Abstract
After experimental infection of squirrel monkeys (Saimiri sciureus) with human T-cell leukemia virus type 1 (HTLV-1)-infected cells, the virus is transcribed only transiently in circulating blood, spleen, and lymph nodes. Stable disappearance of viral expression occurs at 2 to 3 weeks after inoculation. This coincides with the development of the anti-HTLV-1 immune response and persistent detection of the provirus in peripheral blood mononuclear cells (PBMCs). In this study, the HTLV-1 replication pattern was analyzed over time in PBMCs and various organs from two HTLV-1-infected squirrel monkeys. Real-time quantitative PCR confirmed that PBMCs and lymphoid organs constitute the major reservoirs for HTLV-1. The PCR amplification of HTLV-1 flanking sequences from PBMCs evidenced a pattern of clonal expansion of infected cells identical to that observed in humans. Dissemination of the virus in body compartments appeared to result from cellular transport of the integrated provirus. The circulating proviral burden increased as a function of time in one animal studied over a period of 4 years. The high proviral loads observed in the last samples resulted from the accumulation of infected cells via the extensive proliferation of a restricted number of persistent clones on a background of polyclonally expanded HTLV-1-positive cells. Therefore, HTLV-1 primary infection in squirrel monkeys is a two-step process involving a transient phase of reverse transcription followed by persistent multiplication of infected cells. This suggests that the choice of the target for blocking HTLV-1 replication might depend on the stage of infection.
Human T-cell leukemia virus type 1 (HTLV-1), the first human pathogenic retrovirus isolated, is the etiologic agent of a malignant CD4 lymphoproliferation (adult T-cell leukemia/lymphoma [ATLL]) and of a chronic progressive neuromyelopathy (tropical spastic paraparesis/HTLV-1-associated myelopathy [TSP/HAM]) (16, 43). Furthermore, this virus has been associated, to a lesser extent, with the development of a variety of inflammatory diseases including uveitis (37), arthritis (46), polymyositis (33), Gougerot-Sjögren syndrome (50), alveolitis (35), and infective dermatitis (28) in areas where the virus is endemic. Roughly 3 to 5% of the 15 million to 25 million HTLV-1-infected individuals throughout the world will develop TSP/HAM or ATLL, depending on certain unknown cofactors.
The HTLV-1 proviral load is particularly elevated in patients with TSP/HAM, with up to one-fifth of peripheral blood mononuclear cells (PBMCs) harboring HTLV-1 (18, 27). The load in asymptomatic carriers is generally lower but can be as high as 1/20 of PBMCs (47, 54). The problem is that considerable viral replication is needed in order to attain such high proviral loads. Such a situation should lead to extensive sequence diversity, but of all retroviruses, those of the HTLV/bovine leukemia virus group are the most stable genetically (17). This conundrum was resolved by the finding that HTLV-1-bearing T cells underwent clonal expansion in vivo in all symptomatic and asymptomatic carriers (5, 6, 55).
Animal models of HTLV-1 infection have been developed in order to study host-virus interactions, virus transmission, the natural history of infection, and the pathogenesis of HTLV-1-associated diseases. Such models are also essential for testing candidate vaccines. Chronic HTLV-1 infection has been obtained in both rabbit (49) and rat (23) models. In addition, several species of nonhuman primates have also been found to be susceptible to HTLV-1 infection. For instance, marmosets (Callithrix jacchus) are infected orally by the milk of HTLV-1 carrying women (56), while protection against infection with recombinant HTLV-1 Env protein has been obtained in cynomolgus macaques (Macaca fascicularis) (22).
The squirrel monkey (Saimiri sciureus), a New World nonhuman primate that is free of simian T-cell leukemia virus, is susceptible to experimental infection with either syngeneic or allogeneic HTLV-1-immortalized cells (25, 26, 40). As in human subjects, experimental infection leads to proviral expression, persistence, and humoral and cellular immune responses. A recent study investigating primary HTLV-1 infection in this model (26) demonstrated that after experimental infection, the provirus was transcribed only transiently in the circulating blood, spleen, and lymph nodes. The stable disappearance of viral expression, as evidenced by tax/rex reverse transcriptase PCR and in situ hybridization analyses, occurred about 2 weeks postinoculation; it coincided with the development of the anti-HTLV-1 humoral response and was followed by the persistent detection of the provirus in PBMCs (26).
Together with the low level of HTLV-1 genetic drift demonstrated in the squirrel monkey (25), these data led us to hypothesize that primary HTLV-1 infection consists of a first transient step of reverse transcription and viral expression, followed by a second prolonged phase of persistent clonal expansion of HTLV-1-bearing T cells. To investigate this question and better characterize the pattern of HTLV-1 replication at the late stage of experimental infection in the squirrel monkey, we measured the proviral load and assessed the clonality pattern of HTLV-1-infected cells over time and in different anatomic sites in two experimentally infected animals.
Study design.
