Abstract
Using HLA-peptide tetrameric complexes, we isolated human T-cell lymphotrophic virus type 1 Tax peptide-specific CD8+ T cells ex vivo. Antigen-specific amino acid motifs were identified in the T-cell receptor Vβ CDR3 region of clonally expanded CD8+ T cells. This result directly confirms the importance of the CDR3 region in determining the antigen specificity in vivo.
Human T-cell lymphotropic virus type 1 (HTLV-1) (23, 26) infection is closely associated with a slowly progressive neurologic disease called HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (10, 21). Only a minority of HTLV-1-infected individuals develop HAM/TSP, by mechanisms that are incompletely understood (14). In HTLV-1-infected individuals carrying the HLA-A2 allele, the HLA-A2-restricted CD8+ T-cell response is primarily directed to the Tax11-19 peptide (LLFGYPVYV) (22). Since Tax11-19-specific CD8+ T cells have the potential to produce proinflammatory cytokines (15) whereas possession of the HLA-A2 allele was associated with protection against HAM/TSP as well as a lower proviral load (13), it remains unclear whether antigen-specific CD8+ T cells contribute to the inflammatory and demyelinating processes of HAM/TSP or whether the dominant effect of such cells in vivo is protective against disease (13).
The great diversity in the T-cell response results from the large number of different V, D, and J elements in the germ line and additional clone-specific diversity at the V-D and D-J junctions (β-chain) or the V-J junction (α-chain) (9). It is well established that the amino acid sequence within the Vβ CDR3 region is critical for antigen recognition (3, 9). However, these findings are based upon T-cell clones grown in vitro; thus, inadvertent in vitro selection could not be excluded. To overcome this problem, we have exploited tetrameric major histocompatibility complex (MHC)-peptide complexes (1, 20) along with magnetic cell sorting to purify HTLV-1 Tax11-19-specific T cells directly from HLA-A2-positive HTLV-1-infected individuals, in order to characterize immunodominant Tax11-19-specific CD8+ T lymphocytes directly from HAM/TSP patients and asymptomatic carriers (AC) (12, 13). Specific binding of the HLA-A2/Tax11-19 tetramer has been previously demonstrated; thus, there was no detectable staining of peripheral blood mononuclear cells (PBMC) from HLA-A2-positive or HLA-A2-negative healthy subjects or HLA-A2-negative HAM/TSP patients (12, 13). We also confirmed that all of these HLA-A2/Tax11-19 tetramer-positive cells were CD8+ positive (data not shown). Class I MHC tetramer-binding cells can show a range of functions, including cytotoxicity and cytokine production (20). Furthermore, the CD8+ T-cell response to a single peptide (Tax11-19) might not be representative of the host's T-cell response to the virus. However, the observation that the frequency of HLA-A2–Tax11-19 tetramer-positive CD8+ T cells correlated negatively with the percentage of CD4+ cells in infected individuals is consistent with the proposal that a significant proportion of these tetramer-binding CD8+ cells are cytotoxic in vivo (12).
Fresh PBMC from Afro-Caribbean United Kingdom residents (HAM1, HAM2, AC1, and AC2) were obtained by Histopaque-1077 (Sigma) density gradient centrifugation, washed twice in RPMI 1640 with 10% fetal calf serum and resuspended in phosphate-buffered saline with 10% fetal calf serum. Two Japanese PBMC samples (HAM3 and HAM4) were stored in liquid nitrogen until use. All subjects carried the HLA-A*0201 allele, defined by PCR as previously described (13). Positive selection for CD8+ T cells was done by incubating the PBMC with anti-CD8 MACS beads (Miltenyi Biotec Ltd., Bisley, Surrey, United Kingdom) for 15 min at 4°C. Tax11-19-specific cells were positively selected using antiphycoerythrin MACS beads (Miltenyi Biotec Ltd.) for 15 min at 4°C following phycoerythrin-conjugated HLA-A*0201/Tax11-19 tetramer staining for 25 min at 37°C. The purities of tetramer-positive cells and CD8+ T cells were greater than 95 and 98%, respectively, by flow cytometric analysis (data not shown). First-strand cDNA was generated from 105 enriched CD8+ and HLA-A*0201/Tax11-19 tetramer-positive T cells with a High Pure mRNA extraction kit (Boehringer Mannheim, Mannheim, Germany) and a first-strand cDNA synthesis kit (Boehringer Mannheim) in a total volume of 42 μl. One microliter of the first-strand cDNA was subjected to 35 cycles of reverse transcription-PCR (RT-PCR) in which the reaction mixtures contained 20 pmol of one of a panel of 24 T-cell receptor (TCR) Vβ-specific primers and 20 pmol of a reverse primer specific for the TCR β constant region, of which 3 pmol had been end labeled with 6-carboxyfluorescein (6-FAM; PE Applied Biosystems). The sequences of the specific primers were as previously described (5). Preliminary experiments for quantification of the Vβ RT-PCR indicated that 35-cycle amplification is in the exponential phase of amplification for all Vβ transcripts. The semiquantitative PCR results were expressed as follows: percent Vβ = 100 × [(intensity of a Vβ-specific band)/(sum of intensity of all Vβ-specific bands)].
