Abstract
The neocortex varies in size and complexity among mammals due to the tremendous variability in the number and diversity of neuronal subtypes across species. The increased cellular diversity is paralleled by the expansion of the pool of neocortical progenitors and the emergence of indirect neurogenesis during brain evolution. The molecular pathways that control these biological processes and are disrupted in neurological disorders remain largely unknown. Here we show that the transcription factors BRN1 and BRN2 have an evolutionary conserved function in neocortical progenitors to control their proliferative capacity and the switch from direct to indirect neurogenesis. Functional studies in mice and ferrets show that BRN1/2 act in concert with NOTCH and primary microcephaly genes to regulate progenitor behavior. Analysis of transcriptomics data from genetically modified macaques provides evidence that these molecular pathways are conserved in non-human primates. Our findings thus demonstrate that BRN1/2 are central regulators of gene expression programs in neocortical progenitors critical to determine brain size during evolution.
Subject terms: Neural stem cells, Molecular neuroscience
How the diversity and number of neuronal subtypes across species drives neocortical size and complexity remains incompletely understood. Here authors demonstrate BRN1/2 act in concert with NOTCH and primary microcephaly genes to regulate evolutionarily conserved neocortical progenitor behavior across species and determine brain size.
Introduction
The mammalian neocortex differs vastly in size and complexity between species1,2. This cellular diversity is associated with differences in the complexity of the progenitor pools that generate cortical neurons2–6, yet the mechanisms that lead to an increase in brain size during evolution and are disrupted in disease remain largely unknown. Defects in progenitor behavior have been associated to diverse neurodevelopmental disorders including microcephaly7–9. Mutations in genes linked to primary microcephaly, the most severe form of the disease, affect molecular pathways that regulate cell-cycle and progenitor proliferation7,9. Thus, an understanding of cortical progenitor diversity and of the mechanisms by which these progenitors self-renew and differentiate is critical to understand brain evolution and the defects in progenitor behavior that lead to neurological and psychiatric disorders. Cortical progenitors have been broadly divided into two classes named apical progenitors (APs) that undergo mitosis in the ventricular zone (VZ), and basal progenitors (BPs) that undergo mitosis in the subventricular zone (SVZ)10–14. APs engage in two modes of neurogenesis termed direct and indirect neurogenesis. During direct neurogenesis, APs divide asymmetrically to self-renew and to generate one neuron, while during indirect neurogenesis, APs divide asymmetrically to self-renew and generate a BP that then gives rise to neurons10–14. Indirect neurogenesis generates neurons for all cortical layers but is the predominant neurogenic mode that produces upper layer projection neurons (ULNs)11,12,15–18. The number and complexity of ULNs have dramatically increased in gyrencephalic brains, and indirect neurogenesis is thus intricately linked to neocortex expansion during brain evolution3,5,19,20. Two transcriptional regulators that might be involved in the control of gene expression programs that regulate cell proliferation and the neurogenic mode used by cortical progenitors are the POU-homeobox-domain-containing octamer-binding transcription factors BRN1 (POU3F3) and BRN2 (POU3F2). These transcription factors are expressed in the developing mouse cortex in APs and BPs21–23. In mice harboring null alleles for both Brn1 and Brn2, the generation of ULNs is severely affected thus leading to microcephaly21,22. Based on in utero electroporation studies, it has been proposed that murine BRN1/2 are critical to regulate the transition from early- to mid-neurogenesis23. Nevertheless, recent studies suggest that the regulation of BRN2 expression and function differs between mice and monkeys24. Expression of murine BRN2 has been observed in progenitors during the switch to late neurogenesis, but in cynomolgus monkeys BRN2 is already expressed in early progenitors where it regulates the behavior of neuronal progenitors and is required for normal cortical development21–24. However, the molecular and cellular mechanisms by which BRN1/2 regulate progenitor behavior across species is not well understood.
Here, we reveal a central function for BRN1 and BRN2 in establishing transcriptional programs that determine the proliferative capacity and neurogenic mode of cortical progenitors to regulate the production of ULNs and thus determine brain size in lissencephalic and gyrencephalic brains. Notably, BRN1 and BRN2 regulate the expression of large numbers of primary microcephaly genes thus implicating transcriptional deregulation as an important determinant of the molecular pathogenesis of microcephaly.
Results
BRN1/2 regulate the competence of neocortical progenitors to generate ULNs
In Brn1/2-null mice, the generation of ULNs is severely affected21,22. However, effects on brain size could not be evaluated because Brn1/2-null mice die at birth when the formation of cortical layers is still in progress. In addition, the molecular and cellular mechanisms by which BRN1/2 regulate neuronal specification are not well understood. Therefore, we generated mice carrying a floxed Brn1 allele (Brn1fl) and crossed them with mice carrying a floxed Brn2 allele (Brn2fl)25 to generate Brn1fl/fl;Brn2fl/fl mice. We then crossed Brn1fl/fl;Brn2fl/fl mice with Emx1-Cre mice26 on a Brn1fl/+;Brn2fl/+ background to generate mice lacking Brn1/2 in progenitors for excitatory neurons of the dorsal telencephalon (Brn1/2-cKO mice; Supplementary Fig. 1). Brn1/2-cKO mice were viable. We confirmed by immunohistochemistry that inactivation of Brn1 and Brn2 expression followed the pattern of CRE expression and was observed in the mutant mice already at E11.5 (Supplementary Fig. 1b–e and 1h) as expected from the onset of CRE expression in Emx1-Cre mice around E10.526. Compared to control littermates, the cortex of Brn1/2-cKO mice at postnatal day (P) 13 was reduced in thickness (Fig. 1a, b), had reduced cell numbers (Fig. 1c), and showed abnormal layering with cortical heterotopias (Fig. 1a). Defects in brain size were caused by aberrant generation of ULNs. Accordingly, Brn1/2-cKO mice lacked the corpus callosum, which is largely formed by axonal projections of ULNs (Supplementary Fig. 2a, b), and instead had an increased number of subcortical projections (Supplementary Fig. 2c, d). In the somatosensory cortex of Brn1/2-cKO mice, the expression of UL markers RORβ and CUX1 was absent with a concomitant increase in the numbers of cells expressing the deep layer (DL) markers TLE4 and CTIP2 (Supplementary Fig. 2e–h). The increase in DL neurons (DLNs) should have preserved at least some of the DL callosal projections but instead we observed an abnormal ventral misrouting of L1+ projections in Brn1/2-cKO mice (Supplementary Fig. 2b—open arrowhead), suggesting that axon guidance was affected in these neurons. Additionally, the number of glial cells was dramatically increased in Brn1/2-cKO mice at P13 (Supplementary Fig. 3)21,22.
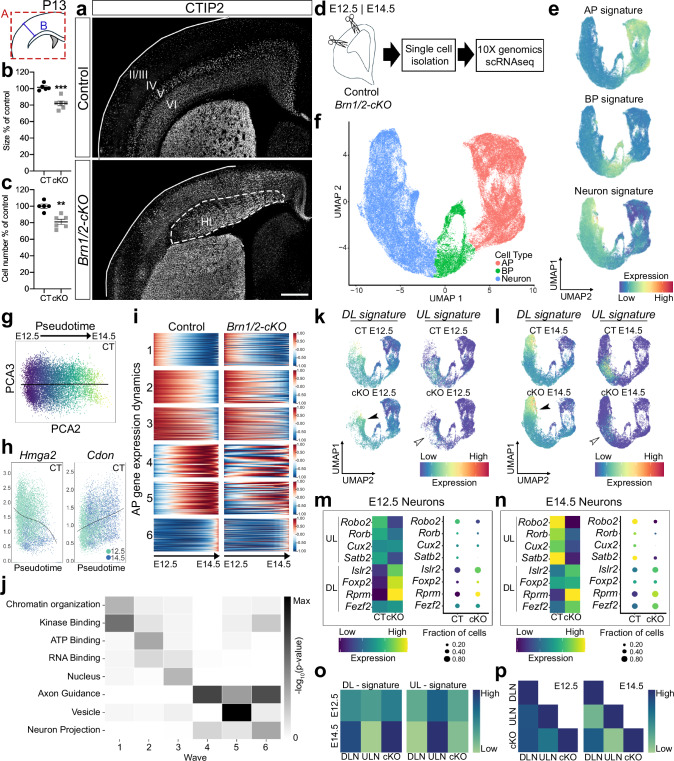
Fig. 1. BRN1/2 regulate the competence of neocortical progenitors to generate ULNs.
a Control (CT) and Brn1/2-cKO (cKO) brains analyzed by CTIP2 immunolabeling at P13. Lines represent the limits of the cortical plate (CP); dashed lines outline cortical heterotopia (Ht). Cortical size (b) and total cell number (c) in control and Brn1/2-cKO cortices at P13 (n = 5 CT, n = 6 cKO mice; two-sided unpaired t-test: (b) p = 0.0004; (c) p = 0.0015). d 10X genomics single-cell RNA sequencing (scRNA-seq) in control and Brn1/2-cKO mice at E12.5 and E14.5. e UMAP of gene signatures for apical progenitors (AP), basal progenitors (BP) and neurons. f UMAPs from control and Brn1/2-cKO cortices at E12.5 and E14.5 by cell type. g Principal component analysis (PCA) of control AP transcriptional identity organization along the pseudotime axis. h Expression of Hmga2 and Cdon in control APs along the pseudotime axis. i Cluster analysis of the gene expression dynamics for the APs along the pseudotime axis in control and Brn1/2-cKO mice (1–6 represent the different transcriptomic waves along the pseudotime axis previously described by Telley et al.32; Supplementary Data 1). j Some of the most relevant gene ontology (GO) terms defining the transcriptomic waves along the pseudotime presented in i. UMAP of deep layer (DL) and upper layer (UL) gene signature in the scRNAseq datasets from control and Brn1/2-cKO cortices at E12.5 (k) and E14.5 (l). Empty arrowheads point to reduced expression of UL gene signature in mutants, arrowheads to increased expression of DL gene signature. Expression of the indicated cortical UL and DL marker genes in control and Brn1/2-cKO neurons at E12.5 (m) and E14.5 (n). o Average DL and UL signature score in DL and UL neurons (DLNs and ULNs) and total cKO neurons at E12.5 and E14.5. p Correlation of DL and UL marker gene expression among DLNs, ULNs and total cKO neurons at E12.5 and E14.5. Values are mean ± SEM; **p < 0.01, ***p < 0.001; Scale bars: 500 µm. Source data are provided as a Source Data file.
Prior to gliogenesis, cortical progenitors generate neurons for different cortical layers in sequential order. DLNs are generated prior to ULNs, which only begin to emerge in mice around E14.527. This temporal order is largely driven by temporal changes in the competence of progenitors to generate distinct neuronal subtypes28–34. In addition, some progenitors for ULNs are already present at E12.5 but they begin to differentiate only after E14.535–38. It has been proposed that BRN1/2 in mice are critical to regulate the transition from early- to mid-neurogenesis21–23,39, while BRN2 in monkeys already acts in early progenitors24. In contrast to earlier findings, we observed BRN2 expression in murine cortical progenitors already at E11.5 (Supplementary Fig. 1f, g) suggesting that BRN1/2 affect progenitor behavior in mice at earlier time points than previously thought. To test this hypothesis, we inactivated Brn1/2 using timed in utero electroporation (IUE) of pCAG-CRE in Brn1fl/fl;Brn2fl/fl mice at E12.5 and E14.5 (Supplementary Fig. 4a). We co-expressed pCAG-RFP to identify electroporated cells. Relative to controls, BRN1/2-deficient RFP+ cells settled in deeper layers regardless of the time point when IUEs were performed (Supplementary Fig. 4b, c, e and f). In addition, experimental manipulations at both ages led to a reduction of BRN1/2-deficient RFP+ cells expressing UL markers with a concomitant increase in the number of BRN1/2-deficient RFP+ cells expressing DL markers (Supplementary Fig. 4b, d, e and g). Similar results were obtained when we injected EdU into pregnant mice at E12.5 and E14.5 and followed the cell-fate of the labeled progenitors by analyzing the co-labeling of EdU+ cells with DL and UL markers at P13 (Supplementary Fig. 4h, i, j, l and n). The EdU+ cells labeled at E12.5 and E14.5 also lost their preference for DLs or ULs, respectively and were distributed more broadly throughout cortical layers of Brn1/2-cKO mice at P13 (Supplementary Fig. 4i, j, k and m), supporting a role of BRN1/2 in neuronal migration. Altogether, these findings suggest that BRN1/2 cell-autonomously regulate the competence of progenitors to generate ULNs starting at early stages of cortical development.
