter with rtTA-responsive tetO elements, we performed transient transfection studies with a variety of LTR-luc constructs (K. Verhoef et al., manuscript in preparation). We varied the number of tetO motifs (2, 4, 6, or 8) that were inserted upstream of the three Sp1 binding sites of the HIV-1 LTR promoter (Fig. 1C). We also tested constructs with or without the two upstream NF-κB elements. The two promoters that provided most robust dox-inducible transcriptional activation were selected for insertion into the HIV-1 genome, and these LTRs are schematically depicted in Fig. 1C. They will be referred to as the K mutant (2 NF-κB + 8 tetO + 3 Sp1 sites) and the S mutant (6 tetO + 3 Sp1 sites). Although insertion into the U3 region of the 3′ LTR will be sufficient to produce a mutant progeny, we also introduced the tetO motifs in the 5′ LTR to generate molecular clones of which the initial round of gene expression in transfected cells is also dox dependent. Thus, both LTRs were modified, and this was done in the wild-type (W) and mutant (Y) Tat backgrounds, resulting in four HIV-rtTA constructs: KWK, KYK, SWS, and SYS. All HIV-rtTA molecular clones have the TAR inactivation and rtTA insertion in common, but they differ in the status of the tat gene and the type of tetO insert (summarized in Table 1). The virus variant KWK is most wild type-like because it maintained the NF-κB sites and a wild-type Tat protein, and SYS is the most minimal HIV-rtTA version.
TABLE 1.
HIV-rtTA constructs
| HIV-rtTA variant | Promoter configurationa | tat gene type | TAR type |
|---|---|---|---|
| KWK | □□▴▴▴▴▴▴▴▴○○○ | Wild type | Mutant |
| KYK | □□▴▴▴▴▴▴▴▴○○○ | Mutant | Mutant |
| SWS | ▴▴▴▴▴▴○○○ | Wild type | Mutant |
| SYS | ▴▴▴▴▴▴○○○ | Mutant | Mutant |
□, NF-κB; ▴, tetO; ○, Sp1.
HIV-rtTA replicates in a dox-dependent manner.
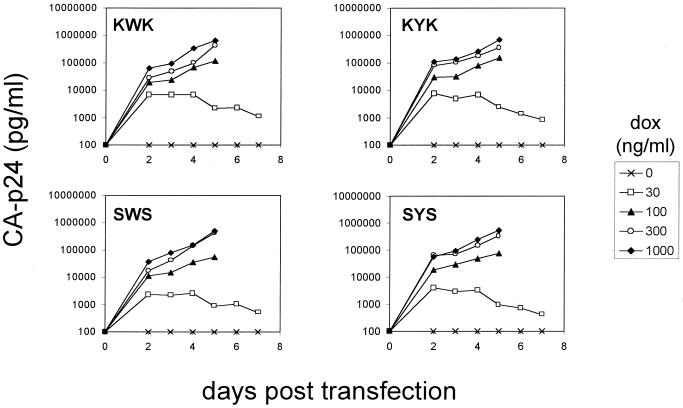
The four pLAI plasmids were individually transfected into the SupT1 T-cell line to test for their replication capacity. The culture was maintained at various dox levels, and virus replication was monitored by measuring the amount of CA-p24 produced in the culture medium (Fig. 2). In the presence of optimal dox levels (1,000 ng/ml), we measured profound replication of all four HIV-rtTA viruses. No virus replication was observed in the absence of dox, indicating that replication is strictly dependent on the inserted Tet system. The Tet system is ideally suited to modulate the level of transcriptional activation in a stepwise manner by changing the amount of dox (4). Indeed, replication of the HIV-rtTA viruses can also be modulated at suboptimal concentrations of the inducing dox reagent (Fig. 2). A progressive reduction in replication rates of all four rtTA-viruses was observed at 300 and 100 ng of dox per ml, and virus replication was nearly abolished at 30 ng/ml.
