Abstract
The glycoprotein precursor (G1/G2) gene of tomato spotted wilt virus (TSWV) was expressed in BHK cells using the Semliki Forest virus expression system. The results reveal that in this cell system, the precursor is efficiently cleaved and the resulting G1 and G2 glycoproteins are transported from the endoplasmic reticulum (ER) to the Golgi complex, where they are retained, a process that could be blocked by tunicamycin. Expression of G2 alone resulted in transport to and retention in the Golgi complex, albeit less efficient, suggesting that G2 contains a Golgi retention signal. G1 alone was retained in the ER, irrespective of whether it contained the precursor's signal sequence or its own N-terminal hydrophobic sequence. Coexpression of G1 and G2 from separate gene constructs resulted in rescue of efficient G1 transport, as the proteins coaccumulated in the Golgi complex, indicating that their interaction is essential for proper targeting to this organelle. The results demonstrate that transport and targeting of the plant TSWV glycoproteins in mammalian BHK cells are strikingly similar to those of animal-infecting bunyavirus glycoproteins in mammalian cells. The observations are likely to reflect the dual tropism of TSWV, which replicates both in its plant host and in its animal (thrips) vector.
Among the Bunyaviridae, Tomato spotted wilt virus (TSWV) is unique in its ability to infect plants rather than animals (6, 8, 9, 10, 11, 28). Like the other bunyaviruses, TSWV particles have a membrane envelope that contains virally encoded glycoproteins, a feature quite uncommon for plant-infecting viruses but rather typical among animal viruses. This led to the suggestion that an ancestral animal-infecting bunyavirus may have evolved into the plant-infecting tospoviruses, of which TSWV is the type species (11). TSWV also replicates in its animal (thrips) vector (39, 43), indicating the dual tropism of this “shuttle” virus, which has to be able to produce virus particles in both plant and animal cells. The presence of the membrane glycoproteins is essential for the virus's ability to replicate alternately in its plant host and its thrips vector (42). This is illustrated by the observation that when the thrips transmission cycle is bypassed by repeated mechanical inoculation of plants, mutants are generated that infect plants but can no longer be transmitted by thrips (42). This feature correlates with the loss of the envelope in these mutants (14, 31, 40). Apparently, the insect transmission cycle guarantees the maintenance of an intact envelope, because infection of the thrips vector is dependent on it and thus selective for it, while for infection of plants the envelope is dispensable.
The formation of the enveloped virus particles is strongly regulated by the viral glycoproteins. They generally accumulate independently at a particular cellular membrane by targeted transport through the secretory pathway, to facilitate the interaction with the viral nucleocapsids and the initiation of budding (reviewed in references 12, 29, and 35). For animal-infecting bunyaviruses, this accumulation site was determined to be the Golgi system (reviewed in references 25 and 30).
The morphogenesis of enveloped TSWV particles has recently been studied in a plant cell system, Nicotiana rustica protoplasts (17), and appeared to be a unique process, very distinct from the morphogenesis of animal-infecting bunyaviruses (16). During infection of plant cells, the TSWV structural proteins, including the glycoproteins, accumulate at the Golgi system, a feature also observed during animal bunyavirus maturation (for Uukuniemi virus [20, 21]). Subsequently, however, doubly enveloped virus particles are formed as a result of wrapping of glycoprotein-containing Golgi cisternae around nucleocapsids in the cytoplasm. In a later stage, these doubly enveloped particles fuse with each other and with endoplasmic reticulum (ER) membranes, giving rise to mature, singly enveloped particles, clustered inside large membrane sacks where they accumulate and await uptake by thrips for transmission to other plants (16). In contrast, animal-infecting bunyaviruses produce singly enveloped particles by direct budding of nucleocapsids into the lumen of glycoprotein-containing Golgi cisternae, without the formation of doubly enveloped particles as intermediates (reviewed in references 7, 12, 25, and 35). These particles are then excreted in order to infect neighboring cells.
Obviously, the rigid cell wall of plants prevents the excretion of plant viruses from the cell, which explains the need to regulate cell-to-cell transport of infectious nucleocapsid structures through plasmodesmata (19, 36). The cell wall also dictates the accumulation of virus particles within the plant cell, illustrating an adapted vector transmission mechanism for plant-infecting TSWV. Assuming that TSWV has evolved from an ancestral animal bunyavirus, an intriguing question is in what way have TSWV glycoproteins changed to adapt to the distinct morphogenesis pathway in plants while maintaining the ability to replicate and produce virus particles in the animal vector?
In this study we addressed this question by expressing the TSWV glycoproteins in mammalian cells, using the Semliki Forest virus expression system, and studying their intracellular trafficking and accumulation behavior. In this way it could be verified whether the TSWV glycoproteins still contain the general transport and targeting signals characteristic of the glycoproteins of the animal-infecting bunyavirus ancestor, or whether the molecular features of TSWV glycoproteins have changed to meet the specific prerequisites for infection of its plant host.
MATERIALS AND METHODS
Cell culture.
Baby hamster kidney (BHK-21) cells were maintained at 37°C with 5% CO2 in Glasgow minimal essential medium (MEM; (Life Technology) supplemented with 10% fetal calf serum, tryptose phosphate broth, pencillin (100 U/ml), and streptomycin (100 μg/ml).
Antisera.