To confirm this hypothesis and to determine the mechanisms by which BRN1/2 regulate neuronal specification, we performed 10X genomics single cell RNA sequencing (scRNAseq) on cortical cells isolated from Brn1/2-cKO and wild-type littermates at E12.5 and E14.5 (Fig. 1d). Cells were classified into cell types using cluster analysis and marker gene enrichment (Supplementary Fig. 5a). We then focused our analysis on APs, BPs and excitatory neurons (Fig. 1e, f; Supplementary Fig. 5b, c). In Brn1/2-cKO and controls, cell numbers were comparable (Supplementary Fig. 5d) and the proportions of AP cells decreased while the number of neurons increased between E12.5 to E14.5 (Supplementary Fig. 5d).
Telley et al. (2019) previously reported that APs undergo temporal transcriptional changes (wave 1–6) during their developmental progression32. We selected the most reproducible genes representing each transcriptional wave and observed in wild-type mice similar gene expression changes along the pseudotime axis as reported32 (Fig. 1g-j; Supplementary Fig. 5e; 1 to 6 represent the different transcriptomic waves along the pseudotime axis previously described by Telley et al.32 to reflect the temporal progression of APs and are characterized by the gene ontology (GO) terms highlighted in Fig. 1j; full gene list is available in Supplementary Data 1). In contrast, the transcriptional waves were severely disrupted in APs from Brn1/2-cKO mice (Fig. 1i; Supplementary Data 1). The transcriptional profile of E14.5 Brn1/2-cKO APs was more similar to the transcriptional profile of E12.5 control APs with a higher and more prolonged expression of the characteristic E12.5high genes and a failure to express high levels of the characteristic E14.5high genes (Fig. 1i; Supplementary Fig. 5f-j; Supplementary Data 1). We also compared gene expression programs of excitatory neurons from the cortex of control and Brn1/2-cKO mice. The expression of UL signature genes was decreased in Brn1/2-cKO neurons, while the expression of DL signature genes was increased (Fig. 1k-n; Supplementary Fig. 6a, b, Supplementary Data 2). Furthermore, despite the abnormal transcriptional profile of Brn1/2-cKO total neurons (Supplementary Data 2), their average DL and UL signature score was closely related to the control DLNs signature score (Fig. 1o) and their gene expression strongly correlated with DL marker gene expression of control DLNs at E12.5 and E14.5 (Fig. 1p). GO terms that significantly changed in Brn1/2-cKO neurons included axon guidance and neuron projections (Supplementary Fig. 6c) consistent with the axonal projection defects that we observed in the mutants at later developmental time points (Supplementary Figs. 2 and 6d). Pioneer neurons and glia guidepost cells were present in Brn1/2-cKO mice at P0 indicating the axonal projection defects observed in the mutants are primarily related to the abnormal expression of axon guidance-associated genes in these neurons at E14.5 (Supplementary Fig. 6b,d and e; Supplementary Data 2). We conclude that BRN1/2 are critical for the normal regulation of temporal gene expression changes in APs that characterize their progressive restriction in fate potential to progress from the production of DLNs to ULNs.
BRN1/2 regulate cell proliferation and the timing of cell cycle exit
To further explore the role of Brn1/2 in progenitor function, we determined the proportion of progenitors in different stages of the cell cycle by analyzing the scRNAseq data using TRIcycle (Transferable Representation and Inference of cell cycle; Fig. 2a). This method defines cell cycle phase based on the expression of reference genes characteristic for different stages of the cell cycle40: (i) G2/M, cells that are preparing to divide (G2) or already have divided (M, mitosis); (ii) S, cells that are synthesizing DNA; (iii) G1/G0, cells that are either waiting to re-enter the cell cycle (G1) or are exiting the cell cycle (G0). The number of progenitors in S-G2/M was reduced in Brn1/2-cKO mice and the number of progenitors in G1/G0 was increased (Fig. 2b). The differences were detected at E12.5 but were more pronounced at E14.5.
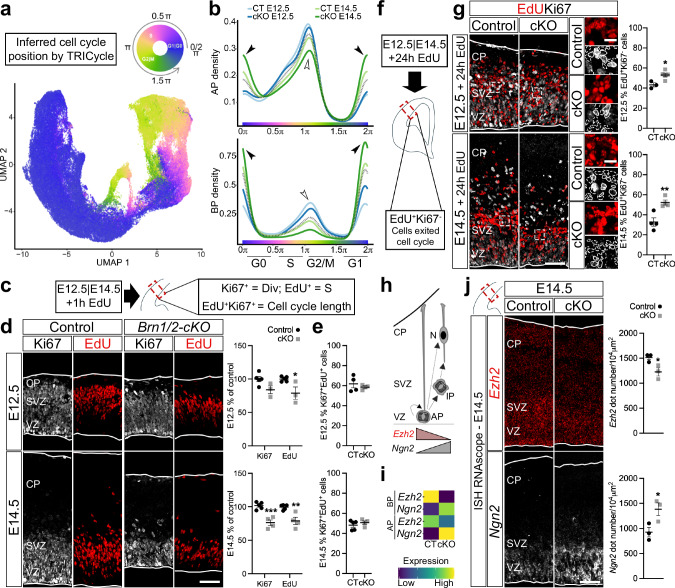
Fig. 2. BRN1/2 regulate cell cycle exit.
a UMAPs from control (CT) and Brn1/2-cKO (cKO) cortices at E12.5 and E14.5 by TRIcycle (Transferable Representation and Inference of cell cycle) analysis40. b Cell cycle phase analysis from control and Brn1/2-cKO apical progenitors (AP) and basal progenitors (BP) at E12.5 and E14.5 represented as cell density (Wilcoxon rank sum test: AP-E12.5 p = 0.085; AP-E14.5 p = 1.84e−15; BP-E12.5 p = 1.95e-06; BP-E14.5 p < 2.2e−16). Empty arrowheads point to reduced S-G2/M state in mutants, arrowheads to increased G1/G0 state. c, f Schematic of the experimental strategy. E12.5 and E14.5 cortices were analyzed by EdU and Ki67 immunolabeling 1 h (c) or 24 h (f) after intraperitoneal injection of EdU. d EdU (red) and Ki67 (grey) immunolabeling in control and Brn1/2-cKO after 1 h EdU injection at E12.5 and E14.5 (E12.5: n = 5 CT, n = 3 cKO mice; E14.5: n = 5 CT, n = 4 cKO mice; two-sided unpaired t-test: E12.5 Ki67 p = 0.1212, E12.5 EdU p = 0.0253, E14.5 Ki67 p = 0.0008, E14.5 EdU p = 0.0053). e EdU labeling index in control and Brn1/2-cKO after 1 h EdU injection at E12.5 and E14.5 (E12.5: n = 4 CT, n = 3 cKO mice; E14.5: n = 5 CT, n = 4 cKO mice; two-sided unpaired t-test: E12.5 p = 0.5296, E14.5 p = 0.4063). g EdU (red) and Ki67 (grey) immunolabeling in control and Brn1/2-cKO after 24 h EdU injection at E12.5 and E14.5 (E12.5: n = 3 CT, n = 5 cKO mice; E14.5: n = 4 mice/group; two-sided unpaired t-test: E12.5 p = 0.0135, E14.5 p = 0.0071). Boxed area at higher magnification on the right. Lines and dashed lines circulating the cells show expression or absence of Ki67, respectively. h Schematic of progenitors dividing and differentiating into neurons. i ScRNAseq expression of Ezh2 and Ngn2 at E14.5 in control and Brn1/2-cKO APs and BPs. j RNAscope for Ezh2 (red) and Ngn2 (grey) in control and Brn1/2-cKO cortices at E14.5 (n = 3 mice/group; two-sided unpaired t-test: Ezh2-p = 0.0357, Ngn2-p = 0.0436). Low and top lines represent the limits of the ventricular zone (VZ) and cortical plate (CP), respectively. SVZ Subventricular Zone, IP Intermediate Progenitors, N Neurons. Values are mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001; Scale bars: 50 µm (lower magnification), 10 µm (higher magnification). Source data are provided as a Source Data file.
To analyze the extent to which progenitor behavior was altered in Brn1/2-cKO mice and to independently validate the TRIcycle data, we injected EdU into pregnant mice at E12.5 and E14.5 and analyzed EdU incorporation into DNA 1 h later (Fig. 2c). Although the number of cortical progenitors (Supplementary Fig. 7a) and the general marker of dividing cells, Ki67 (Fig. 2d), were only significantly reduced in Brn1/2-cKO mice at E14.5, the number of EdU+ cells was reduced in Brn1/2-cKO mice already at E12.5 (Fig. 2d) consistent with the reduced number of progenitors in S-G2/M we observed in mutant mice by TRIcycle analysis.
Next, since cell cycle length was unaltered in Brn1/2-cKO mice at E12.5 and E14.5 (Fig. 2e), we analyzed cell cycle exit by injecting EdU into pregnant mice at E12.5 and E14.5 and analyzing the number of EdU+/Ki67- cells 24 h later (Fig. 2f). The number of EdU+/Ki67- cells was significantly increased in Brn1/2-cKO mice at E12.5 and E14.5 (Fig. 2g). We conclude that in Brn1/2-cKO mice, neocortical progenitors are proliferating less and exit the cell cycle faster. Consistent with this phenotype, the expression of numerous cell cycle-associated genes was significantly changed in the Brn1/2-cKO progenitors at E12.5 and at E14.5 (Supplementary Fig. 7b, c; Supplementary Data 3). These genes included Ezh2 and Ngn2, which play important roles at crucial cell cycle transition points13,41–43. High levels of Ezh2 are associated with proliferation of cortical progenitors, while high levels of Ngn2 are associated with cell cycle exit (Fig. 2h)13,41–43. The expression of Ezh2 and Ngn2 was significantly reduced and increased, respectively, in APs and BPs from Brn1/2-cKO mice at E14.5 (Fig. 2i; Supplementary Data 3). We confirmed this result by ISH RNAscope for Ezh2 and Ngn2 (Fig. 2j). In conclusion, in the absence of BRN1/2 the proliferative capacity of progenitors is reduced, and they undergo precocious neurogenesis.
BRN1/2 regulate the balance between direct and indirect neurogenesis via NOTCH signaling
Since ULNs are absent in Brn1/2-cKO mice, we wondered if BRN1/2 might determine the balance between direct and indirect neurogenesis. We combined TRIcycle with the analysis of the expression of Tbr2 and MKi67 (Figs. 2a and 3b). Tbr2 is expressed in IPs and early differentiating neurons, while MKi67 specifically labels proliferating cells. MKi67+ progenitors are thus actively dividing and among these, Tbr2+/MKi67+ defines the indirect neurogenesis branch33. In contrast, MKi67- cells expressing markers for early differentiation neurons such as Neurod2 labels primarily cells undergoing direct neurogenesis but also a small proportion of quiescent BPs and early neurons generated from IPs (Fig. 3c)33. The ratio between these two groups allowed us to infer the proportion of cells going through direct and indirect neurogenesis. The proportion of cells going through indirect neurogenesis was significantly decreased in Brn1/2-cKO mice at E12.5 and E14.5 (Fig. 3d). These findings were confirmed by TBR2 and Ki67 immunostaining (Fig. 3e) as well as by EdU incorporation analysis in TBR2+ cells (Supplementary Fig. 8a).
