Abstract
Chemical modification of DNA is a common strategy to improve the properties of oligonucleotides, particularly for therapeutics and nanotechnology. Existing synthetic methods essentially rely on phosphoramidite chemistry or the polymerization of nucleoside triphosphates but are limited in terms of size, scalability, and sustainability. Herein, we report a robust alternative method for the de novo synthesis of modified oligonucleotides using template-dependent DNA ligation of shortmer fragments. Our approach is based on the fast and scaled accessibility of chemically modified shortmer monophosphates as substrates for the T3 DNA ligase. This method has shown high tolerance to chemical modifications, flexibility, and overall efficiency, thereby granting access to a broad range of modified oligonucleotides of different lengths (20 → 120 nucleotides). We have applied this method to the synthesis of clinically relevant antisense drugs and ultramers containing diverse modifications. Furthermore, the designed chemoenzymatic approach has great potential for diverse applications in therapeutics and biotechnology.
Subject terms: DNA, DNA, DNA
Chemical modification of DNA is a common strategy to improve the properties of oligonucleotides. Herein, the authors report a robust method based on the template-dependent DNA ligation of shortmer fragments by DNA ligases, which is compatible with numerous chemical modifications and applicable to short (~20 nt) and longer (<100 nt) sequences.
Introduction
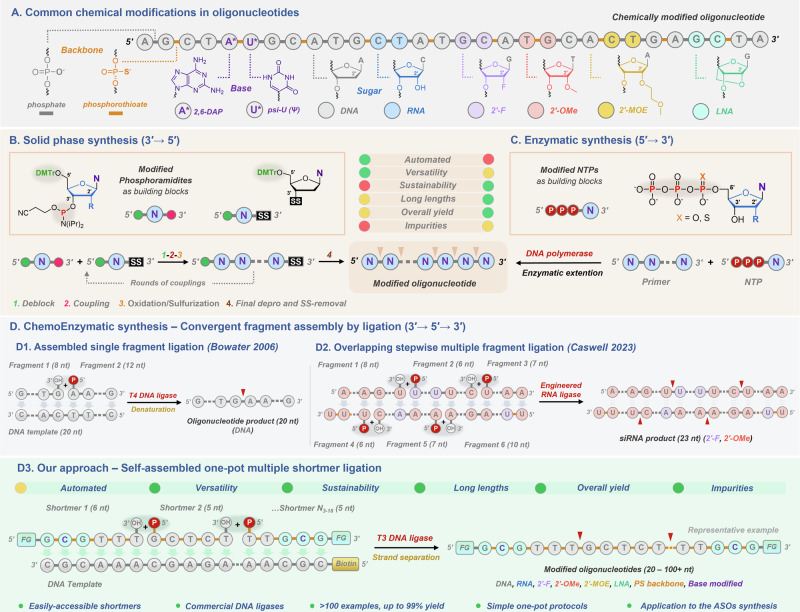
The intrinsic properties of DNA, particularly the high degree of programmability mediated by the Watson-Crick base pairing, have propelled synthetic oligonucleotides into the forefront of numerous applications. For instance, the self-assembly of thousands of short DNA sequences can be harnessed to create intricate 2D and 3D nanomaterials1 and computing2,3 devices. In addition, computing digital information into DNA sequences represents a potentially more powerful storage medium than existing silicon-based technologies4,5. When combined with chemical modifications (Fig. 1A), DNA and RNA can be converted into highly potent therapeutic agents, as highlighted by the recent advent of mRNA-based vaccines and antisense oligonucleotides (ASOs)6–9. This large and increasing demand for oligonucleotides needs to be supported by robust, efficient, cost-affordable, and sustainable synthetic methods10. However, our capacity for reading (i.e., sequencing) still outperforms that of writing (i.e., synthesizing) DNA11. This discrepancy mainly stems from the lack of alternative de novo DNA synthesis methods. Indeed, most synthetic oligonucleotides are produced by solid-phase oligonucleotide synthesis (SPOS) in which activated phosphoramidite building blocks are assembled by iterative synthetic cycles on an immobilized nucleoside (Fig. 1B)12. Application of this versatile and robust method allows for the efficient, large-scale (up to 10 kg batches) synthesis of short to moderately long (5–80 nt) and often heavily modified oligonucleotides6,13–17. Nonetheless, despite these attractive features, SPOS is impinged by low sustainability, truncated byproduct formation, and low overall yields, especially for long sequences. Consequently, its application is restricted to rather shorter sequences of modified oligonucleotides11,18,19. Alternatively, oligonucleotides can be produced by biocatalytic approaches20–24 mainly based on the polymerization of nucleotides catalyzed by polymerases (Fig. 1C)25–36. While polymerase-based methods do not display size limitation, control of the position, number, and complexity of modified nucleotides is very limited.
Fig. 1. Overview of existing synthetic protocols and of the co-polymerization of shortmer fragments.
A Common chemical modifications in oligonucleotides. B Overview of solid phase synthesis. C Schematic representation of enzymatic synthesis. D Alternative chemoenzymatic synthesis based on convergent fragment assembly by ligation. D1 Reported assembled single fragment ligation. D2 Reported overlapping stepwise multiple fragment ligation. D3 Our approach is based on self-assembled one-pot multiple-fragment ligation.
Chemoenzymatic strategies based on the convergent fragment assembly using ligation represent an alternative synthetic approach that could lift the limitations present in existing methods leading to better versatility and sustainability, higher yields, and overall purity (Fig. 1D). Even though several ligation methods have been reported they are restricted either: (1) by the length of assembled fragments (there is little advantage of using fragments of moderate size (10–20 not); (2) by the nature and diversity of chemical modifications accepted by ligases; (3) by the number and order of shortmers that can be assembled, and hence by the length and complexity of modified oligonucleotides that can be produced; (4) by the strict requirement of using engineered ligases; (5) to the combination of above-mentioned factors limiting the process efficiency and versatility (Fig. 1D1, D2)37–40.
Taken together, the main aim of this work was to address these limitations by developing a general, rather than specific, approach using easily accessible shortmers and DNA ligases. Therefore, we present an approach that permits the co-ligation of multiple chemically modified pentamers in a one-pot manner with commercially available ligases and is thus readily accessible to most laboratories (Fig. 1D3).
Our approach represents an alternative method for the de novo synthesis of chemically modified oligonucleotides which can produce short (~ 20 nt) therapeutic antisense oligonucleotides equally well as longer (> 100 nt) sequences decorated with modifications at selected positions of the nucleotidic scaffold.
Results
Design of the method
Despite impressive catalytic efficiencies41 of around 10 µM−1·s−1 (equating to the insertion of up to a few hundred nucleotides per second42), DNA polymerases struggle with substrates consisting of altered sugar motifs, which are not only found in most therapeutic oligonucleotides but also commonly used in chemoenzymatic approaches for de novo synthesis of DNA. On the other hand, DNA ligases have been reported to be rather tolerant to the presence of both base-43–47 and sugar modifications even in short sequences38,39,48 which constitute the basis of our approach. Nonetheless, most existing ligation methods rely on using scarcely modified oligonucleotides of moderate size (10–20 nt), and these are already impacted by some of the aforementioned limitations (vide supra). Instead, we based our method on the co-ligation of short, 5’-phosphorylated pentanucleotides49 containing a broad variety of chemically modified nucleotides. These shortmers are readily produced in high yields and purities by SPOS, and the concomitant templated ligation produces modified oligonucleotides in high yields, overall purity, and without size restrictions.
Scope of the ligase-mediated synthesis of modified oligonucleotides
In order to evaluate the possibility of constructing short, chemically modified oligonucleotides with ligases, we designed primer-template duplexes formed by a 5’-FAM-labeled DNA primer P1 and DNA templates of different lengths (1x−22 nt, 2x−27 nt, 4x−32 nt, 14x−87 nt;) (Fig. 2). In this design, the duplexes can accommodate either one or multiple shortmers in ligase-catalyzed reactions. The shortmer fragments (5 nt) consisted of the random sequence (5’-TAATT-3’)39 and were equipped with 5’-phosphate moieties (Fig. 2). In order to evaluate the compatibility of ligases with chemical modifications as well as the influence of the position of the modifications on the outcome of the reactions, we prepared a small subset library of pentanucleotides 1–23 containing one or multiple chemical modifications at all positions of the nucleotidic scaffold. More specifically, we considered typical sugar modifications present most short therapeutic oligonucleotides (2’-OH, 2’-F, 2’-OMe, 2’-MOE, LNA) 3–15, an altered phosphate backbone (phosphorothioate) 16–18, modified nucleobases (2-amino-dA, 5-octadiynyl-dU) 19, or a combination of multiple modification patterns 20–23. The presence of a 5’-FAM label on the DNA primer facilitated the monitoring of the reaction progress by gel electrophoresis PAGE (20%) followed by phosphorimaging and quantification of the resulting products (Fig. 2A–E).