Antibodies against TSWV glycoproteins G1 and G2 were raised by immunization of rabbits with purified fragments of G1 (hydrophilic amino acid sequence encoded by nucleotides 3012 to 2122, numbered from the 5′ end of the viral M RNA [18]) and G2 (hydrophilic amino acid sequence encoded by nucleotides 4563 to 3928, numbered from the 5′ end of the viral M RNA), expressed in Escherichia coli using the pET11t system (Novagen) according to Kormelink et al. (19). Monoclonal antibody 2B6 against G1 was a gift from Guenter Adam and was described by Adam et al. (1). Antisera against purified TSWV and against the TSWV-encoded N protein were produced as described by de Ávila et al. (5). Monoclonal antibodies against the intermediate compartment p58 protein were kindly provided by J. Saraste and were described by Saraste et al. (33) and Saraste and Svensson (34). The Golgi stack marker anti-p58 was produced by Sigma. The chemical ER marker DiOC6 was purchased from Molecular Probes Inc.
Construction of recombinants.
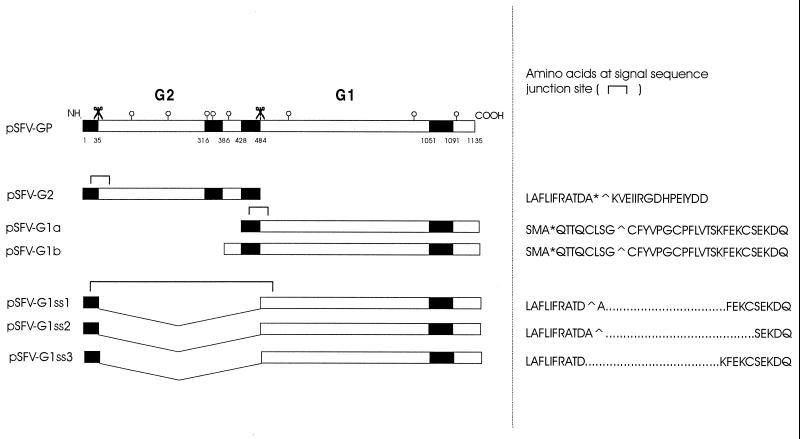
The pSFV1 vector (Gibco-BRL, Life Technology Inc.), containing an NruI linearization site (designated pSFV1-N), was used for cloning and expression in BHK cells. The BamHI site of the multiple cloning site was used to insert an original cDNA fragment of the G1/G2 precursor gene of TSWV, for which the nucleotide sequence has been determined (18). Mutants of the precursor were produced by PCR using specific primers containing a start codon at the 5′ end and a stop codon at the 3′ end of the gene fragment, flanked by a BamHI restriction site for feasible cloning into the SFV1-N vector. The mutant fragments were first cloned into a pGEM-T (Promega) or pSK(−) (Stratagene) vector and verified by sequencing prior to subcloning into the BamHI site of the SFV1-N vector. Recombinants in which the signal sequence of the N terminus (amino acids 1 to 35) of the precursor (18) was linked in frame to the G1 coding sequence (from amino acid 486; see Fig. 1) were produced using a modified ExSite (Stratagene) mutagenesis procedure as follows. cDNA encompassing the glycoprotein precursor open reading frame (ORF) was cloned into the BamHI site of pSKδ, a modified pSK(−) vector which lacked part of the multiple cloning linker from restriction sites SmaI to HincII, including ClaI. This construct was digested with ClaI, which cuts the precursor once, in the G2 sequence. Two PCR primers were engineered, one annealing to the 3′ end of the N-terminal signal sequence of the precursor and extending upstream, and one annealing at the beginning of the G1 coding sequence and extending downstream. With these primers, PCR was performed on the ClaI-cut pSKδGP template, using the proofreading PCR polymerase Elongase (Gibco-BRL), resulting in a product in which the G2 sequence is deleted from the precursor. The PCR product was ligated in the presence of T4 polymerase and deoxynucleoside triphosphates and then cut with ClaI again to linearize any remaining wild-type G1/G2 sequences. After transformation, several clones were selected and sequenced. Positive clones were identified, and mutated ORFs were excised from pSKδ using BamHI and cloned into BamHI-cut pSFV-N vector. Figure 1 shows all investigated recombinants schematically.
FIG. 1.
Schematic representation of Semliki Forest virus constructs described in this article. Solid rectangles indicate hydrophobic sequences (signal sequence/transmembrane domain), and open circles stand for potential N-glycosylation sites. Scissor symbols indicate possible (signal peptidase) cleavages in the precursor. Amino acid sequences around the putative signal sequence junction sites are indicated, where an asterisk indicates a cleavage site predicted by the von Heijne (41) algorithm, and a circumflex (∧) indicates a cleavage site predicted by the Jagla algorithm (unpublished data).
Semliki Forest virus expression system.
The system was first described by Liljeström and Garoff (24). We used transfection of in vitro-capped RNA transcripts of the constructs. To this end, the recombinant vectors were linearized using NruI, cleaned of RNase activity by treatment with proteinase K, and subsequently transcribed in the presence of SP6 RNA polymerase and cap analogs. RNA products were checked by electrophoresis in a 1% agarose gel.