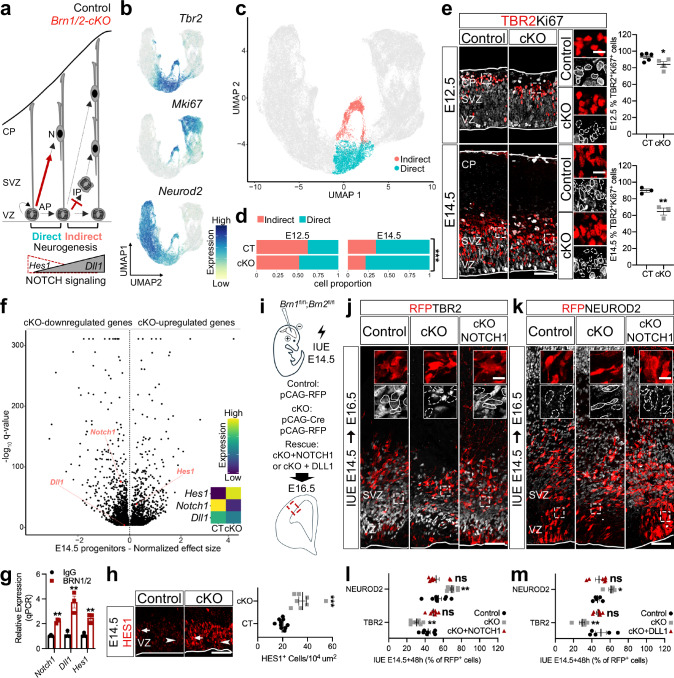
Fig. 3. BRN1/2 regulate the switch from direct to indirect neurogenesis via NOTCH signaling.
a Schematic of progenitor’s neurogenic mode in control (black) and Brn1/2-cKO (red). b UMAP of scRNAseq Tbr2, MKi67 and Neurod2 expression. c UMAP of cells going through indirect and direct neurogenesis in control (CT) and Brn1/2-cKO (cKO) at E12.5 and E14.5. d Proportion of cells going through indirect and direct neurogenesis by age and genotype (Pearson’s Chi-squared test: E12.5-p = 2.212e−11, E14.5-p < 2.2e−16). e TBR2 (red) and Ki67 (grey) immunolabeling in control and Brn1/2-cKO cortical sections at E12.5 and E14.5 (E12.5: n = 5 CT, n = 4 cKO mice; E14.5: n = 3 mice/group; two-sided unpaired t-test: E12.5-p = 0.05, E14.5-p = 0.006). Boxed area at higher magnification on the right. f Volcano plot: differentially expressed genes (DEG) between Brn1/2-cKO progenitors and controls at E14.5 highlighting NOTCH signaling-associated genes (Monocle3 VGAM test; SD = 0.15; q < 0.05; Supplementary Data 3). g ChIP-qPCR analysis of BRN1/2 binding to the indicated promoters/enhancers at E14.5 (n = 3/condition; two-sided unpaired t-test: Notch1-p = 0.0004, Dll1-p = 0.0066, Hes1-p = 0.0042). h HES1 (red) immunolabeling in the ventricular zone (VZ) of control and Brn1/2-cKO cortical sections at E14.5 (n = 10 CT, n = 8 cKO mice; two-sided unpaired t-test: p < 0.0001). i In utero electroporation (IUE) in Brn1fl/fl;Brn2fl/fl mice at E14.5. Cell identities of the control, Brn1/2-cKO and Brn1/2-cKO + NOTCH1 condition immunolabeled for RFP (red), TBR2 (j, grey) and NEUROD2 (k, grey) at E16.5. Boxed area: higher magnification in inserts. RFP+TBR2+ and RFP+NEUROD2+ at E16.5 in the control, Brn1/2-cKO, Brn1/2-cKO + NOTCH1 (l) and Brn1/2-cKO + DLL1 (m) condition (NOTCH1(l): n = 8 CT, n = 8 cKO, n = 7 cKO+NOTCH1; DLL1(m): n = 5 CT, n = 5 cKO, n = 6 cKO+NOTCH1; one-way ANOVA-Dunnett’s multiple comparisons test: NOTCH1-TBR2-p < 0.0001, F2,20 = 20.50; NOTCH1-NEUROD2-p = 0.001, F2,21 = 9.702; DLL1-TBR2-p = 0.0048, F2,13 = 8.294; DLL1-NEUROD2-p = 0.0098, F2,13 = 6.748). Low and top lines represent the limits of the VZ and cortical plate (CP), respectively. Lines and dashed lines outline cells expressing or lacking expression of the indicated marker, respectively. SVZ Subventricular Zone, IP Intermediate Progenitors, N Neurons. Values are mean ± SEM; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: 50 µm (lower magnification), 10 µm (higher magnification). Source data are provided as a Source Data file.
Similar results were obtained when we used CRE to acutely inactivate Brn1/2 by IUE of Brn1fl/fl;Brn2fl/fl mice at E12.5 or E14.5 and analyzed neurogenesis at E14.5 or E16.5 (Supplementary Fig. 8b and Fig. 3i:cKO). We co-expressed pCAG-RFP to identify electroporated cells. The number of mutant RFP+ cells going through indirect neurogenesis was significantly reduced (Supplementary Fig. 8c and Fig. 3j, l:cKO), while the number of mutant RFP+/NEUROD2+ cells was significantly increased consistent with a higher rate of direct neurogenesis (Supplementary Fig. 8c and Fig. 3k, l:cKO). Since indirect neurogenesis is reduced already at early stages of corticogenesis, we reasoned that the generation of CUX2+ BPs, a subgroup of early generated BPs that is fated to generate ULNs35,44,45, might be affected in Brn1/2-cKO mice. Indeed, the proportion of Cux2+ BPs was significantly reduced in Brn1/2-cKO mice at E14.5 (Supplementary Fig. 8d-f), a finding that we confirmed by quantifying Cux2 mRNA expression by ISH RNAscope in TBR2+ cells (Supplementary Fig. 8g). Consistent with the depletion of the CUX2+ BPs fated to generate ULNs in Brn1/2-cKO mice, scRNAseq analysis revealed that several UL markers were reduced, while some DL markers were increased in BPs from Brn1/2-cKO mice at E14.5 (Supplementary Fig. 8h; Supplementary Data 3).
Expression levels of HES1 and the NOTCH ligand DLL1 in neural progenitors oscillate with opposite phases, which is critical to regulate the proliferation and differentiation of these cells (Supplementary Fig. 9a)46,47. In addition, high levels of Hes1 and low levels of Dll1 are associated with direct neurogenesis (Fig. 3a)6,48. Since, BRN1/2 suppress the activity of the Hes1 promoter and cooperate with MASH1 to activate expression of Dll123,49, we wondered whether defects in Hes1 and Dll1 expression might explain the defects in indirect neurogenesis in Brn1/2-cKO mice. scRNAseq analysis revealed that Hes1 was dramatically overexpressed in Brn1/2-cKO progenitors at E14.5 (Fig. 3f; Supplementary Data 3). In contrast, expression of Notch1 and Dll1 were significantly reduced (Fig. 3f; Supplementary Data 3). We confirmed these findings by ISH RNAscope (Supplementary Fig. 9b). In addition, the direct binding of BRN1/2 to the Ensembl-predicted regulatory regions of Notch1, Dll1 and Hes1 was confirmed by BRN1/2 Chromatin Immunoprecipitation at E14.5 (ChIP-qPCR; Fig. 3g), suggesting that these genes are direct targets for BRN1/2.
Given the cross-regulatory interactions between HES1 and DLL146,47, we expected that the amplitude of oscillatory gene expression of NOTCH signaling pathway components might be affected in Brn1/2-cKO mice. We analyzed EdU incorporation and the expression of HES1 during cell cycle in control and Brn1/2-cKO mice 90 minutes (90’; S + G2 phase), 8 h (early G1) or 14 h (late G1) after EdU injection into pregnant mice at E14.5 (Supplementary Fig. 9c). Although the typical HES1 oscillations still occurred in Brn1/2-cKO progenitors (Supplementary Fig. 9c), levels of HES1 were vastly increased compared to controls at any time during the cell cycle (Fig. 3h; Supplementary Fig. 9c). Even lowest levels of HES1 in mutants never fell below highest levels in controls (Fig. 3h; Supplementary Fig. 9c). We conclude that BRN1/2 play a crucial role in establishing normal expression patterns of NOTCH signaling pathway components in progenitors.
To provide direct evidence for a causal relationship between perturbations in NOTCH signaling and the changes in indirect neurogenesis in Brn1/2-cKO mice, we co-expressed pCAG-CRE to inactivate Brn1/2 and pCS2-NOTCH1 to overexpress NOTCH1 by IUE of Brn1fl/fl;Brn2fl/fl mice at E14.5 and analyzed neurogenesis at E16.5 (Fig. 3i). We co-expressed pCAG-RFP to identify electroporated cells. While NOTCH1 overexpression did not change the rate of indirect neurogenesis (RFP+/TBR2+ cells) and neuronal production (RFP+/NEUROD2+ cells) in wild-type (Supplementary Fig. 9d-f), it restored levels of indirect neurogenesis and neuronal production in the Brn1/2-cKO condition to control levels (Fig. 3j-l). Significantly, overexpression of DLL1 in Brn1fl/fl;Brn2fl/fl mice phenocopied the effect of NOTCH1 overexpression (Fig. 3m; Supplementary Fig. 9g-i), which is consistent with the idea that an imbalance in DLL1-dependent NOTCH1 signaling in progenitors is responsible for the defects in indirect neurogenesis in Brn1/2-cKO mice (Fig. 3a).
BRN1/2 are required to maintain the neuronal progenitor pool through the regulation of microcephaly-associated genes
Increases in brain size during evolution are accompanied by a disproportionate increase in ULNs compared to DLNs, and the production of ULNs is frequently affected in microcephaly patients20. Because ULNs are massively affected by defects in BRN1/2 function and the brain of Brn1/2-cKO mice is vastly more microcephalic than is typical for most microcephaly mouse models, we wondered whether BRN1/2 are required for the expression and function of sets of genes linked to microcephaly. ScRNAseq analysis revealed that the expression of a large number of genes that when mutated cause microcephaly in humans was significantly decreased in Brn1/2-cKO progenitors (Fig. 4a; Supplementary Data 3). These included 11 classical primary microcephaly genes: Cdk5rap2 (MCPH3), Knl1 (MCPH4), Aspm (MCPH5), Cenpj (MCPH6), Stil (MCPH7), Cep135 (MCPH8), Cdk6 (MCPH12), Sass6 (MCPH14), Mfsd2a (MCPH15), Cit (MCPH17) and Copb2 (MCPH19)7. We confirmed these findings for Aspm and Cdk6 by ISH RNAscope at E14.5 (Supplementary Fig. 10a).
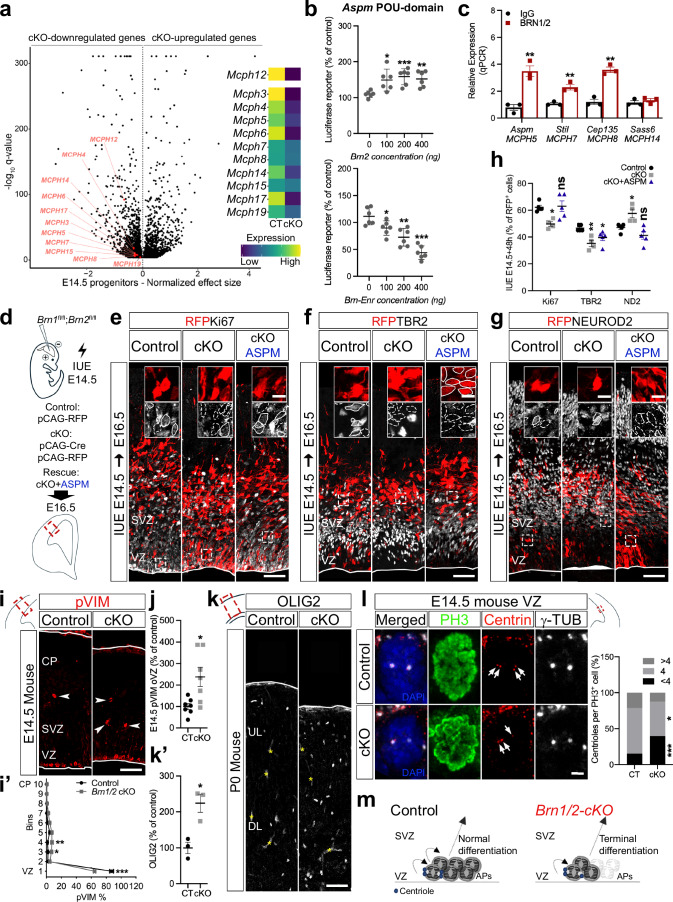
Fig. 4. BRN1/2 are required for the expression of microcephaly-associated genes and maintenance of the neuronal progenitor pool.