S1657 and S1491, two 6-year-old male squirrel monkeys from the primate breeding center at the Pasteur Institute of French Guyana, were inoculated intravenously with 5 × 107 EVO/798 (animal S1491) or EVO/1540 (animal S1657) HTLV-1-transformed monkey cells as previously detailed (26). Humoral and cellular immune responses against HTLV-1 antigens were observed in both monkeys (26), and HTLV-1 provirus was also detected in PBMCs by PCR amplification using gag and tax primers. S1657 was sacrificed at a late stage of experimental infection (26 months after inoculation), whereas S1491 was followed up over a period of 4 years postinfection. DNA was extracted from the two inoculated cell lines, from the PBMCs of both animals, and from 12 organs derived from S1657. HTLV-1 proviral load was measured by an accurate and reproducible quantitative PCR method using a dual-labeled fluorogenic probe (ABI PRISM 7700 sequence detection system). Standard curves for the albumin and HTLV-1 tax genes were generated using DNA extracted from HTLV-1 negative PBMCs for the former and an HTLV-1 plasmid for the latter. It was assumed that 10 ng of high-molecular-weight DNA contained 3,000 copies of the albumin gene. The primer set for HTLV-1 tax gene was PXF (5′-GAAACCGTCAAGCACAGCTT-3′, positions 7163 to 7182) and PXR (5′-TCTCCAAACACGTAGACTGGGT-3′, positions at 7385 to 7364). The primer set for the albumin gene was 5′-GCTGTCATCTCTTGTGGGCTGT-3′ (positions 16283 to 16304) and 5′-ACTCATGGGAGCTGCTGGTTC-3′ (positions 16442 to 16421) (nucleotide coordinates are numbered according to the albumin GenBank reference [HUMALBGC]) (36). The TaqMan probe consisted of an oligonucleotide with 5′-reporter dye and 3′-quencher dye. The fluorescent reporter dye, 6-carboxyfluorescein, was covalently linked to the 5′ end of the oligonucleotide. The reporter was quenched by 6-carboxy-tetramethylrhodamine at the 3′ end. Probes for the HTLV-1 tax and albumin genes were PXT (5′-TTCCCAGGGTTTGGACAGAGTCTTCT-3′, positions 7331 to 7355) and ALB (5′-CCTGTCATGCCCACACAAATCTCTCC-3′, positions 16340 to 16366), respectively. TaqMan amplification was carried out in reaction volumes of 50 μl, using the TaqMan PCR core reagent kit. Each reaction mixture contained 1× TaqMan buffer, primer (300 nmol/liter) and the corresponding fluorescent probe (200 nmol/liter), MgCl2 (3.5 mmol/liter), dATP, dCTP, and dGTP (each at 200 μmol/liter), dUTP (400 μmol/liter), 1.25 U of AmpliTaq Gold, and 0.5 U of AmpErase uracil N-glycosylase. Each sample was analyzed in triplicate, using 500 ng of DNA per reaction. Thermal cycling was initiated with 2 min of incubation at 50°C, followed by a first denaturation step of 10 min at 95°C and then 45 cycles of 15 s at 95°C and 1 min at 58°C (for tax) or 60°C (for albumin). The 7700 sequence detection system software automatically determines the threshold cycle value (Ct), i.e., the threshold cycle at which fluorescence is first detected above background, and infers the starting copy number in each sample. The p4.39 HTLV-1 plasmid, kindly provided by T. Astier-Gin (42), was used to establish the calibration curve for tax. Plasmid concentrations from 5 × 100 to 5 × 104 molecules were mixed with 0.5 μg of HTLV-1-negative genomic DNA, an equivalent of 75,000 cells, and then amplified. The lower limit of detection, i.e., the lowest plasmid concentration having a Ct of ≤45, was 5 copies per 0.5 μg of DNA, which corresponded to 10 copies per 1.5 × 105 PBMCs. The clonality of HTLV-1-infected cells was assessed by the sensitive quadruplicate ligation-mediated PCR (LMPCR) method as described elsewhere (5–8, 30, 32).
The mean proviral copy number of the 16 samples with a positive signal was 750 per μg of DNA (150,000 cell equivalents) (range, 15 to 3,334; median, 61.5; standard error of the mean, 293; mean coefficient of variance, 9.3%). The clonality of infected cells was therefore analyzed by LMPCR, which is the most appropriate method for assessing the amplification of HTLV-1 flanking sequences derived from samples with low proviral loads (52). Briefly, DNA was digested with NlaIII in 1× NlaIII buffer for 3 h at 37°C. Digestion was controlled by gel electrophoresis. DNA was phenol-chloroform extracted and ethanol precipitated. Digested DNA was ligated with BIO1 primer (30, 53) using T4 DNA ligase. This was followed by two phenol-chloroform extractions and precipitation. Ligated DNA was amplified for 100 cycles using the BIO2 primer alone (30). Conditions were 1× Stoffel DNA polymerase buffer, 1.5 mM MgCl2, 50 pmol BIO2, 150 μM each deoxynucleoside triphosphate, and 10 U of Stoffel fragment of Taq DNA polymerase (Perkin-Elmer Cetus) in a final volume of 85 μl. Twenty-five microliters of 1× PCR buffer containing deoxynucleoside triphosphates and primers was loaded in a 750-μl tube, and an Ampliwax PCR Gem 100 (Cetus) was added to each tube. After wax layer formation by incubation at 75°C for 10 min and cooling at room temperature for 15 min, 60-μl aliquots of the remaining reagent and ligated products were loaded. Thermal cycling parameters were 94°C for 10 min; 100 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 2 min; and a final elongation step of 10 min at 72°C. Ten microliters of this linear PCR mixture was used in a classical PCR amplification using the BIO3-BIO4 primer pair (31, 55). Amplification conditions were as before, with 40 pmol of each primer and 2.5 U of Taq polymerase in a final volume of 100 μl. Thermal cycling parameters were 94°C for 10 min; 35 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 1 min; and a final elongation step of 10 min at 72°C.