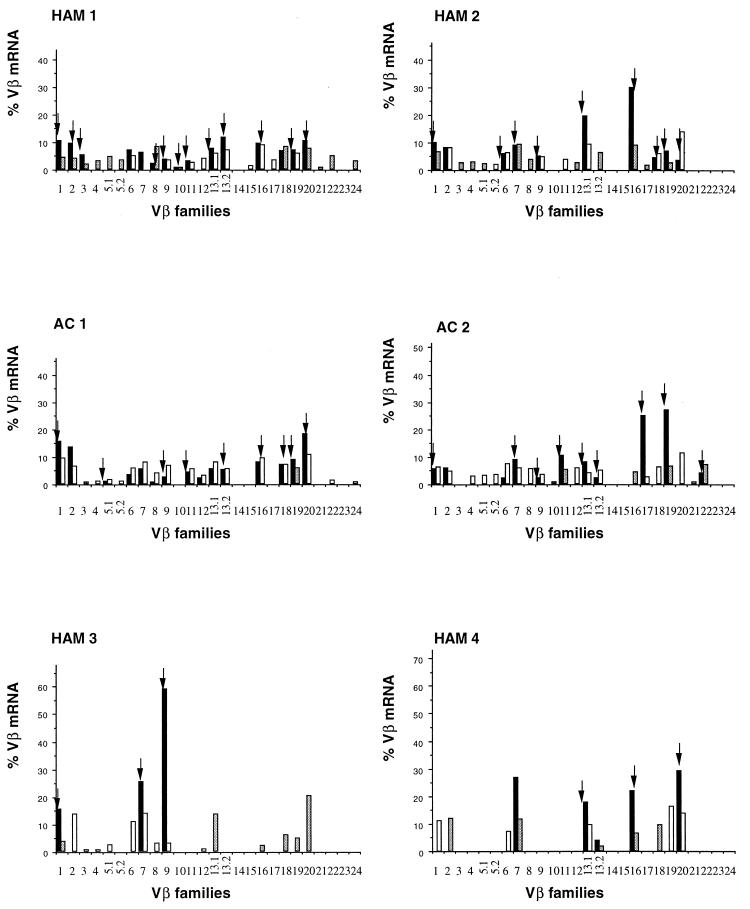
The TCR gene usage of HLA-A2–Tax11-19 tetramer-positive T cells and CD8+ T cells from all subjects is shown in Fig. 1. The T-cell repertoire of HLA-A2–Tax11-19 tetramer-positive T cells is composed of a diverse set of T-cell receptors, in contrast to that of previously reported cultured Tax-specific cytotoxic T lymphocytes (CTLs) (7, 24). The apparently lower diversity of Vβ usage in frozen and thawed samples from two patients (HAM3 and HAM4) may be due to the fact that mRNA recovery was not as efficient as that from fresh samples. As shown in Fig. 1, in freshly isolated Tax11-19-specific cDNA, we detected 10 to 15 different TCR Vβ bands in each patient by using 26 Vβ-specific primer pairs. In contrast to these results, previous workers reported that cultured and cloned anti-Tax CTLs expressed a very limited number of TCR V gene families (7, 24). TCR diversity might be reduced by in vitro selection, for example, for rapidly growing T-cell clones or for clones that resist activation-induced cell death (19). It is possible that certain TCR Vβ bands were amplified from contaminating non-Tax11-19-specific T cells. However, the unusually high frequency of oligoclonal proliferation (10 to 58%) (Table 1) and the selection of a single CDR3 length variant by the tetramer from a minority population in PBMC (Fig. 2b, lower panel) suggest that such contamination played a minor role. Furthermore, our results are consistent with those of Bieganowska et al. (2), who found that Tax11-19–HLA-A2 tetramer-binding cells bound 10 out of 20 anti-TCR Vβ monoclonal antibodies.