a Volcano plot: DEG between Brn1/2-cKO and control progenitors at E14.5 highlighting primary microcephaly genes (Monocle3 VGAM test; SD = 0.15; q < 0.05; Supplementary Data 3). b Luciferase reporter-activity of Aspm when co-expressed with Brn2 or Brn-Enr (n = 6/condition; two-sided unpaired t-test: Brn2: 100-p = 0.0128, 200-p = 0.0006, 400-p = 0.0015; Enr: 100-p = 0.0268, 200-p = 0.0018, 400-p = 0.0001). c ChIP-qPCR analysis of BRN1/2 binding to the indicated promoters/enhancers at E14.5 (n = 3/condition; two-sided unpaired t-test: Aspm-p = 0.0038, Stil-p = 0.0053, Cep135-p = 0.0011, Sass6-p = 0.4289). d In utero electroporation (IUE) in Brn1fl/fl;Brn2fl/fl mice at E14.5. Cell identities of the control, Brn1/2-cKO and Brn1/2-cKO + ASPM condition immunolabeled for RFP (red), Ki67 (e, grey), TBR2 (f, grey) and NEUROD2 (g, grey) at E16.5. Boxed area: higher magnification in inserts. Lines and dashed lines outlining cells expressing or lacking expression of the indicated marker, respectively (n = 5 CT, n = 4 cKO, n = 5 cKO+ASPM mice; one-way ANOVA-Dunnett’s multiple comparisons test: Ki67-p = 0.0132, F2,11 = 6.582; TBR2-p = 0.0055, F2,11 = 8.681; NEUROD2-p = 0.0033, F2,11 = 10.02). i pVIM immunolabeling in control and Brn1/2-cKO mice at E14.5. Arrowheads: pVIM+ cells outside of the VZ (oVZ). i’ Distribution of pVIM+ cells in control (CT) and Brn1/2-cKO (cKO) cortices (n = 7 mice/group; two-way ANOVA-Šídák’s multiple comparisons test: 1-p < 0.0001, 3-p = 0.0403, 4-p = 0.0025, F9,120 = 25.73). j pVIM+ cells in control and Brn1/2-cKO oVZ (n = 7 mice/group; two-sided unpaired t-test: p = 0.0122). k, k’ OLIG2 immunolabeling in cortical sections of control and Brn1/2-cKO mice at P0 (n = 3 mice/group; two-sided unpaired t-test: p = 0.0140). l Centriole number/PH3+ cell in the ventricular zone (VZ) of control and Brn1/2-cKO at E14.5 (n = 3 mice/group; two-sided unpaired t-test: <4-p = 0.0002, 4-p = 0.0383, >4-p = 0.1018). Arrows indicate centrioles. m Schematic of centrosome function and progenitor cell differentiation in control (black) and Brn1/2-cKO (red). Low and top lines represent the limits of the VZ and cortical plate (CP), respectively. Yellow asterisks indicate auto-fluorescent blood vessels. SVZ Subventricular Zone, UL Upper Layers, DL Deep Layers, APs Apical Progenitors. Values are mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: 50 µm (lower magnification; e-g, i, and k), 10 µm (higher magnification; e-g), 2 µm (l). Source data are provided as a Source Data file.
To study the regulation of Aspm by BRN1/2, we cloned the Ensembl-predicted regulatory region of Aspm containing POU-domain binding sites in a luciferase reporter vector and tested its transcriptional activity in absence or presence of pCAG-Brn2 or pCAG-Brn-DBD-EnR, a dominant negative inhibitor of BRN1/2 function23,49. Co-expression with BRN2 increased Aspm-luciferase activity while co-expression with Brn-DBD-EnR inhibited Aspm-luciferase activity (Fig. 4b) indicating BRN1/2 directly regulates the expression of Aspm. The direct binding of BRN1/2 to the Ensembl-predicted regulatory regions of Aspm was confirmed by ChIP-qPCR at E14.5 (Fig.4c; Supplementary Fig. 10b). To understand the extent to which Aspm contributes to the changes of progenitor behavior in Brn1/2-cKO mice, we co-expressed pCAG-CRE to inactivate Brn1/2 and pBlue-hASPM to overexpress ASPM by IUE of Brn1fl/fl;Brn2fl/fl mice at E14.5 and analyzed neurogenesis at E16.5 (Fig. 4d). We co-expressed pCAG-RFP to identify electroporated cells. While ASPM overexpression did not rescue the rate of indirect neurogenesis (RFP+/TBR2+ cells; Fig. 4f, h), it restored the levels of proliferation/cell cycle exit (RFP+/Ki67+ cells) and neuronal production (RFP+/NEUROD2+ cells) in the Brn1/2-cKO condition to control levels (Fig. 4e, g and h). These results indicate that BRN1/2-dependent regulation of Aspm expression and function primarily controls the proliferation and cell cycle exit of progenitor cells without major contribution to the levels of indirect neurogenesis that are mainly regulated by the levels of NOTCH signaling in these cells.
Knock-out of ASPM in ferret, a small carnivore characterized by an expanded and gyrencephalic neocortex, phenocopies the microcephaly defect in human patients50. To gain insights on the cellular mechanisms controlled by BRN1/2-dependent regulation of Aspm expression, we compared the cortical phenotype of Brn1/2-cKO mice and ASPMKO ferrets. In ASPMKO ferrets, APs detached prematurely from the VZ resulting in increased number of terminal divisions and premature differentiation towards the glial lineage (Supplementary Fig. 10c-e and h-j)50. Although the distribution of TBR2+Ki67+ cells within the neocortex was broadened including localization of cells outside of the VZ (Supplementary Fig. 10f), the levels of indirect neurogenesis were not affected in ASPMKO ferrets at E35 (Supplementary Fig. 10g), consistent with the IUE results where ASPM overexpression failed to rescue the balance between direct and indirect neurogenesis in Brn1/2-cKO mice. Similar to ASPMKO ferrets, APs in Brn1/2-cKO mice were displaced at E14.5 (Fig. 4i, j) and the number of glial cells was also increased at P0 (Fig. 4k). In addition, the number of neuronal progenitors declined more rapidly throughout embryonic development in Brn1/2-cKO mice compared to controls (Supplementary Fig. 10k) and glial numbers were further increased at P13 (Supplementary Fig. 3a).
To further determine if there is a precocious shift from neurogenesis to gliogenesis in Brn1/2-cKO mice comparable to ASPMKO, we injected EdU into pregnant mice at E12.5 and E14.5 and followed the cell-fate of the labeled progenitors by analyzing the co-labeling of EdU+ cells with the glia markers SOX9 and OLIG2 at P13 (Supplementary Fig. 3b). Numbers of EdU+SOX9+ and EdU+OLIG2+ cells were significantly increased in Brn1/2-cKO mice at P13 (Supplementary Fig. 3c-f), which is indicative of precocious gliogenesis happening in Brn1/2-cKO mice during early embryonic development. We obtained similar results when we inactivated Brn1/2 in the developing neocortex of Brn1fl/+;Brn2fl/+ by IUE of pCAG-CRE at E14.5 followed by the quantification of the numbers of GFP+OLIG2+ cells at P13 (Supplementary Fig 3g-i).
Since Aspm and most of the microcephaly-associated genes altered in Brn1/2-cKO progenitors encode centrosome proteins7, we tested how the absence of BRN1/2 affects centrosome function in progenitors. The expression of genes essential for centrosome function were significantly reduced in Brn1/2-cKO progenitors at E14.5 (Supplementary Fig. 10l; Supplementary Data 3) and centriole duplication was disrupted (Fig. 4l) indicating that defects in Aspm expression, and possibly in other microcephaly genes like Stil and Cep135, contribute to the cortical phenotype of Brn1/2-cKO mice (Fig. 4m).
BRN1/2 function is conserved across phylogeny
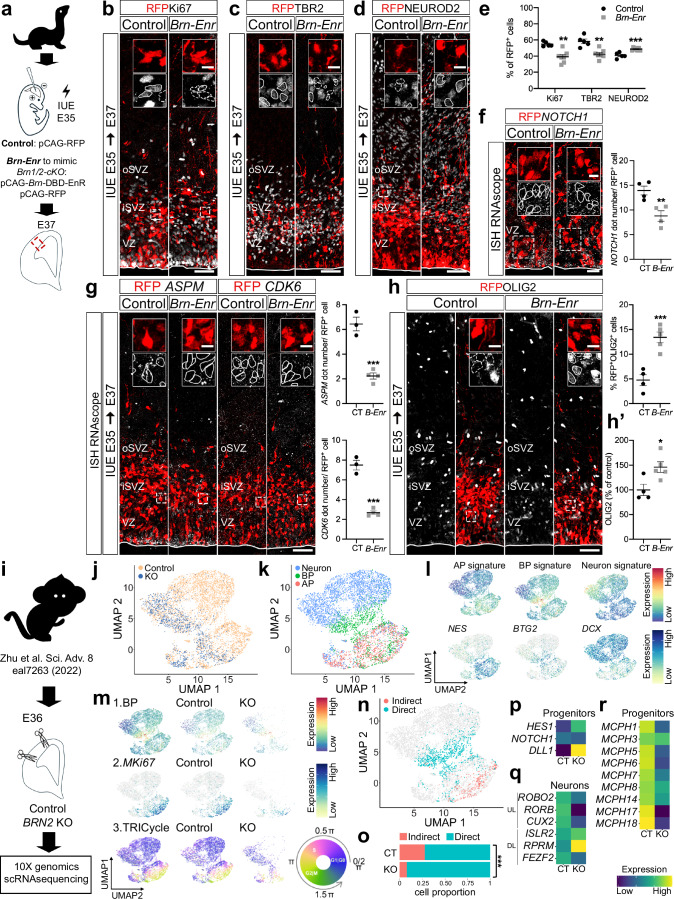
In BRN2KO monkeys, the neurogenic behavior of cortical progenitors is similarly altered as in Brn1/2-cKO mice24. To test if the mechanisms by which BRN1/2 regulate progenitor behavior are evolutionary conserved, we first inhibited BRN1/2 activity in wild-type ferrets at E35 by IUE with pCAG-Brn-DBD-EnR (Fig. 5a)23,49. 48 h after Brn-DBD-EnR expression, the number of dividing progenitors (RFP+/Ki67+) was significantly reduced, indicative of faster cell cycle exit compared to controls (Fig. 5b, e). The number of cells going through indirect neurogenesis (RFP+/TBR2+) was significantly reduced (Fig. 5c, e), while neuronal output (RFP+/NEUROD2+ cells) was increased (Fig. 5d, e). Additionally, the expression of Notch1 was reduced in the cortex of ferrets expressing Brn-DBD-EnR (Fig. 5f-f’). Thus, our results in ferrets mirror those in mice, providing evidence for a conserved role for BRN1/2 in cortical progenitors of lissencephalic and gyrencephalic brains. In further support of this hypothesis, in the cortex of ferrets expressing Brn-DBD-EnR the expression of Aspm and Cdk6 was also reduced (Fig. 5g), resulting in an increase of RFP+/OLIG2+ cells and total OLIG2+ cells (Fig. 5h, h’), indicating a precocious shift from neurogenesis to gliogenesis in ferrets in absence of BRN1/2 activity.
Fig. 5. BRN1/2 function is conserved across mammalian species.
a In utero electroporation (IUE) in wild-type ferrets at E35. Cell identities of the control (CT) and Brn-Enr (B-Enr) condition immunolabeled for RFP (red), Ki67 (b, grey), TBR2 (c, grey) and NEUROD2 (d, grey) at E37 (n = 4 CT, n = 7 Brn-Enr ferrets; two-sided unpaired t-test: Ki67-p = 0.0014, TBR2-p = 0.0012, NEUROD2-p = 0.0004). f NOTCH1 (grey) expression in RFP+ cells of the electroporated ventricular zone (VZ) (n = 4 ferrets/group; two-sided unpaired t-test: p = 0.0099). g ASPM or CDK6 (grey) expression in RFP+ cells of the electroporated cortex (n = 3 CT, n = 4 Brn-Enr ferrets; two-tailed unpaired t-test: ASPM-p = 0.0006, CDK6-p = 0.0001). h RFP+ cells expressing OLIG2 (gray) in the electroporated cortex (n = 4 CT, n = 5 Brn-Enr ferrets; two-sided unpaired t-test: p = 0.0009). h’ Total OLIG2+ cells in the electroporated cortex (n = 4 CT, n = 5 Brn-Enr ferrets; two-sided unpaired t-test: p = 0.0226). i ScRNAseq DATA from E36 cortex of control (CT) and BRN2KO (KO) cynomolgus monkey24 re-analyzed using the same pipeline we used for the analysis in mice. UMAP of scRNAseq from control and BRN2KO by genotype (j) and cell type (k). l UMAP of gene signatures for apical progenitors (AP; e.g., NES), basal progenitors (BP; e.g., BTG2) and neurons (e.g., DCX) in control and BRN2KO. m UMAP of BP signature (1), MKi67 expression (2) and TRIcycle score (3) in control and BRN2KO. n UMAP of cells going through indirect and direct neurogenesis in control and BRN2KO. o Proportion of cells going through indirect and direct neurogenesis by genotype (Pearson’s Chi-squared test: p = 4.454e-12). p Expression of NOTCH signaling-associated genes in control and BRN2KO progenitors. q Expression of the indicated cortical upper layer (UL) and deep layer (DL) neuronal marker genes in control and BRN2KO neurons. r Expression of primary microcephaly DEG in BRN2KO progenitors compared to controls. Boxed areas: higher magnification in inserts. Lines and dashed lines outline cells expressing or not expressing the indicated marker, respectively. iSVZ Inner Subventricular Zone, oSVZ Outer Subventricular Zone. Values are mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: 50 µm (lower magnification), 10 µm (higher magnification). Source data are provided as a Source Data file.
