Abstract
The virion host shutoff (vhs) protein of herpes simplex virus (HSV) triggers global shutoff of host protein synthesis and accelerated turnover of host and viral mRNAs during HSV infection. As well, it induces endoribonucleolytic cleavage of RNA substrates when produced in a rabbit reticulocyte lysate (RRL) in vitro translation system. The vhs1 point mutation (Thr 214→Ile) eliminates vhs function during virus infection and in transiently transfected mammalian cells and was therefore previously considered to abolish vhs activity. Here we demonstrate that the vhs1 mutant protein induces readily detectable endoribonuclease activity on RNA substrates bearing the internal ribosome entry site of encephalomyocarditis virus in the RRL assay system. These data document that the vhs1 mutation does not eliminate catalytic activity and raise the possibility that the vhs-dependent endoribonuclease employs more than one mode of substrate recognition.
Herpes simplex virus (HSV) is a large enveloped DNA virus that replicates in the nuclei of infected mammalian cells. Like many other viruses, HSV inhibits host cell protein synthesis as a key element of its strategy of reprogramming the cellular biosynthetic machinery. HSV-induced host shutoff can be divided into two phases, which occur at early and intermediate times postinfection, respectively. Early shutoff involves disruption of preexisting polysomes and rapid degradation of host mRNAs (6, 7, 9, 10, 15–18, 20, 26). Substantial data indicate that the virion host shutoff (vhs) protein, a tegument protein encoded by HSV gene UL41, is both necessary and sufficient for the early shutoff effect (8, 11, 12, 19–22).
Although the mechanism of vhs action remains to be precisely defined, several lines of evidence strongly indicate that it acts as an RNase. First, cytoplasmic extracts prepared from HSV-infected cells and extracts of partially purified HSV virions contain a vhs-dependent RNase activity (13, 14, 23, 26), and this activity is inhibited by anti-vhs antibodies (26). Second, vhs induces endoribonucleolytic cleavage of a variety of reporter mRNAs when it is expressed as the only HSV protein in a rabbit reticulocyte lysate (RRL) in vitro translation system (4, 5, 26). Third, vhs displays weak but significant amino acid sequence similarity to the fen-1 family of nucleases that are involved in DNA replication and repair in eukaryotes and archaebacteria (3), and human fen-1 has recently been shown to cleave both RNA and DNA substrates (24). Although these data argue that vhs is an integral and required component of the vhs-dependent endoribonuclease, they do not exclude the possibility that one or more cellular factors are also required for activity.
Much of our current knowledge about the role of the UL41 gene product in host shutoff has emerged from studies of the vhs1 mutant isolate of HSV type 1 strain KOS (20). This mutant harbors a single base change in the UL41 open reading frame that converts amino acid residue 214 from threonine to isoleucine (12, 15). The mutation is located in one of the regions of strongest homology to the fen-1 family of nucleases (3), and previous studies have suggested that it completely inactivates vhs function. The vhs1 mutation abolishes early shutoff of host protein synthesis during HSV infection (20) and eliminates vhs activity in transient cotransfection assays (12, 19). In addition, the mutant protein fails to trigger accelerated RNA turnover in in vitro systems derived from extracts of HSV-infected cells and partially purified virions, and it displays little if any activity on the mRNA encoding the α subunit of the signal recognition particle (SRPα mRNA) in the RRL in vitro assay system (4, 14, 23, 26).
Previous work from this laboratory has shown that an internal ribosome entry site (IRES) derived from encephalomyocarditis virus (EMCV) or poliovirus acts to strongly target vhs-dependent RNA cleavage events to a narrow zone located immediately 3′ of the IRES (5). During further studies of this effect, we obtained preliminary evidence that the vhs1 mutant form of vhs displays significant activity on substrates bearing the EMCV IRES (data not shown). This observation was both interesting and surprising, because it raised the possibility that the vhs1 mutation does not completely inactivate the vhs-dependent endoribonuclease. We therefore undertook studies designed to more carefully assess the activity of the vhs1 protein in the RRL assay system.