BHK cells were seeded in 80-cm2 tissue culture flasks. Subconfluent cell monolayers obtained in 1 to 2 days were detached using a trypsin-EDTA solution (Life Technologies). The cells were centrifuged for 5 min at 900 rpm and washed once with PBS-0 (138 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.7 mM KH2PO4 [pH 7.3]). The cell pellet was resuspended carefully in 800 μl of PBS-Ca/Mg (PBS-0 with 0.89 mM CaCl2 · 2H2O and 0.5 mM MgCl2 · 6H2O), and 10 to 50 μg of RNA transcript was added. Electroporation was performed in a Bio-Rad electroporator by two consecutive pulses at 850 V, 25 μF, and 200 Ω. This resulted in a time constant of about 0.8 ms at each pulse. Transfected cells were added to 15 to 20 ml of culture medium in 80-cm2 culture flasks, and from this, samples were taken for immunofluorescence, which were seeded on thin microscopic coverslips in six-well plates. Cells were incubated at 37°C and 5% CO2 for 6 to 21 h. In some experiments tunicamycin (5 μg/ml) was added 1 h after transfection, after which cells were incubated as mentioned above. Cycloheximide was added in some experiments to a concentration of 50 μg/ml 6 h after incubation, after which the incubation was continued for 2 to 3 h before harvesting.
Immunofluorescence microscopy.
Coverslips with attached cells were washed with PBS-0 and fixed with ice-cold methanol for 5 to 10 min or with 4% paraformaldehyde for 20 to 30 min. In the latter case, cells were permeabilized with 0.1% Triton X-100 for 5 min when proteins were to be detected within the cell. Permeabilization was omitted when surface-expressed proteins were to be detected. After fixation, cells were washed with PBS-0 and blocked for at least 30 min with PBS-0 containing 5% bovine serum albumin (BSA). Poly- or monoclonal antisera were diluted in PBS-0 containing 1% BSA and incubated for 1 h at room temperature. After several washes with PBS-0, goat or swine anti-rabbit or mouse immunoglobulin secondary antibodies conjugated to fluorescein isothiocyanate (FITC), tetramethylrhodamine isothiocyanate, or aminomethylcoumarin acid (AMCA) were incubated with the cells for 45 to 60 min at room temperature. Procedures were repeated for double labelings with a different antiserum and fluorescent probe, and at the end of the procedure the slides were washed with PBS-0 overnight. Direct labeling of ER and Golgi membranes was performed using the lectins concanavalin A and wheat germ agglutinin (WGA), respectively, coupled to AMCA or FITC (both from Molecular Probes). ER membranes were also stained using the chemical DiOC6 (Molecular Probes). Preparations were examined and photographed in a Leitz fluorescence microscope.
Western blotting.
Transfected BHK cells were harvested by trypsin treatment from 80-cm2 tissue culture flasks, mostly at 21 h after transfection. Cells were pelleted by centrifugation at 900 rpm for 5 min and washed once or twice with PBS-0 to remove traces of medium. The pellet was resuspended in PBS-0 with a protease inhibitor cocktail (Complete; Boehringer, Mannheim, Germany), sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (22) was added, and the samples were boiled for 3 to 10 min. Samples were frozen at −20°C for later use, or 20 μl was immediately applied to a 10% polyacrylamide gel. After electrophoresis, the gel was blotted onto an Immobilon polyvinylidene difluoride membrane (Millipore) using a semidry blotter (Bio-Rad).
Immunodetection.
Immunoblot analysis using alkaline phosphatase detection was carried out as described by Kikkert et al. (17).
RESULTS
Expressed wild-type TSWV glycoprotein precursor is processed and transported to and retained in the Golgi system.
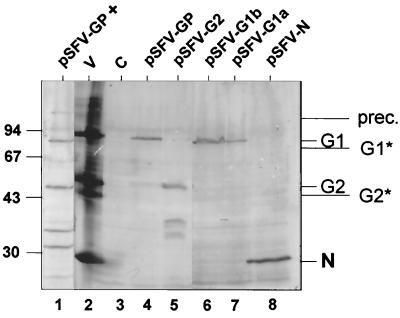
Expression, processing, and intracellular targeting of G1 and G2 in BHK cells containing RNA transcripts from pSFV-GP (Fig. 1) were analyzed by Western blotting and by indirect immunofluorescence. Western blots (Fig. 2) indicated efficient cleavage between G1 and G2, since no glycoprotein precursor molecules (∼127 kDa) were detected (Fig. 2, lane 4). The G1 and G2 glycoproteins both comigrated with the ones from purified TSWV particles isolated from Nicotiana rustica plants (Fig. 2, lanes 2 and 4 to 6), suggesting that the G1/G2 glycoprotein precursor is cleaved in a similar manner in animal and plant cells.
FIG. 2.
Western blots using antiserum against TSWV particles, showing the expression (21 h.p.t.) of the TSWV glycoproteins (pSFV-GP), G2 alone (pSFV-G2), G1 alone (pSFV-G1a and pSFV-G1b), and N (pSFV-N) in BHK cells. Expression of the TSWV glycoproteins in the presence of tunicamycin is shown in lane pSFV-GP + G1* and G2* indicate unglycosylated forms of G1 and G2, and prec. is the glycoprotein precursor protein. The positions of size markers are indicated on the left (in kilodaltons). V, purified TSWV particles; C, control (mock-transfected) cells.
