Summary
Background
Deployment and access to state-of-the-art precision medicine technologies remains a fundamental challenge in providing equitable global cancer care in low-resource settings. The expansion of digital pathology in recent years and its potential interface with diagnostic artificial intelligence algorithms provides an opportunity to democratize access to personalized medicine. Current digital pathology workstations, however, cost thousands to hundreds of thousands of dollars. As cancer incidence rises in many low- and middle-income countries, the validation and implementation of low-cost automated diagnostic tools will be crucial to helping healthcare providers manage the growing burden of cancer.
Methods
Here we describe a low-cost ($230) workstation for digital slide capture and computational analysis composed of open-source components. We analyze the predictive performance of deep learning models when they are used to evaluate pathology images captured using this open-source workstation versus images captured using common, significantly more expensive hardware. Validation studies assessed model performance on three distinct datasets and predictive models: head and neck squamous cell carcinoma (HPV positive versus HPV negative), lung cancer (adenocarcinoma versus squamous cell carcinoma), and breast cancer (invasive ductal carcinoma versus invasive lobular carcinoma).
Findings
When compared to traditional pathology image capture methods, low-cost digital slide capture and analysis with the open-source workstation, including the low-cost microscope device, was associated with model performance of comparable accuracy for breast, lung, and HNSCC classification. At the patient level of analysis, AUROC was 0.84 for HNSCC HPV status prediction, 1.0 for lung cancer subtype prediction, and 0.80 for breast cancer classification.
Interpretation
Our ability to maintain model performance despite decreased image quality and low-power computational hardware demonstrates that it is feasible to massively reduce costs associated with deploying deep learning models for digital pathology applications. Improving access to cutting-edge diagnostic tools may provide an avenue for reducing disparities in cancer care between high- and low-income regions.
Funding
Funding for this project including personnel support was provided via grants from NIH/NCIR25-CA240134, NIH/NCIU01-CA243075, NIH/NIDCRR56-DE030958, NIH/NCIR01-CA276652, NIH/NCIK08-CA283261, NIH/NCI-SOAR25CA240134, SU2C (Stand Up to Cancer) Fanconi Anemia Research Fund – Farrah Fawcett Foundation Head and Neck Cancer Research Team Grant, and the European UnionHorizon Program (I3LUNG).
Keywords: Machine learning, Digital pathology, Cancer diagnostics, Open-source, Low-cost microscope, Global health, Precision oncology
Research in context.
Evidence before this study
We searched PubMed and Google Scholar with the terms: (“low cost” OR “mobile” OR “portable”) AND (“microscope”) AND/OR (“digital pathology” OR “machine learning” OR “deep learning”) on June 24, 2022 and repeated this search on Aug 29, 2023. Papers published in English that discussed low-cost microscopy with or without machine learning or deep learning modules were identified. There is significant literature that involves the development of deep learning-based diagnostic and clinical tools more generally, and several papers describe devices for mobile and lower cost microscopy. A small number of papers discuss using deep learning with low-cost microscopy however no papers were found that validate existing deep learning models for histopathologic cancer subtyping on low cost and open-source hardware and software.
Added value of this study
This work intends to show that it is possible to combine existing deep learning methods with exclusively low-cost, open-source tools while preserving model accuracy. Our findings serve as a proof of concept that lower resolution pathology images can be used for deep learning models and that high-performing deep learning models can be run successfully on low-cost computational hardware.
Implications of all the available evidence
We show that it is possible to use deep learning models with digital pathology image capture workstations one thousand times less expensive than some institutional slide scanners. Given the potential for automation to increase diagnostic bandwidth in a wide range of clinical settings, more resources should be devoted to careful analysis and eventual implementation of methods that reduce cost and improve access to precision cancer care.
Introduction
The global burden of cancer is increasing as mortality associated with communicable diseases, starvation, and war declines. In the past, most cancer cases and cancer-related deaths occurred in higher income countries. However, the demographics of cancer are shifting. Incidence of cancer in low Human Development Index (HDI) countries is projected to double between 2008 and 2030 and increase by 81% in middle HDI countries.1 Advances in cancer care disproportionately benefit people in high HDI countries: fewer cancer-related gains in life expectancy are seen in low versus high HDI countries.1 Adapting advanced cancer diagnostics currently used in high-resource settings for broader application may help address some of the inequities perpetually seen in cancer care. Cancer diagnostics and treatment decision-making worldwide rely on pathological analysis with hematoxylin and eosin (H&E) stained tumor biopsy sections and molecular tests, but access to extensive molecular testing remains limited by cost. As the demographics of disease change and a growing number of people are diagnosed with cancers that could be better treated with advanced diagnostics, failure to close the gap in diagnostic precision will result in significant mortality.2
Digital pathology involves the acquisition and analysis of digital histopathological images in place of conventional microscopy.3,4 Specific advantages of digital pathology in resource-limited settings include the ability to share images for remote collaboration, the easy acquisition of large amounts of analyzable data, faster diagnosis, reduced burden of collecting and storing physical glass slides, and decreased costs achieved by lessening the amount of human pathologist review needed to make a diagnosis.4,5 As digitized whole slide images (WSIs) of histopathologic slides become widely available, computer vision and machine learning methods in digital pathology have the potential to assist with automating diagnostic processes. Deep learning (DL), a subdomain of machine learning that uses neural networks to identify patterns and features in complex datasets,6 can automate diagnostic workflows and reduce costs while providing the same information that human pathologists identify in histologic images.3 DL algorithms can analyze higher-order image characteristics to discern histologic and clinically actionable features, such as survival, treatment response, and genetic alterations.7,8
DL algorithms have the potential to automate tasks, thereby reducing personnel needs, equipment, and costs associated with precision cancer care. This may be especially useful in low- and middle-income countries where the cancer burden is growing quickly relative to available healthcare resources. Governments and corporations invest extensive resources into oncologic research to develop advanced diagnostics and targeted therapies, yet implementation barriers limit the ability of these advances to reduce the global burden of cancer.
