Abstract
This study aims to detect Escherichia coli which encodes beta-lactamase Cefotaxime (blaCTX-M) gene from the reproductive tract of Bali cattle with repeat breeder cases. This research was conducted from June to August 2021 using 16 Bali cattle with repeat breeder cases. The reproductive fluids were taken using a plastic sheet gun which was inserted into a Brain Heart Infusion medium, isolated in eosin-methylene blue agar (EMB) and identified using biochemical tests. Antibiotic susceptibility testing of E. coli was carried out using the disc diffusion method. Double-disk approximation test was used to screen the presence of E. coli which produces Extended-spectrum beta-lactamase. The polymerase chain reaction (PCR) method was used to detect the blaCTX-M gene of E. coli and sequences of the blaCTX-M gene were phylogenetically analyzed. The research results obtained three E. coli isolates from 16 reproductive tract fluids of Bali cattle. Antibiotic sensitivity tests showed that 100% of E. coli was resistant to penicillin G and oxytetracycline. 66.66% of E. coli was resistant to cefotaxime (CTX) and gentamicin, and 33.33% of E. coli was resistant to tetracycline. Escherichia coli isolates that were resistant to penicillin and CTX showed positive results in the double-disk approximation test. The results of E. coli detection using PCR showed that three E. coli isolates encoded the blaCTX-M gene located at 370 bp on gel electrophoresis. The results of the phylogenetic analysis showed that E. coli from the reproductive tract of Bali cattle was related to E. coli that encoded blaCTX-M-14 isolated from humans.
Key Words: Bali cattle, blaCTX-M, Escherichia coli, Repeat breeder
Introduction
West Nusa Tenggara province, which consists of the islands of Lombok and Sumbawa, is one of the provinces that is rich in people's livestock and supplies national beef cattle. Data from the West Nusa Tenggara province Central Statistics Agency until 2019 stated that the total cattle population in West Nusa Tenggara was 1, 234, 357 heads.1 West Nusa Tenggara province is a national meat supplier with a 1000-cow village program.2 Problems that are often found on people's farms in West Nusa Tenggara province, especially Lombok island, are cases of reproductive disorders that are treated using antibiotics. Previous research data on repeat breeders shows that the incidence rate of repeat breeder cases in Bali cattle in the Pringsewu Regency, Indonesia was 19. 85%.3
The incidence of cases of reproductive disorders such as the repeat breeder case cannot be separated from the use of antibiotics. Uncontrolled antibiotic treatment causes the bacteria in the reproductive tract to become resistant to the antibiotics used. The beta-lactam class of antibiotics is one of the antibiotics that is often used in animal husbandry and has given rise to resistance genes.4,5 The use of beta-lactam antibiotics has given rise to bacteria that can code for resistance genes that have been found in animals and humans. One of the resistance genes widely studied by world researchers is the cefotaxime-munich (CTX-M). Extended-Spectrum beta-Lactamase (ESBL) gene found in Escherichia coli produceESBL other than the Temoneira (TEM) gene.