FIG. 1.
RT-PCR analysis of expression of TCR Vβ transcripts in CD8+ T cells and Tax11-19 tetramer-positive cells from HTLV-1-infected individuals. RT-PCR products were separated on a 1% agarose gel and visualized with ethidium bromide. The relative amounts of Vβ transcripts in CD8+ T cells (white bars) and Tax11-19 tetramer-positive cells (black bars) were calculated with Genescan software. Hatched bars indicate CD8+ T cells with a mono- or oligoclonal spectratype; arrows indicate Tax11-19 tetramer-positive cells with a mono- or oligoclonal spectratype.
TABLE 1.
TCR Vβ oligoclonality in the CD8+ T cells from HTLV-1-infected individualsa
| Patient | No. of:
|
Frequency of oligoclonal Vβ (%) | |
|---|---|---|---|
| Oligoclonal Vβ segments | Positive Vβ segments | ||
| HAM1 | 12 | 24 | 50.0 |
| HAM2 | 11 | 19 | 57.9 |
| HAM3 | 6 | 15 | 40.0 |
| HAM4 | 5 | 10 | 50.0 |
| AC1 | 2 | 20 | 10.0 |
| AC2 | 5 | 22 | 22.7 |
CD8+ samples that showed marked expansions of CDR3 segments of a certain length within the given Vβ were defined as oligoclonal by spectratyping analysis when a single peak contained >50% of the total area under the spectratype curve for that Vβ family.
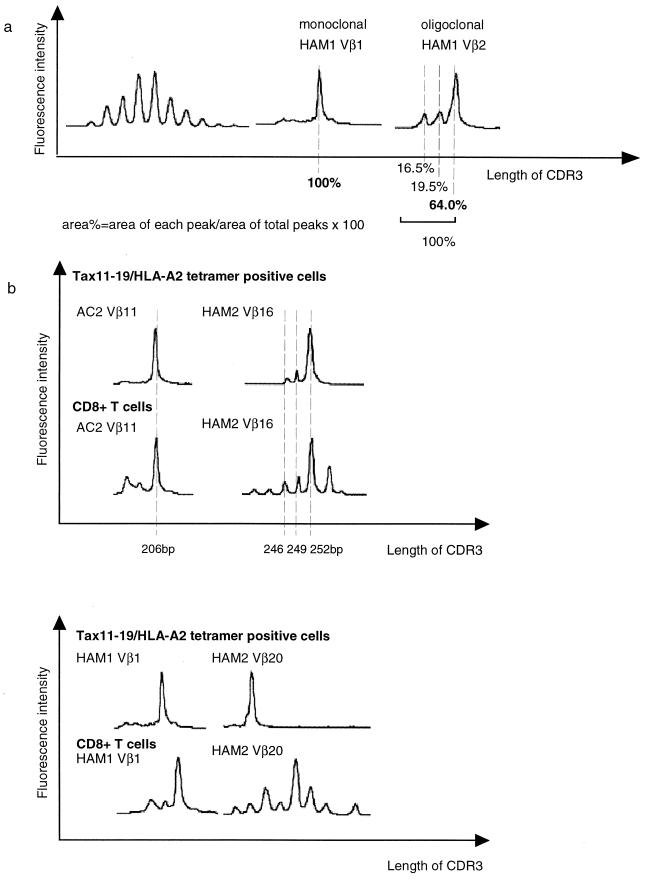
FIG. 2.
CDR3 length profiles for TCR transcripts in Tax11-19 tetramer-positive cells and CD8+ T cells from HTLV-1-infected individuals. TCR Vβ transcripts were reverse transcribed and amplified using Vβ- and Cβ-specific primers. A 6-FAM dye-labeled Cβ-specific primer was used to visualize the amplified products. CDR3 length is in base pairs of Vβ-Cβ elongation reaction products. (a) (Left) Typical spectratype profile of Gaussian distribution found in CD8+ T cells. Each Vβ generally had five to seven peaks with one or two dense bands in the middle portion. (Center and right) Monoclonal and oligoclonal spectratypes observed in CD8+ T cells. There are marked expansions of CDR3 segments of a certain length within the given Vβ. We defined such skewed spectratype bands as oligoclonal when >50% of the area within a V segment or family was calculated for the highest peak band. (b) Profiles of the Vβ11 spectratypes of AC2, Vβ16 of HAM2, Vβ1 of HAM1, and Vβ20 of HAM2. Fluorescence intensities were calculated with Genescan software (reflecting the number of clones with a particular CDR3 length).