Currently, state-of-the-art DL-based methods for digital pathology diagnostics fail to integrate their technology with the hardware and computing resources commonly found in lower-resource settings. As of October 2023, only four FDA-approved commercial AI technologies for digital pathology exist, and no open-source technologies have gained approval.9 Purchasing commercial software, computing resources, and WSI scanners needed for available AI pathology tools costs hundreds of thousands of dollars, making them inaccessible to low resource settings. By contrast, open-source tools are free to use, transparent, adaptable, and can be integrated into existing workflows. Validation of digital pathology DL methods is typically performed with high-cost equipment in high-income countries, although some work is being done to examine how these methods might be translated to lower-resource settings.10, 11, 12, 13, 14 An open-source pipeline would allow local users to contribute to device development and modify hardware or software components to fit variable clinical needs.
To overcome the cost barrier in digital pathology analysis, we compiled a fully integrated set of open-source, low-cost hardware and software components to compare achievable DL model performance against high-cost methods. Several studies have shown that digital images from diagnostic glass slides can be captured with low-cost equipment at sufficient resolution for DL model analysis.11,14, 15, 16, 17, 18 We constructed a workflow consisting of entirely open-source resources that integrates images captured with low-cost hardware, low-cost computing equipment, and publicly available DL models, producing an end-to-end low-cost digital histology deep learning analysis pipeline (Fig. 1a).19 We developed and tested a USD$230 platform for image capture of histopathological slides and used an open-source DL pipeline run on a USD$55 Raspberry Pi computer to classify tissue samples from three distinct datasets. We show that model performance with images captured with low-cost equipment is comparable to that of images captured with gold standard high-cost equipment, and we validate our pipeline for automated digital pathology biomarker-based classification of head and neck squamous cell carcinoma (HNSCC), lung cancer, and breast cancer subtypes.
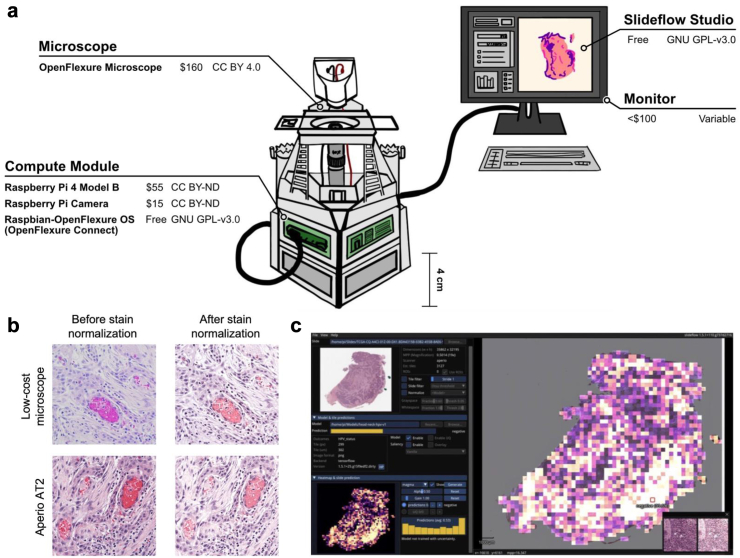
Fig. 1.
Open-source workflow. (a) Low-cost, open-source digital pathology workstation. All hardware (OpenFlexure Microscope, Raspberry Pi 4 Model B, Raspberry Pi camera module, monitor) and software (OpenFlexure Connect, Raspberry Pi OS, Slideflow) components with their costs and licenses are shown. (b) Open-source user interface for interactive visualization and generation of model predictions. Slideflow can be used to deploy a variety of trained models for digital pathology image classification, generating predictions for both partial-slide and whole-slide images. Predictions can be rendered for whole slides (rendered as a heatmap, as shown) or focal areas (rendered as individual tiles, as shown in the bottom right corner). The Slideflow user interface has been optimized for both x86 and low-power ARM-based devices. The above screenshot displays a heatmap of a WSI prediction, captured on the Raspberry Pi 4B. (c) Effects of computational stain normalization. Stain normalization increases visual similarity between the images captured by the Aperio AT2 slide scanner and the low-cost OpenFlexure device.
Methods
Data were collected to train DL models to perform a set of distinct pathologic classification tasks. To study the feasibility of leveraging existing DL methods within a low-cost, end-to-end, open-source pipeline, we selected DL models that mirrored published or pre-print data, including human papillomavirus (HPV) status in HNSCC as well as lung cancer and breast cancer subtypes.20, 21, 22 We collected publicly available histology images, which were captured using an Aperio ScanScope or other Aperio slide scanner and stored in SVS format from The Cancer Genome Atlas (TCGA). Tissue samples in these TCGA datasets originate from patients who live primarily in the United States and Europe.23 To perform external validation on models trained with data from TCGA, we collected WSIs from the University of Chicago Medical Center (UCMC), in accordance with University of Chicago IRB protocol 20-0238. For TCGA and UCMC datasets, pathologists identified and annotated tumor regions of interest within each WSI using QuPath. Patient demographics in the UCMC dataset reflected those in the TCGA datasets.