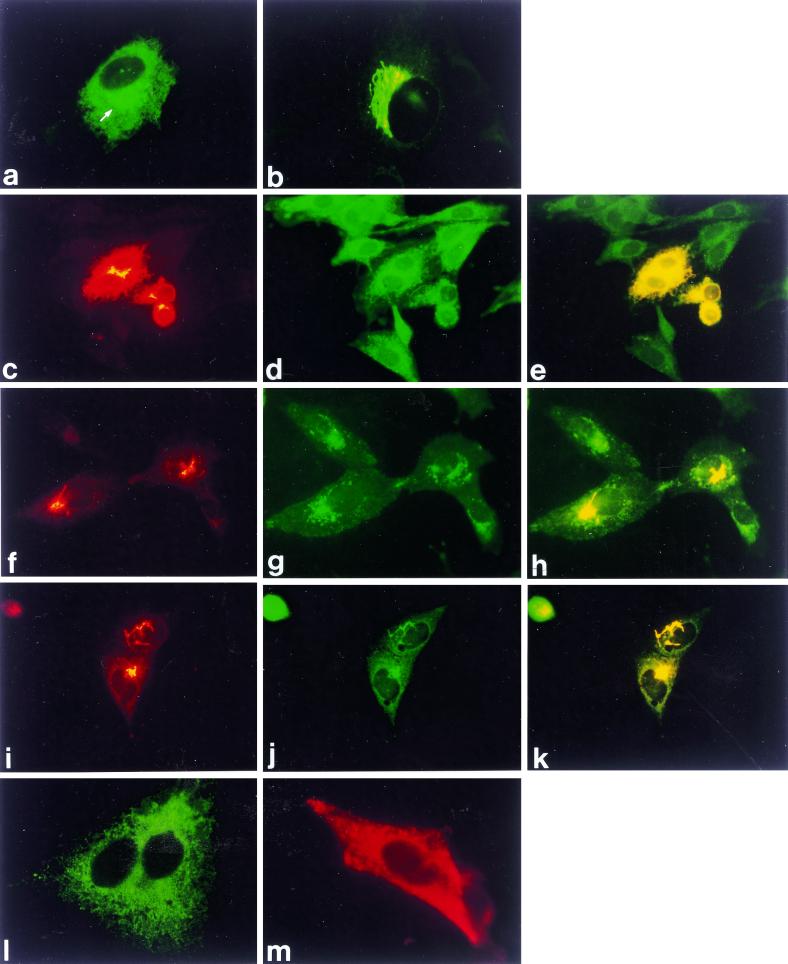
The subcellular location of G1 and G2 in BHK cells was determined using polyclonal antiserum raised against intact TSWV particles (containing G1/G2 glycoproteins) or against the separately E. coli-expressed glycoproteins, by means of indirect immunofluorescence microscopy. Identical results were obtained using the three different sera, indicating that G1 and G2 colocalize and are probably closely associated during transport (Fig. 3; only data for anti-TSWV are shown). At 6 posttransfection (h.p.t.), a typical picture was observed, predominated by an extensive reticular pattern covering virtually the whole cell, combined with a significant perinuclear signal (Fig. 3a). Upon treatment with cycloheximide (data not shown) or after prolonged incubation times (21 h.p.t.) (Fig. 3b), the reticular signal largely vanished and only the perinuclear signal remained. Using the ER markers concanavalin A (not shown) and DiOC6 (Fig. 3c, d, and e), the reticular signal could be identified as ER, while the Golgi marker WGA identified the perinuclear signal to represent the Golgi system (Fig. 3f, g, and h). The anti-p58 Golgi stack marker confirmed this localization (Fig. 3i, j, and k). The intermediate compartment marker anti-p58 gave primarily a punctate pattern throughout the cell, which showed no clear colocalization with perinuclear G1 and G2 expressed from the precursor (data not shown). These results strongly suggest that the TSWV glycoproteins are transported from the ER to the Golgi system and are then retained in the Golgi stacks. Upon labeling of the surface of G1/G2-expressing cells, no signal was detected (data not shown), suggesting that G1 and G2 are not transported to the cell surface in BHK cells.
FIG. 3.
Immunofluorescence analysis of G1 and G2 expression from transcripts from pSFV-GP (a to l) and of N expression from transcripts from pSFV-N (m). (a) At 6 h.p.t. with anti-TSWV; arrow indicates perinuclear signal. (b) At 21 h.p.t. with anti-TSWV. (c) At 6 h.p.t. with anti-TSWV. (d) Same cell treated with ER marker DiOC6. (e) Merge (double exposure) of c and d. (f) At 21 h.p.t. with anti-TSWV. (g) Same cell treated with Golgi marker WGA-FITC. (h) Merge of f and g. (i) At 21 h.p.t. with anti-TSWV. (j) Same cell treated with Golgi stack marker anti-p58. (k) Merge of i and j. (l) Expression in the presence of the glycosylation inhibitor tunicamycin at 21 h.p.t. with anti-TSWV. (m) At 21 h.p.t. with anti-N. Yellow areas in c, f, and i indicate overexposure of the film to clearly visualize additional (ER) localization elsewhere in the cell.
When the N-glycosylation inhibitor tunicamycin (37, 38) was added during expression of the precursor, the staining pattern of the G1/G2 proteins at 21 h.p.t. was reticular only (Fig. 31), indicating that tunicamycin affects the exit of G1 and G2 from the ER. As shown in Western blots (Fig. 2, lane 1), this was not caused by complete inhibition of precursor cleavage. Faster-migrating G1 and G2 proteins were produced in the presence of tunicamycin, presumably corresponding to the unglycosylated forms of the proteins. In addition, some uncleaved precursor protein was observed as well as some smaller immunoreactive polypeptides likely representing degradation products (Fig. 2, lane 1). These findings may indicate that the glycosylation of G1/G2 is important for proper transport out of the ER, for stability of the proteins, and apparently for efficient cleavage of the precursor protein in BHK cells.
