Abstract
Background
Our study aims to perform functional exploration and drug prediction of programmed cell death (PCD)-related biomarkers in lung adenocarcinoma (LUAD).
Methods
UCSC-Xena obtained LUAD-related genes. DESeq2 screened PCD-specific differentially expressed genes (DEGs), and these DEGs were intersected with genes identified by weighted gene co-expression network analysis (WGCNA) to pinpoint the key genes. KOBAS-i was used for enrichment analysis. String and GeneMania were used to construct protein interaction networks and gene-gene interaction networks, respectively. Using two machine learning algorithms to screen for key genes, and taking the intersection as biomarkers, validating via receiver operating characteristic (ROC) and in vitro experiments. Building a diagnostic model with a nomogram. Construct transcription factor (TF) regulatory network. CIBERSORT was used for immune infiltration analysis. Enrichr predicts targeted drugs and AutodockTools simulates molecular docking.
Results
120 hub genes related to PCD were identified, and an intersection of these genes with DEGs yielded 10 key genes, which were enriched in apoptosis-related pathways. Further machine learning screening of these genes led to the selection of 7 genes, among which 6 genes (FGR, LAPTM5, SIRPA, TLR4, ZEB2, and NLRC4) exhibited significant differences upon ROC validation, ultimately serving as biomarkers, in vitro experiments also confirmed. A nomogram demonstrated their excellent diagnostic performance. These six biomarkers are correlated with the infiltration status of most immune cells, suggesting that they affect LUAD through the immune system. TF regulation analysis identified the upstream miRNAs. Finally, drug prediction yielded three potential drugs: Lenvatinib, methadone, and trimethoprim.
Conclusion
PCD-related biomarkers in LUAD were explored, which may contribute to further understanding on PCD in LUAD.
Keywords: Lung adenocarcinoma, Programmed cell death, Transcriptome sequencing, Machine learning
1. Introduction
Lung adenocarcinoma (LUAD) is one of the deadliest cancers around the world due to its late diagnosis and high heterogeneity [1]. According to research estimates, there will be as many as 234,580 new cases of lung and bronchus cancer in the United States in 2024, with 125,070 deaths. The probability of men suffering from lung and bronchus cancer is as high as 6.3 %, while that of women is 5.9 % [2]. Existing studies have identified the risk factors of LUAD, tobacco, for instance, which is the main etiological factor [3]. Localized and early-stage LUAD patients undergo standard surgery, while the overwhelming majority of LUAD patients are typically diagnosed at an advanced stage, receiving conventional treatments such as combined radiation and chemotherapy, unfortunately experiencing a high mortality rate [4]. Recent years have witnessed the discovery of cancer driver genes and their functions in predicting the effects of targeted therapies, which leads to the development of methods aiming at the diagnosis and treatment of LUAD [5]. Since altered driver genes are strongly associated with the prognosis of patients with LUAD, it would arouse additional interest to delve into the molecular mechanisms of LUAD at the micro level [6,7].
In addition, great attention has been paid on the cell death mechanisms and their significance in combating LUAD [8]. Since the 60s–70s of the last century, systematic studies on cell death have commenced, which has been one of the most rapid developing areas of biomedicine currently [9]. The removal of functionally dispensable or potentially neoplastic cells is driven by the programmed cell death (PCD) pathways, several types of which have been described already, like apoptosis, necroptosis, pyroptosis, and ferroptosis [10,11]. PCD has been demonstrated to play either pro-tumor or anti-tumor role partly depending on the released intracellular contents [12]. When it comes to LUAD, a PCD-related model based on machine learning has been established to predict the prognosis and immunotherapy responses [13]. Another study has explored PCD patterns to develop novel gene signature to predict the prognosis of LUAD patients, where 9-gene signature based on PCD-related genes has been identified [14]. Besides, LASSO algorithm and multiple cox regression have been applied to establish a PCD-related gene prognostic model in The Cancer Genome Atlas (TCGA) cohort, which has been suggested to provide guidance for immunotherapy in LUAD patients [15]. Hence, it will become the goal of our study to delve into the specific involvement of PCD in LUAD so as to develop relevant targeted therapies.
Transcriptome sequencing (RNA-seq) has been commonly applied to assess differential expression in case-control studies and is a powerful tool for the identification of alternative splicing, which can also provide qualitative (RNA sequence) and quantitative (RNA abundance) analyses of either targeted mRNA transcripts or the complete transcriptome of a specific tissue [16,17]. Recent progresses in the RNA-seq workflow, from sample preparation to sequencing platforms to bioinformatics analysis, has enabled the opportunity to interpret different physiological and pathological conditions [18]. Further, RNA sequencing has provided an effective approach in the detection of diverse types of cancers, thus shedding lights on developing the effective treatments [19]. When it comes to LUAD, RNA-seq has contributed to the identification of cuproptosis, disulfidptosis/ferroptosis-related prognostic signature [20,21]. Combining these evidences above, we have the reason to believe PCD indeed plays a key role in LUAD and our study aims to reveal relevant PCD-related biomarkers and carry out functional exploration and drug prediction on these identified biomarkers using RNA-seq (Detailed scheme was shown in Supplementary Fig. 1). By initiating this study, we can provide additional insights on the PCD and its involvement in LUAD.
2. Material and methods
2.1. Data source
TCGA dataset: The count and FPKM data in LUAD were downloaded from UCSC-Xena (https://xena.ucsc.edu/). Following the sorting process, only 01A tumor sample (n = 510) and 11A peri-tumor sample (n = 58) were retained for further analyses.
GEO dataset: The LUAD dataset GSE118370 were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/) and 6 pairs of LUAD and normal tissues were obtained.
PCD-related genes (PCD-RGs): a total of 1548 PCD-RGs were downloaded based on a previous study [22].
2.2. Analytical methods
Differential gene expression analysis: DESeq 2 package was applied to sort the differentially expressed genes (DEGs) at the thresholds of |log2FC| ≥ 1 and padj <0.05 [23].
Dataset scoring: ssGSEA package was used to score the enrichment of sample in different gene sets [24].
Weighted correlation network analysis (WGCNA): WGCNA package was used to sort key genes as needed [25]. In detail, the soft threshold and the hub modules were determined, following which the strongest positive correlation was selected for additional analysis via calculating the Pearson correlation coefficient. Then the gene significance (GS) score for the genes’ traits and module membership (MM) in the hub modules were measured and genes were screened using GS score ≥0.2 and |MM| ≥ 0.8 as the thresholds.