Next, we analyzed available raw scRNAseq data from BRN2KO monkeys at E3624 using identical criteria as employed in our analysis of scRNAseq from mice (Fig. 5i-k). We identified APs, BPs and neurons (Fig. 5l) and used the BP gene expression signature, the expression profile of MKi67 and TRIcycle analysis (Fig. 5m1-3), to identify cells going through direct and indirect neurogenesis (Fig. 4n). Similar to our findings in mice, the proportion of cells going through indirect neurogenesis was significantly reduced in BRN2KO monkeys (Fig. 5o). Expression of NOTCH1 was reduced while HES1 was dramatically increased in BRN2KO progenitors (Fig. 5p; Supplementary Data 4). Unlike in mice, DLL1 expression was increased in BRN2KO progenitors (Fig. 5p; Supplementary Data 4). Effects on the expression of DLL1 might not be observed in monkeys because BRN1 may compensate in part for loss of BRN2. Alternatively, the regulation of NOTCH signaling pathways in monkeys might reflect differences between species. Interestingly, the expression of some UL markers was already reduced in the BRN2KO neurons at E36 (Fig. 5q; Supplementary Data 4). In addition, the expression of 9 classical microcephaly genes (MCPH1, CDK5RAP2 (MCPH3), ASPM (MCPH5), CENPJ (MCPH6), STIL (MCPH7), CEP135 (MCPH8), SASS6 (MCPH14), CIT (MCPH17) and WDFY3 (MCPH18))7 was reduced in BRN2KO progenitors at E36 (Fig. 5r; Supplementary Data 4) mimicking the results in Brn1/2-cKO mice.
Overall, our findings reveal a striking similarity in the defects caused by perturbations in BRN1/2 function in mice, ferrets and monkeys, supporting a model in which BRN1/2 have evolutionarily conserved functions to regulate neocortical progenitor behavior in lissencephalic and gyrencephalic animals.
Discussion
The molecular mechanisms that regulate the behavior of cortical progenitors in the mammalian clade and thus control brain size are not well understood. We show here that BRN1/2 drive the transcriptional programs in neocortical progenitors of mice, ferrets and monkeys that determine the timing of their differentiation and their total output of ULNs. Loss of BRN1/2 in progenitors leads to the deregulation of the expression of genes that regulate cell cycle progression, NOTCH signaling and centrosome function, thus coordinating the expression of sets of genes that affect different aspects of progenitor behavior.
We show that BRN1/2 are expressed in cortical progenitors of mice already at E11.5 during lower layer neurogenesis. BRN2 in monkeys is also expressed in cortical progenitors from the earliest stages of neurogenesis24, suggesting evolutionary conservation in the regulation of BRN2 expression. Consistent with the BRN1/2 expression pattern, we demonstrate that BRN1/2 act in progenitors during the earliest stages of cortical development to regulate their transcriptional programs, thus leading to defects in cell proliferation, precocious cell cycle exit, and altered balance of direct and indirect neurogenesis.
Our functional studies demonstrate that deregulation of NOTCH signaling contributes to defects in indirect neurogenesis and the production of ULNs observed in Brn1/2-cKO mice, which is consistent with previous studies demonstrating the crucial function of NOTCH signaling for regulating the neurogenic mode of cortical progenitors6,48. We show that BRN1/2 directly bind to the predicted regulatory regions of Notch1, Dll1 and Hes1 promoters/enhancers. Consistent with this finding, we observed increased levels of Hes1 and reduced levels of Dll1 and Notch1 in Brn1/2-cKO mice. Expression of NOTCH signaling components in wild-type mice is tightly regulated by feedback mechanisms that establish their oscillatory expression and the amplitude of these oscillations is crucial for balanced levels of progenitor’s proliferation and differentiation46,47. Oscillations at higher levels than normal have been shown to inhibit proliferation51,52. In Brn1/2-cKO mice, the lowest HES1 expression levels during cell cycle are higher than peak levels of HES1 in wild-type, consistent with the proliferative defects in the mutant mice. Also, Dll1 and Hes1 are intricately linked to the neurogenic mode of cortical progenitors, where low levels of Dll1 are associated with reduced indirect neurogenesis6, while high levels of Hes1 are associated with high levels of direct neurogenesis48, which is consistent with the phenotype of Brn1/2-cKO mice. BRN1/2 functional defects are restored when we overexpress NOTCH1 or DLL1 to override the loss of BRN1/2 function, indicating that NOTCH signaling components regulate the neurogenic mode of cortical progenitors downstream of BRN1/2.
In addition to the transcriptional changes in cell cycle- and NOTCH signaling-associated genes, the expression of large numbers of microcephaly-associated genes is altered in Brn1/2-cKO mice. In addition, we show that BRN1/2 directly bind and regulate the transcriptional activity of Aspm, a gene essential for normal centrosome function. Centrosomes have crucial roles in the organization of microtubules and regulate cell polarity, the formation of primary cilia and mitotic spindle assembly53. We found that centrosome function is disrupted in Brn1/2-cKO progenitors. Consistent with the deregulation of the expression of microcephaly genes as shown by perturbation of ASPM function50, defects in centrosome function result in precocious cell cycle exit in Brn1/2-cKO progenitors and accelerated depletion of neuronal progenitors during progressive stages of neurogenesis, contributing to the absence of ULNs, increased gliogenesis and reduced brain size.
The Brn1/2-cKO proliferation defects but not the levels of indirect neurogenesis are restored when we overexpress ASPM to override the loss of BRN1/2 function, indicating that BRN1/2-dependent regulation of Aspm expression and function primarily controls the proliferation and cell cycle exit of progenitor cells without major contribution to the levels of indirect neurogenesis that are mainly controlled by the tightly regulated NOTCH signaling by BRN1/2 in these cells.
While our findings provide evidence for an evolutionary conserved role for BRN1/2 regulation of transcriptional programs that control progenitor behavior, it remains to be established to what extent the function of these transcriptional regulators has been modified during brain evolution to produce differences in brain size. For example, disruption of BRN2 function alone affects cortical development in monkeys24 but not in mice21,22, suggesting that redundancy of BRN1 and BRN2 function is not universally conserved. In addition, recent studies have shown that during evolution, enhancers that control neuronal gene expression have been altered by mutations that affect predominantly binding sites for a limited set of transcription factors, including BRN1/254. Microcephaly-associated genes are included in the list of genes that contain enhancer elements enriched in BRN1/2 binding sites54. Changes to regulatory regions of the genome that affect the transcriptional landscape are thus likely to be intricately linked to adaptive changes crucial for neocortical expansion and affected in microcephaly in humans.
Methods
All the experiments reported in this study adhered to the ethical regulations and guidelines of the National Institute of Health Guide for Care and Use of Laboratory Animals. The experiments involving mice were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine under the study protocol numbers: MO16M273, MO19M255 and MO22M217. The experiments involving ferrets were also approved by the Animal Care and Use Committee at Johns Hopkins University School of Medicine under the study protocol number: FE21M265.
Mice
All animal experiments adhered to the guidelines of the National Institute of Health and were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine. Mice were maintained on a 14 h light/10 h dark cycle. Both male and female mice were used, and no obvious differences between the sexes were noted. All mice were group-housed in pathogen-free facilities with regulated temperature and humidity and given ad libitum access to food and water. All mice used were seemingly free of infection, health abnormalities or immune system deficiencies and were employed independently of their gender. None of the mice used had been used for previous experiments. The date of the vaginal plug detection was designated E0.5, and the date of birth P0. Emx1-Cre [B6.129S2-Emx1tm1(cre)Krj]26 and Brn2 conditional knockout [Brn-2fl/fl]25 mice have been described. Brn1 conditional knockout mouse was generated in collaboration with Janelia Research Campus - Howard Hughes Medical Institute. The embryonic stem cells (ESCs) were designed to insert loxP sites flanking the coding sequence of Brn1 gene, with upstream elements including a neomycin-resistance cassette (PGK-neo) and lacZ reporter flanked by two FRT sites. ESCs were transplanted into mouse blastocysts to produce transgenic mice. Heterozygous F1 mice (Brn1flox-neo/+) were mated with FLPe mice to remove the PGK-neo cassette and lacZ reporter. The resulting offspring were subsequently mated to C57BL/6 J mice to remove the FLPe transgene. C57BL/6 J wild type mice were obtained from Charles River Laboratories. Brn1fl/fl and Brn2fl/fl mice were crossed and the resulting Brn1fl/fl;Brn2fl/fl mice were crossed with heterozygous Emx1-Cre mice. Animals with no CRE-recombination were used as control littermates in the scRNAseq experiments. Animals with no CRE-recombination or heterozygous for one or the two genes (Brn1 and Brn2) were used as control littermates in the different histological experiments. We used these mice interchangeably because they were indistinguishable from each other and from wild-type mice.
Ferrets
All procedures adhered to the guidelines of the National Institute of Health and were approved by the Animal Care and Use Committee at Johns Hopkins University. Experiments were performed in female ferrets (Mustela putoris furo, Marshall BioResources) with normal immune status. Animals were housed in a 16 h light/8 h dark cycle. Ferrets were not involved in previous studies. The ASPMKO brain sections50 were obtained through the collaboration with Dr. Byoung-Il Bae, Dr. Richard S. Smith and Dr. Christopher A. Walsh.
Animal Euthanasia
We used the most humane methods for euthanasia following the recommendations of the panel report on Euthanasia of the American Veterinary Medical Association and the guidelines of the National Institute of Health Guide for Care and Use of Laboratory Animals. Different methods were used for mice of different ages, according to the experiments that needed to be performed: neonatal and adult mice—isoflurane inhalation followed by cervical dislocation; newborn mice—chilling and decapitation; embryos 9–19 days post coitum—chilling and decapitation; newborn to adult mice—cardiac perfusion under full anesthesia. For tissue collection in ferrets, pregnant females were first anesthetized with ketamine (40 mg/kg IM), supplemented with isoflurane (1–3%), before euthanizing them with an overdose of Euthasol (0.26 mL/kg intracardially or IV). Removal of fetuses only began after cessation of heart beat and breathing was confirmed for the jill. In the unlikely case that live fetuses were discovered after administration of the euthanasia solution, an additional dose of Euthasol was administered through the uterine artery and fetuses were decapitated before removing them from the mother.