FIG. 2.
dox-controlled replication of the HIV-rtTA viruses. The SupT1 T-cell line was electroporated with 10 μg of the indicated molecular clones, and cells were cultured without or with an increasing concentration of dox (0 to 1,000 ng/ml). Some of the cultures were stopped at day 5 because of massive HIV-induced syncytium formation and cell death. Virus production was measured by CA-p24 ELISA on culture supernatant samples.
The SupT1 cells were killed within 1 week by massive virus-induced syncytia, and the peak CA-p24 production is similar to what we regularly observe in infections with the wild-type LAI virus. Nevertheless, the HIV-rtTA variants have significantly reduced replication capacity compared to wild-type HIV-1 (results not shown). We managed to passage all four HIV-rtTA viruses as cell-free inocula onto fresh, uninfected T cells, and a spreading infection was sustained for at least 5 weeks (five passages). The combined results of several independent replication assays, either started by transfection or infection, indicated that there are modest but reproducible differences in the replication efficiency of the four HIV-rtTA constructs. The most efficient replication was measured for the KWK variant that maintained both the NF-κB sites and a functional tat gene, but the other variants were not much delayed. As expected, the minimal SYS virus replicated most weakly, but this variant also induced a spreading infection. From these experiments the following ranking order of replication was apparent: LAI (wild type) ≫ KWK > KYK, SWS > SYS.
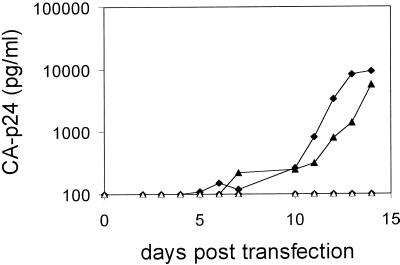
Putative HIV-rtTA vaccine viruses should be able to replicate in primary cells. The LAI molecular clone used in these studies represents a primary isolate that is able to efficiently infect primary cells (43, 52), but a complication of our design is that we removed the nef gene that contributes to optimal virus replication in primary cells (18). We transfected pooled PBMCs by means of electroporation with the KWK and KYK molecular clones and measured CA-p24 production in the culture supernatant for up to 2 weeks (Fig. 3). Both HIV-rtTA variants, with or without a functional tat gene, replicated in the presence of 1,000 ng of dox per ml, whereas no replication was detectable without dox. These results demonstrate that the HIV-rtTA viruses can replicate in primary cells, despite the absence of the nef function.
FIG. 3.
dox-dependent replication of HIV-rtTA viruses in primary cells. PBMCs were electroporated with the KWK (diamonds) and KYK (triangles) constructs (20 μg), and the cultures were maintained without (open symbols) or with (1,000 ng/ml) (solid symbols) dox. Fresh uninfected cells were added immediately after transfection and at day 6 postinfection. Virus production was measured by CA-p24 ELISA on culture supernatant samples.
Turning virus replication on and off in a reversible manner.
Subsequent tests were performed with the SWS virus in SupT1 infections (Fig. 4). First, we repeated the dox response experiment. Efficient virus replication was observed at 1,000 ng of dox per ml, but 100 ng of dox per ml was not sufficient to support a spreading infection in this more sensitive replication assay (Fig. 4A). We next analyzed virus replication kinetics when dox was added 3 days after infection of the cells (Fig. 4B). This resulted in a delay of virus production of approximately 3 days. In the absence of dox, the HIV-rtTA virus can still infect cells, reverse transcribe its RNA genome, and integrate the DNA into the host genome. In other words, the provirus form can be established, but replication is blocked at the level of viral transcription. These latently infected cells will remain present in the culture and can be activated by dox after 3 days to produce new, infectious particles.