Lung cancer dataset preparation
941 digitized WSIs of H&E tissue samples from patients with lung cancer of known subtype were collected in SVS format from TCGA for model training. 472 samples from this training dataset were classified as lung adenocarcinoma and 469 were classified as squamous cell carcinoma. We performed external validation of the lung classification model using a UCMC dataset with ten lung cancer histopathology slides. Of the patients in the validation cohort, five had known lung adenocarcinoma and five had known squamous cell carcinoma.
Breast cancer dataset preparation
852 digitized WSIs of H&E tissue samples from patients with breast cancer of known subtype were collected in SVS format from TCGA for model training. 187 tissue samples from this training dataset were known invasive lobular carcinoma and 665 were known invasive ductal carcinoma. We performed external validation of the breast cancer classification model using a UCMC dataset consisting of ten breast cancer histopathology slides. Within this validation dataset, five patients had known invasive lobular carcinoma and five had known invasive ductal carcinoma.
HPV status HNSCC dataset preparation
For DL model training using retrospective data, 472 digitized WSIs of H&E tissue samples from patients with HNSCC and a known HPV status were collected in SVS format from TCGA. Among the TCGA training cohort, 52 patients were classified as HPV positive and 407 were classified as HPV negative. For model testing, we used a UCMC external validation dataset consisting of ten histopathology glass slides collected from patients with HNSCC and a known HPV status. Of these patients, five had known HPV positive and five had known HPV negative cancers.
Current standard imaging hardware and acquisition
For baseline “gold standard” image capture in the validation dataset, we used an Aperio AT2 ($250,000) digital pathology microscope slide scanner for image acquisition at 40× magnification in SVS format. As part of slide processing, 299 × 299 pixel image tiles are extracted from WSIs at 10× magnification (0.5 microns-per-pixel).
Open-source microscopy manufacture and image acquisition
To assess a cost-efficient alternative to professional grade microscopes, we modified the open-source OpenFlexure Microscope v6 design and assembled it with a fused deposition modeling 3D-printer (Creality CR-10s). The device includes optics, illumination, and stage modules (Supplementary Fig. S1). Our model of the device differs from the published OpenFlexure design in that it excludes the motorized stage. We used manual rather than motorized stage actuators to maximize stage mobility and attached the optics and illumination modules to the stage piece with the system of rails in the OpenFlexure design, securing the parts with M3 hex head screws. This microscope was then paired with a low-cost Raspberry Pi Model 3 (USD$55) and associated Camera Module 2 (USD$15) for image capture.19,24
After manual calibration, effective optical magnification was determined to be 0.4284 microns per pixel. The Raspbian-OpenFlexure operating system provided the software needed to visualize the glass slide on a monitor as well as capture and save images. Visual landmarks were used to locate the region of tissue for image capture, and images from the low-cost microscope were saved in JPEG format on-device and used for subsequent deep learning predictions. As a part of image preprocessing, these partial-slide images were segmented into individual tiles and resized to 299 × 299 pixels at 10× magnification (0.5 microns-per-pixel).
Histopathologic image processing
To assess the impact of low-versus high-cost image capture methods, we produced two sets of scanned histopathologic images for the UCMC validation cohorts: 1) WSIs from a clinical-grade microscope, and 2) partial-slide images from the low-cost microscope. Slides from the validation dataset were first scanned as WSIs with the clinical-grade microscope. Partial-slide images from this same dataset were then captured using the low-cost microscope, as the low-cost microscope is not yet capable of automated WSI capture. Regions of interest containing strongly predictive morphological features were selected based on predictive heatmap images generated by Slideflow. This partial-slide, proof-of-concept data capture method enabled us to demonstrate that histopathological images acquired using low-cost equipment can be accurately classified with open-source deep learning algorithms. We captured 150 images of morphologically informative regions at 10× magnification using OpenFlexure Connect software on the Raspberry Pi, which also accommodates a 40× lens for digital pathology tasks requiring higher optical magnification.19 From each of the slides in our validation datasets (10 HNSCC slides, 10 lung cancer slides, 10 breast cancer slides), we captured five partial-slide images. For both whole-slide (Aperio AT2) and partial-slide (low-cost microscope) images, DL predictions were generated on smaller image tiles (tile width 299 × pixels and 10 × magnification), and final slide-level predictions were calculated by averaging tile predictions. For the HNSCC dataset, we also carried out image capture of unguided, randomly chosen tissue regions with the low-cost microscope to further characterize the concordance between the low- and high-cost predictions. All image processing, including Reinhard-Fast stain normalization (Fig. 1b), was performed using Slideflow version 1.2.3.25
Deep learning models
Prior work has shown successful DL classification of H&E stained WSIs of lung cancer as adenocarcinoma versus squamous cell carcinoma, breast cancer as invasive ductal or invasive lobular carcinoma, and HNSCC as HPV positive or negative.5,21,22 For this study, we trained DL classification models for HPV positive versus negative status in TCGA HNSCC cases, lung adenocarcinoma versus squamous cell carcinoma from TCGA lung cancer cases, and breast lobular versus ductal carcinoma in the TCGA breast cancer cohort. A validation set of ten slides from UCMC (five from each class) was then used for validation of each model.