On the other hand, no restriction to certain Vβ gene segments has been described for TCR usage of cultured and cloned Tax peptide-specific CTLs (7, 24). This diverse TCR Vβ repertoire might result either from HLA-peptide-driven clonal selection or from antigen-independent, TCR-independent HTLV-1 infection-induced expansion of T-cell clones without particular Vβ gene restriction. To rule out this possibility, we investigated the clonality and sequence diversity of each different TCR Vβ band derived from RT-PCR.
First, we carried out CDR3 size spectratyping of each TCR Vβ PCR product as described previously (11). Two microliters of the final PCR mixture was electrophoresed through a 5% polyacrylamide sequencing gel, and the resulting bands were quantified by fluorescence detection on an automated sequencer (model 377A; PE Applied Biosystems) using Genescan software (PE Applied Biosystems). In CD8+ T cells, each Vβ generally had five to seven length variants with one or two dense bands in the middle of the spectratype, consistent with a Gaussian distribution (Fig. 2a, left panel). But in some HLA-A2–Tax11-19 tetramer-positive T cells and CD8+ T cells, there were marked expansions of CDR3 segments of a certain length within the given Vβ (Vβ1 and Vβ2 spectratypes of HAM1). We defined such skewed spectratype bands as “oligoclonal” when a single peak contained >50% of the total area under the spectratype curve for that Vβ family (Fig. 2a, right panel). Hatched bars shown in Fig. 1 indicate the TCR Vβ segments that were found to be oligoclonal, and Table 1 summarizes the number of oligoclonal CD8+ T cells. Clonal expansion of CD8+ T cells was frequent and widespread across Vβ families, in both asymptomatic carriers and patients with HAM/TSP, consistent with previous observations (5). This suggests that these cells have recently encountered the viral antigen in vivo, in the course of chronic infection. The frequency of oligoclonal spectratypes was significantly higher in HAM/TSP patients than in asymptomatic carriers (P = 0.0079, Student's t test). This may be due to the higher antigen load in HAM/TSP patients.
The length of the expanded CD8+ Vβ11 CDR3 in AC2 was exactly the same as the Vβ11 single peak of Tax11-19-specific cells on the same gel. This was also observed in the Vβ16 spectratype of HAM2 (Fig. 2b, upper panel). On the other hand, in the Vβ1 spectratype of HAM1 and the Vβ20 spectratype of HAM2, a minor peak of CDR3 length in circulating CD8+ T cells was expanded in Tax11-19-specific T cells (Fig. 2b, lower panel). These findings indicate that the expanded Tax11-19-specific T cells account for a high proportion of certain Vβ families in the circulating CD8+ T-cell repertoire.
Next, to determine whether these clonally expanded CD8+ T cells in mixed PBMC were identical with the HLA-A2–Tax11-19 tetramer-binding cells, we subcloned and sequenced both CD8-positive- and tetramer-positive-cell-derived RT-PCR products from two infected individuals (Vβ11 of AC2 and Vβ16 of HAM2) (Fig. 2b, upper panel). As shown in Table 2, the major clonotypes seen in the CD8+ T cells were exactly the same as those seen in Tax11-19 tetramer-positive cells in both infected individuals. Moreover, the size of the dominant CDR3 in Table 2 corresponded to the length of the major spectratype peaks shown in Fig. 3b. These data indicate that some in vivo clonally expanded CD8+ T cells in HTLV-1-infected individuals are indeed HTLV-1 Tax11-19 specific. We also sequenced the Vβ chain CDR3 regions (between Vβ and Jβ) of the HLA-A2–Tax11-19-specific T cells which showed a discrete spectratype peak. Most samples that showed a single peak spectratype had identical sequence at the nucleotide level throughout its CDR3, reflecting clonal expansion (Table 3).
TABLE 2.