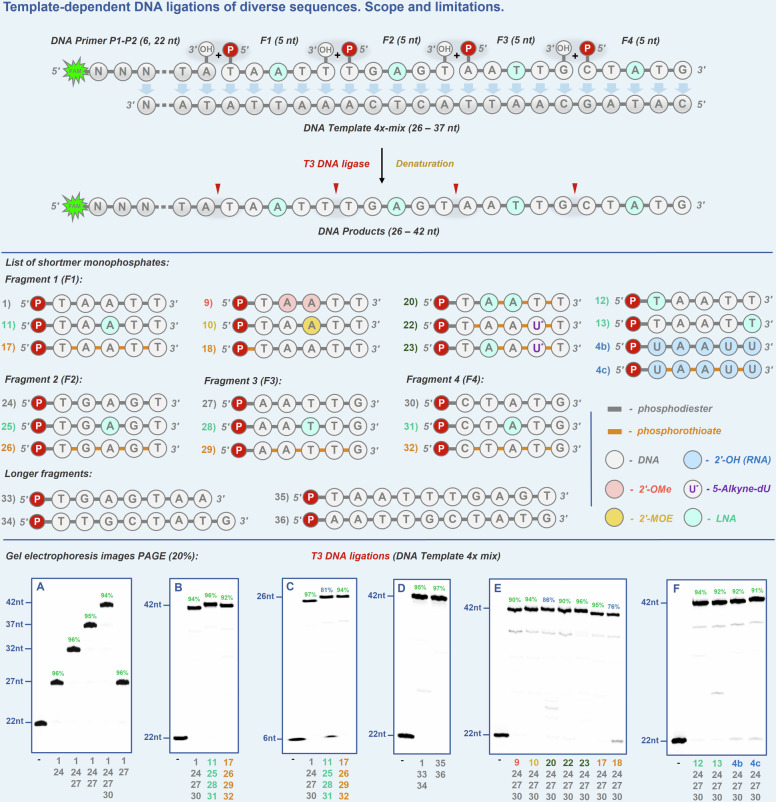
Fig. 2. DNA ligations of repetitive sequences.
Scope and limitations. Gel electrophoresis images PAGE (20%): A Screening of commercial DNA ligases. B–E DNA ligation reactions of shortmer (5 nt) fragments 1–23 using T3 DNA ligase, 5’-FAM labeled DNA primer P1 (22 nt) on complementary DNA templates of different lengths (1x-22 nt, 2x-27 nt, 4x-47 nt, 14x-87 nt). (-) – negative control in the absence of DNA ligase.
In addition to various chemical modifications, we also evaluated the capacity of various, commercially available DNA ligases (i.e. T4, Hi-T4TM, T3, and T7) to catalyze the ligation reactions of natural shortmer 1 with the primer using DNA templates 1x (22 nt) and 2x (27 nt) (Fig. 2A and Supplementary Fig. 2A). Most of the DNA ligases (T4, Hi-T4TM, and T3) showed excellent activity yielding the desired products (27–32 nt) with high conversion efficiencies (82–96%) except for the T7 DNA ligase which was reluctant to catalyze the reaction (7–10% of product formation). Considering the high compatibility of the T3 DNA ligase with base-modified fragments and the high conversion yields obtained with fragments 1–2343–47, we decided to further explore the substrate scope of the reaction with this enzyme.
To do so, we first optimized the reaction conditions for ligation reactions with the unmodified shortmer 1 by modulating different parameters such as ratios of oligonucleotide components (i.e. primer, template, shortmer), temperature, and reaction time (Supplementary Fig. 2O). Suitable conditions (resulting in high conversions) were obtained by using a slight excess of DNA template and shortmer compared to primer, with reaction times of over 6 hours at 16 °C. With these conditions, we evaluated the scope and limitations of the method by running the ligation reactions of all monophosphate shortmers 1–23 on DNA templates of different lengths (1x, 2x, 4x) (Supplementary Fig. 2B–D).
From these first studies, we learned that: 1) 5’/3’-bisphosphorylated shortmers (substrate 2) are only added once since the 3’-phosphate moiety acts as an efficient blocking group (but could be deprotected with alkaline phosphatase28); 2) substrates containing either 2’-OH or 2’- F sugar modifications (substrates 3–7), LNA units in the internal positions (substrates 11, 14-15), phosphorothioate modifications at all but the last phosphodiester or at all linkages, respectively (substrates 16-17), nucleobase-modified substrate 19, substrates 22-23 (containing all modification types), are generally well-tolerated by the T3 DNA ligase, leading to full-length products with good to high conversions on either of the DNA templates; 3) substrates containing internal 2’-OMe and 2’-MOE modifications (8–10), LNA as first or last nucleotide (substrates 12-13), equipped with a 5’-α-monophosphorothioate unit (substrate 18), substrates 20-21 (with different modification patterns) showed limited substrate tolerance by the T3 DNA ligase leading to mostly lower reactivities and only truncated products (Supplementary Fig. 2B–D). Hence, due to the low reactivities of some of the substrates, we undertook another optimization campaign. To do so, we had to take into more detailed consideration the properties of such short DNA sequences (namely, base pairing efficiency, low Tm values, bulky modifications), and we considered several parameters to be further optimized: (1) lower reaction temperatures to ensure more efficient annealing on the template; (2) addition of crowding agents like polyethylene glycol (PEG) and DMSO in order to improve the close proximity and efficiency of the alignment of the shortmer fragments on templates; (3) addition of other metal cofactors (e.g., Mn2+, Ca2+, or Co2+) along with Mg2+ (present in the reaction buffer) to increase flexibility and tolerance of bulkier modifications; (4) optimization of the amount of ATP cofactor (present in the reaction buffer) to avoid potential side reactions such as adenylation50. Consequently, by varying the above-mentioned parameters, we identified reaction conditions (4 °C reaction temperature, the addition of PEG8000 (20%), DMSO (10%), MnCl2 (1 mM final), 20x less ATP (50 µM final)) which significantly improved the reactivity and substrate tolerance. Indeed, under these optimized reaction conditions, most of the evaluated chemical modifications were well-tolerated with excellent conversions to the desired full-length products by ligating multiple, consecutive shortmer substrates 1–23 on DNA templates 1x, 2x, 4x (Fig. 2B–D and Supplementary Fig. 2B–D). Nonetheless, some of the substrates (8–10) bearing bulkier 2’-sugar modifications (2’-OMe, MOE) struggled to yield full-length products, as well as those (12-13) having unfavorable C3’-endo sugar conformations at the beginning and the end of the sequence. It is also worth mentioning that even though reactions with substrate 18 led to high conversions, some desulfurization (PS to PO) could be detected by LCMS analysis. Importantly, shortmer substrates 21–23 containing all combinations of chemical modifications (i.e., sugar, phosphate, and nucleobase) in a single fragment acted as excellent substrates for the DNA ligase (Fig. 2B–D).
Next, we evaluated the possibility of producing longer modified oligonucleotides by this method, and we tested the repetitive DNA ligation of several substrates bearing various modifications on a longer DNA template 14x allowing for the successive, one-pot incorporation of up to 14 pentanucleotide fragments. Under the optimized conditions, we could produce full-length (92 nt) products with good conversions (Fig. 2E). Finally, the chemical nature of all reaction products (Fig. 2A–E) was confirmed by LCMS analysis after running the large-scale analytical reactions (Supplementary Tables 1–4). Taken together, DNA ligation of repetitive sequences offers the possibility of efficiently introducing multiple chemical modifications at user-defined positions by simply changing the nature of the pentanucleotide fragments hence overcoming the challenges associated with classical strategies such as primer extension (PEX) reactions.
