Abstract
RNA exon editing is a therapeutic strategy for correcting disease-causing mutations by inducing trans-splicing between a synthetic RNA molecule and an endogenous pre-mRNA target, resulting in functionally restored mRNA and protein. This approach enables the replacement of exons at the kilobase scale, addresses multiple mutations with a single therapy, and maintains native gene expression without changes to DNA. For genes larger than 5 kb, RNA exon editors can be delivered in a single vector despite AAV capacity limitations because only mutated exons need to be replaced. While correcting mutations by trans-splicing has been previously demonstrated, prior attempts were hampered by low efficiency or lack of translation in preclinical models. Advances in synthetic biology, next-generation sequencing, and bioinformatics, with a deeper understanding of mechanisms controlling RNA splicing, have triggered a re-emergence of trans-splicing and the development of new RNA exon editing molecules for treating human disease, including the first application in a clinical trial (this study was registered at ClinicalTrials.gov [NCT06467344]). Here, we provide an overview of RNA splicing, the history of trans-splicing, previously reported therapeutic applications, and how modern advances are enabling the discovery of RNA exon editing molecules for genetic targets unable to be addressed by conventional gene therapy and gene editing approaches.
Keywords: MT: RNA and epigenetic editing Special Issue, RNA exon editing, trans-splicing, spliceosome, gene therapy, rare diseases
Graphical abstract
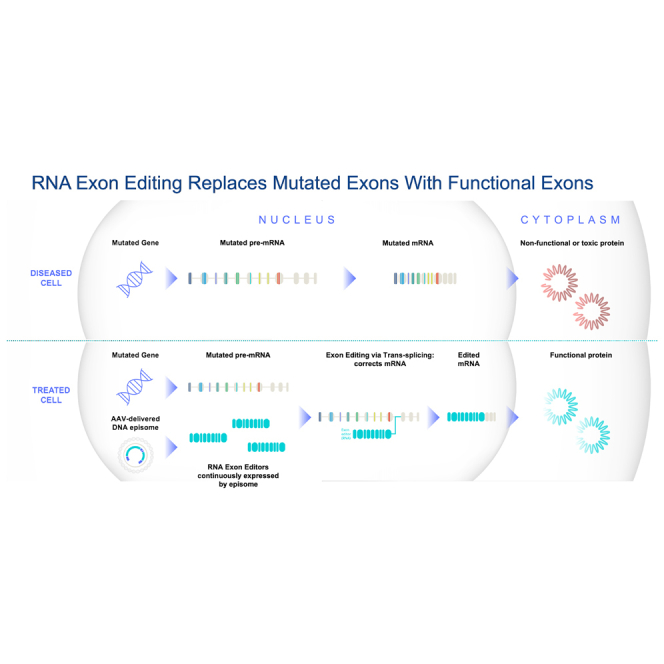
RNA exon editing is a promising therapeutic approach that corrects genetic mutations by replacing faulty RNA segments with functional ones, overcoming limitations of conventional gene replacement and other gene editing strategies.
Introduction to RNA splicing in cis and trans
RNA splicing is a ubiquitous process in the post-transcriptional modification of pre-mRNA in eukaryotic cells that enables the removal of introns, joining of exons, and formation of mature mRNA. The major spliceosome complex that controls most RNA splicing is a dynamic ribonucleoprotein (RNP) complex comprising five small nuclear RNAs (U1, U2, U4, U5, and U6 snRNAs) and numerous associated proteins, collectively forming small nuclear RNPs (snRNPs) that assemble in a highly coordinated process onto pre-mRNA.1 In brief, following transcription, assembly of the spliceosome begins with the U1 snRNP binding to the 5′ splice site of the intron (GU sequence) and the U2 snRNP binding to the branchpoint sequence (BPS) (usually an adenine nucleotide located within the intron) located upstream of the 3′ splice site (AG sequence). Next, a preassembled U4/U6-U5 tri-snRNP complex joins the spliceosome, creating what is referred to as the B complex. Several RNA-RNA and protein rearrangements occur, displacing U1 and U4 snRNP, forming the catalytically active C complex. The catalytic processing first involves a transesterification reaction, where the 2′-OH of the branch site nucleotide attacks the 5′ splice site, forming a lariat structure. The second transesterification reaction involves the 3′-OH of the upstream exon attacking the 3′ splice site, thereby joining the exons and releasing the intron lariat. After this splicing event, the spliceosome disassembles, and the RNA and protein components can be recycled and used for additional splicing reactions. While this is a basic summary of RNA splicing, several review articles provide more detail into structural and molecular mechanisms of the spliceosome complex and role in gene regulation and proteomic diversity.1,2,3
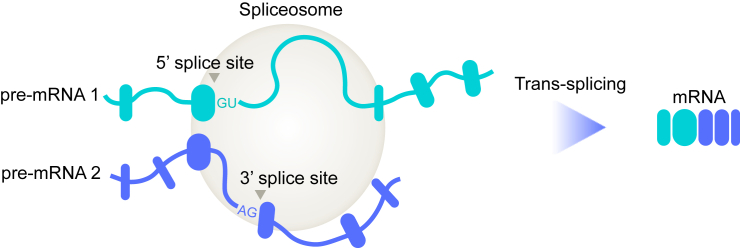
Splicing in trans is another naturally occurring process where exons from two separate pre-mRNA molecules are joined together by the spliceosome complex, resulting in a chimeric mRNA (Figure 1). The first report of RNA trans-splicing occurring in mammalian cells came in 1995 based on experimental observation of a novel small T antigen transcript.4 Since then, multiple examples of natural trans-splicing in several vertebrates have been observed.5 Moreover, synthetic trans-splicing constructs (which we call RNA exon editors) can be designed to potentially treat human diseases by editing pre-mRNA.6,7,8 RNA exon editing corrects genetic diseases by replacing exons containing the relevant variants with functional ones from an introduced synthetic RNA exon editor. In addition, RNA exon editing can be used to introduce toxic genes selectively into cancers cells, leading to targeted cell death. Additional applications of RNA exon editing are described further below.
Figure 1.
Spliceosome-mediated RNA trans-splicing
Simple illustration depicting trans-splicing between two pre-mRNA transcripts mediated by the spliceosome.
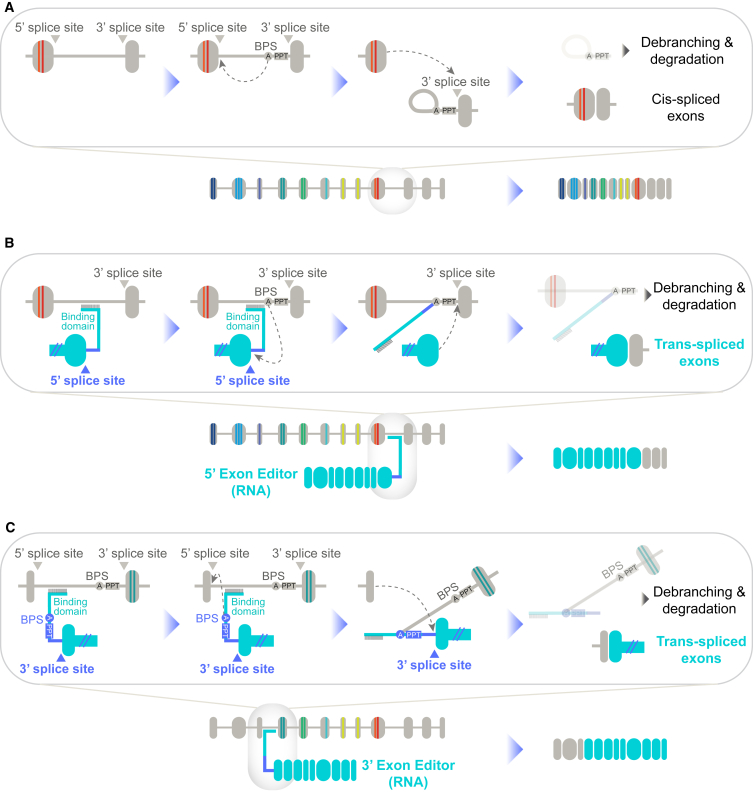
The process of RNA exon editing by trans-splicing follows a mechanism similar to cis-splicing (Figure 2), allowing for molecular insights related to cis-splicing to be applied to the design of RNA exon editors. In cis-splicing, the step I catalytic reaction mediated by the spliceosomal C complex occurs by the 2′-OH group of a bulged nucleotide at the BPS initiating a nucleophilic attack on the 5′ phosphate of the 5′ splice site, leading to the formation of the free 5′ exon and the lariat intermediate. This is followed by the catalytically active step II spliceosomal C∗ complex, where exon ligation occurs via the nucleophilic attack by the 3′-OH group of the free 5′ exon onto the 5′ phosphate of the 3′ exon. This reaction generates a ligated exon product and the intron lariat that is subject to debranching and degradation. The 5′ trans-splicing reaction involves a 5′ exon editor that contains a 5′ mRNA replacement coding sequence (CDS) and a binding domain that base pairs with an endogenous intron of the target pre-mRNA. In the catalytically active step I spliceosomal C complex, the 2′-OH group of the branchpoint nucleotide in the pre-mRNA attacks the 5′ phosphate of the 5′ splice site in the exon editor, leading to the formation of the free 5′ replacement CDS and the Y-branch intermediate. This is followed by the catalytically active step II spliceosomal C∗ complex, where exon ligation occurs via the nucleophilic attack by the 3′-OH group of the free 5′ replacement CDS onto the 5′ phosphate of the 3′ exon. This reaction generates a ligated exon product that is a fusion of the exon editor CDS with the native exon. The released Y-branch intermediate is subject to debranching and degradation. For 3′ trans-splicing, a 3′ exon editor that contains a binding domain that base pairs with an endogenous intron of the target pre-mRNA and a 3′ replacement CDS is introduced. In the catalytically active step I spliceosomal C complex, the 2′-OH group of the branchpoint adenosine in the exon editor attacks the 5′ phosphate of the 5′ splice site in the upstream native exon, leading to the formation of the free 5′ exon and the Y-branch intermediate. In the catalytically active step II spliceosomal C∗ complex, exon ligation occurs via the nucleophilic attack by the 3′-OH group of the free 5′ exon onto the 5′ phosphate of the 3′ replacement CDS. This reaction generates a ligated exon product that is a fusion of the native exon with the exon editor CDS. The released Y-branch intermediate is subject to debranching and degradation.