CDR3 amino acid sequences of HLA-A2–Tax11-19 tetramer-positive cells and oligoclonally proliferated CD8+ T cells with the same Vβa
| Subject and cell type | Vβ gene segment | Vβ | N-D-N | Jβ | Cβ | No. of sequences/totalb |
|---|---|---|---|---|---|---|
| HAM2 | ||||||
| Tetramer+ | Vβ16 | YFCASS | QEAVSM | NYGYTFGSGTRLTVV (JB1.2) | EDLNK | 8/11 |
| YFCASS | QEKDM | NTEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 3/11 | ||
| CD8+ | Vβ16 | YFCASS | QEAVSM | NYGYTFGSGTRLTVV (Jβ1.2) | EDLNK | 12/37 |
| YFCASS | QEKDM | NTEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 6/37 | ||
| YFCASS | QDWRTDLN | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 4/37 | ||
| YFCAS | TIYTGIGQG | TEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 3/37 | ||
| YFCASS | QETGF | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 3/37 | ||
| YFCASS | HLAQGAYP | GNTIYFGEGTWLTVV (Jβ1.3) | EDLNK | 2/37 | ||
| YFCASS | LGRG | YNEQFFGPGTRLTVL (Jβ2.1) | EDLKN | 1/37 | ||
| YFCASS | LAGV | EQFFGPGTRLTVL (Jβ2.1) | EDLKN | 1/37 | ||
| YFCASS | QVSGA | KLFFGSGTQLSVL (Jβ1.4) | EDLNK | 1/37 | ||
| YFCASS | QVP | GELFFGEGSRLTVL (Jβ2.2) | EDLKN | 1/37 | ||
| YFCASS | QGRGTGLK | NEKLFFGSGTQLSVL (Jβ1.4) | EDLNK | 1/37 | ||
| YFCASS | QETSS | DTQYFGPGTRLTVL (Jβ2.3) | EDLKN | 1/37 | ||
| YFCASS | LSY | SNQPQHFGDGTRLSIL (Jβ1.5) | EDLNK | 1/37 | ||
| AC2 | ||||||
| Tetramer+ | Vβ11 | YLCASS | GEDSTF | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 17/17 |
| CD8+ | Vβ11 | YLCASS | GEDSTF | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 21/25 |
| YLCASS | EYGG | TGELFFGEGSRLTVL (Jβ2.2) | EDLKN | 1/25 | ||
| YLCAS | TQTGIA | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 1/25 | ||
| YLCASS | GTGEVA | PLHFGNGTRLTVT (Jβ1.6) | EDLNK | 1/25 | ||
| YLCASS | EPGEW | NEQFFGPGTRLTVL (Jβ2.1) | EDLKN | 1/25 |
Bold type indicates CDR3 sequence of oligoclonally proliferated CD8+ T cells.
Number of indicated CDR3 sequences derived from bacterial colonies containing PCR-amplified DNA per total number of colonies sequenced.
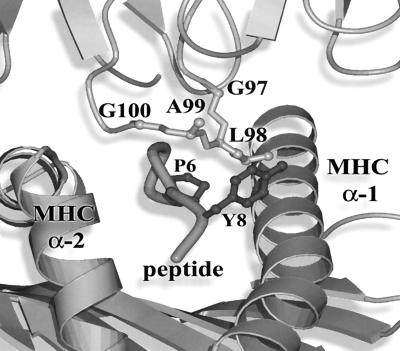
FIG. 3.
Crystal structure illustration of the A6 TCR HLA-A2–Tax11-19. One corner of the apex of the Vβ CDR3 loop (GLAG) inserts into a hydrophobic pocket formed by the alpha-1 helix of HLA-A2 and the side chain of the Tyr residue at position 8 in the Tax 11-19 peptide. The hydrophobic Leu residue at position 98 on the TCR Vβ loop makes several strong interactions with both the Tax peptide and HLA-A2. Leu is strongly favored (34 of 38 clones) at this position in the Vβ13.1-containing clonally expanded CD8+ T cells that bind the HLA-A2–Tax11-19 tetramer. The illustration was created by using MolScript (6) and Raster3D (18).
TABLE 3.
