Abstract
The complete nucleotide sequence of the genome segment 4 (S4) of Bombyx mori cytoplasmic polyhedrosis virus (BmCPV) was determined. The 3,259-nucleotide sequence contains a single long open reading frame which spans nucleotides 14 to 3187 and which is predicted to encode a protein with a molecular mass of about 130 kDa. Western blot analysis showed that S4 encodes BmCPV protein VP3, which is one of the outer components of the BmCPV virion. Sequence analysis of the deduced amino acid sequence of BmCPV VP3 revealed possible sequence homology with proteins from rice ragged stunt virus (RRSV) S2, Nilaparvata lugens reovirus S4, and Fiji disease fijivirus S4. This may suggest that plant reoviruses originated from insect viruses and that RRSV emerged more recently than other plant reoviruses. A chimeric protein consisting of BmCPV VP3 and green fluorescent protein (GFP) was constructed and expressed with BmCPV polyhedrin using a baculovirus expression vector. The VP3-GFP chimera was incorporated into BmCPV polyhedra and released under alkaline conditions. The results indicate that specific interactions occur between BmCPV polyhedrin and VP3 which might facilitate BmCPV virion occlusion into the polyhedra.
Cytoplasmic polyhedrosis viruses (CPVs) belong to the genus Cypovirus in the family Reoviridae (13, 36). These viruses produce large proteinaceous occlusion bodies called polyhedra in the cytoplasms of infected midgut epithelial cells of a wide range of insects (2–4, 14, 34). The polyhedra are the result of the crystallization of a virus-encoded protein, polyhedrin, late during the viral infection, and many virus particles are occluded into the polyhedra (4, 36). One of the functions of these polyhedra is to protect the virions from hostile environmental conditions during horizontal transmission of the disease (2, 4). The polyhedra are highly resistant to both nonionic and ionic detergents and to solubilization at neutral pH. Another function of the polyhedra is to ensure the delivery of virus particles to the target intestinal cells. Here the polyhedra are dissolved by the strongly alkaline pH of the insect midgut, thereby releasing the virions and allowing the infection to proceed.
The CPV genome is composed of 10 discrete equimolar double-stranded RNA (dsRNA) segments (S1 to S10) (36). Based on the variations in the electrophoretic migration patterns of the genomic dsRNA segments, 14 types of CPV have been identified (5, 23, 32, 33). The polyhedrin has a molecular mass ranging from 27 to 31 kDa, and the smallest genome segment encodes the polyhedrin. The polyhedrin genes of type 1 Bombyx mori CPV (BmCPV) and type 5 CPVs, including Euxoa scandens CPV (EsCPV), Orgyia pseudotsugata CPV, and Heliothis armigera CPV, were cloned, and the nucleotide sequences were determined (1, 7, 8, 26). No similarities were found in the DNA sequences of the polyhedrin gene of two distinct virus types, type 1 (BmCPV) and type 5 (EsCPV). The amino acid sequences of BmCPV and EsCPV polyhedrins, however, show weak homology in three regions. In particular, the hydrophilic profiles and predicted secondary structures of both BmCPV and EsCPV polyhedrins show some similarities, mainly in the amino-terminal half of the polypeptides (4).
Virus particles of BmCPV are composed of VP1 (151 kDa), VP2 (142 kDa), VP3 (130 kDa), VP4 (67 kDa), and VP5 (33 kDa) (31). The in vitro labeling of BmCPV with 125I indicated that VP1 and VP3 were outer components (20). Recently, it was reported that the BmCPV particle has a single shell capsid and that there are two sizes of protrusions on the capsid shell (12). The coding assignments of dsRNA segments for BmCPV were determined by in vitro translation studies with rabbit reticulocytes (22). The nucleotide sequences of S6 and S7, which encode BmCPV VP4 and VP5, and those of S8 and S9, which encode two nonstructural proteins, have been determined (9–11). From the results of in vitro translation studies, it was speculated that the BmCPV outer components VP1 and VP3 are encoded by S1 and S4, respectively; however, these two dsRNA segments have not been characterized.
Here, the complete nucleotide sequence of S4, which is thought to encode an outer component, BmCPV VP3, is reported and possible evolutionary relationships of BmCPV and other members of the family Reoviridae are examined. While it is known that the shape of BmCPV polyhedra and the crystallization pattern of the polyhedrin are susceptible to mutations in amino acid sequence (14, 15, 16, 29), little is known about the specific interactions between the CPV polyhedrin and other viral components that control virion occlusion. Therefore, a VP3 mutant containing green fluorescent protein (GFP) at the C-terminal region was constructed and expressed together with BmCPV polyhedrin by using a baculovirus expression vector. The green fluorescence was used to determine whether the chimeric protein is incorporated in the BmCPV polyhedra and released under alkaline conditions.
MATERIALS AND METHODS
Virus and cells.
BmCPV strain H was originally described by Hukuhara and Midorikawa (15). The Spodoptera frugiperda cell line IPLB-Sf21-AE (Sf21) was maintained in tissue culture flasks in TC-100 medium (GIBCO/BRL) with 10% fetal bovine serum. The recombinant virus AcCP-H, which produces cubic polyhedra, was used in this study (27).