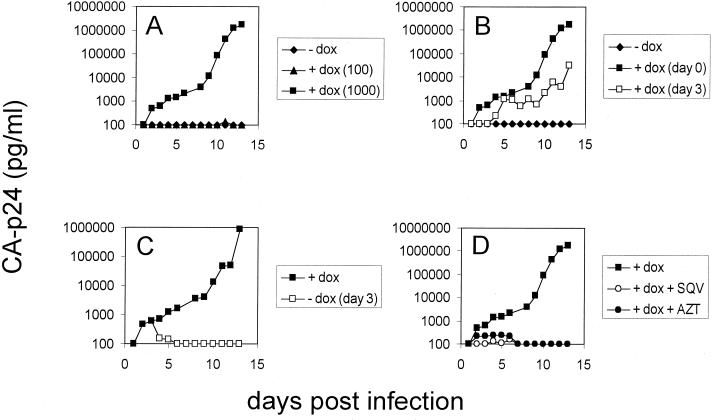
FIG. 4.
Replication of HIV-rtTA can be turned on and turned off. These experiments were performed with the SWS virus, but similar experiments have been performed with the other HIV-rtTA variants. We used the SWS virus (2,200 ng of CA-p24) to infect 6 × 106 SupT1 cells at day 0. (A) Replication potential with 0, 100, and 1,000 ng of dox per ml. (B) Effect of delayed addition of dox (1,000 ng/ml) at day 3 after infection. (C) Infected cells were grown in 1,000 ng of dox per ml for 3 days, at which point the cells were washed and incubated in the absence or presence of dox. (D) Infected cells were maintained in the presence of dox and either the Pro inhibitor SQV (200 nM) or the RT inhibitor AZT (1 μM).
A key feature of the Tet system is that it provides reversible regulation. To test this, SupT1 cells were infected with the SWS virus and cultured in the presence of dox (Fig. 4C). At day 3, the cells were washed to remove extracellular dox and resuspended in medium either with or without dox. Indeed, replication can be stopped abruptly by the removal of dox. These combined results confirm that replication of the HIV-rtTA virus is absolutely dependent on dox, and the level of virus replication can be strictly controlled in a graded and reversible manner.
Gene expression characteristics of the HIV-rtTA variants.
We next set out to accurately measure the level of gene expression and virus production of the HIV-rtTA variants compared with the wild-type LAI construct. These experiments were performed by transient transfection of C33A cells, which allows virus production without replication due to the absence of appropriate receptors for virus entry. The results are summarized in Table 2. We measured low virus production for all HIV-rtTA constructs in the absence of dox, ranging from 4,850 to 11,600 pg of CA-p24 per ml. By comparing the KWK and SWS versus KYK and SYS constructs, it is clear that the wild-type Tat protein makes a modest, twofold contribution to gene expression without dox. This basal activity is 0.8 to 2.1% of the virus production measured by the wild-type LAI plasmid, which is an excellent value compared with the “leakiness” measured in transient LTR-luc assays (7 to 10%; results not shown). Profound dox induction was measured for all constructs, ranging from 47- to 120-fold induction over the level of basal activity. In fact, virus production levels (455,000 to 580,000 pg of CA-p24 per ml) are similar to that of LAI, indicating that gene expression and virion production are efficiently executed by the HIV-rtTA variants. Comparable results were recently presented for SIV constructs with a similarly modified LTR promoter (55). Consistent with the replication ranking order of the HIV-rtTA variants, SYS produced the least amount of virus.
TABLE 2.
Transient HIV-rtTA production in C33A and SupT1 cells
| Cell type and HIV-rtTA variant | HIV-rtTA production (pg of CA-p24 per ml)a
|
|||||
|---|---|---|---|---|---|---|
| −CMV-rtTA
|
+CMV-rtTA
|
|||||
| −dox | +dox | Fold induction | −dox | +dox | Fold induction | |
| C33A cells | ||||||
| KWK | 11,600 | 545,000 | 47 | 91,500 | 715,000 | 7.8 |
| SWS | 11,350 | 560,000 | 49 | 49,500 | 560,000 | 11.3 |
| KYK | 4,850 | 580,000 | 120 | 47,500 | 500,000 | 10.5 |
| SYS | 5,950 | 455,000 | 76 | 32,500 | 520,000 | 16 |
| LAI | 635,000 | 520,000 | 0.8 | |||
| SupT1 cells | ||||||
| KWK | 110 | 54,000 | 491 | |||
| SWS | 30 | 65,000 | 2,167 | |||
| KYK | 10 | 39,000 | 3,900 | |||
| SYS | 20 | 7,800 | 390 | |||
Production was measured for the variants with (+) or without (−) dox and CMV-rtTA as indicated.