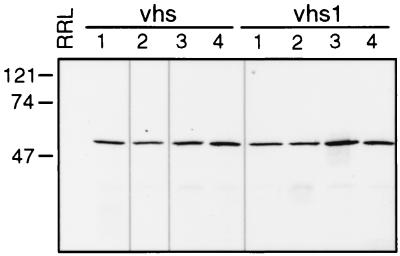
The vhs1 in vitro translation vector pSP6vhs1 (4) was first subcloned by retransforming Escherichia coli, to eliminate possible contamination with wild-type vhs sequences. Following verification of the vhs1 mutation by DNA sequence analysis, DNA obtained from a single transformed colony was amplified and used for all experiments described in this report. Multiple RRL in vitro translation reactions were then programmed with vhs mRNA generated from pSP6vhs (4) and pSP6vhs1 to generate wild-type and mutant vhs, respectively. Template DNA was linearized with EcoRI, and transcriptions were done for 1 h at 37°C in a 20-μl reaction mixture containing 1 μg of template, 1 U of SP6 RNA polymerase, 0.5 mM cap primer 7mG(5′)ppp(5′)G (Amersham Pharmaceutical), 2 U of RNase inhibitor (Gibco BRL), and 0.5 mM GTP, CTP, ATP, and UTP. The DNA template was then digested with 10 U of RNase-free DNase I (Boehringer Mannheim) at 37°C for 15 min; the RNA product was recovered, precipitated with ethanol, and dissolved in RNase-free water. Approximately 2 μg of the vhs mRNA was then used to program each of several 40-μl aliquots of RRL (Promega) using the manufacturer's procedure. Reaction mixtures were incubated at 30°C for 60 min in the presence of [35S]methionine (NEN Life Science Products). Blank RRL controls were generated in the same way except that no mRNA was added to the translation reaction. A 2-μl aliquot of each reaction mixture was then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the remainder of each reaction mixture was stored at −70°C for future use (4). Following autoradiography, pairs of translation reactions containing comparable amounts of wild-type and mutant protein were identified (Fig. 1); these were then thawed and used for the activity assays depicted in Fig. 2, 3, 5, and 6. This approach ensured that most of the activity comparisons presented in this report were performed with wild-type and mutant vhs preparations that were generated in parallel and were closely matched for vhs protein content.
FIG. 1.
vhs protein used in activity assays. vhs and vhs1 mRNAs were each used to program four RRL in vitro translation reactions in the presence of [35S]methionine. RRL control (lane RRL) was generated as above except that no mRNA was added to the translation reaction. The products of each translation reaction were then analyzed by SDS-PAGE. Numbers to the left indicate the sizes (in kilodaltons) of protein markers. The samples of wild-type and vhs1 mutant labeled 1, 2, 3, and 4 were used for the activity assays depicted in Fig. 2, 3, 5, and 6, respectively.
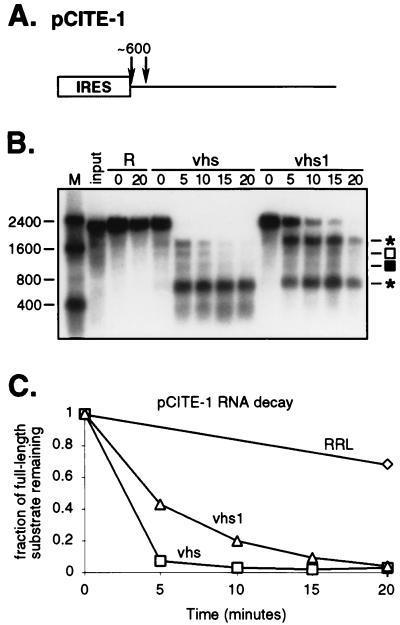
FIG. 2.
Activities of wild-type and mutant vhs on pCITE-1 RNA. (A) Diagram of the 2.3-kb pCITE-1 RNA, indicating the location of the IRES and approximate locations of the sites of preferential initial cleavage by wild-type vhs. (B) Internally labeled pCITE-1 RNA was added to control RRL (R) and RRL containing pretranslated wild-type or vhs1 mutant vhs, as indicated. RNA extracted at the indicated times (minutes) was resolved on a 1% agarose–1.8% formaldehyde gel and transferred to a GeneScreen Plus membrane. Numbers to the left indicate the sizes (in nucleotides) of RNA markers (lane M). Asterisks indicate the 3′ and 5′ products of IRES-directed cleavage (ca. 1,800 and 600 nt, respectively); open and closed squares indicate the ca. 1,500- and 1,000-nt fragments. (C) The data displayed in panel B, quantified by phosphorimager analysis.