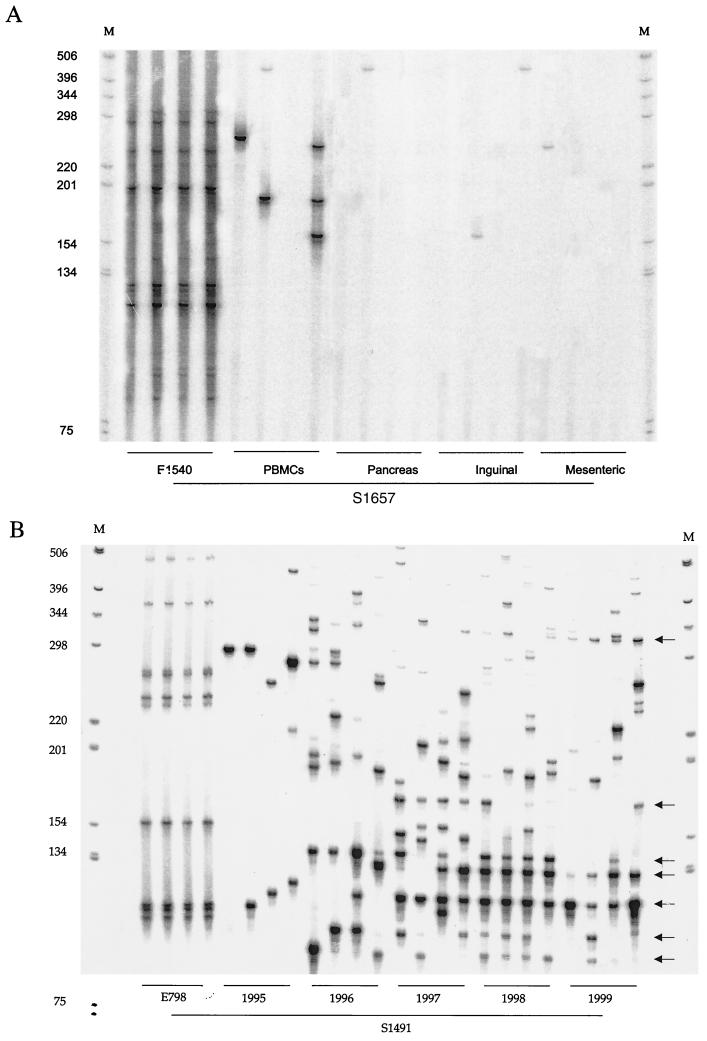
HTLV-1 does not integrate in a specific region of the cellular genome (31). In addition, the absolute LMPCR detection threshold (the value below which a signal is never detected) is ∼20 copies (8); therefore, each signal obtained after runoff analysis corresponded to a cluster of at least 20 proviruses sharing the same integration site and thus derived from a single HTLV-1-infected progenitor. A stochastic element has previously been evidenced in the detection of HTLV-1 integration sites by LMPCR; signals detected one, two or three, and four times after four LMPCR experiments correspond to 50, 100, and more than 500 copies, respectively, per μg of DNA (8). Quadruplicate experiments and runoff analyses of LMPCR products were performed as previously described (5–8, 30, 32). The clonality pattern of HTLV-1-infected cells is represented in Fig. 1. Table 1 shows the proviral load and number of detected clones in each of the 18 samples studied.
FIG. 1.
Quadruplicate LMPCR analysis of HTLV-1 integration sites in DNA samples from experimentally infected squirrel monkeys. DNA was submitted to quadruplicate LMPCR; amplified products were subjected to runoff analysis with an HTLV-1 3′ long terminal repeat-specific oligonucleotide. Run-off products were resolved on a sequencing gel. M, molecular weight marker (positions are indicated in base pairs). (A) E1540 cell line, PBMCs and organs from animal S1657; (B) E798 cell line, PBMCs from animal S1491 collected over 4 years with 1-year intervals (abundant persistent clones are identified by arrows).
TABLE 1.
Proviral load and infected cell clonality in PBMCs and various organs from two squirrel monkeysa
| Monkey | Sample | No. of proviral copies/μg of DNA | No. of HTLV-1- infected clones after 4 expts | No. of clones ≥1/1,500 PBMCs |
|---|---|---|---|---|
| 1491 | PBMC | 2,153 | 21 | 5 |
| 1657 | PBMC | 274 ± 32.3 | 6 | 0 |
| Spleen | 40 ± 7.2 | 0 | 0 | |
| Mesenteric LN | 75 ± 8.4 | 1 | 0 | |
| Submaxillary LN | 22 ± 2.11 | 0 | 0 | |
| Axillary LN | 20 ± 2.14 | 0 | 0 | |
| Inguinal LN | 110 ± 12.9 | 2 | 0 | |
| Bone marrow | 20 ± 3.6 | 0 | 0 | |
| Salivary gland | 20 ± 2.2 | 0 | 0 | |
| Lung | ≤10 | 0 | 0 | |
| Pancreas | 48 ± 2.24 | 1 | 0 | |
| Intestine | 13 ± 1.5 | 0 | 0 | |
| Cervical spinal cord | ≤10 | 0 | 0 | |
| Lumbar spinal cord | 15 ± 1.6 | 0 | 0 |
Clonal expansion of HTLV-1-infected PBMCs in squirrel monkeys.
Results from quadruplicate analyses of the two monkey cell lines evidenced patterns similar to those observed with human HTLV-1 cell lines (Fig. 1) (5–8, 30, 32). PBMCs from monkey 1657 were analyzed 26 months postinfection, at which time the circulating proviral load was 274 of 150,000 PBMCs (Table 1). Figure 1 shows that six distinct clones of cells harboring an integrated HTLV-1 provirus were detected after quadruplicate LMPCR (4 × 0.5 μg). All of these clones were detected in a single sampling (clonal frequency of about 1/3,000). This corresponded to a calculated proviral load of ∼300 copies in 150,000 PBMCs, which was consistent with the value obtained after quantitative PCR (Table 1). Figure 1A shows that the six clones detected in the PBMCs from monkey 1657 were absent from the EVO/1540 cell line, demonstrating that they corresponded to newly infected cells rather than to persistent expanded allogeneic cells. Figure 1B shows that a typical pattern of clonal expansion of newly infected cells also characterized the PBMCs of monkey 1491. Taken together, these results indicate that in squirrel monkeys, the circulating HTLV-1 proviral burden results mainly from the clonal expansion of newly HTLV-1 infected PBMCs.
Cellular transport of the HTLV-1 provirus in lymphoid and nonlymphoid organs.