Purification of MAbs.
An anti-BmCPV monoclonal antibody (MAb) was purified from ascites fluid of mice inoculated intraperitoneally with MAb-producing S11 hybridoma cells by using a MAb Trap GII affinity chromatography kit (Amersham Pharmacia Biotech) according to the manufacturer's recommendations (24).
cDNA synthesis.
Virus dsRNA was recovered from purified virus, and the fourth segment (S4) was isolated by electrophoresis in a low-melting-point agarose gel (SeaPlaque GTG; FMC). For the synthesis of cDNA, two primers (5′GATCGCGGCCGCAGTAATTTCCACCATG3′ for the plus strand, containing a NotI site, which is underlined, and 5′GATCGGATCCGGCTAACGTTTCC3′ for the minus strand, containing a BamHI site, which is underlined) were constructed on the basis of the terminal RNA sequence of S4 (18). S4 cDNA was synthesized by using the primers and a Timeserver cDNA synthesis kit (Amersham Pharmacia Biotech.). The resultant cDNA was amplified by PCR by using ExTaq polymerase (Takara). The PCR products were digested with NotI and BamHI, ligated into the NotI-BamHI site of pBlueScript II (SK+), and transformed into Escherichia coli JM109 (Toyobo). Five clones were used for the sequence analysis.
Sequence analysis.
Deletion mutants were made from each recombinant plasmid containing full-length segment 4 cDNA by using a deletion kit (Takara). The deletion-containing recombinant plasmids were prepared by standard techniques and sequenced with an ABI Prism terminator cycle sequencing kit (PE Applied Biosystems) and a PE Applied Biosystems model 373A automated sequencer.
Construction of recombinant baculoviruses.
S4 cDNA was cut out with NotI and BamHI and ligated into the NotI-BamHI site of a baculovirus transfer vector, pVL1392 (PharMingen), resulting in recombinant transfer vector pAcVP3. The cDNA fragment was excised from pAcVP3 by digestion with BglII and SalI (bases 2964 to 2999 in the nucleotide sequence of S4) and ligated into the BglII-SalI site of pEGFPN2 (Clontech). A chimeric gene consisting of the GFP gene and S4 cDNA could then be liberated by digestion with NotI and ligated into the dephosphorylated NotI site of the pVL1392 transfer vector. Also, the GFP gene insert from pEGFPN2 was excised with BamHI and NotI and ligated into the BamHI-NotI site of pVL1393 (PharMingen). The recombinant transfer vector containing S4 cDNA and GFP gene was named pAcVP3/GFP, and that containing only the GFP gene was named pAcGFP. Sf21 cells were transfected with 5 μg of the resulting recombinant transfer vector and 0.5 μg of linearized Autographa californica nuclear polyhedrosis virus (AcNPV) DNA (Baculogold baculovirus DNA; PharMingen). A recombinant AcNPV rescued by the transfer vector was isolated and plaque purified. Recombinant baculoviruses AcVP3, AcVP3/GFP, and AcGFP were obtained from pAcVP3, pAcVP3/GFP, and pAcGFP, respectively.
Expression of the recombinant proteins in Sf21 cells.
The Sf21 cells (106 cells per 35-mm plate) were inoculated with the recombinant virus at 5 PFU/cell. For double infection with AcVP3/GFP and AcCP-H or AcGFP and AcCP-H, each virus was also used at 10 PFU/cell. After incubation for 1 h at room temperature, the inoculum was removed and replaced with 2 ml of TC-100 medium containing 10% fetal bovine serum. After incubation at 27°C for 4 days, the infected cells were subjected to Western blot analysis and measurement of green fluorescence.
Purification of polyhedra and virions.
The Sf21 cells (108 cells) infected with the recombinant AcNPV were collected, washed with phosphate-buffered saline (PBS; 20 mM NaH2PO4, 20 mM Na2HPO4, 150 mM NaCl [pH 7.2]), and homogenized with a blender on ice. The homogenates were washed with 1% Tween 20, and polyhedra were partially purified by cycles of differential centrifugation. Further purification was performed by discontinuous sucrose density gradient centrifugation in 1.5 to 2.2 M sucrose at 50,000 × g for 45 min (25). Then polyhedra were recovered with syringes, washed with PBS, and collected by centrifugation at 15,000 × g for 10 min. BmCPV virions were purified as described previously (11).
Western blotting.
Purified BmCPV virions and infected Sf21 cells were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled, and separated by SDS–10% PAGE under reducing conditions as described by Laemmli (19). After electrophoresis, proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad) by using a Transblot cell (Bio-Rad). The membranes were blocked with 1% gelatin in PBS and incubated with purified anti-BmCPV MAb which was 100-fold diluted in PBS containing 0.05% Triton X-100 (PBST). After a washing with PBST, the membrane was incubated with 5,000-fold-diluted goat anti-mouse immunoglobulin G conjugated with horseradish peroxidase (Bio-Rad) in PBST. The POD Immunostrain set (Wako) was used as protein weight markers.
Measurement of fluorescence by GFP.
