Abstract
The Hsp90 chaperone cycle catalyzes the final activation step of several important eukaryotic proteins (Hsp90 “clients”). Although largely a functional form of Hsp90, an Hsp90-Gal4p DNA binding domain fusion (Hsp90-BD) displays no strong interactions in the yeast two-hybrid system, consistent with a general transience of most Hsp90-client associations. Strong in vivo interactions are though detected when the E33A mutation is introduced into this bait, a mutation that should arrest Hsp90-client complexes at a stage where the client is stabilized, yet prevented from attaining its active form. This E33A mutation stabilized the two-hybrid interactions of the Hsp90-BD fusion with ∼3% of the Saccharomyces cerevisiae proteome in a screen of the 6,000 yeast proteins expressed as fusions to the Gal4p activation domain (AD). Among the detected interactors were the two stress-activated mitogen-activated protein (MAP) kinases of yeast, Hog1p and Slt2p (Mpk1p). Column retention experiments using wild-type and mutant forms of Hsp90 and Slt2p MAP kinase, as well as quantitative measurements of the effects of stress on the two-hybrid interaction of mutant Hsp90-BD and AD-Slt2p fusions, revealed that Hsp90 binds exclusively to the dually Thr/Tyr-phosphorylated, stress-activated form of Slt2p [(Y-P,T-P)Slt2p] and also to the MAP kinase domain within this (Y-P,T-P)Slt2p. Phenotypic analysis of a yeast mutant that expresses a mutant Hsp90 (T22Ihsp82) revealed that Hsp90 function is essential for this (Y-P,T-P)Slt2p to activate one of its downstream targets, the Rlm1p transcription factor. The interaction between Hsp90 and (Y-P,T-P)Slt2p, characterized in this study, is probably essential in this Hsp90 facilitation of the Rlm1p activation by Slt2p.
Hsp90 is an essential and abundant molecular chaperone of eukaryotic cells. It orchestrates the chaperone cycle that catalyzes the final activation step of several of their key signaling and regulatory proteins (targets hereafter designated Hsp90 “clients”). Rather than acting at an early stage of protein folding, Hsp90 binds these clients when they are already substantially folded, so as to induce the structural changes and protein associations whereby they attain their full activity. A number of cochaperone proteins assist in these events, becoming associated with the Hsp90-based complex at different steps of the multistage chaperone cycle (reviewed in references 8, 45, 46, 48, 50, and 71).
One of the best-understood events of client activation catalyzed by this chaperone cycle is the process whereby certain steroid hormone receptors (SHRs) become competent for steroid binding. Here the cochaperone p60/Hop is present in the early-stage Hsp90-SHR complexes, prior to the attainment of the steroid-activatable state. These early complexes then progress to later-stage, more mature Hsp90-SHR complexes, where the SHR is competent for activation though the binding of the steroid. In these later-stage complexes, certain other cochaperones (p23 and also immunophilins such as Cyp40 and FKBP52) have replaced p60/Hop in the Hsp90-based complex (48). In some instances the association of a cochaperone with the Hsp90-client complex may be specific for a particular class of client. Thus, Hsp90 complexes with protein kinases often contain p50/Cdc37p, whereas those with SHRs generally do not (48, 59).
Proteins tend to be identified as Hsp90 clients on an ad hoc basis, as workers identify their protein of interest as having Hsp90 binding properties and an activity that is susceptible to highly selective inhibitors of Hsp90 function (notably the antibiotics geldanamycin and radicicol). Hsp90 is frequently also identified by mass spectrometry as a component of multiprotein complexes (63). No in vivo screen for identifying Hsp90 interactors has yet been developed. In this report we describe a modification of the yeast two-hybrid system that allows such a screen. One of the novel interactions discovered in this way, that between Hsp90 and a mitogen-activated protein (MAP) kinase, is characterized in detail. Kinases of the MAP kinase class constitute the terminal kinases of phosphorylation cascades controlling cell proliferation, differentiation, stress responses, and cell survival (27, 60, 61). These important protein kinases are not generally considered Hsp90 clients, though certain kinases modestly related to conventional MAP kinases are known to bind Hsp90 (39). Nevertheless, through protein binding studies and genetic analysis, we revealed that the function of the Hsp90 chaperone is essential for one stress-activated MAP kinase, Slt2p (Mpk1p), to activate a downstream target. Hsp90 binding by this Slt2p is selective for the dually Tyr/Thr-phosphorylated, stress-activated form of the MAP kinase.
MATERIALS AND METHODS
Construction of “bait” constructs comprising mutant versions of the Hsp90-BD fusion.
Saccharomyces cerevisiae expressing point mutant forms of yeast Hsp82 with a C-terminal Gal4p binding domain (BD) extension (mutant Hsp82-BD fusions) were constructed by a slight modification of the procedure used to make the strains expressing a native, wild-type Hsp82-BD fusion (see reference 38 for details). Instead of using the wild-type HSP82 gene as a template in the first of two sequential PCRs, well-characterized mutant hsp82 alleles from an earlier study were employed (49). Strain PJ69-4α (MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) (20) was then transformed with the product of the second PCR and NcoI-digested pBDC, so as to generate the gene encoding the mutant Hsp82-BD fusion by homologous recombination within the yeast (38). Transformants were initially selected by plating on dropout agar medium (DO) lacking tryptophan (3) and then checked for the presence of the correct fusion gene by colony PCR (using the same primers as in the second round of the above PCR amplification) and by Western blotting (using an anti-Gal4p BD antiserum [Clontech]).
Two-hybrid screening of protein interactions.
Baits in PJ69-4α were checked for self-activation by plating onto DO minus tryptophan and histidine and containing increasing concentrations (0 to 8 mM) of 3-amino-1,2,4-triazole (3-AT). Having ensured, from the absence of appreciable growth, that the introduced mutations did not lead to self-activation, these baits in PJ69-4α were mated to PJ69-4a (20) transformants from an earlier study (37), the latter expressing fusions of the Gal4p activation domain (AD) to known Hsp90 system cochaperones and Hsp90 clients. They were also mated to a previously described (64) 16-plate, 384-well format array of yeast AD-protein fusions carried in strain PJ69-4a. All replications and inoculations were carried out using the 96- or 384-pin replicators of a Biomek 2000 Laboratory Automation Workstation, with movements programmed using the BioWorks Version software (Beckman).
After mating, PJ69-4 diploids were selected by plating onto DO minus leucine and tryptophan. Screening for protein-protein interactions was by the pinning of these diploids onto DO lacking leucine, tryptophan, and histidine and supplemented with increasing concentrations (0 to 20 mM) of 3-AT. Growth on these plates was scored after 4, 10, and 16 days at 30°C. As with the earlier screens of the AD-protein fusion array (37, 64), two identically performed matings of each bait to the array were conducted.
Measurements of interaction-responsive lacZ expression.
Automated measurement of β-galactosidase activity due to the basal and stress-induced expression of the interaction-responsive, GAL7 promoter-regulated lacZ gene of PJ69-4 was done essentially as described previously(37); data shown (see Fig. 3 and 4) are the means and standard deviations of eight assays. The control cells were those containing pBDC (38) lacking any gene insert and the pOAD-derived plasmid for the expression of the AD fusion, since the basal lacZ expression in this system is generally due to the AD-protein fusion, and cells containing the vector for Hsp82-BD expression and empty pOAD display an even lower basal lacZ expression that is unaffected by stress (37).
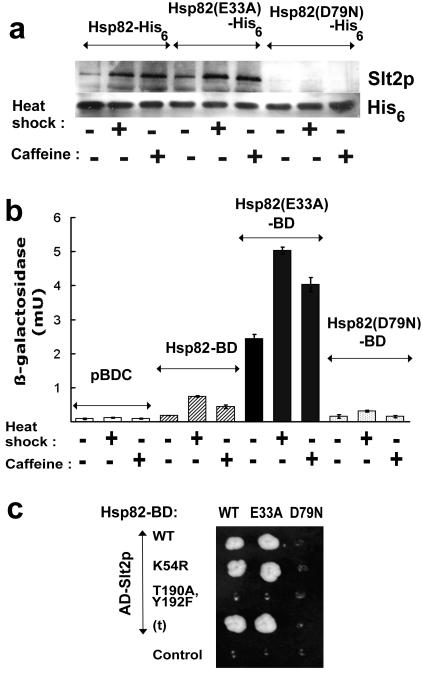
FIG. 3.
Effects of heat shock and caffeine stress on interaction of Hsp82 with the Slt2p MAP kinase. (a) Analysis of the level of Hsp82-His6-bound Slt2p in extracts from unstressed (25°C), heat shocked (from 25 to 39°C for 1 h), and caffeine-treated (8 mM for 1 h) cells expressing either a wild-type, an E33A mutant, or a D79N mutant Hsp82-His6. The Western blot was probed with anti-Slt2p and anti-His6 antisera. (b) Quantitation of the strength of Hsp82-BD-AD-Slt2p interaction (measurements of interaction-responsive lacZ expression in strain PJ694), showing that the Hsp82-BD-AD-Slt2p interaction in the two-hybrid system is moderately reinforced by stress, strongly reinforced by the E33A mutation, which inhibits the ATPase reaction of Hsp82, but abolished by the D79N mutation, inhibiting ATP/ADP binding by the chaperone. The control cells (pBDC) expressed AD-Slt2p but contained empty pBDC, since basal lacZ expression in this system is generally due to the AD fusion (37). (c) 3-AT-resistant growth of the same PJ694 cells expressing the wild-type (WT) or the E33A or D79N mutant form of Hsp82-BD in conjunction with either a wild-type AD-Slt2p fusion (WT), a nonphosphorylatable (T190A,Y192F) or phosphorylatable, yet kinase-dead (K54R), mutant form of this AD-Slt2p, or a C terminally truncated AD-Slt2p [AD-Slt2(t)p] (t). Growth was for 16 days at 30°C on DO minus leucine, tryptophan, and histidine and supplemented with 2 mM 3-AT.