Extended spectrum beta-lactamase is an enzyme produced by E. coli bacteria and has increased activity in hydrolyzing beta-lactam antibiotics, especially oxyimino-cephalosporins.6 Enterobacteriaceae including E. coli producing ESBLs type CTX-M have been found in people living in rural areas in Thailand with a prevalence of 65.70% from 417 fecal samples.7 Escherichia coli which can carry two ESBL genes with the CTX-M type has also been found in cattle on farms in Florida.8 The genes encoding CTX-M-1 and CTX-M-9 in E. coli have been successfully isolated in 8.60% of 220 cattle feces samples at slaughterhouses in Bogor.9 Escherichia coli encoding the CTX-M gene has also been found in cattle feces and the environment in Peninsular Malaysia.10 Meanwhile, on Lombok island, there were only reports of E. coli which was resistant to beta-lactam antibiotics isolated from the feces of Bali cattle with reproductive disorders. Previous research reported that E. coli could be isolated from Bali cattle that were repeat breeders.11 Previous research data showed that isolated E. coli which was resistant to beta-lactam antibiotics such as penicillin G, cefotaxime (CTX) and oxytetracycline from the feces of Bali cattle on community farms on Lombok island.12
Research data states that there are many cases of repeat breeders in Bali cattle on Lombok island, if they are kept by people's farms who are not aware of the dangers of using antibiotics. This have the potential for cases of antimicrobial resistance (AMR) to arise. One of the factors causing the increase in cases of AMR is the lack of awareness and understanding of the people of West Nusa Tenggara regarding the dangers of AMR. Surveillance on how AMR occurs on people's livestock in West Nusa Tenggara is urgently needed to raise awareness among farmers about the dangers of resistant microbes. The lack of surveillance of resistance development is one of the key causes of the increase in cases of AMR.13
Cases of repeat breeders in Bali cattle on Lombok island, if treated with poorly controlled antibiotics, are very likely to cause the emergence of E. coli encoding the beta-lactamase Cefotaxime (blaCTX-M) gene. Escherichia coli which codes for the blaCTX-M gene can be released from the reproductive tract fluids of Bali cattle so that it can spread to animals, humans and the environment which can potentially increase cases of antimicrobial resistance. Results of previous research stated that E. coli produceing ESBL had the potential to increase cases of AMR in animals and humans because the gene encoding AMR can be spread horizontally to other bacteria in their microenvironment.14 Therefore, research on molecular-based detection of the presence of E. coli which codes for the blaCTX-M gene in Bali cattle that are repeat breeders on Lombok island is very important to anticipate the spread of this bacteria.
This study aims to detect E. coli which encodes the blaCTX-M gene from the reproductive tract of Bali cattle with repeat breeder cases which had implications for increasing cases of AMR, considering that AMR caused deaths around 700,000 people, and by 2050 this number is expected to increase to around 10 million deaths each year.15
Materials and Methods
Study design. This research was a cross-sectional descriptive study carried out from June to August 2021 on people's farms located on Lombok island, West Nusa Tenggara Province with sample testing carried out at the Health Testing and Calibration Laboratory of West Nusa Tenggara province accredited by the National Accreditation Committee. This research used 16 Bali cattle whose reproductive fluid samples were taken from 30 animals suffering from repeat breeder cases. The criteria for Bali cattle suffering from breeder cases were female Bali cattle with normal heat cycles and periods that mated two or more times with fertile males or inseminated with the semen of fertile males but were not pregnant.16
Isolation and identification of E. Coli . The fluid collection was carried out by veterinarians from the Faculty of Veterinary Medicine, Universitas Pendidikan Mandalika who have professional qualifications. The reproductive fluids of Bali cattle with repeat breeder cases were collected using a sterile artificial insemination gun coated with a plastic sheet which was inserted into the reproductive tract of Bali cattle.17 The tip of the plastic sheet was cut 2.00 - 3.00 cm and immediately placed in the Brain Heart Infusion (Merck, Darmstadt, Germany) broth and immediately taken to the Health Testing and Calibration Laboratory of West Nusa Tenggara province and incubated for 24 hours at 37.00 ˚C. The samples were then planted in Eosin Methylene Blue (EMB; Merck) media and incubated for 24 hr at 37.00 ˚C to grow E. coli. The E. coli that grew were then identified by Gram staining and biochemical tests. The biochemical tests included sulfide indole motility, fermentation of lactose, glucose, fructose and mannitol performed using basic laboratory procedures in clinical bacteriology.18