Figure 2.
Mechanism of cis- and trans-splicing
(A) RNA cis-splicing involves the splicing and removal of introns to generate a mRNA transcript, involving two steps of transesterification reactions to form a lariat intermediate and the ligated exon product. The exons are indicated as gray ovals, and the intron is depicted as a gray line. (B) 5′ trans-splicing involves a 5′ exon editor that contains 5′ replacement CDS (teal ovals) and a binding domain that base-pairs with the endogenous intron of the target pre-mRNA (gray). Transesterification reaction between the branchpoint nucleotide in the pre-mRNA and the 5′ splice site in the exon editor, followed by a second transesterification reaction generates the Y-branch intermediate and a ligated exon product that is a fusion of the exon editor CDS with native exon. (C) 3′ trans-splicing involves a 3′ exon editor that contains a binding domain that base-pairs with the endogenous intron of the target pre-mRNA and 3′ replacement CDS (teal ovals). Transesterification between the branchpoint nucleotide in the exon editor and the 5′ splice site in the upstream native exon, followed by a second transesterification reaction generates the Y-branch intermediate and a ligated exon product that is a fusion of the native exon with the exon editor CDS.
Before reviewing potential therapeutic applications of RNA exon editing explored to date, we provide a brief overview of the discovery of RNA trans-splicing in nature.
Discovery of RNA trans-splicing
The discovery of RNA trans-splicing was a groundbreaking event in the field of molecular biology. The first observations occurred in the early 1980s, when researchers studying a group of parasitic protozoa called trypanosomes noticed the presence of a common 5′ spliced leader (SL) sequence present in the mRNA that was not present in the genomic sequence of the same gene.9,10,11 Further analysis revealed that this 1.3-kb “mini-exon” sequence is tandemly repeated about 200 times throughout the trypanosome genome. This discovery was significant because it showed that exons from different pre-mRNA molecules could be joined together, unlike the previously known cis-splicing, where only exons from the same pre-mRNA were spliced together. While SL trans-splicing is absent in vertebrates,12 SL RNAs can be accurately trans-spliced in mammalian cells in vivo and in vitro.13
Around the same time as the discovery of SL trans-splicing, Sharp and colleagues provided some of the first in vitro experimental evidence elucidating the basic molecular and chemical processes governing RNA trans-splicing.14 Radiolabeled RNA substrates derived from adenovirus 2 were prepared by in vitro transcription, with constructs containing additional short 38-nt complementary sequences at the 3′ and 5′ ends. The RNA substrates were then annealed, and the association of the two RNAs was assayed by electrophoresis on non-denaturing gels before and after incubation in nuclear extracts. The authors concluded that the two substrate RNAs in the trans-splicing reaction are associated by base pairing of complementary sequences and are not joined by ligation prior to splicing. During these studies, it was also observed that trans-splicing of two RNA molecules with no significant complementarity can occur (albeit at a much lower efficiency), and it was suggested that secondary structures, not sequence complementary within intervening sequences, may affect splicing reactions.
In the years to come, RNA trans-splicing was observed in chloroplasts15 and nematodes.16 Vandenberghe et al. first reported the discovery of SL trans-splicing in chordates in the early 2000s in the ascidian Ciona intestinalis.17 As ascidians are the closest relatives to vertebrates, this observation supported an ancestral origin of trans-splicing and hinted that different types of RNA trans-splicing may naturally occur in high-order species.5 Moreover, computational analysis by Dandekar and Sibbald in 1990 predicted trans-splicing to be a potentially widespread phenomenon, including in vertebrates.18 Indeed, work from Sullivan and colleagues identified that alternative processing of androgen-binding protein RNA transcripts in fetal rat liver was the result of trans-splicing.19 A cDNA transcript containing the ABP-SHBG gene (exons 1–5) and the histidine decarboxylase gene, localized to rat chromosomes 10 and 3, respectively, were joined at the splice junctions from the separate transcripts. A few years later Eul et al. identified the occurrence of trans-splicing between two SV40 transcripts, utilizing a cryptic 5′ splice donor site in the second exon and an upstream small T antigen 3′ splice acceptor site, to generate the T1 mRNA molecule.4 Currently, several studies and high-throughput RNA analyses continue to expand the number of known trans-splicing event in vertebrates,20,21 and the field continues to investigate these events with respect to underlying mechanisms.
Although trans-splicing has been established in mammals, the underlying mechanisms that induce trans-splicing remain to be elucidated. A seminal finding related to chromosomal translocation in cancer identified rearrangements that led to premature transcriptional termination, resulting in early terminated RNA molecules with an unsaturated splice donor site that can trans-splice and give rise to chimeric fusion RNAs. For example, using nested PCR experiments in combination with cloning and sequencing, Kowarz et al. were able to demonstrate a predominant intergenic trans-splicing product of transcripts deriving from an early terminated MLL gene with the AF4 gene in healthy individuals (in-frame fusion of MLL exon 9 with the AF4 exon 4).22 Whether chimeric trans-spliced RNAs serve a functional purpose or are part of the random noise of molecular machinery found in nature is yet to be determined.
Therapeutic applications of RNA exon editing
Recently several RNA editing technologies such as adenosine deaminase acting on RNA(ADAR)-based RNA editing, ribozyme-mediated trans-splicing, or Cas-mediated mRNA trans-splicing have progressed toward the clinic, but here, we describe RNA trans-splicing and its therapeutic applications, focusing specifically on spliceosome-mediated trans-splicing (Table 1). As illustrated in Figure 2, RNA exon editing allows for the replacement of large stretches of exons. RNA exon editors can be designed to rewrite multiple contiguous exons, enabling the treatment of diseases that have high mutational variance in the target gene with a single therapeutic construct. In contrast, using a base editing approach to treat such diseases would require developing a different therapeutic for each mutation. In addition, the technology allows for the control of precise endogenous gene expression levels by interacting specifically with pre-mRNA, an advantage for the treatment of diseases that involve narrow gene dosage indices. Stated differently, the expression levels of the pre-mRNA target serve as a maximum setpoint for the expression of any trans-spliced mRNA product generated, thereby limiting any complications due to overexpression. When expressed via an adeno-associated virus (AAV) vector, RNA exon editors provide the durability of a one-time gene therapy treatment, without the use of foreign enzymes, avoiding the risks associated with direct DNA editing or gene replacement therapies. Moreover, RNA exon editors can be designed for expression by a single AAV, providing an important advantage over many DNA editing technologies, which require dual vectors for in vivo delivery.
Table 1.