Enrichment analysis: KOBAS-i (http://kobas.cbi.pku.edu.cn/) was chosen to perform the GO/KEGG enrichment analysis of relevant DEGs [26].
Protein-protein interaction (PPI) and gene-gene interaction (GGI) networks: String (https://cn.string-db.org/) and GeneMania (http://genemania.org) were used to construct the PPI and GGI networks.
Machine learning: Glmnet and e1071 packages were applied for machine learning and sorting of LUAD-related genes [27].
Receiver operating characteristics (ROC) curve analysis: LUAD-related genes were then subjected to ROC analysis using pROC package [28].
Analysis of immune checkpoint expression: We calculated the scores of 79 immune checkpoint related genes using ssGSEA.
Immune infiltration analysis: the enrichment scores of 22 types of immune cells in each sample were quantified via CIBERSORT package [29]. Specifically, the samples with P-value >0.05 were removed and only 507 cases were retained. The results were then categorized based on the tumor and control group and the immune cells infiltration as visualized using a boxplot. Additionally, the correlation in the feature genes and the immune cells infiltration was determined and the corresponding results were seen in a heatmap.
Nomogram and the transcription factor (TF) regulatory networks: rms package was applied to plot the nomogram based on the relevant biomarkers [30]. Relevant TFs to the biomarkers were downloaded from hTFtarget (https://guolab.wchscu.cn/hTFtarget/#!/) and those with No. of dataset ≥2 in LUAD were retained to construct the TF regulatory network. In the meantime, the upstream microRNAs (miRNAs) targeting these biomarkers were retrieved from ENCORI (https://rnasysu.com/encori/) with those clipExpNum ≥2 collected for the analysis. Then, the TF regulatory network was created in cytoscape (version 3.8.0).
Drug prediction and molecular docking: Enrichr package was selected to predict the drug candidates in DSigDB database [31]. Based on the prediction on drug candidates, PubChem (https://pubchem.ncbi.nlm.nih.gov/) was employed to download the 3D structure. Drugs, if lacked the 3D structure, would be traced downwards according to the P-value. The energy of 3D structure of predicted drugs was minimized via ChemBioOffice and the crystal structure of receptor proteins was retrieved from Uniprot (https://www.uniprot.org/).
Prior to the molecular docking, all molecules were dehydrated, hydrogenated, and then subjected to the removal of small molecules. AutodockTools was then used for the molecular docking of drug candidates and genes of interest. Only the results with binding energy < −5 kcal/mol were visualized in PyMOL, and the parameters for molecular docking were listed in Supplementary Table 1.
2.3. Cell culture and transfection
Human bronchial epithelial cell line BEAS-2B (BNCC359274) and LUAD cell line A549 (BNCC337696) were procured from BeiNa Culture Collection (Beijing, China) and cultured in 90 % high-glucose Dulbecco's modified Eagle's medium (C3260, Solarbio Lifesciences, Beijing, China) or Ham's F–12K medium (C3340, Solarbio Lifesciences) supplemented with 10 % fetal bovine serum (S9020, Solarbio Lifesciences). All cells were incubated in an incubator at 37 °C with 5 % CO2.
For the silencing of TLR4 via liposome transfection, the corresponding transfection assay kit Lipofectamine 2000 transfection reagent (11668027, Invitrogen, Carlsbad, CA) and the small interfering RNA against TLR4 (target sequence: 5′-TTCCTAATATTACTTATCAATGC-3′) and the negative control with scramble target sequence were ordered and applied for transfection.
2.4. Wound healing assay
The transfected LUAD cells A549 were cultured in a 6-well plate at the density of 1 × 105 cells/well until growing completely confluent. An artificial scratch was then made on the monolayers of these cells using a 10 μL pipette tip, following which cells were continued to be cultured in the non-serum medium at 37 °C. Relevant cell images at 0 and 24 h were visualized under Eclipse Ts2R microscope (Nikon, Tokyo, Japan) and the wound closure (%) was accordingly calculated.
2.5. Transwell assay
Transwell chambers (pore: 8 μm, 3422, Corning, Inc., Corning, NY) pre-coated with 50 μL Matrigel (356234, Corning, Inc.) were applied and put on the 24-well plates beforehand. Hereafter, the transfected A549 cells were transferred to the upper chamber containing 200 μL serum-depleted medium at a density of 1 × 105 cells/well, while the corresponding lower chamber was supplemented with 700 μL complete medium. Following the 24-h culture, cells invaded to the lower chamber were fixed with 4 % paraformaldehyde (P1110, Solarbio Lifesciences) for 30 min and stained with 0.1 % crystal violet staining solution (G1063, Solarbio Lifesciences) for 15 min. The same optical microscope in wound healing assay was applied to visualize the invaded cells and the number of invaded cells was quantified hereafter.
2.6. Quantitative real-time PCR analysis of mRNA expression
The total RNAs from BEAS-2B and A549 cells were extracted via TriZol assay kit (15596026, Invitrogen) and were reversely transcribed into the complementary DNA (cDNA) using a first-strand cDNA synthesis kit (1708890, Bio-Rad Laboratories, Hercules, CA). The PCR was then initiated with the help of SYBR Green Supermix (1708880, Bio-Rad Laboratories) in CFX384 qPCR System (1855484, Bio-Rad Laboratories) at the following parameters: 95 °C for 2 min, and 40 repeated cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. The relative mRNA expression level was finally calculated with the method 2−ΔΔct with GAPDH as the normalization control [32]. The primers involved were all listed in Supplementary Table 2 for reference.
2.7. Statistical analyses
R software (version 4.1.1) was applied as needed for relevant bioinformatics analyses. Wilcoxon test was used to compare the data of two groups. The ROC curve was applied in this study and a larger area under the ROC curve (AUC) was indicative of a better predictive performance. Meanwhile, all data were expressed as mean ± standard deviation based on three independent tests, and SPSS (version 17.0, SPSS, Inc., Chicago, IL) was applied to compare the data of two groups using student's t-test. The data were statistically significant when P-value was lower than 0.05.