Weakly-supervised, tile-based DL model training was performed using a convolutional neural network with an Xception-based architecture, ImageNet pretrained weights, and two hidden layers of width 1024 with dropout of 0.1 after each hidden layer, using the package Slideflow as previously described.26 In this training schema, tiles are extracted from whole-slide images and receive a ground-truth label inherited from the corresponding slide. Slide-level predictions are aggregated across tiles via average pooling. During training, tiles received data augmentation with flipping, rotating, JPEG compression, and Gaussian blur. Model training was performed with 1 epoch and 3-fold cross-validation, using the Adam optimizer, with a learning rate of 10−4. Models were trained using Slideflow version 1.2.3 with the Tensorflow backend (version 2.8.2) in Python 3.8.25 All hyperparameters are listed in Supplementary Table S1. For the lung cancer subtyping endpoint, the publicly available “lung-adeno-squam-v1” model was used, with its training strategy and validation performance previously reported.27,28
Computational resources
DL models often require central processing units (CPUs) and graphical processing units (GPUs) with sufficient computational power to generate predictions. DL classification of the images captured using the Aperio AT2 microscope with Slideflow was carried out with an NVIDIA Titan RTX GPU (USD$3000) that is representative of the high-cost computational resources currently used for automated analysis of WSIs. To demonstrate the feasibility of using low-cost computational hardware to generate DL predictions, the images captured using the low-cost microscope were classified using Slideflow hosted on a USD$55 Raspberry Pi 4B computer with a Broadcom BCM2711, Quad core Cortex-A72 (ARM v8) 64-bit SoC @ 1.5 GHz processor and 4 GB of RAM.
User interface
Slideflow includes an open-source graphical user interface (GUI) that allows for interactive visualization of model predictions from histopathological images (Fig. 1c, Supplementary Video S1). Slideflow Studio, the Slideflow GUI, can flexibly accommodate a variety of trained models for digital pathology image classification and can generate predictions from both Raspberry Pi camera capture, partial-slide images, and whole-slide images. We optimized this interface for deployment on low-memory, ARM-based edge devices including the Raspberry Pi, and used the interface to generate predictions for all images captured on the low-cost microscope. The user interface utilizes a Python wrapper of Dear Imgui for GUI rendering, pyimgui.29,30 The interface supports displaying and navigating both partial-slide and whole-slide images (JPEG, SVS, NDPI, MRXS, TIFF) with panning and zooming. Slide images are read using VIPS and rendered using OpenGL.31 Loading and navigating a WSI utilizes <2 GB of RAM and provides a smooth experience, rendering slides at an average of 18 frames per second while actively panning and zooming. Models trained in both PyTorch and Tensorflow can be loaded, allowing focal predictions of specific areas of a slide or whole-slide predictions and heatmaps of an entire image. All necessary preprocessing, including optional stain normalization, is encoded in model metadata and performed on-the-fly. Predictions can also be generated for WSIs, rendering a final slide-level prediction, and displaying tile-level predictions as a heatmap. We implemented and optimized a low-memory mode for this interface to support future whole-slide predictions on Raspberry Pi hardware to ensure flexibility such that new versions of the device can capture WSIs. In low memory mode, partial-slide and whole-slide predictions are generated using a batch size of 4 without multiprocessing. Active CPU cooling is recommended for systems generating WSI predictions due to high thermal load associated with persistent CPU utilization.
Statistical analysis
Deep learning model performance was assessed at patient- and tile-level using confusion matrices and area under the receiver operating curve (AUROC). Chi-squared tests were used to determine whether the differences between low- and high-cost AUROC were statistically significant. Additionally, for each classification model, a correlation coefficient was calculated to assess the degree of concordance between model output-based numerical prediction values for images captured with low-cost microscope versus those captured with Aperio AT2. A strong association between the numerical prediction values generated by the two pipelines for each tissue sample supports the proof-of-concept that our DL algorithms can feasibly be implemented using less expensive image capture methods.
Results
Benchmarking DL predictions on the Raspberry Pi
We benchmarked DL inference speed on the low-cost microscope using 24 model architectures, four different tile sizes (71 × 71, 128 × 128, 256 × 256, and 299 × 299 pixels), and a variety of batch sizes (Supplementary Table S2). Deep learning benchmarks were performed using the Tensorflow backend of Slideflow. A recognizable, Xception-based architecture (Xception at 299 × 299 pixels) allowed predictions at 1.04 images/second. At all tile sizes, the fastest architecture was MobileNet, allowing 4.64 img/sec at 299 × 299 pixels, 6.30 img/sec at 256 × 256 pixels, 16.72 img/sec at 128 × 128 pixels, and 28.50 img/sec at 71 × 71 pixels.
All raw images captured at 10× magnification using the low-cost microscope exhibited some degree of blur, color distortion, and/or spherical aberration. Using the whole-slide user interface, focal predictions from a model trained on 299 × 299 pixel images could be generated at approximately 1 image per second, utilizing 2.2 GB of RAM on average. WSI predictions at 10× magnification required an average of 15 min for a typical slide with ∼0.8 cm2 tumor area and utilized all 4 GB of RAM. The swap file size needed to be increased to 1 GB to generate WSI predictions. Thermal throttling was observed during WSI predictions when using standard passive CPU cooling, suggesting that prediction speed might improve with active cooling.