FIG. 4.
Interaction between Hsp82 and Slt2p is abolished by mutations that prevent dual Thr190/Tyr192 phosphorylation of Slt2p. (a) Ni-NTA-retained Hsp82 in unstressed (25°C), heat shocked (from 25 to 39°C for 1 h), and caffeine-treated (8 mM for 1 h) cells of the p82aslt2Δ strain. Cultures expressed either the wild-type Slt2-His6, a nonphosphorylatable (T190A,Y192F mutant) Slt2-His6, or a phosphorylatable, yet catalytically dead (K54R mutant), Slt2p-His6 as their sole form of Slt2p MAP kinase. Since the cells also expressed just the Hsp82 isoform of yeast Hsp90, the protein detected by the anti-yeast Hsp90 antibody is entirely this isoform. (b) lacZ expression measurement of the basal and stress-induced two-hybrid interactions of the Hsp82-BD bait with a wild-type AD-Slt2p fusion and with nonphosphorylatable (T190A,Y192F) and phosphorylatable, yet kinase-dead (K54R), mutant forms of this AD-Slt2p The control cells (pBDC) expressed wild-type AD-Slt2p but contained the empty pBDC vector.
Protein binding studies.
TRP1 vectors for TDH1 promoter-regulated expression of a C terminally His6-tagged wild-type Hsp82 (Hsp82-His6) and also an E33A or D79N mutant form of this Hsp82-His6 were constructed by PCR amplifying the appropriate wild-type or mutant HSP82 genes from the plasmid DNAs of an earlier study (43) (primer sequences available on request). Products of these PCRs were then digested with BamHI plus SalI, prior to insertion into BamHI-plus-XhoI-cleaved pG1 (55).
A URA3 vector for MET25 promoter-regulated expression of a C terminally His6-tagged Slt2p (Slt2-His6) was constructed by first PCR amplifying the SLT2 gene from yeast chromosomal DNA, using primers GGACTAGTATGGCTGATAAGATAGAGAGG and CCATCGATCTAATGATGATGATGATGATGACCAGGTTCGTCAGCTGGATCATGCCA (restriction sites underlined; His-tag sequence in bold). This was then inserted as a SpeI/ClaI fragment into pUT36 (pUT36 being pUG36 (http://www.mips.biochem.mpg.de/proj/yeast/info/tools/hegemann/gfp/html) with the XbaI fragment containing the green fluorescent protein gene excised). Plasmids for Slt2(T190A,Y192F)-His6 and Slt2(K54R)-His6 expression were derived from this vector using the QuickChange Mutagenesis kit (Stratagene), the introduced mutations being confirmed by sequencing. These Slt2-His6, Slt2(T190A,Y192F)-His6, and Slt2(K54R)-His6 expression vectors were then inserted into p82aslt2Δ (a strain constructed by kanMX4 cassette deletion of the chromosomal SLT2 gene of p82a (40). The MKK1S386P gene was also inserted into pUT36, but as an XbaI/XhoI fragment PCR amplified from the vector described by Watanabe et al. (65).
Preparation of total yeast protein extracts was as described previously (43), the different forms of Slt2p-His6 or Hsp82-His6 in these extracts being isolated using nickel-nitrilotriacetate (Ni-NTA) chromatography. Western blot analysis of yeast Hsp90, Slt2p, and Sba1p used, as the primary antibody, rabbit polyclonal antisera raised against these proteins. Analysis of His6- tagged proteins used a mouse tetra-His antibody (QIAGEN). The secondary antibody was horseradish peroxidase-anti-rabbit or -anti-mouse immunoglobulin G (Amersham) diluted 2,000-fold. Analysis of the active form of Slt2p used an anti-(Thr202/Tyr204)-p44/42 MAP kinase antiserum (New England Biolabs) which specifically recognizes the dually Thr190/Tyr192-phosphorylated Slt2p in yeast extracts (34). Enhanced chemiluminescence reagents (Amersham) were used for detection. Measurements of Rlm1p transcription factor activity used a YIL117c promoter-lacZ reporter plasmid (22).
RESULTS
A genomic screen for two-hybrid interactions strongly reinforced by the E33A mutation in an Hsp90-BD bait fusion.
The yeast two-hybrid system detects in vivo protein-protein association as the reconstitution of transcription factor activity, the DNA binding domain (BD) and the activation domain (AD) of this transcription factor being expressed on different protein fusions. Reconstitution of an active transcription factor requires these BD- and AD-bearing fusions (“bait” and “prey,” respectively) to associate noncovalently in the yeast nucleus (3). In proteome-wide screens it is generally an interaction-responsive HIS3 gene that is monitored (64), an activity scored as growth over 4 to 16 days in the absence of histidine and the presence of 3-AT (an inhibitor of the HIS3 product). Growth under these conditions requires a high, sustained activation of the HIS3 gene and therefore a relatively long-lived, stable association of the “bait” and “prey” fusions. It is unlikely that the in vivo associations of Hsp90 with its clients could be readily detected by such a system, as these tend to be rather transient (involving a chaperone cycle thought to take place over a time scale of minutes (52, 57). Indeed, two-hybrid bait fusions comprising the Gal4p BD positioned at the C terminus of either isoform of yeast Hsp90 (Hsp82-BD or Hsc82-BD) display just a few, weak interactions when screened against the complete yeast proteome expressed as fusions to the Gal4p AD (37, 38). To detect Hsp90 interactors by an in vivo screen of this nature, it would probably be necessary to increase very considerably the length of time that these are in association with Hsp90 (i.e., to slow the chaperone cycle at a stage where the client is both stable and chaperone bound).
It is thought that the ATPase reaction of Hsp90 constitutes the rate-limiting step of the Hsp90 chaperone cycle, this ATP turnover rate therefore determining the length of time a client is bound by Hsp90 (35, 49, 69). We therefore investigated if it might be possible to stabilize the two-hybrid associations of an Hsp82-BD fusion by the mutation in this fusion of glutamate 33, the residue acting as a general base in the ATPase reaction of Hsp90 (51). Such a mutation should inhibit the final ATPase step of the chaperone cycle but allow this cycle to progress to a late-stage complex where the client is stabilized in association with the ATP-bound Hsp90 (43).
Strain PJ694α was engineered to express an E33A mutant form of Hsp82-BD, [Hsp82(E33A)-BD; see Materials and Methods]. Also constructed was an Hsp82(D79N)-BD fusion where the sole amino acid of the N-terminal domain of yeast Hsp82 that directly contacts the bound ATP/ADP is mutated, leading to loss of ADP/ATP binding by the chaperone (43). As neither mutation caused the Hsp82-BD fusion to self-activate, PJ694α cells expressing either Hsp82(E33A)-BD, Hsp82(D79N)-BD, or the control (Hsp82-BD) fusion, were mated to cells of the opposite mating type (PJ694a) that express AD fusions to different cochaperones or AD fusions to some of the known Hsp90 clients of yeast (AD-Ste11p [31], AD-Gcn2p [10], and AD-Hap1p [29]). The resultant diploids were next replicated onto DO lacking tryptophan, leucine, or histidine but containing either 4 or 8 mM 3-AT, and the plates then incubated at 30°C.
The presence of the E33A mutation in the Hsp82-BD fusion generated strong two-hybrid interactions with most of these cochaperones and known Hsp90 clients, as shown by growth of the yeast in the presence of 4 mM 3-AT (Fig. 1). In contrast, the D79N mutation generated no such effect (only the AD-Gcn2p interaction appearing to be reinforced by D79N in this initial screen [Fig. 1], a result not investigated further). Since this preliminary analysis had indicated that the E33A mutation was reinforcing Hsp82-BD interactions to a sufficient extent to allow a genomic screen for the in vivo binding partners of Hsp90, Hsp82(E33A)-BD and the control Hsp82-BD fusion were next introduced into the previously described 16-plate, 384-colony format array of colonies of the opposite mating type, each of which expresses a different AD-yeast protein fusion (64) (see Materials and Methods). Following mating, the cells were pinned onto DO minus tryptophan or leucine, thereby selecting the diploids expressing both Hsp82(E33A)-BD (or the control Hsp82-BD) and an AD-protein fusion. These diploid colonies were next transferred to DO minus tryptophan, leucine, or histidine but containing 4, 8, or 20 mM 3-AT. Then, after 4, 8, and 16 days of growth at 30°C, the colonies scoring positive for HIS3 gene activation were identified from their growth at distinct positions on the arrays. As self-activators can arise in two-hybrid screens by spontaneous mutation of the interactor fusions (3), the mating of each bait to the genomic array of AD-yeast protein fusions was performed in duplicate, the positive interacting partners being scored as the “double hits” identified in both of the identically executed screens.