We then explored the possibility of synthesizing oligonucleotides with more diverse sequence compositions ligating the shortmers F1–F4 to the 5’-FAM-labeled DNA primers P1–P2 on the complementary DNA templates 4x-mix (Fig. 3). First, we used the optimized conditions to add the unmodified shortmers 1, 24, 27, 30 to the primer P1 in a stepwise manner to show the possibility of controlled synthesis of desired lengths with one single template, and to exclude the possibility of template-independent cross-ligation reactions (Fig. 3A). Next, we carried out mixed ligation experiments with the unmodified substrates 1, 24, 27, 30, the LNA-containing substrates 11, 25, 28, 31, and substrates 17, 26, 29, 32 equipped with phosphorothioates along with longer (P1, 22 nt) and shorter (P2, 6 nt) DNA primers. All these T3 DNA ligation reactions resulted in excellent conversions to the desired products (Fig. 3B, C). Our method is mainly based on the ligation of pentanucleotide fragments, nonetheless, for the construction of specific oligonucleotides of lengths differing from multiples of five, fragments of other lengths would be required51. Hence, we evaluated the possibility of adjusting our method to slightly longer oligomers. To do so, we carried out two separate DNA ligation reactions with primer P1 using shortmers 1, 33, 34 (5 nt, 7 nt, 8 nt, respectively) and 35, 36 (10 nt each) (Fig. 3D). All reactions proceeded with excellent conversions and full-length products could be identified without the formation of truncated products, suggesting that the length of the shortmer does not negatively impact the outcome of the reaction. Since these reactions with unmodified shortmers proceeded very well, we rationalized that a mixture of unmodified and modified fragments could be employed, to incorporate the least reactive of the modified pentanucleotide substrates identified previously. Hence, we carried out co-ligation reactions with modified pentanucleotides 9, 10, 17, 18, 20, 22, 23 along with unmodified substrates 24, 27, and 30 using the 5’-FAM-labeled DNA primer P1 (Fig. 3E).
Fig. 3. DNA ligations of diverse sequences. Scope and limitations.
Gel electrophoresis images PAGE (20%): A Study of stepwise ligation of natural shortmers (5 nt) to DNA primer P1 (22 nt) on DNA template 4x-mix (37 not). B Ligation of diverse natural and modified (LNA, phosphorothioate) shortmers (5 nt) to DNA primer P1 (22 not). C Ligation of diverse natural and modified (LNA, phosphorothioate) shortmers (5 nt) to DNA primer P2 (6 nt) on DNA template 4x-mixS (26 not). D Ligation of diverse natural shortmers of different length (5–10 not). E Ligation of diverse natural and variously modified (2’-OMe, 2’-MOE, phosphorothioate, NB) shortmers (5 not); (-) – negative control in the absence of DNA ligase. F Ligation of diverse natural and variously modified (LNA, RNA, RNA-phosphorothioate) shortmers (5 nt); (-) – negative control in the absence of DNA ligase.
In the same manner, we performed the co-ligation of shortmers 12–13 (LNA as first or last nucleotide) and fully RNA and RNA-phosphorothioate shortmers 4b-c (Fig. 3F). Gratifyingly, all reactions proceeded with high conversions toward the expected full-length products with marginal truncated product formation, suggesting that with a careful design in mixed ligation, even highly modified DNA or RNA, XNA shortmers can be incorporated into DNA with good efficiency. The integrity of all ligated DNA products (Fig. 3A–E) was confirmed by LCMS analysis after running the large-scale analytical reactions (Supplementary Table 4).
Having explored the scope and limitations of the method, we next explored different methods for isolation of the single-stranded product stemming from the ligation protocol. Thus, we first tested two methods based on the template digestion using an RNA instead of a DNA template followed by RNase H treatment52 and using a 5’-phosphorylated DNA template followed by λ-exonuclease digestion53. Unfortunately, the RNA template was not compatible with the DNA ligase, and the use of a 5’-phosphorylated DNA template resulted in the formation of longer side-products and reduced/absence of λ-exonuclease digestion (Supplementary Fig. 3G). Nevertheless, we then successfully applied the use of a 5’-biotinylated template and magnetic separation using Streptavidin magnetic beads (see the following sections, Supplementary Protocol for magnetic separation, and Supplementary Fig. 3G)54.
Synthesis of long and highly modified oligonucleotides
Having established a method for the production of modified DNA oligonucleotides by co-ligating short fragments to a primer on a template, we next set out to evaluate its compatibility with practical applications. In this context, we wanted to assess the possibility of applying this method to the synthesis of ultralong (> 100 nt), natural and modified oligonucleotide sequences. To fulfill this goal, we considered two different yet complementary general strategies: (1) ligating multiple diverse shortmer fragments F1-F18 (5 nt) directly on an ultramer DNA template TU (107 nt) produced by phosphoramidite chemistry; (2) ligation of two long fragments (VL and IL, each of 79 nt) with a shorter DNA splint (TL, 31 nt)55 (Fig. 4A, B).
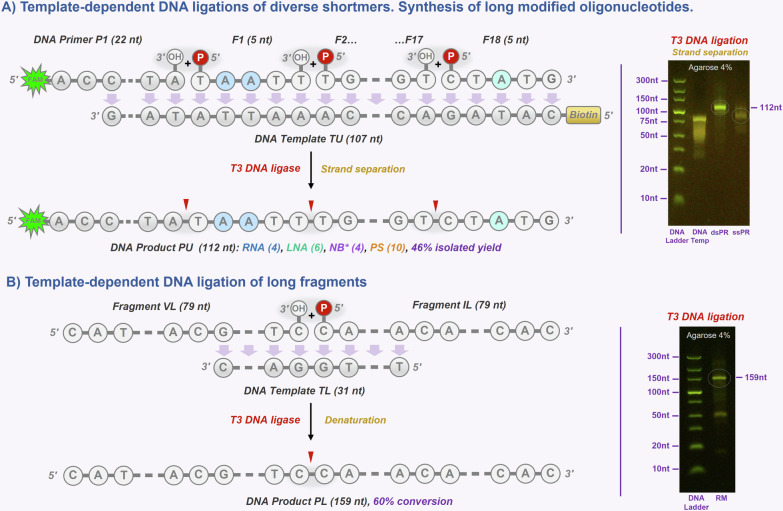
Fig. 4. Synthesis of long and modified oligonucleotides.
Gel electrophoresis images Agarose (4%). A DNA ligations of multiple diverse shortmer fragments (5 nt) F1–F18 to DNA primer P1 on ultramer DNA template TU (107 nt). B DNA ligation of long fragments VL-IL (79 nt) on shorter DNA template TL (31 nt).
We started with programmed ligation of multiple unmodified and modified shortmer fragments F1–F18 (5 nt) on ultramer DNA template TU (107 nt). We wanted to demonstrate that our approach is compatible with the diversity of chemically modified shormers as well as to show that the desired product could be recovered using biotinylated template TU (Fig. 4A). Using gel electrophoresis (Agarose 4%), we monitored the formation of the product PU (dsDNA, 112 nt) before and after the strand separation (ssDNA, 112 nt) that resulted in satisfactory 46% isolated yield (Supplementary Fig. 4A). The nature of the product was also confirmed by LCMS analysis (Supplementary Table 6).
At the same time, we wanted to demonstrate that our ligation approach is compatible with longmer assembly using the same setup that could lead to the advantageous production of DNA templates (e.g., for ligation, PEX) and ultralong modified sequences (> 150 nt, overcoming the limitation of SPOS). Therefore, we ligated two long fragments VL and IL (79 nt each) on a short DNA template TL (31 not), leading to the formation of ultramer DNA product PL (159 nt) with a good 60% conversion (Fig. 4B). Product formation was analyzed by agarose (4%) gel electrophoresis and further confirmed by LCMS analysis (Supplementary Fig. 4B and Supplementary Table 7). Efficient product isolation could be achieved by either magnetic separation from biotinylated template56, denaturing HPLC, electrophoresis, or membrane filtration.