In ATLL, cellular transport of HTLV-1 as a provirus has been documented in various anatomic sites such as the skin, lymph nodes, and central nervous system (7, 34). It was recently observed that in patients with TSP/HAM, the virus could cross the blood-brain barrier via its host cell (4). During early HTLV-1 infection of squirrel monkeys, PBMCs, spleens, and lymph nodes serve as virus reservoirs (26). In addition, other organs could harbor proviral sequences in the late stage of experimental infection. Indeed, PCR amplification experiments with gag- and tax-specific primers were previously found to give a positive signal in animal 1657 when the DNA from the spleen, lymph nodes, bone marrow, salivary gland, lung, pancreas, intestine, and spinal cord was analyzed. To assess the distribution of HTLV-1 proviral sequences in these body compartments, we analyzed the proviral load and clonality pattern of HTLV-1 infected cells in 12 organs from monkey 1657. As shown in Table 1, the circulating proviral load in this animal was up to more than seven times higher than that of other organs analyzed. In addition, the mean proviral load of lymphoid organs was about twice as high as that of other sites displaying a positive signal: 47.8 versus 24 copies per μg, respectively. The clonal distribution of HTLV-1 integration sites was evidenced in two lymph nodes and in the pancreas. Given the sensitivity of the LMPCR, it appears that almost all the proviral sequences from these three organs were distributed among the four detected clones. As shown in Fig. 1, the signal at ∼420 bp observed for the pancreas and inguinal lymph node was also detected in the corresponding PBMCs. Similarly, the band at ∼160 bp observed for the inguinal lymph node was also present in the PBMCs. An additional band at ∼250 bp was also present in both PBMCs and the mesenteric lymph node. All of the organs tested were histologically normal. These results suggest that the four detected clones corresponded to infiltration of these organs by infected lymphoid cells. The proviral copy number of the nine remaining organs was below the LMPCR detection threshold.
Persistent clonal expansion with accumulation of infected cells within clones increases the proviral load over time.
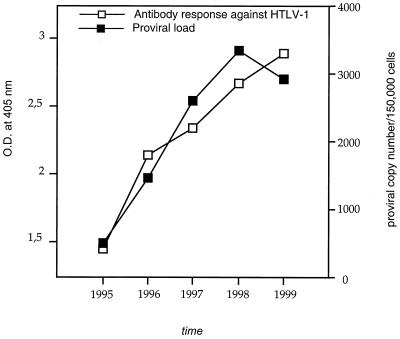
Clonal stability with fluctuating abundance of persistent clones characterizes HTLV-1-infected malignant or nonmalignant cells in humans (6, 12, 14, 32). The fact that 10 to 80% of human PBMCs from HTLV-1 infected individuals are capable of expressing Tax in vivo (20) suggests that such clonal stability results from the persistent proliferation of infected lymphocytes driven by the positive effect of Tax on cell cycling (13, 41, 45, 48). To assess the pattern of HTLV-1 replication in squirrel monkeys more precisely, the temporal stability of infected clones was examined in blood samples collected from animal S1491 on four occasions over a period of 4 years with 1-year intervals. Circulating clones of infected cells were evidenced in every sample, i.e., 3 months to 4 years after experimental infection, and the frequency of abundant clones (those detected more than once and having a clonal frequency of ≥1/1,500 PBMCs) increased as a function of time (Fig. 1B). In this experiment, numerous circulating HTLV-1-positive clones persisted over time in PBMCs. In addition, quadruplicate experiments revealed that there was a fluctuation in the clonal frequency of these persistent clones (Fig. 1B). For example, proportions of the clone corresponding to the signals at ∼140 bp were <1/3,000 in 1995, ≥1/300 in 1996, 1/1,500 in 1997, ≥1/300 in 1998, and 1/3,000 in 1999. Such fluctuation was clearly observed for other clones. However, as shown in Fig. 1B, the abundance of persistent circulating clones was higher in the last samples than in those collected early after experimental infection. The mean proviral loads of persistent clones, as calculated from their mean frequency of detection at each point, in samples collected in 1995, 1996, 1997, 1998, and 1999 were 25, 67, 117, 316, and 131, respectively. Real-time quantitative PCR analysis of the proviral load was assessed over time in animal S1491. As shown in Fig. 2, both the level of the anti-HTLV-1 antibody response, as measured by enzyme immunoassay (Cobas Core Anti-HTLV-I/II EIA; Roche, Basel, Switzerland), and the circulating HTLV-1 proviral copy number increased as a function of time. The proviral loads were found to correlate with both the intensity of the antibody response (Fig. 2; P = 0.005, R ∼ 0.9429, Spearman rank correlation) and the frequency of abundant clones (i.e., those having a clonal frequency higher than 1/300) (P = <10−4, R ∼ 0.99, Spearman rank correlation) rather than with the overall number of clones (P = 0.22, R ∼ 0.67).
FIG. 2.
Temporal evolution of circulating HTLV-1 proviral load and anti-HTLV-1 antibody titers in animal S1491. The first sample was collected 3 months after experimental infection. For the proviral load, the mean viral copy number obtained after three experiments is given at each time point. For the five samples, the standard deviation ranged from 3.3 to 11.8 and the mean coefficient of variance was 6%. O.D., optical density.
HTLV-1 replicates in squirrel monkeys as in humans.