Vβ CDR3 sequences of TCR amplified from A2 tetramer-positive cells
| Subject | Vβ gene segment | Vβ | N-D-Na | Jβ | Cβ | No. of sequences/totalb |
|---|---|---|---|---|---|---|
| HAM1 | Vβ1 | YFCASS | VSDTT | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 15/15 |
| HAM3 | Vβ1 | YFCASS | VSDTT | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 15/15 |
| AC2 | Vβ1 | YFCASS | PSGIAY | TGELFFGEGSRLTVL (Jβ2.2) | EDLKN | 11/11 |
| HAM2 | Vβ13.1 | YFCASS | HPLAGA | NEQFFGPGTRLTVL (Jβ2.1) | EDLKN | 12/15 |
| YFCASS | SPGTGE | ETQYFGPGTRLLVL (Jβ2.5) | EDLKN | 2/15 | ||
| YFCASS | HPLAGV | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 1/15 | ||
| HAM4 | Vβ13.1 | YFCAS | NTGLAGVG | EQFFGPGTRLTVL (Jβ2.1) | EDLKN | 8/12 |
| YFCASS | WGLRGPP | GYTFGSGTRLTVV (Jβ1.2) | EDLNK | 4/12 | ||
| AC2 | Vβ13.1 | YFCASS | PGLAG | TGELFFGEGSRLTVL (Jβ2.2) | EDLKN | 4/11 |
| YFCAS | RPGLAGAKD | EQYFGPGTRLTVT (Jβ2.7) | EDLKN | 2/11 | ||
| YFCASS | YPLAGV | NEQFFGPGTRLTVL (Jβ2.1) | EDLKN | 2/11 | ||
| YFCA | GRQDH | NSPLHFGNGTRLTVT (Jβ1.6) | EDLNK | 2/11 | ||
| YFCAS | RPGLAGGVD | EQFFGPGTRLTVL (Jβ2.1) | EDLKN | 1/11 | ||
| HAM1 | Vβ16 | YFCASS | QDPLLQGAR | TEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 11/11 |
| HAM2 | Vβ16 | YFCASS | QEAVSM | NYGYTFGSGTRLTVV (Jβ1.2) | EDLNK | 8/11 |
| YFCASS | QEKDM | NTEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 3/11 | ||
| AC1 | Vβ16 | YFCASS | QDFAGNLN | NEQFFGPGTRLTVL (Jβ2.1) | EDLKN | 14/15 |
| YFCASS | HAQGG | YNSPLHFGNGTRLTVT (Jβ1.6) | EDLNK | 1/15 | ||
| HAM1 | Vβ20 | YLCAW | SRGG | TEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 10/13 |
| YLCAW | REGTFG | NQPQHFGDGTRLSIL (Jβ1.5) | EDLNK | 3/13 | ||
| HAM2 | Vβ20 | YLCAW | SVQR | TEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 17/17 |
| HAM4 | Vβ20 | YLCAW | RTGF | NYGYTFGSGTRLTVV (Jβ1.2) | EDLNK | 11/11 |
Bold type indicates conserved amino acid motifs.
Number of indicated CDR3 sequences derived from bacterial colonies containing PCR-amplified DNA per total number of colonies sequenced.
We wished to test the hypothesis that chronic stimulation by the Tax antigen in vivo leads to the selection of specific sequences in the TCR Vβ chain. Previous evidence, from X-ray analysis (4, 8) and from site-directed mutagenesis (17) of TCR Vβ residues, showed that certain TCR Vβ residues make specific contact with side chains in the antigenic peptide. Indeed, although the Vβ rearrangements of the HLA-A2–Tax11-19-specific T cells varied between infected individuals, shared motifs consisting of four amino acids (P/G-L-A/R-G) were found within Vβ13.1-Dβ-Jβ junctional regions of HLA-A2–Tax11-19-specific T cells (Table 3). The P/G-L-A/R-G motif was present in the Vβ13.1-Dβ-Jβ junctional region of 34 out of 38 transcripts from Vβ13.1 PCR products of HLA-A2–Tax11-19-specific CD8+ T cells with oligoclonal spectratypes (Table 3). This motif is also present in the Tax11-19-specific CTL clone A6, which was generated from a patient with HAM/TSP (24). To clarify the significance of individual amino acid residues in the P/G-L-A/R-G motif, we used the known X-ray crystallographic structure of the A6 TCR complex (8). The interactions between the Vβ CDR3 GLAG motif and the MHC peptide complex are shown in Fig. 3.