Establishment of an rtTA-tetO autoregulatory loop.
The previous results indicate that gene expression and replication of HIV-rtTA are strictly dependent on dox. The excellent performance of the HIV-rtTA viruses is novel due, at least in part, to the use of the rtTA-S2 reagent, which was shown to exhibit no measurable basal activity in the uninduced state (48). Moreover, we have placed rtTA expression under control of an rtTA-regulated LTR promoter, a situation that mimics the natural autoregulatory loop of the TAR-Tat axis. This means that both the activity of rtTA and its synthesis are dox dependent. Thus, only minute amounts of rtTA protein will be present in the absence of dox, resulting in an extremely low basal level of gene expression, and consequently a more-profound dox induction. In other Tet-controlled gene expression systems, the tTA or rtTA protein is produced in a constitutive manner from a second locus, e.g., the CMV-rtTA plasmid, which causes a significant level of gene activation in the off state.
We next designed an experiment to critically test whether an autoregulatory loop is established in HIV-rtTA. We mimicked the regular Tet system by cotransfection of HIV-rtTA with CMV-rtTA. The latter plasmid will produce a constitutive level of rtTA protein (even in the absence of dox), which is expected to enhance the level of virus production in the uninduced state. This is indeed what we observed (Table 2). The uninduced level of virus production was increased 5- to 10-fold with CMV-rtTA. The results in Table 2 also indicate that additional synthesis of rtTA protein from the cotransfected CMV-rtTA plasmid does not increase the level of virus production in the presence of dox, indicating that all HIV-rtTA constructs are able to produce saturating amounts of rtTA trans-activator.
Due to increased basal expression levels in cotransfections with CMV-rtTA, we measured only 8- to 16-fold dox induction levels. In fact, these results are very similar to the results obtained with the corresponding LTR-luc constructs (K. Verhoef et al., manuscript in preparation). These LTR-luc transfection experiments also revealed that higher dox induction levels (up to 80-fold) can be obtained in the T-cell line SupT1. Although we currently do not understand this effect, the responsiveness of the Tet system has been reported to differ in different cell types (27). We therefore tested all four HIV-rtTA variants in SupT1 cells. The transfected cells were cultured with or without dox, and virus production was measured at 2 days posttransfection. The latter point is critical because SupT1 cells support virus replication, which will eventually disturb the transient expression data. However, we have demonstrated previously that replication does not contribute to virus production measured at day 2 (15), and the transient virus production results are summarized in Table 2. Indeed, a more profound dox effect was measured in SupT1 cells, an effect ranging from 390- to 3,900-fold induction for the different HIV-rtTA constructs. These values are somewhat inaccurate because of the extremely low level of virus production in the uninduced state, which hardly exceeds the cutoff value of the CA-p24 assay. However, it is obvious that dox inducibility is 5- to 10-fold more profound in SupT1 cells than in C33A cells. The combined effects of the autoregulatory loop established in HIV-rtTA and the T-cell-specific augmentation of the dox response result in rather dramatic induction levels. In SupT1 cells, an extremely low basal level of virus production is measured, which is estimated to be approximately 0.03 to 0.2% of the dox-induced state.
Safety issues.