Since GFP is stable between pH 5 to 12 and polyhedra dissolve at pHs over 10.0, green fluorescence from GFP was measured before and after the dissolution of polyhedra, in order to determine whether the chimeric protein consisted of VP3 and whether GFP was incorporated into polyhedra. Sf21 cells (107 cells per 75-cm2 flask) were inoculated with AcVP3/GFP and AcCP-H at 10 PFU/cell each, and a second set of Sf21 cells were inoculated with AcGFP and AcCP-H at 10 PFU/cell each. Polyhedra were collected from the infected Sf21 cells 4 days postinfection (p.i.) at 27°C. The polyhedra were purified as described above and suspended in 1 ml of distilled water, 50 mM acetate buffer (CH3COOH-CH3COONa, pH 4.0), and 50 mM carbonate buffer (Na2CO3-NaHCO3, pH 11.0). The pH of the polyhedron suspension in acetate buffer was adjusted to 6.0, 10.0, and 12.5 by the addition of 5 N NaOH. After the incubation at 30°C for 30 min, GFP-mediated fluorescence levels were measured by excitation at 475 nm and emission at 510 nm using an F-2000 fluorescence spectrophotometer (Hitachi). The protein concentration was determined as described by Lowry et al. (21) with bovine serum albumin as a standard, and the level of green fluorescence was reported as a relative amount.
Sf21 cells were seeded into 35-mm plates at 106 cells per plate. Cells were inoculated with AcVP3/GFP at 20 PFU/cell or with AcVP3/GFP and AcCP-H at 10 PFU/cell each and viewed 4 days p.i. using an Olympus photomicroscope equipped for fluorescence microscopy.
Nucleotide sequence accession number.
The nucleotide sequence data reported here for segment 4 of BmCPV strain H will appear in the GenBank database with accession no. AB041008.
RESULTS
Sequencing of BmCPV S4.
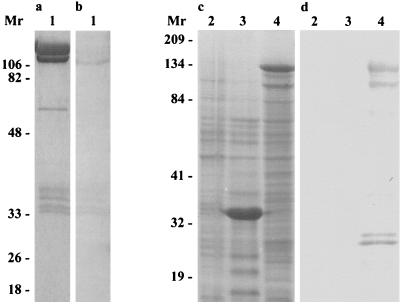
McCrae and Mertens have reported that BmCPV VP3, which is one of the outer components of the virus particle, is encoded by S4 (22). The cDNAs of BmCPV S4 were synthesized and cloned into pBlueScript II. To minimize the sequencing errors for PCR amplification, the nucleotide sequence of each clone was carefully determined by repetition in both the forward and reverse directions. The complete nucleotide sequence of gene S4, corresponding to the plus strand (mRNA sense strand), and the deduced amino acid sequence are shown in Fig. 1. S4 consisted of 3,259 nucleotides and possessed a single, long open reading frame (ORF) starting with the ATG codon (bases 14 to 16) and terminating with a TAA stop codon (bases 3185 to 3187), encoding a protein of 1,057 amino acids with a deduced molecular mass of 130 kDa. MAbs raised against BmCPV VP3 were used for Western blot analyses. The antibody reacted with the 130-kDa protein band of VP3 from the purified virions (Fig. 2). The uninfected Sf21 cells and those infected with AcNPV and AcVP3 were examined 4 days p.i. by Western blot analyses. The major protein band appearing at a molecular mass of 130 kDa from Sf21 cells infected with AcVP3 reacted with the MAb. BmCPV VP3 was thus confirmed to be encoded by S4 (Fig. 2). Although the other protein bands from AcVP3-infected Sf21 cells also reacted with the MAb, they were supposed to be degraded products of the expressed 130-kDa protein.
FIG. 1.
Complete nucleotide and deduced amino acid sequences of BmCPV S4.
FIG. 2.
Immunoblot analysis of VP3 with anti-VP3 antibody. Sf21 cells were infected with AcVP3 and collected 4 days after infection. Purified BmCPV virions and the infected Sf21 cells were lysed in SDS-PAGE sample buffer. Each sample was loaded on a SDS–10% polyacrylamide gel and subjected to electrophoresis. Proteins were detected by Coomassie brilliant blue staining (a and c) and Western blot analysis (b and d). Lane 1, Purified BmCPV virions; lane 2, uninfected Sf21 cells; lane 3, AcNPV-infected Sf21 cells; lane 4, AcVP3-infected Sf21 cells. Molecular weights are in thousands.