As a control, transcripts from a construct containing a DNA copy of the nucleoprotein (N) gene were introduced into BHK cells, and, as expected, a 29-kDa protein was observed (Fig. 2, lane 8). Immunofluorescence showed a clustered staining pattern of nucleoprotein in the cytoplasm (Fig. 3m), as observed earlier in infection of protoplasts (17).
G2 can reach the Golgi complex on its own.
To investigate the trafficking and location of G1 and G2 separately, different deletion mutants of the precursor were produced encompassing either G2 or G1 sequences alone. As the precursor cleavage sites have not been mapped precisely, the C terminus of G2 and the N terminus of G1 are not known for TSWV. The pSFV-G2 construct (Fig. 1), meant to produce mature G2, included the hydrophobic domain contained within amino acids 428 through 484. Transfection of mRNA from construct pSFV-G2 produced a protein comigrating with the G2 species from virus particles and reacting with antiserum against G2 (not shown) and against purified virus (Fig. 2, lane 5). Also, some faster-migrating products were found in Western blot analysis, most probably representing degradation products of the protein. At 6 h.p.t., immunofluorescence analysis showed a reticular staining pattern in about 50% of the transfected cells (Fig. 4a), while in the remaining cells an additional perinuclear fluorescence was observed (Fig. 4b). The percentage of cells with perinuclear signal increased after prolonged (up to 21 h) incubation times. Costaining with ER and Golgi markers showed that the reticular signal represented again the ER and the perinuclear signal represented the Golgi complex (Fig. 4c to h). The results suggest that transport of G2 alone to the Golgi apparatus is occurring, albeit less efficiently than when coexpressed with G1 from the precursor, since a considerable part of G2 stayed in the ER. Addition of cycloheximide to the cells showed that much of the G2 signal was gradually lost during this treatment (not shown), suggesting degradation of G2 protein in the ER and the Golgi complex. Cell surface staining for G2 did not show an immunofluorescence signal in any of the G2 expression experiments (data not shown), indicating that G2 alone is not transported to the cell surface.
FIG. 4.
Immunofluorescence analysis of G2 expression from transcripts from pSFV-G2. (a) At 6 h.p.t. with anti-G2. (b) At 21 h.p.t. with anti-G2; arrows indicate perinuclear signal. (c) At 6 h.p.t. with anti-G2. (d) Same cells treated with ER marker DiOC6. (e) Merge (double exposure) of c and d. (f) At 21 h.p.t. with anti-G2. (g) Same cells treated with Golgi marker WGA-FITC. (h) Merge of f and g. Yellow areas in c and f indicate overexposure of the film as explained in the legend to Fig. 3.
G1 can be inserted into the ER membrane by its own signal sequence or that of the precursor, but is not transported to the Golgi.
Several different mutants encompassing G1 were produced, since the native N terminus of G1 is not precisely known, nor is it known whether G1 carries its own functional signal sequence or whether the signal sequence of the precursor is used to direct both G2 and G1 into the ER.
pSFV-G1a and pSFV-G1b (Fig. 1) contain the hydrophobic region between amino acid residues 428 and 484, which has been suggested to act as an internal signal sequence (18), while another set of constructs, pSFV-G1ss1, pSFV-G1ss2, and pSFV-G1ss3, contain the sequence for the signal peptide of the N terminus of the G2-G1 precursor (amino acids 1 to 35) attached in frame to the G1 ORF, i.e., lacking the sequence encoding the hydrophobic region of amino acids 428 to 484 (Fig. 1). The latter three constructs differed slightly in the junction area and were tested to obtain insight into the prerequisites for signal peptidase cleavage of these chimeras (Fig. 1).
Construct pSFV-G1a, which contained the putative internal signal sequence at its N terminus, produced a protein comigrating with G1 from virus particles, although expression was not very high (Fig. 2, lane 7). Construct pSFV-G1b produced a protein of exactly the same size as well (Fig. 2, lane 6), although it contained an extra hydrophilic sequence (residues 386 to 428) N terminal of the putative signal sequence. Expression of G1 in BHK cells from transcripts from pSFV-G1ss1 and pSFV-G1ss2 also resulted in a protein of the same size as G1 from purified TSWV (data not shown). Immunofluorescence analysis showed an ER staining for pSFV-G1a, pSFV-G1b, pSFV-G1ss1 (Fig. 5), and pSFV-G1ss2 (not shown). Even after prolonged incubation times, when ER staining could still be detected, no Golgi signal was observed for any of the G1 constructs. The pSFV-G1ss3 construct did not result in detectable protein levels in Western blots or immunofluorescence analysis (not shown).
FIG. 5.
Immunofluorescence analysis of G1 expression from transcripts from (a) pSFV-G1a, (d) pSFV-G1b, and (g) pSFV-G1ss1. All were taken at 21 h.p.t. with anti-G1. (b, e, and h) Corresponding cells treated with the DiOC6 ER marker. (c, f, and i) Merge (double exposures) of corresponding panels.
Coexpression of G1 and G2 from separate constructs results in rescue of transport to the Golgi system.