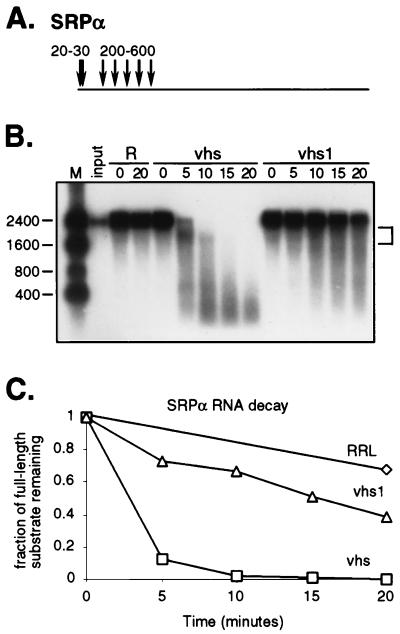
FIG. 3.
Activities of wild-type and mutant vhs on SRPα RNA. (A) Diagram of the 2.4-kb RNA substrate, indicating the approximate location(s) of the sites of preferential initial cleavage by wild-type vhs. (B) Internally labeled SRPα RNA was added to control RRL (R) and RRL containing pretranslated wild-type or vhs1 mutant vhs, as indicated. The reaction products were analyzed as for Fig. 2. Numbers to the left indicate the sizes (in nucleotides) of RNA markers (lane M). The bracket indicates the mobility of several discrete 3′ products (1,800 to 2,200 nt). (C) The data displayed in panel B, quantified by phosphorimager analysis.
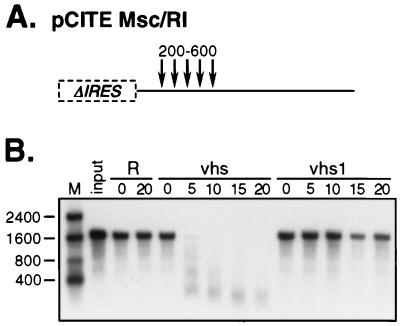
FIG. 5.
Activities of wild-type and mutant vhs on pCITE Msc/RI RNA. (A) Diagram of the 1.7-kb transcript, indicating the IRES deletion and approximate locations of the sites of preferential initial cleavage by wild-type vhs. (B) Internally labeled pCITE Msc/RI RNA was added to control RRL (R) and RRL containing pretranslated wild-type or vhs1 mutant vhs, as indicated. The reaction products were analyzed as for Fig. 2. Numbers to the left indicate the sizes (in nucleotides) of RNA markers (lane M).
FIG. 6.
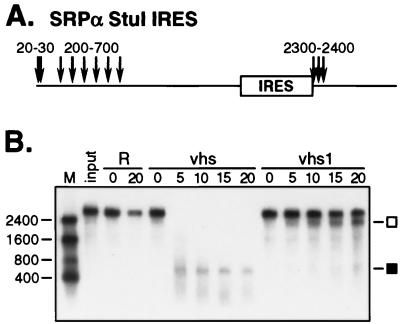
Activities of wild-type and mutant vhs on SRPα StuI IRES RNA. (A) Structure of the 3-kb RNA and approximate locations of the sites of preferential initial cleavage by wild-type vhs. (B) Internally labeled SRPα StuI IRES RNA was added to control RRL (R) and RRL containing pretranslated wild-type or vhs1 mutant vhs, as indicated. The reaction products were analyzed as for Fig. 2. Numbers to the left indicate the sizes (in nucleotides) of RNA markers (lane M). The open and closed squares indicate the ca. 2,300- and 700-nt fragments arising through IRES-directed cleavage.