Summary of published therapeutic applications of RNA trans-splicing
| Target | Disease | Year | 5′ or 3′ design | Delivery method | In vitro/in vivo | Reference |
|---|---|---|---|---|---|---|
| AFP | HCC or HPV-16 | 2018 | 5′, 3′ | transfection | in vitro | Poddar et al.23 |
| Apolipoprotein A1 | hemophilia A | 2009 | 3′ | injected minicircle | in vivo | Wang et al.24 |
| β-globin | sickle cell disease, β-thalassemia | 2017 | 5′ | transfection, lentivirus | in vitro | Uchida et al.25; Kierlin-Duncan et al.26 |
| 2007 | 5′ | transfection | in vitro | |||
| β hCG6 | cancer | 1999 | 3′ | transfection, electroporation | in vitro, in vivo | Puttaraju et al.8 |
| CD22 | pediatric B lineage ALL | 2015 | 3′ | transfection of polypeptide nanoparticles | in vitro, in vivo | Uckun et al.27,28 |
| 2015 | 3′ | transfection | in vitro | |||
| CD40L | X-linked immunodeficiency with HIGM1 | 2004 | 3′ | transfection, lentivirus | in vitro, in vivo | Tahara et al.29 |
| CEP290 | LCA10 | 2018 | 5′ | AAV | in vitro, in vivo | Dooley et al.30 |
| COL17A1 | EB | 2011 | IER | transfection | in vitro | Koller et al.31; Dallinger et al.32 |
| 2003 | 3′ | transfection | in vitro | |||
| COL7A1 | EB | 2023 | 3′ | minicircle in liposome | in vitro | Koller et al.33; Liemberger et al.34; Mayr et al.35; Peking et al.36,37; Huttner et al.38; Tockner et al.39; Murauer et al.40,41 |
| 2022 | 5′ | retrovirus | in vitro | |||
| 2017 | 3′ | lentivirus | in vitro, ex vivo | |||
| 2016 | IER | retrovirus | in vitro | |||
| 2016 | 3′ | retrovirus | in vitro | |||
| 2016 | 5′ | transfection, minicircle gene gun | in vitro, in vivo | |||
| 2015 | 3′ | transfection | In vitro | |||
| 2013 | 3′ | retrovirus | In vitro | |||
| 2011 | 3′ | retrovirus | In vitro | |||
| Ct-OATP1B3 | cancer | 2018 | 3′ | transfection, retrovirus | in vitro, in vivo | Sun et al.42 |
| CFTR | cystic fibrosis | 2009 | segmental | transfection, AAV | in vitro | Liu et al.43; Mansfield et al.44,45; Song et al.46; Liu et al.47 |
| 2005 | 3′ | AAV | in vitro | |||
| 2003 | 5′ | transfection | in vitro | |||
| 2002 | 3′ | recombinant adenovirus | in vitro, in vivo | |||
| 2000 | 3′ | transfection | in vitro | |||
| DNA-PKcs | SCID | 2007 | 3′ | nucleofection | in vitro | Zayed et al.48 |
| DMPK | DM1 | 2009 | 3′ | transfection | in vitro | Chen et al.49 |
| DNM2 | Autosomal dominant centronuclear myopathy | 2016 | 5′, 3′ | transfection, AAV | in vitro, in vivo | Trochet et al.50 |
| DYSF | Dysferlinopathies | 2015 | 3′ | lentivirus, AAV | in vitro, in vivo | Philippi et al.51; Monjaret et al.52 |
| 2014 | 3′ | transfection, AAV | in vitro, in vivo | |||
| Dystrophin | DMD | 2013 | 3′ | transfection, lentivirus, AAV | in vitro, in vivo | Lorain et al.53,54 |
| 2010 | IER | transfection | in vitro | |||
| FVIII | hemophilia A | 2003 | 3′ | transfection, AAV | in vitro, in vivo | Chao et al.55 |
| GNE | GNE myopathy | 2014 | 3′ | transfection, AAV | in vitro | Tal-Goldberg et al.56 |
| HIV | HIV | 2017 | 5′, 3′ | transfection | in vitro | Ingemarsdotter et al.57 |
| HTT | HD | 2017 | 5′ | transfection, lentivirus | in vitro | Rindt et al.58,59 |
| 2012 | 5′ | transfection, lentivirus | in vitro | |||
| IgG | 2015 | segmental | transfection | in vitro | Shang et al.60 | |
| KRT14 | EB simplex | 2019 | 5′ | retrovirus | ex vivo | Liemberger et al.34; Peking et al.61; Wally et al.62 |
| 2018 | 5′ | transfection | in vitro | |||
| 2010 | 5′ | transfection | in vitro | |||
| LMNA | L-CMD | 2018 | 5′ | transfection, AAV | in vitro, in vivo | Azibani et al.63 |
| MAPT | Tauopathies | 2018 | 3′ | lentivirus | in vivo | Avale et al.64; Espindola et al.65; Lacovich et al.66; Rodriguez-Martin et al.67,68 |
| 2017 | 3′ | lentivirus | in vitro | |||
| 2013 | 3′ | lentivirus | in vitro, in vivo | |||
| 2009 | 3′ | transfection | in vitro | |||
| 2005 | 3′ | transfection | in vitro | |||
| MMP9 | EB-SCC | 2011 | 3′ | transfection | in vitro | Gruber e al.69 |
| Mybpc3 | HCM | 2017 | 5′, 3′ | AAV | in vitro | Prondzynski et al.70; Mearini et al.71 |
| 2013 | 5′ | AAV | in vitro, in vivo | |||
| PLEC1 | EB simplex with muscular dystrophy | 2008 | 5′ | transfection, retrovirus | in vitro | Wally et al.72 |
| p53 | cancer | 2015 | 3′ | transfection, adenovirus | in vitro, in vivo | He et al.73,74 |
| 2015 | 3′ | transfection, adenovirus | in vitro, in vivo | |||
| RET protooncogene | 2001 | 3′ | transfection | in vitro | Kikumori et al.75 | |
| RHO | autosomal dominant retinitis pigmentosa | 2015 | 3′ | transfection, AAV | in vitro, in vivo | Berger et al.76 |
| STX1A1 | cancer | 2005 | segmental | adenovirus | in vitro, in vivo | Nakayama et al.77 |
| SLCO1B3 | EB-SCC | 2013 | 3′ | transfection, retrovirus | in vitro | Gruber et al.78 |
| SMN | SMA | 2011 | 3′ | transfection | in vitro, in vivo | Coady et al.79,80,81; Shababi and Lorson82; Shababi et al.83 |
| 2011 | 3′ | transfection | in vivo | |||
| 2010 | 3′ | transfection | in vivo | |||
| 2008 | 3′ | transfection | in vitro, in vivo | |||
| 2007 | 3′ | transfection, AAV | in vitro | |||
| SYN1 | 2018 | 3′ | transfection, lentivirus, AAV | in vitro, in vivo | Davidsson et al.84 | |
| TTN | titinopathies | 2014 | 3′ | transfection, AAV | in vitro, in vivo | Monjaret et al.52 |
AAV, adeno-associated virus; AFP, α-fetoprotein; ALL, acute lymphoblastic leukemia; β hCG6, β-subunit of human chorionic gonadotropin gene 6; CD40L, CD40 ligand; CFTR, cystic fibrosis transmembrane conductance regulator; COL17A1, collagen 17A1; COL7A1, type VII collagen; Ct-OATP1B3, cancer-type organic anion transporting polypeptide 1B3; DM1, dystrophia myotonica type 1; DMD, Duchenne muscular dystrophy; DMPK, dystrophia myotonica protein kinase; DNA-PKcs, DNA protein kinase catalytic subunit; DNM2, dynamin 2; DYSF, dysferlin; EB-SCC, epidermolysis bullosa-associated squamous cell carcinoma; FVIII, factor VIII; HCC, hepatocellular carcinoma; GNE, UDP-N-acetylglucosamine 2-epimerase/N-acetyl-mannosamine kinase; HCM, hypertrophic cardiomyopathy; HD, Huntington disease; HIGM1, hyper-immunoglobulin M1; HPV-16, human papillomavirus type 16; HTT, Huntingtin; IgG, immunoglobulin G; IER, internal exon replacement; KRT14, keratin 14; LCA10, Leber congenital amaurosis type 10; L-CMD, LMNA-related congenital muscular dystrophy; LMNA, lamin; MAPT, microtubule-associated protein; MMP9, matrix metalloproteinase-9; PLEC1, plectin; RHO, rhodopsin; SCID, severe combined immune deficiency; SMA, spinal muscular atrophy; SMN, survival motor neuron; STX1A1, shigatoxin1A1; SYN1, synapsin 1; TTN, titin.
RNA exon editing has been considered a potential therapeutic option for many years, with reviews detailing the history of progress in terms of on-target efficiencies.7 Table 1 provides a comprehensive overview of preclinical therapeutic applications of RNA trans-splicing to date. However, comparing the reported efficiency numbers of these studies is difficult due to their varying experimental designs and data analyses. A more reliable way to track the history of the field and its progress to date is examining specific case studies.