3. Results
3.1. WGCNA analysis
GSEA is a technique applied to assess the distribution trend of genes from a predefined gene set in a list ranked with phenotypic correlation, so as to determine the contribution to the relevant phenotypes. ssGSEA, an extension of GSEA calculating the enrichment score of each sample paired with the specific gene set, was applied in this study to calculate the enrichment of sample in PCD-RGs [24]. Accordingly, the evidently higher ssGSEA score was seen in the normal tissue (Fig. 1A).
Fig. 1.
Results of WGCNA analysis. (A) Quantified results of ssGSEA score on LUAD tissue (n = 510) and control tissue (n = 58) in LUAD. (B) The procedures sorting on the optimal soft threshold. (C) Gene dendrogram based on the topological overlap, together with the assigned modules colors. (D) Correlation between the traits and the modules (the numbers without the brackets were the correlation coefficient and those within the brackets were the P-values). (E) The gene-significance-module membership plot of MEgreen module.
WGCNA is a method used to describe genes’ association patterns in different samples, which identifies gene sets with high covariation and candidate biomarkers or therapeutic targets based on the endogeneity of the specific gene set and the relationship between the gene set and the phenotype. The results of ssGSEA score were applied as traits for the WGCNA analysis on the training set. The soft threshold interception height was set at 0.9 and the optimal soft threshold (power) was 4 (Fig. 1B). The minimum number of genes in the module and the high cut were adjusted to be 200 and 0.2, respectively. 19 modules were finally obtained (Fig. 1C) and the correlation heatmaps based on these modules were then drawn and MEgreen, the module with the highest correlation, was screened and selected as the specific modules containing 2582 genes (Fig. 1D). Additionally, 120 Hubgenes were sorted with GS score ≥0.2 and |MM| ≥ 0.8 (Fig. 1E).
3.2. DEGs analysis
The relevant data from TCGA were applied in this phase of study to determine the DEGs in different samples. In accordance with the results, a total of 1505 PCD-related genes were sorted out using the data from TCGA and DESeq2 package was then used to determine the differential gene expressions in LUAD and normal tissues. Then the DEGs were sorted at the thresholds of |log2FC| ≥ 1 and padj <0.05 and displayed in a Volcano plot (Fig. 2A). Accordingly, 333 DEGs were obtained, including 217 up-regulated genes and 116 down-regulated genes. A heatmap was then drawn based on the top 20 up-regulated and down-regulated DEGs (Fig. 2B).
Fig. 2.
DEGs analysis. (A) Volcano plot displaying the DEGs (n = 333) based on the data from TCGA. Each dot annotates a gene, and the left and right parts represents the down-regulated and up-regulated DEGs, respectively. (B) Heatmap based on the top 20 up-regulated and down-regulated DEGs in tumor and normal tissue. (C) DEGs (n = 333) and Hubgene(n = 120) obtained by WGCNA were intersected to obtain 10 key genes. (D) Locations of the intersected PCD-related genes on the chromosome. The outermost circle in the figure is chromosome, the middle gene line represents the position of the gene on the chromosome, and the inner circle represents the DEGs, with red representing upregulation and blue representing downregulation.
Subsequently, these DEGs were intersected with the Hubgenes from WGCNA. Based on the Venn diagram, 10 common PCD-related genes were obtained and their location on the chromosome was mapped (Fig. 2C–D). These 10 genes are considered to be key genes related to PCD.
3.3. Gene enrichment analysis and gene-gene network construction
GO and KEGG enrichment analyses were then carried out on the 10 common PCD-related genes using KOBAS-i. In accordance with the results on GO enrichment analysis, these genes were evidently enriched in apoptotic signaling pathway and neutrophil degranulation (Fig. 3A), and those from KEGG enrichment analyses have suggested that these genes were most significantly enriched in osteoclast differentiation (Fig. 3B).
Fig. 3.
Gene enrichment analysis and gene-gene network construction. (A–B) GO and KEGG enrichment analyses were then carried out on the 10 common PCD-related genes using KOBAS-i. (C–D) Constructed PPI network (C) and GGI network (D) of these 10 PCD-related genes using String and GeneMania.
Relevant gene-gene network on these 10 PCD-related genes was constructed using String (Fig. 3C) and GeneMania (Fig. 3D), and the relevant PPI and GGI networks were visible.
3.4. Machine learning on the identification of feature genes
Glmnet package was applied for the LASSO regression analysis on the 10 PCD-related genes and the results were taken at the lowest mean squared error (lambda.min = 0.0028). 7 featured genes were obtained (Fig. 4A–B). In the meantime, e1071 package was also applied for the support vector machines recursive feature elimination (SVM-RFE) analysis on these 10 PCD-related genes. The feature genes were recursively eliminated and the model with the least error was selected, which included 10 feature genes (Fig. 4C).
Fig. 4.
Machine learning results. (A–B) Results on LASSO regression on the training set. (C) Relationship between generalization error and the characteristics in LUAD. (D) Venn diagram showing the common feature genes from LASSO regression test and SVM-RFE.
Then these feature genes from Glmnet and e1071 packages were intersected in a Venn diagram and 7 common genes were collected, including FGR, NLRC4, TLR4, LAPTM5, CYBB, ZEB2 and SIRPA.
3.5. Validation on the efficacy of feature genes in LUAD
The ROC curves of these feature genes in the training set and the testing sets were then plotted to quantify the AUC, which was used to test the efficacy of these common feature genes in LUAD. The AUC of these common feature genes in the training set (TCGA dataset) and the testing set (GSE118370) was quantified and presented in Fig. 5A–B, respectively, where it was seen that the AUC of all these genes was all higher than 0.7 (Fig. 5A–B).
Fig. 5.
Validation on the efficacy of feature genes in LUAD. (A–B) ROC curve on the feature genes in the training set (A) and testing set (B). (C–D) Box plot on the expression levels of feature genes in the training set (C) and testing set (D).
Hereafter, the expression levels of these 7 genes in both the training set and the testing set were also visualized. Considering the significant difference on the expression levels of FGR, NLRC4, TLR4, LAPTM5, ZEB2 and SIRPA in these two datasets, these 6 genes were applied biomarkers for the current study. A nomogram was then plotted to additionally determine the diagnostic efficacy of these 6 genes (Fig. 6A). Then the ROC curve and the calibration curve on the nomogram were drawn, where an AUC of 0.989 and the mean absolute error of 0.012 were seen (Fig. 6B–C). These results were indicative of a satisfying predictive efficacy of the nomogram.