We first compared the activities of wild-type and mutant vhs on a 2.3-kb transcript which bears the EMCV IRES at its 5′ end (pCITE-1 RNA [Fig. 2A]). Uncapped, internally labeled pCITE-1 RNA was generated by in vitro transcription as previously described (5) and then added to RRL containing pretranslated vhs. Following incubation at 30°C, RNA extracted at various time points was analyzed by electrophoresis on a 1% agarose-formaldehyde gel. Previous work has demonstrated that the initial sites of vhs-induced cleavage of pCITE-1 RNA are clustered immediately downstream of the IRES (diagrammed in Fig. 2A). These IRES-directed cleavage events give rise to two relatively discrete early degradation products: a ca. 600-nucleotide (nt) 5′ fragment that contains the IRES, and a ca. 1,800-nt 3′ fragment. The 5′ fragment is stable over the course of the reaction, while the 3′ fragment is subject to further decay. We found that wild-type vhs generated the predicted 600- and 1,800-nt products (Fig. 2B). However, we also detected additional products of ca. 1,500 and 1,000 nt early during the reaction, and these declined in abundance as the reaction proceeded. The 1,500- and 1,000-nt fragments were not noted in our earlier study, and we have not yet determined their structure; it is possible that they represent degradation intermediates generated from the 1,800-nt product. The vhs1 mutant protein also induced degradation of the pCITE-1 RNA (Fig. 2B), leading to production of the same degradation intermediates as wild-type vhs. However, the reaction proceeded more slowly, and the 1,800-nt 3′ fragment was substantially more stable than in the presence of wild-type vhs. The data presented in Fig. 2B, quantified by phosphorimager analysis and plotted in Fig. 2C, demonstrate that the vhs1 mutant protein displays reduced but significant activity on pCITE-1 RNA.
We next reexamined the activities of wild-type and mutant vhs on SRPα RNA, a transcript that lacks an IRES (Fig. 3). Elgadi et al. (4) have shown that the most prominent sites of initial cleavage of this RNA are nonrandomly clustered over the 5′ quadrant of the transcript (Fig. 3A), leading to early production of relatively discrete sets of 5′ and 3′ degradation intermediates of ca. 200 to 600 nt and 1,800 to 2,200 nt, respectively. Both sets of intermediates are then further degraded as the reaction proceeds. We observed a similar pattern in reactions containing wild-type vhs (Fig. 3B). In contrast, reactions containing the vhs1 mutant protein did not display readily detectable quantities of these early 5′ and 3′ degradation intermediates. However, the RNA substrate was somewhat less stable than in control RRL and gave rise to a heterogeneous set of degradation products that migrated as a broad smear during gel electrophoresis. These data suggest that the vhs1 protein displays very weak activity on SRPα RNA in the RRL assay (see also Fig. 4). However, this activity was not always detected in repeated trials (reference 5 and additional data not shown), indicating that it is close to the lower detection limit of our assay system.
FIG. 4.
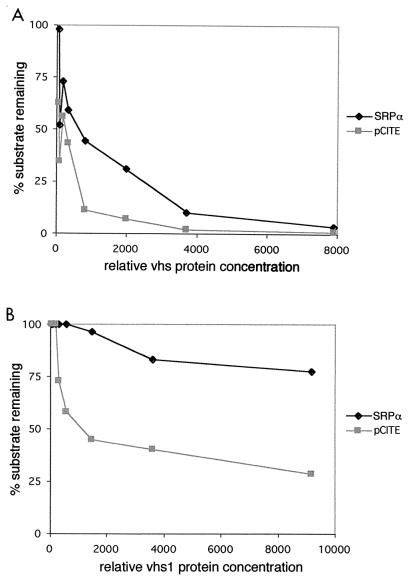
Effects of dilution on activities of wild-type and mutant vhs. [35S]methionine-labeled wild-type (A) and vhs1 mutant (B) vhs generated by in vitro translation were serially diluted in RRL. Aliquots of each dilution were assayed for vhs protein content by SDS-PAGE and phosphorimager analysis and then tested for activity on internally labeled pCITE-1 and SRPα RNA as for Fig. 2 and 3. Reaction mixtures were incubated for 4 min. The activity data were quantified by phosphorimager analysis and plotted against the relative amount of vhs protein present in each reaction.
The reactions depicted in Fig. 2 and 3 contained similar quantities of vhs protein, and the substrate concentrations were identical. pCITE-1 RNA decayed significantly more rapidly than SRPα RNA in the reactions containing vhs1 (compare Fig. 2 and 3), suggesting that pCITE-1 RNA serves as a preferred substrate for this mutant form of vhs. We were unable to determine whether pCITE-1 RNA also serves as a preferred substrate for wild-type vhs in these experiments, because both substrates were almost fully degraded at the earliest time point analyzed. As one approach to answering this question, we compared the activities of serially diluted samples containing wild-type and vhs1 mutant protein on both RNAs. 35S-labeled wild-type and mutant vhs were generated by in vitro translation as described above. Following translation, reactions were serially diluted in RRL; 2-μl aliquots of each dilution were analyzed by SDS-PAGE as for Fig. 1, and the relative amount of vhs protein in each sample was quantified by phosphorimager analysis; 5-μl aliquots of each dilution were then combined with internally labeled pCITE-1 or SRPα RNA, and incubated for 4 min at 30°C; the RNA products were extracted and analyzed by gel electrophoresis as described above. The proportion of undigested substrate was determined for each dilution by phosphorimager analysis and plotted against the relative amount of vhs protein present in the corresponding reaction (Fig. 4). The results of this experiment confirmed that the vhs1 protein displays substantially greater activity on pCITE-1 RNA than on SRPα RNA. For example, the concentration of vhs1 that led to 50% decay of pCITE-1 RNA had virtually no effect on the SRPα substrate over the 4-min time course of the experiment (Fig. 4B). This experiment additionally provided evidence that the wild-type vhs protein also displays greater activity on pCITE-1 RNA than on SRPα RNA, although in this case the difference was not as apparent.