Overall, these results demonstrate the versatility of the DNA ligation strategy and its applicability for the synthesis of long (> 100 nt), natural, and modified oligonucleotides, a daunting task for most existing chemical or biocatalytic methods. Shortmer approach will be more practical for the production of longmers roughly up to 150 nt in length. Consequently, longmer assembly will be more practical to access longer (> 150 nt) templates. Moreover, both approaches could be combined towards ultralong modified sequences (e.g., 300–500 nt).
Synthesis of therapeutic antisense oligonucleotides
Finally, we set out to demonstrate the compatibility of our chemoenzymatic approach with the preparation of antisense oligonucleotides (ASOs) and their conjugates. As a proof-of-principle, we evaluated the possibility of synthesizing the first FDA-approved antisense drug Fomivirsen (21 nt, all linkages are phosphorothioates), as well as various conjugates and LNA gapmer analog (Fig. 5A–F).
Fig. 5. Synthesis of therapeutic antisense oligonucleotide and its conjugates.
A Synthesis of Fomivirsen. B Synthesis of Fomivirsen FAM conjugate. C Synthesis of Fomivirsen FG (NH2, N3) conjugates. D Synthesis of Fomivirsen Fatty acid (C16:0) conjugate. E Synthesis of Fomivirsen Cholesterol-TEG conjugate. F Synthesis of Fomivirsen LNA gapmer analog.
In this context, our synthetic strategy was based on the simultaneous ligation of four shortmer fragments F1–F4 on the complementary 5’-biotinylated DNA template TF-Bio (21 nt) to isolate the ssDNA Fomivirsen products by magnetic separation on Streptavidin beads. We started the synthesis of Fomivirsen by searching optimal reaction conditions using 5’-FAM-labeled fragment F1 which enables a facile visualization and quantification of the reaction products by gel electrophoresis. Once again, as demonstrated above, the addition of crowding agents (PEG, DMSO) was essential to improve the initial yield of 30% obtained with standard conditions to 85%. Finally, when the reaction mixtures were supplemented with Mn2+ as an additional cofactor, the ligation reaction led to > 95% conversion of the expected products (Supplementary Fig. 6A). In addition, since all phosphodiester linkages were modified to the corresponding phosphorothioates, we investigated the reactivity of the 5’-monophosphorothioate-containing fragments F2–F4. To do so, we carried out a competitive reactivity study between these modified fragments and the corresponding monophosphate analogs (Supplementary Fig. 6B). This analysis revealed that even by lowering the reaction temperature to 4 °C, conversion of the 5’-monophosphate fragments to products was >95% while only 30% for the corresponding 5’-monophosphorothioate shortmers reacted. Nonetheless, even though the reactivity of 5’-monophosphorothioated fragments was lower compared to that of unmodified fragments, we demonstrated that it was still possible to reach high conversions to the expected fully modified oligonucleotide products by comprehensive multi-parametric optimization without requiring the assistance of enzyme engineering. Consequently, with optimized reaction conditions at hand, we performed the synthesis and isolation of Fomivirsen and its corresponding 5’-FAM conjugate that could be used for imaging and cellular uptake studies (Fig. 5A, B). We then synthesized two Fomivirsen conjugates bearing 5’-terminal reactive amino and azide functional groups that are readily available for post-functionalization via amide coupling or click chemistry (Fig. 5C). In addition, we demonstrated that our ligation approach is also compatible with pre-functionalized shortmers at both 5’ and 3’ ends. Thus, we prepared two examples of lipid conjugates (with a fatty acid at the 5’- and сholesterol-TEG at the 3’-position) for the improved cellular uptake of ASOs (Fig. 5D, E). Finally, we also synthesized the Fomivirsen LNA gapmer analog to show the compatibility of our ligation approach with the production of gapmers (Fig. 5F).
The isolated yields for the products after magnetic separation were mostly in the 60–70% range, with purities around 60–90%, as confirmed by LCMS analysis (Supplementary Table 8). The majority of the impurities stem from a PS to PO conversions which have many origins including small PO impurities (~ 5%) in starting fragments after phosphoramidite synthesis57,58, conversion during LCMS analysis, and potentially due the ligation reaction conditions.
In addition, using the example of Fomivirsen, we also demonstrated that our shortmer co-ligation approach is suitable for using longer templates. Therefore, we performed ligation on long natural DNA template TF-3x (63 nt) with repetitive sequence leading to 3 equivalents of Fomivirsen with good 77% conversion as detected by gel electrophoresis analysis (Supplementary Fig. 8A, B). By taking advantage of the size difference (63 nt for template vs. 21 nt for the product) this approach could be applied for qualitative product isolation by either denaturing HPLC, electrophoresis, or membrane filtration. It is also worth mentioning that the synthesis of antisense drugs by ligation strategy could be potentially compatible with the use of self-priming templates and restriction endonuclease as demonstrated by the Lovelock group for polymerases26.
Discussion
We have developed a general chemoenzymatic approach for the synthesis of natural and modified oligonucleotides based on the assembly of easily accessible short, 5’-phosphorylated DNA oligomers that does not require engineered enzyme or specific equipment and hence is readily available to most laboratories. In order to evaluate the scope and limitations of this method, we have tested a large variety of chemically modified shortmer monophosphate (5 nt) fragments in template-dependent DNA ligation reactions using commercial T3 DNA ligase under thoroughly optimized conditions. Overall, we have found that the DNA ligation method displays a high tolerance for chemical modifications at all three levels of the nucleotidic scaffold, including the reactive phosphate group, as first and terminal nucleotides are directly involved in ligation. Moreover, even fully modified shortmer fragments can be incorporated into DNA.
Importantly, this method not only demonstrated high flexibility in terms of the nature of chemical modifications but also in terms of the product size that can be achieved, since we were capable not only of producing shorter oligonucleotides similar to that of clinically relevant ASOs but also larger fragments (> 100 nt) which might be directly amenable to address biological questions such as the three-dimensional structure and dynamics of nucleosomes59. It is also worthwhile mentioning that longer (> 100 nt) modified sequences are difficult to obtain by standard phosphoramidite-based methods, especially with high overall yield and purity. In addition, our ligation method demonstrates great flexibility in terms of the length and sequence combinations of the shortmers, which is an additional important perquisite for the construction of a wide range of sequences of interest (shortmers from the portfolio can be easily manipulated).
This synthetic methodology also displays a high atom and enzyme economy as fewer functional groups per nucleotide are involved in the ligation reactions. This atom and cost economy can yet be further improved in the future by using optimized automated SPOS or employing manual solid-phase synthesis of shortmers, which requires fewer equivalents of phosphoramidites and shunts the need for some synthetic steps leading to better overall sustainability18,33. The enzyme economy and atom economy are also in stark contrast to the classical enzymatic extension reactions in which nucleoside triphosphates are used in (often large) excess together with DNA polymerases, whereas in our approach, every fifth nucleotide is ligated, and we use nearly stoichiometric amounts of pentamers. Importantly, extending DNA primers with five nucleotide long blocks rather than single nucleotides (triphosphates or phosphoramidites) also facilitates purification efforts since truncated side-products, if they arise, will be easier to remove. However, a more comprehensive technical study will be crucial in the future to compare the sustainability of our ligase-based method, traditional SPOS, and polymerase-catalyzed enzymatic synthesis and to identify parameters that can be improved.
It is important to mention that this approach does not represent a substitute for SPOS but rather a complementary alternative method. Also, as for most methods, there are some limitations to this approach that will need to be improved in the future. For instance, we have not evaluated the possibility of producing long (> 100 nt) fully-modified oligonucleotides, and some modifications (e.g., 2’-MOE) are only moderately well-tolerated. In this context, the use of alternative60 or engineered ligases61 will certainly represent a necessary future step to further improve our methodology. For instance, by further increasing the reactivity and flexibility while ligating the fragments containing terminal 5’-phosphorothioate shortmers, fully or highly modified shortmer sequences towards short or yet longer (> 150 nt) target sequences. Another necessary future step to further improve the practical aspect of the method lies in template recovery and recycling. As we demonstrated, longer repetitive templates can be efficiently used and we are currently working on that in our laboratory as well as on the use of self-priming templates. Lastly, engineered ligases will be paramount for the production of fully modified oligonucleotides containing more complex, XNA-like modifications such as Hexitol Nucleic Acids (HNA), Peptide Nucleic Acids (PNA), or Phosphorodiamidate Morpholino Oligomers (PMO).