One representative example of RNA exon editing in a monogenic disease comes from the work of Dooley et al. on the CEP290 gene as it relates to mutations causing Leber congenital amaurosis type 10 (LCA10).30 In this disease, mutations can be found throughout the CEP290 gene, which is 7.4 kb and therefore too large for traditional gene augmentation therapy using AAV. In this study, the authors utilized a model in which a mutation in CEP290 intron 26 introduces a cryptic splice donor site leading to the inclusion of an intronic region as an exon (exon X). This creates a premature stop codon, and the gene is effectively knocked down. The authors used a pre-mRNA trans-splicing molecule targeting intron 26 downstream of exon X to replace the upstream exons with wild-type exon sequences that remove the unintended exon, thereby restoring the full-length CEP290 transcript. The optimal targeting site in the intron was determined by performing a binding domain screen using a GFP mini-gene reporter system. The 5′ half of the GFP CDS was cloned into the reporter trans-splicing molecule, while the remaining 3′ GFP CDS was inserted downstream of the CEP290 target intron in a custom mini-gene plasmid, allowing for the reconstitution of the GFP open reading frame and the detection of GFP fluorescence signal upon successful trans-splicing. A series of trans-splicing molecules with different binding domains were co-transfected into HEK293T cells with the CEP290 mini-gene, and the optimal target region for further testing was identified based on the percentage of GFP+ cells. Using transient transfection of the top-performing trans-splicing molecule into HEK293T cells, the authors tested editing of endogenous CEP290 transcripts by specifically measuring the trans-spliced RNA product using a qPCR-based assay that detects the codon-optimized CDS present in the trans-spliced edited RNA. Next, the trans-splicing molecule was packaged into AAV2.7m8 and tested in an engineered mouse model by performing subretinal injections at 15 days old. qPCR analyzing the edited trans-spliced junction from RNA in the mouse retina 30 days after treatment showed a population of the correct trans-spliced product. However, protein detection probing an epitope tag on the expected trans-spliced product was not observed. Despite the various challenges associated with both non-spliced editor and delivery to mice retina, it is clear in this paper that editing the CEP290 transcript using trans-splicing is possible both in vitro and in vivo. This example also demonstrates some advantages of the technology: trans-splicing corrects the native transcript rather than simply regulating its expression and can cover both alleles for diseases like LCA10, where mutations are often compound heterozygous.
Therapeutic applications of trans-splicing have been described in contexts beyond monogenic disorders. An early study by Garcia-Blanco and colleagues demonstrated the applicability of RNA exon editing targeting highly abundant transcripts to express a therapeutic protein of interest.24 The authors developed 3′ exon editors that target the highly expressed albumin pre-mRNA and contain the CDS of therapeutic proteins of interest (human apoA-I protein, HPV-E7-specific single-chain monoclonal antibody, or factor VIII). Successful trans-splicing would produce an edited mRNA containing the ATG start codon and signal peptide from the albumin locus fused to the CDS provided by the exon editor. In mouse studies, the authors demonstrated the production of trans-spliced RNA and functional protein in vivo. Importantly, treatment of hemophilia A mice with an exon editor that targets the albumin pre-mRNA and encodes factor VIII demonstrated the expression of functional protein resulting from edited pre-mRNA and phenotypic correction in the treated mice. This strategy of targeting highly abundant transcripts can provide beneficial levels of therapeutic protein even with modest levels of trans-splicing efficiency while providing cell- and tissue-specific expression.
RNA exon editing approaches have also been proposed in the context of suicide gene therapy to induce cell death in tumor cells. Cancer cell-specific induction of cell death can be achieved by RNA exon editors that encode a cytotoxic gene and target tumor-specific pre-mRNAs. By utilizing the intrinsic ability of RNA exon editing to provide gene specificity, successful trans-splicing would result in cancer cell-specific expression of the cytotoxic protein and reduce unwanted side effects associated with suicide gene therapy. An example of such an approach has been described utilizing the herpes simplex virus 1 thymidine kinase (HSV-tk)-ganciclovir suicide gene system, in which the HSV-tk protein phosphorylates and leads to the eventual cytotoxic activity of the prodrug ganciclovir.42,69,78 Sun et al. engineered a 3′ exon editor that targets the cancer-type organic anion transporting polypeptide 1B3 (Ct-OATP1B3) pre-mRNA, a transcript that is expressed in a highly cancer-specific manner.42 The authors demonstrated that RNA exon editor expression results in the generation of Ct-OATP1B3/HSV-tk edited RNA and its subsequent translation to a fusion protein that renders cells susceptible to ganciclovir treatment in colorectal cancer cell lines and in vivo in a xenograft mouse model. This approach to using RNA exon editing to induce cancer cell-specific cell death shows promise in targeting and eliminating tumor cells if challenges in delivering RNA exon editors to tumor cells can be overcome in vivo. Taken together, such studies highlight the potential for RNA exon editor approaches that are broadly applicable to not only monogenic disorders but also inflammatory, infectious disease or oncologic conditions.57
Why now?
Despite being experimentally confirmed for multiple decades, only recently has RNA trans-splicing been considered a viable therapeutic modality. This is largely due to technological progress enabling efficient screening both experimentally and computationally, along with a deeper understanding of underlying spliceosome biology. Together, these advances may provide powerful clinical benefits to patients. Here, we discuss various improvements that have opened the door to identifying more potent RNA exon editors.
A key advantage for scientists seeking potent RNA exon editors today is the ability to accurately screen large libraries to identify candidate molecules and test component-specific improvements using modern synthetic biology and computational tools. Products such as oligo pools for large-scale cloning and droplet digital PCR for accurate and benchmarked expression measurement have taken the screening process from small scale and manual to large scale and automated.85,86,87,88 Larger screening libraries allow for a level of granularity in RNA diversity that has not been previously interrogated for trans-splicing molecules. These advances have allowed RNA-based approaches to start properly searching the vast nucleotide space for more efficient molecules. High-throughput and cost-efficient screening aside, generating reliable readout for such efforts has also been improved dramatically by advances in next-generation sequencing approaches such as hybrid capture and long-read sequencing,89,90,91,92 which give researchers many options for assessing the efficiency of an RNA exon editor. Being able to detect trans-spliced product directly is simply more achievable than ever before, and the same technological advances that make direct detection possible also facilitate examination of any potential off-target events, a key assessment for ensuring patient safety and drug efficacy. Although delivery of molecules continues to pose a significant challenge, efficiency improvements in the production of research-grade AAV allow for direct screening of molecules in their packaged forms, leading to identification of RNA exon editors in more relevant cell types and tissues.
RNA secondary structure is still considered one of the most important factors in RNA molecular design, and more advanced methods for determining the secondary structure of molecules have been gaining considerable traction, including selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) and dimethyl sulfate mutational profiling with sequencing (DMS-MaPseq).93,94,95,96 These methods predict RNA structures with single-nucleotide resolution in vitro or in-cell by the chemical probing of 2′-OH at conformationally flexible RNA nucleotides for SHAPE, or methylation of unpaired adenines and cytosines by DMS, followed by mutational profiling to identify reactive positions in the RNA. It is important to note that computationally deducing the folding of the RNA is key to achieving a more accurate and timely prediction of RNA secondary structure. Assessing RNA exon editors and target pre-mRNA for functional structure is of critical importance when designing and offers important insight into the behavior of certain elements. Hence, access to methods that can interrogate RNA secondary structures offers the potential for highly efficient screening processes and may expand our ability to design more efficient and specific molecules. Of note, the role of RNA structure of the target pre-mRNA and the structure-function relationship of cis-splicing remains to be understood, and RNA structure-probing methods should provide insights into both target pre-mRNA and exon editor activity. Such computational approaches have improved significantly over the last few decades to provide accurate understanding and prediction of splice junctions within RNA molecules97 and the secondary structure associated with those sequences.98,99,100,101 These computational techniques reduce the screening of molecules with high failure probability and provide insight into potential structural mechanisms of various sequence elements.
In addition to technological advances, our biological understanding of the cis-splicing process has guided the engineering of molecules that might result in higher trans-splicing efficiencies. Important work has been done in the area of group II intron splicing. These introns are thought to be the ancestors of spliceosomal introns and self-catalyze their excision, mirroring the spliceosome with a lariat formation.102 Experiments performed to understand the dynamics of group II introns have shown that they exhibit many structural properties similar to those of the spliceosomal machinery, including the interaction of the 5′ splice site with the branch helix and the recognition of the branchpoint.103,104 Understanding the chemistry of naturally occurring splicing reactions has shed light on elements and strategies that can increase the potency of exon editors by engineering their interaction with the spliceosome.
These advances have enabled the development of RNA exon editors that have the potential to treat patients with monogenic disorders such as Stargardt disease.105 Stargardt disease is an autosomal recessive disorder that is caused by mutations in ABCA4, a gene too large to package in AAV and within which disease-causing mutations occur throughout all 50 exons of the gene. These two features mean that Stargardt disease cannot be addressed by conventional gene therapies or gene editing approaches. ACDN-01, an exon editor that can replace multiple exons in ABCA4 at a time, has the potential to treat up to 70% of the patients carrying ABCA4 mutations and is now in a phase 1/2 trial (STELLAR; this trial was registered at ClinicalTrials.gov [NCT06467344]), demonstrating the translational viability of this therapeutic strategy.