Immunohistochemistry
Embryonic brains were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 2–4 h at 4 °C. Postnatal mice were transcardially perfused with 20 ml ice-cold 4% PFA using a peristaltic pump at a rate of 2 ml/min. Brains were removed from the skull and postfixed in 4% PFA overnight at 4 °C. Embryonic brains were cryopreserved with 30% sucrose, embedded in Tissue-Tek O.C.T. compound (#4583, SAKURA) and frozen at −80 °C. They were then sectioned coronally at 16 µm with a cryostat (CM3050 S, Leica). Postnatal brains were embedded in 3% low melting point agarose (#R0801, Thermo Fisher Scientific) in PBS and sectioned coronally or sagittally at 60 µm with a vibrating microtome (VT1200S, Leica). For immunohistochemistry, brain sections were permeabilized in PBS containing 0.2% Triton X-100 and blocked in 10% goat serum for 1 h at room temperature. Brain slices were then incubated with primary antibodies in blocking solution overnight (or 48 h depending on the thickness of the section and antibody used) at 4 °C and subsequently incubated with the appropriate secondary antibodies diluted in blocking solution for 2 h at room temperature. To label all cell nuclei, the fluorescent nuclear dye DAPI (1 μg/ml, #D9542, Sigma-Aldrich) was included with the secondary antibody solution. The sections were mounted with ProLongTM Gold (#P36930, Thermo Fisher Scientific). Primary antibodies used were: anti-L1 (1:500, rat monoclonal, MAB5272 Millipore Sigma); anti-NRP1 (1:500, goat polyclonal, AF566 R&D systems); anti-TLE4 (1:500, rabbit polyclonal, ab64833 Abcam); anti-CTIP2 (1:1000, rat monoclonal, ab18465 Abcam); anti-RORβ (1:500, mouse monoclonal, PP-N7927-00 R&D systems); anti-CUX1 (1:1000, rabbit polyclonal, 11733-1-AP Proteintech); anti-GFP (1:500, chicken polyclonal, GFP-1020 Aves Labs Inc.); anti-BRN2 (1:1000, rabbit polyclonal, PA5-30124 Thermo Fisher Scientific or 1:1000, rabbit monoclonal, 12137 Cell Signaling); anti-BRN1 (1:1000, rabbit polyclonal, sc-6028-R Santa Cruz Biotechnology); anti-CRE (1:200, mouse monoclonal, MAB3120 Millipore Sigma); anti-NeuN (1:500, mouse monoclonal, MAB377 Millipore Sigma); anti-Ki67 (1:500, rabbit polyclonal, ab15580 Abcam or 1:500, rat monoclonal, 14-5698-82 Thermo Fisher Scientific); anti-RFP (1:1000, rat monoclonal, 5F8 ChromoTek or 1:1000, rabbit polyclonal, ab62341 Abcam); anti-Pax6 (1:250, mouse monoclonal, MA1-109 Thermo Fisher Scientific); anti-TBR2 (1:500, rat monoclonal, 14-4875-82 Thermo Fisher Scientific or 1:500, rabbit polyclonal, ab183991 Abcam); anti-NEUROD2 (1:500, rabbit polyclonal, ab104430 Abcam); anti-HES1 (1:1000, rabbit polyclonal, R. Kageyama gift46); anti-pVIM (1:500, mouse monoclonal, D076-3 MBL); anti-OLIG2 (1:500, rabbit polyclonal, AB9610 Millipore Sigma); anti-SOX9 (1:1000, rabbit monoclonal, ab185966 Abcam); anti-GFAP (1:1000, chicken polyclonal, AB5541 Millipore Sigma); anti-PH3 (1:1000, rat monoclonal, ab10543 Abcam); anti-γ-TUBULIN (1:500, goat polyclonal, A. Holland gift55); anti-Centrin (1:500, rabbit polyclonal, A. Holland gift56). Secondary antibodies used were: Alexa 555, 647, 488 anti-rat (A21434; A21247, A11006), anti-rabbit (A21430; A21246) and anti-mouse (A21425; A21237); Alexa 488 anti-rabbit (A11070); Alexa 488 and 647 anti-chicken (A11039; A21449); anti-rabbit HRP (A27036); all diluted 1:500 and all from Thermo Fisher Scientific. Antigen retrieval57 was performed for all immunohistochemistry at embryonic ages and to stain for OLIG2 and RORβ postnatally. The method used here was a heat-induced citrate method57. HES1 immunostaining was done using a horseradish peroxidase (HRP)-secondary antibody and an additional tyramide signal amplification (TSA) step. Therefore, after antigen retrieval, the brains sections were treated with 3% hydrogen peroxide to block HRP-unspecific binding. After the secondary antibody, the sections were washed and incubated with TSA solution (#NEL744001KT, Akoya Biosciences) overnight at 4 °C. The sections were then mounted with ProLongTM Gold (P36930, Thermo Fisher Scientific).
RNAscope multiplex in situ hybridization
Brain sections were processed for multiplex fluorescent in situ hybridization RNAscope58 following the manufacturer’s instructions (#323110, Advanced Cell Diagnostics). Gene-specific probes: Fezf2 (313301-C2), Satb2 (413261), Ezh2 (446611-C2), Ngn2 (417291-C2), Cux2 (469551-C3), Notch1 (404641), Dll1 (425071-C2), Hes1 (417701), Aspm (427881) and Cdk6 (570091-C2). When we combined RNAscope with immunohistochemistry, the sections were briefly washed in 1X PBS after the last conjugation step and the immunohistochemistry protocol was followed as above.
EdU injections and labeling
EdU (5-ethynyl-2´-deoxyuridine; #A10044, Thermo Fisher Scientific) was diluted at 10 mg/ml in 0.9% NaCl (#114-055-101, Quality Biological) and administered in pregnant females at the specified embryonic pregnancy stage at 50 mg/Kg body weight. For proliferation and cell cycle length analysis, a single intraperitoneal EdU injection was administered at E12.5 or E14.5, embryos were fixed after 1 h and the number of EdU+ cells and the percentage of Ki67+ cells labeled with EdU were quantified. For cell cycle exit calculation, a single intraperitoneal EdU injection was administered at E12.5 or E14.5, embryos were fixed after 24 h and the percentage of EdU+ cells labeled with Ki67 was calculated. For cell-fate analysis, a single intraperitoneal EdU injection was administered at E12.5 or E14.5, and the number of EdU+ cells expressing the indicated cellular markers were analyzed at P13. For HES1 expression analysis during different phases of cell cycle, EdU was injected at E14.5, embryos were fixed after 90 min (S + G2), 8 h (early G1) or 14 h (late G1) and the percentage of EdU+ cells labeled with HES1 was calculated. EdU was labeled using Click-iT® EdU imaging kits with alexa 555 or 647 following the manufacturer’s instructions (#C10638 or #C10340, Thermo Fisher Scientific). Briefly, brain sections were post-fixed with 3.7% formaldehyde and permeabilized with 0.5% Triton X-100 in PBS. The Click-iT® reaction cocktail was then added, and the sections incubated in the dark for 30 minutes at room temperature. After the washes, the brain sections were stained for the different proteins of interest using the immunohistochemistry protocol previously described.
Expression constructs
pCAG-mRFP (plasmid #32600, Addgene) was deposited by Anna-Katerina Hadjantonakis59. pCAG-CRE (plasmid #13775, Addgene) was generated by Connie Cepko60. pCS2 NOTCH1 Full Length-6MT (pCS2-Notch1-FL, plasmid #41728, Addgene) was deposited by Raphael Kopan61. pCAG-Brn-DBD-EnR (plasmid #19714, Addgene) and pCAG-Brn2 (plasmid #19711, Addgene) were generated by Connie Cepko62. pCMV6 Delta1 Full Length Myc-DDK was obtained from Origene (pCMV6-Dll1-FL, #MR226161). pBlue-hASPM(WT) was generated by Byoung-Il Bae.
In utero electroporation in mice
The protocol used here was adapted from Saito and Nakatsuji63. Briefly, timed pregnant mice were continuously anesthetized with 2% isoflurane and 5 mg/kg carprofen in saline (OstiFen Injection Sterile Injectable Solution, 50 mg/ml, VetOne) was injected subcutaneously for pain relief. DNA combination of interest (total 2 µg/μl) with 1/10 volume of 1% Fast Green (#F7252, Sigma-Aldrich) in TE buffer was injected into the embryos’ lateral ventricles. Sterile saline solution (37 °C) was constantly applied to moisturize the embryos. The embryo’s head was held using tweezers with disk electrodes (3 mm diameter electrode disk, CUY650P3, Sonidel Limited) with the positive electrode contacting the dorsolateral region of the injected side (for selective targeting of the somatosensory cortex). For electroporation of embryonic day 12.5 (E12.5) embryos, five 50 ms pulses of 30 V were delivered and for electroporation of E14.5 embryos, five 80 ms pulses of 40 V were delivered using an electroporation system (ECM 830 BTX, Harvard Apparatus). Upon completion of the injection and electroporation of all desired embryos, the uterine horns were carefully inserted back into the abdominal cavity filled with saline solution (37 °C). The surgical incision was closed, and the females were closely monitored for the first hours post-surgery. Embryos were left to develop in utero for the indicated time or females were allowed to give birth and pups were euthanized at the indicated age.
In utero electroporation in ferrets
The protocol used here followed the established method described by Kawasaki et al.64. Briefly, time pregnant ferrets were anesthetized with ketamine, and their body temperature was monitored and maintained using a bair hugger warmer. Anesthesia was then continued with isoflurane at 1–2%. Respiratory rate was closely observed, and analgesic solution was injected subcutaneously (Buprenorphine, 0.001 mg/Kg). DNA combination of interest (total 2 µg/μl) with 1/10 volume of 1% Fast Green (#F7252, Sigma-Aldrich) in TE buffer was injected into the embryos’ lateral ventricles. Sterile saline solution (37 °C) was constantly applied to moisturize the embryos. The embryo’s head was held using tweezers with disk electrodes (5 mm diameter electrode disk CUY650P5, Sonidel Limited) with the positive electrode contacting the dorsolateral region of the injected side (for selective targeting of the somatosensory cortex). For E35 electroporation five 50 ms pulses of 100 V were delivered using an electroporation system (ECM 830 BTX, Harvard Apparatus). Upon completion of the injection and electroporation of all desired embryos, the uterine horns were carefully inserted back into the abdominal cavity filled with saline solution (37 °C). The surgical incision was closed, and the females were closely monitored for the first hours post-surgery. The embryos were left to develop in utero for 48 h.
Luciferase assay
Ensembl-predicted regulatory region including the POU domain binding site on Aspm promoter/enhance (sequence from 5’-GAAAAAGTGGGCAGTAACTCGC-3’ to 5’-CAACCTTTCCCTGAGGACGATC-3’) was synthesized by Twist Bioscience and cloned into pGL3-Basic Luciferase Reporter vector (E1751, Promega) using the Gibson assembly method. 150 ng of both pRL-CMV and pGL3-Aspm vectors were co-transfected with 0, 100, 200, 400, or 800 ng of either pCAG-Brn2 vector or pCAG-Brn-DBD-EnR. HEK 293 cells (293 [HEK-293] - CRL-1573™, ATCC) were seeded into 96-well plates at 4,000 cells per well and transfected the following day with PEI Prime linear polyethylenimine transfection reagent (919012-100MG, Sigma) at a 1:3 ratio of DNA:PEI. Cells were harvested 48 h after transfection and luciferase activity was measured using the Dual-Glo Luciferase Assay system (E2920, Promega) on a FlexStation® 3 plate reader. Firefly luciferase activity was normalized to Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP-qPCR)
The interaction of BRN1/2 with the indicated genes was studied by ChIP-qPCR in E14.5 wild-type brains using the ChIP assay kit following the manufacturer’s instructions (Millipore). Immunoprecipitation was performed with 5 µg of anti-BRN1/2 (PA5-30124, Thermo Fisher Scientific) or rabbit IgG (2729S, Cell Signaling) as a negative control. This BRN1/2 antibody recognizes both BRN1 and BRN2 protein sequence. Quantitative PCR (qPCR) was performed with 5 ng of ChIP-enriched genomic DNA in a 20 µl reaction using PowerTrack SYBR Green Master Mix (A46012, Thermo Scientific) with readout on a QuantStudio 6 Flex System (Thermo Scientific) using standard cycling parameters. ΔΔCt was calculated using Gapdh as a housekeeping gene control. Relative expression was calculated using the equation: relative expression =2−ΔΔCt. Three technical replicates of three independent biological replicates were run to account for assay variance. Primer sequences used for qPCR are listed in Supplementary Table 1. The expression of Zic1 was used as a positive control65.
Single cell RNA sequencing
Tissue preparation and single cell RNA sequencing: Cortex of E12.5 and E14.5 embryos was dissected and dissociated into single cells using the Papain Dissociation System (#LK0031500, Worthington Biochemical). Individual collected cells were processed for scRNAseq using. Cell counts and viability were determined using the Cell Countess II with Trypan Blue. A maximum volume of 43.3 µl/sample was used for processing to target up to 10,000 cells. Cells were combined with RT reagents and loaded onto 10X Next GEM Chip G along with 3’ v3.1 gel beads. The NextGEM protocol was run on the 10X Chromium Controller to create GEMs, composed of a single cell, gel bead with unique barcode and UMI primer, and RT reagents. 100 µl of emulsion is retrieved from the chip and incubated (45 min at 53 °C, 5 min at 85 °C, cool to 4 °C), generating barcoded cDNA from each cell. The GEMs were broken using Recovery Agent and cDNA cleaned, following manufacturer’s instructions using MyOne SILANE beads. cDNA was amplified for 11 cycles (3 min at 98 °C, 11 cycle: 15 sec at 98 °C, 20 sec at 63 °C, 1 min at 72 °C; 1 min at 72 °C, cool to 4 °C). Samples were cleaned using 0.6X SPRIselect beads. Quality control (QC) was completed using Qubit and Bioanalyzer to determine size and concentrations. 10 µl of amplified cDNA was carried into library prep. Fragmentation, end repair and A-tailing were completed (5 min at 32 °C, 30 min at 65 °C, cool to 4 °C), and samples were cleaned up using double sided size selection (0.6X, 0.8X) with SPRIselect beads. Adaptor ligation (15 min at 20 °C, cool to 4 °C), 0.8X cleanup and amplification were performed by PCR using unique i7 index sequences. Libraries underwent a final cleanup using double sided size selection (0.6X, 0.8X) with SPRIselect beads. Library QC was performed using Qubit, Bioanalyzer and KAPA library quantification qPCR kit. Libraries were sequenced on the Illumina NovaSeq 6000 using v1.5 kits, targeting 50 K reads/cell, at read lengths of 28 (R1), 8 (i7), 91 (R2). Demultiplexing and FASTQ generation was completed using Illumina’s BaseSpace software. Data were aligned to the mouse reference genome (refdata-gex-mm10-2020-A) publicly available at the 10X genomics’ website using the Cell Ranger pipeline. Although we started with seven Brn1/2-cKO and seven wild-type littermates of both sexes the genotype for one of the wild-type samples could not be confirmed and therefore this sample was removed from the analysis.