Lung cancer subtype model performance with open-source pipeline
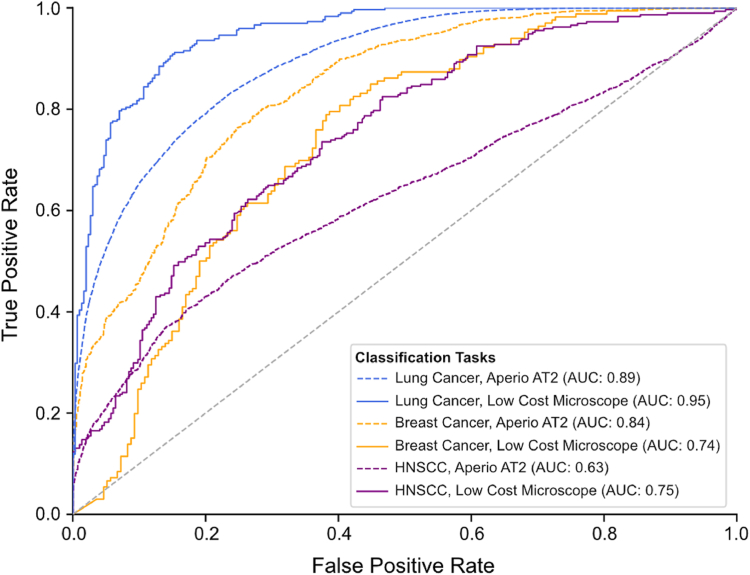
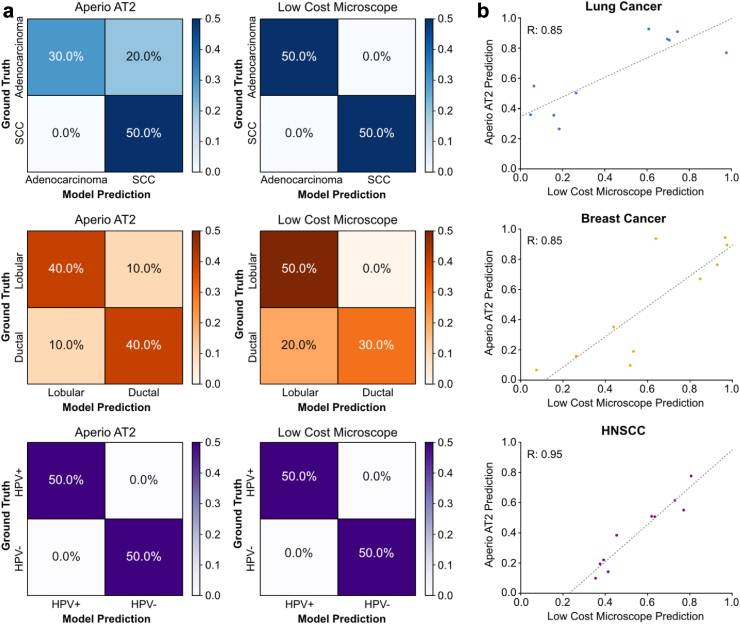
With tumor samples from ten patients in the lung cancer validation dataset, the patient-level and tile-level AUROCs for the model tested on images captured with the high-cost Aperio AT2 were 0.7 and 0.89, respectively. The model tested on images captured by the low-cost microscope performed with a patient-level and tile-level AUROC of 1.0 and 0.95 (Fig. 2). A chi-squared test showed no statistically significant difference in the proportion of correctly predicted tiles extracted from images captured and analyzed by the low-versus high-cost workstations (X2 (1, Nlow-cost = 595, Nhigh-cost = 29,532) = 0.363, p = 0.5468). Confusion matrices of model predictions on images captured with Aperio AT2 and the low-cost microscope are shown in Fig. 3a. Predictions made on images from the UCMC external validation cohort captured by the Aperio AT2 and the low-cost microscopes had a correlation coefficient of 0.85 (Fig. 3b).
Fig. 2.
AUROC of model performance for images captured on high- versus low-cost image acquisition and computational hardware. AUROC compares tile-level accuracy between high-cost Aperio AT2 and low-cost OpenFlexure image acquisition hardware. Model accuracy in terms of AUROC was maintained when the low-cost OpenFlexure device was used for image capture instead of the gold standard Aperio AT2 slide scanner, with chi-squared tests showing no statistically significant differences in model performance between low- and high-cost methods for any of the cancer subtypes (Lung cancer: X2 (1, Nlow-cost = 595, Nhigh-cost = 29532) = 0.363, p = 0.5468; Breast cancer: X2 (1, Nlow-cost = 360, Nhigh-cost = 1947) = 0.286, p = 0.5929; HNSCC: X2 (1, Nlow-cost = 587, Nhigh-cost = 5932) = 0.629, p = 0.4276).
Fig. 3.
Comparing model accuracy for high- versus low-cost image acquisition and computational hardware. (a) Confusion matrices showing patient-level accuracy when predictions were made using images captured with the Aperio AT2 or OpenFlexure device. Model accuracy at the patient level with the low-cost image capture and analysis pipeline was equal to or greater than accuracy with the high-cost hardware for image capture and analysis. (b) Correlation between DL numerical predictions made on images captured by Aperio AT2 versus OpenFlexure device. Strong and very strong correlation coefficients (Lung cancer R = 0.85; Breast cancer R = 0.85; HNSCC R = 0.95) imply that the underlying analyses involved in model prediction are similar whether the images are captured by the Aperio AT2 slide scanner or by the OpenFlexure device.
Breast cancer subtype model performance with open-source pipeline
With the ten patients’ slides in the breast cancer validation dataset, the patient-level and tile-level AUROCs for the classification model tested on images captured with Aperio AT2 were 0.84 and 0.84 respectively, and the model tested on images captured by low-cost microscope performed with a patient-level and tile-level AUROC of 0.80 and 0.74 (Fig. 2). A chi-squared test showed no statistically significant difference in the proportion of correctly predicted tiles extracted from images captured and analyzed by the low-versus high-cost workstations ((X2 (1, Nlow-cost = 360, Nhigh-cost = 1947) = 0.286, p = 0.5929)). Confusion matrices of DL model predictions on images captured with Aperio AT2 and low-cost microscope are shown in Fig. 3a. For the breast cancer dataset, predictions made on images from the UCMC external validation cohort captured by the Aperio AT2 and the low-cost microscopes had a correlation coefficient of 0.85 (Fig. 3b).