Future directions
Despite recent technology advances that have expanded the understanding of RNA biology, several improvements will help bring RNA exon editing approaches to their full potential: the ability to predict the function and efficacy of RNA molecules in silico, deeper understanding of the dynamics of trans-splicing relative to the transcription of the target gene, further understanding of how the spliceosome defines introns and exons, and correct subcellular localization of RNA exon editors.
The ability to predict the function and efficacy of RNA molecules in silico will improve steadily. Secondary structure prediction software has been rapidly advancing with the rise of more complex machine learning models.100,106 Structurally aware in silico RNA gene therapy design could dramatically improve efficiencies and expand therapeutic approaches, such as RNA switches or the design of binding sites that take advantage of the unique structure of the target gene.
Furthermore, a deeper mechanistic understanding of trans-splicing would guide the design and discovery of elements that could enhance the efficiency of exon editors. For example, blocking of endogenous splice sites in the target pre-mRNA by antisense oligonucleotides (ASOs) or antisense RNAs has conferred improved trans-splicing by favoring trans- over cis-splicing.33,79,82 For 3′ exon editing, competitive inhibition of a downstream splice site in the target pre-mRNA was shown to enhance trans-splicing efficiency in SMN2 transcripts79,82 and in a minigene system for COL7A1 trans-splicing.33 For 5′ exon editing, using a minigene reporter system for KRT14 trans-splicing, Liemberger et al. demonstrated that co-delivery of ASOs that directly interfere with the competing 5′ splice site or ASOs that block the splicing of an upstream intron exhibit improved trans-splicing efficiencies.34 These studies highlight that trans-splicing reactions occur in direct competition with cis-splicing; however, the mechanisms by which the above-mentioned ASOs enhance trans-splicing efficiencies or how trans-splicing competes with cis-splicing are not well understood. Key questions that remain include understanding how the exon editor might be primed for interaction with the spliceosome and elucidating at which spliceosome assembly step the RNA exon editor intersects with the spliceosome assembly dynamics of cis-splicing.
Understanding the dynamics of trans-splicing in relation to transcription of the target gene could also provide insight into the feasibility of trans-splicing into a given target or intron selection for a target of interest. Mechanistic insight into how the transcriptional rate of a gene, splicing rates of introns, and/or intron splicing order might influence trans-splicing could guide designs of RNA exon editors. Several recent studies have highlighted the coordinated and predetermined nature of intron splicing across multiple introns,107,108,109,110 where the intron splicing order influences splicing fidelity across long pre-mRNAs. There is accumulating evidence, moreover, that splicing does not occur in the order of transcription, and the defined order of splicing is conserved across cell types.107 The regulatory mechanism that defines the order of splicing is not fully understood; however, spliceosomal components and RNA secondary structure likely play a role in this process. These regulatory mechanisms could be an important factor to consider for RNA exon editor designs.
Further understanding how the spliceosome defines introns and exons could inform future RNA exon editor designs. In organisms that have short (<100 nt) introns, splicing predominantly occurs through intron definition, where splice site pairing occurs across an intron. In vertebrates that have an abundance of long introns and short exons, introns are predominantly spliced through exon definition, where interactions across an exon define the splice sites (i.e., between the upstream 3′ splice site and the downstream 5′ splice site) before reorganizing takes place to form interactions across the intron and complete splicing.2,111,112 Transcriptome-wide examination of this mechanism proposed that there is a much stronger exon definition compared to intron definition in mammals.112 The orchestrated nature of pre-mRNA splicing mediated by intron/exon definition could, therefore, be used to advantage in the design of efficient exon editors. For example, modifications that strengthen the exon definition in 3′ exon editors could enhance the association of U2 snRNP with the branchpoint sequence, enabling the “activation” of the trans-splicing reaction.
Ensuring correct subcellular localization is yet another consideration that could lead to improved RNA exon editor performance. Recent studies on nuclear speckles, which are ideal subcellular targeting locations for therapies that act on the pre-mRNA, are of significance in this regard. Nuclear speckles are biomolecular condensates that are enriched in splicing factors (reviewed by Ilik and Aktas113), and recent models propose splicing to occur at the phase-separated nuclear speckle interface. This model is supported by experimental evidence, including spatial studies of active spliceosomal components and the positioning of introns and exons of transcripts.114 Elucidation of nuclear speckle function could guide improvements to molecular design whether that be via targeting, specific recruitment sequences, or retention elements.
Finally, while outside the scope of this review, we are encouraged by recent progress in the delivery of gene therapy in tissue-specific ways, a major roadblock preventing many novel therapeutic payloads from clinical translation.
Conclusion
RNA exon editing uses synthetic RNA molecules to replace defective exons in a target pre-mRNA, leading to the production of functional protein. This approach allows for kilobase-scale editing without the need for exogenous enzymes, overcoming the limitations of other gene therapy and gene editing approaches. RNA exon editing can address multiple mutations in a single gene, enables precise control of gene expression, and avoids the potential risks associated with altering DNA directly. The therapeutic applications for RNA exon editing are vast, and initial preclinical and clinical efforts are primarily focused on monogenic disease and targeting cancer cell death. The remaining challenges to the development of successful RNA exon editing technology include efficient delivery of the RNA editing molecule to target cells, and maximizing both efficiency and specificity of the trans-splicing reaction. Advancements in synthetic biology, computational technology, and our understanding of splicing mechanisms are paving the way for this promising therapeutic approach to genetic medicine, which has the potential to help patients in need, many with no other options.
Acknowledgments
None.
Author contributions
A.D., C.D., D.T., K.B., and R.D.B. contributed to writing this article. A.D. prepared Figures 1 and 2. D.T. prepared Table 1.
Declaration of interests
All authors are shareholders of Ascidian Therapeutics.
References
- 1.Nagasawa C.K., Garcia-Blanco M.A. Early Splicing Complexes and Human Disease. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241411412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Conti L., Baralle M., Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 3.Wan R., Bai R., Zhan X., Shi Y. How Is Precursor Messenger RNA Spliced by the Spliceosome? Annu. Rev. Biochem. 2020;89:333–358. doi: 10.1146/annurev-biochem-013118-111024. [DOI] [PubMed] [Google Scholar]
- 4.Eul J., Graessmann M., Graessmann A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J. 1995;14:3226–3235. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei Q., Li C., Zuo Z., Huang C., Cheng H., Zhou R. Evolutionary Insights into RNA trans-Splicing in Vertebrates. Genome Biol. Evol. 2016;8:562–577. doi: 10.1093/gbe/evw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Blanco M.A. Messenger RNA reprogramming by spliceosome-mediated RNA trans-splicing. J. Clin. Invest. 2003;112:474–480. doi: 10.1172/JCI19462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong E.M., Ingemarsdotter C.K., Lever A.M.L. Therapeutic applications of trans-splicing. Br. Med. Bull. 2020;136:4–20. doi: 10.1093/bmb/ldaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puttaraju M., Jamison S.F., Mansfield S.G., Garcia-Blanco M.A., Mitchell L.G. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 9.Boothroyd J.C., Cross G.A. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5' end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 10.Van der Ploeg L.H., Liu A.Y., Michels P.A., De Lange T., Borst P., Majumder H.K., Weber H., Veeneman G.H., Van Boom J. RNA splicing is required to make the messenger RNA for a variant surface antigen in trypanosomes. Nucleic Acids Res. 1982;10:3591–3604. doi: 10.1093/nar/10.12.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milhausen M., Nelson R.G., Sather S., Selkirk M., Agabian N. Identification of a small RNA containing the trypanosome spliced leader: a donor of shared 5' sequences of trypanosomatid mRNAs? Cell. 1984;38:721–729. doi: 10.1016/0092-8674(84)90267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douris V., Telford M.J., Averof M. Evidence for multiple independent origins of trans-splicing in Metazoa. Mol. Biol. Evol. 2010;27:684–693. doi: 10.1093/molbev/msp286. [DOI] [PubMed] [Google Scholar]