To test whether the separately translated G1 and G2 proteins were able to complement each other so that efficient trafficking of both glycoproteins to the Golgi complex would be restored, transcripts from pSFV-G2 were transfected together with transcripts from pSFV-G1b or pSFV-G1ss1. For immunofluorescence analysis of these cotransfections, a polyclonal antiserum against G2 was used in combination with a monoclonal antiserum against G1 (2B6; kindly provided by G. Adam, University of Hamburg, Hamburg, Germany). This anti-G1 monoclonal serum detects only Golgi-localized G1 and does not react with G1 localized in the ER (shown in Fig. 6a, b, and c). Cells that expressed both G1 (from either pSFV-G1b or pSFV-G1ss1) and G2 (from pSFV-G2) at 21 h.p.t. (Fig. 6d and g) gave a signal using the monoclonal against G1, whereas cells expressing only G1 (which is ER localized) did not (not shown). This indicates that G1 is transported to the Golgi when coexpressed with G2 (Fig. 6d to i). Detection of G1 in coexpressing cells using the polyclonal serum against G1 (which also detects ER-localized G1) showed little G1 staining associated with ER in these cells (data not shown), confirming efficient transport of G1 in the presence of G2.
FIG. 6.
(a to c) Expression of G1 and G2 from transcripts from pSFV-GP at 6 h.p.t. using antisera (a) 2B6 (monoclonal against Golgi-localized G1) and (b) anti-G2 (polyclonal). (c) Merge of a and b. (d to i) Immunofluorescence analysis of coexpression of pSFV-G2 with pSFV-G1b (d, e, and f) or pSFV-G1ss1 (g, h, and i) at 21 h.p.t. (d and g) 2B6 monoclonal against G1; (e and h) anti-G2; (f and i) merge (double exposures) of corresponding panels.
Cotransfection of G1 and G2 also rescued the impaired transport of G2 when expressed alone, as concluded from the reduced ER signal of G2 in coexpressing cells (Fig. 6e and h).
DISCUSSION
As a first step toward detailed understanding of the molecular processes underlying the maturation of TSWV glycoproteins and the subsequent assembly of virus particles in both plant and animal cells, we used the Semliki Forest virus expression system to study these glycoproteins in animal cells.
The results show that, as for the animal-infecting bunyaviruses, the TSWV glycoproteins themselves contain information necessary and sufficient for their transport to and retention in the Golgi system of mammalian cells. As was found for the animal-infecting bunyaviruses Uukuniemi virus (27, 32) and Bunyamwera virus (23), the N-terminal protein G2 of the TSWV glycoprotein precursor could be transported to the Golgi system on its own, though with decreased efficiency, and apparently contains a Golgi retention signal. The C-terminal protein G1 expressed on its own was unable to leave the ER and thus seems to require an interaction with G2 in order to be transported out of the ER. This was illustrated in cotransfections, where the interaction of G2 with G1 from separate RNAs resulted in the transport of both glycoproteins to the Golgi system.
G1 and G2 are glycoproteins, presumably acquiring one or two N-linked oligosaccharide side chains at the predicted sites in their luminal (i.e., N-terminal) domains (see Fig. 1). When glycosylation was inhibited by treatment with tunicamycin, precursor cleavage still occurred, though less efficiently (Fig. 2, lane 2), but the proteins were unable to leave the ER. Presumably, the absence of N-linked glycans results in aberrant folding of the proteins, which leads to hampered transport from the ER to the Golgi complex, as observed. The same was also found for Uukuniemi virus proteins lacking their N-linked glycans (20).
The TSWV glycoproteins apparently do not reach the plasma membrane, since they were undetectable by cell surface immunofluorescence staining. This indicates that the proteins are tightly retained in the Golgi complex. We cannot, however, rule out that a fraction of the proteins, too small to be detectable, escape to the plasma membrane but are continually retrieved to the Golgi complex, as has been demonstrated for some resident Golgi membrane proteins.
Expression of different TSWV G1 constructs in BHK cells suggests that the hydrophobic sequence encompassing amino acids 428 to 484 can function as a signal sequence for G1, since it is able to guide G1 into the ER. The same was found for the Gc glycoprotein of other bunyaviruses (reviewed in reference 30) and also for the glycoprotein E1 of Semliki Forest virus (13). TSWV G1 can be targeted to the ER by attachment of the precursor protein signal peptide to its sequence as well. A number of such constructs were produced (Fig. 1), since we were interested in the prerequisites for efficient cleavage of these chimeric molecules. Apparently, omitting a few of the N-terminal residues of G1, as in pSFV-G1ss2 (Fig. 1), did not affect the ER targeting. However, when the putative last residue of the signal sequence itself was missing, as in pSFV-G1ss3 (Fig. 1), no protein could be detected. Using the von Heijne algorithm (41) and a new algorithm based on a computer neural network (Jagla et al., personal communication of unpublished results), cleavage sites could indeed be predicted for all constructs produced except pSFV-G1ss3 (Fig. 1). This may account for aberrant targeting of G1 generated from transcripts obtained from pSFV-G1ss3 and result in an unstable protein product, as observed.
Further research is needed to map the region(s) in the TSWV G2 sequence that is necessary for its Golgi retention in mammalian cells. This issue has already been investigated for Punta Toro virus (26) and Uukuniemi virus (2). For these viruses, the signal was found to be located in the cytoplasmic tail of GN, close to the transmembrane anchor. However, no sequence homology was found for these and other Golgi retention signals, suggesting that Golgi retention signals in bunyavirus glycoproteins are based on the conformation of the protein rather than on a primary sequence motif.