HNSCC HPV status model performance with open-source pipeline
With one slide from each of the ten patients in our HNSCC validation dataset, the AUROC for the model tested with images captured by Aperio AT2 and low-cost microscopes were 0.84 and 0.88 at the patient-level and 0.63 and 0.75 at the tile-level, respectively (Fig. 2). A chi-squared test showed no statistically significant difference in the proportion of correctly predicted tiles extracted from images captured and analyzed by the low-versus high-cost workstations (X2 (1, Nlow-cost = 587, Nhigh-cost = 5932) = 0.629, p = 0.4276). Fig. 3A shows confusion matrices of predictions made when images were captured using the Aperio AT2 or the low-cost microscope. When comparing predictions made on images from the UCMC external validation cohort captured by the Aperio AT2 versus low-cost microscopes, the HNSCC HPV status model predictions had a correlation coefficient of 0.95 (Fig. 3b). For the HNSCC dataset, we analyzed an alternative method for image capture with the low-cost device, in which model predictions were made from unguided, randomly chosen tissue regions. These results how high-concordance between model predictions for the images captured on the low-and high-cost devices and are reported in Supplementary Fig. S3.
Discussion
Open-source technology ensures access to artificial intelligence-based diagnostic tools among a wider range of providers and communities. It also allows users globally to contribute to the development of clinical tools that serve the needs of their patient populations. Our results serve as proof-of-concept that it is possible to preserve model accuracy while dramatically reducing the cost of image acquisition and computational hardware. We show the feasibility of using low-cost, open-source hardware for the image acquisition and computational steps required to apply machine learning methods to digital pathology and cancer diagnostics.
The DL models used to classify lung cancer, breast cancer, and HNSCC images in this study were trained with similar methods used in previously published analyses.32,33 Model performance was maintained with an open-source, 3D-printed microscope for image acquisition despite it costing orders of magnitude less than the clinical-grade microscopes currently used. Despite the presence of color distortion and blur prior to image normalization and lower resolution, images captured using the low-cost microscope and the Raspberry Pi camera were classified by our DL model as accurately as images captured on a clinical-grade microscope. Model accuracy was maintained whether the task involved predicting breast cancer subtype, lung cancer subtype, or HNSCC HPV status, supporting the idea that significant reduction in resolution associated with reduced hardware costs does not hinder model performance and can be applied to histopathologic cancer diagnostics more broadly.
Model accuracy can be optimized by exploring strategies to computationally augment lower-quality images or modify the training dataset such that the model can be trained on images whose quality reflects those captured by low-cost devices. We used established algorithms and training paradigms, but significant gains in accuracy and computational efficiency may be possible through further model development. Emerging methods for computational image normalization, including CycleGAN and other generative algorithms, could be useful in this context. In addition, Slideflow not only applies stain normalization to images prior to analysis, but also computationally alters the training dataset by introducing random blur and JPEG compression augmentation. It will be important to determine the influence of these techniques on model performance.
There are several limitations to this study, including the small quantity of validation data, the limited number of models tested, and the intentional selection of highly informative regions of tissue for image capture to test the low-cost device. We have not accounted for site or batch effects, out of distribution data, or domain shift. We chose a limited number of outcomes to evaluate in this analysis, and subsequent studies are planned to explore whether other common clinical biomarkers, such as breast cancer receptor status and microsatellite instability, can be identified and used to make accurate predictions with lower-resolution images such as those captured by the low-cost device.8 Further testing should be performed to validate this process for multiclass models, which are highly relevant for many digital pathology applications. Additionally, it is crucial that DL models are trained on datasets that are representative of the intended patient populations.9 Since the TCGA data used for model training came primarily from patients in the United States and Europe, exploring how dataset demographics affect model performance prior to the deployment of these technologies in clinical settings is essential. Further open-source software and support for biorepositories in low- and middle-income countries is essential to building more robust, diverse, and globally representative datasets.34,35
Design improvements in the low-cost microscope to improve image quality, reduce costs, and ease construction and transport are ongoing. Parts of the device are currently 3D printed, which might limit accessibility and ease of assembly. In resource-constrained areas, designs that adapt existing equipment to accommodate digital pathology and DL technologies may be preferable to an additional piece of distinct equipment that would need to be purchased. In general, the development of low-cost equipment for image capture in low-resource clinical settings expands the collaborative potential associated with telepathology such that providers in low- and high-resource settings can consult with each other when there is diagnostic uncertainty.
The success of this open-source workstation demonstrates that there are opportunities to reduce hardware costs while maintaining predictive accuracy. Although this analysis is limited in scope, the results are promising. Successful implementation of low-cost digital pathology and DL pipelines will increase access to precision cancer care in lower-resource clinical settings.
Contributors
A.T.P. conceived the study. A.T.P., D.C., S.K., and J.M.D. designed the experiments. J.M.D. wrote the software and designed the algorithms for the lung and head and neck cancer models. J.M.D. and D.C. performed the experiments and analyzed data with support from S.K. and S.R. F.M.H. designed the algorithm for the breast cancer model. J.S. performed histological analysis and pathologist review. J.R.M. and A.S. designed, prototyped, and built the low-cost microscope. D.C., J.M.D., and E.C.D. wrote the manuscript. D.C. and J.M.D. verified the underlying data. All authors contributed to general discussion and manuscript revision, and all authors read and approved the final version of the manuscript.