The finding that HIV-rtTA has an extremely low level of virus expression in the absence of dox seems the ideal situation concerning the safety of a vaccine strain. We performed some additional assays to address safety aspects. First, we screened for leaky virus replication in the absence of dox in prolonged cultures. For instance, the SupT1 cultures that were transfected with the four different HIV-rtTA constructs (Fig. 2) were maintained up to day 170, but no virus production was measured. Similarly, no replicating virus was observed in primary cell cultures without dox (Fig. 3). Second, HIV-rtTA virus spreading in SupT1 cultures could be “turned off” effectively by the removal of dox (see, e.g., Fig. 4C for the SWS virus), without any sign of virus production. Third, we analyzed the sensitivity of the HIV-rtTA virus to antiretroviral drugs that are in current clinical use. Because we did not alter the basic set of viral genes in HIV-rtTA, including the genes encoding protease (Pro) and reverse transcriptase (RT), these viruses are expected to remain fully sensitive to well-known drugs that target these essential enzymes. As shown in Fig. 4D, replication of the dox-dependent SWS virus can be inhibited efficiently either by 3′-azido-3′-deoxythymidine (AZT; a nucleoside RT inhibitor) or saquinavir (SQV; a Pro inhibitor). These experiments may be viewed as the first safety tests of these putative vaccine strains.
DISCUSSION
We have incorporated the Tet regulatory system in the HIV-1 genome such that virus transcription and replication can now be controlled from the outside by addition of a nontoxic inducer molecule such as dox. Specifically, we constructed replicating HIV-1 variants with inactivating mutations in both arms of the Tat-TAR axis through replacement with the rtTA-tetO elements of the Tet system. Replication experiments in a T-cell line and primary cells demonstrate that we have successfully designed dox-dependent HIV-1 variants. Replication of these designer HIV-rtTA viruses is regulatable in a graded and reversible manner. Although leakiness, that is, residual activity of the rtTA activator in the absence of dox, has been a problem in some applications of the rtTA system, we have not observed any virus replication in the absence of dox. This may be due, at least in part, to the superior performance of the improved rtTA-S2 protein that was used in these experiments (48). Another possibility is that expression of the rtTA trans-activator in the viral context is fully dependent on the presence of dox. Indeed, we demonstrated that an autoregulatory loop is established that resembles the natural situation with the TAR-Tat axis. This mechanism restricts leakiness or dox-independent replication, thereby providing an additional safety feature.
The HIV-rtTA viruses have some unique properties that make them ideal reagents for a variety of biological experiments. The most obvious application for such a virus is in the field of live-attenuated vaccines, and a similar approach may be used to put control over other retroviral pathogens (e.g., HIV-2 and human T-cell leukemia virus type 1), pararetroviruses (e.g., hepatitis B virus), or DNA viruses (e.g., herpesvirus or adenovirus). The HIV-rtTA viruses will improve the current generation of live-attenuated HIV-1 variants as potential vaccine strains because the conditional replication provides a unique safety feature. Of the variants constructed in this study, the SYS variant has the most minimal “genotype”: TAR− Tat− ΔU3 ΔNF-κB Δnef. However, it should be possible to further delete some of the “accessory” genes such as vpr and vpu. The absence of functional Tat seems particularly attractive because this viral protein is known to have several adverse effects on the host cell.
It is anticipated that HIV-rtTA vaccine viruses will be able to induce a protective immune response by dox-induced replication, after which replication can be turned off by the withdrawal of dox, such that the virus will remain nonpathogenic. In case a booster vaccination is required to mount an optimal immune response, replication can be switched on transiently at later times by dox. Experiments in mice indicate that the expression of a transgene can be regulated by simply adding or removing dox from the drinking water (35). Our tissue culture experiments also indicate that the level of virus replication can be fine-tuned by varying the dox level. We demonstrate that the HIV-rtTA viruses are inhibited by antiviral drugs such as the RT inhibitor AZT and the Pro inhibitor SQV. These viruses will await extensive replication tests to verify their genetic stability, followed by animal tests with homologous SIV constructs to screen for their pathogenic potential and their ability to induce a protective immune response.
Because the TAR RNA and tat gene may have become nonessential parts of the HIV-rtTA genome, these elements may be “free” to evolve. If these transcriptional elements have no other function in the viral replication cycle, one would predict that they would eventually be lost either by deletion or by accumulation of point mutations. This would further reduce the likelihood of a wild-type-like reversion, thereby making the vaccine strain more safe. However, the situation may be more complex since additional roles have been proposed for both motifs (reviewed in reference 6). This is most obvious for the TAR motif, which is part of the R (repeat) region that is critical in strand transfer during reverse transcription. But TAR has also been reported to contribute to RNA packaging in virion particles. The Tat protein has also been implicated in nontranscriptional roles, e.g., during mRNA translation and the process of reverse transcription (13, 26, 28, 44). Prolonged culture experiments and the analysis of revertant viruses may provide more insight into some of these possibilities.