- 13.Bruzik J.P., Maniatis T. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992;360:692–695. doi: 10.1038/360692a0. [DOI] [PubMed] [Google Scholar]
- 14.Konarska M.M., Padgett R.A., Sharp P.A. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. doi: 10.1016/s0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- 15.Koller B., Fromm H., Galun E., Edelman M. Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell. 1987;48:111–119. doi: 10.1016/0092-8674(87)90361-8. [DOI] [PubMed] [Google Scholar]
- 16.Nilsen T.W. Trans-splicing in nematodes. Exp. Parasitol. 1989;69:413–416. doi: 10.1016/0014-4894(89)90191-4. [DOI] [PubMed] [Google Scholar]
- 17.Vandenberghe A.E., Meedel T.H., Hastings K.E. mRNA 5'-leader trans-splicing in the chordates. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dandekar T., Sibbald P.R. Trans-splicing of pre-mRNA is predicted to occur in a wide range of organisms including vertebrates. Nucleic Acids Res. 1990;18:4719–4725. doi: 10.1093/nar/18.16.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan P.M., Petrusz P., Szpirer C., Joseph D.R. Alternative processing of androgen-binding protein RNA transcripts in fetal rat liver. Identification of a transcript formed by trans splicing. J. Biol. Chem. 1991;266:143–154. [PubMed] [Google Scholar]
- 20.Frenkel-Morgenstern M., Gorohovski A., Lacroix V., Rogers M., Ibanez K., Boullosa C., Andres Leon E., Ben-Hur A., Valencia A. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013;41:D142–D151. doi: 10.1093/nar/gks1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herai R.H., Yamagishi M.E.B. Detection of human interchromosomal trans-splicing in sequence databanks. Brief. Bioinform. 2010;11:198–209. doi: 10.1093/bib/bbp041. [DOI] [PubMed] [Google Scholar]
- 22.Kowarz E., Merkens J., Karas M., Dingermann T., Marschalek R. Premature transcript termination, trans-splicing and DNA repair: a vicious path to cancer. Am. J. Blood Res. 2011;1:1–12. [PMC free article] [PubMed] [Google Scholar]
- 23.Poddar S., Loh P.S., Ooi Z.H., Osman F., Eul J., Patzel V. RNA Structure Design Improves Activity and Specificity of trans-Splicing-Triggered Cell Death in a Suicide Gene Therapy Approach. Mol. Ther. Nucleic Acids. 2018;11:41–56. doi: 10.1016/j.omtn.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Mansfield S.G., Cote C.A., Jiang P.D., Weng K., Amar M.J.A., Brewer B.H., Jr., Remaley A.T., McGarrity G.J., Garcia-Blanco M.A., Puttaraju M. Trans-splicing into highly abundant albumin transcripts for production of therapeutic proteins in vivo. Mol. Ther. 2009;17:343–351. doi: 10.1038/mt.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchida N., Washington K.N., Mozer B., Platner C., Ballantine J., Skala L.P., Raines L., Shvygin A., Hsieh M.M., Mitchell L.G., Tisdale J.F. RNA Trans-Splicing Targeting Endogenous beta-Globin Pre-Messenger RNA in Human Erythroid Cells. Hum. Gene Ther. Methods. 2017;28:91–99. doi: 10.1089/hgtb.2016.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kierlin-Duncan M.N., Sullenger B.A. Using 5'-PTMs to repair mutant beta-globin transcripts. RNA. 2007;13:1317–1327. doi: 10.1261/rna.525607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uckun F.M., Mitchell L.G., Qazi S., Liu Y., Zheng N., Myers D.E., Song Z., Ma H., Cheng J. Development of Polypeptide-based Nanoparticles for Non-viral Delivery of CD22 RNA Trans-splicing Molecule as a New Precision Medicine Candidate Against B-lineage ALL. EBioMedicine. 2015;2:649–659. doi: 10.1016/j.ebiom.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uckun F.M., Qazi S., Ma H., Reaman G.H., Mitchell L.G. CD22DeltaE12 as a molecular target for corrective repair using RNA trans-splicing: anti-leukemic activity of a rationally designed RNA trans-splicing molecule. Integr. Biol. 2015;7:237–249. doi: 10.1039/c4ib00221k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahara M., Pergolizzi R.G., Kobayashi H., Krause A., Luettich K., Lesser M.L., Crystal R.G. Trans-splicing repair of CD40 ligand deficiency results in naturally regulated correction of a mouse model of hyper-IgM X-linked immunodeficiency. Nat. Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- 30.Dooley S.J., McDougald D.S., Fisher K.J., Bennicelli J.L., Mitchell L.G., Bennett J. Spliceosome-Mediated Pre-mRNA trans-Splicing Can Repair CEP290 mRNA. Mol. Ther. Nucleic Acids. 2018;12:294–308. doi: 10.1016/j.omtn.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koller U., Wally V., Mitchell L.G., Klausegger A., Murauer E.M., Mayr E., Gruber C., Hainzl S., Hintner H., Bauer J.W. A novel screening system improves genetic correction by internal exon replacement. Nucleic Acids Res. 2011;39:e108. doi: 10.1093/nar/gkr465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dallinger G., Puttaraju M., Mitchell L.G., Yancey K.B., Yee C., Klausegger A., Hintner H., Bauer J.W. Development of spliceosome-mediated RNA trans-splicing (SMaRT) for the correction of inherited skin diseases. Exp. Dermatol. 2003;12:37–46. doi: 10.1034/j.1600-0625.2003.120105.x. [DOI] [PubMed] [Google Scholar]
- 33.Koller U., Hainzl S., Kocher T., Hüttner C., Klausegger A., Gruber C., Mayr E., Wally V., Bauer J.W., Murauer E.M. Trans-splicing improvement by the combined application of antisense strategies. Int. J. Mol. Sci. 2015;16:1179–1191. doi: 10.3390/ijms16011179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liemberger B., Pinon Hofbauer J., Wally V., Arzt C., Hainzl S., Kocher T., Murauer E.M., Bauer J.W., Reichelt J., Koller U. RNA Trans-Splicing Modulation via Antisense Molecule Interference. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19030762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr E., Ablinger M., Lettner T., Murauer E.M., Guttmann-Gruber C., Piñón Hofbauer J., Hainzl S., Kaiser M., Klausegger A., Bauer J.W., et al. 5'RNA Trans-Splicing Repair of COL7A1 Mutant Transcripts in Epidermolysis Bullosa. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peking P., Koller U., Duarte B., Murillas R., Wolf S., Maetzig T., Rothe M., Kocher T., García M., Brachtl G., et al. An RNA-targeted therapy for dystrophic epidermolysis bullosa. Nucleic Acids Res. 2017;45:10259–10269. doi: 10.1093/nar/gkx669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peking P., Koller U., Hainzl S., Kitzmueller S., Kocher T., Mayr E., Nyström A., Lener T., Reichelt J., Bauer J.W., Murauer E.M. A Gene Gun-mediated Nonviral RNA trans-splicing Strategy for Col7a1 Repair. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.3. [DOI] [PubMed] [Google Scholar]
- 38.Huttner C., Murauer E.M., Hainzl S., Kocher T., Neumayer A., Reichelt J., Bauer J.W., Koller U. Designing Efficient Double RNA trans-Splicing Molecules for Targeted RNA Repair. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tockner B., Kocher T., Hainzl S., Reichelt J., Bauer J.W., Koller U., Murauer E.M. Construction and validation of an RNA trans-splicing molecule suitable to repair a large number of COL7A1 mutations. Gene Ther. 2016;23:775–784. doi: 10.1038/gt.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murauer E.M., Koller U., Hainzl S., Wally V., Bauer J.W. A reporter-based screen to identify potent 3' trans-splicing molecules for endogenous RNA repair. Hum. Gene Ther. Methods. 2013;24:19–27. doi: 10.1089/hgtb.2012.180. [DOI] [PubMed] [Google Scholar]
- 41.Murauer E.M., Gache Y., Gratz I.K., Klausegger A., Muss W., Gruber C., Meneguzzi G., Hintner H., Bauer J.W. Functional correction of type VII collagen expression in dystrophic epidermolysis bullosa. J. Invest. Dermatol. 2011;131:74–83. doi: 10.1038/jid.2010.249. [DOI] [PubMed] [Google Scholar]
- 42.Sun Y., Piñón Hofbauer J., Harada M., Wöss K., Koller U., Morio H., Stierschneider A., Kitamura K., Hashimoto M., Chiba K., et al. Cancer-type organic anion transporting polypeptide 1B3 is a target for cancer suicide gene therapy using RNA trans-splicing technology. Cancer Lett. 2018;433:107–116. doi: 10.1016/j.canlet.2018.06.032. [DOI] [PubMed] [Google Scholar]