Data sharing statement
Whole-slide images from The Cancer Genome Atlas (TCGA) lung adenocarcinoma project are publicly available at https://portal.gdc.cancer.gov/projects/TCGA-LUSC, and images from the lung squamous cell project are available at https://portal.gdc.cancer.gov/projects/TCGALUAD. Whole-slide images from TCGA Head and Neck Squamous Cell Carcinoma project are publicly available at https://portal.gdc.cancer.gov/projects/TCGA-HNSC. Whole-slide images from TCGA Breast Invasive Carcinoma project are publicly available at https://portal.gdc.cancer.gov/projects/TCGA-BRCA. The training datasets from University of Chicago analyzed during the current study are not publicly available due to patient privacy obligations but are available from the corresponding author on reasonable request. Data can only be shared for non-commercial academic purposes and will require institutional permission and a data use agreement.
Code availability
Slideflow and Slideflow Studio are available at https://github.com/slideflow/slideflow, via the Python Package Index (PyPI), and Docker Hub (https://hub.docker.com/r/jamesdolezal/slideflow). Slideflow is licensed with Apache License, Version 2.0. Deep learning analyses were performed using Slideflow 1.5.4 with Tensorflow 2.11 as the primary deep learning package.
Declaration of interests
A.T.P. reports no competing interests for this work, and reports personal fees from Prelude Therapeutics Advisory Board, Elevar Advisory Board, AbbVie consulting, Ayala Advisory Board, and stock options ownership in Privo Therapeutics, all outside of submitted work. J.M.D. is Founder/CEO of Slideflow Labs Inc, a digital pathology startup company founded in April 2024; he reports no financial interests related to the contents of this manuscript. S.R. is CSO of Slideflow Labs, owns stock/stock options in Slideflow Labs, and reports no competing interests for this work. F.M.H. reports receiving grants from the NIH/NCI, the Cancer Research Foundation, and the Department of Defense Breast Cancer Research Program and has no competing interests for this work. J.N.K. reports no competing interests for this work. He receives consulting fees from Owkin, DoMore Diagnostics, Panakeia, Scailyte, and Histofy, honoraria from AstraZeneca, Bayer, Eisai, MSD, BMS, Roche, Pfizer, and Fresenius, and reports owning stock/stock options in StratifAI GmbH. M.G. reports no competing interests for this work, and reports personal financial support from AstraZeneca, Abion, Merck Sharp & Dohme International GmbH, Bayer, Bristol-Myers Squibb, Boehringer Ingelheim Italia S.p.A, Celgene, Eli Lilly, Incyte, Novartis, Pfizer, Roche, Takeda, Seattle Genetics, Mirati, Daiichi Sankyo, Regeneron, Merck, Blueprint, Janssen, Sanofi, AbbVie, BeiGenius, Oncohost, and Medscape, Gilead, and Io Biotech.
Acknowledgements
We would like to acknowledge the Human Tissue Resource Center at the University of Chicago Medical Center for their support in this work.
This work was supported in part by from NIH/NCI R25-CA240134, NIH/NCI U01-CA243075, NIH/NIDCR R56-DE030958, NIH/NCI R01-CA276652, NIH/NCI K08-CA283261, NIH/NCI-SOAR 25CA240134, SU2C (Stand Up to Cancer) Fanconi Anemia Research Fund – Farrah Fawcett Foundation Head and Neck Cancer Research Team Grant, and the European Union Horizon Program (I3LUNG).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105276.
Appendix ASupplementary data
References
- 1.Fidler M.M., Bray F., Soerjomataram I. The global cancer burden and human development: a review. Scand J Public Health. 2018;46(1):27–36. doi: 10.1177/1403494817715400. [DOI] [PubMed] [Google Scholar]
- 2.Chen S., Cao Z., Prettner K., et al. Estimates and projections of the global economic cost of 29 cancers in 204 countries and territories from 2020 to 2050. JAMA Oncol. 2023;9(4):465–472. doi: 10.1001/jamaoncol.2022.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Rojo M. International clinical guidelines for the adoption of digital pathology: a review of technical aspects. Pathobiology. 2016;83(2–3):99–109. doi: 10.1159/000441192. [DOI] [PubMed] [Google Scholar]
- 4.Jahn S.W., Plass M., Moinfar F. Digital pathology: advantages, limitations and emerging perspectives. J Clin Med. 2020;9(11) doi: 10.3390/jcm9113697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kacew A.J., Strohbehn G.W., Saulsberry L., et al. Artificial intelligence can cut costs while maintaining accuracy in colorectal cancer genotyping. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.630953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamamoto R., Suvarna K., Yamada M., et al. Application of artificial intelligence technology in oncology: towards the establishment of precision medicine. Cancers. 2020;12(12) doi: 10.3390/cancers12123532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bera K., Schalper K.A., Rimm D.L., Velcheti V., Madabhushi A. Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard F.M., Dolezal J., Kochanny S., et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat Commun. 2021;12(1):4423. doi: 10.1038/s41467-021-24698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good machine learning practice for medical device development: guiding principles. 2021. https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles [Google Scholar]