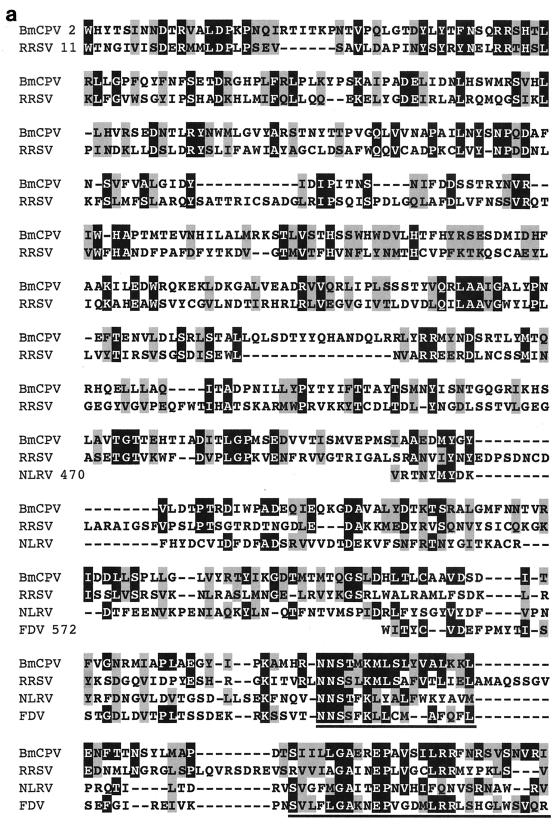
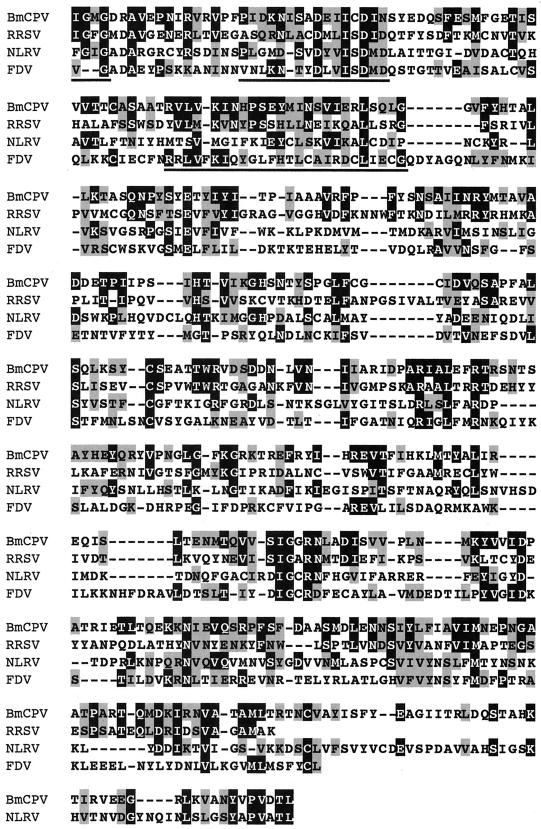
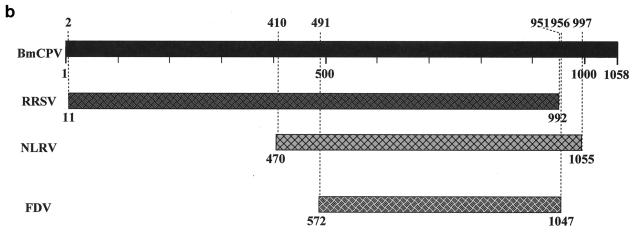
Comparison of the amino acid sequence encoded by BmCPV S4 with GenBank and EMBL databases using BLAST or FASTA programs revealed some similarity with plant reoviruses. Figure 3a shows 22% identity and 39% similarity with an S2-encoded P2 protein of rice ragged stunt virus (RRSV), a member of the genus Oryzavirus (38), 17% identity and 33% similarity with an S4-encoded 130-kDa protein of Nilaparvata lugens reovirus (NLRV), a putative member of the genus Fijivirus (28), and 17% identity and 36% similarity with an S4-encoded protein of Fiji disease fijivirus (FDV), a member of the genus Fijivirus (37). There appears to be little sequence similarity between reoviruses of different genera (13). Nevertheless, database analysis of the amino acid sequence of BmCPV VP3 showed similarities with those encoded by RRSV S2, NLRV S4, and FDV S4. For RRSV, the homologous region covered almost the full length of the amino acid sequence (residues 2 to 951 in BmCPV and residues 11 to 992 in RRSV). The homology of BmCPV to NLRV and FDV was less evident and observed only in the carboxy-terminal half (residues 470 to 1055 for NLRV and residues 572 to 1047 for FDV) (Fig. 3b). These results might indicate that BmCPV is more closely related to RRSV than to the other two viruses and that these two viruses split more recently. According to the estimation of the rate of nucleic acid substitution (2.2 × 10−3 substitutions/site/year) in the case of the genus Orbivirus in the family Reoviridae (17), the divergence of BmCPV and RRSV can be estimated to have occurred as recently as about 180 years ago. However, CPVs are extremely stable in the environment and can survive for long periods without replication in insects, and it is supposed that they spent many years without changing significantly in terms of their RNA sequences. Therefore, some caution must be exercised in interpretation of the data and in any speculation of a date for divergence of two genera within the Reoviridae.
FIG. 3.
(a) Multiple sequence alignment of homologous proteins from BmCPV S4, RRSV S2, NLRV S4, and FDV S4. Black and shaded regions represent identical and similar amino acids, respectively. The highly conserved regions of the amino acid sequences are underlined. RRSV S2, accession no. AF020335; NLRV S4, D49696; FDV S4, AF049705. (b) Locations of the homologous regions in the amino acid sequences between BmCPV S4, RRSV S2, NLRV S4, and FDV S4.
