Abstract
Eukaryotic translation initiation factor 4E (eIF4E) binds to the m7GTP of capped mRNAs and is an essential component of the translational machinery that recruits the 40S small ribosomal subunit. We describe here the identification and characterization of two eIF4E homologues in an ancient protist, Giardia lamblia. Using m7GTP-Sepharose affinity column chromatography, a specific binding protein was isolated and identified as Giardia eIF4E2. The other homologue, Giardia eIF4E1, bound only to the m2,2,7GpppN structure. Although neither homologue can rescue the function of yeast eIF4E, a knockdown of eIF4E2 mRNA in Giardia by a virus-based antisense ribozyme decreased translation, which was shown to use m7GpppN-capped mRNA as a template. Thus, eIF4E2 is likely the cap-binding protein in a translation initiation complex. The same knockdown approach indicated that eIF4E1 is not required for translation in Giardia. Immunofluorescence assays showed wide distribution of both homologues in the cytoplasm. But eIF4E1 was also found concentrated and colocalized with the m2,2,7GpppN cap, 16S-like rRNA, and fibrillarin in the nucleolus-like structure in the nucleus. eIF4E1 depletion from Giardia did not affect mRNA splicing, but the protein was bound to Giardia small nuclear RNAs D and H known to have an m2,2,7GpppN cap, thus suggesting a novel function not yet observed among other eIF4Es in eukaryotes.
Most eukaryotic mRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) are modified posttranscriptionally in the nucleus at their 5′ end by addition of a 7-methylguanosine (m7G) cap linked by a 5′-5′-triphosphate bridge to the first transcribed residue (11, 45). The cap of snRNAs and snoRNAs is then further methylated at its N2 position in the cytoplasm or nucleus to yield an N2,N2,7-methylguanosine (m2,2,7G) cap (41). The m7GpppN-cap structure is required for recruitment of mRNA by the translational machinery, whereas the m2,2,7GpppN-cap plays crucial roles in gene expression, such as mRNA splicing, methylation, pseudouridylation, and rRNA processing and ribosome assembly (41).
The m7GpppN-cap of mRNA is recognized by eukaryotic initiation factor 4E (eIF4E), which interacts also with scaffold protein eIF4G (13, 30). The latter binds to an ATP-dependent RNA helicase, eIF4A, and ribosome-bound eIF3 to recruit the 40S small ribosomal subunit to the 5′ end of an mRNA for translation initiation (14, 16, 20, 24). eIF4E is not only a translation initiation factor but also a protein that modulates the overall rate of translation and the mRNA selectivity of the translation apparatus (11, 13). The functional importance of eIF4E is illustrated by the lethality of eif4e gene disruption in Saccharomyces cerevisiae (3). Both the function and structure of eIF4E have been conserved throughout evolution; human and yeast homologues are 31% identical at the amino acid level, and human eIF4E can rescue eif4e gene disruption in S. cerevisiae (3). In spite of the variable N and C termini among the eIF4Es, a core of about 170 amino acids within the eIF4E molecule has been apparently well conserved among all eukaryotes (28, 32, 49). Deletion analysis of eIF4E in S. cerevisiae showed that this core region alone is sufficient for cap recognition (49). The three-dimensional structures of cap analogue-bound eIF4Es from mice (28), humans (48), and S. cerevisiae (29) have been resolved. They each contain an eight-stranded antiparallel β-sheet on top of three α-helices in a cupped-hand shape. Two Trp residues, located within a narrow cavity inside the concave surface, hold the guanine residue of the cap analogue through π-π stacking interaction (4, 28, 29, 51), which is further stabilized by the hydrogen bonds among the purine base, the polypeptide backbone, and a conserved Glu residue. A third Trp residue recognizes the N7-methyl group of the cap structure, thus contributing to the specificity of cap binding (43).
Recently, multiple eIF4E homologues have been reported in mammals (43), Drosophila melanogaster (25), Caenorhabditis elegans (18, 21), plants (44), and Schizosaccharomyces pombe (38). This multiplicity could reflect simple redundancy but may also suggest more complex roles for the eIF4E isoforms beyond mere involvement with translation initiation. The issue was partially addressed in C. elegans, in which the existence of several eIF4E-like proteins was attributed to the necessity of recruiting mRNAs with different caps (7- and 2,2,7-methylguanosine) to the initiation complex (18, 21). In another case, one of the two eIF4Es in zebra fish, eIF4E-1B, cannot bind to the m7GpppN-cap structure and plays no apparent role in translation initiation. Though it shares 66% identity with human prototypical eIF4E and possesses a conserved cap-binding domain, its function remains unknown (19). In most other cases, the biological significance in multiple eIF4E-like proteins remains unclear. To distinguish the prototypical eIF4E protein in the initiation complex from the other eIF4E homologues in humans, the former has been renamed eIF4E-1 (20).
Giardia lamblia, a parasitic protozoan, is one of the deeply branched and most primitive eukaryotes based on phylogenetic analysis of its small rRNA, as well as many other proteins (1, 17, 42, 46). Within its small 12-Mb genome, most of the genes reported so far do not contain introns (36). Their transcripts have exceedingly short 5′ untranslated regions (5′-UTRs), ranging from 0 to 14 nucleotides, and similarly short 3′-UTRs of 10 to 30 nucleotides (2). In a previous study, we demonstrated that translation of an mRNA in Giardia could initiate efficiently from the first initiation codon located only 1 nucleotide downstream from the m7GpppN-cap structure (26). When the 5′-UTR between the cap and AUG was lengthened beyond 9 nucleotides, translation initiation was decreased drastically. This is in contrast to what were observed in yeast and mammalian systems, in which a minimum of 20 nucleotides are required in the 5′-UTR to allow optimal ribosome scanning and prevention of scanning leakiness (22, 23). This interesting discrepancy between Giardia and higher eukaryotes suggests the presence of a unique and perhaps much simpler protein synthetic machinery in Giardia that may not depend on ribosome scanning but recognize the simple structure of cap-AUG as a sufficient signal for translation initiation. The precise cap structure in Giardia RNAs has, however, not yet been determined, even though m7GpppN-capped mRNA introduced into the cells appeared to express well (26). Eight m2,2,7GpppN-capped snRNA species were also identified in Giardia (35). Two of them, snRNA D and snRNA H, shared some common features with snoRNAs and were associated with fibrillarin, which suggested their location in the nucleolus and their potential role in rRNA processing and ribosome maturation (12, 34, 35).
To begin elucidating the structure of translation initiation machinery in Giardia, we describe in this report the identification and characterization of two eIF4E homologues. eIF4E2 binds to m7GpppN-cap and is apparently responsible for translation initiation as a functional equivalent of human eIF4E-1, whereas eIF4E1 binds to the m2,2,7GpppN-capped snoRNA-like molecules and is not involved with translation initiation. The protein is associated with a nucleolus-like structure in Giardia and could perform the function of a snoRNA cap-binding protein for rRNA processing and ribosome biogenesis (15).
MATERIALS AND METHODS
Construction of recombinant expression plasmids.
Two pairs of primers were designed to amplify the two Giardia eIF4E homologues by reverse transcription (RT)-PCR. 5′-GGCATATGACCGACTACTCGCTCCCG-3′ (NdeI site underlined) and 5′-GGAAGCTTTCACCGTGAGCGTTTGTGCGTTCG-3′ (Hind III site underlined) were used for amplifying the 654-bp eIF4E1 cDNA encoding a 218-amino-acid 25-kDa protein, whereas 5′-GGCATATGGAGGACATTATACTTGACG-3′ (NdeI site underlined) and 5′-GGAAGCTTTCATGGCTGGATGTGGAACATGC-3′ (HindIII site underlined) were employed for synthesizing the 504-bp eIF4E2 cDNA coding for a 168-amino-acid 20-kDa protein. The two full-length cDNA fragments were cloned into pGEM-T easy vector, released with NdeI/HindIII digestion, and inserted into the pET28b expression vector (Novagen). The recombinant plasmids pET4E1 and pET4E2 thus produced were verified by DNA sequencing and transformed into the cells of Escherichia coli BL21(DE3)/pLysS for expression.