Fig. 6.
Construction of the diagnostic model. (A) Nomogram on the 6 biomarkers (FGR, NLRC4, TLR4, LAPTM5, ZEB2 and SIRPA) in LUAD. (B) Plotted ROC curve of the nomogram. (C) Plotted calibration curve of the nomogram.
3.6. Measurement of immune cell infiltration and analysis of immune checkpoints
The immune cells infiltration in the training sets was then calculated using the CIBERSORT package. The results of the immune infiltration analysis revealed significant differences in the expression of B cells naive, Plasma cells, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), NK cells resting, Monocytes, Macrophages M0, Macrophages M1, Macrophages M2, Dendritic cells resting, Dendritic cells activated, Mast cells resting, Eosinophils, and Neutrophils between the control group and the cancer group (Fig. 7A). Notably, the six biomarkers identified in this study exhibited a significant negative correlation with the expression of B cells naive, Plasma cells, T cells follicular helper, T cells regulatory (Tregs), and NK cells activated, while displaying a notable positive correlation with the expression of NK cells resting, Monocytes, Macrophages M0, Macrophages M2, Mast cells resting, Eosinophils, and Neutrophils (Fig. 7B). The analysis revealed a high degree of consistency between the immune cells significantly associated with the six biomarkers and those exhibiting marked expression differences in tumor cells, indicating that these biomarkers exert an influence on the onset of LUAD primarily through modulating immune pathways.
Fig. 7.
Determination on the immune cell infiltration. (A) Differentially immune cells infiltration in both LUAD tumor tissue and normal tissue. (B) Correlation on the 6 biomarkers and the immune cells infiltration. *P ≤ 0.05, *P ≤ 0.01, *P ≤ 0.001, *P ≤ 0.0001.
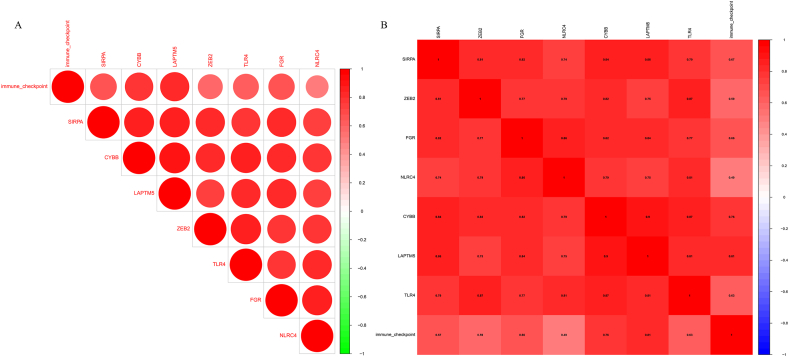
The analysis of 79 immune checkpoints significantly revealed a strong positive correlation (P < 0.05) between seven specific genes (FGR, NLRC4, TLR4, LAPTM5, CYBB, ZEB2 and SIRPA) and these immune checkpoints, suggesting that these genes may play a crucial role in the immune evasion mechanisms of tumor cells (Fig. 8).
Fig. 8.
Immune checkpoint analysis results. (A) ssGSEA calculated scores for 79 immune checkpoint related genes. (B) The analysis of correlation between immune checkpoints and seven PCD genes (FGR, NLRC4, TLR4, LAPTM5, CYBB, ZEB2 and SIRPA) indicates that the higher the numerical value, the stronger the correlation.
3.7. Plotting of the TF regulatory network
Then we set out to plot the relevant TF regulatory network to ascertain the specific mechanisms underlying the effects of these 5 genes in LUAD (NLRC4 was removed due to 0 TF predicted). As per the illustration in the methods on sorting the relevant TFs and miRNAs, 3, 4, 4, 1, and 6 TFs were predicted for FGR, LAPTM5, SIRPA, TLR4 and ZEB2, respectively. In the meantime, the upstream miRNAs targeting these 5 genes were also predicted using ENCORI, and the number of miRNAs targeting these genes was 2, 39, 80, 27 and 63, respectively. The relevant miRNAs and the TFs of these genes were then plotted in a TF regulatory network (Fig. 9).
Fig. 9.
Plotted TF regulatory network on the biomarkers (NLRC4 was removed due to the zero number of TF predicted).
3.8. Drug prediction and molecular docking
Enrichr is a comprehensive database for gene set enrichment analysis, which contains 180,184 annotated gene sets from 102 gene set databanks. The obtained 6 genes were applied as the gene sets and the enrichment analysis was then implemented with Enrichr and DSigDB was selected as the database to predict the targeted drugs (Table 1).
Table 1.
LUAD-related biomarkers and parts of targeted drugs.
| Term | P.value | Odds.Ratio | Combined.Score | Genes |
|---|---|---|---|---|
| valproic acid HL60 DOWN | 1.79E-05 | 103.1354 | 1127.555 | FGR; ZEB2; SIRPA |
| trichostatin A HL60 DOWN | 0.000374 | 36.02593 | 284.2487 | FGR; ZEB2; SIRPA |
| vorinostat HL60 DOWN | 0.00043 | 34.32509 | 266.1062 | FGR; ZEB2; SIRPA |
| Trimethoprim BOSS | 0.001327 | 52.39418 | 347.1037 | NLRC4; TLR4 |
| Retinoic acid CTD 00006918 | 0.002155 | 18.50576 | 113.6231 | FGR; ZEB2; LAPTM5; SIRPA; TLR4 |
According to the results from molecular docking, the 7JT9 protein of FGR and 2Z62 protein of TLR4 were selected. Based on the results of Pymol, the binding energy of FGR to Lenvatinib was −6.48 kcal/mol and the hydrogen binding sites were located on the amino acids of MET-90 and LEU-95 (Fig. 10A). Meanwhile, the binding energy of TLR4 to Methadone and trimethoprim was −6.49 kcal/mol and −5.78 kcal/mol, and the hydrogen binding sites were located at the amino acids of ALA-139 (for Methadone) and PRO-202, LEU-204, MET-201 and LEU-198 (for trimethoprim). The length of hydrogen bond was all lower than 3.5 Å (Fig. 10A–C, Table 2). Due to their good binding stability, we choose Lenvatinib, methadone, and trimethoprim as potential candidate drugs.