Comparison of the nucleotide sequences encoding the highly conserved amino acid sequences shown in Fig. 3a (BmCPV S4, RRSV S2, NLRV S4, and FDV S4) yielded the following results. Between BmCPV S4 and RRSV S2, the levels of identity of the first and the second nucleotides in the codons were 50 and 76%, respectively. The third nucleotide, having a higher degree of freedom, showed a much lower level of homology, only 25%. In the case of NLRV S4 and FDV S4, comparison of the first and the second nucleotides in the codons showed similar homologies of about 55% and the identity of the third nucleotide was about 35%. The nonrandom distribution of nucleotide exchanges was shown in the codons of ORFs of BmCPV S4 and RRSV S4. In conjunction with the amino acid sequence data, these findings might be a good example of the continuing divergence in viral evolution, which takes place under the functional constraint of maintaining the necessary capsid properties.
Expression of a chimeric gene consisting of GFP and VP3 genes.
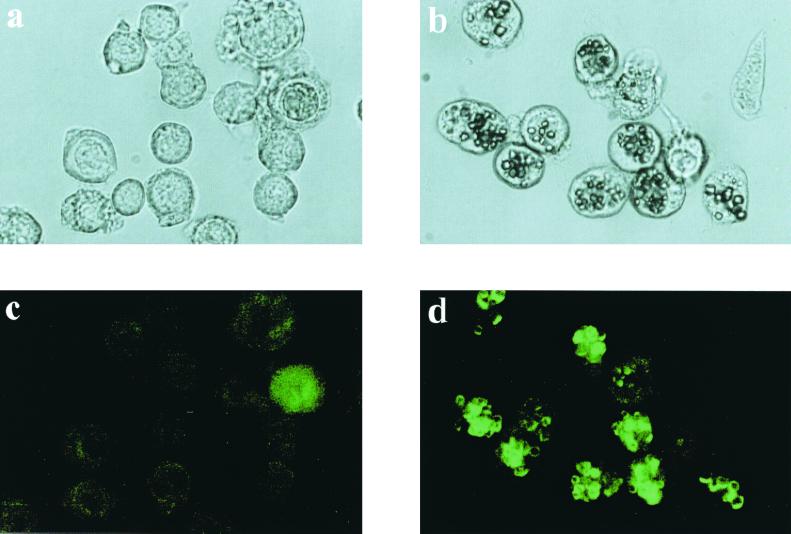
Light microscopy of Sf21 cells coinfected with AcVP3/GFP and AcCP-H showed that the green fluorescence was located around BmCPV polyhedra. Infection with AcVP3/GFP only, on the other hand, resulted in a dispersed green fluorescence in the cytoplasm (Fig. 4). This localized green fluorescence around BmCPV polyhedra suggested that specific interactions occur between BmCPV polyhedrin and VP3. However, it is unknown whether the VP3-GFP chimeric protein is occluded in BmCPV polyhedra and whether the specific interactions facilitate BmCPV virion occlusion into polyhedra.
FIG. 4.
Light (a and b) and fluorescence (c and d) micrographs of BmCPV polyhedra and VP3-GFP chimera. Sf21 cells were infected with AcVP3/GFP (a and c) or AcCP-H and AcVP3/GFP (b and d).
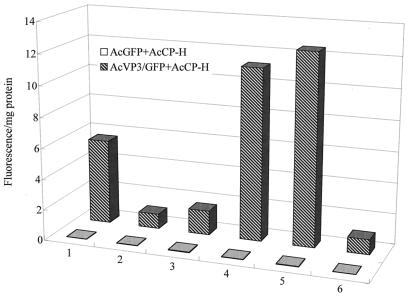
The green fluorescence of the polyhedron suspension prepared from double infection with AcVP3/GFP and AcCP-H completely disappeared in acetate buffer at pH 4. After the pH was increased, the green fluorescence was observed again (Fig. 5). There was no change in the appearance of BmCPV polyhedra produced by AcCP-H in a solution below pH 10; however, when the pH increased above 10, the polyhedra were dissolved more rapidly and the green fluorescence was intensified (Fig. 5). This effect could be due to the release of the VP3-GFP chimera embedded in the polyhedra by the dissolving polyhedra. No fluorescence was detected in polyhedra obtained from cells infected with both AcGFP and AcCP-H, indicating that the chimeric protein occlusion was initiated by specific interactions between BmCPV polyhedrin and VP3.
FIG. 5.
Measurement of green fluorescence of the VP3-GFP chimera in BmCPV polyhedra. Polyhedra obtained by double infection with AcGFP and AcCP-H or with AcVP3/GFP and AcCP H were resuspended in distilled water, acetate buffer (pH 4.0), and carbonate buffer (pH 11.0). After resuspension in acetate buffer, the pH was adjusted to 6.0, 10.0, and 12.5 by the addition of 5 N NaOH. Fluorescence intensity was represented as fluorescence emission at 510 nm per milligram of protein which was illuminated with 475-nm light. Column 1, distilled water; column 2, pH 4; column 3, pH 6; column 4, pH 10; column 5, pH 11; column 6, pH 12.5.