Expression and purification of G. lamblia eIF4Es from transformed E. coli.
A colony of the transformed E. coli cells was cultured overnight in 50 ml of LB medium with 50 μg/ml kanamycin at 37°C. The culture was transferred into 1 liter of LB medium and incubated until the optical density at 600 nm reached ∼0.8. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to 200 μM, and the cells were incubated for another 2 h. The cells were harvested by centrifugation and suspended in lysis buffer (100 mM NaH2PO4, 10 mM Tris-Cl, 8 M urea, pH 8.0) at a ratio of 5 ml of buffer per g of wet cells. The C-terminally His-tagged recombinant protein was purified from the lysate under denatured conditions as suggested by the manufacturer (QIAGEN). Eluents thus collected were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot assay using mouse anti-His tag monoclonal antibody (1:1,000 dilution). Purified eIF4E1 and eIF4E2 (10 mg of each) were used to raise polyclonal antibodies in rabbits at Animal Pharm Services. The antisera thus obtained were titrated against the recombinant antigens by enzyme-linked immunosorbent assay and purified via two-step affinity chromatography using the ImmunoPure rProtein A immunoglobulin G (IgG) Plus Orientation column and the Immobilized E. coli lysate column (Pierce). The purified antibodies were titrated by enzyme-linked immunosorbent assay and used for subsequent immunoblotting and immunofluorescence assays.
m7GTP-Sepharose affinity column chromatography.
Trophozoites of Giardia strains WB and WBI (WB strain infected with giardiavirus) in 1.4-liter fresh cultures (50) were cooled on ice for 20 min, harvested at 3,000 × g for 30 min, washed in phosphate-buffered saline (PBS) (137 mM NaCl, 8 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), and suspended in 10 ml of buffer A (20 mM HEPES [pH 7.4], 150 mM KCl, 1 mM EDTA, 2 mM dithiothreitol) with two complete proteinase inhibitor tablets (Roche) added per 50 ml of buffer A. The cells were sonicated on ice at about 200 W for 3 min with 5-s bursts and 15-s intervals. The lysates were centrifuged for 20 min at 10,000 × g, and the supernatant was mixed with 1 ml of the m7GTP-Sepharose 4B slurry (0.5 ml of beads; Amersham), which was prewashed with 10 column volumes (CV) of water and then 10 CV of buffer A. The mixture was incubated at 4°C overnight and packed into a column. The flowthrough was collected, and the column was washed with 20 CV of buffer A and 20 CV of buffer A plus 0.1 mM GTP. Protein still attached to the column was eluted with 1 ml of buffer A plus 0.1 mM m7GTP four consecutive times.
Protein identification by MALDI-TOF mass spectrometry.
Proteins in the final elute were separated in SDS-PAGE, and the gel was stained with E-Zinc as instructed by the manufacturer (Invitrogen). The specific protein band at about 20 kDa was cut off the gel and destained with the destaining buffer (Invitrogen). After reduction and alkylation, the protein band was treated with 12.5 μg/ml trypsin (Promega) at 37°C overnight. The digest, cleaned with ZipTipC18, was analyzed with a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry instrument (Voyager DE-STR mass spectrometer; Applied Biosystems). Molecular weights of individual peptide peaks were collected to BLAST the National Center for Biotechnology Information genome database using MS-FIT of the ProteinProspector program, version 4.0.5, from the University of California San Francisco (5).
Western blot analyses.
Electrotransfer of protein was carried out at 80 V in 10% (vol/vol) methanol-25 mM Tris-Cl (pH 7.4)-192 mM glycine at 4°C for 1 h. Polyvinylidene difluoride membranes were blocked with 5% milk in TBST buffer (50 mM Tris-Cl [pH 8.0], 200 mM NaCl, 0.1% Tween 20) and incubated with mouse monoclonal anti-c-myc IgG (1:5,000 dilution) and rabbit polyclonal anti-Giardia eIF4E1 or eIF4E2 antibodies. Horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies (1:10,000 dilution; Amersham) were then added prior to washing and visualization with enhanced chemiluminescence (Amersham).
Cap-binding specificity assay.
A 200-bp DNA fragment containing the T7 promoter and an AAGAAACC stretch before the ATG initiation codon was digested from pGiaLuc (26) with EcoRI/BstXI and gel purified. The fragment was used as the template for in vitro synthesis of the 178-nucleotide RNA molecule with T7 RNA polymerase in the presence of 12 mM m7G(5′)ppp(5′)G or 12 mM m2,2,7G(5′)ppp(5′)G as suggested by the manufacturer (Ambion). The product was purified by phenol-chloroform extraction and ethanol precipitation and examined for integrity by electrophoresis in a 1.5% agarose gel. The concentration of each mRNA sample was estimated at 260 nm in a Beckman DU7 spectrophotometer.
The RNA sample (3 μg) in 50 μl of TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA) was labeled with biotin using the BrightStar Psoralen-Biotin nonisotopic labeling kit in accordance with the instructions of the manufacturer (Ambion). The excess biotin was removed by n-butanol extraction, and the two biotinylated RNA samples were used in the in vitro pulldown experiments described as follows.
Constructs pGBK4E1 and pGBK4E2 with T7 promoters and HA or c-myc located at the 5′ ends of the eIF4E1 and eIF4E2 open reading frames, respectively, were each used as a template in the rabbit reticulocyte TnT quick coupled transcription-translation system (Promega) for in vitro synthesis of the corresponding proteins. The reaction mixtures (5 μl of eIF4E1 and 20 μl of eIF4E2) were each incubated with 1 μg of the biotinylated RNA sample described above in 500 μl of the binding buffer (50 mM HEPES-KOH [pH 7.6], 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 5%[vol/vol] glycerol) at 4°C for 60 min and centrifuged at 12,000 rpm for 3 min to remove any aggregated material. Preblocked streptavidin-Sepharose beads (Novagen) were added to the supernatant, and the mixture was incubated at 4°C for 60 min with gentle shaking. The beads were washed twice with the binding buffer, and the bound proteins were eluted off the beads with 200 μM GTP, m7GTP, or m2,2,7GpppG. Aliquots of the collected fractions were analyzed by SDS-PAGE.
Complementation in S. cerevisiae.
The pGADT7 vector (Invitrogen) was converted to pGADT7′ by removing the AD domain and the hemagglutinin (HA) tag peptide domain. pGADT7′ was used to construct the expression cassette of Giardia eIF4E1 and eIF4E2 in yeast. The two cDNAs were removed from pET4E1 and pET4E2 with NdeI/XhoI digestion and inserted into pGADT7′. Yeast eIF4E in pYCP33-supex2 (a kind gift from John McCarthy of UMIST) was removed by NdeI/EcoRI and inserted into pGADT7′ as the positive control. The recombinant constructs were verified by restriction enzyme digestion and DNA sequencing.
The S. cerevisiae yTHC strain YSC1180-7428777 (Open Biosystems) was grown in liquid YPD broth containing 200 μg/ml Geneticin to be made competent for transformation. Cells transformed with the construct were plated on −Leu and Geneticin plates and incubated at 30°C for 3 days. Cell colonies were selected, and the cells from individual colonies were used in various dilutions to inoculate plates with 10 μg/ml doxycycline and incubated at 30°C for 2 days.
Ribozyme knockdown of gene expression in G. lamblia.
Fragments (∼400 bp) of the 5′ ends of the Giardia eIF4E1 and eIF4E2 cDNAs were each inserted into the pC631neo plasmid in which neo was used to substitute for pac in the pC631pac plasmid (40). A hammerhead ribozyme sequence was introduced into the constructs by site-directed mutagenesis using two pairs of primers (7, 8, 33): 4E1RZ1 (CGAGGAGCTCTGGCGTGTTTCGTCCTCACGGACTCATCAGGTCGATAACATCCC) plus 4E1RZ2 (GGGATGTTATCGACCTGATGAGTCCGTGAGGACGAAACACGCCAGAGCTCCTCG) and 4E2RZ1 (GCTATCAGTTTTTGCGTTTCGTCCTCACGGACTCATCAGGGCCCAACGTTATTGAC) plus 4E2RZ2 (GTCAATAACGTTGGGCCCTGATGAGTCCGTGAGGACGAAACGCAAAAACTGATAGC).