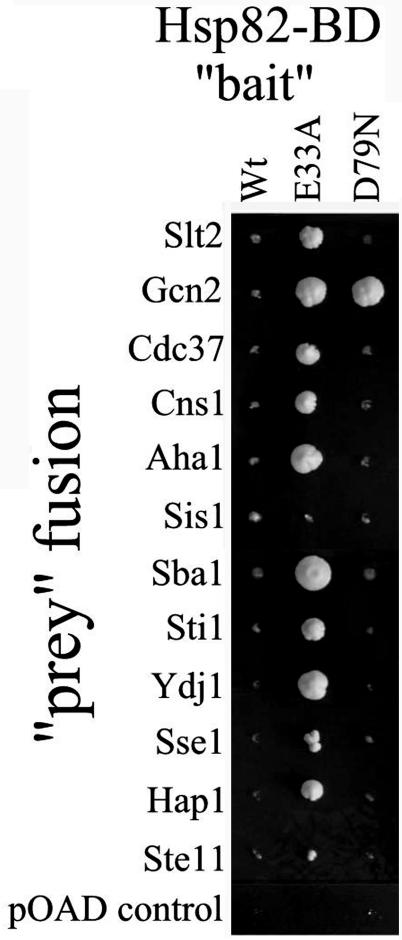
FIG. 1.
Effects of the E33A and D79N mutations in an Hsp82-BD bait fusion on two-hybrid interaction of this fusion with selected Hsp90 system cochaperones and clients. Growth was for 12 days at 30°C on DO lacking leucine, tryptophan, and histidine and supplemented with 4 mM 3-AT.
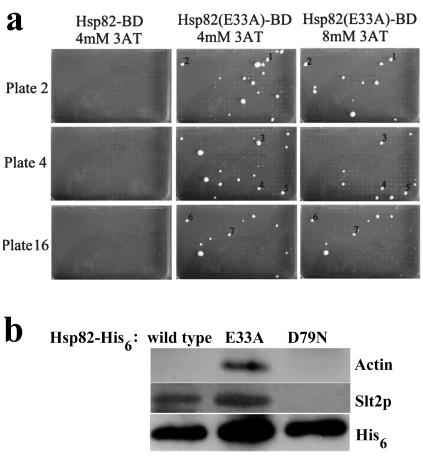
Figure 2a shows three sample plates from the 16-plate array of AD-fusion expressor cells (64) following their mating to PJ694α cells expressing either the control fusion (Hsp82-BD) or the E33A mutant fusion [Hsp82(E33A)-BD] and their subsequent growth (now as diploids expressing both AD and BD fusions) in the presence of 3-AT. Note that only a few of the 384 colonies on each plate are exhibiting 3-AT-resistant growth dependent upon the presence of the E33A mutation in the bait fusion. In total, some 177 proteins (∼3% of the total yeast proteome) were identified as potential Hsp82 interactors on the basis of such strong stabilization of their two-hybrid interaction by the E33A mutation in the Hsp82-BD fusion. These are listed in Table 1, together with their apparent interaction strengths with the Hsp82(E33A)-BD bait (based on 3-AT-resistant growth) and their annotated functions in SGD (www.yeastgenome.org).
FIG. 2.
The presence of the E33A mutation in the Hsp82-BD bait fusion stabilizes certain interactions with the library array of yeast AD-protein fusions. (a) Three sample 384-colony format plates of the 16-plate library array (64), containing either the Hsp82-BD or the Hsp82(E33A)-BD baits, as indicated. The numbered interactors with the latter are as follows: 1, ACT1; 2, YCL056c; 3, OLE1; 4, NUP57; 5, SLT2; 6, PDR3; and 7, ESC5. Growth was for 16 days at 30°C on DO minus leucine, tryptophan, and histidine and supplemented with 4 mM or 8 mM 3-AT. (b) Analysis of Ni-NTA-retained material in extracts from unstressed PJ694 cells expressing either Hsp82-His6, Hsp82(E33A)-His6, or Hsp82(D79N)-His6. The Western blot was probed with anti-actin, anti-Slt2p, and anti-His6 antisera.
TABLE 1.
Two-hybrid interactors with the Hsp82(E33A)-BD bait fusion
| ORFc | Gene | Inta | Function of gene productb |
|---|---|---|---|
| Known or putative Hsp90 system cochaperones | |||
| YDR214w | AHA1 | 2 | Nonessential cochaperone which binds the middle domain of Hsp90 (36) and activates the Hsp90 ATPase (44) |
| YDR168w | CDC37 | 1 | Essential Hsp90 system cochaperone; believed to target protein kinases to the Hsp90 chaperone machine (32) |
| YBR155w | CNS1 | 1 | Essential Hsp90 system cochaperone |
| YKL117w | SBA1 | 2 | Nonessential Hsp90 system cochaperone; binds the N-terminal Hsp90 domain in mature Hsp90-client complexes, probably assisting client protein release (69) |
| YOR027w | STI1 | 1 | Nonessential stress-induced protein that interacts with both Hsp70 and Hsp90 through its TPR domains (72) |
| YOR035c | SHE4 | 3 | Nonessential protein required for mother cell-specific expression of homothallic switching endonuclease; equivalent of the UNC45 myosin-specific chaperone, of metazoans (2); though fungal forms of She4p lack the Hsp90-interacting TPR domain of UNC45 (16), the yeast She4p still binds Hsp90 (S. H. Millson, unpublished data) |
| YPL106c | SSE1 | 1 | Nonessential form of Hsp110, a member of a subgroup of Hsp70 family chaperones (56) |
| YCR060w | TAH1 | 2 | Nonessential protein with TPR repeats and similarity to Sti1p |
| YNL064c | YDJ1 | 2 | DnaJ homolog; activator of the ATPase of Hsp70 |
| Other stress proteins, chaperones, chaperonins | |||
| YDL143w | CCT4 | 1 | Component of chaperonin-containing T complex (TCP ring complex, TRiC), required for the assembly of actin and tubulins in vivo |
| YOL053c-a | DDR2 | 1 | Stress protein induced by DNA damage, heat shock, osmotic shock, and oxidative stress |
| YOR020c | HSP10 | 1 | Mitochondrial chaperonin that cooperates with Hsp60p; homolog of E. coli GroES |
| YDR171w | HSP42 | 2 | Heat shock protein with similarity to Hsp26p; involved in restoration of the cytoskeleton during mild stress |
| YLR259c | HSP60 | 1 | Mitochondrial chaperonin that cooperates with Hsp10p; homolog of E. coli GroEL |
| Cytoskeleton | |||
| YFL039c | ACT1 | 2 | Actin, critical for cell polarization, endocytosis, and other cytoskeletal functions |
| YNL079c | TPM1 | 2 | Tropomyosin, coiled-coil protein localized to actin cables |
| YHR107c | CDC12 | 2 | Component of 10-nm filaments of mother-bud neck (septin); involved in cytokinesis. |
| Protein kinases | |||
| YDR283c | GCN2 | 2 | Kinase that phosphorylates translation initiation factor eIF2; known Hsp90 client protein (10) |
| YLR113w | HOG1 | 2 | MAP kinase of the high-osmolarity signal transduction pathway |
| YGR040w | KSS1 | 1 | MAP kinase involved in signal transduction pathways that control filamentous growth and pheromone response |
| YHR030c | SLT2 | 2 | MAP kinase of the cell wall integrity (hypotonic stress-activated) pathway |
| YLR362w | STE11 | 1 | MAP kinase kinase kinase of the Hog1p and Kss1p MAP kinase pathways; known Hsp90 client protein (31) |
| YHR135c | YCK1 | 1 | Casein kinase I isoform |
| Known or putative transcriptional activators | |||
| YLR256w | HAP1 | 2 | Heme-responsive Zn(2)-Cys(6) binuclear cluster domain transcription factor; redox-sensing regulator of gene expression; known Hsp90 client protein (29) |
| YBL005w | PDR3 | 2 | Zn(2)-Cys(6) binuclear cluster domain transcription factor; regulator of genes of pleiotropic drug resistance |
| YCR029c-a | RIM1 | 1 | Zinc finger single-stranded DNA-binding protein; required for mitochondrial genome maintenance |
| YKR064w | YKR064w | 1 | Zn(2)-Cys(6) binuclear cluster domain protein with similarity to transcription factors |
| Known or putative regulators of transcriptional silencing or telomere organizations | |||
| YJL076w | ESC5 | 2 | Involved in chromatin silencing, has similarity to Tof2p |
| YMR263w | SAP30 | 2 | Subunit of a histone deacetylase complex |
| YDL042c | SIR2 | 1 | NAD-dependent histone deacetylase; involved in maintenance of silencing at HMRd, HMLd, and telomeres |
| YOR229w | WTM2 | 1 | Transcriptional modulator protein involved in meiotic regulation and silencing |
| Protein import or export from the nucleus | |||
| YJR074w | MOG1 | 2 | Involved in nuclear protein import; potential Cdc28p substrate |
| YJR132w | NMD5 | 3 | Beta-karyopherin; involved in nuclear import |
| YGR119c | NUP57 | 2 | Nuclear pore protein (nucleoporin) that acts in a complex with Nic96p, Nup49p, and Nsp1p |
| Ras superfamily small GTPases | |||
| YOR101w | RAS1 | 1 | GTP-binding protein involved in regulation of cyclic AMP pathway; homolog of mammalian proto-oncogene ras |
| YML001w | YPT7 | 2 | GTP-binding protein of the Rab family (Ras superfamily) with a role in protein transport between endosome-like compartments |
| YER031c | YPT31 | 2 | GTP-binding protein of the Rab family (Ras superfamily) required in the secretory pathway at the stage of formation of trans-Golgi vesicles |
| YNL093w | YPT53 | 2 | GTP-binding protein of the Rab family (Ras superfamily) involved in endocytosis and transport of proteins to the vacuole |
| Other proteins of vesicle trafficking | |||