DISCUSSION
Members of the Reoviridae are grouped into nine genera, Orthoreovirus, Rotavirus, Orbivirus, Cypovirus, Phytoreovirus, Fijivirus, Oryzavirus, Coltivirus, and Aquareovirus, and have a multipartite genome consisting of 10 to 12 linear segments of dsRNA (13). The natural hosts of these viruses include vertebrates, invertebrates, and plants, but the virulence they exhibit toward these hosts differs widely. CPVs belong to the genus Cypovirus and possess 10 segmented dsRNAs (S1 to S10) (36). In this study the complete nucleotide sequence of BmCPV S4 encoding BmCPV VP3 was determined. BmCPV S4 consists of 3,259 bp and contains a single large ORF encoding a product of 1,057 amino acids.
Multiple sequence alignments of the amino acid sequences of proteins encoded by BmCPV S4, RRSV S2, NLRV S4, and FDV S4 seem to confirm the previous hypothesis that insect reoviruses are closely related to plant reoviruses and that plant reoviruses originated from insect reoviruses (30). According to the rate of nucleic acid substitution estimated for bluetongue virus (Orbivirus), BmCPV and RRSV could have diversified more recently than expected. RRSV infects rice and is transmitted exclusively by an insect, the brown plant hopper N. lugens (38). Although this virus replicates in both rice and its insect vector, BmCPV replicates only in the midgut epithelial cells of the silkworm. The recent emergence of RRSV relative to other plant reoviruses and the dramatically expanded host range of RRSV, from insects to plants, illustrate the possibility that a future virus may arise from members of the Reoviridae. The cultivation of rice started about 10 thousand years ago, and sericulture, invented about 5 thousand years later, took place near rice fields. An insect reovirus developed in the silkworms used for sericulture. The increased development of sericulture and rice cultivation facilitated the evolution of this virus.
Other occluded viruses that are pathogenic for insect hosts include the CPV group, the baculovirus group (nucleopolyhedrovirus and granulovirus), and the entomopoxvirus group. The occlusion body protein of the baculovirus group is very similar in size to CPV polyhedrin; however, there is no homology between the amino acid sequences of the two types of polyhedrins. The maintenance of occlusion body proteins in such a diverse group of viruses indicates the importance of polyhedrins in the life cycles of these viruses. The polyhedrin forms a protective crystal around the virus particles, and it resists solubilization except under strongly alkaline conditions similar to those found in the insect midgut. These properties allow the virus to remain viable for many years outside the insect host.
It has been hypothesized that in nuclear polyhedrosis viruses, virion occlusion and polyhedral growth are initiated by specific interactions between polyhedrin molecules and the virion envelope (6). However, little is known about the specific interactions between CPV polyhedrin and the viral capsid protein. Iodination of BmCPV virion and analysis of the labeled polypeptides by SDS-PAGE indicate that VP1 and VP3 are outer components of BmCPV. VP3 was thus selected to investigate interaction with BmCPV polyhedrin in the occlusion of virus particles into the polyhedra. Examination by microscopy of cells infected with AcVP3/GFP and AcCP-H under irradiation by long-wavelength UV light revealed clusters of very intense green fluorescence around BmCPV polyhedra. Infection with only AcVP3/GFP resulted in a dispersed green fluorescence in the cytoplasm. This suggests that there is indeed a specific interaction between BmCPV polyhedrin and VP3; however, it is not known whether the chimeric protein consisting of VP3 and GFP is actually incorporated into the polyhedra. Since UV light is unable to penetrate the polyhedra (a reason why the occluded virus particles in the polyhedra remain viable for long periods under normal environmental conditions), it is difficult to excite the GFP that is integrated together with the VP3. Purified polyhedra, obtained by double infection with AcVP3/GFP and AcCP-H, show a weak green fluorescence when suspended in distilled water and excited with UV light. This fluorescence might be due to GFP on the surface of the polyhedra. GFP is known to be stable in a broad pH range (pH 5 to 12) but is rapidly inactivated at pH values below 5 or above 12. After the polyhedra were resuspended in acetic acid buffer at pH 4, the green fluorescence disappeared. When the pH in this polyhedron suspension was increased beyond 10, a very strong green fluorescence was again detected and then disappeared at pH 12.5. There was no change in the appearance of the polyhedra in a solution below pH 10. The polyhedron suspension at pH 10 was clear, and examination by microscopy revealed that the polyhedra were completely dissolved. Polyhedra obtained by a double infection with AcGFP and AcCP-H and subjected to the same treatment yielded no green fluorescence under any condition. These results indicate that most of the chimeric protein consisting of VP3 and GFP was occluded in the polyhedra and that specific interactions must occur between BmCPV polyhedrin and VP3 to allow this occlusion. The extent to which these interactions might facilitate BmCPV virion occlusion and occlusion body assembly has yet to be determined.