Single cell RNA sequencing analysis: Briefly, the 10x data were manually aggregated to create a comprehensive data set and subjected to additional QCing. Raw counts were aggregated together for input into the Monocle3 R/Bioconductor platform66. The raw counts were first scaled by the estimated size factor to normalize the total UMI difference of each cell. The normalized counts were then log transformed after applying a pseudo count. For dimensionality reduction, the log-transformed counts were zero centered with unit variance. Batch correction was done using the aligned_cds function within Monocle3 for the purpose of visualization67. Dimensionality reduction and visualization for the aggregate 10x data were performed using Uniform Manifold Approximation and Projection (UMAP)68 for all cells passing QC. Briefly the first 50 principle components of the log10(CPT + 1) was used as input for the monocle3 implementation of the UMAP algorithm with the following additional parameters: max_components = 3, metric =”cosine”. Initial cell clusters were called using the louvain algorithm with manual annotation using marker genes. The progenitor and neuronal lineages were subset from the main data using this initial clustering. A new embedding was learned for this subset and cell with ambiguous identity were removed. Although we sequenced seven Brn1/2-cKO and seven wild-type littermates of both sexes (total cells = 161,000) the genotype for one of the wild-type samples could not be confirmed and therefore this sample was removed from the subsequent analysis (total cells = 149,712). AP (28% of cells), BP (6.7% of cells), Neurons (31% of cells), DL and UL signatures scores were calculated using the AddModuleScore function from Seurat with cell type signatures from Telley et al.31,32 and refined the combination of the results of the clustering and the module Scores with additional marker genes31–34,69. The log-transformed counts were projected into cell-cycle latent space pretrained by the TRIcycle package40. The cell cycle position of each cell was inferred by its polar angle in the cell-cycle latent space ranging from 0 to 2 pi. The final cell-cycle phase was assigned considering both the cell cycle position and the expression of certain maker genes. Final cell types were called using a combination of cluster analysis and cell marker genes31–34. All differential expression tests were performed across all expressed genes using the Monocle3 VGAM likelihood ratio test with batch and conditions included in the model formula66. Additional statistical tests were carried out in R/Bioconductor. Unpaired t-test was used to compare the total cell types per age and genotype. Wilcoxon rank sum test was used for TRIcycle cell cycle phase analysis and Pearson’s Chi-squared test for given probabilities was used for cell proportion analysis. The heatmaps and dotplots were generated with ggplot2 using the row normalized nonzero means. The signature score heatmaps were generated by averaging the DL signature and UL signature scores within control DLNs, ULNs, and all Brn1/2-cKO neurons at each age. To generate the correlation plots, the control ULNs, the control DLNs, and all Brn1/2-cKO neurons at each age were pseudo-bulked and the Pearson correlation between each group was calculated within each age. The DE genes were selected using the following thresholds: standard error <0.15, q value 0.001 for neurons, AP and BP, and q value 0.05 for all progenitors (AP and BP together). The ribosomal and mitochondrial genes were removed. GO analysis was performed by the Enrichr API from Gseapy using the filtered DE lists. The terms of interest were selected for visualization from the significant terms with adjusted q value less than 0.05. The embedding of cynomolgus monkey data was generated by scanpy. Cells with less than 5500 UMIs were excluded. The UMAP was generated with the first 15 principle components and the min_dist of 0.3. AP, BP, UL, and DL signatures scores were calculated using the “score_genes” function. The TRIcycle and DE gene analysis used the same setting as described above. The DE genes were selected by the following criteria: standard error 0.15, q value 0.001 for neurons and q value 0.05 for all progenitors (AP and BP together).
Transcriptional waves along pseudotime projection: Only AP cells were selected to learn the pseudotime projection for cell maturity. First, we performed the dimensionality reduction on the control cells. Since one of the sequencing batches (Batch 1) failed to show temporal difference in the latent space, we removed this batch from the following pseudotime analysis. In the latent space of the remaining control data, the second and third principal components best describe the temporal difference and explain a significant amount of variance. Therefore, we fitted a principal curve based on these two PCs using prinPY and projected the cells back into the fitted trajectory to define the maturity pseudotime. Similar to the analysis of the different transcriptomic waves along the pseudotime axis previously described by Telley et al.32, the side with the highest expression of Sox2 and Hmga2 was selected as the beginning and the pseudotime score was normalized between 0 to 1. For the Brn1/2-cKO cells, we used KDtree to search for their nearest neighbor among control cells in the latent space and assigned the Brn1/2-cKO cells with the same pseudotime score as its nearest control neighbor. We then visualized the previously described temporal wave genes using Seaborn heatmap. The cells were aligned according to their pseudotime score and the heatmap was smoothed by the UnivariateSpline. We have confirmed that the control and Brn1/2-cKO cells from Batch 1 showed similar average age-related expression pattern compared with the other batches within the same genotype for the vast majority of the genes used to define the transcriptional waves. The Fisher’s Exact test was used to measure the enrichment of the selected GO terms in each wave and the heatmap was colored by the -log10(p-value) from the statistical test.
Imaging
All immunohistochemistry and ISH RNAscope data were acquired in an equivalent latero-medial level of the cortex from at least 3 independent animals per condition or genotype. Images were captured using a Zeiss LSM 800 confocal microscope. Maximum intensity projections were generated in ImageJ or Imaris software. In all fluorescence microscopy figures, different channels of image series were represented in pseudocolor, and contrast and brightness were adjusted manually using ImageJ or Adobe Photoshop software. All markers were counterstained with DAPI to allow visualization of overall cellular density.
Histological analysis
A series of z-stack (depth of 10 μm) confocal tiled images were used for cell quantification; 150 μm-wide images (embryonic brains) or 300 μm-wide images (postnatal brains) comprising the entire extension of the cortex were analyzed. All quantification were made in an equivalent latero-medial level of the cortex from at least 3 independent animals per condition or genotype. For cortical thickness quantifications, P13 postnatal brains were sectioned coronally at 60 µm and sections sequentially collected over 10 wells. Each well had a full representation of the brain. Three equivalent midbrain sections were imaged and quantified per mice per genotype. The midbrain sections were classified in reference to their proximity to the ventricle, hippocampus, and corpus callosum/midline (1st section ~Interaural 3.46 mm; Bregma −0.34 mm; 2nd section ~Interaural 2.58 mm; Bregma −1.22 mm; 3rd section ~Interaural 1.74 mm; Bregma −2.06 mm). The number of cells expressing the markers of interest was quantified in radial sections of the same latero-medial level (determined as mentioned above) and adjusted per area. For the IUE analysis, to divide the cortex into longitudinal bins, a rectangle was drawn from the border of the white matter with layer VI to the pial surface. This rectangle was then divided into ten equal bins and the number of cells within each bin was quantified. To quantify ISH-RNAscope data, all punctate dots or all punctate dots in RFP+ cells were counted in 50 μm-wide images comprising the entire extension of the cortex. Notch1 mRNA levels in the ferret in utero electroporated cells were quantified in all RFP+ cells in the area comprising 50 µm from the VZ. For the centriole number analysis, a z-stack of 10 sections (0.5μm each) was selected for all analyzed cells and the number of centrioles overlapping or in the very close proximity of the g-TUBULIN staining were counted. Since Brn1/2-cKO show a 20% reduction of total cell number and cortical thickness, the quantifications were normalized to the cortical thickness of each analyzed section in controls and mutants.
Statistical analysis
Statistical analyses of the immunohistochemistry and ISH-RNAscope experiments were done using GraphPad Prism software. Statistically significant differences were assessed by Student’s unpaired t-test or one-way analysis of variance (one-way ANOVA-Dunnett’s multiple comparisons test; FDFn, DFd (DFn: degrees of freedom in the numerator; DFd: degrees of freedom in the denominator)), comparing two or more groups, respectively. To compare the distribution of RFP+ cells in the different bins of the cortex for the two different conditions we used two-way analysis of variance (two-way ANOVA-Šídák’s multiple comparisons test; FDFn, DFd). P < 0.05 was considered a significant difference. All values represent individual animals mean ± standard error of the mean (SEM). The statistical test used, and the statistical significance are indicated in figure legends. Statistical analyses of the scRNAseq experiments were done using Monocle 3 and R/Bioconductor. Monocle3 VGAM likelihood ratio test was used for the differential expressed gene analysis. Unpaired t-test was used to compare the total cell types per age and genotype. Wilcoxon rank sum test was used for TRIcycle cell cycle phase analysis and Pearson’s Chi-squared test for given probabilities was used for cell proportion analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank members of the Müller and Kolodkin laboratories for suggestions, reagents and technical help; Alex Kolodkin, Jakub Ziak and Cristina Gil-Sanz for critical comments on the manuscript; Caiying Guo for help in generating the Brn1 floxed mice; Amanda Maxwell, Caroline Garrett and Eric Hutchinson for help with ferrets; Michele Pucak for imaging assistance; Linda Orzolek and Tyler J. Creamer for help with scRNAseq; Anna-Katerina Hadjantonakis, Connie Cepko and Raphael Kopan for plasmids; Ryoichiro Kageyama for the HES1 antibody; Andrew Holland for the γ-TUBULIN and Centrin antibodies. We are grateful to Michelle Monroe, Tajma Smith, Kaiping Zhang, Femi Cleola Villamor and Trinity Walker for assistance with mouse maintenance and genotyping. National Institutes of Health grant R01HG012357 and RF1MH121539 (UM); Brain Research Foundation grant BRFSG-2022-02 (B.-I.B).
Author contributions
Conceptualization: S.B., U.M.; methodology: S.B., G.S.O’.B., Y.X., L.G., K.J.N., R.V., U.M.; investigation: S.B., Y.X., J.P.L., G.S.O’.B., U.M.; validation: S.B., Y.X., J.P.L., G.S.O’.B., U.M.; visualization: S.B.; formal analysis: S.B., Y.X., J.P.L., G.S.O’.B., U.M.; data curation: S.B., Y.X., J.P.L., G.S.O’.B.; resources: K.J.N., B.-I.B, R.S.S., C.A.W., U.M.; supervision: U.M.; project administration: U.M.; funding acquisition: U.M.; writing - original draft: S.B., U.M.; writing - review & editing: S.B., Y.X., J.P.L., L.G., K.J.N., R.V., B.-I.B., R.S.S., C.A.W., G.S.O’.B., U.M.
Peer review
Peer review information
Nature Communications thanks Tianqing Li and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw single cell RNA sequencing datasets generated in this study have been deposited in the GEO repository database under the accession code GSE229129. Raw single cell RNA sequencing datasets for the monkey data24 are deposited in the SRA repository database under the accession code SRP366952. All the other data generated in this study are provided in the Supplementary Information and Supplementary Data files. Source data are provided with this paper.
Material availability
All data are available in the main text or the supplementary information. Correspondence and requests for materials should be addressed to U.M.
Code availability
The code for the scRNAsequencing analysis presented in this manuscript is available on: 10.5281/zenodo.12582808 (10.5281/zenodo.12582809).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Soraia Barão, Email: sbarao@jhmi.edu.
Ulrich Müller, Email: umuelle3@jhmi.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52443-x.