The two final constructs, pC631neo4E1RZ and pC631neo4E2RZ, together with pC631neo as a negative control, were each linearized by NdeI and transcribed in vitro with T7 RNA polymerase as instructed by the manufacturer (Ambion). The RNA thus synthesized, containing an antisense sequence of eIF4E1 or eIF4E2 plus an inserted hammerhead ribozyme, was electroporated into Giardia WBI trophozoites by a previously described procedure (26, 52). G418 (200 μg/ml) was added to the cell culture for selection of transfectants. When cells grew to confluency, G418 was increased stepwise to 4 mg/ml and the incubation continued until confluency was reached again each time. The efficiency in knocking down the target mRNA was examined by semiquantitative RT-PCR, and the growth of transfectants was monitored in three independent experiments and presented with calculated standard deviations.
Semiquantitative RT-PCR.
Total RNA was extracted from antisense RNA-ribozyme-transfected Giardia trophozoites, and the concentration of each RNA sample was measured spectrophotometrically. An equal quantity of RNA was used as the template in a semiquantitative one-step RT-PCR (Invitrogen) using primers of corresponding genes as instructed by the manufacturer. DNA fragments resulting from RT-PCR and identified in agarose gel were quantitated by laser densitometer tracing.
In vitro transcription, RNA transfection of Giardia, and luciferase assay. The luciferase-containing plasmid pGiaLuc mentioned above (26) was linearized with SacII and used as the template for in vitro synthesis of the luciferase mRNA as suggested by the manufacturer (Ambion) in the presence of 15 mM GTP, 12 mM m7G(5′)ppp(5′)G, or 12 mM m2,2,7G(5′)ppp(5′)G. The products were purified by phenol-chloroform extraction and ethanol precipitation and examined for integrity by electrophoresis in 1.5% agarose gel. The concentration of each mRNA sample was estimated by its absorbance at 260 nm in a Beckman DU7 spectrophotometer, and 20 μg of each in vitro transcript was introduced into Giardia WBI trophozoites by electroporation (26). The luciferase activity thus expressed in Giardia was assayed 4 h after electroporation as previously described (26).
Pulldown of Giardia snRNAs by GST-fused eIF4E1.
The Giardia eIF4E1 gene was cloned into pGEX4T-3 vector between BamHI and NotI restriction sites, and the recombinant plasmid, pGEX4E1, confirmed by DNA sequencing, was transformed into E. coli BL21(DE3) cells. The eIF4E1 protein fused with glutathione S-transferase (GST) at its N terminus was overexpressed and purified following the instruction of the manufacturer (Amersham). Four micrograms of the fusion protein was incubated with 40 μg of Giardia total RNA for 1 h. Glutathione-Sepharose 4B beads (50 μl), thoroughly washed with PBS and preincubated with 1 mg of yeast tRNA and 1 mg of salmon sperm DNA, were incubated with the protein-RNA mixture for 30 min at room temperature. After a thorough wash, the beads were eluted with glutathione and the eluate was extracted with phenol and precipitated. One-tenth of the RNA sample was used for semiquantitative RT-PCR assay using primers RNA D5 (5′-GTCCTAGACGCGTCCTGGGATAATGC-3′) and RNA D3 (5′-AAGGACTATAGGGGCGGTGATTAGGC-3′) for snRNA D, primers RNA H5 (5′-CTGCCTCTCCTGAGGCAGATG-3′) and RNA H3 (5′-GAATTCAGAATACGACAAACTTCG-3′) for snRNA H, and primers RNA J5 (5′-AATGTAGCGAACCCACGCGCAAGC-3′) and RNA J3 (5′-ATTAAGTAAGGAAGGCTCGGACATCC-3′) for snRNA J.
Immunofluorescence assays.
Giardia trophozoites cultivated in vitro were harvested, washed with PBS three times, and fixed in 4% paraformaldehyde in PBS for 30 min. The cells were then washed three times in PEM buffer (100 mM PIPES, 2 mM EGTA, 0.1 mM EDTA, 1 mM MgSO4, pH 6.9), permeabilized with 0.1% Triton X-100 for 5 min, and washed another three times with PEM buffer. The fixed cells were blocked in blocking buffer (2% bovine serum albumin and 0.1% Triton X-100 in PBS) for 60 min at room temperature and incubated with the following primary antibodies for another 60 min: purified anti-eIF4E1 rabbit polyclonal antibodies (1:100 final dilution), purified anti-eIF4E2 rabbit polyclonal antibodies (1:100 final dilution), mouse monoclonal antibody against yeast fibrillarin (1:500 final dilution), and mouse monoclonal antibody against the m2,2,7G cap structure (1:100 final dilution). Alexa Fluor 488- or 555-conjugated secondary donkey antibodies against mouse or rabbit IgG (1:500 dilution; Invitrogen) were then applied to the cells and incubated for another 60 min at room temperature. Slides were mounted in the presence of 1 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) and examined under an Olympus 1X70 microscope equipped with bright-field and epifluorescence optics. The images were acquired with the MetaVue software.
RNA fluorescence in situ hybridization.
Giardia trophozoites were harvested, suspended in PBS, placed on coverslips pretreated with 0.5% poly-l-lysine, and incubated at 37°C for 30 min to allow the trophozoites to adhere. The cells were then fixed in 4% paraformaldehyde for 30 min at room temperature and washed with PBS and 2× SSC (300 mM NaCl, 30 mM sodium citrate) each for 5 min. Triton X-100 (0.5%) was added and incubated for 15 min at room temperature, and the cells were dehydrated in 70% and 100% ethanol and denatured in 70% formamide in 2× SSC at 70°C for 2 min and dehydrated again in 70% ethanol (−20°C) and 100% ethanol each for 5 min. After a further treatment with 1 μl/ml of proteinase K at 37°C for 30 min, the cells were washed with 70% and 100% ethanol for 5 min at −20°C and air dried.
A 30-mer oligonucleotide, 5′-CACCTACGGATACCTTGTTACGACTTCTCC-3′, which is complementary to the nucleotide 1403 to 1432 sequence of the Giardia 16S-like rRNA, was synthesized by Operon with fluorescein isothiocyanate (FITC) labeling at the 5′ end. Ten picomoles of the labeled probe was mixed with 10 μg of salmon DNA and 10 μg of S. cerevisiae tRNA, dried, suspended in 10 μl of 100% formamide, denatured at 95°C for 10 min, and placed immediately on ice. An equal volume of hybridization buffer (4× SSC, 20% dextran sulfate, 4 mg/ml bovine serum albumin) was added. The cells were hybridized with the probe overnight at 37°C and washed in 2× SSC with 50% formamide at 37°C for 30 min, followed by 2× SSC at 37°C and 1× SSC at room temperature for 30 min. The coverslips were mounted and examined using an Olympus 1X70 microscope equipped with bright-field and epifluorescence optics. The images were acquired with the MetaVue software.
RESULTS
Presence of two eIF4E homologues in G. lamblia.
Homology searches using human eIF4E-1, 4E-HP, and S. cerevisiae eIF4E protein sequences revealed in the Giardia genome database two homologous genes encoding two eIF4E isoforms. These two genes were designated Giardia eIF4E1 and eIF4E2.