ACKNOWLEDGMENTS
We acknowledge support from Enhancement of Center of Excellence, Special Coordination Funds for Promoting Science and Technology, Science and Technology Agency, Japan. K.I. was supported by the Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Arella M, Lavallée C, Belloncik S, Furuichi Y. Molecular cloning and characterization of cytoplasmic polyhedrosis virus polyhedrin and a viable deletion mutant gene. J Virol. 1988;62:211–217. doi: 10.1128/jvi.62.1.211-217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belloncik S. Cytoplasmic polyhedrosis viruses—Reoviridae. Adv Virus Res. 1989;37:173–209. [PubMed] [Google Scholar]
- 3.Belloncik S. Interactions of cytoplasmic polyhedrosis viruses with insects. Adv Insect Physiol. 1996;26:235–295. [Google Scholar]
- 4.Belloncik S, Mori H. Cypoviruses. In: Miller L K, Ball L A, editors. The insect viruses. New York, N.Y: Plenum Publishing Corporation; 1998. pp. 337–369. [Google Scholar]
- 5.Belloncik S, Liu J, Su D, Arella M. Identification and characterization of a new cypovirus, type 14, isolated from Heliothis armigera. J Invertebr Pathol. 1996;67:41–47. [Google Scholar]
- 6.Eason J E, Hice R H, Johnson J J, Federici B A. Effects of substituting granulin or a granulin-polyhedrin chimera for polyhedrin on virion occlusion and polyhedral morphology in Autographa californica multinucleocapsid nuclear polyhedrosis virus. J Virol. 1998;72:6237–6243. doi: 10.1128/jvi.72.7.6237-6243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fossiez F, Belloncik S, Arella M. Virology 169:462–465. 1989. Nucleotide sequence of the polyhedrin gene of Euxoa scandens cytoplasmic polyhedrosis virus (EsCPV) [DOI] [PubMed] [Google Scholar]
- 8.Galinski S M, Yu Y, Heminway B R, Beaudreau G S. Analysis of the C polyhedrin genes from different geographical isolates of a type 5 cytoplasmic polyhedrosis virus. J Gen Virol. 1994;75:1969–1974. doi: 10.1099/0022-1317-75-8-1969. [DOI] [PubMed] [Google Scholar]
- 9.Hagiwara K, Matsumoto T. Nucleotide sequences of genome segments 6 and 7 of Bombyx mori cypovirus 1, encoding the viral structural proteins V4 and V5, respectively. J Gen Virol. 2000;81:1143–1147. doi: 10.1099/0022-1317-81-4-1143. [DOI] [PubMed] [Google Scholar]
- 10.Hagiwara K, Tomita M, Kobayashi J, Miyajima S, Yoshimura T. Nucleotide sequence of Bombyx mori cytoplasmic polyhedrosis virus segment 8. Biochem Biophys Res Commun. 1998;247:549–553. doi: 10.1016/s0006-291x(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 11.Hagiwara K, Tomita M, Nkai K, Kobayashi J, Miyajima S, Yoshimura T. Determination of the nucleotide sequence of Bombyx mori cytoplasmic polyhedrosis virus segment 9 and its expression in BmN cells. J Virol. 1998;72:5762–5768. doi: 10.1128/jvi.72.7.5762-5768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill C L, Booth T F, Prasad B V V, Grimes J M, Mertens P P C, Sutton G C, Stuart D I. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 13.Holmes I H, Boccardo G, Estes M K, Furuichi M K, Hoshino Y, Joklik W K, McCrae M, Mertens P P C, Milne R G, Samal K S K, Shikata E, Winton J R, Uyeda I, Nuss D L. Family Reoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer; 1994. pp. 208–239. [Google Scholar]
- 14.Hukuhara T, Bonami J R. Reoviridae. In: Adams J R, Bonami J R, editors. Atlas of invertebrate viruses. Boca Raton, Fla: CRC Press; 1992. pp. 394–430. [Google Scholar]
- 15.Hukuhara T, Midorikawa M. Pathogenesis of cytoplasmic polyhedrosis in the silkworm. In: Compans R W, Bishop D H L, editors. double-stranded RNA viruses. New York, N.Y: Elsevier Biomedical; 1983. pp. 405–414. [Google Scholar]
- 16.Ikeda K, Nakazawa H, Alain R, Belloncik S, Mori H. Characterizations of natural and induced polyhedrin gene mutants of Bombyx mori cytoplasmic polyhedrosis viruses. Arch Virol. 1998;143:241–248. doi: 10.1007/s007050050283. [DOI] [PubMed] [Google Scholar]
- 17.Kowalik T F, Li J K. Bluetongue virus evolution: sequence analyses of the genomic S1 segments and major core protein VP7. Virology. 1991;181:749–755. doi: 10.1016/0042-6822(91)90911-t. [DOI] [PubMed] [Google Scholar]