Fig. 10.
Results on the molecular docking. (A) Molecular docking of FGR to Lenvatinib and the hydrogen binding sites located on the amino acids of MET-90 and LEU-95. (B) Molecular docking of TLR4 to Methadone and the hydrogen binding sites located on the amino acid of ALA-139. (C) Molecular docking of TLR4 to trimethoprim and the hydrogen binding sites located on the amino acids of PRO-202, LEU-204, MET-201 and LEU-198.
Table 2.
Binding energy from molecular docking.
| Compound CID | Molecula_name | Gene_name | PDB_ID | Energy (kcal/mol) |
|---|---|---|---|---|
| 9823820 | Lenvatinib | FGR | 7JT9 | −6.48 |
| 4095 | methadone | TLR4 | 2Z62 | −6.49 |
| 5578 | trimethoprim | TLR4 | 2Z62 | −5.78 |
3.9. In-vitro validation using LUAD cells
Then we sought to determine the involvement of these obtained 6 biomarkers in LUAD and relevant qPCR assay was implemented to calculate their expression levels in LUAD cells A549 and human bronchial epithelial cells BEAS-2B, revealing the higher expression of FGR (Fig. 11A, P < 0.001), LAPTM5 (Fig. 11B, P < 0.0001), SIRPA (Fig. 11C, P < 0.001), TLR4 (Fig. 11D, P < 0.001) and ZEB2 (Fig. 11E, P < 0.001) yet lower expression of NLRC4 in LUAD cells A549 (Fig. 11F, P < 0.001).
Fig. 11.
In-vitro validation using LUAD cells. (A–F) Quantified expression levels of 6 biomarkers FGR (A), LAPTM5 (B), SIRPA (C) TLR4 (D), ZEB2 (E) and NLRC4 (F) in LUAD cells A549 and bronchial epithelial cells BEAS-2B via qPCR. (G) Effects of TLR4 silencing on the migration of LUAD cells A549 tested via wound healing assay. (H) Effects of TLR4 silencing on the invasion of LUAD cells A549 tested via Transwell assay. All experimental data of independent triplicates were expressed as mean ± standard deviation. ***P < 0.001; ****P < 0.0001.
Hence, we determined the effects of TLR4 silencing on the metastasis of LUAD cells in vitro using wound healing and Transwell assays, and it was clear that the silencing of TLR4 led to the diminished number of both migrated and invaded cells (Fig. 11G–H, P < 0.001). These results suggest that these 6 biomarkers, particularly TLR4, play a key role in LUAD, highlighting their potential as therapeutic targets.
4. Discussion
Recent years have witnessed the progress made in the treatment of lung cancer, and relevant therapies, for instance, targeted therapy, immunotherapy, and multidisciplinary treatment models, have been applied in clinical practice [33]. However, the reliable biomarkers capable of predicting the sensitivity to immunotherapy are still limited, necessitating further exploration. Our current study, while not directly focusing on PCD-related biomarkers in LUAD from the angles of functional exploration and relevant drug candidates, has illuminated the potential significance of FGR, NLRC4, TLR4, LAPTM5, ZEB2 and SIRPA as biomarkers worthy of further investigation. Further exploration has indicated the correlation between these biomarkers and the immune cells infiltration and immune checkpoint expression, hinting the roles of these biomarkers in LUAD. Besides, in addition to the relevant TF regulatory network of these biomarkers, the molecular docking results have confirmed the binding of TLR4 and FGR, two candidates of interest, to Lenvatinib, Methadone and trimethoprim. These results have therefore contributed to another insight on developing relevant biomarkers in LUAD.
The analysis of omics is becoming increasingly important for the advancement of disease research [34]. Our current study was initiated following the intersection of Hub genes in WGCNA and the DEGs in LUAD, and a total of 10 common genes were retrieved, which were proven to be enriched in apoptosis-related pathways and Osteoclast differentiation, to our surprise. Then these 10 common genes were subjected to machine learning via glmnet and e1071 packages. The 7 feature genes were taken following the intersection on the results from the analysis of these packages, including FGR, NLRC4, TLR4, LAPTM5, CYBB, ZEB2 and SIRPA. FGR is a tyrosine kinase gene and has been identified to be related to immune cell infiltration in LUAD [35], it may affect the occurrence of LUAD by influencing the immune response in the TME. NLRC4 has been recognized as a pyroptosis-related gene in a pan-cancer analysis including LUAD [36], which is a crucial regulator of the inflammatory signaling pathway in macrophages and is essential for the development of protective CD4+ and CD8+ T cells that produce IFN-γ [37], and its abnormal expression affects the immune response of LUAD. TLR4 has been selected as the representative of immunogenic cell death (ICD)-related genes and its stimulation represses the growth of LUAD (based on an animal xenograft model) [38]. LAPTM5 is one of the genes related to myeloid dendritic cell (which predicted the prognosis and efficacy of immunotherapy) in LUAD [39]. The protein encoded by LAPTM5 plays a role in the integrity of lysosomal membrane, and its abnormal expression may affect the activity and function of immune cells [40]. CYBB is one of the ferroptosis-related gene signature (FRGS) genes which have been identified to predict the clinical outcome in LUAD [41]. ZEB2, a transcriptional E-cadherin repressor, has been recognized as an epithelial-to-mesenchymal transition (EMT) promoter in LUAD [42]. SIRPA participates in the induction of tumor immune suppressive environment and promotes the growth of lung cancer [43]. The correlation analysis of immune checkpoints also confirms that most of these genes influence the occurrence and development of LUAD by impacting immune expression. Moreover, the results of ROC curves have also obtained high AUC values (all >0.7), demonstrating the effectiveness of these common characteristic genes in LUAD.
Previous studies have demonstrated that TLR4 can serve as a potential biomarker for LUAD(43), as its overexpression in tumor cells and the microenvironment may contribute to cancer progression. Chronic inflammation induced by the TLR4 pathway facilitates the development of various malignancies [44]. Additionally, in the process of molecular docking conducted in this study, the 2Z62 protein of TLR4 was used as a docking site. Therefore, TLR4 was ultimately selected as the silenced gene in this research. The relevant validation using LUAD cells in vitro has demonstrated the abnormally higher expression of FGR, LAPTM5, SIRPA, and TLR4, ZEB2 yet lower expression of NLRC4 in LUAD. These evidences further supported the involvement of these 6 genes in LUAD. And indicate that silencing of the TLR4 gene plays a significant role in effectively suppressing the occurrence of LUAD, which strongly confirms that TLR4 promotes the development of LUAD, consistent with previous research findings [45].