Antibiotic sensitivity test. The identified E. coli was then tested for sensitivity to antibiotics to determine its resistance to antibiotics using disk diffusion (Oxoid, Basingstoke, UK) on Mueller Hinton Agar using the Kirby-Bauer method. The colonies of E. coli from EMB media were put into a tube containing 0.90% NaCl and homogenized until they reached the McFarland standard of 0.50. Escherichia coli suspension and the standard of 0.50. Then, was swabbed on Mueller Hinton Agar (Oxoid) planted with several antibiotics. Antibiotics planted on Mueller Hinton Agar (Oxoid) included penicillin G 10.00 U (Oxoid), oxytetracycline 30.00 µg (Oxoid), gentamicin 10.00 µg (Oxoid), tetra-cycline 30.00 µg (Oxoid) and CTX 30.00 µg (Oxoid), and then incubated for 24 hr at 37.00 ˚C. Sensitivity to antibiotics was carried out by measuring the diameter of the inhibition zone formed. Penicillin and CTX antibiotics were used in this study because these antibiotics are beta-lactam antibiotics. Cefotaxime is a third-generation cephalosporin antibiotic that is used as an indication of the presence of ESBL-producing bacteria. The antibiotic oxytetracycline was used in this research because it has been used in the treatment of large animals in either short-acting or long-acting form, while tetracycline was used in this research as a comparison to oxytetracycline because tetracycline is usually used in humans. Gentamicin was used in this study because gentamicin has been used for reproductive disorders such as metritis in combination with prostaglandin F2 alpha.19 The screening test for the presence of ESBL uses a double-disk approach test method. The antibiotics used in the double-disk approximation test method included CTX 30.00 µg and amoxicillin-clavulanate (AMC) 30.00 µg. This research used E. coli American Type Culture Collection (ATCC) 25,922 as a negative control. Interpretation of the results of the E. coli sensitivity test to antibiotics was carried out by measuring the diameter of the inhibition zone formed. The zone of inhibition formed was measured using a caliper. Assessment of the sensitivity of E. coli to antibiotics was categorized into sensitive, intermediate and resistant based on the standards of the Clinical and Laboratory Standards Institute.20
Detection of blaCTX-M gene. Molecular detection of the blaCTX-M gene in Escherichia coli was preceded by extraction of E. coli DNA using the QIAprep® Spin Miniprep Kit procedure (Qiagen, Hilden, Germany). The purity and DNA concentration was estimated using a spectrophotometer. Molecular detection of the blaCTX-M gene from E. coli was carried out using a polymerase chain reaction (PCR) assay by a specific primer. The specific primer was designed based on NCBI reference sequence: KX669628.1. The reagents for PCR were done with a total volume of 51.00 μL consisting of 25.00 μL PCR mix Dream Taq HS (Thermo Fisher Scientific, Waltham, USA), 1.50 μL primer, 20.00 μL ddH2O, and 3.00 μL extracted DNA of E. coli as a template. The mixture was processed in the Biorad i-cycler PCR machine (BioRad, Hercules, USA). The specific primers were used in this study for blaCTX-M genes forward (blaCTX-MF) 5' CTTTATGCGCAGACGAGTGC -3' and blaCTX-M genes reverse (blaCTX-MR) 5' CGTATTG CCTTTGAGCCACG -3' with an amplicon of 579 bp.21 The PCR conditions used were as follows: Predenaturation temperature at 95.00 ˚C for 3 min, denaturation at 94.00 ˚C for 30 sec, annealing at 57.00 ˚Cfor 30 sec, and elongation at 72.00 ˚C for 1 min and post- elongation at 72.00 ˚C for 5 min with 35 cycles. The PCR amplicon from the blaCTX-M gene was electrophoresed on 2.00% agarose gel. The results of electrophoresis were read using the BioRad Gel Imager. This research used E. coli isolate type 25,922 of ATCC as a negative control. Sequencing of PCR results was carried out at Genetica Science Indonesia. The data obtained from the research results were analyzed descriptively in pictures including the results of electrophoresis of the gene encoding blaCTX-M from E.coli isolates. The phylogenetic tree analysis was carried out to analyze the relationship of the gene encoding blaCTX-M of E. coli isolates with various E. coli encoding blaCTX-M in GenBank® using MEGA Software (version 10.0; Biodesign Institute, Tempe, USA).