The HIV-1 TAR-Tat axis was successfully replaced by the tetO-rtTA system, and the latter elements appear to have become essential viral functions. This may preclude the spontaneous loss of these exogenous elements by simple deletion, thereby enhancing the genetic stability of vaccine strains based on HIV-rtTA. On the other hand, the current HIV-rtTA viruses do replicate suboptimally. The advantage of working with HIV is that even if a poorly replicating virus is identified, the error-prone nature of the RT enzyme will allow for the generation and outgrowth of faster-replicating variants by a method termed forced evolution (7, 37). This evolutionary refinement of the initial designer HIV-rtTA viruses provides a powerful genetic method to identify modified forms of the E. coli-derived rtTA protein and/or the tetO sites that are better suited for their transcription function in a eukaryotic background. Consistent with this idea, we recently observed improved replication of the HIV-rtTA viruses in long-term infection experiments, apparently without repair of the Tat-TAR axis.
ACKNOWLEDGMENTS
We thank Rainer Loew and Maz Hasan for providing several reagents of the Tet system. We thank Wim van Est for excellent artwork.
Research within the Berkhout laboratory is sponsored by the Dutch AIDS Fund (AIDS Fonds, Amsterdam, The Netherlands), the Dutch Cancer Society (KWF, Amsterdam, The Netherlands), the Technology Foundation (STW, Utrecht, The Netherlands), and the National Institutes of Health (NIH, Bethesda, Md.). G.M. was supported by EMBO and HFSP fellowships, and K.V. was supported by a short-term EMBO fellowship.
REFERENCES
- 1.Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet. 1995;345:1342–1344. doi: 10.1016/s0140-6736(95)92540-6. [DOI] [PubMed] [Google Scholar]
- 2.Baba T W, Liska V, Khimani A H, Ray N B, Dailey P J, Penninck D, Bronson R, Greene M F, McClure H M, Martin L N, Ruprecht R M. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 3.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude Essink B B, van Kuilenburg A B P, Van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Baron U, Gossen M, Bujard H. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv Pharmacol. 2000;48:29–73. doi: 10.1016/s1054-3589(00)48003-8. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout B, Das A T. Functional analysis of RNA signals in the HIV-1 genome by forced evolution. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 249–275. [Google Scholar]
- 8.Berkhout B, Jeang K T. Transactivation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol. 1989;63:5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout B, Klaver B. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 1993;21:5020–5024. doi: 10.1093/nar/21.22.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J Mol Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 11.Berkhout B, Verhoef K, van Wamel J L B, Back N K T. Genetic instability of live attenuated human immunodeficiency virus type 1 vaccine strains. J Virol. 1999;73:1138–1145. doi: 10.1128/jvi.73.2.1138-1145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–204. [PubMed] [Google Scholar]
- 13.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 14.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 15.Das A T, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA(3Lys) J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das A T, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 18.de Ronde A, Klaver B, Keulen W, Smit L, Goudsmit J. Natural HIV-1 NEF accelerates virus replication in primary human lymphocytes. Virology. 1992;188:391–395. doi: 10.1016/0042-6822(92)90772-h. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 Tat protein binds trans-activating-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyer W B, Ogg G S, Demoitie M-A, Jin X, Geczy A F, Rowland-Jones S L, McMichael A J, Nixon D F, Sullivan J S. Strong human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte activity in Sydney blood bank cohort patients infected with nef-defective HIV type 1. J Virol. 1999;73:436–443. doi: 10.1128/jvi.73.1.436-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng S, Holland E C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 22.Fisher J, Goff S P. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 23.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 24.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenough T C, Sullivan J L, Desrosiers R C. Declining CD4 T-cell counts in a person infected with nef-deleted HIV-1. N Engl J Med. 1999;340:236–237. doi: 10.1056/NEJM199901213400314. [DOI] [PubMed] [Google Scholar]