Overall, we envision that the ligation of short, chemically modified oligonucleotides will improve our capacity to produce short therapeutic and longer oligonucleotides in a high yield and purity, easy-to-implement, and versatile one-pot approach that represents a valid alternative to existing methods.
Methods
Chemicals, oligonucleotides, and ligases
All modified and unmodified oligonucleotides including primers and templates were purchased from Integrated DNA Technologies and Microsynth. All DNA ligases (T4, Hi-T4TM, T3 and T7) were purchased from New England BioLabs.
Protocols for T3 DNA ligations of repetitive sequences
Optimized conditions for T3 DNA ligations on DNA templates 1x, 2x, 4x, 14x (analytical scale, 10 µL)
DNA Primer P1 10 µM (1.0 µL), DNA ligase bufferOPTI 10X (1.0 µL), PEG8000-50% (4.0 µL), DMSO (1.0 µL), T3 DNA ligase (0.5 µL). For DNA template 1x 10 µM (1.5 µL), DNA shortmers 1–23 10 µM (1.5 µL) were used. For DNA template 2x 10 µM (1.5 µL), DNA shortmers 1–23 20 µM (1.5 µL) were used. For DNA template 4x 10 µM (1.5 µL), DNA shortmers 1–23 40 µM (1.5 µL) were used. For DNA template 14x 10 µM (1.5 µL), DNA shortmers 1, 4, 6, 14, 17, 19 100 µM (2.0 µL) were used. DNA ligase bufferOPTI 1X composition: 66 mM Tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 50 µM ATP, 7.5% polyethylene glycol (PEG8000), pH 7.6 at 25. Reaction mixtures were incubated at 4 °C for 12 h. Reaction mixtures were then subjected to denaturing gel electrophoresis (PAGE 20%), followed by phosphorimaging and quantification analysis using Typhoon phosphorimager with the ImageQuantTL software (v.10.2) from CytivaTM.
Optimized conditions for T3 DNA ligations on DNA templates 1x, 2x, 4x, 14x (large scale analytical, 100 µL)
DNA Primer P1 100 µM (5.0 µL), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). For DNA template 1x 100 µM (7.5 µL), DNA shortmers 1–23 100 µM (7.5 µL) and water (10.0 µL) were used. For DNA template 2x 100 µM (7.5 µL), DNA shortmers 1–23 100 µM (15.0 µL) and water (2.5 µL) were used. For DNA template 4x 100 µM (7.5 µL), DNA shortmers 1–23 200 µM (15.0 µL) and water (2.5 µL) were used. For DNA template 14x 100 µM (7.5 µL), DNA shortmers 1, 4, 6, 14, 17, 19 700 µM (15.0 µL) and water (2.5 µL) were used. DNA ligase bufferOPTI 1X composition: 66 mM Tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 50 µM ATP, 7.5% polyethylene glycol (PEG8000), pH 7.6 at 25. Reaction mixtures were incubated at 4 °C for 12 h. The large-scale analytical reaction mixture was diluted with water to 500 µL final volume. Then washed and concentrated using an Amicon® Ultra centrifugal filter (3 kDa). Resulted DNA concentrate was additionally precipitated with ethanol, dried (typically 80–90% overall recovery), and used in LCMS analysis.
Protocols for T3 DNA ligations of diverse sequences
Optimized reaction conditions for T3 DNA ligation on DNA template 4x-mix and 4x-mixS (analytical scale, 10 µL)
DNA Primer P1/P2 10 µM (1.0 µL), DNA template 4x-mix/4x-mixS 10 µM (1.5 µL), DNA ligase bufferOPTI 10X (1.0 µL), PEG8000-50% (4.0 µL), DMSO (0.5 µL), T3 DNA ligase (0.5 µL), the mix of DNA shortmers 1, 24, 27, 30 30 µM (0.5 µL each), or shortmers 11, 25, 28, 31 30 µM (0.5 µL each), or shortmers 17, 26, 29, 32 30 µM (0.5 µL each), or shortmers 1, 33, 34 30 µM (0.5 µL each), or shortmers 35, 36 15 µM (1.0 µL each), or shortmers (9 or 10, 17, 18, 20, 22, 23), 24, 27, 30 30 µM (0.5 µL each). DNA ligase bufferOPTI 1X composition: 66 mM Tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 50 µM ATP, 7.5% polyethylene glycol (PEG8000), pH 7.6 at 25. Reaction mixtures were incubated at 4 °C for 12 h. Reaction mixtures were then subjected to denaturing gel electrophoresis (PAGE 20%), followed by phosphorimaging and quantification analysis using Typhoon phosphorimager with the ImageQuantTL software (v.10.2) by CytivaTM.
Optimized reaction conditions for T3 DNA ligation on DNA template 4x-mix and 4x-mixS (large-scale analytical, 115 µL)
DNA Primer P1/P2 100 µM (5.0 µL), DNA template 4x-mix/4x-mixS 100 µM (7.5 µL), DNA ligase bufferOPTI 10X (11.5 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (11.0 µL), mix of DNA shortmers 1, 24, 27, 30 100 µM (7.5 µL each), or shortmers 11, 25, 28, 31 100 µM (7.5 µL each), or shortmers 17, 26, 29, 32 100 µM (7.5 µL each), or shortmers 1, 33, 34 100 µM (7.5 µL each), or shortmers 35, 36 100 µM (7.5 µL each), or shortmers (9 or 10, 17, 18, 20, 22, 23), 24, 27, 30 100 µM (7.5 µL each). DNA ligase bufferOPTI 1X composition: 66 mM Tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 50 µM ATP, 7.5% polyethylene glycol (PEG8000), pH 7.6 at 25. Reaction mixtures were incubated at 4 °C for 12 h. The large-scale analytical reaction mixture was diluted with water to 500 µL final volume. Then washed and concentrated using an Amicon® Ultra centrifugal filter (3 kDa). Resulted DNA concentrate was additionally precipitated with ethanol, dried (typically 80–90% overall recovery), and used in LCMS analysis.
Synthesis of long and modified oligonucleotides
T3 DNA ligation of multiple shortmers for ultramer PU synthesis
Reaction conditions for T3 DNA ligation on DNA template TU (large-scale analytical, 200 µL)
DNA Primer P1 100 µM (5.0 µL), DNA template TU 100 µM (6.0 µL), Mix of DNA shortmers: 4 100 µM (15.0 µL), 25 100 µM (15.0 µL), 28 100 µM (7.5 µL), 31 100 µM (15.0 µL), 37 100 µM (15.0 µL), 38 100 µM (7.5 µL), 39 100 µM (7.5 µL), 40 100 µM (7.5 µL), 41 100 µM (15.0 µL), 42 100 µM (7.5 µL), 43 100 µM (7.5 µL), 44 100 µM (7.5 µL), 45 100 µM (7.5 µL). StickTogetherTM DNA ligase buffer 10X (20.0 µL), water (19.0 µL), T3 DNA ligase (15.0 µL). StickTogetherTM DNA ligase buffer 1X composition: 66 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, 1 mM ATP, 7.5% polyethylene glycol (PEG8000), pH 7.6 at 25 °C. Reaction mixture was incubated at 4 °C for 12 h.
Product PU isolation by magnetic separation
The reaction mixture obtained with biotinylated template TU was diluted with water to 500 µL final volume. Then washed and concentrated using an Amicon® Ultra centrifugal filter (10 kDa) (> 90% overall recovery). The reaction mixture was then subjected to magnetic separation using the general protocol (vide infra).
Protocol for magnetic separation
The T3 DNA ligation reaction mixture (~ 500 pmol of product) was subjected to magnetic separation from 5’-biotinylated template TU (107 nt) using streptavidin-coated magnetic beads (SMB).
Prior to the magnetic separation, the ligation mixture was washed and concentrated using an Amicon® (10 kDa) centrifugal filter, then distributed in three Eppendorf tubes (3 × 50 µL).
Streptavidin magnetic beads (SMB) (3 × 150 µL) were pre-washed 3 × 200 µL with binding buffer TEN100 (10 mM Tris, 1 mM EDTA, 100 mM NaCl, pH 7.5).
Next, the ligation mixture (3 × 50 µL) was combined with pre-washed (SMB) (3 × 200 µL) and mixed on a rotating wheel (speed 20) at room temperature for one hour.