Results
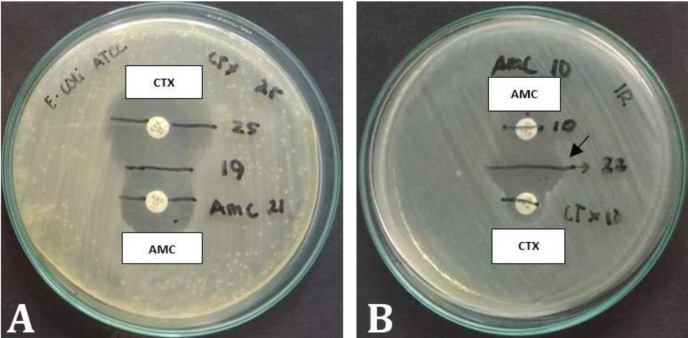
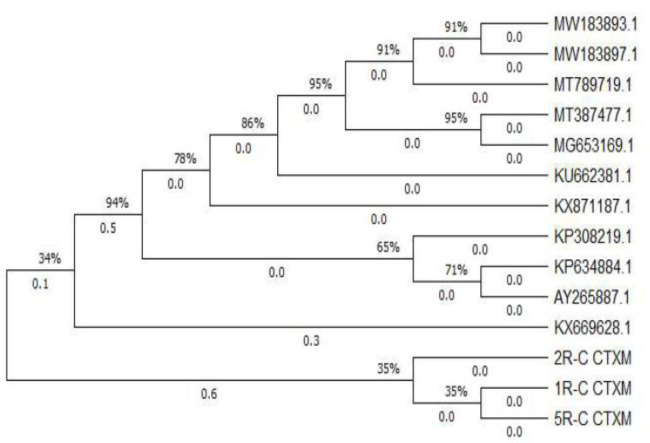
The results of isolation of E. coli on EMB Agar ) medium obtained three (18.75%) of 16 reproductive tract fluid samples of Bali cattle. The results of the research on the sensitivity test to antibiotics on three isolated E. coli showed that 100% of E. coli was resistant to penicillin G and oxytetracycline. 66.66 % of E. coli was resistant to CTX and gentamicin, and 33.33 % of E. coli was resistant to tetracycline. This research also found that 66.66% of E. coli were resistant to penicillin and CTX (Table 1). The results of the screening test for the presence of ESBL in E. coli which was resistant to penicillin and CTX using the double-disk approach test method showed positive results with the presence of an expansion of the CTX disc diameter zone around the AMC disc (Fig. 1). The results of 2.00% agarose electrophoresis from the CTX-M gene PCR product of E. coli showed that for the blaCTX-M gene, three samples were positive, namely samples No. 1, 2, and 3 at position 370 bp in the agarose electrophoresis band for each sample (Fig. 2). The phylogenetic results of the three E. coli isolates that were positive for the blaCTX-M gene in this study when compared to the ESBL-producing E. coli in NCBI using the Neighbor-Joining method showed that the E. coli blaCTX-M gene from the Bali cattle reproductive tract was located in one group (Fig. 3). Figure 3 shows that E. coli CTX-M genes from the reproductive tract of Bali cattle (CTX-M_R1, R2, and R5) are located in one group. The E. coli group from the reproductive tract of Bali cattle (CTX-M_R1, R2, and R5) was related to the E. coli producing ESBL gene blaCTX-M which is used as a reference from GenBank® data with code KX669628.1. The code KX669628.1 is E. coli which encodes blaCTX-M-14. genes originating from humans.
Table 1.
The result of the antibiotics sensitivity test of isolated Escherichia coli, based on inhibition zone diameter (mm).
| No. | Cefotaxime | Penicillin | Oxytetracycline | Gentamycin | Tetracycline |
|---|---|---|---|---|---|
| 1 | 12.00 R | 0.00 R | 0.00 R | 0.00 R | 12.00 I |
| 2 | 40.00 S | 0.00 R | 0.00 R | 18.00 S | 16.00 S |
| 3 | 10.00 R | 0.00 R | 0.00 R | 0.00 R | 0.00 R |
S: Susceptible, I: intermediate, and R: resistant.
Fig. 1.
Double-disk approximation synergy test on Escherichia coli. A) E. coli, American type culture collection (ATCC®) 25,922, and B) E. coli sample. AMC: Amoxicillin-clavulanate and CTX: cefotaxime. Arrow shows the synergy.
Fig. 2.