Our results surprisingly indicate that the TSWV glycoproteins contain transport and retention characteristics that are functional in mammalian cells and resemble those of animal-infecting bunyavirus glycoproteins very closely. These observations could be interpreted as a strong confirmation of the putative evolution of an animal-infecting ancestral bunyavirus into the plant-infecting tospoviruses. However, it is unlikely that such detailed molecular features would be conserved if they did not have a function in the infection cycle of TSWV. Therefore, these features probably reflect the ability of TSWV to replicate in its animal thrips vector, in which the formation of particles may be very homologous to the process observed for other bunyaviruses in mammalian and insect vector cells. Furthermore, the literature increasingly indicates that cellular transport and retention signals are not only conserved among closely related organisms but are similar if not identical for all eukaryotes (reviewed in references 4 and 15). This would suggest that the molecular features underlying the behavior of TSWV glycoproteins in mammalian cells may also function during maturation in plant cells. In particular, accumulation in the Golgi system was also observed during TSWV infection of plant cells (16) and may thus be regulated by the same molecular signals as in mammalian cells. The differences in the subsequent formation of particles in plant and animal cells could be the result of extraregulatory signals acquired by the TSWV (glyco)proteins to meet the prerequisites of the plant host.
ACKNOWLEDGMENTS
We are extremely grateful to Gert-Jan Godeke of Utrecht University in The Netherlands for assistance with the SFV expression system. Christina Spiropoulou is thanked for sending us her Sigma Golgi marker anti 58K, and Agneta Andersson is thanked for sending us the IC marker anti 58K. Guenter Adam of the University of Hamburg sent us the monoclonal 2B6 against G1, for which we are very grateful. We furthermore thank Cecile van Woensel and Bart Hesselink for excellent technical assistance and Claire Pacot-Hiriart for preparing useful constructs in preparation for this work.
REFERENCES
- 1.Adam G, Peters D, Goldbach R W. Serological comparison of tospovirus isolates using polyclonal and monoclonal antibodies: Proceedings of the International Symposium on Tospoviruses and Thrips of Floral and Vegetable Crops. Acta Hortic (Wageningen) 1996;431:135–158. [Google Scholar]
- 2.Andersson A M, Melin L, Bean A, Pettersson R F. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J Virol. 1997;71:4717–4727. doi: 10.1128/jvi.71.6.4717-4727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson A M, Melin L, Persson R, Rashperger E, Wikström L, Pettersson R F. Processing and membrane topology of the spike proteins G1 and G2 of Uukuniemi virus. J Virol. 1997;71:218–225. doi: 10.1128/jvi.71.1.218-225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Peled M, Bassham D C, Raikhel N V. Transport of proteins in eucaryotic cells: more questions ahead. Plant Mol Biol. 1996;32:223–249. doi: 10.1007/BF00039384. [DOI] [PubMed] [Google Scholar]
- 5.De Ávila A C, de Haan P, Smeets M L L, de O. Resende R, Kormelink R, Kitajima E W, Goldbach R W, Peters D. Distinct levels of relationships between tospovirus isolates. Arch Virol. 1993;128:211–227. doi: 10.1007/BF01309435. [DOI] [PubMed] [Google Scholar]
- 6.Elliot R M. Molecular biology of the Bunyaviridae. J Gen Virol. 1990;71:501–522. doi: 10.1099/0022-1317-71-3-501. [DOI] [PubMed] [Google Scholar]
- 7.Elliot R M, editor. The bunyaviridae. New York, N.Y: Plenum Press; 1996. [Google Scholar]
- 8.Francki R I B, Fauquet C M, Knudson D L, Brown F. Classification and nomenclature of viruses: fifth report of the International Committee on Taxonomy of Viruses. Archives of Virology, Supplementum 2. Vienna, Austria: Springer-Verlag; 1991. [Google Scholar]
- 9.German T L, Ullman D E, Moyer J W. Tospoviruses: diagnosis, molecular biology, phylogeny and vector relationships. Annu Rev Phytopathology. 1992;30:315–348. doi: 10.1146/annurev.py.30.090192.001531. [DOI] [PubMed] [Google Scholar]
- 10.Goldbach R, Kormelink R, de Haan P, de O. Resende R, de Ávila A, van Poelwijk F, van Lent J, Wijkamp I, Prins M, Peters D. Tomato spotted wilt virus: genome organization, transmission and symptom induction. In: Bills D D, Kung S-D, editors. Biotechnology and plant protection: viral pathogenesis and disease resistance. River Edge, N.J: World Scientific; 1995. pp. 297–311. [Google Scholar]
- 11.Goldbach R, Peters D. Molecular and biological aspects of tospoviruses. In: Elliott R M, editor. The bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 129–157. [Google Scholar]
- 12.Griffiths G, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto K, Erdei S, Keränen S, Saraste J, Kääriäinen L. Evidence for a separate signal sequence for the carboxy-terminal envelope glycoprotein E1 of Semliki Forest virus. J Virol. 1981;38:34–40. doi: 10.1128/jvi.38.1.34-40.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ie T S. A sap transmissable, defective form of tomato spotted wilt virus. J Gen Virol. 1982;59:387–392. [Google Scholar]
- 15.Kermode A R. Mechanisms of intracellular protein transport and targetting in plant cells. Crit Rev Plant Sci. 1996;15:285–423. [Google Scholar]