- 18.Kuchino Y, Nishimura S, Smith R E, Furuichi Y. Homologous terminal sequences in the double-stranded RNA genome segments of cytoplasmic polyhedrosis virus of the silkworm Bombyx mori. J Virol. 1982;44:538–543. doi: 10.1128/jvi.44.2.538-543.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lewandowski L J, Traynor B L. Comparison of the structure and polypeptide composition of three double-stranded ribonucleic acid-containing viruses (Diplornaviruses): cytoplasmic polyhedrosis virus, wound tumor virus, and reovirus. J Virol. 1972;10:1053–1070. doi: 10.1128/jvi.10.5.1053-1070.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.McCrae M A, Mertens P P C. In vitro translation studies on and RNA coding assignments for cytoplasmic polyhedrosis viruses. In: Compans R W, Bishop D H L, editors. Double-stranded RNA viruses. New York, N.Y: Elsevier Biomedical; 1983. pp. 35–41. [Google Scholar]
- 23.Mertens P P C, Crook N, Rubinstein R, Pedley S, Payne C. Cytoplasmic polyhedrosis virus classification by electropherotype: validation by serological analyses and agarose gel electrophoresis. J Gen Virol. 1989;70:173–185. doi: 10.1099/0022-1317-70-1-173. [DOI] [PubMed] [Google Scholar]
- 24.Mike A, Ohwaki M, Fukada T, Miyajima S. Preparation of monoclonal antibodies to the Bombyx mori cytoplasmic polyhedrosis virus. J Seric Sci Jpn. 1984;53:77–80. [Google Scholar]
- 25.Mori H, Kawase S. Alkaline protease in cytoplasmic polyhedra of the silkworm, Bombyx mori (Lepidoptera: Bombycidae) Appl Entomol Zool. 1983;18:342–350. [Google Scholar]
- 26.Mori H, Minobe Y, Sasaki T, Kawase S. Nucleotide sequence of the polyhedrin gene of Bombyx mori cytoplasmic polyhedrosis virus A strain with nuclear localization of polyhedra. J Gen Virol. 1989;70:1885–1888. doi: 10.1099/0022-1317-70-7-1885. [DOI] [PubMed] [Google Scholar]
- 27.Mori H, Ito R, Nakazawa H, Sumida M, Matsubara F, Minobe Y. Expression of Bombyx mori cytoplasmic polyhedrosis virus polyhedrin in insect cells by using a baculovirus expression vector, and its assembly into polyhedra. J Gen Virol. 1993;74:99–102. doi: 10.1099/0022-1317-74-1-99. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima N, Koizumi M, Watanabe H, Noda H. Complete nucleotide sequence of the Nilaparvata lugens reovirus: a putative member of the genus Fijivirus. J Gen Virol. 1996;77:139–146. doi: 10.1099/0022-1317-77-1-139. [DOI] [PubMed] [Google Scholar]
- 29.Nakazawa H, Kendirgi F, Belloncik S, Ito R, Takagi S, Minobe Y, Higo K, Sumida M, Matsubara F, Mori H. Effect of mutations on the intracellular localization of Bombyx mori cytoplasmic polyhedrosis virus polyhedrin. J Gen Virol. 1996;77:147–153. doi: 10.1099/0022-1317-77-1-147. [DOI] [PubMed] [Google Scholar]
- 30.Nault L R. Transmission biology, vector specificity and evolution of planthopper transmitted plant viruses. In: Denno R F, Perfect T J, editors. Planthoppers. New York, N.Y: Chapman & Hall; 1994. pp. 429–448. [Google Scholar]
- 31.Payne C C, Kalmakoff J. Biochemical properties of polyhedra and virus particles of the cytoplasmic polyhedrosis virus of Bombyx mori. Intervirology. 1974;4:354–364. doi: 10.1159/000149870. [DOI] [PubMed] [Google Scholar]
- 32.Payne C C, Rivers C F. A provisional classification of cytoplasmic polyhedrosis viruses based on the size of the RNA genome segments. J Gen Virol. 1976;33:71–85. doi: 10.1099/0022-1317-33-1-71. [DOI] [PubMed] [Google Scholar]
- 33.Payne C C, Piasecka-Serafin M, Pilley B. The properties of two recent isolates of cytoplasmic polyhedrosis viruses. Intervirology. 1977;8:155–163. doi: 10.1159/000148890. [DOI] [PubMed] [Google Scholar]
- 34.Payne C C. Cytoplasmic polyhedrosis viruses. In: Davidson E W, editor. Pathogenesis of invertebrate microbial diseases. Totowa, N.J: Allanheld, Osmun & Co. Publishers; 1981. pp. 61–100. [Google Scholar]
- 35.Payne C C, Rubinstein R, Crook N E, Mertens P P C. Serological and molecular studies of variation in cytoplasmic polyhedrosis viruses. In: Compans R W, Bishop D H L, editors. Double-stranded RNA viruses. New York, N.Y: Elsevier Biomedical; 1983. pp. 253–262. [Google Scholar]
- 36.Payne C C, Mertens P P C. Cytoplasmic polyhedrosis viruses. In: Joklik W K, editor. The Reoviridae. New York, N.Y: Plenum; 1983. pp. 425–504. [Google Scholar]
- 37.Soo H M, Handley J A, Maugeri M M, Burns P, Smith G R, Dale J L, Harding R M. Molecular characterization of Fiji disease fijivirus genome segment 9. J Gen Virol. 1998;79:3155–3161. doi: 10.1099/0022-1317-79-12-3155. [DOI] [PubMed] [Google Scholar]
- 38.Upadhyaya N M, Yang M, Kositratana W, Ghosh A, Waterhouse P M. Molecular analysis of rice ragged stunt oryzavirus segment 9 and sequence conservation among isolates from Thailand and India. Arch Virol. 1995;140:1945–1956. doi: 10.1007/BF01322684. [DOI] [PubMed] [Google Scholar]