It seemed possible that the marked difference in activity of the vhs1 protein on pCITE-1 versus SRPα RNA reflected the presence of the EMCV IRES in the former substrate. We tested this hypothesis, by examining the effect of deleting the IRES (construct pCITE Msc/RI RNA, ca. 1.7 kb [Fig. 5A]). Previous work has shown that vhs-induced decay of pCITE Msc/RI RNA proceeds similarly to that of SRPα RNA, in that the sites of initial cleavage are nonrandomly clustered over the 5′ quadrant of the transcript (Fig. 5A) (5). The vhs1 protein displayed little, if any, activity on this substrate; in contrast, wild-type vhs induced rapid decay (Fig. 5B). Thus, deletion of the IRES rendered the pCITE-1 transcript refractory to degradation by the vhs1 protein (compare Fig. 2 and 5).
Taken in combination, the foregoing results suggested that the vhs1 protein retains significant IRES-directed cleavage activity. As an additional test of this hypothesis, we tested whether the vhs1 protein was capable of inducing cleavage downstream of the EMCV IRES when the IRES was transplanted onto a heterologous RNA. pSRPα StuI IRES is a derivative of pSRPα which bears the EMCV IRES element inserted into the unique StuI site in SRPα RNA (5). Using 5′-labeled RNA, Elgadi and Smiley (5) have shown that the transplanted IRES efficiently targets vhs activity, resulting in a complex pattern of initial cleavage events. Specifically, some of the substrate RNA molecules are initially cleaved at sites clustered over the 5′ quadrant of the transcript (in the same fashion as unmodified SRPα RNA), while others are initially cleaved immediately downstream of the IRES (Fig. 6A). If, as suggested by data presented above, the vhs1 protein retains significant IRES-directed activity but is otherwise severely impaired, then pSRPα StuI IRES RNA presumably would be cleaved predominantly or exclusively downstream of the transplanted IRES. Such IRES-directed events would give rise to stable products of ca. 2,300 and 700 nt, representing the 5′ and 3′ segments of the RNA, respectively. Our results (Fig. 6B) confirmed this prediction. In contrast, wild-type vhs rapidly degraded the substrate into low-molecular-weight products. As expected, no stable 2,300-nt 5′ fragment accumulated (previous work has shown that this early product is subject to rapid further decay in reactions containing wild-type vhs [5]). However, we observed a stable ca. 700-nt product which may correspond to either the 3′ end of the RNA (5) or the excised IRES element (which is refractory to vhs-induced attack [5]). Further experiments are required to clarify the nature of this product. The results of this experiment provided further evidence that the readily detectable endoribonuclease activity of the vhs protein is IRES dependent.
Overall, the results presented in this report establish that the vhs1 point mutation does not abolish the activity of the vhs-dependent endoribonuclease. Thus, if the vhs polypeptide indeed forms an integral component of the endoribonuclease, then it follows that the mutation does not completely inactivate the active site of the enzyme. Although this finding at first glance may seem inconsistent with the fact that the vhs1 mutation is located in a region of strong similarity to fen-1 nucleases, the threonine residue that is altered by the mutation is not itself conserved in fen-1 nucleases and is therefore likely not essential for activity. Our data also provide strong evidence that the vhs1 mutant form of vhs retains significant IRES-directed activity but is essentially inactive on RNA substrates that lack an IRES. One interesting possibility is that wild-type vhs normally utilizes both IRES-dependent and IRES-independent modes of substrate recognition, and the vhs1 mutation selectively impairs the latter. If so, then screening other mutant forms of vhs may help to unveil which regions of vhs participate in each mode of substrate recognition and thus clarify the mechanisms involved. Alternatively, it is possible that the apparently selective activity of the mutant protein simply mirrors an inherent preference of wild-type vhs for IRES-bearing substrates (Fig. 4A). According to this hypothesis, the mutation coordinately reduces (but does not eliminate) activity on all substrates, and our inability to clearly detect activity on substrates lacking an IRES simply reflects limitations of our assay system. Distinguishing between these possibilities will require detailed kinetic analysis of the activity of wild-type and mutant vhs on a variety of substrates. However, such analyses are very difficult to perform with the present RRL assay system (data not shown).