- 43.Liu X., Luo M., Zhang L.N., Yan Z., Zak R., Ding W., Mansfield S.G., Mitchell L.G., Engelhardt J.F. Spliceosome-mediated RNA trans-splicing with recombinant adeno-associated virus partially restores cystic fibrosis transmembrane conductance regulator function to polarized human cystic fibrosis airway epithelial cells. Hum. Gene Ther. 2005;16:1116–1123. doi: 10.1089/hum.2005.16.1116. [DOI] [PubMed] [Google Scholar]
- 44.Mansfield S.G., Clark R.H., Puttaraju M., Kole J., Cohn J.A., Mitchell L.G., Garcia-Blanco M.A. 5' exon replacement and repair by spliceosome-mediated RNA trans-splicing. RNA. 2003;9:1290–1297. doi: 10.1261/rna.5101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansfield S.G., Kole J., Puttaraju M., Yang C.C., Garcia-Blanco M.A., Cohn J.A., Mitchell L.G. Repair of CFTR mRNA by spliceosome-mediated RNA trans-splicing. Gene Ther. 2000;7:1885–1895. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- 46.Song Y., Lou H.H., Boyer J.L., Limberis M.P., Vandenberghe L.H., Hackett N.R., Leopold P.L., Wilson J.M., Crystal R.G. Functional cystic fibrosis transmembrane conductance regulator expression in cystic fibrosis airway epithelial cells by AAV6.2-mediated segmental trans-splicing. Hum. Gene Ther. 2009;20:267–281. doi: 10.1089/hum.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X., Jiang Q., Mansfield S.G., Puttaraju M., Zhang Y., Zhou W., Cohn J.A., Garcia-Blanco M.A., Mitchell L.G., Engelhardt J.F. Partial correction of endogenous DeltaF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotechnol. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- 48.Zayed H., Xia L., Yerich A., Yant S.R., Kay M.A., Puttaraju M., McGarrity G.J., Wiest D.L., McIvor R.S., Tolar J., Blazar B.R. Correction of DNA protein kinase deficiency by spliceosome-mediated RNA trans-splicing and sleeping beauty transposon delivery. Mol. Ther. 2007;15:1273–1279. doi: 10.1038/sj.mt.6300178. [DOI] [PubMed] [Google Scholar]
- 49.Chen H.Y., Kathirvel P., Yee W.C., Lai P.S. Correction of dystrophia myotonica type 1 pre-mRNA transcripts by artificial trans-splicing. Gene Ther. 2009;16:211–217. doi: 10.1038/gt.2008.150. [DOI] [PubMed] [Google Scholar]
- 50.Trochet D., Prudhon B., Jollet A., Lorain S., Bitoun M. Reprogramming the Dynamin 2 mRNA by Spliceosome-mediated RNA Trans-splicing. Mol. Ther. Nucleic Acids. 2016;5:e362. doi: 10.1038/mtna.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Philippi S., Lorain S., Beley C., Peccate C., Précigout G., Spuler S., Garcia L. Dysferlin rescue by spliceosome-mediated pre-mRNA trans-splicing targeting introns harbouring weakly defined 3' splice sites. Hum. Mol. Genet. 2015;24:4049–4060. doi: 10.1093/hmg/ddv141. [DOI] [PubMed] [Google Scholar]
- 52.Monjaret F., Bourg N., Suel L., Roudaut C., Le Roy F., Richard I., Charton K. Cis-splicing and translation of the pre-trans-splicing molecule combine with efficiency in spliceosome-mediated RNA trans-splicing. Mol. Ther. 2014;22:1176–1187. doi: 10.1038/mt.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorain S., Peccate C., Le Hir M., Griffith G., Philippi S., Précigout G., Mamchaoui K., Jollet A., Voit T., Garcia L. Dystrophin rescue by trans-splicing: a strategy for DMD genotypes not eligible for exon skipping approaches. Nucleic Acids Res. 2013;41:8391–8402. doi: 10.1093/nar/gkt621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorain S., Peccate C., Le Hir M., Garcia L. Exon exchange approach to repair Duchenne dystrophin transcripts. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao H., Mansfield S.G., Bartel R.C., Hiriyanna S., Mitchell L.G., Garcia-Blanco M.A., Walsh C.E. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- 56.Tal-Goldberg T., Lorain S., Mitrani-Rosenbaum S. Correction of the Middle Eastern M712T mutation causing GNE myopathy by trans-splicing. NeuroMolecular Med. 2014;16:322–331. doi: 10.1007/s12017-013-8278-2. [DOI] [PubMed] [Google Scholar]
- 57.Ingemarsdotter C.K., Poddar S., Mercier S., Patzel V., Lever A.M.L. Expression of Herpes Simplex Virus Thymidine Kinase/Ganciclovir by RNA Trans-Splicing Induces Selective Killing of HIV-Producing Cells. Mol. Ther. Nucleic Acids. 2017;7:140–154. doi: 10.1016/j.omtn.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rindt H., Tom C.M., Lorson C.L., Mattis V.B. Optimization of trans-Splicing for Huntington's Disease RNA Therapy. Front. Neurosci. 2017;11:544. doi: 10.3389/fnins.2017.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rindt H., Yen P.F., Thebeau C.N., Peterson T.S., Weisman G.A., Lorson C.L. Replacement of huntingtin exon 1 by trans-splicing. Cell. Mol. Life Sci. 2012;69:4191–4204. doi: 10.1007/s00018-012-1083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shang Y., Tesar D., Hötzel I. Modular protein expression by RNA trans-splicing enables flexible expression of antibody formats in mammalian cells from a dual-host phage display vector. Protein Eng. Des. Sel. 2015;28:437–444. doi: 10.1093/protein/gzv018. [DOI] [PubMed] [Google Scholar]
- 61.Peking P., Breitenbach J.S., Ablinger M., Muss W.H., Poetschke F.J., Kocher T., Koller U., Hainzl S., Kitzmueller S., Bauer J.W., et al. An ex vivo RNA trans-splicing strategy to correct human generalized severe epidermolysis bullosa simplex. Br. J. Dermatol. 2019;180:141–148. doi: 10.1111/bjd.17075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wally V., Brunner M., Lettner T., Wagner M., Koller U., Trost A., Murauer E.M., Hainzl S., Hintner H., Bauer J.W. K14 mRNA reprogramming for dominant epidermolysis bullosa simplex. Hum. Mol. Genet. 2010;19:4715–4725. doi: 10.1093/hmg/ddq405. [DOI] [PubMed] [Google Scholar]
- 63.Azibani F., Brull A., Arandel L., Beuvin M., Nelson I., Jollet A., Ziat E., Prudhon B., Benkhelifa-Ziyyat S., Bitoun M., et al. Gene Therapy via Trans-Splicing for LMNA-Related Congenital Muscular Dystrophy. Mol. Ther. Nucleic Acids. 2018;10:376–386. doi: 10.1016/j.omtn.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avale M.E., Rodríguez-Martín T., Gallo J.M. Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum. Mol. Genet. 2013;22:2603–2611. doi: 10.1093/hmg/ddt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Espindola S.L., Damianich A., Alvarez R.J., Sartor M., Belforte J.E., Ferrario J.E., Gallo J.M., Avale M.E. Modulation of Tau Isoforms Imbalance Precludes Tau Pathology and Cognitive Decline in a Mouse Model of Tauopathy. Cell Rep. 2018;23:709–715. doi: 10.1016/j.celrep.2018.03.079. [DOI] [PubMed] [Google Scholar]
- 66.Lacovich V., Espindola S.L., Alloatti M., Pozo Devoto V., Cromberg L.E., Čarná M.E., Forte G., Gallo J.M., Bruno L., Stokin G.B., et al. Tau Isoforms Imbalance Impairs the Axonal Transport of the Amyloid Precursor Protein in Human Neurons. J. Neurosci. 2017;37:58–69. doi: 10.1523/JNEUROSCI.2305-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez-Martin T., Anthony K., Garcia-Blanco M.A., Mansfield S.G., Anderton B.H., Gallo J.M. Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing. Hum. Mol. Genet. 2009;18:3266–3273. doi: 10.1093/hmg/ddp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez-Martin T., Garcia-Blanco M.A., Mansfield S.G., Grover A.C., Hutton M., Yu Q., Zhou J., Anderton B.H., Gallo J.M. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: implications for tauopathies. Proc. Natl. Acad. Sci. USA. 2005;102:15659–15664. doi: 10.1073/pnas.0503150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gruber C., Gratz I.K., Murauer E.M., Mayr E., Koller U., Bruckner-Tuderman L., Meneguzzi G., Hintner H., Bauer J.W. Spliceosome-mediated RNA trans-splicing facilitates targeted delivery of suicide genes to cancer cells. Mol. Cancer Ther. 2011;10:233–241. doi: 10.1158/1535-7163.MCT-10-0669. [DOI] [PubMed] [Google Scholar]
- 70.Prondzynski M., Krämer E., Laufer S.D., Shibamiya A., Pless O., Flenner F., Müller O.J., Münch J., Redwood C., Hansen A., et al. Evaluation of MYBPC3 trans-Splicing and Gene Replacement as Therapeutic Options in Human iPSC-Derived Cardiomyocytes. Mol. Ther. Nucleic Acids. 2017;7:475–486. doi: 10.1016/j.omtn.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mearini G., Stimpel D., Krämer E., Geertz B., Braren I., Gedicke-Hornung C., Précigout G., Müller O.J., Katus H.A., Eschenhagen T., et al. Repair of Mybpc3 mRNA by 5'-trans-splicing in a Mouse Model of Hypertrophic Cardiomyopathy. Mol. Ther. Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wally V., Klausegger A., Koller U., Lochmüller H., Krause S., Wiche G., Mitchell L.G., Hintner H., Bauer J.W. 5' trans-splicing repair of the PLEC1 gene. J. Invest. Dermatol. 2008;128:568–574. doi: 10.1038/sj.jid.5701152. [DOI] [PubMed] [Google Scholar]