| YLR078c | BOS1 | 2 | Synaptobrevin (v-SNARE) homolog involved in endoglasmic reticulum (ER)-to-Golgi transport |
| YAL042w | ERV46 | 2 | Protein localized to Sec31p-coated vesicles |
| YOR036w | PEP12 | 2 | Syntaxin homolog (t-SNARE) involved in Golgi-to-vacuole transport |
| YKL006c-a | SFT1 | 2 | v-SNARE required for vesicle traffic between Golgi compartments |
| YOL018c | TLG2 | 1 | Syntaxin homolog (t-SNARE) involved in endocytosis and maintenance of resident proteins in the trans-Golgi network |
| YDR084c | TVP23 | 2 | Integral membrane protein localized to vesicles along with the v-SNARE Tlg2p |
| YJR044c | VPS55 | 2 | Vacuolar protein sorting; involved in Golgi-to-vacuole targeting |
| YGL161c | YIP5 | 2 | Protein that interacts with Rab GTPases; possible role in vesicle-mediated transport |
| Known or putative membrane transporters | |||
| YBR132c | AGP2 | 2 | General amino acid permease |
| YGR289c | AGT1 | 2 | General alpha-glucoside permease with similarity to maltose permeases and other members of the sugar permease family |
| YPR128c | ANT1 | 1 | Protein with similarity to ADP/ATP carrier proteins and members of the mitochondrial carrier family (MCF) |
| YNL065w | AQR1 | 2 | Member of major facilitator superfamily (MFS) multidrug resistance (MFS-MDR) protein family |
| YER119c | AVT6 | 3 | Vacuolar transporter; exports aspartate and glutamate from the vacuole |
| YDR046c | BAP3 | 3 | Branched-chain amino acid permease, valine transporter |
| YOL137w | BSC6 | 2 | MFS protein of unknown function. |
| YBR290w | BSD2 | 2 | Copper ion transporter of the ER; metal homeostasis |
| YEL063c | CAN1 | 2 | Permease for arginine, lysine, and histidine |
| YJR152w | DAL5 | 2 | Allantoate and ureidosuccinate permease, MFS superfamily member |
| YNL125c | ESBP6 | 2 | Protein with similarity to mammalian monocarboxylate transporters MCT1 and MCT2 |
| YAL022c | FUN26 | 3 | Nucleoside transporter with broad nucleoside selectivity |
| YHR094c | HXT1 | 2 | Low-affinity hexose transporter and member of sugar permease family; induced by high glucose |
| YHR096c | HXT5 | 2 | Strong similarity to hexose transporters; member of sugar permease family |
| YFL011w | HXT10 | 2 | Hexose transporter; member of sugar permease family |
| YNR072w | HXT17 | 2 | Similarity to hexose transporters, including Hxt13p, Hxt16p, Hxt6p, Hxt7p, and Lgt1p |
| YNL268w | LYP1 | 2 | Lysine-specific permease, high affinity |
| YDL247w | MPH2 | 2 | Alpha-glucoside permease |
| YGR055w | MUP1 | 2 | High-affinity methionine permease |
| YHL036w | MUP3 | 2 | Low-affinity methionine permease |
| YJL117w | PHO86 | 3 | Phosphate permease |
| YOR348c | PUT4 | 2 | Proline and gamma-aminobutyrate permease |
| YDL199c | YDL199c | 3 | Similarity to members of the sugar permease family |
| YIL120w | YIL120w | 2 | Member of MFS-MDR protein family |
| YKL174c | YKL174c | 2 | Similarity to Hnm1p and other permeases |
| YMR155w | YMR155w | 2 | Unknown function, MFS superfamily member |
| YPL264c | YPL264c | 2 | Unknown function, MFS superfamily member |
| YPR003c | YPR003c | 2 | Member of the sulfate permease subfamily of MFS superfamily |
| Miscellaneous | |||
| YCR048w | ARE1 | 2 | Acyl coenzyme A (CoA):sterol acyltransferase (sterol-ester synthetase) |
| YNR019w | ARE2 | 2 | Acyl-CoA:sterol acyltransferase (sterol-ester synthetase) |
| YHR018c | ARG4 | 2 | Argininosuccinate lyase; arginine biosynthesis |
| YPR201w | ARR3 | 2 | Arsenic resistance protein |
| YJL170c | ASG7 | 2 | Protein expressed only in cells of mating type a |
| YBR128c | ATG14 | 2 | Required for autophagy |
| YPR171w | BSP1 | 2 | Binding protein of synaptojamin polyphosphoinositide phosphate domain |
| YLR438w | CAR2 | 2 | Ornithine aminotransferase |
| YIL003w | CFD1 | 3 | Putative P-loop ATPase localized in cytoplasm |
| YLL018c | DPS1 | 2 | Aspartyl-tRNA synthetase, cytosolic |
| YLR284c | EHD1 | 1 | Enoyl-CoA hydratase, peroxisomal |
| YHR123w | EPT1 | 2 | sn-1,2-Diacylglycerol ethanolaminephosphotransferase |
| YFL022c | FRS2 | 1 | Phenylalanyl-tRNA synthetase, beta subunit, cytoplasmic |
| YEL042w | GDA1 | 1 | Guanosine diphosphatase of Golgi membrane |
| YHR183w | GND1 | 3 | 6-Phosphogluconate dehydrogenase |
| YPL059w | GRX5 | 1 | Mitochondrial glutaredoxin |
| YJR069c | HAM1 | 1 | Protein controlling 6-N-hydroxylaminopurine sensitivity and mutagenesis |
| YDR224c | HTB1 | 2 | Histone H2B |
| YDL125c | HNT1 | 2 | Member of the HIT family of Zn-binding proteins and the histidine triad nucleotide-binding protein family |
| YPR033c | HTS1 | 1 | Histidyl-tRNA synthetase, mitochondrial and cytoplasmic |
| YOL065c | INP54 | 2 | Inositol polyphosphate 5-phosphatase |
| YOL101c | IZH4 | 2 | Membrane protein involved in zinc metabolism |
| YDL131w | LYS21 | 2 | Homocitrate synthase, involved in lysine metabolism |
| YDL078c | MDH3 | 1 | Malate dehydrogenase, peroxisomal |
| YKL001c | MET14 | 2 | Adenosine-5′-phosphosulfate 3′-phosphotransferase; sulfate assimilation pathway |
| YIL014w | MNT3 | 2 | Alpha-1,3-mannosyltransferase |
| YNL110c | NOP15 | 2 | Constituent of 66S preribosomal particles |
| YGL055w | OLE1 | 3 | Stearoyl-CoA desaturase (delta-9 fatty acid desaturase); synthesis of unsaturated fatty acids |
| YOL147c | PEX11 | 2 | Peroxisomal biogenesis |
| YBR035c | PDX3 | 2 | Pyridoxine (pyridoxamine) phosphate oxidase |
| YHR037w | PUT2 | 1 | Delta-1-pyrroline-5-carboxylate dehydrogenase |
| YOR347c | PYK2 | 1 | Pyruvate kinase, glucose-repressed isoform |
| YCR106w | RDS1 | 2 | Regulator of drug sensitivity |
| YOL120c | RPL18A | 2 | Ribosomal protein L18A |
| YPL152w | RRD2 | 1 | Protein involved in rapamycin sensitivity |
| YDL244w | THI13 | 1 | Thiamine synthesis |
| YER100w | UBC6 | 2 | Ubiquitin-conjugating enzyme anchored in the ER membrane, with catalytic activity facing into cytoplasm |
| YBR273c | UBX7 | 2 | Ubiquitin regulatory X domain-containing protein; interacts with Cdc48p |
| YKL035w | UGP1 | 2 | UDP-glucose pyrophosphorylase (UTP-glucose-1-P uridylyltransferase) (UGPase) |
| YBR130c | SHE3 | 2 | Required for mother cell-specific expression of HO |
| YER118c | SHO1 | 3 | Osmosensor in the Hog1p MAP kinase; high-osmolarity signal transduction pathway |
| YML066c | SMA2 | 1 | Spore membrane assembly |
| YJR159w | SOR1 | 1 | Sorbitol dehydrogenase |
| YMR101c | SRT1 | 1 | cis-prenyltransferase involved in dolichol synthesis |
| YIR011c | STS1 | 1 | Protein that when overexpressed restores protein transport and rRNA stability to a sec23 mutation |
| YPL048w | TEF3 | 1 | Translation elongation factor EF-1gamma |
| YNL131w | TOM22 | 2 | Component of mitochondrial outer membrane receptor complex; required for protein import and cell viability |
| YOR332w | VMA4 | 2 | Vacuolar H-ATPase hydrophilic (27-kDa) subunit |
| YOR083w | WHI5 | 2 | Cell size regulation of passage through start and commitment to cell division |
| YLR070c | XYL2 | 2 | Xylitol dehydrogenase |
| YJL114w | YJL114w | 1 | TyA Gag protein; main structural constituent of virus-like particles |
Strength of interaction. 1, 2, and 3 indicate growth of the cells containing this interactor and the Hsp82(E33A)-BD bait to 4 mM, 8 mM, or 20 mM 3-AT, respectively.