The data presented here show an elevated HTLV-1 proviral load in PBMCs and lymphoid organs that results from the persistent clonal expansion of infected cells. This indicates that HTLV-1 replication at the late stage of experimental infection in squirrel monkeys is identical to that observed in humans (5, 6, 12, 14, 32, 52, 53). Indeed, such cell-associated proviral multiplication helps explain the low level of HTLV-1 genetic drift demonstrated in squirrel monkeys (25). At this stage of infection, quantitative PCR measurement of the proviral load confirms that PBMCs and lymphoid organs correspond to the major reservoirs for HTLV-1. A similar distribution of infected cells was previously reported for the rabbit model after experimental infection with lethally irradiated HTLV-1-infected cells from patients with ATLL or TSP/HAM (29). It is of note that the circulating proviral loads observed in experimentally infected squirrel monkeys (reference 26 and this study) are in the same range as those measured by quantitative competitive PCR in rabbit inoculated with irradiated cell lines expressing a wild-type molecular clone of HTLV-1 (2, 3). As for ATLL and TSP/HAM, two diseases in which there is cellular transport of the provirus in various anatomic sites (6, 7, 34), PCR amplification of proviral flanking sequences integrated in the DNA from squirrel monkeys suggests that dissemination of the virus in body compartments results from the same pathway. Persistent clonal expansion characterizes HTLV-1 replication in humans (6, 12, 14), and we previously showed that the number of abundant clones increased with age among asymptomatic carriers (5). The present study addresses for the first time the temporal evolution of both the proviral loads and clonality patterns of infected PBMCs in an asymptomatic monkey. Figure 1B shows that some clones of infected cells persist over time. Interestingly, we observed an accumulation of infected cells via persistent clonal expansion, which accounted for an increase of the circulating proviral load over time. The clonality pattern of infected PBMCs differed significantly between the two animals. S1657 harbored a relatively low proviral load together with the polyclonal expansion of only six circulating clones. By contrast, animal S1491 displayed, at the same time postinoculation, an elevated proviral load correlated with, and therefore resulting from, the persistent proliferation of a restricted number of abundant clones on a background of polyclonally expanded cells. This mode of infected cell proliferation is reminiscent of the HTLV-1 replication pattern observed in ATLL (7, 30, 32) and suggests that monkey S1491 presents an equivalent of the pre-ATLL stage (10). Prolonged follow-up of this animal will permit us to determine whether this replication pattern is associated with the development of malignancy.
The two-step nature of HTLV-1 primary infection in squirrel monkeys.
The results presented here strongly support the hypothesis that HTLV-1 primary infection is a two-step process: a transient first step of reverse transcription and integration characterized by a burst of viral expression, followed by the persistent multiplication of infected cells that account for the dissemination of the virus in the organism.
ATLL is a disease with long latency in the development of which the immune status of the infected individual plays an important role. Clinical observation and experimental investigation have shown that a decrease in the host cellular immunity against HTLV-1 led to malignant transformation (11, 19, 24, 51). Furthermore, additional cofactors such as parasitic or viral superinfections have been found to increase the risk for asymptomatic carriers to develop ATLL (1, 11, 15, 19, 39, 44). The finding that the persistent clonal expansion of HTLV-1-bearing cells precedes ATLL suggests that in ATLL (7), tumor cells may originate in a clonally expanding nonmalignant cell, presumably through the acquisition of subsequent mutations in genes such as p53 or p16 (9, 21). The results presented here suggest that squirrel monkeys constitute a promising animal model for assessing the impact of putative ATLL cofactors on the clonality of infected cells in vivo.
Finally, the two-step nature of HTLV-1 replication over time suggests that two specific approaches might be used to inhibit HTLV-1 replication in vivo. First, blocking the horizontal route of virus multiplication with drugs such as reverse transcriptase inhibitors may be suitable for the treatment of primary infection, i.e., during the 2 to 3 weeks that follow contamination. By contrast, such an approach might be inappropriate in the second phase of the infection, which is characterized by vertical dissemination of the virus via clonal expansion. However, at this stage, blocking the proliferation of infected T cells may help to reduce the proviral load, which is associated with the risk of developing HTLV-1-associated diseases (18, 27, 38, 47, 54).
Acknowledgments
We thank G. de Thé for initiating the HTLV-1 program in squirrel monkeys, A. Gessain for helpful discussion, and P. Wattre and collaborators who kindly welcomed us in their laboratories for DNA extraction, digestion, ligation, and PCR. We also thank Marie-Dominique Reynaud for assistance.