As shown in Fig. 3, one corner of the apex of the Vβ CDR3 loop (GLAG) inserts into a hydrophobic pocket formed by the alpha-1 helix of HLA-A2 and the side chain of the Tyr residue at position 8 in the Tax11-19 peptide. In this CDR3 loop, the Leu residue at position 98 is distinguished by the extent of its interactions with the Tax peptide, i.e., hydrophobic interactions with Pro 6 and Tyr 8 as well as with Gln 72 of the alpha-1 helix of HLA-A2. Thus, the leucine residue in the conserved CDR3 motif is very likely to have particular importance for the interaction between the HLA-A2–Tax11-19 peptide complex and TCR. Since Leu and Gly residues are strongly favored (34 of 38 clones) at this position in the Vβ13.1-containing clonally expanded HLA-A2–Tax11-19 tetramer-positive cells, it seems likely that these CDR3 residues play a critical role in recognition of the HLA-A2–Tax11-19 complex in vivo. On the other hand, the side chain of the Arg at position 99, instead of Ala in the structure, could be readily accommodated without any disruption to interactions in the TCR HLA-A2–Tax11-19 peptide complex. We therefore suggest that the neighboring residues in the motif, particularly those at positions 97 and 99, which do not themselves make major interactions with HLA-A2 or the peptide, may simply allow the hydrophobic residue at position 98 to make optimal interactions with the complex. The Vβ1-Dβ-Jβ junctional sequence derived from two unrelated HAM/TSP patients (HAM1, from Japan, and HAM3, of Afro-Caribbean origin) shared an identical motif (VSDTT) (Table 3). Finally, a third motif consisting of a Q residue followed by an acidic residue (D or E) was identified in the Vβ16-Dβ-Jβ junctional regions of oligoclonally proliferating tetramer-binding cells in three subjects (Table 3). These results suggest that the observed amino acid motifs are the result of in vivo selection by the combination of particular MHC and peptide complexes rather than the result of proliferation of randomly infected and activated T cells in vivo.
The observation that HLA-A*02 alleles are associated with a reduced proviral load and protection against HAM/TSP suggests that HLA-A*02-restricted anti-Tax CTLs are protective in southern Japan (13). However, recently reported findings show that anti-HTLV CTLs also have the potential to produce proinflammatory cytokines (15) and that anti-HTLV CTLs are found in cerebrospinal fluid at a higher frequency than in peripheral blood in some HAM/TSP patients. Since our present data have shown that the same junctional sequence motifs were present in both HAM/TSP patients and asymptomatic carriers, there is no simple correlation between fine TCR specificity and disease manifestation in HTLV-1 infection. Recently it has been reported that the concentration of antigen required to elicit gamma interferon secretion by CD8+ T cells is greater than that required for target cell lysis (25). On the other hand, the frequency of gamma interferon-positive cells is positively correlated with the proviral load in HAM/TSP patients but not in asymptomatic carriers (16). These findings suggest that the anti-HTLV-1 CTLs are protective in a subject with a strong CTL response to Tax (an asymptomatic carrier), whereas the anti-HTLV-1 CTLs in a patient with a weak response to Tax (HAM/TSP patient) contribute to inflammation, because individuals with strong CTL responses to Tax maintain a low equilibrium concentration of the Tax protein, whereas in those with weak CTL responses the equilibrium concentration of Tax exceeds the threshold needed to elicit proinflammatory cytokines (1a).
Nucleotide sequence accession numbers.
All TCR sequence data have been deposited in EMBL-GenBank-DDBJ under accession numbers AB044099 to AB044135.
Acknowledgments
We thank the staff and blood donors of Kagoshima University Hospital and St. Mary's Hospital. We also thank Nathan Zaccai and Yvonne Jones (Wellcome Trust Centre for Human Genetics, Oxford, United Kingdom) for crystal structure illustration and Graham Ogg (John Radcliffe Hospital, Oxford, United Kingdom) for providing Tax11-19–HLA-A2 tetramers.
This study was supported by the Program for Promotion of Fundamental Studies in Health Science of the Organization for Pharmaceutical Safety and Research (OPSR) (Japan) and the Wellcome Trust (United Kingdom).