References
- 1.Lodato, S. & Arlotta, P. Generating Neuronal Diversity in the Mammalian Cerebral Cortex. Annu. Rev. Cell Dev. Biol.31, 699–720 (2015). 10.1146/annurev-cellbio-100814-125353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrell, V. & Reillo, I. Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol.72, 955–971 (2012). 10.1002/dneu.22013 [DOI] [PubMed] [Google Scholar]
- 3.Kalebic, N. & Huttner, W. B. Basal Progenitor Morphology and Neocortex Evolution. Trends Neurosci.43, 843–853 (2020). 10.1016/j.tins.2020.07.009 [DOI] [PubMed] [Google Scholar]
- 4.Pinson, A. & Huttner, W. B. Neocortex expansion in development and evolution—from genes to progenitor cell biology. Curr. Opin. Cell Biol.73, 9–18 (2021). 10.1016/j.ceb.2021.04.008 [DOI] [PubMed] [Google Scholar]
- 5.Pebworth, M. P., Ross, J., Andrews, M., Bhaduri, A. & Kriegstein, A. R. Human intermediate progenitor diversity during cortical development. Proc. Natl Acad. Sci. USA118, 1–10 (2021). 10.1073/pnas.2019415118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cárdenas, A. et al. Evolution of Cortical Neurogenesis in Amniotes Controlled by Robo Signaling Levels. Cell174, 590–606.e21 (2018). 10.1016/j.cell.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayaraman, D. et al. The genetics of primary microcephaly. Annu Rev. Genomics Hum. Genet19, 177–200 (2018). 10.1146/annurev-genom-083117-021441 [DOI] [PubMed] [Google Scholar]
- 8.Villalba, A., Götz, M. & Borrell, V. The regulation of cortical neurogenesis. Curr. Top. Dev. Biol.142, 1–66 (2021). 10.1016/bs.ctdb.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 9.Klingler, E., Francis, F., Jabaudon, D. & Cappello, S. Mapping the molecular and cellular complexity of cortical malformations. Science (1979)371, eaba4517 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S. & Kriegstein, A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature409, 714–720 (2001). 10.1038/35055553 [DOI] [PubMed] [Google Scholar]
- 11.Haubensak, W., Attardo, A., Denk, W. & Huttner, W. B. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc. Natl Acad. Sci. USA101, 3196–3201 (2004). 10.1073/pnas.0308600100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noctor, S. C., Martínez-Cerdeño, V., Ivic, L. & Kriegstein, A. R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci.7, 136–144 (2004). 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- 13.Miyata, T. et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development131, 3133–3145 (2004). 10.1242/dev.01173 [DOI] [PubMed] [Google Scholar]
- 14.Hansen, D. V., Lui, J. H., Parker, P. R. L. & Kriegstein, A. R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature464, 554–561 (2010). 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- 15.Tarabykin, V., Stoykova, A., Usman, N. & Gruss, P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development128, 1983–1993 (2001). 10.1242/dev.128.11.1983 [DOI] [PubMed] [Google Scholar]
- 16.Kowalczyk, T. et al. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb. Cortex19, 2439–2450 (2009). 10.1093/cercor/bhn260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasistha, N. A. et al. Cortical and clonal contribution of Tbr2 expressing progenitors in the developing mouse brain. Cereb. Cortex25, 3290–3302 (2015). 10.1093/cercor/bhu125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huilgol, D. et al. Direct and indirect neurogenesis generate a mosaic of distinct glutamatergic projection neuron types in cerebral cortex. Neuron111, 2557–2569.e4 (2023). 10.1016/j.neuron.2023.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reillo, I., De Juan Romero, C., García-Cabezas, M. Á. & Borrell, V. A Role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex21, 1674–1694 (2011). 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- 20.Molnár, Z. et al. New insights into the development of the human cerebral cortex. J. Anat.235, 432–451 (2019). 10.1111/joa.13055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugitani, Y. et al. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev.16, 1760–1765 (2002). 10.1101/gad.978002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEvilly, R. J., Ortiz de Diaz, M., Schonemann, M. D., Hooshmand, F. & Rosenfeld, M. G. Transcriptional regulation of cortical neuron migration by POU domain factors. Science295, 1528–1532 (2002). 10.1126/science.1067132 [DOI] [PubMed] [Google Scholar]
- 23.Dominguez, M. H., Ayoub, A. E. & Rakic, P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb. Cortex23, 2632–2643 (2013). 10.1093/cercor/bhs252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, X. et al. BRN2 as a key gene drives the early primate telencephalon development. Sci. Adv.8, 1–17 (2022). 10.1126/sciadv.abl7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaegle, M. et al. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev.17, 1380–1391 (2003). 10.1101/gad.258203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci.22, 6309–6314 (2002). 10.1523/JNEUROSCI.22-15-06309.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greig, L. C., Woodworth, M. B., Galazo, M. J., Padmanabhan, H. & Macklis, J. D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci.14, 755–769 (2013). 10.1038/nrn3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SK, M. & CE, K. Cell cycle dependence of laminar determination in developing neocortex. Science254, 282–285 (1991). 10.1126/science.254.5029.282 [DOI] [PubMed] [Google Scholar]
- 29.Frantz, G. D. et al. Restriction of Late Cerebral Cortical Progenitors to an Upper-Layer Fate. Neuron17, 55–61 (1996). 10.1016/S0896-6273(00)80280-9 [DOI] [PubMed] [Google Scholar]
- 30.Desai, A. R. & McConnell, S. K. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development127, 2863–2872 (2000). 10.1242/dev.127.13.2863 [DOI] [PubMed] [Google Scholar]
- 31.Telley, L. et al. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science351, 1443–1446 (2016). 10.1126/science.aad8361 [DOI] [PubMed] [Google Scholar]
- 32.Telley, L. et al. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science364, eaav2522 (2019). 10.1126/science.aav2522 [DOI] [PubMed] [Google Scholar]
- 33.Moreau, M. X., Saillour, Y., Cwetsch, A. W., Pierani, A. & Causeret, F. Single-cell transcriptomics of the early developing mouse cerebral cortex disentangle the spatial and temporal components of neuronal fate acquisition. Development148, dev197962 (2021). 10.1242/dev.197962 [DOI] [PubMed] [Google Scholar]
- 34.Di Bella, D. J. et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature595, 554–559 (2021). 10.1038/s41586-021-03670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer, C., Tiveron, M. C., Bodmer, R. & Cremer, H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb. Cortex14, 1408–1420 (2004). 10.1093/cercor/bhh102 [DOI] [PubMed] [Google Scholar]
- 36.Franco, S. J. et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science337, 746–749 (2012). 10.1126/science.1223616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gil-Sanz, C. et al. Lineage Tracing Using Cux2-Cre and Cux2-CreERT2 Mice. Neuron86, 1091–1099 (2015). 10.1016/j.neuron.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Moreno, F. & Molnár, Z. Subset of early radial glial progenitors that contribute to the development of callosal neurons is absent from avian brain. Proc. Natl Acad. Sci. USA112, E5058–E5067 (2015). 10.1073/pnas.1506377112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahr, S. K. et al. A Translational Repression Complex in Developing Mammalian Neural Stem Cells that Regulates. Neuronal Specif. Neuron97, 520–537.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Zheng, S. C. et al. Universal prediction of cell-cycle position using transfer learning. Genome Biol.23, 1–27 (2022). 10.1186/s13059-021-02581-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertrand, N., Castro, D. S. & Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci.3, 517–530 (2002). 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- 42.Britz, O. et al. A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex16, i138–i151 (2006). 10.1093/cercor/bhj168 [DOI] [PubMed] [Google Scholar]
- 43.Chou, R. H., Yu, Y. L. & Hung, M. C. The roles of EZH2 in cell lineage commitment. Am. J. Transl. Res3, 243–250 (2011). [PMC free article] [PubMed] [Google Scholar]
- 44.Nieto, M. et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J. Comp. Neurol.479, 168–180 (2004). 10.1002/cne.20322 [DOI] [PubMed] [Google Scholar]
- 45.Cubelos, B. et al. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb. Cortex18, 1758–1770 (2008). 10.1093/cercor/bhm199 [DOI] [PubMed] [Google Scholar]
- 46.Shimojo, H., Ohtsuka, T. & Kageyama, R. Oscillations in Notch Signaling Regulate Maintenance of Neural Progenitors. Neuron58, 52–64 (2008). 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 47.Imayoshi, I. et al. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science342, 1203–1208 (2013). 10.1126/science.1242366 [DOI] [PubMed] [Google Scholar]
- 48.Borrell, V. et al. Slit/Robo Signaling Modulates the Proliferation of Central Nervous System Progenitors. Neuron76, 338–352 (2012). 10.1016/j.neuron.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castro, D. S. et al. Proneural bHLH and Brn Proteins Coregulate a Neurogenic Program through Cooperative Binding to a Conserved DNA Motif. Dev. Cell11, 831–844 (2006). 10.1016/j.devcel.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 50.Johnson, M. B. et al. Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size letter. Nature556, 370–375 (2018). 10.1038/s41586-018-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marinopoulou, E. et al. HES1 protein oscillations are necessary for neural stem cells to exit from quiescence. iScience24, 103198 (2021). 10.1016/j.isci.2021.103198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baek, J. H., Hatakeyama, J., Sakamoto, S., Ohtsuka, T. & Kageyama, R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development133, 2467–2476 (2006). 10.1242/dev.02403 [DOI] [PubMed] [Google Scholar]
- 53.Saade, M., Morin, X., Onorati, M., Wilsch-Bräuninger, M. & Huttner, W. B. Primary Cilia and Centrosomes in Neocortex Development. Front. Neurosci.15, 755867 (2021). 10.3389/fnins.2021.755867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, S., Hannenhalli, S. & Ovcharenko, I. De novo human brain enhancers created by single-nucleotide mutations. Sci. Adv.9, eadd2911 (2023). 10.1126/sciadv.add2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine, M. S. et al. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev. Cell40, 313–322.e5 (2017). 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moyer, T. C. & Holland, A. J. PLK4 promotes centriole duplication by phosphorylating STIL to link the procentriole cartwheel to the microtubule wall. Elife8, e46054 (2019). 10.7554/eLife.46054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang, X., Falls, D. L., Li, X., Lane, T. & Luskin, M. B. Antigen-retrieval procedure for bromodeoxyuridine immunolabeling with concurrent labeling of nuclear DNA and antigens damaged by HCl pretreatment. J. Neurosci.27, 5837–5844 (2007). 10.1523/JNEUROSCI.5048-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, F. et al. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagnostics14, 22–29 (2012). 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long, J. Z., Lackan, C. S. & Hadjantonakis, A. K. Genetic and spectrally distinct in vivo imaging: Embryonic stem cells and mice with widespread expression of a monomeric red fluorescent protein. BMC Biotechnol.5, 1–11 (2005). 10.1186/1472-6750-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuda, T. & Cepko, C. L. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl Acad. Sci. USA104, 1027–1032 (2007). 10.1073/pnas.0610155104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature393, 382–386 (1998). 10.1038/30756 [DOI] [PubMed] [Google Scholar]
- 62.Kim, D. S., Matsuda, T. & Cepko, C. L. A core paired-type and POU homeodomain-containing transcription factor program drives retinal bipolar cell gene expression. J. Neurosci.28, 7748–7764 (2008). 10.1523/JNEUROSCI.0397-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saito, T. & Nakatsuji, N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol.240, 237–246 (2001). 10.1006/dbio.2001.0439 [DOI] [PubMed] [Google Scholar]
- 64.Kawasaki, H., Iwai, L. & Tanno, K. Rapid and efficient genetic manipulation of gyrencephalic carnivores using in utero electroporation. Mol. Brain5, 24 (2012). 10.1186/1756-6606-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urban, S. et al. A Brn2-Zic1 axis specifies the neuronal fate of retinoic-acid-treated embryonic stem cells. J. Cell Sci.128, 2303–2318 (2015). 10.1242/jcs.168849 [DOI] [PubMed] [Google Scholar]
- 66.Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature566, 496–502 (2019). 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol.36, 421–427 (2018). 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.37, 38–47 (2019). 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- 69.Molyneaux, B. J. et al. DeCoN: Genome-wide analysis of invivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron85, 275–288 (2015). 10.1016/j.neuron.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw single cell RNA sequencing datasets generated in this study have been deposited in the GEO repository database under the accession code GSE229129. Raw single cell RNA sequencing datasets for the monkey data24 are deposited in the SRA repository database under the accession code SRP366952. All the other data generated in this study are provided in the Supplementary Information and Supplementary Data files. Source data are provided with this paper.
All data are available in the main text or the supplementary information. Correspondence and requests for materials should be addressed to U.M.
The code for the scRNAsequencing analysis presented in this manuscript is available on: 10.5281/zenodo.12582808 (10.5281/zenodo.12582809).