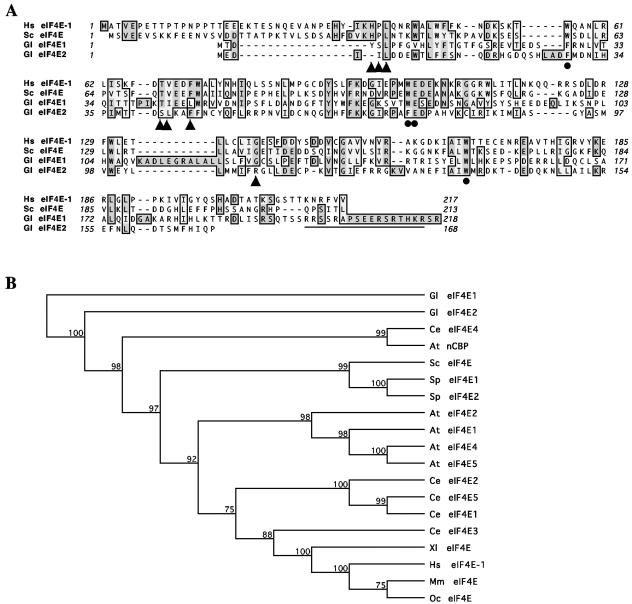
The sequence of Giardia eIF4E1 encodes a putative protein of 218 amino acids with an estimated molecular mass of 25,129 Da. It shares 13% identity and 29% similarity with human eIF4E-1 (Fig. 1A) and contains four out of the eight conserved Trp residues in human eIF4E-1 and three of the other four residues are two Tyr and one Phe. For the two Trp residues (Trp56 and Trp102) in human eIF4E-1 that are known to hold the guanine moiety of the cap, Trp56 is replaced with Phe whereas Trp102 remains unchanged at the corresponding positions in Giardia eIF4E1. The conserved Glu103 in human eIF4E-1 that stabilizes the interaction with the cap and a third Trp residue (Trp166 in human eIF4E-1) that recognizes the N7-methyl group of the cap structure remain also conserved in Giardia eIF4E1. Thus, in spite of the relatively poor sequence identity with human eIF4E-1, Giardia eIF4E1 could still function as a cap-binding protein. There is also in Giardia eIF4E1 a basic amino acid-rich sequence, RRSSRAPSEERSRTHKR, at the C terminus (Fig. 1A), which was predicted to be a bipartite nuclear localization signal, suggesting that eIF4E1 may locate and even function in the nucleus.
FIG. 1.
Alignment and phylogeny of eIF4Es from Giardia and other organisms. (A) Alignment of Giardia eIF4Es with human eIF4E-1 and S. cerevisiae eIF4E. The predicted amino acid sequences of Giardia eIF4E1 and eIF4E2 were aligned with those of human eIF4E-1 and S. cerevisiae eIF4E using the CLUSTAL W algorithm (47). Identical residues are shaded in dark gray, and related residues are in light gray. Residues with dots underneath are essential for cap binding, whereas triangles indicate residues essential for interaction with eIF4G in humans and yeast (27). The sequences predicted to be the bipartite nucleus localization signal in Giardia eIF4E1 is underlined. (B) Phylogeny of eIF4Es. The selected eIF4Es were analyzed using unweighted-pair group method using average linkages distance matrix methods from the MacVector software. The bootstrap tree was shown with 1,000 distance replicates performed. Abbreviated sequence identifiers are as follows: At, A. thaliana; Ce, C. elegans; Gl, G. lamblia; Hs, Homo sapiens; Mm, Mus musculus; Sc, S. cerevisiae; Sp, S. pombe; Xl, Xenopus laevis.
The Giardia eIF4E2 gene (Fig. 1A) encodes a putative protein of 168 amino acids with a molecular mass of 19,874 Da. It is also quite divergent from other eIF4Es, with only 14% identity and 27% similarity with human eIF4E-1. However, Trp56 and Trp102 in human eIF4E-1 are substituted only by Phe, whereas Glu103 and Trp166 are conserved in Giardia eIF4E2. This protein is thus likely capable of binding to the cap as well. There is no apparent nuclear targeting signal in this protein, which could mean that it cannot enter the nucleus and remains and performs its function in the cytoplasm of Giardia (see below).
Alignment of the two Giardia eIF4E amino acid sequences with each other showed that they are not similar to each other at all except for some of the conserved residues in the 170-amino-acid core. Even there, there are only 8% identity and 20% similarity (Fig. 1A), suggesting that though both proteins may be able to bind to the cap structure, they may perform distinctive biological functions in Giardia.
Phylogenetic analysis revealed that the two Giardia eIF4Es are highly divergent from all the other eIF4Es (Fig. 1B). While eIF4E1 is the farthest removed from the others, eIF4E2 shows a closer relationship only to C. elegans eIF4E4 and Arabidopsis thaliana nCBP. It verifies the fact that Giardia is one of the most deeply branched eukaryotes and raises the interest in identifying the precise functions of these two proteins in Giardia.
cDNAs containing the complete open reading frame sequences of eIF4E1 and eIF4E2 were amplified by RT-PCR from Giardia mRNAs. The two recombinant proteins, each tagged with a hexahistine peptide at the C terminus, were overexpressed in transformed E. coli, purified under denaturing conditions (see Fig. S1 in the supplemental material), and used to raise antibodies in rabbits (see Materials and Methods).
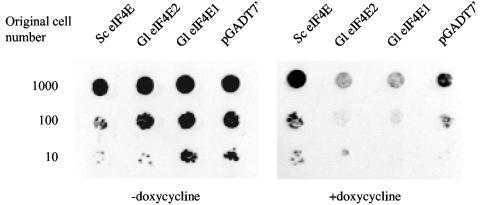
Neither Giardia homologue can complement an eIF4E knockout in S. cerevisiae.
Since human eIF4E-1 is capable of complementing the eIF4E function in S. cerevisiae (3) in spite of the sequence divergence (31% identity), we tested the two homologues from Giardia for complementing the function of yeast eIF4E (3). In S. cerevisiae yTHC strain YSC1180-7428777, the promoter of endogenous eIF4E was replaced with a tetracycline-titratable promoter. Thus, the expression of endogenous eIF4E can be switched off by doxycycline, forcing dependence of cell growth on an ectopic expression of another functional eIF4E. cDNAs of the two Giardia eIF4Es, as well as yeast eIF4E, were each cloned into pGADT7′, which has a LEU selection marker and allows insert expression from the constitutive ADH1 promoter. Following transformation and selection for transformants in leucine-deficient medium, cell cultures were applied to doxycycline-containing plates in serial dilutions. The outcome (Fig. 2) indicates that neither eIF4E1 nor eIF4E2 from Giardia is capable of rescuing the growth of yeast lacking endogenous eIF4E. The failure could be attributed to the significant sequence divergence between Giardia and yeast eIF4Es (18% identify for eIF4E1 and 15% for eIF4E2). It could also raise doubt about whether the two homologues perform a function in Giardia similar to that of yeast eIF4E in S. cerevisiae.
FIG. 2.
Giardia eIF4Es are incapable of complementing the function of S. cerevisiae eIF4E. S. cerevisiae yTHC strain YSC1180-7428777 was transformed with the Leu-selectable vector pGADT7′ containing cDNAs encoding yeast eIF4E, Giardia eIF4E1, and Giardia eIF4E2, respectively. Following selection on the −Leu plate under G418 selection, cells from individual colonies were serially diluted and applied to synthetic complete medium plates lacking leucine but containing doxycycline.
Purification and identification of cap-binding proteins from G. lamblia.
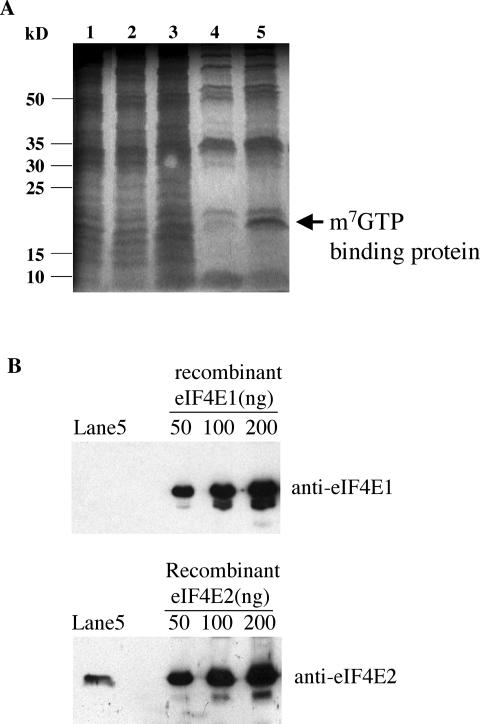
With the identification of two eIF4E homologues in the Giardia genome database and expression of their protein products in E. coli, it will be important to verify if these two proteins are also expressed in Giardia and perform the anticipated cap-binding function.