Immune cells are the crucial component of the tumor microenvironment (TME) which affect the proliferation, metastasis, and prognosis of cancers, and their interaction with tumor cells exhibits a dynamic and complex nature [[46], [47], [48]]. The infiltration of immune cells in the TME is a key factor in cancer prognosis, since increased immune cells infiltration into tumors is related to the improved survival and predicts the response to immune therapies [49,50]. By analyzing the immune infiltration results in this study, it was found that the immune cells that showed significant positive correlations with the six biomarkers were primarily concentrated in NK cells, Monocytes, Mast cells, Eosinophils, and Neutrophils. NK cells, a subtype of innate immune cells, can rapidly recognize and kill tumor cells [51], and have been proven as biomarkers for LUAD in previous studies [52]. Tumor-associated macrophages play pivotal roles in tumorigenesis, regulation of neovascularization, immunosuppression, and tumor metastasis [53]. Mast cells, as tissue-resident bone marrow cells existing throughout the connective tissues in the body, contain coarse granules and potent inflammatory mediators [54]. Their infiltration into tumors is associated with lymphangiogenesis and poor prognosis in LUAD [55]. Eosinophils, known as granulocytic white blood cells, differentiate from myeloid progenitors in the bone marrow [56]. Studies have revealed increased eosinophil counts in tumor-associated tissues [57]. Neutrophils are major immune cells, and transcriptome analysis shows that tumor-associated neutrophils from cancer patients significantly differ from those from healthy individuals [58]. The above evidence all demonstrates that these immune cells are closely related to the development of cancer, particularly LUAD, suggesting that the six biomarkers identified in this study exert their influence on LUAD by modulating immune cells within the TME, which is consistent with the results obtained from the previous analysis of the six biomarkers.
Further, TF regulatory network and molecular docking were implemented to evaluate the specific mechanisms underlying the effects of these feature PCD-related genes in LUAD. Recent studies on TF regulatory network in LUAD have already revealed some key TFs in LUAD, like SOX10, SPIB, NR4A2, FOXD1, ELF5, HOXA5, KLF5, ESRRA, SREBF1 and REL [59]. Here, the predicted results from hTFtarget have mapped out POLR2A, EP300, SMAD3, SMAD4, CEBPB, CTCF, SPI1, and FOXA2 as the relevant TFs of these feature PCD-related genes in LUAD. Additional prediction on the relevant upstream of miRNAs were predicted and the TF regulatory network was plotted. Future studies, hence, will continue to explore such interaction between the PCD-related feature genes and relevant TFs or miRNAs. Besides, the molecular docking results have proven Lenvatinib (a multi-targeted tyrosine kinase inhibitor [60]), methadone (a potential antineoplastic compound known to affect TLR4 expression in lung cancer [61]) and trimethoprim (an antibiotic which exhibits anti-tumor efficacy in vitro and in vivo [62]) as the potential drug candidates. Future studies will continue to explore the interaction and consolidate the results generated in this study.
In the field of cancer treatment, targeted therapy and immunotherapy, as two major breakthroughs in recent years, are gradually reshaping the landscape of cancer treatment. However, while demonstrating their enormous potential, these two cutting-edge therapies also face non-negligible problems and challenges. For targeted therapy, although it brings hope to many patients by precisely attacking specific molecules or proteins of cancer cells, the limited availability of biomarkers has become a significant bottleneck [63]. On the other hand, immunotherapy, which utilizes the body's own immune system to fight cancer, has demonstrated unique efficacy, but the complexity of the TME poses numerous obstacles to its effective implementation [64]. Against this backdrop, this study's exploration of biomarkers is expected to uncover new biomarkers closely related to immune infiltrating cells, potentially expanding the scope of targeted therapy, increasing the patient population that can benefit, and potentially providing new targets and strategies for optimizing immunotherapy.
Nonetheless, there are some other shortcomings that should be noticed in this study. For instance, the cohort for the validation on the efficacy of these key PCD-related genes in LUAD should be expanded, and the association between the expression levels of these genes and the prognosis of patients has not been expounded. In the meantime, our in-vitro validation focuses on TLR4 only, while those for the remaining genes have been missing. Our future studies, accordingly, will continue to explore this topic and complete the results concluded in this study.
5. Conclusion
In short, this study explores the PCD-related biomarkers in LUAD based on their functions and relevant drug candidates. It is hoped that this research may contribute to a further understanding of the involvement of PCD in LUAD and other malignancies, as well as providing new avenues for the treatment of LUAD.
Funding
The authors declare that they have receive no funding.
Availability of data and materials
The public dataset used in this study is available in GSE118370 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118370).
Ethnic statement
None.