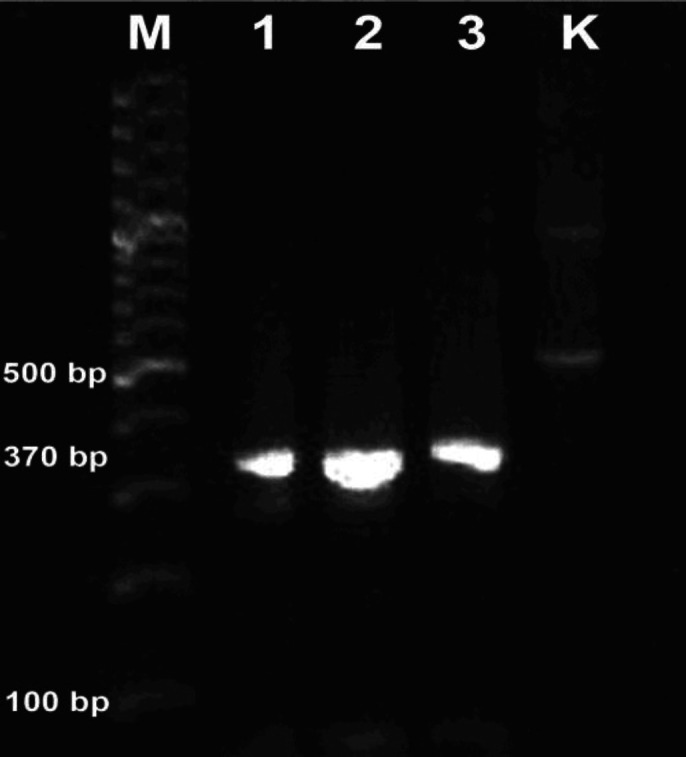
The 2.00% of agarose gel electrophoresis shows three samples in No. (1-3) of E. coli encode beta-lactamase Cefotaxime gene. Lane M is the DNA marker, the sample is in lanes 1-3, and lane K is E. coli, American type culture collection (ATCC®) 25,922.
Fig. 3.
Phylogenetic analysis of the beta-lactamase Cefotaxime gene on E. coli samples with data from GenBank®. 1R-C CTXM , 2R-C CTXM, 5R-C CTXM: Sample of E. coli; (KX669628.1, KU662381.1, MT789719.1, MT387477.1, MG653169.1, KX871187.1, AY265887.1, KP634884.1, KP308219.1, MW183893.1 dan MW183897.1: Data reference in GenBank®.
Discussion
The results of the isolation of E. coli on EMB Agar medium obtained 18.75% of 16 reproductive tract fluid samples of Bali cattle. This research was consistent with research conducted by Aminuddi et al. which found E. coli in the reproductive tract of Bali cattle that were repeat breeders. The incidence of E. coli in the reproductive tract of Bali cattle that experienced repeat breeders was almost the same as research which stated that E. coli was detected in 25.00% of Bali cattle that experienced repeat breeders.22
The results of the present research showed that E. coli isolated from the reproductive tract of Bali cattle showed resistance to penicillin G, oxytetracycline, CTX, gentamicin and tetracycline, indicating that various antibiotics were given to Bali cattle in repeat breeders whose management had received little attention. Poorly controlled administration of antibiotics will cause E. coli bacteria to adapt and mutate, including the gene encoding ESBL found on chromosomes and plasmids. The results of the present study were in agreement with the statement that the AMR profile of E. coli almost reflected the use of antimicrobials in animals for food production.23
The results of the screening test in the present study for the phenotypic test using double-disk diffusion showed that the AMC disk diffused into the agar and inhibited beta-lactamase around the CTX disk. Previous research data stated that the expansion of the ceftazidime disk zone around the AMC disk was interpreted as a positive result.24 The results of the double-disk diffusion test with a 5.00 mm increase in zone diameter for the antimicrobial agent tested with clavulanic acid versus the zone when tested alone can be suspected that E. coli produces ESBL. Escherichia coli which was isolated from the reproductive tract of Bali cattle in the present study produced ESBLs. This event was caused by the adaptation of E. coli to the administration of beta-lactam antibiotics used in treating reproductive disorders. One of the bacterial efforts to avoid the pressure of beta-lactam antibiotics is by producing enzymes that can hydrolyze beta-lactam anti-biotics known as ESBL. These results were consistent with a previous research that found that beta-lactam antibiotics were one of the antibiotics most frequently used in animal husbandry and gave rise to resistance genes.4
The PCR results of the CTX-M gene in E. coli isolated from the reproductive tract of Bali cattle in this study was located at position 370 bp in the agarose band which was lower when compared to several studies. The results of previous research found that blaCTX-M isolated from E. coli from the urine of dairy cows with uterine infections in India was at 580 bp.25 The results of other research also found that the blaCTX-M gene of E. coli in cattle was located at around 400 bp.10 These results showed that the molecular size of the blaCTX-M gene of E. coli samples was lower because there were variations in the position of the blaCTX-M gene of E. coli on the agarose gel electrophoresis in several research results because there were several factors that influenced the electrophoresis results. These factors included the size of the DNA molecule, concentration of the agarose gel, DNA conformation, voltage, presence of DNA dye and composition of the electrophoresis buffer.