This work was supported by grants from the Association pour la Recherche sur le Cancer (ARC), the Fondation contre la Leucémie, the comité départemental du Rhône de la Ligue Nationale Contre le Cancer, and the Association Virus Cancer Prévention (VCP). F.M. was supported by funds from the Ministère de l'Enseignement Supérieur et de la Recherche and from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Agape P, Copin M C, Cavrois M, Panelatti G, Plumelle Y, Ossondo-Landeau M, Quist D, Grossat N, Gosselin B, Fenaux P, Wattel E. Implication of HTLV-I infection, strongyloidiasis, and P53 overexpression in the development, response to treatment, and evolution of non-Hodgkin's lymphomas in an endemic area (Martinique, French West Indies) J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:394–402. doi: 10.1097/00042560-199904010-00011. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht B, Collins N D, Newbound G C, Ratner L, Lairmore M D. Quantification of human T-cell lymphotropic virus type 1 proviral load by quantitative competitive polymerase chain reaction. J Virol Methods. 1998;75:123–140. doi: 10.1016/s0166-0934(98)00087-1. [DOI] [PubMed] [Google Scholar]
- 3.Bartoe J T, Albrecht B, Collins N D, Robek M D, Ratner L, Green P L, Lairmore M D. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J Virol. 2000;74:1094–1100. doi: 10.1128/jvi.74.3.1094-1100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavrois M, Gessain A, Gout O, Wain-Hobson S, Wattel E. Common human T cell leukemia virus type 1 (HTLV-1) integration sites in cerebrospinal fluid and blood lymphocytes of patients with HTLV-1-associated myelopathy/tropical spastic paraparesis indicate that HTLV-1 crosses the blood-brain barrier via clonal HTLV-1-infected cells. J Infect Dis. 2000;182:1044–1050. doi: 10.1086/315844. [DOI] [PubMed] [Google Scholar]
- 5.Cavrois M, Gessain A, Wain-Hobson S, Wattel E. Proliferation of HTLV-1 infected circulating cells in vivo in all asymptomatic carriers and patients with TSP/HAM. Oncogene. 1996;12:2419–2423. [PubMed] [Google Scholar]
- 6.Cavrois M, Leclercq I, Gout O, Gessain A, Wain-Hobson S, Wattel E. Persistent oligoclonal expansion of human T-cell leukemia virus type 1-infected circulating cells in patients with tropical spastic paraparesis/HTLV-1 associated myelopathy. Oncogene. 1998;17:77–82. doi: 10.1038/sj.onc.1201906. [DOI] [PubMed] [Google Scholar]
- 7.Cavrois M, Wain-Hobson S, Gessain A, Plumelle Y, Wattel E. Adult T-cell leukemia/lymphoma on a background of clonally expanding human T-cell leukemia virus type-1-positive cells. Blood. 1996;88:4646–4650. [PubMed] [Google Scholar]
- 8.Cavrois M, Wain-Hobson S, Wattel E. Stochastic events in the amplification of HTLV-I integration sites by linker-mediated PCR. Res Virol. 1995;146:1779–1784. doi: 10.1016/0923-2516(96)80578-4. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Chadburn A, Inghirami G, Gaidano G, Knowles D M. Structural and functional analysis of oncogenes and tumor suppressor genes in adult T-cell leukemia/lymphoma shows frequent p53 mutations. Blood. 1992;80:3205–3216. [PubMed] [Google Scholar]
- 10.Chen Y X, Ikeda S, Mori H, Hata T, Tsukasaki K, Momita S, Yamada Y, Kamihira S, Mine M, Tomonaga M. Molecular detection of pre-ATL state among healthy HTLV-1 carriers in an endemic area of Japan. Int J Cancer. 1995;60:798–801. doi: 10.1002/ijc.2910600612. [DOI] [PubMed] [Google Scholar]
- 11.D'Incan M, Combemale P, Verrier B, Garin D, Audoly G, Brunot J, Desgranges C, Flechaire A. Transient adult T-cell leukemia/lymphoma picture during varicella infection in an HTLV-1 carrier. Leukemia. 1994;8:682–687. [PubMed] [Google Scholar]
- 12.Etoh K, Tamiya S, Yamaguchi K, Okayama A, Tsubouchi H, Ideta T, Mueller N, Takatsuki K, Matsuoka M. Persistent clonal proliferation of human T-lymphotropic virus type I-infected cells in vivo. Cancer Res. 1997;57:4862–4867. [PubMed] [Google Scholar]
- 13.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 14.Furukawa Y, Fujisawa J, Osame M, Toita M, Sonoda S, Kubota R, Ijichi S, Yoshida M. Frequent clonal proliferation of human T-cell leukemia virus type 1 (HTLV-1)-infected T cells in HTLV-1-associated myelopathy (HAM-TSP) Blood. 1992;80:1012–1016. [PubMed] [Google Scholar]
- 15.Gabet A, Mortreux F, Talarmin A, Plumelle Y, Leclercq I, Leroy A, Gessain A, Wattel E. High circulating proviral load with oligoclonal expansion of HTLV-1 bearing T cells in HTLV-1 carriers with strongyloidiasis. Oncogene. 2000;19:4954–4960. doi: 10.1038/sj.onc.1203870. [DOI] [PubMed] [Google Scholar]
- 16.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;2:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 17.Gessain A, Gallo R C, Franchini G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol. 1992;66:2288–2295. doi: 10.1128/jvi.66.4.2288-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gessain A, Saal F, Gout O, Daniel M T, Flandrin G, de-The G, Peries J, Sigaux F. High human T-cell lymphotropic virus type I proviral DNA load with polyclonal integration in peripheral blood mononuclear cells of French West Indian, Guianese, and African patients with tropical spastic paraparesis. Blood. 1990;75:428–433. [PubMed] [Google Scholar]
- 19.Hanabuchi S, Ohashi T, Koya Y, Kato H, Takemura F, Hirokawa K, Yoshiki T, Yagita H, Okumura K, Kannagi M. Development of human T-cell leukemia virus type 1-transformed tumors in rats following suppression of T-cell immunity by CD80 and CD86 blockade. J Virol. 2000;74:428–435. doi: 10.1128/jvi.74.1.428-435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanon E, Hall S, Taylor G P, Saito M, Davis R, Tanaka Y, Usuku K, Osame M, Weber J N, Bangham C R. Abundant tax protein expression in CD4+ T cells infected with human T-cell lymphotropic virus type I (HTLV-I) is prevented by cytotoxic T lymphocytes. Blood. 2000;95:1386–1392. [PubMed] [Google Scholar]