CRediT authorship contribution statement
Xugang Zhang: Writing – review & editing, Writing – original draft, Supervision, Software, Resources, Project administration, Formal analysis, Data curation, Conceptualization. Taorui Liu: Visualization, Supervision, Resources, Project administration, Methodology. Ying Hao: Validation, Supervision, Resources, Project administration, Formal analysis, Data curation. Huiqin Guo: Visualization, Software, Project administration, Methodology, Investigation, Formal analysis. Baozhong Li: Writing – original draft, Visualization, Supervision, Project administration, Methodology, Formal analysis, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36616.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Seguin L., Durandy M., Feral C.C. Lung adenocarcinoma tumor origin: a guide for personalized medicine. Cancers. 2022;14(7) doi: 10.3390/cancers14071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Giaquinto A.N., Jemal A. Cancer statistics, 2024. CA: a cancer journal for clinicians. 2024;74(1) doi: 10.3322/caac.21820. [DOI] [PubMed] [Google Scholar]
- 3.Ortega M.A., Navarro F., Pekarek L., Fraile-Martínez O., García-Montero C., Saez M.A., et al. Exploring histopathological and serum biomarkers in lung adenocarcinoma: clinical applications and translational opportunities. Int. J. Oncol. 2022;61(6) doi: 10.3892/ijo.2022.5444. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spella M., Stathopoulos G.T. Immune resistance in lung adenocarcinoma. Cancers. 2021;13(3) doi: 10.3390/cancers13030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J.W., Li M. Molecular typing of lung adenocarcinoma with computed tomography and CT image-based radiomics: a narrative review of research progress and prospects. Transl. Cancer Res. 2021;10(9):4217–4231. doi: 10.21037/tcr-21-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E., et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Guan J.L., Zhong W.Z., An S.J., Yang J.J., Su J., Chen Z.H., et al. KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann. Surg Oncol. 2013;20(4):1381–1388. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 8.Denisenko T.V., Budkevich I.N., Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117. doi: 10.1038/s41419-017-0063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopeina G.S., Zhivotovsky B. Programmed cell death: past, present and future. Biochem. Biophys. Res. Co. 2022;633:55–58. doi: 10.1016/j.bbrc.2022.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Bedoui S., Herold M.J., Strasser A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 2020;21(11):678–695. doi: 10.1038/s41580-020-0270-8. [DOI] [PubMed] [Google Scholar]
- 11.Kari S., Subramanian K., Altomonte I.A., Murugesan A., Yli-Harja O., Kandhavelu M. Programmed cell death detection methods: a systematic review and a categorical comparison. Apoptosis. 2022;27(7–8):482–508. doi: 10.1007/s10495-022-01735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Hong M., Li Y., Chen D., Wu Y., Hu Y. Programmed cell death tunes tumor immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.847345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Wang Y., Chen J., Xia Y., Huang Y. A programmed cell death-related model based on machine learning for predicting prognosis and immunotherapy responses in patients with lung adenocarcinoma. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1183230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Z., Wang Y., Yu Y., Fu F., Zhang Y. Exploring diverse programmed cell-death patterns to develop a novel gene signature for predicting the prognosis of lung adenocarcinoma patients. J. Thorac. Dis. 2024;16(2):911–923. doi: 10.21037/jtd-23-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y., Ouyang W., Wang Z., Huang H., Ou Q., Lin R., et al. A comprehensive analysis of programmed cell death-associated genes for tumor microenvironment evaluation promotes precise immunotherapy in patients with lung adenocarcinoma. J. Personalized Med. 2023;13(3) doi: 10.3390/jpm13030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H., Huang A.Y., Wang L.K., Yoon A.J., Renteria G., Eskin A., et al. Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet. Med. 2020;22(3):490–499. doi: 10.1038/s41436-019-0672-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuksin M., Morel D., Aglave M., Danlos F.X., Marabelle A., Zinovyev A., et al. Applications of single-cell and bulk RNA sequencing in onco-immunology. European journal of cancer (Oxford, England : 1990) 2021;149:193–210. doi: 10.1016/j.ejca.2021.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Kukurba K.R., Montgomery S.B. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015;2015(11):951–969. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ergin S., Kherad N., Alagoz M. RNA sequencing and its applications in cancer and rare diseases. Mol. Biol. Rep. 2022;49(3):2325–2333. doi: 10.1007/s11033-021-06963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W., Qu H., Ma X., Li L., Wei Y., Wang Y., et al. Identification of cuproptosis and immune-related gene prognostic signature in lung adenocarcinoma. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1179742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X., Deng Z., Li Z., Ma T., Li G., Zhang C., et al. Leveraging a disulfidptosis/ferroptosis-based signature to predict the prognosis of lung adenocarcinoma. Cancer Cell Int. 2023;23(1):267. doi: 10.1186/s12935-023-03125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin H., Abulaiti A., Maimaiti A., Abulaiti Z., Fan G., Aili Y., et al. Integrated machine learning survival framework develops a prognostic model based on inter-crosstalk definition of mitochondrial function and cell death patterns in a large multicenter cohort for lower-grade glioma. J. Transl. Med. 2023;21(1):588. doi: 10.1186/s12967-023-04468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abazeed M.E., Adams D.J., Hurov K.E., Tamayo P., Creighton C.J., Sonkin D., et al. Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res. 2013;73(20):6289–6298. doi: 10.1158/0008-5472.CAN-13-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bu D., Luo H., Huo P., Wang Z., Zhang S., He Z., et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317. doi: 10.1093/nar/gkab447. w25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J., Liang C., Li J. A signal recognition particle-related joint model of LASSO regression, SVM-RFE and artificial neural network for the diagnosis of systemic sclerosis-associated pulmonary hypertension. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M., Gao Y., Yuan Y., Liu X., Wang Y., Li L., et al. A comprehensive study of clinicopathological and genetic features of neuronal intranuclear inclusion disease. Neurol. Sci. 2023;44(10):3545–3556. doi: 10.1007/s10072-023-06845-2. [DOI] [PubMed] [Google Scholar]
- 31.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTMethod. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Wang M., Herbst R.S., Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 2021;27(8):1345–1356. doi: 10.1038/s41591-021-01450-2. [DOI] [PubMed] [Google Scholar]
- 34.Shahrajabian M.H., Sun W.L. Survey on multi-omics, and multi-omics data analysis, integration and application. Curr. Pharm. Anal. 2023;19(4):267–281. [Google Scholar]