In the phylogenetic analysis of the blaCTX-M gene of E. coli isolates from the reproductive tract of Bali cattle that experienced repeat breeding, there was a relationship with the code KX699628.1, which was E. coli strain EC271 ESBL blaCTX-M-14 isolated from humans. Escherichia coli encoding the blaCTX-M-14 gene in the reproductive tract of Bali cattle could come from humans and the environment.
Escherichia coli contamination in the reproductive tract of Bali cattle could occur through livestock keepers or staff who did not pay attention to hygiene in treating reproductive tract disorders such as postpartum treatment, artificial insemination and other cases of reproductive disorders. Previous research results stated that the predisposing factors causing repeat breeders of cattle were the result of bacterial infections when treating cases of dystocia, retained placenta, uterine prolapse, and ovarian disorders.26 Infection of the uterus can be caused by environmental conditions during childbirth, however, placental retention is the most important predisposition.27 Previous research data stated that E. coli which coded for the beta-lactamase gene could come from human feces and urine because of the discovery of E. coli which coded for the beta-lactamase Temoneira (blaTEM) gene from the reproductive tract related to E. coli strain U-10 TEM.28
This research detected E. coli blaCTX-M from the reproductive tract of Bali cattle with repeat breeder cases on Lombok island that was related to the E. coli strain EC271 blaCTX-M-14 isolated from humans. The data from this research could be used as a basis for study and prevention with a one-health approach in the spread of E. coli encoding blaCTX-M which has the potential to increase the incidence of AMR.
Acknowledgments
The authors are grateful to the Faculty of Veterinary Medicine, Universitas Pendidikan Mandalika, the Institute of Biosciences, and the Faculty of Veterinary Medicine, Universitas Brawijaya, and the Health Laboratory of Testing and Calibration of West Nusa Tenggara province for collaborating in sampling and testing that permitted and facilitated this research.
Conflict of interest
No conflict of interest in this research
References
- 1.BPS-Statistics of Nusa Tenggara Barat Province. Nusa TenggaraBarat provine. CV: Maharani. 2020:241–242. [Google Scholar]
- 2.Mashur M, Bilad MR, Kholik K, et al. The sustainability and development strategy of a cattle feed bank: a case study. Sustainability. 2022;14(13):7989. [Google Scholar]
- 3.Juliana A, Hartono M, Suharyati S. Repeat breeder of Bali cattles in Pringsewu regency [Indonesian] J Ilmiah Peternakan Terpadu . 2015;3(2):42–47. [Google Scholar]
- 4.Huang L, Xu Y, Xu J, et al. Dissemination of antibiotic resistance genes (ARGs) by rainfall on a cyclic economic breeding livestock farm. Int Biodeterior Biodegradation. 2019;138:114–121. [Google Scholar]
- 5.Qian X, Gu J, Sun W, et al. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J Hazard Mater. 2018;344:716–722. doi: 10.1016/j.jhazmat.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Bradford PA. Extended-spectrum beta-lactamase in the 21st century: characterization epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luvsansharav UO, Hirai I, Nakata A, et al. Prevalence of and risk factors associated with faecal carriage of CTX-M β-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother. 2012;67(7):1769–1774. doi: 10.1093/jac/dks118. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Teng L, DiLorenzo N, et al. Prevalence and molecular characteristics of extended-spectrum and AmpC β-Lactamase producing Escherichia coli in grazing beef cattle. Front Microbiol. 2019;10:3076. doi: 10.3389/fmicb.2019.03076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudarwanto MB, Lukman DW, Latif H, et al. CTX-M producing Escherichia coli isolated from cattle feces in Bogor slaughterhouse, Indonesia. Asian Pac J Trop Biomed. 2016;6(7):605–608. [Google Scholar]
- 10.Kamaruzzaman EA, Abdul Aziz S, Bitrus AA, et al. Occurrence and characteristics of extended-spectrum β-lactamase-producing Escherichia coli from dairy cattle, milk, and farm environments in Peninsular Malaysia. Pathogens. 2020;9(12):1007. doi: 10.3390/pathogens9121007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aminuddi SP, Alpian SB, Dita P, et al. Identification of gram-negative bacteria of Bali cattle with repeat breeding cases on East Lombok, West Nusa Tenggara Province. J Phys Conf Ser. 2020;1430(1):012013 . [Google Scholar]
- 12.Kholik K, Munawaroh M, Saputra MR, et al. Antibiotic resistance in Escherichia coli isolated from feces of Bali cattle with reproductive disorders. Jurnal Biodjati. 2021;6(2):303–311. [Google Scholar]
- 13.Chokshi A, Sifri Z, Cennimo D, et al. Global contributors to antibiotic resistance. J Glob Infect Dis. 2019;11(1):36–42. doi: 10.4103/jgid.jgid_110_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Roux F, Blokesch M. Eco-evolutionary dynamics linked to horizontal gene transfer in vibrios. Ann Rev Microbiol. 2018;(72):89–110. doi: 10.1146/annurev-micro-090817-062148. [DOI] [PubMed] [Google Scholar]
- 15.O'Neill J. Tackling drug-resistant infections globally: Final report and recommendations 2016. [Accessed June 19, 2024]. Available at: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf.