Lysate of Giardia trophozoites was subjected to m7GTP-Sepharose affinity column chromatography. Column eluates were fractionated by SDS-PAGE and visualized with E-Zinc staining (Fig. 3A). One protein band with an estimated molecular mass of 20 kDa was eluted with m7GTP and tentatively designated a cap-binding protein (Fig. 3A, lane 5). The gel containing this protein band was removed, digested with trypsin, and subjected to MALDI-TOF mass spectrometric analysis (see Fig. S2 in the supplemental material). A database search using the peptide masses thus obtained resulted in identification of a single match, eIF4E2, with 46% of its sequence covered by the identified peptides. There is thus little doubt that eIF4E2 is expressed in Giardia and that it binds to the m7GpppG-cap structure.
FIG. 3.
Purification of Giardia eIF4E2 by m7GTP-Sepharose affinity chromatography and identification by Western blotting. (A) Cell lysate from 2 liters of Giardia cell culture was applied to an m7GTP-Sepharose column and eluted. Proteins in collected fractions (lanes: 1, total protein; 2, flowthrough; 3, column wash; 4, column washed with GTP; 5, column eluted with m7GTP) were separated by SDS-PAGE and visualized by E-zinc staining. (B) The eluted fraction from an m7GTP-Sepharose affinity column separated by SDS-PAGE (Fig. 2A, lane 5) was transferred onto a polyvinylidene difluoride membrane and subjected to immunoblotting with purified rabbit polyclonal antibodies against Giardia eIF4E1 and eIF4E2, respectively. Purified recombinant Giardia eIF4Es (see Fig. S1 in the supplemental material) were included in various quantities as a control.
From the same m7GTP-Sepharose affinity column chromatography, eIF4E1 was, however, not identified in the eluent (Fig. 3A), which was confirmed by subsequent Western blot analysis (Fig. 3B). The results suggest that eIF4E1 may be either not expressed in Giardia trophozoites at all or incapable of binding to the m7GpppN-cap structure. Unfortunately, both eIF4E1 and eIF4E2 were undetectable in a Western blot from a crude lysate of 107 Giardia trophozoites (data not shown). It was not until the subsequent immunofluorescence assays that the expression of both proteins in the trophozoites was verified (see below). eIF4E1 is thus present in Giardia but most likely incapable of binding to m7GTP.
Distinctive cap-binding specificities of eIF4E1 and eIF4E2.
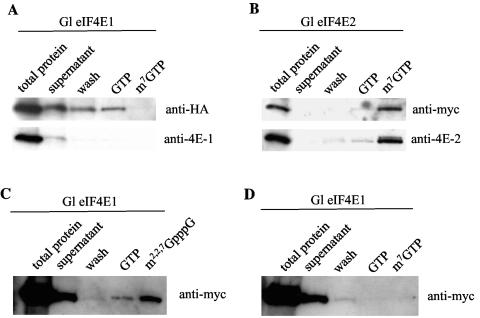
The cap-binding properties of HA-tagged eIF4E1 and c-myc-tagged eIF4E2, both synthesized from the rabbit TnT quick coupled transcription-translation reticulocyte system, were further examined by m7GTP affinity column chromatography. Effluents from the column were examined by Western blot analysis. The results indicated that eIF4E1 was not retained by the column (Fig. 4A), whereas eIF4E2 was retained and eluted only by m7GTP (Fig. 4B).
FIG. 4.
Binding of Giardia eIF4E1 and eIF4E2 to different cap structures. (A) HA-tagged eIF4E1 eluted through a m7GTP-Sepharose column was analyzed by immunoblotting (Fig. 2B), and the result demonstrated no detectable binding between the protein and the cap. (B) c-myc-tagged eIF4E2 applied to an m7GTP-Sepharose column showed that it was bound to the cap structure. (C) c-myc-tagged eIF4E1 was mixed with a biotinylated RNA species capped with m2,2,7GpppG and subjected to a pulldown experiment with streptavidin-agarose beads. The results from the subsequent immunoblotting indicated that the protein was pulled down with the beads and was released only by washing with m2,2,7GpppG. (D) In a similar experiment, c-myc-tagged eIF4E1 was not brought down by the m7GpppG-capped RNA.
To probe into the possibility that eIF4E1 may bind to a different cap structure such as m2,2,7GpppN, known to exist in Giardia (35), c-myc-tagged eIF4E1 was examined in another binding assay with RNA molecules of different caps added as potential substrates. The RNA molecules were biotinylated for pulldown by streptavidin-Sepharose beads. The data in Fig. 4C and D show that while eIF4E1 cannot be pulled down by the m7GpppG-capped RNA, it is coprecipitated with the m2,2,7GpppN-capped RNA. The protein is thus likely an m2,2,7GpppN-binding protein. Testing of eIF4E2 in a similar experiment showed that it was pulled down by m7GpppN-cap but not by m2,2,7GpppN-cap (data not shown).
Only eIF4E2 plays an essential function in protein synthesis in Giardia.
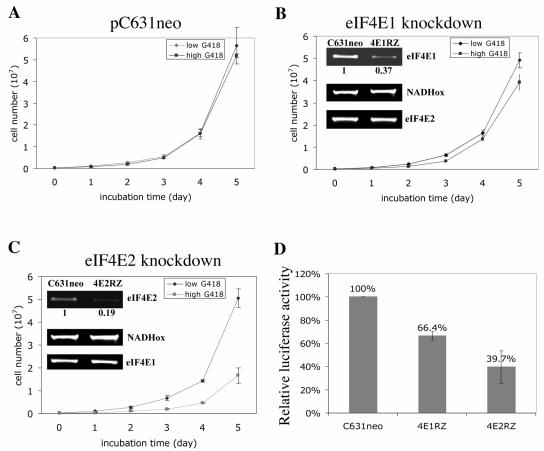
To elucidate whether eIF4E1, eIF4E2, or both perform an essential function in Giardia, an antisense-ribozyme strategy with a viral vector, which has been proven efficient for specific gene knockdowns in Giardia (7, 8, 33), was utilized. An in vitro transcript of the antisense sequence from a fragment of eIF4E1 or eIF4E2 cDNA with an inserted hammerhead ribozyme sequence was introduced into Giardia trophozoites, multiplied by the giardiavirus RNA replicating machinery in the cells, and enriched under drug pressure. Semiquantitative RT-PCR and densitometer tracing showed that in the eIF4E1 knockdown experiment, the mRNA of eIF4E1 was reduced to 37% of the original level, while that of eIF4E2 remained relatively unchanged (Fig. 5B, inset). For the eIF4E2 knockdown, the eIF4E2 mRNA was decreased to 19% whereas eIF4E1 mRNA remained unaffected (Fig. 5C inset). Growth of the eIF4E1 knockdown cells reached about 80% of the control (Fig. 5B), whereas eIF4E2 knockdown resulted in only 30% cell growth (Fig. 5C). These outcomes suggest that each protein may perform certain role in Giardia trophozoites, but eIF4E2 appears to play a more essential function in cell growth.
FIG. 5.
Effects on cell growth and cap-dependent translation of knocking down the eIF4Es in Giardia. Giardia WBI trophozoites transfected with in vitro transcripts from the empty vector pC631neo (A), the Giardia eIF4E1 antisense-ribozyme construct pC631neo4E1RZ (B), and the Giardia eIF4E2 antisense-ribozyme construct pC631neo4E2RZ (C) were incubated at 37°C in culture medium containing G418. Cell growth was monitored daily for up to 5 days. The insets show the intracellular mRNA levels estimated by semiquantitative RT-PCR after 1 day of incubation. RT-PCR quantitation of NADH oxidase mRNA (NADHox) was included as a control. (D) A luciferase transcript capped with m7GpppG was electroporated into the ribozyme-transfected cells mentioned above. The luciferase activities thus expressed were monitored 4 h thereafter and are presented with the activity in pC631neo-transfected cells designated as 100%. RLU, relative light units.