- 35.Chen Q., Ma J., Wang X., Zhu X. Identification of prognostic candidate signatures by systematically revealing transcriptome characteristics in lung adenocarcinoma with differing tumor microenvironment immune phenotypes. Aging. 2022;14(11):4786–4818. doi: 10.18632/aging.204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan M., Ai M., Du K., Song J., Wang B., Lin J., et al. Pyroptosis relates to tumor microenvironment remodeling and prognosis: a pan-cancer perspective. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1062225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X.Y., Zhou J., Li Z.N., Chen X.Q., Wei Q.H., Chen K.D., et al. A Novel Pyroptosis-Related Prognostic Signature for Lung Adenocarcinoma: Identification and Multi-Angle Verification. Front. Genet. 2023;14 doi: 10.3389/fgene.2023.1160915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Y., Li Y., Long S., Xu Y., Liu X., Sun Z., et al. Comprehensive analysis of the immunogenic cell death-related signature for predicting prognosis and immunotherapy efficiency in patients with lung adenocarcinoma. BMC Med. Genom. 2023;16(1):184. doi: 10.1186/s12920-023-01604-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X., Shang X., Li J., Zhang S. The prognosis and immune checkpoint blockade efficacy prediction of tumor-infiltrating immune cells in lung cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.707143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X.Z., Shang X.Q., Li J., Zhang S.J. The Prognosis and Immune Checkpoint Blockade Efficacy Prediction of Tumor-Infiltrating Immune Cells in Lung Cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.707143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S., Wu C., Ma D., Hu Q. Identification of a ferroptosis-related gene signature (FRGS) for predicting clinical outcome in lung adenocarcinoma. PeerJ. 2021;9 doi: 10.7717/peerj.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Y., Shetty A., Jiang F. Integrated analysis of miRNAs and DNA methylation identifies miR-132-3p as a tumor suppressor in lung adenocarcinoma. Thoracic cancer. 2020;11(8):2112–2124. doi: 10.1111/1759-7714.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B., Pan L., Chen M., Ma Y., Gao J., Tang D., et al. SIRP-alpha-IL-6 axis induces immunosuppressive macrophages in non-small-cell lung cancer. Biochem. Biophys. Res. Co. 2023;682:386–396. doi: 10.1016/j.bbrc.2023.10.035. [DOI] [PubMed] [Google Scholar]
- 44.Chen C.Y., Kao C.L., Liu C.M. The cancer prevention, anti-inflammatory and anti-oxidation of bioactive phytochemicals targeting the TLR4 signaling pathway. Int. J. Mol. Sci. 2018;19(9):2729. doi: 10.3390/ijms19092729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashani B., Zandi Z., Pourbagheri‐Sigaroodi A., Bashash D., Ghaffari S.H. The role of toll‐like receptor 4 (TLR4) in cancer progression: a possible therapeutic target? J. Cell. Physiol. 2021;236(6):4121–4137. doi: 10.1002/jcp.30166. [DOI] [PubMed] [Google Scholar]
- 46.Lin X., Tian C., Pan F., Wang R. A novel immune-associated prognostic signature based on the immune cell infiltration analysis for hepatocellular carcinoma. Oncologie. 2024;26(1):91–103. [Google Scholar]
- 47.Guo H., Hou Y., Wang C., Ding J. How to optimize the immune checkpoint blockade therapy for cancers? Oncologie. 2024 [Google Scholar]
- 48.Feng J., Zhang J., Chen Y. Prognostic value of a glycolysis and cholesterol synthesis related gene signature in osteosarcoma: implications for immune microenvironment and personalized treatment strategies. Oncologie. 2024;26(2):301–310. [Google Scholar]
- 49.Kohli K., Pillarisetty V.G., Kim T.S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2022;29(1):10–21. doi: 10.1038/s41417-021-00303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melssen M.M., Sheybani N.D., Leick K.M., Slingluff C.L. Barriers to immune cell infiltration in tumors. J. ImmunoTher. 2023;11(4) doi: 10.1136/jitc-2022-006401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moretta L. NK cell-mediated immune response against cancer. Surgical Oncology. 2007;16:3–5. doi: 10.1016/j.suronc.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 52.Song P., Li W., Guo L., Ying J., Gao S., He J. Identification and Validation of a Novel Signature Based on NK Cell Marker Genes to Predict Prognosis and Immunotherapy Response in Lung Adenocarcinoma by Integrated Analysis of Single-Cell and Bulk RNA-Sequencing. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.850745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cendrowicz E., Sas Z., Bremer E., Rygiel T.P. The role of macrophages in cancer development and therapy. Cancers. 2021;13(8):1946. doi: 10.3390/cancers13081946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L., Chen J., Cheng T., Yang H., Li H., Pan C. Identification of the key genes and characterizations of tumor immune microenvironment in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) J. Cancer. 2020;11(17):4965–4979. doi: 10.7150/jca.42531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang B.C., Gao J., Wang J., Rao Z.G., Wang B.C., Gao J.F. Tumor-associated macrophages infiltration is associated with peritumoral lymphangiogenesis and poor prognosis in lung adenocarcinoma. Med. Oncol. 2011;28(4):1447–1452. doi: 10.1007/s12032-010-9638-5. [DOI] [PubMed] [Google Scholar]
- 56.Hogan S.P., Rosenberg H.F., Moqbel R., Phipps S., Foster P.S., Lacy P., et al. Eosinophils: biological properties and role in health and disease. J Clinical Experimental Allergy. 2008;38(5):709–750. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- 57.Ghaffari S., Rezaei N. Eosinophils in the tumor microenvironment: implications for cancer immunotherapy. J. Transl. Med. 2023;21(1):551. doi: 10.1186/s12967-023-04418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaul M.E., Fridlender Z.G. Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 2019;16(10):601–620. doi: 10.1038/s41571-019-0222-4. [DOI] [PubMed] [Google Scholar]
- 59.Huang F., Xue F., Wang Q., Huang Y., Wan Z., Cao X., et al. Transcription factor-target gene regulatory network analysis in human lung adenocarcinoma. J. Thorac. Dis. 2023;15(12):6996–7012. doi: 10.21037/jtd-23-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng P., Xia Y., Shen X., Lu H., Chen Y., Xu C., et al. Combination of TrxR1 inhibitor and lenvatinib triggers ROS-dependent cell death in human lung cancer cells. Int. J. Biol. Sci. 2024;20(1):249–264. doi: 10.7150/ijbs.86160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaseghi G., Rashidi N., Zare N., Ghasemi F., Pourhadi M., Rafiee L., et al. Effects of methadone on the toll-like receptor 4 expression in human non-small cell lung carcinoma A549 cell line using in-silico and in vitro techniques. Adv. Biomed. Res. 2022;11:122. doi: 10.4103/abr.abr_97_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren B.X., Li Y., Li H.M., Lu T., Wu Z.Q., Fu R. The antibiotic drug trimethoprim suppresses tumour growth and metastasis via targeting Snail. Br. J. Pharmacol. 2022;179(11):2659–2677. doi: 10.1111/bph.15763. [DOI] [PubMed] [Google Scholar]
- 63.Vanneman M., Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer. 2012;12(4):237–251. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye F., Dewanjee S., Li Y., Jha N.K., Chen Z.-S., Kumar A., et al. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol. Cancer. 2023;22(1):105. doi: 10.1186/s12943-023-01805-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public dataset used in this study is available in GSE118370 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118370).