Then, (SMB) was collected on the magnet using a magnetic separation rack (S1506S, NEB), then washed with wash buffer TEN1000 (2 × 300 µL) (10 mM Tris, 1 mM EDTA, 1 M NaCl, pH 7.5) and water (2 × 300 µL).
Next, (SMB) were re-suspended with water (3 × 100 µL), and NaOH 1 M (3 × 10 µL) was added for denaturation.
Then (SMB) were rapidly collected on the magnet, and the aqueous solutions with ssDNA product were combined and neutralized by dropwise addition of HCl 100 mM.
Finally, the aqueous solution with ssDNA product was washed and concentrated using an Amicon® (10 kDa) centrifugal filter for further analysis.
The resulting ultramer product PU (ssDNA, 112 nt) was verified by gel electrophoresis (Agarose 4%) (Supplementary Fig. 4A). The concentration was measured by NanoDrop to estimate the yield (230 pmol, 46%) and the purity was verified by LCMS analysis (Supplementary Table 6).
T3 DNA ligation of long sequences for ultramer PL synthesis
Reaction conditions for T3 DNA ligation on DNA Template TL (large-scale analytical, 100 µL)
Fragment VL 100 µM (5.0 µL), DNA template TL 100 µM (5.0 µL), Fragment IL 100 µM (7.5 µL), StickTogetherTM DNA ligase buffer 2X (50.0 µL), water (22.5 µL), T3 DNA ligase (10.0 µL). The reaction mixture was incubated at 16 °C over 6 h. StickTogetherTM DNA ligase buffer 1X composition: 66 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, 1 mM ATP, 7.5% polyethylene glycol (PEG6000), pH 7.6 at 25 °C. The reaction mixture was incubated at 16 °C for 6 h. The large-scale analytical reaction mixture was diluted with water to 500 µL final volume. Then washed and concentrated using an Amicon® Ultra centrifugal filter (10 kDa). The resulting DNA concentrate was additionally precipitated with ethanol and dried (90% overall recovery). The concentration was measured by UV spectroscopy on a NanoDrop, and the purity was verified by LCMS analysis (Supplementary Table 7).
Synthesis of Fomivirsen antisense drug, its analogs and conjugates
Synthesis on biotinylated template TF-Bio followed by magnetic separation
A) Synthesis of Fomivirsen (large-scale analytical, 100 µL)
Shortmer 47 100 µM (5.0 µL), DNA template TF-Bio 100 µM (5.0 µL), Shortmers 48, 49, 50 100 µM (7.0 µL each), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). The reaction mixture was incubated at 4 °C for 24 h.
B) Synthesis of Fomivirsen FAM conjugate (large scale analytical, 100 µL):
Shortmer 46 100 µM (5.0 µL), DNA template TF-Bio 100 µM (5.0 µL), Shortmers 48, 49, 50 100 µM (7.0 µL each), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). Reaction mixtures were incubated at 4 °C for 24 h.
C) Synthesis of Fomivirsen Amino/Azide FG conjugates (large-scale analytical, 100 µL):
Shortmer 51/52 100 µM (5.0 µL), DNA template TF-Bio 100 µM (5.0 µL), Shortmers 48, 49, 50 100 µM (7.0 µL each), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). Reaction mixtures were incubated at 4 °C for 24 h.
D) Synthesis of Fomivirsen Fatty acid conjugate (large-scale analytical, 100 µL):
Shortmer 53 100 µM (5.0 µL), DNA template TF-Bio 100 µM (5.0 µL), Shortmers 48, 49, 50 100 µM (7.0 µL each), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). Reaction mixtures were incubated at 4 °C for 24 h.
E) Synthesis of Fomivirsen Cholesterol-TEG conjugate (large-scale analytical, 100 µL):
Shortmer 47 100 µM (5.0 µL), DNA template TF-Bio 100 µM (5.0 µL), Shortmers 48, 49, 54 100 µM (7.0 µL each), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). Reaction mixtures were incubated at 4 °C for 24 h.
F) Synthesis of Fomivirsen LNA gapmer (large-scale analytical, 100 µL)
Shortmer 55 100 µM (5.0 µL), DNA template TF-Bio 100 µM (5.0 µL), Shortmers 48, 49, 56 100 µM (7.0 µL each), DNA ligase bufferOPTI 10X (10.0 µL), PEG8000-50% (40.0 µL), DMSO (10.0 µL), T3 DNA ligase (10.0 µL). Reaction mixtures were incubated at 4 °C for 24 h.
Fomivirsen products isolation by magnetic separation
Large-scale analytical reaction mixture using a biotinylated template was diluted with water to 500 µL final volume. Then washed and concentrated using an Amicon® Ultra centrifugal filter (3 kDa) (> 90% overall recovery). The reaction mixture was then subjected to magnetic separation using the general protocol (see below).
General protocol for magnetic separation of Fomivirsen and analogs
The T3 DNA ligation reaction mixture of Fomivirsen or its analogs (~ 500 pmol of product) was subjected to magnetic separation from 5’-biotinylated template TF-Bio (21 nt) using streptavidin magnetic beads (SMB).
Prior to magnetic separation, the ligation mixture was washed and concentrated using an Amicon® (3 kDa) centrifugal filter, then distributed in three Eppendorf tubes (3 × 50 µL).
Streptavidin magnetic beads (SMB) (3 × 150 µL) were pre-washed with binding buffer TEN100 (3 × 200 µL) (10 mM Tris, 1 mM EDTA, 100 mM NaCl, pH 7.5).
Next, the ligation mixture (3 × 50 µL) was combined with pre-washed (SMB) (3 × 200 µL) and mixed on a rotating wheel (speed 20) at room temperature for one hour.
Then (SMB) was collected on the magnet using a magnetic separation rack (S1506S, NEB), then washed with wash buffer TEN1000 (2 × 300 µL) (10 mM Tris, 1 mM EDTA, 1 M NaCl, pH 7.5) and water (2 × 300 µL).
Next, (SMB) were re-suspended with water (3 × 100 µL), and NaOH 1 M (3 × 10 µL) was added for denaturation.
Then, (SMB) was rapidly collected on the magnet, and the aqueous solutions with the ssDNA product were combined and neutralized by dropwise addition of 100 mM of HCl.
Finally, the aqueous solution with ssDNA product was washed and concentrated using an Amicon® (3 kDa) centrifugal filter for further analyses.
The nature of the resulting Fomivirsen products (ssDNA, 21 nt) was verified by gel electrophoresis (Agarose 4%) (Supplementary Fig. 7). The concentration was measured by NanoDrop to estimate the yield, and the purity was verified by LCMS analysis (Supplementary Table 8).
Statistics and reproducibility
All analytical scale experiments were independently repeated at least three times before scaling up. For experiments where representative gel electrophoresis images are shown (both in the manuscript and supplementary information files), the results were consistent across three independent replicates.
LCMS analysis of enzymatic reactions
LCMS settings and method
Chromatographic separations for LC-MS experiments were performed on a Thermo Scientific™ Vanquish™ Flex Binary UHPLC system (Thermo Fisher Scientific, Reinach, Switzerland).
The column used for all separations was a Waters Aquity Premier BEH C18 Peptide 2.1*50 mm 1.7 µm 300 Å (Waters (CH) AG, Baden-Dättwil, Switzerland). The solvents were A: 15 mM Amylamine (ALDRICH, W424201 (inhouse re-distilled), CAS 110-58-7, SIGMA-ALDRICH CHEMIE GMBH (CH), Buchs Switzerland) and 50 mM 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP, ACROS ORGANICS, ACR14754 (99.5 + %, PURE), CAS 920-66-1, ThermoFisher Scientific, Reinach Switzerland) in water (Milli-Q® IQ 7000, Millipore, Merck & Cie, Schaffhausen, Switzerland) and B: methanol/acetonitrile (9/1;v/v) both gradient grade. Both solvents contained 1 µM Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA), SIGMA, E5134-100G (99.0–101.0%, titration)), CAS 6381-92-6, SIGMA-ALDRICH CHEMIE GMBH (CH), Buchs Switzerland, helping to suppress metal adducts forming in the mass spectrometer. A flow rate of 400 μL min−1 was applied, and the column compartment was held at 80 °C. The gradient system was A vs B, starting at 5%B with a rise to 20%B within 3 min and reaching 35%B at 25.5 min, followed by a short step to 100% B before coming back to the initial 5% B for reinjection.