These ribozyme-transfected cell lines were also used to monitor the effect of knocking down eIF4E1 or eIF4E2 on m7GpppN-cap-dependent translation initiation in Giardia. The m7GpppG-capped in vitro transcript of firefly luciferase was electroporated into the antisense ribozyme-transfected cells, and the luciferase activity thus expressed in the cells was assayed (33). The results indicated that the translation initiation in eIF4E1 knockdown cells reached 66.4% of the control, whereas luciferase expression in eIF4E2-depleted cells was reduced to 39.7% of the wild-type control (Fig. 5D). These data suggest that eIF4E2, but not eIF4E1, could be the cap-binding protein in the translation initiation complex of Giardia.
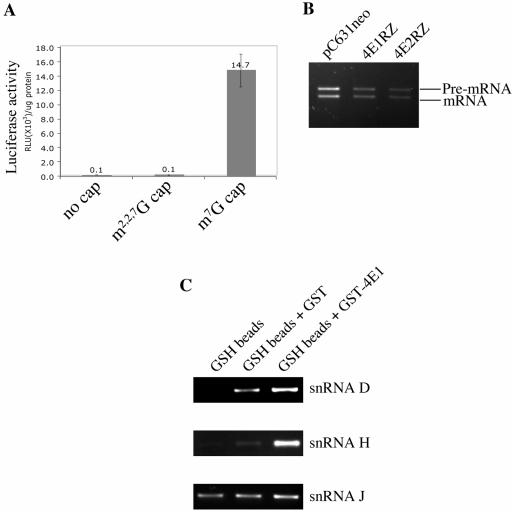
To verify if translation initiation in Giardia is mediated by m7GpppN-cap, m2,2,7GpppN-cap or no cap at all, a firefly luciferase transcript without a cap or with the m7GpppG-cap or m2,2,7GpppG-cap structure was electroporated into Giardia trophozoites. No luciferase activity could be detected from the cells transfected with the transcript without a cap or with the m2,2,7GpppG cap structure (see Fig. 7A), but cells transfected with the m7GpppG-capped luciferase transcript expressed a significant level of luciferase activity (Fig. 6A), indicating that m7GpppN-cap is most likely the cap of Giardia mRNAs and functions in translation initiation. The observation also agrees with the previous findings that eIF4E2 is most likely the cap-binding protein involved in translation initiation (Fig. 5D) and that it binds to the m7GpppG-cap structure (Fig. 3 and 4B).
FIG. 7.
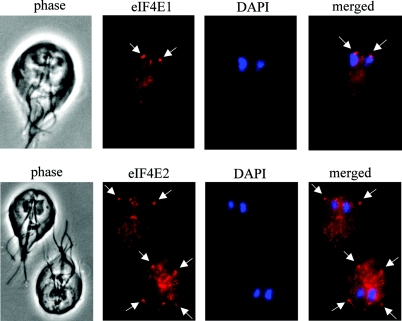
Immunofluorescence localization of eIF4Es in Giardia. Giardia WB cells stained with DAPI were further stained with rabbit polyclonal antibodies against Giardia eIF4E1 (top panel) and eIF4E2 (bottom panel). The stained cells were examined with a fluorescence microscope. Arrows point to concentrated localizations of the protein.
FIG. 6.
Functional analysis of Giardia eIF4E1. (A) Firefly luciferase in vitro transcripts with different cap structures were used to transfect Giardia trophozoites. The luciferase activity thus expressed was assayed 4 h after electroporation. Each value was derived from three independent electroporation experiments. (B) Total RNA was extracted from each of the knockdown cell lines described in the legend to Fig. 5 1 day after incubation in the presence of 4 mg/ml G418. These RNA samples were used as templates for semiquantitative RT-PCR to monitor the pre-mRNA and mature mRNA of 2Fe-2S ferredoxin in the eIF4E-depleted cells. (C) Giardia total RNA was incubated with the GST-eIF4E1 fusion protein and pulled down by glutathione-Sepharose 4B beads. Results from RT-PCR amplification indicate pulldown of snRNA D and snRNA H by GST-eIF4E1, whereas the uncapped snRNA J is not.
This identification of the function of eIF4E2 left the function of eIF4E1 still unclear, except that it binds to the m2,2,7GpppG-cap (Fig. 4C) and is thus most likely associated with the snRNAs or snoRNAs in Giardia (35). snRNAs are known to play important roles in RNA splicing (6, 40, 41). An intron-containing gene, encoding a 2Fe-2S ferredoxin, was recently identified in Giardia (36), which prompted us to look into the potential involvement of eIF4E1 in converting the pre-mRNA into the mature mRNA of this particular gene. Using RT-PCR, the ratios of pre-mRNA to mature mRNA of 2Fe-2S ferredoxin in the wild-type, eIF4E1, and eIF4E2 knockdown cells of Giardia (Fig. 5) were compared. The results (Fig. 6B) showed no apparent change in the ratios of pre-mRNA to mature mRNA from the three cell lines, suggesting that eIF4E1 does not function in mRNA splicing in Giardia. The snRNAs with the m2,2,7GpppN cap structure identified so far in Giardia, snRNA D and snRNA H, are associated with fibrillarin and share some common features with snoRNAs rather than snRNAs (35). To verify if eIF4E1 could be associated with snRNA D and snRNA H, N-terminally GST-tagged eIF4E1 was overexpressed and purified from transformed E. coli, incubated with Giardia total RNA, and pulled down by glutathione Sepharose 4B beads. Results from semiquantitative RT-PCR (Fig. 6C) indicated that m2,2,7GpppN-capped RNA D and RNA H were indeed pulled down with eIF4E1, but not snRNA J, which is without an m2,2,7GpppN cap structure (Fig. 6C). eIF4E1 is thus most likely bound to the m2,2,7GpppN-capped snRNAs in Giardia.
Localization of the two eIF4E proteins in Giardia trophozoites.
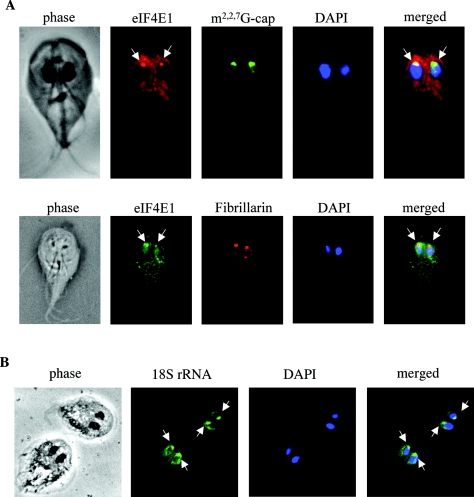
As the two eIF4Es bind to different cap structures and may perform distinctive functions in Giardia, we asked whether this might be reflected in their intracellular distribution. Immunofluorescence assays using purified rabbit polyclonal antibodies against eIF4E1 and eIF4E2 revealed very different patterns of distribution for the two proteins (Fig. 7). Although both proteins were identified in the cytoplasm of Giardia, eIF4E1 also showed a concentrated presence in a nucleolus-like structure in the nucleus (Fig. 7, top panel). To verify this observation, two more monoclonal antibodies against the m2,2,7GpppN-cap structure and the nucleolus marker fibrillarin (37) were used in the immunofluorescence assay and showed that eIF4E1 was indeed colocalized with the cap and fibrillarin in the nucleolus-like structures in both nuclei of Giardia (Fig. 8A). The nucleolus-like structure was mostly located at the anterior end of the nucleus. Its identity was further verified in an RNA fluorescence in situ hybridization study using FITC-conjugated oligonucleotide complementary to the 3′ end of Giardia 16S-like rRNA molecule. The results indicated the presence of the 16S-like rRNA in the nucleolus-like organelles at the anterior ends of the nuclei (Fig. 8B) similar to those structures containing eIF4E1, m2,2,7GpppN-cap, and fibrillarin (Fig. 7, top panel, and 8A).
FIG. 8.