REFERENCES
- 1.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 1a.Asquith B, Bangham C R M. The role of cytotoxic T lymphocytes in human T-lymphotropic virus type 1 infection. J Theor Biol. 2000;207:65–79. doi: 10.1006/jtbi.2000.2156. [DOI] [PubMed] [Google Scholar]
- 2.Bieganowska K, Hollsberg P, Buckle G J, Lim D G, Greten T F, Schneck J, Altman J D, Jacobson S, Ledis S L, Hanchard B, Chin J, Morgan O, Roth P A, Hafler D A. Direct analysis of viral-specific CD8+ T cells with soluble HLA-A2/Tax11-19 tetramer complexes in patients with human T cell lymphotropic virus-associated myelopathy. J Immunol. 1999;162:1765–1771. [PubMed] [Google Scholar]
- 3.Davis M M, Bjorkman P J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 4.Ding Y H, Smith K J, Garboczi D N, Utz U, Biddison W E, Wiley D. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 5.Eiraku N, Hingorani R, Ijichi S, Machigashira K, Gregersen P K, Monteiro J, Usuku K, Yashiki S, Sonoda S, Osame M, Hall W W. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotrophic virus type 1-infected individuals. J Immunol. 1998;161:6674–6680. [PubMed] [Google Scholar]
- 6.Esnouf R M. An extensive modified version of MolScript that includes greatly enhanced colouring capabilites. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa K, Mori M, Ohta N, Ikeda H, Shida H, Furukawa K, Shiku H. Clonal expansion of CD8+ cytotoxic T lymphocytes against human T cell lymphotropic virus type 1 (HTLV-1) genome products in HTLV-1 associated myelopathy/tropical spastic paraparesis patients. J Clin Investig. 1994;94:1830–1839. doi: 10.1172/JCI117532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Structure of the complex between human T cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 9.Garcia K C, Degan M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. An alpha-beta T cell receptor structure at 2.5A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. [Google Scholar]
- 10.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calende A, De The G. Antibodies to human T-lymphotropic virus type-1 in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 11.Gorski J, Yassai M, Zhu X, Kissella B, Keever C, Flomenberg N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. J Immunol. 1994;152:5109–5119. [PubMed] [Google Scholar]
- 12.Hanon E, Hall S, Taylor G P, Saito M, Davis R, Tanaka Y, Usuku K, Osame M, Weber J N, Bangham C R M. Abundant Tax protein expression in CD4+ T cells infected with human T-cell lymphotropic virus type-1 (HTLV-1) is prevented by cytotoxic T lymphocytes. Blood. 2000;95:1386–1392. [PubMed] [Google Scholar]
- 13.Jeffery K J M, Usuku K, Hall S E, Matsumoto W, Taylor G P, Procter J, Bunce M, Ogg G S, Welsh K I, Weber J N, Lloyd A L, Nowak M A, Nagai M, Kodama D, Izumo S, Osame M, Bangham C R M. HLA alleles determine human T-lymphotropic virus-I (HTLV-1) proviral load and the risk of HTLV-1-associated myelopathy. Proc Natl Acad Sci USA. 1999;96:3848–3853. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan J E, Osame M, Kubota H, Igata A, Nishitani H, Maeda Y, Khabbaz R F, Janssen R S. The risk of development of HTLV-1 associated myelopathy/tropical spastic paraparesis among persons infected with HTLV-1. J Acquir Immune Defic Syndr. 1990;3:1096–1101. [PubMed] [Google Scholar]
- 15.Kubota R, Kawanishi T, Matsubara H, Manns A, Jacobson S. Demonstration of human T lymphotropic virus type I (HTLV-1) Tax-specific CD8+ lymphocytes directly in peripheral blood of HTLV-1-associated myelopathy/tropical spastic paraparesis patients by intracellular cytokine detection. J Immunol. 1998;161:482–488. [PubMed] [Google Scholar]
- 16.Kubota R, Kawanishi T, Matsubara H, Manns A, Jacobson S. HTLV-1 specific IFN-gamma+ CD8+ lymphocytes correlate with the proviral load in peripheral blood of infected individuals. J Neuroimmunol. 2000;102:208–215. doi: 10.1016/s0165-5728(99)00175-7. [DOI] [PubMed] [Google Scholar]
- 17.Manning T C, Schlueter C J, Brodnicki T C, Parke E A, Speir J A, Garcia K C, Teyton L, Wilson I A, Kranz D M. Alanine scanning mutagenesis of an αβ T cell receptor: mapping the energy of antigen recognition. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- 18.Merrit E A, Bacon D J. Raster 3D photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 19.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J D, Zajac A J, Miller J D, Slansky J, Ahmed R. Counting antigen-specific CD8+ T cells: a re-evaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 20.Ogg G S, McMichael A J. HLA-peptide tetrameric complexes. Curr Opin Immunol. 1998;10:393–396. doi: 10.1016/s0952-7915(98)80110-6. [DOI] [PubMed] [Google Scholar]
- 21.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-1 associated myelopathy, a new clinical entity. Lancet. 1986;i:1031. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 22.Pique C, Connan F, Levilain J-P, Choppin J, Dokhelar M-C. Among all human T-cell leukemia virus type I proteins, Tax, polymerase, and envelope proteins are predicted as preferential targets for the HLA-A2-restricted cytotoxic T-cell response. J Virol. 1996;70:4919–4926. doi: 10.1128/jvi.70.8.4919-4926.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poiesz B J, Ruscetti R W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Utz U, Banks D, Jacobson S, Biddison W E. Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: evidence for oligoclonal expansion. J Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valitutti S, Muller S, Dessing M, Lanzavecchia A. Different responses are elicited in cytotoxic T lymphocytes by different levels of T cell receptor occupancy. J Exp Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]