It has been suggested that vhs may destabilize mRNAs by inducing a limited number of endoribonucleolytic cleavages, which predispose the transcript to further decay through cellular mRNA surveillance systems (19, 23, 26) in a fashion analogous to the mechanisms that regulate the stability of some cellular mRNAs (for examples, see references 1 and 2). However, our data provide strong evidence that vhs is required not only for initial endoribonucleolytic cleavage events but also for subsequent degradation to low-molecular-weight products, at least in the RRL system. Thus, although the vhs1 protein is able to perform IRES-directed cleavage of pCITE-1 RNA, the resulting 3′ product is much more stable than in reactions containing wild-type vhs. The implication is that wild-type vhs actively induces further decay of the products of IRES-directed cleavage.
It will be interesting to determine if the vhs1 mutant protein is capable of destabilizing IRES-bearing mRNAs in vivo. If so, vhs1 may offer an approach to studying the biological function of the small minority of cellular mRNAs that bear an IRES. We suspect that further studies of the IRES-directed activity of vhs1 and other mutant forms of vhs will clarify the mechanism of vhs action.
Acknowledgments
We thank Rob Maranchuk for superb technical assistance and Kim Ellison for valuable advice and criticism.
This research was supported by a grant from the Medical Research Council of Canada.
REFERENCES
- 1.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown B D, Zipkin I D, Harland R M. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 1993;7:1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- 3.Doherty A J, Serpell L C, Ponting C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgadi M M, Hayes C E, Smiley J R. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elgadi M M, Smiley J R. Picronavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenwick M L, Clark J. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J Gen Virol. 1982;61:121–125. doi: 10.1099/0022-1317-61-1-121. [DOI] [PubMed] [Google Scholar]
- 7.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 8.Fenwick M L, Everett R D. Inactivation of the shutoff gene (UL41) of herpes simplex virus type 1 and 2. J Gen Virol. 1990;71:2961–2967. doi: 10.1099/0022-1317-71-12-2961. [DOI] [PubMed] [Google Scholar]
- 9.Fenwick M L, Owen S A. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J Gen Virol. 1988;69:2869–2877. doi: 10.1099/0022-1317-69-11-2869. [DOI] [PubMed] [Google Scholar]
- 10.Fenwick M L, Walker M J. Suppression of synthesis of cellular macromolecules by herpes simplex virus. J Gen Virol. 1978;41:37–51. doi: 10.1099/0022-1317-41-1-37. [DOI] [PubMed] [Google Scholar]
- 11.Fenwick M L, Everett R D. Transfer of UL41, the gene controlling virion-associated host cell shutoff, between different strains of herpes simplex virus. J Gen Virol. 1990;71:411–418. doi: 10.1099/0022-1317-71-2-411. [DOI] [PubMed] [Google Scholar]
- 12.Jones F E, Smibert C A, Smiley J R. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J Virol. 1995;67:4863–4871. doi: 10.1128/jvi.69.8.4863-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karr B M, Read G S. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology. 1999;264:195–204. doi: 10.1006/viro.1999.9986. [DOI] [PubMed] [Google Scholar]
- 14.Krikorian C R, Read G S. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate early (alpha) mRNA. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pak A S, Everly D N, Knight K, Read G S. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 20.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate-early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 23.Sorenson C M, Hart P A, Ross J. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 1991;19:4459–4465. doi: 10.1093/nar/19.16.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens A. Endonucleolytic cleavage of RNA at 5′ endogenous stem structures by human flap endonuclease 1. Biochem Biophys Res Commun. 1998;251:501–508. doi: 10.1006/bbrc.1998.9499. [DOI] [PubMed] [Google Scholar]
- 25.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]