Colocalization of Giardia eIF4E1 with m2,2,7GpppN cap, fibrillarin, and 16S-like rRNA. (A) Giardia WB cells stained with rabbit polyclonal antibody against eIF4E1, mouse monoclonal antibody against m2,2,7GpppN cap (top), and mouse monoclonal antibody against yeast fibrillarin (bottom). (B) Giardia WB trophozoites were fixed and stained with an FITC-conjugated 30-mer oligonucleotide complementary to the 3′ end of the Giardia 16S-like rRNA. Concentrated stains were visible in the nucleolus-like structures at the anterior ends of both nuclei.
The cytoplasmic distribution of eIF4E2 was punctate in appearance, with four concentrated spots located at the conjunctions where flagella emerged across the cell membrane of Giardia (Fig. 7, bottom panel). The potential significance in this punctate appearance of eIF4E2 remains unclear.
DISCUSSION
In this report we showed that there are two eIF4E homologues in Giardia. eIF4E1 carries a putative bipartite nuclear localization signal and was found concentrated in a nucleolus-like structure in the nucleus of Giardia. It does not bind to m7GpppN-cap and plays no apparent role in translation initiation in Giardia. But it binds to the m2,2,7GpppN-cap, is colocalized with it in the nucleolus-like structure, and interacts with the snRNA molecules with an m2,2,7GpppN-cap structure. These snRNAs represent an abundant, evolutionarily ancient group of noncoding RNAs among the eukaryotes (3, 44). They perform highly diverse functions ranging from methylation and pseudouridylation of RNA and nucleolytic processing of rRNAs to synthesis of telomeric DNA. eIF4E1 could be involved in some or all of these activities in Giardia. Its apparent lack of involvement in mRNA splicing (Fig. 6B) agrees with the fact that it is colocalized with the cap only in the nucleolus-like structure and is most likely a snoRNA binding protein. Giardia eIF4E1 may thus play important roles in rRNA and ribosome maturation in the nucleolus (10). To the best of our knowledge, this is the first case in which an eIF4E homologue has been found to bind to m2,2,7GpppN-capped snoRNA-like molecules in a nucleolus-like structure instead of m7GpppN-capped mRNA for translation initiation. The only protein identified so far that interacts specifically with m2,2,7GpppN-cap but not m7GpppN-cap, Snurportin1, functions as an snRNP-specific nuclear import receptor (15). However, Giardia eIF4E1 shares little sequence homology with that protein.
The structural basis for the specific binding between eIF4E1 and m2,2,7GpppN-cap is a little more difficult to envision. The five eIF4E variants found in C. elegans have eIF4E-3 and eIF4E-4 binding only to the m7GpppN-cap, whereas eIF4E-1, eIF4E-2, and eIF4E-5 bind to both m7GpppN- and m2,2,7GpppN-cap (21). Molecular dynamic simulation suggested that the width and depth of the cap-binding cavity were smaller in eIF4E-3 than in eIF4E-5 (31). Site-directed mutations of eIF4E-5 aimed at reducing the size of its cap-binding cavity resulted in a mutant binding only to m7GpppN-cap. This well-derived structural basis for selective cap binding cannot, however, explain the specific binding to only the trimethyl-cap by Giardia eIF4E1. In view of the vastly divergent amino acid sequence of eIF4E1 compared with those of the other eIF4Es, it is possible that a different structural basis may underlie the binding specificity of this Giardia protein, which will have to wait for the resolution of its three-dimensional structure.
Giardia eIF4E2 binds only to m7GpppN-cap and is most likely involved in translation initiation in Giardia. The fact that only transcripts with the m7GpppN-cap are translated in Giardia has further confirmed this conclusion. This is the first translation initiation protein identified in Giardia thus far.
By the knowledge from other eukaryotes, His37, Pro38, Val69, Trp73, Leu131, Glu132, and Leu135 in eIF4E (by human eIF4E-1 numbering) are involved in interacting with the scaffold protein eIF4G and 4E-BPs (27). Val69 and Trp73 are located in a conserved sequence (S/T)V(E/D)(E/D)FW in which substitution of the third residue E in yeast eIF4E with either A or D reduced the interaction with eIF4G (39). A comparison between Giardia eIF4E2 and human eIF4E-1 indicated that residues corresponding to His37, Pro38, and Leu131 are missing from the Giardia protein. Val69 is replaced by Leu and Trp73 is substituted by Phe, whereas Glu132 and Leu135 remain unchanged. The conserved sequence (S/T)V(E/D)(E/D)FW is changed to SLKAFF (Fig. 1). These drastic changes in the Giardia protein raise the possibility that either the eIF4G homologue in Giardia is also altered significantly in its eIF4E-binding domain or it does not exist. The composition of the initiation complex in Giardia could be radically different from those in the higher eukaryotes.
Using various domains in human eIF4G1 that are known to bind to the mRNA, poly(A)-binding protein, eIF4E, eIF4A, eIF3, and mitogen-activated protein kinase signal-integrating kinase 1 (4) as individual queries in mining the Giardia genome data bank, a 410-amino-acid protein was identified (accession number EAA40087). It has a 73-amino-acid stretch at the C terminus bearing 28.8% sequence identity with a portion of the eIF4A-binding domain in eIF4G1. The other binding domains are, however, all missing from this protein. It is dubious whether this protein is a bona fide initiation factor performing the function of a scaffold in the translation initiation complex of Giardia. An eIF4A homologue (EAA36655) was, however, identified and shown to function in translation initiation in Giardia (unpublished data), whereas homologues of eIF4B and eIF4H could not be found in Giardia. These two proteins are the RNA-binding factors thought to promote eIF4A to unwind the RNA secondary structure near the 5′ end of the mRNA presumably required for ribosome scanning (26). Their absence may verify our previous observation (26) that translation initiation in Giardia may not include ribosome scanning but leaves the structure of the initiation complex even more obscure.
The Archaea, which are the closest relatives to the nuclear components of eukaryotes, have uncapped mRNA with very short or no 5′-UTR (9). They also possess a regiment of initiation factors that are clearly homologous to those in the eukaryotes that participate in identifying the initiation codon. In Methanobacterium jannaschii, the presence of eIF1, eIF1A, eIF2, and eIF4A was verified, but eIF2B, eIF4B, eIF4E, eIF4G, eIF4H, and eIF5 were not found (9). This similar deficiency of initiation factors between Archaea and Giardia suggests a simple initiation complex shared by the two with the additional presence of cap-binding protein eIF4E2 in the latter. The composition of this relatively simple complex in Giardia will be further pursued by monitoring protein-protein interactions in future studies.
Supplementary Material
Acknowledgments
We thank John McCarthy at UMIST, Manchester, United Kingdom, for his kind gift of yeast plasmid pYCP33-supex2 and John Gross at UCSF for helpful discussion and critical reading of the manuscript.