For mass spectrometric data acquisition, a Thermo Scientific™ Fusion Lumos™ Hybrid Quadrupole-Orbitrap mass spectrometer equipped with a heated electrospray ionization-II (HESI-II) probe in a standard Thermo Scientific™ Ion Max™ ion source (Thermo Fisher Scientific, San José, CA, USA) was used. Data acquisition was performed with Thermo Scientific™ Xcalibur 4.5. HR LC-MS measurements were performed under Thermo Scientific™ Xcalibur™ Orbitrap Fusion Lumos Tune Application 3.5. The mass range was 600–2000 Da at a resolution of 120 K. In addition, the full DAD but also a 260 nm UV trace were acquired.
MS raw files were exported to ThermoFisher Scientific BioPharma Finder (BPF) 5.1 software and analyzed using the ReSpect algorithm via the sliding window deconvolution feature either against the full structure of the compounds or just against the molecular mass. All data evaluation was done based on the Thermo Scientific BioPharma Finder User Guide Software Version 5.1, XCALI-98492 Revision A, July 2022.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors acknowledge Institut Pasteur, CNRS grant PEPR MolecularXiv (ANR-22-PEXM-0002), and Roche for financial support via its Technology, Innovation and Science program. Yannick Rondelez (CNRS/ESPCI Laboratory Gulliver) and Anthony Genot (CNRS/University of Tokyo) for fruitful discussions.
Author contributions
Conceptualization was done by M.H., K.P., F.S., S.H., S.B., and N.S. Methodology was done by M.H. and N.S. Validation was carried out by N.S. and M.H. Formal analysis was done by all authors. The investigation was carried out by N.S. and A.S. Resources were provided by K.P., S.H., and M.H. Data were curated by N.S., A.S., and M.H. The original draft was written by N.S. and M.H. Review and editing of the draft was done by all authors. Supervising the project were M.H., S.H., and K.P. Project administration was done by M.H. Funding was acquired by K.P., S.H., and M.H.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The Supplementary Information provides experimental procedures and characterization data. Correspondence and requests for materials should be addressed to M.H.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52141-8.
References
- 1.Zhan, P. et al. Recent advances in DNA origami-engineered nanomaterials and applications. Chem. Rev.123, 3976–4050 (2023). 10.1021/acs.chemrev.3c00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lv, H. et al. DNA-based programmable gate arrays for general-purpose DNA computing. Nature622, 292–300 (2023). 10.1038/s41586-023-06484-9 [DOI] [PubMed] [Google Scholar]
- 3.Kieffer, C., Genot, A. J., Rondelez, Y. & Gines, G. Molecular computation for molecular classification. Adv. Biol.7, 2200203 (2023). 10.1002/adbi.202200203 [DOI] [PubMed] [Google Scholar]
- 4.Doricchi, A. et al. Emerging approaches to DNA data storage: challenges and prospects. ACS Nano16, 17552–17571 (2022). 10.1021/acsnano.2c06748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gervasio, J. H. D. B. et al. How close are we to storing data in DNA? Trends Biotechnol.42, 156–167 (2024). 10.1016/j.tibtech.2023.08.001 [DOI] [PubMed] [Google Scholar]
- 6.McKenzie, L. K., El-Khoury, R., Thorpe, J. D., Damha, M. J. & Hollenstein, M. Recent progress in non-native nucleic acid modifications. Chem. Soc. Rev.50, 5126–5164 (2021). 10.1039/D0CS01430C [DOI] [PubMed] [Google Scholar]
- 7.Egli, M. & Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res.51, 2529–2573 (2023). 10.1093/nar/gkad067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov.20, 817–838 (2021). 10.1038/s41573-021-00283-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi, Y. et al. Chemically modified platforms for better RNA therapeutics. Chem. Rev.124, 929–1033 (2024). 10.1021/acs.chemrev.3c00611 [DOI] [PubMed] [Google Scholar]
- 10.Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods11, 499–507 (2014). 10.1038/nmeth.2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoose, A., Vellacott, R., Storch, M., Freemont, P. S. & Ryadnov, M. G. DNA synthesis technologies to close the gene writing gap. Nat. Rev. Chem.7, 144–161 (2023). 10.1038/s41570-022-00456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaucage, S. L. & Caruthers, M. H. Deoxynucleoside phosphoramidites–A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett.22, 1859–1862 (1981). 10.1016/S0040-4039(01)90461-7 [DOI] [Google Scholar]
- 13.Kawamoto, Y., Wu, Y., Takahashi, Y. & Takakura, Y. Development of nucleic acid medicines based on chemical technology. Adv. Drug Deliv. Rev.199, 114872 (2023). 10.1016/j.addr.2023.114872 [DOI] [PubMed] [Google Scholar]
- 14.Sandahl, A. F. et al. On-demand synthesis of phosphoramidites. Nat. Commun.12, 2760 (2021). 10.1038/s41467-021-22945-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Giesen, K. J. D., Thompson, M. J., Meng, Q. & Lovelock, S. L. Biocatalytic synthesis of antiviral nucleosides, cyclic dinucleotides, and oligonucleotide therapies. JACS Au3, 13–24 (2023). 10.1021/jacsau.2c00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, C. et al. Fully automated fast-flow synthesis of antisense phosphorodiamidate morpholino oligomers. Nat. Commun.12, 4396 (2021). 10.1038/s41467-021-24598-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, R. A., Märcher, A., Pedersen, K. N. & Gothelf, K. V. Insertion of chemical handles into the backbone of DNA during solid-phase synthesis by oxidative coupling of amines to phosphites. Angew. Chem. Int. Ed.62, e202305373 (2023). 10.1002/anie.202305373 [DOI] [PubMed] [Google Scholar]
- 18.Andrews, B. I. et al. Sustainability challenges and opportunities in oligonucleotide manufacturing. J. Org. Chem.86, 49–61 (2021). 10.1021/acs.joc.0c02291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obexer, R., Nassir, M., Moody, E. R., Baran, P. S. & Lovelock, S. L. Modern approaches to therapeutic oligonucleotide manufacturing. Science384, eadl4015 (2024). 10.1126/science.adl4015 [DOI] [PubMed] [Google Scholar]
- 20.Liu, L., Huang, Y. & Wang, H. H. Fast and efficient template-mediated synthesis of genetic variants. Nat. Methods20, 841–848 (2023). 10.1038/s41592-023-01868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blümler, A., Schwalbe, H. & Heckel, A. Solid-phase-supported chemoenzymatic synthesis of a light-activatable tRNA derivative. Angew. Chem. Int. Ed.61, e202111613 (2022). 10.1002/anie.202111613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buller, R. et al. From nature to industry: Harnessing enzymes for biocatalysis. Science382, eadh8615 (2023). 10.1126/science.adh8615 [DOI] [PubMed] [Google Scholar]
- 23.McGorman, B. et al. Enzymatic synthesis of chemical nuclease triplex-forming oligonucleotides with gene-silencing applications. Nucleic Acids Res.50, 5467–5481 (2022). 10.1093/nar/gkac438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aitken, H. R. M., Wright, T. H., Radakovic, A. & Szostak, J. W. Small-molecule organocatalysis facilitates in situ nucleotide activation and RNA copying. J. Am. Chem. Soc.145, 16142–16149 (2023). 10.1021/jacs.3c04635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verardo, D. et al. Multiplex enzymatic synthesis of DNA with single-base resolution. Sci. Adv.9, eadi0263 (2023). 10.1126/sciadv.adi0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moody, E. R., Obexer, R., Nickl, F., Spiess, R. & Lovelock, S. L. An enzyme cascade enables production of therapeutic oligonucleotides in a single operation. Science380, 1150–1154 (2023). 10.1126/science.add5892 [DOI] [PubMed] [Google Scholar]