- 16.Kikkert M, van Lent J, Storms M, Bodegom P, Kormelink R, Goldbach R. Tomato spotted wilt virus particle morphogenesis in plant cells. J Virol. 1999;73:2288–2297. doi: 10.1128/jvi.73.3.2288-2297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikkert M, Van Poelwijk F, Storms M, Kassies W, Bloksma H, van Lent J, Kormelink R, Goldbach R. A protoplast system for studying tomato spotted wilt virus infection. J Gen Virol. 1997;78:755–763. doi: 10.1099/0022-1317-78-7-1755. [DOI] [PubMed] [Google Scholar]
- 18.Kormelink R, de Haan P, Meurs C, Peters D, Goldbach R. The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J Gen Virol. 1992;73:2795–2804. doi: 10.1099/0022-1317-73-11-2795. [DOI] [PubMed] [Google Scholar]
- 19.Kormelink R, Storms M, van Lent J, Peters D, Goldbach R. Expression and subcellular localization of the NSm protein of tomato spotted wilt virus (TSWV), a putative movement protein. Virology. 1994;200:56–65. doi: 10.1006/viro.1994.1162. [DOI] [PubMed] [Google Scholar]
- 20.Kuismanen E, Bång B, Hurme M, Pettersson R F. Uukuniemi virus maturation: immunofluorescence microscopy with monoclonal glycoprotein-specific antibodies. J Virol. 1984;51:137–146. doi: 10.1128/jvi.51.1.137-146.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuismanen E, Hedman K, Saraste J, Pettersson R F. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol Cell Biol. 1982;2:1444–1458. doi: 10.1128/mcb.2.11.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lapin D F, Nakitare G W, Palfreyman J W, Elliott R M. Localization of bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J Gen Virol. 1994;75:3441–3451. doi: 10.1099/0022-1317-75-12-3441. [DOI] [PubMed] [Google Scholar]
- 24.Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka Y, Chen S Y, Compans R W. Bunyavirus protein transport and assembly. Curr Top Microbiol Immunol. 1991;169:161–180. doi: 10.1007/978-3-642-76018-1_6. [DOI] [PubMed] [Google Scholar]
- 26.Matsuoka Y, Chen S Y, Holland C E, Compans R W. Molecular determinants of Golgi retention in the Punta Toro G1 protein. Arch Biochem Biophys. 1996;336:184–189. doi: 10.1006/abbi.1996.0547. [DOI] [PubMed] [Google Scholar]
- 27.Melin L, Persson R, Andersson A, Bergström A, Rönnholm R, Pettersson R. The membrane glycoprotein G1 of Uukuniemi virus contains a signal for localization to the Golgi complex. Virus Res. 1995;36:49–66. doi: 10.1016/0168-1702(95)00006-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mumford R A, Barker I, Wood K R. The biology of the tospoviruses. Ann Appl Biol. 1996;128:159–183. [Google Scholar]
- 29.Petterson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–104. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 30.Pettersson R F, Melin L. Synthesis, assembly, and intracellular transport of Bunyaviridae membrane proteins. In: Elliott R M, editor. The bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 159–188. [Google Scholar]
- 31.Resende R de O, de Haan P, de Avila A C, Kitajima E W, Koermelink R, Goldbach R, Peters D. Generation of envelope and defective RNA mutants of tomato spotted wilt virus by mechanical passage. J Gen Virol. 1991;72:2375–2383. doi: 10.1099/0022-1317-72-10-2375. [DOI] [PubMed] [Google Scholar]
- 32.Rönnholm R. Localization to the Golgi complex of Uukuniemi virus glycoproteins G1 and G2 expressed from cloned cDNAs. J Virol. 1992;66:4525–4531. doi: 10.1128/jvi.66.7.4525-4531.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saraste J, Palade G E, Farquhar M G. Antibodies to rat pancreas Golgi subfractions: identification of a 58 kD cis-Golgi complex. Semin Cell Biol. 1987;105:2021–3030. doi: 10.1083/jcb.105.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saraste J, Svensson K. Distribution of intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- 35.Stephens E B, Compans R W. Assembly of animal viruses at cellular membranes. Annu Rev Microbiol. 1988;42:489–516. doi: 10.1146/annurev.mi.42.100188.002421. [DOI] [PubMed] [Google Scholar]
- 36.Storms M, Kormelink R, Peters D, van Lent J, Goldbach R. The non-structural protein NSm of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology. 1995;214:485–493. doi: 10.1006/viro.1995.0059. [DOI] [PubMed] [Google Scholar]
- 37.Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot. 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 38.Tkacz J S, Lampen J O. Tunicamycin inhibition of polyisoprenol N-acetylglucosaminyl pyrophosphate formation in calf liver microsomes. Biochem Biophys Res Commun. 1975;65:248–257. doi: 10.1016/s0006-291x(75)80086-6. [DOI] [PubMed] [Google Scholar]
- 39.Ullman D E, German T L, Sherwood J L, Westcot D M, Cantone F E. Tospovirus replication in insect vector cells: immuno-cytochemical evidence that the non structural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology. 1993;83:456–463. [Google Scholar]
- 40.Verkleij F N, Peters D. Characterization of a defective form of tomato spotted wilt virus. J Gen Virol. 1983;64:677–682. [Google Scholar]
- 41.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wijkamp I. Virus-vector relationships in the transmission of tospoviruses. Ph.D. thesis. Wageningen, The Netherlands: Wageningen University; 1996. [Google Scholar]
- 43.Wijkamp I, van Lent J, Kormelink R, Goldbach R, Peters D. Multiplication of tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J Gen Virol. 1993;74:341–349. doi: 10.1099/0022-1317-74-3-341. [DOI] [PubMed] [Google Scholar]