- 21.Hatta Y, Hirama T, Miller C W, Yamada Y, Tomonaga M, Koeffler H P. Homozygous deletions of the p15 (MTS2) and p16 (CDKN2/MTS1) genes in adult T-cell leukemia. Blood. 1995;85:2699–2704. [PubMed] [Google Scholar]
- 22.Ibuki K, Funahashi S I, Yamamoto H, Nakamura M, Igarashi T, Miura T, Ido E, Hayami M, Shida H. Long-term persistence of protective immunity in cynomolgus monkeys immunized with a recombinant vaccinia virus expressing the human T cell leukaemia virus type I envelope gene. J Gen Virol. 1997;78:147–152. doi: 10.1099/0022-1317-78-1-147. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro N, Abe M, Seto K, Sakurai H, Ikeda H, Wakisaka A, Togashi T, Tateno M, Yoshiki T. A rat model of human T lymphocyte virus type I (HTLV-I) infection. 1. Humoral antibody response, provirus integration, and HTLV-I-associated myelopathy/tropical spastic paraparesis-like myelopathy in seronegative HTLV-I carrier rats. J Exp Med. 1992;176:981–989. doi: 10.1084/jem.176.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenks P J, Barrett W Y, Raftery M J, Kelsey S M, van-der-Walt J D, Kon S P, Breuer J. Development of human T-cell lymphotropic virus type I-associated adult T-cell leukemia/lymphoma during immunosuppressive treatment following renal transplantation. Clin Infect Dis. 1995;21:992–993. doi: 10.1093/clinids/21.4.992. [DOI] [PubMed] [Google Scholar]
- 25.Kazanji M, Moreau J P, Mahieux R, Bonnemains B, Bomford R, Gessain A, de The G. HTLV-I infection in squirrel monkeys (Saimiri sciureus) using autologous, homologous, or heterologous HTLV-I-transformed cell lines. Virology. 1997;231:258–266. doi: 10.1006/viro.1997.8528. [DOI] [PubMed] [Google Scholar]
- 26.Kazanji M, Ureta-Vidal A, Ozden S, Tangy F, de Thoisy B, Fiette L, Talarmin A, Gessain A, de The G. Lymphoid organs as a major reservoir for human T-cell leukemia virus type 1 in experimentally infected squirrel monkeys (Saimiri sciureus): provirus expression, persistence, and humoral and cellular immune responses. J Virol. 2000;74:4860–4867. doi: 10.1128/jvi.74.10.4860-4867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kira J, Koyanagi Y, Yamada T, Itoyama Y, Goto I, Yamamoto N, Sasaki H, Sakaki Y. Increased HTLV-I proviral DNA in HTLV-I-associated myelopathy: a quantitative polymerase chain reaction study. Ann Neurol. 1991;29:194–201. doi: 10.1002/ana.410290214. . (Erratum, 29:363.) [DOI] [PubMed] [Google Scholar]
- 28.LaGrenade L, Hanchard B, Fletcher V, Cranston B, Blattner W. Infective dermatitis of Jamaican children: a marker for HTLV-I infection. Lancet. 1990;336:1345–1347. doi: 10.1016/0140-6736(90)92896-p. [DOI] [PubMed] [Google Scholar]
- 29.Lairmore M D, Roberts B, Frank D, Rovnak J, Weiser M G, Cockerell G L. Comparative biological responses of rabbits infected with human T-lymphotropic virus type I isolates from patients with lymphoproliferative and neurodegenerative disease. Int J Cancer. 1992;50:124–130. doi: 10.1002/ijc.2910500125. [DOI] [PubMed] [Google Scholar]
- 30.Leclercq I, Cavrois M, Mortreux F, Hermine O, Gessain A, Morschhauser F, Wattel E. Oligoclonal proliferation of human T-cell leukaemia virus type 1 bearing T cells in adult T-cell leukaemia/lymphoma without deletion of the 3′ provirus integration sites. Br J Haematol. 1998;101:500–506. doi: 10.1046/j.1365-2141.1998.00743.x. [DOI] [PubMed] [Google Scholar]
- 31.Leclercq I, Mortreux F, Cavrois M, Leroy A, Gessain A, Wain-Hobson S, Wattel E. Host sequences flanking the human T-cell leukemia virus type 1 provirus in vivo. J Virol. 2000;74:2305–2312. doi: 10.1128/jvi.74.5.2305-2312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leclercq I, Mortreux F, Morschhauser F, Duthilleul P, Desgranges C, Gessain A, Cavrois M, Vernant J P, Hermine O, Wattel E. Semiquantitative analysis of residual disease in patients treated for adult T-cell leukaemia/lymphoma (ATLL) Br J Haematol. 1999;105:743–751. doi: 10.1046/j.1365-2141.1999.01389.x. [DOI] [PubMed] [Google Scholar]
- 33.Leon-Monzon M, Illa I, Dalakas M C. Polymyositis in patients infected with human T-cell leukemia virus type I: the role of the virus in the cause of the disease. Ann Neurol. 1994;36:643–649. doi: 10.1002/ana.410360414. [DOI] [PubMed] [Google Scholar]
- 34.Marshall A G, Pawson R, Thom M, Schulz T F, Scaravilli F, Rudge P, Schutz T F. HTLV-I associated primary CNS T-cell lymphoma. J Neurol Sci. 1998;158:2226–2231. doi: 10.1016/s0022-510x(98)00111-7. . (Erratum, 162:210, 1999.) [DOI] [PubMed] [Google Scholar]
- 35.Mattos K, Queiroz C, Pecanha-Martins A C, Publio L, Vinhas V, Melo A. Lymphocyte alveolitis in HAM/TSP patients. Preliminary report. Arq Neuropsiquiatr. 1993;51:134–136. doi: 10.1590/s0004-282x1993000100022. [DOI] [PubMed] [Google Scholar]
- 36.Minghetti P P, Ruffner D E, Kuang W J, Dennison O E, Hawkins J W, Beattie W G, Dugaiczyk A. Molecular structure of the human albumin gene is revealed by nucleotide sequence within q11–22 of chromosome 4. J Biol Chem. 1986;261:6747–6757. [PubMed] [Google Scholar]