Listed where a function has been assigned. The following are the ORFs encoding interactors of unknown function, with the corresponding gene name, where assigned, and the interaction strength of the encoded interactor in parentheses: YAL049c (1); YBR220c (2); YBR227c (1); YCL056c (2); YCR101c (3); YDL012c (2); YDL015c (TSC13) (2); YDL133w (1); YDL180w (1); YDL246c (SOR2) (1); YDL248c (COS7) (2); YDR051c (1); YDR281c (PHM6) (2); YDR319c (2); YDR412w (1); YER156c (2); YHR036w (2); YGR295c (COS6) (2); YIL041w (3); YIL146c (ECM37) (2); YIR030c (DCG1) (1); YIR042c (2); YJL060w (BNA3) (2); YJR083c (ACF4) (3); YJR134c (2); YKL008c (LAC1) (2); YKL069w (2); YKR030w (GMH1) (3); YKR044w (UIP5) (2); YKR067w (1); YLR036c (1); YLR404w (2); YML131w (2); YML132w (COS3) (2); YMR159c (SAP18) (2); YNL100w (1); YNL194c (1); YNL326c (2); YOL026c (MIM1) (1); YOR097c (2); YPL014w (1); YPL109c (1); YPL156c (PRM2) (1); YPL186c (UIP4) (1); YPL188w (POS5) (1); YPL199c (2); YPL246c (RBD2) (1); YPL257w (2); YPR089w (3); YPR115w (2); YPR127w (2); YPR148c (1).
ORF, open reading frame.
Mating type cassettes on chromosome 3.
Confirmation by protein binding that the E33A mutation strengthens interactions of Hsp82 with actin and with Slt2p.
The demonstration that a two-hybrid interaction is stabilized by the E33A mutation in Hsp82-BD can constitute only a first step towards showing the biological relevance of this interaction in terms of a functional Hsp90 complex (see Discussion). It was necessary therefore to validate this approach in studies of direct protein binding. To this end, investigations were conducted into the effects of the E33A and D79N mutations in Hsp82 binding to two proteins that, in the above screen, interacted more strongly with the Hsp82(E33A)-BD bait as compared to the control Hsp82-BD fusion. One, actin, represented a protein with well-established Hsp90-binding properties (41), whereas the other, Slt2p MAP kinase, had not previously been shown to bind Hsp90.
PJ694 cells were transformed with TRP1 plasmid vectors for the expression of C terminally His6-tagged wild-type Hsp82 (Hsp82-His6), as well as E33A and D79N mutant versions of this Hsp82-His6 [Hsp82(E33A)-His6 and Hsp82(D79N)-His6 respectively; see Materials and Methods]. Protein extracts were then prepared from vegetative 25°C, unstressed) cultures of these transformants, the His6-tagged Hsp82 of these extracts being isolated on a nickel-nitrilotriacetate (Ni-NTA) affinity resin. Western blot analysis of the Ni-NTA-retained material revealed that the E33A mutation was reinforcing the Hsp82-His6 association with actin (Fig. 2b), whereas D79N did not generate any such association, thus confirming the results of the two-hybrid screen (Fig. 2a).
Analysis of Slt2p MAP kinase in the same Ni-NTA-retained protein samples indicated that Hsp82-His6 association with this Slt2p was also being reinforced by the E33A mutation but was abolished totally by D79N (Fig. 2b). Some Slt2p binding to the nonmutant Hsp82-His6 was also apparent, consistent with a fairly weak two-hybrid interaction of the nonmutant Hsp82-BD and AD-Slt2p fusions (growth to 2 mM 3-AT [Fig. 3c] but not 4 mM 3-AT [Fig. 2a]).
Caffeine or heat shock generate an increased Hsp82 binding to Slt2p MAP kinase.
Slt2p is one of five MAP kinases of S. cerevisiae and the terminal MAP kinase of a signaling cascade that becomes activated in response to either a weakening of the cell wall or the need for polarized growth at the cell surface. Cell wall integrity is monitored continuously by Rho1p, a GTPase that acts as both a regulatory subunit of the 1,3-β-glucan synthase complex and an activator of protein kinase C (Pkc1p) (see references 13, 21, 24, and 42 for literature). Pkc1p in turn controls the cell integrity MAP kinase cascade, composed of a MAP kinase kinase kinase (Bck1p) (30), a pair of redundant MAP kinase kinases (Mkk1/2p) (19), and the Slt2p stress-activated MAP kinase. There are numerous stress activators of this pathway, notably hypotonic stress (9), high temperature (23, 34), endoplasmic reticulum stress (5), caffeine and vanadate (34), and agents that cause cell wall weakening (12, 26, 53). The pathway is also activated with the polarized growth of budding and mating (73), as well as with the operation of the morphogenesis checkpoint (15).
MAP kinases are characteristically activated through dual threonine/tyrosine phosphorylation of a TXY motif, found within an activation loop some distance from the active site of the enzyme (6, 61). Yeast Slt2p exhibits a basal activity in growing cultures but becomes much more strongly activated by Mkk1/2p-catalyzed phosphorylation of its TEY motif whenever the cell integrity MAP kinase pathway undergoes further stimulation. Heat shock and caffeine are just two of the inducers of this dually Thr190/Tyr192-phosphorylated, active Slt2p (23, 34). To determine how these stresses would affect the Hsp82-Slt2p interaction, extracts were prepared from unstressed and stressed cultures of the above Hsp82-His6-, Hsp82(E33A)-His6-, or Hsp82(D79N)-His6-expressing cells, cultures either in growth at 25°C (unstressed), heat shocked from 25°C to 39°C for 1 h, or treated with 8 mM caffeine for 1 h. The Ni-NTA-retained protein of these extracts was then analyzed by Western blotting. As shown in Fig. 3a, heat or caffeine stress increased Slt2p binding by Hsp82-His6 and Hsp82(E33A)-His6. Stress exposure did not, though, restore any capacity for Slt2p binding to the non-ATP-binding, Hsp82(D79N)-His6 mutant form of the chaperone (Fig. 3a).
PJ694, the two-hybrid strain used for this study, allows quantitative measurements of the effects of in vivo stress on the strength of any two-hybrid interaction through the monitoring of its interaction-responsive, GAL7 promoter-regulated lacZ expression. Such measurements readily lend themselves to automation (37). By this approach the effects of in vivo heat shock and caffeine stress on the Hsp82-BD “bait”-AD-Slt2p “prey” interaction were determined, as were the influences of the E33A and D79N Hsp82-BD mutations on this Hsp82-BD-AD-Slt2p interaction (see Materials and Methods). As shown in Fig. 3b, interaction of the wild-type Hsp82-BD and AD-Slt2p fusions (functional forms of Hsp90 [37] and Slt2p [58] in yeast) increased under conditions of heat or caffeine stress. Interaction was reinforced strongly by the presence of the E33A mutation in the Hsp82-BD fusion but was abolished completely by D79N (a result consistent with the protein binding experiments; Fig. 2b). 3-AT-resistant growth of the same two-hybrid strains (a monitoring of interaction-responsive HIS3 gene activation [64]) provided essentially the same result (Fig. 3c), confirming the importance of the ATP/ADP-interacting D79 residue of the chaperone for any significant interaction with Slt2p.
Hsp82 binds exclusively to the dually Thr190/Tyr192-phosphorylated, stress-activated form of Slt2p MAP kinase.
The above results indicated that Slt2p was interacting with Hsp82 when the latter was in ATP-bound form, the state of the chaperone present in late-stage complexes of the Hsp90 cycle. Slt2p-Hsp82 association was reinforced by heat or caffeine (Fig. 3), conditions of in vivo stress that increase the dual phosphorylation of Slt2p (23, 34). This indicated that Hsp82 might be binding more tightly to, or exclusively to, the dually Thr/Tyr-phosphorylated form of Slt2p, the state corresponding to the active MAP kinase.
Vectors were constructed for the in vivo expression of a functional, C terminally His6-tagged Slt2p (Slt2-His6), as well as two nonfunctional mutant forms of this Slt2-His6 (see Materials and Methods). The mutant forms were a T190A Y192F double mutant (corresponding to conservative substitutions of the TEY motif residues that must become phosphorylated for MAP kinase activity) and K54R (a mutation which generates a phosphorylatable, yet catalytically dead Slt2p [33]). These plasmids for Slt2-His6, Slt2(T190A,Y192F)-His6, and Slt2(K54R)-His6 expression were then transformed into an slt2Δ yeast strain that expresses, at a constant level from the TDH1 promoter, just the Hsp82 isoform of yeast Hsp90 (p82aslt2Δ; see Materials and Methods). Expression of these Slt2-His6 forms in an slt2Δ background eliminated the possibility of heterodimer formation with a native, chromosomally derived Slt2p (MAP kinases can undergo dimerization in response to the structural changes induced by dual Tyr/Thr phosphorylation [28]). Finally, extracts were prepared from these p82aslt2Δ cells expressing Slt2-His6, Slt2(T190A,Y192F)-His6, or Slt2(K54R)-His6 as their sole Slt2p, either in growth at 25°C (unstressed), after a heat shock from 25°C to 39°C for 1 h, or following treatment with 8 mM caffeine for 1 h. The Ni-NTA-retained protein of these extracts was then analyzed by Western blotting.