- 10.Holmström O., Linder N., Kaingu H., et al. Point-of-Care digital cytology with artificial intelligence for cervical cancer screening in a resource-limited setting. JAMA Netw Open. 2021;4(3) doi: 10.1001/jamanetworkopen.2021.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan K., Ceylan Koydemir H., Rivenson Y., et al. Automated screening of sickle cells using a smartphone-based microscope and deep learning. NPJ Digital Med. 2020;3(1):76. doi: 10.1038/s41746-020-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu M.Y., Williamson D.F.K., Chen T.Y., Chen R.J., Barbieri M., Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng. 2021;5(6):555–570. doi: 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar D.S., Kulkarni P., Shabadi N., Gopi A., Mohandas A., Narayana Murthy M.R. Geographic information system and foldscope technology in detecting intestinal parasitic infections among school children of South India. J Fam Med Prim Care. 2020;9(7):3623–3629. doi: 10.4103/jfmpc.jfmpc_568_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naqvi A., Manglik N., Dudrey E., Perry C., Mulla Z.D., Cervantes J.L. Evaluating the performance of a low-cost mobile phone attachable microscope in cervical cytology. BMC Wom Health. 2020;20(1):60. doi: 10.1186/s12905-020-00902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Soto-Montoya H., Voisin M., Valenzuela L.F., Prakash M. Octopi: open configurable high-throughput imaging platform for infectious disease diagnosis in the field. bioRxiv. 2019 doi: 10.1101/684423v1. [DOI] [Google Scholar]
- 16.Salido J., Toledano P.T., Vallez N., et al. MicroHikari3D: an automated DIY digital microscopy platform with deep learning capabilities. Biomed Opt Express. 2021;12(11):7223–7243. doi: 10.1364/BOE.439014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Villena J., Torres J.E., Aguilar C., et al. 3D-Printed portable robotic mobile microscope for remote diagnosis of global health diseases. Electronics. 2021;10(19):2408. [Google Scholar]
- 18.Chen P.C., Gadepalli K., MacDonald R., et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat Med. 2019;25(9):1453–1457. doi: 10.1038/s41591-019-0539-7. [DOI] [PubMed] [Google Scholar]
- 19.Sharkey J.P., Foo D.C.W., Kabla A., Baumberg J.J., Bowman R.W. A one-piece 3D printed flexure translation stage for open-source microscopy. Rev Sci Instrum. 2016;87(2) doi: 10.1063/1.4941068. [DOI] [PubMed] [Google Scholar]
- 20.Kather J.N., Schulte J., Grabsch H.I., et al. Deep learning detects virus presence in cancer histology. bioRxiv. 2019 doi: 10.1101/690206v1. [DOI] [Google Scholar]
- 21.Kanavati F., Toyokawa G., Momosaki S., et al. A deep learning model for the classification of indeterminate lung carcinoma in biopsy whole slide images. Sci Rep. 2021;11(1):8110. doi: 10.1038/s41598-021-87644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couture H.D., Williams L.A., Geradts J., et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer. 2018;4(1):30. doi: 10.1038/s41523-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Steensma J.T., Bailey M.H., Feng Q., Padda H., Johnson K.J. Characteristics of the cancer genome atlas cases relative to U.S. general population cancer cases. Br J Cancer. 2018;119(7):885–892. doi: 10.1038/s41416-018-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins J.T., Knapper J., Stirling J., et al. Robotic microscopy for everyone: the OpenFlexure microscope. Biomed Opt Express. 2020;11(5):2447–2460. doi: 10.1364/BOE.385729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolezal J., Kochanny S., Howard F.M. 2022. Slideflow: a unified deep learning pipeline for digital histology (1.3.1) Zenodo. [Google Scholar]
- 26.Dolezal J.M., Kochanny S., Dyer E., et al. Slideflow: deep learning for digital histopathology with real-time whole-slide visualization. BMC Bioinf. 2024;25(1):134. doi: 10.1186/s12859-024-05758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolezal J.M., Srisuwananukorn A., Karpeyev D., et al. Uncertainty-informed deep learning models enable high-confidence predictions for digital histopathology. Nat Commun. 2022;13(1):6572. doi: 10.1038/s41467-022-34025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolezal J.M., Wolk R., Hieromnimon H.M., et al. Deep learning generates synthetic cancer histology for explainability and education. NPJ Precis Oncol. 2023;7(1):49. doi: 10.1038/s41698-023-00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cython-based Python bindings for dear imgui. GitHub repository; 2023. https://github.com/pyimgui/pyimgui [Google Scholar]
- 30.Dear ImGui: bloat-free graphical user interface for C++ with minimal dependencies. GitHub repository; 2023. https://github.com/ocornut/imgui [Google Scholar]
- 31.Martinez KaC J. VIPS – a highly tuned image processing software architecture. Proc IEEE Int Conf Image Process. 2005;2:574–577. [Google Scholar]
- 32.Howard F.M., Dolezal J., Kochanny S., et al. Integration of clinical features and deep learning on pathology for the prediction of breast cancer recurrence assays and risk of recurrence. NPJ Breast Cancer. 2023;9(1):25. doi: 10.1038/s41523-023-00530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolezal J.M., Kather J.N., Kochanny S.E., et al. Deep learning detects actionable molecular and clinical features directly from head/neck squamous cell carcinoma histopathology slides. Int J Radiat Oncol Biol Phys. 2020;106:1165. [Google Scholar]
- 34.Müller H., Lopes-Dias C., Holub P., et al. BIBBOX, a FAIR toolbox and App Store for life science research. New Biotechnol. 2023;77:12–19. doi: 10.1016/j.nbt.2023.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Anderson D., Bendou H., Kipperer B., Zatloukal K., Müller H., Christoffels A. In: Biobanks in low- and middle-income countries: relevance, setup and management. Sargsyan K., Huppertz B., Gramatiuk S., editors. Springer International Publishing; Cham: 2022. Software tools for Biobanking in LMICs; pp. 137–146. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.