- 73.He X., Liu F., Yan J., Zhang Y., Yan J., Shang H., Dou Q., Zhao Q., Song Y. Trans-splicing repair of mutant p53 suppresses the growth of hepatocellular carcinoma cells in vitro and in vivo. Sci. Rep. 2015;5:8705. doi: 10.1038/srep08705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He X., Liao J., Liu F., Yan J., Yan J., Shang H., Dou Q., Chang Y., Lin J., Song Y. Functional repair of p53 mutation in colorectal cancer cells using trans-splicing. Oncotarget. 2015;6:2034–2045. doi: 10.18632/oncotarget.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kikumori T., Cote G.J., Gagel R.F. Promiscuity of pre-mRNA spliceosome-mediated trans splicing: a problem for gene therapy? Hum. Gene Ther. 2001;12:1429–1441. doi: 10.1089/104303401750298580. [DOI] [PubMed] [Google Scholar]
- 76.Berger A., Lorain S., Joséphine C., Desrosiers M., Peccate C., Voit T., Garcia L., Sahel J.A., Bemelmans A.P. Repair of rhodopsin mRNA by spliceosome-mediated RNA trans-splicing: a new approach for autosomal dominant retinitis pigmentosa. Mol. Ther. 2015;23:918–930. doi: 10.1038/mt.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakayama K., Pergolizzi R.G., Crystal R.G. Gene transfer-mediated pre-mRNA segmental trans-splicing as a strategy to deliver intracellular toxins for cancer therapy. Cancer Res. 2005;65:254–263. [PubMed] [Google Scholar]
- 78.Gruber C., Koller U., Murauer E.M., Hainzl S., Hüttner C., Kocher T., South A.P., Hintner H., Bauer J.W. The design and optimization of RNA trans-splicing molecules for skin cancer therapy. Mol. Oncol. 2013;7:1056–1068. doi: 10.1016/j.molonc.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coady T.H., Baughan T.D., Shababi M., Passini M.A., Lorson C.L. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coady T.H., Lorson C.L. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J. Neurosci. 2010;30:126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coady T.H., Shababi M., Tullis G.E., Lorson C.L. Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol. Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- 82.Shababi M., Lorson C.L. Optimization of SMN trans-splicing through the analysis of SMN introns. J. Mol. Neurosci. 2012;46:459–469. doi: 10.1007/s12031-011-9614-3. [DOI] [PubMed] [Google Scholar]
- 83.Shababi M., Glascock J., Lorson C.L. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum. Gene Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- 84.Davidsson M., Díaz-Fernández P., Torroba M., Schwich O.D., Aldrin-Kirk P., Quintino L., Heuer A., Wang G., Lundberg C., Björklund T. Molecular barcoding of viral vectors enables mapping and optimization of mRNA trans-splicing. RNA. 2018;24:673–687. doi: 10.1261/rna.063925.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hughes R.A., Ellington A.D. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harbor Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kosuri S., Church G.M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuiper B.P., Prins R.C., Billerbeck S. Oligo Pools as an Affordable Source of Synthetic DNA for Cost-Effective Library Construction in Protein- and Metabolic Pathway Engineering. Chembiochem. 2022;23 doi: 10.1002/cbic.202100507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taylor S.C., Laperriere G., Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 2017;7:2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E.M., Brockman W., Fennell T., Giannoukos G., Fisher S., Russ C., et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Samorodnitsky E., Jewell B.M., Hagopian R., Miya J., Wing M.R., Lyon E., Damodaran S., Bhatt D., Reeser J.W., Datta J., Roychowdhury S. Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing. Hum. Mutat. 2015;36:903–914. doi: 10.1002/humu.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh R.R. Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology. Diagnostics. 2022;12 doi: 10.3390/diagnostics12071539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marx V. Method of the year: long-read sequencing. Nat. Methods. 2023;20:6–11. doi: 10.1038/s41592-022-01730-w. [DOI] [PubMed] [Google Scholar]
- 93.Lucks J.B., Mortimer S.A., Trapnell C., Luo S., Aviran S., Schroth G.P., Pachter L., Doudna J.A., Arkin A.P. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siegfried N.A., Busan S., Rice G.M., Nelson J.A.E., Weeks K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gupta P., Rouskin S. In Vivo RNA Structure Probing with DMS-MaPseq. Methods Mol. Biol. 2022;2404:299–310. doi: 10.1007/978-1-0716-1851-6_16. [DOI] [PubMed] [Google Scholar]
- 96.Zubradt M., Gupta P., Persad S., Lambowitz A.M., Weissman J.S., Rouskin S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods. 2017;14:75–82. doi: 10.1038/nmeth.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 98.Do C.B., Woods D.A., Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–e98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 99.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J., Fei Y., Sun L., Zhang Q.C. Advances and opportunities in RNA structure experimental determination and computational modeling. Nat. Methods. 2022;19:1193–1207. doi: 10.1038/s41592-022-01623-y. [DOI] [PubMed] [Google Scholar]
- 101.Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lambowitz A.M., Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harbor Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manigrasso J., Chillón I., Genna V., Vidossich P., Somarowthu S., Pyle A.M., De Vivo M., Marcia M. Visualizing group II intron dynamics between the first and second steps of splicing. Nat. Commun. 2020;11:2837. doi: 10.1038/s41467-020-16741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu L., Liu T., Chung K., Pyle A.M. Structural insights into intron catalysis and dynamics during splicing. Nature. 2023;624:682–688. doi: 10.1038/s41586-023-06746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mullard A. RNA-rewriting candidate moves into the clinic. Nat. Rev. Drug Discov. 2024;23:407–409. doi: 10.1038/d41573-024-00086-4. [DOI] [PubMed] [Google Scholar]
- 106.Zhang H., Zhang C., Li Z., Li C., Wei X., Zhang B., Liu Y. A New Method of RNA Secondary Structure Prediction Based on Convolutional Neural Network and Dynamic Programming. Front. Genet. 2019;10:467. doi: 10.3389/fgene.2019.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Choquet K., Baxter-Koenigs A.R., Dülk S.L., Smalec B.M., Rouskin S., Churchman L.S. Pre-mRNA splicing order is predetermined and maintains splicing fidelity across multi-intronic transcripts. Nat. Struct. Mol. Biol. 2023;30:1064–1076. doi: 10.1038/s41594-023-01035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Drexler H.L., Choquet K., Churchman L.S. Splicing Kinetics and Coordination Revealed by Direct Nascent RNA Sequencing through Nanopores. Mol. Cell. 2020;77:985–998.e8. doi: 10.1016/j.molcel.2019.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim S.W., Taggart A.J., Heintzelman C., Cygan K.J., Hull C.G., Wang J., Shrestha B., Fairbrother W.G. Widespread intra-dependencies in the removal of introns from human transcripts. Nucleic Acids Res. 2017;45:9503–9513. doi: 10.1093/nar/gkx661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takahara K., Schwarze U., Imamura Y., Hoffman G.G., Toriello H., Smith L.T., Byers P.H., Greenspan D.S. Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a COL5A1 acceptor-site mutation that results in abnormal pro-alpha1(V) N-propeptides and Ehlers-Danlos syndrome type I. Am. J. Hum. Genet. 2002;71:451–465. doi: 10.1086/342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ellis J.D., Llères D., Denegri M., Lamond A.I., Cáceres J.F. Spatial mapping of splicing factor complexes involved in exon and intron definition. J. Cell Biol. 2008;181:921–934. doi: 10.1083/jcb.200710051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu L., Das U., Ogunsola S., Xie J. Transcriptome-Wide Detection of Intron/Exon Definition in the Endogenous Pre-mRNA Transcripts of Mammalian Cells and Its Regulation by Depolarization. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231710157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ilik I.A., Aktas T. Nuclear speckles: dynamic hubs of gene expression regulation. FEBS J. 2022;289:7234–7245. doi: 10.1111/febs.16117. [DOI] [PubMed] [Google Scholar]
- 114.Liao S.E., Regev O. Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res. 2021;49:636–645. doi: 10.1093/nar/gkaa1209. [DOI] [PMC free article] [PubMed] [Google Scholar]