- 27.Sabat, N. et al. Towards the controlled enzymatic synthesis of LNA containing oligonucleotides. Front. Chem.11, 1161462 (2023). 10.3389/fchem.2023.1161462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flamme, M. et al. Evaluation of 3 ‘-phosphate as a transient protecting group for controlled enzymatic synthesis of DNA and XNA oligonucleotides. Commun. Chem.5, 68 (2022). 10.1038/s42004-022-00685-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, J. A. et al. Spatially selective electrochemical cleavage of a polymerase-nucleotide conjugate. ACS Synth. Biol.12, 1716–1726 (2023). 10.1021/acssynbio.3c00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichon, M., Levi-Acobas, F., Kitoun, C. & Hollenstein, M. 2’,3’-Protected nucleotides as building blocks for enzymatic de novo RNA synthesis. Chem. Eur. J.30, e202400137 (2024). 10.1002/chem.202400137 [DOI] [PubMed] [Google Scholar]
- 31.Palluk, S. et al. De novo DNA synthesis using polymerase-nucleotide conjugates. Nat. Biotechnol.36, 645–650 (2018). 10.1038/nbt.4173 [DOI] [PubMed] [Google Scholar]
- 32.Wiegand, D. J. et al. Template-independent enzymatic synthesis of RNA oligonucleotides. Nat. Biotechnol. 10.1038/s41587-024-02244-w (2024). [DOI] [PMC free article] [PubMed]
- 33.Sabat, N. et al. Artificial nucleotide codons for enzymatic DNA synthesis. Chem. Commun.59, 14547–14550 (2023). 10.1039/D3CC04933G [DOI] [PubMed] [Google Scholar]
- 34.Tang, L. An enzymatic oligonucleotide synthesizer. Nat. Methods15, 568–568 (2018). 10.1038/s41592-018-0096-x [DOI] [PubMed] [Google Scholar]
- 35.Brunderová, M. et al. Expedient production of site specifically nucleobase-labelled or hypermodified RNA with engineered thermophilic DNA polymerases. Nat. Commun.15, 3054 (2024). 10.1038/s41467-024-47444-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollenstein, M. Enzymatic synthesis of RNA oligonucleotides. Nat. Biotechnol.10.1038/s41587-024-02322-z (2024). [DOI] [PubMed]
- 37.Paul, S. et al. Convergent biocatalytic mediated synthesis of siRNA. ACS Chem. Biol.18, 2183–2187 (2023). 10.1021/acschembio.3c00071 [DOI] [PubMed] [Google Scholar]
- 38.Kestemont, D. et al. XNA ligation using T4 DNA ligase in crowding conditions. Chem. Commun.54, 6408–6411 (2018). 10.1039/C8CC02414F [DOI] [PubMed] [Google Scholar]
- 39.McCloskey, C. M., Liao, J.-Y., Bala, S. & Chaput, J. C. Ligase-mediated threose nucleic acid synthesis on DNA templates. ACS Synth. Biol.8, 282–286 (2019). 10.1021/acssynbio.8b00511 [DOI] [PubMed] [Google Scholar]
- 40.Bullard, D. R. & Bowater, R. P. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J.398, 135–144 (2006). 10.1042/BJ20060313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beard, W. A., Shock, D. D., Vande Berg, B. J. & Wilson, S. H. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem.277, 47393–47398 (2002). 10.1074/jbc.M210036200 [DOI] [PubMed] [Google Scholar]
- 42.Beard, W. A. & Wilson, S. H. Structural insights into the origins of DNA polymerase fidelity. Structure11, 489–496 (2003). 10.1016/S0969-2126(03)00051-0 [DOI] [PubMed] [Google Scholar]
- 43.Chen, Z., Lichtor, P. A., Berliner, A. P., Chen, J. C. & Liu, D. R. Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem.10, 420–427 (2018). 10.1038/s41557-018-0008-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong, D., Yeung, W. & Hili, R. In vitro selection of diversely functionalized aptamers. J. Am. Chem. Soc.139, 13977–13980 (2017). 10.1021/jacs.7b07241 [DOI] [PubMed] [Google Scholar]
- 45.Guo, C., Mahdavi-Amiri, Y. & Hili, R. Influence of linker length on ligase-catalyzed oligonucleotide polymerization. ChemBioChem20, 793–799 (2019). 10.1002/cbic.201800616 [DOI] [PubMed] [Google Scholar]
- 46.Riedl, J., Ding, Y., Fleming, A. M. & Burrows, C. J. Identification of DNA lesions using a third base pair for amplification and nanopore sequencing. Nat. Commun.6, 8807 (2015). 10.1038/ncomms9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oda, Y., Chiba, J., Kurosaki, F., Yamade, Y. & Inouye, M. Additive-free enzymatic phosphorylation and ligation of artificial oligonucleotides with C-nucleosides at the reaction points. ChemBioChem20, 1945–1952 (2019). 10.1002/cbic.201900217 [DOI] [PubMed] [Google Scholar]
- 48.Mann, G. et al. Biocatalytic assembly of chemically modified oligonucleotides. Tetrahedron Lett.93, 153696 (2022). 10.1016/j.tetlet.2022.153696 [DOI] [Google Scholar]
- 49.Horspool, D. R., Coope, R. J. N. & Holt, R. A. Efficient assembly of very short oligonucleotides using T4 DNA Ligase. BMC Res. Notes3, 291 (2010). 10.1186/1756-0500-3-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei, Y. & Hili, R. Structure-activity relationships of the ATP cofactor in ligase-catalysed oligonucleotide polymerisations. Org. Biomol. Chem.15, 2349–2352 (2017). 10.1039/C6OB02792J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei, Y., Washington, J. & Hili, R. Efficiency and fidelity of T3 DNA ligase in ligase-catalysed oligonucleotide polymerisations. Org. Biomol. Chem.17, 1962–1965 (2019). 10.1039/C8OB01958D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ondruš, M., Sýkorová, V. & Hocek, M. Traceless enzymatic synthesis of monodispersed hypermodified oligodeoxyribonucleotide polymers from RNA templates. Chem. Commun.58, 11248–11251 (2022). 10.1039/D2CC03588J [DOI] [PubMed] [Google Scholar]
- 53.Hollenstein, M., Hipolito, C. J., Lam, C. H. & Perrin, D. M. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M 2+). Nucleic Acids Res.37, 1638–1649 (2009). 10.1093/nar/gkn1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kilili, G. K., Tilton, L. & Karbiwnyk, C. M. Letter to the Editor] NaOH concentration and streptavidin bead type are key factors for optimal DNA aptamer strand separation and isolation. BioTechniques61, 114–116 (2016). 10.2144/000114449 [DOI] [PubMed] [Google Scholar]
- 55.Renders, M., Miller, E., Hollenstein, M. & Perrin, D. A method for selecting modified DNAzymes without the use of modified DNA as a template in PCR. Chem. Commun.51, 1360–1362 (2015). 10.1039/C4CC07588A [DOI] [PubMed] [Google Scholar]
- 56.Uhlen, M. Magnetic separation of DNA. Nature340, 733–734 (1989). 10.1038/340733a0 [DOI] [PubMed] [Google Scholar]
- 57.Bergot, B. J. & Egan, W. Separation of synthetic phosphorothioate oligodeoxynucleotides from their oxygenated (phosphodiester) defect species by strong-anion-exchange high-performance liquid chromatography. J. Chromatogr. A599, 35–42 (1992). 10.1016/0021-9673(92)85456-4 [DOI] [Google Scholar]
- 58.Rentel, C. et al. Assay, purity, and impurity profile of phosphorothioate oligonucleotide therapeutics by ion Pair-HPLC-MS. Nucleic Acid Ther.32, 206–220 (2022). 10.1089/nat.2021.0056 [DOI] [PubMed] [Google Scholar]
- 59.Hirashima, S., Park, S. & Sugiyama, H. Evaluation by experimentation and simulation of a FRET pair comprising fluorescent nucleobase analogs in nucleosomes. Chem. Eur. J.29, e202203961 (2023). 10.1002/chem.202203961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lohman, G. J. S. et al. Efficient DNA ligation in DNA–RNA hybrid helices by Chlorella virus DNA ligase. Nucleic Acids Res.42, 1831–1844 (2013). 10.1093/nar/gkt1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanmeert, M. et al. Rational design of an XNA ligase through docking of unbound nucleic acids to toroidal proteins. Nucleic Acids Res.47, 7130–7142 (2019). 10.1093/nar/gkz551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Supplementary Information provides experimental procedures and characterization data. Correspondence and requests for materials should be addressed to M.H.