As shown in Fig. 4a, stresses that lead to an increased dual phosphorylation of Slt2p also cause increased Hsp82 binding by the wild-type (Slt2-His6) and the kinase-dead [Slt2(K54R)-His6] versions of this MAP kinase. In contrast, the mutations rendering this MAP kinase nonphosphorylatable (T190A and Y192F) completely abolished any interaction with Hsp82. Additional evidence that Hsp82 binds only the dually Thr/Tyr-phosphorylated Slt2p was provided by the two-hybrid system, both through a monitoring of GAL1 promoter-regulated HIS3 expression (3-AT-resistant growth; Fig. 3c) and by quantitative measurements of GAL7 promoter-regulated lacZ expression (Fig. 4b). Using PJ694 cells containing the wild-type Hsp82-BD bait fusion and either the AD-Slt2p, the AD-Slt2p(T190A,Y192F), or the AD-Slt2p(K54R) prey fusion, the T190A and Y192F mutations, which render the AD-Slt2p fusion nonphosphorylatable, were found to completely abolish Hsp82-BD-AD-Slt2p interaction (Fig. 3c and 4b). In contrast, the phosphorylatable, yet kinase-dead K54R mutant version of this AD-Slt2p still displayed a stress-reinforced interaction with the Hsp82-BD fusion (Figs. 3c and 4b). Interaction-responsive lacZ expression measurements indicated that this K54R mutant AD-Slt2p was exhibiting a slightly enhanced, stress-induced interaction with Hsp82-BD as compared to the wild-type AD-Slt2p fusion (Fig. 4b).
Slt2p (55.6 kDa) differs from the other MAP kinases of yeast in having an extended C-terminal domain of uncertain function. Its closest relative among mammalian MAP kinases, ERK5/BMK1, also possesses an extended C terminus, and heterologous expression of ERK5 can substantially provide Slt2p function in yeast (A. W. Truman, unpublished). In ERK5 this C-terminal domain is known to contain a transcriptional activator region (25, 68) and a nuclear localization signal (67). Nevertheless, the function of Slt2p in yeast is substantially preserved with loss of a substantial C-terminal region (58). To determine whether Hsp82 interaction is also preserved with the loss of this domain, a C terminally truncated Slt2p (amino acids 1 to 328, essentially just the MAP kinase module) was expressed as an AD fusion [AD-Slt2(t)p] in the two-hybrid system. The Hsp82-BD bait still interacted with this truncated Slt2(t)p (Fig. 3c), showing that the MAP kinase domain of Slt2p is alone sufficient for the two-hybrid interaction with Hsp82.
Analysis of the T22Ihsp82 yeast mutant reveals that Hsp90 chaperone function is essential for Slt2p-mediated stimulation of the Rlm1p transcriptional activator of cell integrity genes.
To search for genetic evidence of whether the action of Slt2p MAP kinase is Hsp90 dependent in vivo, a set of eight hsp82 mutants were investigated for an altered Slt2p-mediated response. These are strains temperature sensitive for growth at 37°C due to point mutations in Hsp82, their sole form of the Hsp90 chaperone (40). They express this Hsp82 to similar levels (47) but are compromised, to variable degrees, in the essential Hsp90 function that this Hsp82 confers (40).
While a low, basal activity of the cell integrity pathway is beneficial for yeast growth, exceptionally high stress-activated MAP kinase activity is generally extremely detrimental (14, 66). We found that one of these hsp82 mutants lacked the detrimental effects of overactive Slt2p-mediated signaling that normally result with the presence of an overactive form of the Mkk1p MAP kinase kinase activator of Slt2p. As shown in Fig. 5a, the expression of this hyperactive MKK1P386 allele (65) was not inhibitory for growth of the T22Ihsp82 mutant, though it was toxic in the isogenic cells expressing the wild-type Hsp82 (strain p82a). T22Ihsp82 is temperature sensitive due to expression of the T22I point mutant form of Hsp82 as its sole Hsp90 (40) (lack of high-temperature growth being one of the phenotypes displayed by Slt2p MAP kinase cascade mutants [13, 34]). This absence of toxic effects of MKK1P386 expression in T22Ihsp82 cells (Fig. 5a) was an indication that the T22I mutant Hsp82 was generating a defect in cell integrity signaling epistatic to Pkc1p, Bck1p, and Mkk1/2p in the Slt2p MAP kinase cascade.
FIG. 5.
Analysis of the T22Ihsp82 mutant phenotype. (a) The MKK1S386P allele is toxic in strain p82a, which expresses wild-type Hsp82 as its sole Hsp90, but not in the T22Ihsp82 mutant, which is isogenic but for expression of the T22I mutant form of this Hsp82. Cells containing either empty vector (pUT36) or a plasmid for MET25 promoter-regulated expression of this overactive MKK1 allele were plated on either methionine-containing or methionine-deficient DO and grown for 2 days at the indicated temperature. (b) Measurement of dually phosphorylated Slt2p [(Y-P,T-P)Slt2p] and total Slt2p levels in unstressed, heat-shocked, or caffeine-stressed cells of the T22Ihsp82 mutant and its corresponding wild type, p82a. (c) Measurements of Rlm1p activity in the same unstressed, caffeine-stressed, or heat-shocked p82a and T22Ihsp82 cells, either growing at 25°C, treated with 8 mM caffeine for 1 h at 25°C, or heat shocked from 25°C to 39°C for 1 h.
Downstream of Mkk1/2p in this signaling cascade is Slt2p, one of whose direct phosphorylation targets is the Rlm1p trans activator of cell wall genes. On this Rlm1p transcription factor, the sites of docking interaction by the dually Thr/Tyr-phosphorylated Slt2p and of activating phosphorylation by this Slt2p are reasonably well characterized (12, 21, 22). Furthermore Rlm1p activity is readily monitored by using a lacZ reporter gene regulated by the Rlm1p-responsive promoter of the YIL117c gene (22). As shown in Fig. 5c, unstressed cells of the above-mentioned T22Ihsp82 mutant were found to possess extremely low basal levels of Rlm1p activity. This mutant also completely lacked the normal increases in this activity that result from stress stimulation of cell integrity MAP kinase signaling (Fig. 5c). Nevertheless, even though the T22Ihsp82 mutant exhibits this pronounced Rlm1p activity defect, its Slt2p still became dually Tyr/Thr phosphorylated in response to either caffeine or heat stress activation of cell integrity signaling (Fig. 5b). Therefore, the compromised Hsp82 function of the T22Ihsp82 mutant, though not abolishing Mkk1/2p-mediated Slt2p kinasing in response to Pkc1p-mediated signaling (Fig. 5b), is preventing any relay of this activating signal to the Rlm1p target of this Slt2p. This effect of the T22Ihsp82 allele is strong genetic evidence for an Hsp90 chaperone requirement in this Rlm1p activation by Slt2p. The physical interaction of Hsp82 and dually Tyr/Thr-phosphorylated Slt2p, identified and characterized in this study (Fig. 2 to 4), is probably essential in this Hsp90 facilitation of Slt2p MAP kinase activity.
The fraction of the total cellular Slt2p existing as the dually Thr190/Tyr192-phosphorylated form was higher in the heat-shocked cells of the T22Ihsp82 mutant as compared to that in the identically stressed control cells expressing the wild-type Hsp82 (Fig. 5b). This might reflect sensing of a weakened cell wall in this mutant (one of the signals for Slt2p pathway activation being sensing of a weakening of cell wall architecture; see above). Alternatively, it might be due to a compromised dephosphorylation of the active Slt2p in T22Ihsp82 mutant cells by the multiple phosphatases that inactivate this MAP kinase in vivo (7, 11, 14).
DISCUSSION
Protein chaperones often engage in rather transient associations with their target proteins, preventing an analysis of these associations by the conventional two-hybrid system. This study though revealed that many associations of Hsp90 can be stabilized in vivo using a mutation that inhibits the essential ATPase reaction of the Hsp90 chaperone cycle. The E33A mutation in the Hsp82-BD two-hybrid bait strongly reinforced the interactions of this bait with about 3% of the yeast proteome, the latter expressed as AD fusions (Table 1). That many of the interactors are known cochaperones or clients of the yeast Hsp90 system (Fig. 1) is a strong indication that this E33A mutant Hsp82-BD bait is allowing the detection of bona fide Hsp90 associations. This screening approach is therefore potentially of use in finding new binding partners of Hsp90, though it is improbable that it could reveal every Hsp90 interactor in the cell (false negatives arise for a large number of reasons in two-hybrid screens) (3). Indeed, while our screen succeeded in identifying some of the known Hsp90 clients of yeast (10, 29, 31), it also failed to identify others (1, 17, 18).
Table 1 lists the protein fusions selected by the E33A mutant Hsp82-BD bait. For most of these, further work is needed to confirm whether or not the two-hybrid association is meaningful in terms of a biologically relevant Hsp90 complex. It is noteworthy that a few other chaperones and chaperonins of yeast were identified as putative Hsp90 interactors (Table 1). They include Hsp60/Hsp10, the functional equivalent of the bacterial GroEL/ES in mitochondria. Since yeast Hsp90 is localized to the cytosol (and probably also the nucleus), the detected interactions (also the interaction with the Grx5p mitochondrial glutaredoxin) might be to the precursor forms of these proteins that exist prior to mitochondrial import, especially since the presence of the Gal4p AD at the N termini of these protein fusions may interfere with the recognition of signal sequences for mitochondrial import. In mammalian systems the Hsp70/Hsp90 chaperone system delivers mitochondrial protein precursors to the preprotein translocase of the outer mitochondrial membrane, but it appears that this process is not Hsp90 dependent in yeast (70). Nevertheless, a component of the multisubunit mitochondrial import receptor, Tom22p, was identified as a potential Hsp90 interactor in this screen.