- 16.Wodaje HB, Mekuria TA. Risk factors of repeat breeding in dairy cattle. Adv Biol Res. 2016;10(4):213–221. [Google Scholar]
- 17.Andriani AI, Madyawati SP, Sabdoningrum EK. Non-specific bacterial profiles in reproductive tract of dairy cattle during artificial insemination. Worlds Vet J. 2021;11(1):110–115. [Google Scholar]
- 18.Vandepitte J, Verhaegen J, Engbaek K, et al. Basic laboratory procedures in clinical bacteriology. 2nd ed. Geneva, Switzerland: World Health Organization; 2003. pp. 45–49. [Google Scholar]
- 19.Melia J, Amrozi A, TumbelakaLigaya LI. Dynamics of the ovaries of endometritis cows treated with gentamicine, flumequine and prostaglandin F2 alpha (PGF2α) analogs intrauterinely [Indonesian] Indonesian J Vet Sci. 2014;8(2):111–115. [Google Scholar]
- 20.CLSI. Performance standards for antimicrobial disk susceptibility tests; approved standard. 11th ed. USA: Clinical and Laboratory Standards Institute: 2012. [Google Scholar]
- 21.Guggiana-Nilo P, Lima CA, Cid-Maldonado N. Escherichia coli strain EC271 extended spectrum beta-lactamase CTX-M-14 (blaCTX-M) gene, blaCTX-M-14 allele. 2016. Available at: https://www.ncbi.nlm.nih.gov/nuccore/KX669628.1/ [Google Scholar]
- 22.Kholik K. Detection of antibiotic resistant in Escherichia coli from the reproductive tract of Bali cattle on smallholder farm. J Biosains Pascasarjana. 2022;24SP(1):44–53. [Google Scholar]
- 23.EFSA panel on biological hazards (BIOHAZ) Scientific opinion on the public health risks of bacterial strains producing extended‐spectrum β‐lactamases and/or AmpC β‐lactamases in food and food‐producing animals. EFSA J. 2011;9(8):2322. [Google Scholar]
- 24.Igwe J, Onaolapo JA, Kachallah M, et al. Molecular characterization of extended spectrum β-lactamase genes in clinical E coli Isolates. J Biomed Sci Eng. 2014;7:276–285. [Google Scholar]
- 25.Agrawal S, Singh AP, Singh R, et al. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolated from postpartum uterine infection in dairy cattle in India. Vet World. 2021;14(1):200–209. doi: 10.14202/vetworld.2021.200-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rafika I, Thasmi CN, Herrialfian H, et al. Isolation and identification of Gram-negative bacteria in uterine Aceh cow with repeat breeding [Indonesian] J Agripet. 2020;20(2):187–192. [Google Scholar]
- 27.Sheldon IM, Williams EJ, Miller AN, et al. Uterine diseases in cattle after parturition. Vet J. 2008;176(1):115–121. doi: 10.1016/j.tvjl.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kholik K, Srianto P, Aulanniam A, et al. Characterization and phylogenetics of beta-lactamase Temoneira gene in Escherichia coli of the Bali cattle on Lombok island, Indonesia. Iraqi J Vet Sci. 2023;37(2):487–493. [Google Scholar]