- 26.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howe J R, Skryabin B V, Belcher S M, Zerillo C A, Schmauss C. The responsiveness of a tetracycline-sensitive expression system differs in different cell lines. J Biol Chem. 1995;270:14168–14174. doi: 10.1074/jbc.270.23.14168. [DOI] [PubMed] [Google Scholar]
- 28.Huang L M, Joshi A, Willey R, Orenstein J, Jeang K T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ilyinskii P O, Daniel M D, Simon M A, Lackner A A, Desrosiers R C. The role of the upstream U3 sequences in the pathogenesis of simian immunodeficiency virus-induced AIDS in rhesus monkeys. J Virol. 1994;68:5933–5944. doi: 10.1128/jvi.68.9.5933-5944.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilyinskii P O, Desrosiers R C. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 1998;17:3766–3774. doi: 10.1093/emboj/17.13.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeeninga R E, Hoogenkamp M, Armand-Ugo M, de Baar M, Verhoef K, Berkhout B. Functional differences between the LTR transcriptional promoters of HIV-1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson R P, Lifson J D, Czajak S C, Stefano Cole K, Manson K H, Glickman R L, Yang J Q, Montefiori D C, Montefioro R C, Wyand M S, Desrosiers R C. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J Virol. 1999;73:4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchhoff F, Greenough T, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 34.Kirchhoff F, Kestler III H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klaver B, Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohman B L, McChesney M B, Miller C J, McGowan E, Joye S M, van Rompay K K, Reay E, Antipa L, Pedersen N C, Marthas M L. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 42.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 44.SenGupta D N, Berkhout B, Gatignol A, Zhou A M, Silverman R H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata R, Siemon C, Czajak S C, Desrosiers R C, Martin M A. Live, attenuated simian immunodeficiency virus vaccines elicit potent resistance against a challenge with a human immunodeficiency virus type 1 chimeric virus. J Virol. 1997;71:8141–8148. doi: 10.1128/jvi.71.11.8141-8148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl-Hennig C, Dittmer U, Nisslein T, Petry H, Jurkiewicz E, Fuchs D, Wachter H, Matz-Rensing K, Kuhn E M, Kaup F J, Rud E W, Hunsmann G. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J Gen Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- 47.Ulich C, Dunne A, Parry E, Hooker C W, Gaynor R B, Harrich D. Functional domains of Tat required for efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1999;73:2499–2508. doi: 10.1128/jvi.73.3.2499-2508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urlinger S, Baron U, Thellmann M, Hasan M T, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Rompay K K A, Spinner A, Otsyula M, McChesney M B, Marthas M L. Attenuated retrovirus vaccines and AIDS. Science. 1995;270:1218. [PubMed] [Google Scholar]
- 50.Verhoef K, Berkhout B. A second-site mutation that restores replication of a Tat-defective human immunodeficiency virus. J Virol. 1999;73:2781–2789. doi: 10.1128/jvi.73.4.2781-2789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 52.Wain-Hobson S, Vartanian J-P, Henry M, Chenciner N, Cheynier R, Delassus S, Pedroza Martins L, Sala M, Nugeyre M T, Guétard D, Klatzmann D, Gluckman J-C, Rozenbaum W, Barré-Sinoussi F, Montagnier L. LAV revisited: origins of the early HIV-1 isolates from Institut Pasteur. Science. 1991;252:961–965. doi: 10.1126/science.2035026. [DOI] [PubMed] [Google Scholar]
- 53.Wei P, Garber M E, Fang S-M, Fisher W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 54.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao Y, Kuwata T, Miura T, Hayami M, Shida H. Dox-dependent SIVmac with tetracycline-inducible promoter in the U3 promoter region. Virology. 2000;269:268–275. doi: 10.1006/viro.2000.0213. [DOI] [PubMed] [Google Scholar]