A few cytoskeletal proteins (actin, tropomyosin, and Cdc12p) and chaperones/cochaperones involved in cytoskeletal function (Cct4p, Hsp42, and She4p) were identified as potential Hsp90 binding partners (Table 1). Hsp90 has long been known to possess actin-binding properties (41). There is also a constant requirement for Hsp90 function in the organization of actin microfilaments, as indicated by an almost immediate delocalization of rhodamine-phalloidin staining and loss of polarized growth in yeast cells treated with Hsp90 inhibitor drugs (S. H. Millson, unpublished observations). Certain Rab/Ras small GTPase protein family members were also apparent Hsp90 interactors (Table 1). This is consistent with the recently discovered role of a Rab-recycling, membrane-associated Hsp90 chaperone complex in operation of the alphaGDP-dissociation inhibitor. The latter acts in the Rab-mediated targeting of vesicles to an acceptor compartment, coordinating the Ca2+-dependent events that trigger the hydrolysis of Rab-bound GTP with the retrieval of the product of this reaction, Rab-GDP, from vesicle membranes to the cytosol (54). Among the other potential Hsp90 interactors were 28 known or potential membrane transporter proteins, including no less than six of the plasma membrane permeases for amino acids (Table 1). This is unexpected, since the two-hybrid system requires BD- and AD-containing fusions to associate noncovalently in the yeast nucleus. Possibly the fusion of these membrane proteins to the Gal4p AD, a domain with a nuclear import signal, has allowed their mislocalization to the nucleus. Only further work will tell if this apparent Hsp90 association with membrane transporters is of biological significance.
The E33A mutation inhibits the final, essential ATPase step of the Hsp90 chaperone cycle and the ensuing release of the activated client protein (43). Client proteins interacting with this mutant Hsp90 should therefore accumulate as a late-stage chaperone complex, stabilized in association with the ATP-bound chaperone. When expressed at normal cellular Hsp90 levels, the E33A mutant Hsp82 cannot provide the essential Hsp90 function in yeast (43). This mutant Hsp82 can though allow very slow growth when highly overexpressed as the sole Hsp90 of yeast cells (unpublished observations). An E33A mutant Hsp82 must therefore be capable of very slow progression through the chaperone cycle. It might be argued that the two-hybrid interactions detected on the basis of this E33A mutation are artifacts of such slow cycle progression and the resultant alterations to the stoichiometry of the different Hsp90 complexes in the yeast. It was necessary therefore to validate this approach for revealing new Hsp90 binding partners by confirming that an interaction reinforced by the E33A mutation involves a hitherto-unidentified Hsp90 client (Fig. 2b to 5).
Just 6 of the 117 yeast protein kinases exhibited a reinforced two-hybrid interaction dependent on the E33A mutation in the Hsp82-BD bait (Table 1). Nevertheless, these included two of the kinases already identified as Hsp90 clients, as well as three of the five MAP kinases of yeast. We were particularly intrigued by the latter finding since, while an Hsp90 dependence has been shown for a number of MAP kinase pathway signaling events (see below), the MAP kinase family kinases that are the targets of this signaling are not generally regarded as Hsp90 clients. Indeed, it would seem that Hsp90 is not required for the activity of many of these kinases (for example, recombinant ERK2 and p38γ can be obtained in active states by Escherichia coli expression in the absence of eukaryotic forms of Hsp90 [4, 27]). We therefore sought to establish whether one of the MAP kinases identified in the screen was Hsp90 binding and Hsp90 dependent in its activity. As shown in Fig. 3 and 4, Slt2p interaction is specific for the ATP-bound form of Hsp82 and the dually Thr190/Tyr192-phosphorylated, stress-activated state of Slt2p, most probably as a late-stage Hsp90 chaperone complex. The phenotype of the T22Ihsp82 mutant indicates, in turn, that Hsp90 function is essential for this dually phosphorylated Slt2p to activate one of its targets, the Rlm1p trans activator of cell wall genes (Fig. 5). It is probable therefore that the Slt2p-Hsp82 interaction, identified and characterized in this study (Fig. 2 to 4), is essential for Slt2p MAP kinase activity.
This Hsp90 requirement in the action of Slt2p may represent the Hsp90 machine participating in the formation of the active Slt2p MAP kinase in response to the structural changes induced in this MAP kinase as a consequence of the activating Thr/Tyr phosphorylation. Alternatively, a phosphorylation-induced dimerization of Slt2p (as in mammalian ERK2 [28]) may be an essential step before Hsp90 (also a dimeric protein) is able to bind to Slt2p and promote its interaction with the MAP kinase docking site (D domain) on Rlm1p. A third possibility is that the Hsp90 function is required for the ability of Slt2p to phosphorylate its targets after the docking interaction. Future work should readily distinguish which of these events in the operation of this client protein require the Hsp90 chaperone.
A number of the signaling events of MAP kinase pathway activation are known to be Hsp90 dependent. In Schizosaccharomyces pombe the Hsp90 cochaperone Cdc37p is involved in Spc1p stress-activated MAP kinase signaling and cdc37 mutations affect both Spc1p level and Spc1p phosphorylation by the Wis1p stress-activated MAP kinase kinase (62). Hsp90 is also needed for the activity of Ste11p, a MAP kinase kinase kinase of S. cerevisiae that signals to no less than two MAP kinases, Kss1p and Hog1p (31). In addition, Hsp90 binding stabilizes mammalian Mok1p, a protein kinase that is structurally moderately related to conventional MAP kinases and activated during spermatogenesis (39). Nevertheless stress-activated MAP kinases, such as the Hog1p and Slt2p of S. cerevisiae, the Sty1p of S. pombe, and the p38 of mammalian systems, have not previously been considered as Hsp90-dependent activities. As a MAP kinase with well-established genetics (5, 9, 12, 15, 23, 26, 34, 53, 73), Slt2p should be an ideal model protein with which to investigate the Hsp90 dependence of protein kinase activity.
ADDENDUM IN PROOF
While this paper was in press, the results of another two-hybrid screen for Hsp90 interactors were published (R. Zhao, M. Davey, Y. C. Hsu, P. Kaplanek, A. Tong, A. B. Parsons, N. Krogan, G. Cagney, D. Mai, J. Greenblatt, C. Boone, A. Emili, and W. A. Houry, Cell 120:715-727, 2005). While both studies have identified some common interactors (e.g., Tah1p, Slt2p), there are also major differences between our findings and the findings of this latest study. These can be largely attributed to the use of different bait fusions in the screens. Zhao et al. used a bait comprising the Ga14p BD positioned at the N terminus of the wild-type Hsp82 sequence, a BD-Hsp82 fusion that cannot provide the essential Hsp90 chaperone function in vivo (38). Our screen, in contrast, relied upon the introduction of an interaction-stabilizing mutation (E33A) in a fusion (Hsp82-BD) where the BD is placed at the Hsp82 C terminus. The latter fusion is substantially functional as an Hsp90 chaperone in vivo (37, 38).
Acknowledgments
We are indebted to G. Cagney, T. Hazbun, S. Fields, K. Ayscough, J. Hegemann, D. Levin, S. Lindquist, K. Matsumoto, M. Molina, and B. Panaretou for their kind assistance in providing materials for this project.
This work was supported by BBSRC grants 31/C13023 and C506721/1.