- 37.Mochizuki M, Tajima K, Watanabe T, Yamaguchi K. Human T lymphotropic virus type 1 uveitis. Br J Ophthalmol. 1994;78:149–154. doi: 10.1136/bjo.78.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai M, Usuku K, Matsumoto W, Kodama D, Takenouchi N, Moritoyo T, Hashiguchi S, Ichinose M, Bangham C R, Izumo S, Osame M. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol. 1998;4:586–593. doi: 10.3109/13550289809114225. [DOI] [PubMed] [Google Scholar]
- 39.Nakada K, Yamaguchi K, Furugen S, Nakasone T, Nakasone K, Oshiro Y, Kohakura M, Hinuma Y, Seiki M, Yoshida M, Matutes E, Catovsky D, Ishii T, Takatsuky K. Monoclonal integration of HTLV-I proviral DNA in patients with strongyloidiasis. Int J Cancer. 1987;40:145–148. doi: 10.1002/ijc.2910400203. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura H, Tanaka Y, Komuro-Tsujimoto A, Ishikawa K, Takadaya K, Tozawa H, Tsujimoto H, Honjo S, Hayami M. Experimental inoculation of monkeys with autologous lymphoid cell lines immortalized by and producing human T-cell leukemia virus type-I. Int J Cancer. 1986;38:867–875. doi: 10.1002/ijc.2910380614. [DOI] [PubMed] [Google Scholar]
- 41.Neuveut C, Low K G, Maldarelli F, Schmitt I, Majone F, Grassmann R, Jeang K T. Human T-cell leukemia virus type 1 Tax and cell cycle progression: role of cyclin D-cdk and p110Rb. Mol Cell Biol. 1998;18:3620–3632. doi: 10.1128/mcb.18.6.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicot C, Astier-Gin T, Edouard E, Legrand E, Moynet D, Vital A, Londos-Gagliardi D, Moreau J P, Guillemain B. Establishment of HTLV-I-infected cell lines from French, Guianese and West Indian patients and isolation of a proviral clone producing viral particles. Virus Res. 1993;30:317–334. doi: 10.1016/0168-1702(93)90099-9. [DOI] [PubMed] [Google Scholar]
- 43.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-I associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 44.Plumelle Y, Gonin C, Edouard A, Bucher B J, Thomas L, Brebion A, Panelatti G. Effect of Strongyloides stercoralis infection and eosinophilia on age at onset and prognosis of adult T-cell leukemia. Am J Clin Pathol. 1997;107:81–87. doi: 10.1093/ajcp/107.1.81. [DOI] [PubMed] [Google Scholar]
- 45.Santiago F, Clark E, Chong S, Molina C, Mozafari F, Mahieux R, Fujii M, Azimi N, Kashanchi F. Transcriptional up-regulation of the cyclin D2 gene and acquisition of new cyclin-dependent kinase partners in human T-cell leukemia virus type 1-infected cells. J Virol. 1999;73:9917–9927. doi: 10.1128/jvi.73.12.9917-9927.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato K, Maruyama I, Maruyama Y, Kitajima I, Nakajima Y, Higaki M, Yamamoto K, Miyasaka N, Osame M, Nishioka K. Arthritis in patients infected with human T lymphotropic virus type I. Clinical and immunopathologic features. Arthritis Rheum. 1991;34:714–721. doi: 10.1002/art.1780340612. [DOI] [PubMed] [Google Scholar]
- 47.Shinzato O, Ikeda S, Momita S, Nagata Y, Kamihira S, Nakayama E, Shiku H. Semiquantitative analysis of integrated genomes of human T-lymphotropic virus type I in asymptomatic virus carriers. Blood. 1991;78:2082–2088. [PubMed] [Google Scholar]
- 48.Suzuki T, Kitao S, Matsushime H, Yoshida M. HTLV-1 Tax protein interacts with cyclin-dependent kinase inhibitor p16INK4A and counteracts its inhibitory activity towards CDK4. EMBO J. 1996;15:1607–1614. [PMC free article] [PubMed] [Google Scholar]
- 49.Taguchi H, Sawada T, Fukushima A, Iwata J, Ohtsuki Y, Ueno H, Miyoshi I. Bilateral uveitis in a rabbit experimentally infected with human T-lymphotropic virus type I. Lab Investig. 1993;69:336–339. [PubMed] [Google Scholar]
- 50.Terada K, Katamine S, Eguchi K, Moriuchi R, Kita M, Shimada H, Yamashita I, Iwata K, Tsuji Y, Nagataki S, Miyamoto T. Prevalence of serum and salivary antibodies to HTLV-1 in Sjögren's syndrome. Lancet. 1994;344:1116–1119. doi: 10.1016/s0140-6736(94)90630-0. [DOI] [PubMed] [Google Scholar]
- 51.Tsurumi H, Tani K, Tsuruta T, Shirato R, Matsudaira T, Tojo A, Wada C, Uchida H, Ozawa K, Asano S. Adult T-cell leukemia developing during immunosuppressive treatment in a renal transplant recipient. Am J Hematol. 1992;41:292–294. doi: 10.1002/ajh.2830410414. [DOI] [PubMed] [Google Scholar]
- 52.Wattel E. Proliferation of HTLV-1 infected cells in vivo: pathogenic implications in leukemogenesis and neuropathogenesis. In: Semmes O J, Hannarskjöld M L, editors. Molecular pathogenesis of HTLV-1, a current perspective. Arlington, Va: ABI Professional Publications; 1999. pp. 139–152. [Google Scholar]
- 53.Wattel E, Cavrois M, Gessain A, Wain-Hobson S. Clonal expansion of infected cells—a way of life for HTLV-1. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl. 1):92–99. doi: 10.1097/00042560-199600001-00016. [DOI] [PubMed] [Google Scholar]
- 54.Wattel E, Mariotti M, Agis F, Gordien E, Le Coeur F F, Prin L, Rouger P, Chen I S, Wain-Hobson S, Lefrere J J. Quantification of HTLV-1 proviral copy number in peripheral blood of symptomless carriers from the French West Indies. J Acquir Immune Defic Syndr. 1992;5:943–946. [PubMed] [Google Scholar]
- 55.Wattel E, Vartanian J P, Pannetier C, Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol. 1995;69:2863–2668. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamanouchi K, Kinoshita K, Moriuchi R, Katamine S, Amagasaki T, Ikeda S, Ichimaru M, Miyamoto T, Hino S. Oral transmission of human T-cell leukemia virus type-I into a common marmoset (Callithrix jacchus) as an experimental model for milk-borne transmission. Jpn J Cancer Res. 1985;76:481–487. [PubMed] [Google Scholar]