This work was supported by grant AI-30475 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, R. D. 2000. The Giardia lamblia genome. Int. J. Parasitol. 30:475-484. [DOI] [PubMed] [Google Scholar]
- 3.Altmann, M., P. P. Muller, J. Pelletier, N. Sonenberg, and H. Trachsel. 1989. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J. Biol. Chem. 264:12145-12147. [PubMed] [Google Scholar]
- 4.Carberry, S. E., R. E. Rhoads, and D. J. Goss. 1989. A spectroscopic study of the binding of m7GTP and m7GpppG to human protein synthesis initiation factor 4E. Biochemistry 28:8078-8083. [DOI] [PubMed] [Google Scholar]
- 5.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 6.Cougot, N., E. van Dijk, S. Babajko, and B. Seraphin. 2004. Cap-tabolism. Trends Biochem. Sci. 29:436-444. [DOI] [PubMed] [Google Scholar]
- 7.Dan, M., A. L. Wang, and C. C. Wang. 2000. Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Mol. Microbiol. 36:447-456. [DOI] [PubMed] [Google Scholar]
- 8.Dan, M., and C. C. Wang. 2000. Role of alcohol dehydrogenase E (ADHE) in the energy metabolism of Giardia lamblia. Mol. Biochem. Parasitol. 109:25-36. [DOI] [PubMed] [Google Scholar]
- 9.Dennis, P. P. 1997. Ancient ciphers: translation in Archaea. Cell 89:1007-1010. [DOI] [PubMed] [Google Scholar]
- 10.Eliceiri, G. L. 1999. Small nucleolar RNAs. Cell. Mol. Life Sci. 56:22-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, S., R. Ghosh, P. Das, and D. Chattopadhyay. 2001. Expression and purification of recombinant Giardia fibrillarin and its interaction with small nuclear RNAs. Protein Expr. Purif. 21:40-48. [DOI] [PubMed] [Google Scholar]
- 13.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 14.Hentze, M. W. 1997. eIF4G: a multipurpose ribosome adapter? Science 275:500-501. [DOI] [PubMed] [Google Scholar]
- 15.Huber, J., U. Cronshagen, M. Kadokura, C. Marshallsay, T. Wada, M. Sekine, and R. Luhrmann. 1998. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17:4114-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17:6940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagaki, Y., and W. F. Doolittle. 2000. Evolution of the eukaryotic translation termination system: origins of release factors. Mol. Biol. Evol. 17:882-889. [DOI] [PubMed] [Google Scholar]
- 18.Jankowska-Anyszka, M., B. J. Lamphear, E. J. Aamodt, T. Harrington, E. Darzynkiewicz, R. Stolarski, and R. E. Rhoads. 1998. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem. 273:10538-10542. [DOI] [PubMed] [Google Scholar]
- 19.Joshi, B., A. Cameron, and R. Jagus. 2004. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 271:2189-2203. [DOI] [PubMed] [Google Scholar]
- 20.Keiper, B. D., W. Gan, and R. E. Rhoads. 1999. Protein synthesis initiation factor 4G. Int. J. Biochem. Cell Biol. 31:37-41. [DOI] [PubMed] [Google Scholar]
- 21.Keiper, B. D., B. J. Lamphear, A. M. Deshpande, M. Jankowska-Anyszka, E. J. Aamodt, T. Blumenthal, and R. E. Rhoads. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275:10590-10596. [DOI] [PubMed] [Google Scholar]
- 22.Kozak, M. 1991. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1:117-125. [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak, M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1:111-115. [PMC free article] [PubMed] [Google Scholar]
- 24.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 25.Lasko, P. 2000. The drosophila melanogaster genome: translation factors and RNA binding proteins. J. Cell Biol. 150:F51-F56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., and C. C. Wang. 2004. Capped mRNA with a single nucleotide leader is optimally translated in a primitive eukaryote, Giardia lamblia. J. Biol. Chem. 279:14656-14664. [DOI] [PubMed] [Google Scholar]
- 27.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3:707-716. [DOI] [PubMed] [Google Scholar]
- 28.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951-961. [DOI] [PubMed] [Google Scholar]
- 29.Matsuo, H., H. Li, A. M. McGuire, C. M. Fletcher, A. C. Gingras, N. Sonenberg, and G. Wagner. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4:717-724. [DOI] [PubMed] [Google Scholar]
- 30.McKendrick, L., V. M. Pain, and S. J. Morley. 1999. Translation initiation factor 4E. Int. J. Biochem. Cell Biol. 31:31-35. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi, H., D. S. Dwyer, B. D. Keiper, M. Jankowska-Anyszka, E. Darzynkiewicz, and R. E. Rhoads. 2002. Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 21:4680-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morino, S., H. Hazama, M. Ozaki, Y. Teraoka, S. Shibata, M. Doi, H. Ueda, T. Ishida, and S. Uesugi. 1996. Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem. 239:597-601. [DOI] [PubMed] [Google Scholar]
- 33.Munagala, N., and C. C. Wang. 2002. The pivotal role of guanine phosphoribosyltransferase in purine salvage by Giardia lamblia. Mol. Microbiol. 44:1073-1079. [DOI] [PubMed] [Google Scholar]
- 34.Narcisi, E. M., C. V. Glover, and M. Fechheimer. 1998. Fibrillarin, a conserved pre-ribosomal RNA processing protein of Giardia. J. Eukaryot. Microbiol. 45:105-111. [DOI] [PubMed] [Google Scholar]
- 35.Niu, X. H., T. Hartshorne, X. Y. He, and N. Agabian. 1994. Characterization of putative small nuclear RNAs from Giardia lamblia. Mol. Biochem. Parasitol. 66:49-57. [DOI] [PubMed] [Google Scholar]
- 36.Nixon, J. E., A. Wang, H. G. Morrison, A. G. McArthur, M. L. Sogin, B. J. Loftus, and J. Samuelson. 2002. A spliceosomal intron in Giardia lamblia. Proc. Natl. Acad. Sci. USA 99:3701-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochs, R. L., M. A. Lischwe, W. H. Spohn, and H. Busch. 1985. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell 54:123-133. [DOI] [PubMed] [Google Scholar]
- 38.Ptushkina, M., K. Berthelot, T. von der Haar, L. Geffers, J. Warwicker, and J. E. McCarthy. 2001. A second eIF4E protein in Schizosaccharomyces pombe has distinct eIF4G-binding properties. Nucleic Acids Res. 29:4561-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ptushkina, M., T. von der Haar, M. M. Karim, J. M. Hughes, and J. E. McCarthy. 1999. Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 18:4068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 41.Ro-Choi, T. S. 1999. Nuclear snRNA and nuclear function (discovery of 5′ cap structures in RNA). Crit. Rev. Eukaryot. Gene Expr. 9:107-158. [DOI] [PubMed] [Google Scholar]
- 42.Roger, A. J., S. G. Svard, J. Tovar, C. G. Clark, M. W. Smith, F. D. Gillin, and M. L. Sogin. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc. Natl. Acad. Sci. USA 95:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rom, E., H. C. Kim, A. C. Gingras, J. Marcotrigiano, D. Favre, H. Olsen, S. K. Burley, and N. Sonenberg. 1998. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J. Biol. Chem. 273:13104-13109. [DOI] [PubMed] [Google Scholar]
- 44.Ruud, K. A., C. Kuhlow, D. J. Goss, and K. S. Browning. 1998. Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J. Biol. Chem. 273:10325-10330. [DOI] [PubMed] [Google Scholar]
- 45.Shuman, S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1-40. [DOI] [PubMed] [Google Scholar]
- 46.Sogin, M. L., J. H. Gunderson, H. J. Elwood, R. A. Alonso, and D. A. Peattie. 1989. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 243:75-77. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomoo, K., X. Shen, K. Okabe, Y. Nozoe, S. Fukuhara, S. Morino, T. Ishida, T. Taniguchi, H. Hasegawa, A. Terashima, M. Sasaki, Y. Katsuya, K. Kitamura, H. Miyoshi, M. Ishikawa, and K. Miura. 2002. Crystal structures of 7-methylguanosine 5′-triphosphate (m7GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem. J. 362:539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasilescu, S., M. Ptushkina, B. Linz, P. P. Muller, and J. E. McCarthy. 1996. Mutants of eukaryotic initiation factor eIF-4E with altered mRNA cap binding specificity reprogram mRNA selection by ribosomes in Saccharomyces cerevisiae. J. Biol. Chem. 271:7030-7037. [DOI] [PubMed] [Google Scholar]
- 50.Wang, A. L., and C. C. Wang. 1986. Discovery of a specific double-stranded RNA virus in Giardia lamblia. Mol. Biochem. Parasitol. 21:269-276. [DOI] [PubMed] [Google Scholar]
- 51.Wieczorek, Z., K. Zdanowski, L. Chlebicka, J. Stepinski, M. Jankowska, B. Kierdaszuk, A. Temeriusz, E. Darzynkiewicz, and R. Stolarski. 1997. Fluorescence and NMR studies of intramolecular stacking of mRNA cap-analogues. Biochim. Biophys. Acta 1354:145-152. [DOI] [PubMed] [Google Scholar]
- 52.Yu, D. C., A. L. Wang, C. H. Wu, and C. C. Wang. 1995. Virus-mediated expression of firefly luciferase in the parasitic protozoan Giardia lamblia. Mol. Cell. Biol. 15:4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.