REFERENCES
- 1.Bali, M., B. Zhang, K. A. Morano, and C. A. Michels. 2003. The Hsp90 molecular chaperone complex regulates maltose induction and stability of the Saccharomyces MAL gene transcription activator Mal63p. J. Biol. Chem. 278:47441-47448. [DOI] [PubMed] [Google Scholar]
- 2.Barral, J. M., A. H. Hutagalung, A. Brinker, F. U. Hartl, and H. F. Epstein. 2002. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295:669-671. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, P. L., and S. Fields. 1997. The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 4.Bellon, S., M. J. Fitzgibbon, T. Fox, H. M. Hsiao, and K. P. Wilson. 1999. The structure of phosphorylated p38gamma is monomeric and reveals a conserved activation-loop conformation. Structure Fold Des. 7:1057-1065. [DOI] [PubMed] [Google Scholar]
- 5.Bonilla, M., and K. W. Cunningham. 2003. Mitogen-activated protein kinase stimulation of Ca(2+) signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 14:4296-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canagarajah, B. J., A. Khokhlatchev, M. H. Cobb, and E. J. Goldsmith. 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90:859-869. [DOI] [PubMed] [Google Scholar]
- 7.Collister, M., M. P. Didmon, F. MacIsaac, M. J. Stark, N. Q. MacDonald, and S. M. Keyse. 2002. YIL113w encodes a functional dual-specificity protein phosphatase which specifically interacts with and inactivates the Slt2/Mpk1p MAP kinase in S. cerevisiae. FEBS Lett. 527:186-192. [DOI] [PubMed] [Google Scholar]
- 8.Csermely, P., T. Schnaider, C. Soti, Z. Prohaszka, and G. Nardai. 1998. The 90kDa molecular chaperone family: structure, function and clinical applications. A comprehensive review. Pharmacol. Ther. 79:1-39. [DOI] [PubMed] [Google Scholar]
- 9.Davenport, K. R., M. Sohaskey, Y. Kamada, D. E. Levin, and M. C. Gustin. 1995. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J. Biol. Chem. 270:30157-30161. [DOI] [PubMed] [Google Scholar]
- 10.Donze, O., and D. Picard. 1999. Hsp90 binds and regulates Gcn2, the ligand-inducible kinase of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 19:8422-8432. (Erratum, 20:1897, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flandez, M., I. C. Cosano, C. Nombela, H. Martin, and M. Molina. 2004. Reciprocal regulation between Slt2 MAPK and isoforms of Msg5 dual-specificity protein phosphatase modulates the yeast cell integrity pathway. J. Biol. Chem. 279:11027-11034. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, R., C. Bermejo, C. Grau, R. Perez, J. M. Rodriguez-Pena, J. Francois, C. Nombela, and J. Arroyo. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279:15183-15195. [DOI] [PubMed] [Google Scholar]
- 13.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, J. S., and D. J. Thiele. 2002. Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J. Biol. Chem. 277:21278-21284. [DOI] [PubMed] [Google Scholar]
- 15.Harrison, J. C., E. S. G. Bardes, Y. Ohya, and D. J. Lew. 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3:417-420. [DOI] [PubMed] [Google Scholar]
- 16.Hutagalung, A. H., M. L. Landsverk, M. G. Price, and H. F. Epstein. 2002. The UCS family of myosin chaperones. J. Cell Sci. 115:3983-3990. [DOI] [PubMed] [Google Scholar]
- 17.Imai, J., M. Maruya, H. Yashiroda, I. Yahara, and K. Tanaka. 2003. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 22:3557-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai, J., and I. Yahara. 2000. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell. Biol. 20:9262-9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie, K., M. Takase, K. S. Lee, D. E. Levin, H. Araki, K. Matsumoto, and Y. Oshima. 1993. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 13:3076-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 22.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781-789. [DOI] [PubMed] [Google Scholar]
- 23.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 24.Kamada, Y., H. Qadota, C. P. Python, Y. Anraku, Y. Ohya, and D. E. Levin. 1996. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 271:9193-9196. [DOI] [PubMed] [Google Scholar]
- 25.Kasler, H. G., J. Victoria, O. Duramad, and A. Winoto. 2000. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 20:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khokhlatchev, A., S. Xu, J. English, P. Wu, E. Schaefer, and M. H. Cobb. 1997. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J. Biol. Chem. 272:11057-11062. [DOI] [PubMed] [Google Scholar]
- 28.Khokhlatchev, A. V., B. Canagarajah, J. Wilsbacher, M. Robinson, M. Atkinson, E. Goldsmith, and M. H. Cobb. 1998. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93:605-615. [DOI] [PubMed] [Google Scholar]
- 29.Lan, C., H. C. Lee, S. Tang, and L. Zhang. 2004. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J. Biol. Chem. 279:27607-27612. [DOI] [PubMed] [Google Scholar]
- 30.Levin, D. E., B. Bowers, C. Y. Chen, Y. Kamada, and M. Watanabe. 1994. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell. Mol. Biol. Res. 40:229-239. [PubMed] [Google Scholar]
- 31.Louvion, J. F., T. Abbas-Terki, and D. Picard. 1998. Hsp90 is required for pheromone signaling in yeast. Mol. Biol. Cell 9:3071-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLean, M., and D. Picard. 2003. Cdc37 goes beyond Hsp90 and kinases. Cell Stress Chaperones 8:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, H., J. Arroyo, M. Sanchez, M. Molina, and C. Nombela. 1993. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 degrees C. Mol. Gen. Genet. 241:177-184. [DOI] [PubMed] [Google Scholar]
- 34.Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:1511-1519. [DOI] [PubMed] [Google Scholar]
- 35.McLaughlin, S. H., H. W. Smith, and S. E. Jackson. 2002. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J. Mol. Biol. 315:787-798. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, P. 2004. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23:1402-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millson, S. H., A. Truman, F. Wolfram, V. King, B. Panaretou, C. Prodromou, L. H. Pearl, and P. W. Piper. 2004. Investigating the protein-protein interactions of the yeast Hsp90 chaperone system by two hybrid analysis; potential uses and limitations of this approach. Cell Stress Chaperones 9:359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millson, S. M., A. Truman, and P. W. Piper. 2003. Vectors for N- or C-terminal positioning of the yeast Gal4p DNA binding or activator domains. BioTechniques 35:60-64. [DOI] [PubMed] [Google Scholar]
- 39.Miyata, Y., Y. Ikawa, M. Shibuya, and E. Nishida. 2001. Specific association of a set of molecular chaperones including HSP90 and Cdc37 with MOK, a member of the MAP kinase superfamily. J. Biol. Chem. 16:16. [DOI] [PubMed] [Google Scholar]
- 40.Nathan, D. F., and S. Lindquist. 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishida, E., S. Koyasu, H. Sakai, and I. Yahara. 1986. Calmodulin-regulated binding of the 90-kDa heat shock protein to actin filaments. J. Biol. Chem. 261:16033-16036. [PubMed] [Google Scholar]
- 42.Nonaka, H., K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno, M. Umikawa, A. Mino, and Y. Takai. 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14:5931-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panaretou, B., C. Prodromou, S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1998. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17:4829-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panaretou, B., G. Siligardi, P. Meyer, A. Maloney, J. K. Sullivan, S. Singh, S. H. Millson, P. A. Clarke, S. Naaby-Hansen, R. Stein, R. Cramer, M. Mollapour, P. Workman, P. W. Piper, L. H. Pearl, and C. Prodromou. 2002. Activation of the ATPase activity of Hsp90 by AHA1 and other co-chaperones. Mol. Cell 10:1307-1318. [DOI] [PubMed] [Google Scholar]
- 45.Pearl, L. H., and C. Prodromou. 2000. Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol. 10:46-51. [DOI] [PubMed] [Google Scholar]
- 46.Picard, D. 2002. Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 59:1640-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piper, P. W., S. H. Millson, M. Mollapour, B. Panaretou, G. Siligardi, L. H. Pearl, and C. Prodromou. 2003. Sensitivity to Hsp90-targeting drugs can arise with mutation to the Hsp90 chaperone, cochaperones and plasma membrane ATP binding cassette transporters of yeast. Eur. J. Biochem. 270:4689-4695. [DOI] [PubMed] [Google Scholar]
- 48.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228:111-133. [DOI] [PubMed] [Google Scholar]
- 49.Prodromou, C., B. Panaretou, S. Chohan, G. Siligardi, R. O'Brien, J. E. Ladbury, S. M. Roe, P. W. Piper, and L. H. Pearl. 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19:4383-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prodromou, C., and L. H. Pearl. 2003. Structure and functional relationships of Hsp90. Curr. Cancer Drug Targets 3:301-323. [DOI] [PubMed] [Google Scholar]
- 51.Prodromou, C., S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90:65-75. [DOI] [PubMed] [Google Scholar]
- 52.Prodromou, C., G. Siligardi, R. O'Brien, D. N. Woolfson, L. Regan, B. Panaretou, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinoso-Martin, C., C. Schuller, M. Schuetzer-Muehlbauer, and K. Kuchler. 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2:1200-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakisaka, T., T. Meerlo, J. Matteson, H. Plutner, and W. E. Balch. 2002. Rab-alphaGDI activity is regulated by a Hsp90 chaperone complex. EMBO J. 21:6125-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schena, M., D. Picard, and K. R. Yamamoto. 1991. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 194:389-398. [DOI] [PubMed] [Google Scholar]
- 56.Shaner, L., A. Trott, J. L. Goeckeler, J. L. Brodsky, and K. A. Morano. 2004. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 279:21992-22001. [DOI] [PubMed] [Google Scholar]
- 57.Smith, D. F., L. Whitesell, and K. Katsanis. 1998. Molecular chaperones: biology and prospects for pharmacological intervention. Pharmacol. Rev. 50:493-514. [PubMed] [Google Scholar]
- 58.Soler, M., A. Plovins, H. Martin, M. Molina, and C. Nombela. 1995. Characterization of domains in the yeast MAP kinase Slt2 (Mpk1) required for functional activity and in vivo interaction with protein kinases Mkk1 and Mkk2. Mol. Microbiol. 17:833-842. [DOI] [PubMed] [Google Scholar]
- 59.Stepanova, L., X. Leng, S. B. Parker, and J. W. Harper. 1996. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 10:1491-1502. [DOI] [PubMed] [Google Scholar]
- 60.Tanoue, T., and E. Nishida. 2002. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol. Ther. 93:193-202. [DOI] [PubMed] [Google Scholar]
- 61.Tanoue, T., and E. Nishida. 2003. Molecular recognitions in the MAP kinase cascades. Cell. Signal. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 62.Tatebe, H., and K. Shiozaki. 2003. Identification of Cdc37 as a novel regulator of the stress-responsive mitogen-activated protein kinase. Mol. Cell. Biol. 23:5132-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 64.Uetz, P., G. Cagney, D. Lockshon, A. Qureshi-Emili, D. Conover, M. Johnston, and S. Fields. 2000. A protein array for genomewide screens of protein-protein interactions. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe, Y., K. Irie, and K. Matsumoto. 1995. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 15:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yaakov, G., M. Bell, S. Hohmann, and D. Engelberg. 2003. Combination of two activating mutations in one HOG1 gene forms hyperactive enzymes that induce growth arrest. Mol. Cell. Biol. 23:4826-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan, C., H. Luo, J. D. Lee, J. Abe, and B. C. Berk. 2001. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J. Biol. Chem. 276:10870-10878. [DOI] [PubMed] [Google Scholar]
- 68.Yang, C. C., O. I. Ornatsky, J. C. McDermott, T. F. Cruz, and C. A. Prody. 1998. Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 26:4771-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young, J. C., and F. U. Hartl. 2000. Polypeptide release by hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 19:5930-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41-50. [DOI] [PubMed] [Google Scholar]
- 71.Young, J. C., I. Moarefi, and F. U. Hartl. 2001. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 154:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young, J. C., W. M. Obermann, and F. U. Hartl. 1998. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J. Biol. Chem. 273:18007-18010. [DOI] [PubMed] [Google Scholar]
- 73.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 15:83-91. [PMC free article] [PubMed] [Google Scholar]