Abstract
Bovine leukemia virus (BLV) is closely associated with the development of B-cell leukemia and lymphoma in cattle. BLV infection has also been studied extensively in an in vivo ovine model that provides a unique system for studying B-cell leukemogenesis. There is no evidence that BLV can directly infect ovine B cells in vitro, and there are no direct data regarding the oncogenic potential of the viral Tax transactivator in B cells. Therefore, we developed ovine B-cell culture systems to study the interaction between BLV and its natural target, the B cell. In this study, we used murine CD154 (CD40 ligand) and γ-chain-common cytokines to support the growth of B cells isolated from ovine lymphoid tissues. Integrated provirus, extrachromosomal forms, and viral transcripts were detected in BLV-exposed populations of immature, rapidly dividing surface immunoglobulin M-positive B cells from sheep ileal Peyer's patches and also in activated mature B cells isolated from blood. Conclusive evidence of direct B-cell infection by BLV was obtained through the use of cloned B cells derived from sheep jejunal Peyer's patches. Finally, inoculation of sheep with BLV-infected cultures proved that infectious virus was shed from in vitro-infected B cells. Collectively, these data confirm that a variety of ovine B-cell populations can support productive infection by BLV. The development of ovine B-cell cultures permissive for BLV infection provides a controlled system for investigating B-cell leukemogenic processes and the pathogenesis of BLV infection.
Bovine leukemia virus (BLV) is a B-lymphotropic retrovirus that is associated with B-cell lymphoproliferative disorders in cattle (2, 22). Infection can either remain asymptomatic or result in persistent lymphocytosis, characterized by an increased number of circulating B lymphocytes and, more rarely, by clonal lymphoid tumors after a long latency period. Under experimental conditions, BLV can infect sheep and induces B-cell neoplasia at very high frequencies (26).
Structurally and functionally, BLV is related to human T-cell lymphotropic viruses type 1 and 2 (HTLV-1 and HTLV-2), which infect T cells and are associated with adult T-cell leukemia, tropical spastic paraparesis, and hairy T-cell leukemia in humans (11, 38, 51). BLV, HTLV-1 and HTLV-2 have a similar genomic organization, encode gene products with biologically similar functions, and share mechanisms of transactivation (10). Aside from the structural genes (gag, pol, and env), the BLV provirus contains a region, X, which encodes at least four auxiliary proteins, Tax, Rex, R3, and G4. Tax and Rex are involved in transcriptional and posttranscriptional regulation of viral expression, respectively, and are essential for viral infectivity in vivo (5, 9). R3 and G4 may play a role in virus propagation in the infected host (48), and G4 was shown to have moderate oncogenic potential in vitro (21).
The BLV provirus integrates into bovine and ovine B cells, and this viral infection may result in B-cell transformation. In cattle, infected cells have been reported as mature, CD5+ surface immunoglobulin M-positive (sIgM+) and CD5+ sIgG+ B cells (27, 45). In sheep, BLV-infected cells from aleukemic animals were found to be either CD5+ or CD5−, Cd11b+ sIgM+ B cells (1, 31, 32, 37, 42, 43), and cells isolated from lymphoma and leukemic tissues were consistently sIgM+ B cells. It has also been suggested that BLV may infect other cell types, including granulocytes, monocytes, and CD8+ T cells (7, 19, 39). However, Mirsky et al. (29) did not obtain convincing evidence that either monocytes or T cells contained BLV provirus.
BLV infection has been studied extensively in an in vivo ovine model using whole blood, blood leukocytes, fresh or cultured lymphocytes from BLV-seropositive animals, and the injection of proviral DNA (26, 33, 40, 41, 49, 50). It is possible to establish tumor-derived B-cell lines from BLV-infected sheep, and these cell lines, with integrated provirus, provide a unique system for studying B-cell leukemogenic processes (23, 41, 42). Furthermore, a variety of cell types have been used to study BLV infection and viral gene expression (6, 7, 28, 37). Both the HTLV-1 and BLV Tax proteins exhibit oncogenic potential, based on immortalization and transformation assays with rat embryonic fibroblasts (35, 46). It has not been possible, however, to perform similar studies with either bovine or ovine B cells due to an inability to culture primary B cells or establish permanent B-cell lines.
Therefore, we sought to develop an ovine B-cell culture system as a model to study the interaction between BLV and its natural target, the B cell. The first culture system used was a pure population of immature, sIgM+ B cells from the lymphoid follicles of sheep ileal Peyer's patches (12). The culture of these ileal Peyer's patch follicular B cells (iPfB cells) is limited by extensive cell death during the first 24 to 48 h after isolation. Cocultivation of iPfB cells with murine CD154 (CD40 ligand) inhibits much of this cell death, induces cytokine responsiveness, and specifically supports B-cell growth (14). Thus, it is possible to maintain pure cultures of sheep B cells for prolonged periods using CD154 coculture in conjunction with γ-chain-common cytokines (13). In this study, we used these culture conditions to maintain the growth of B cells isolated from different ovine lymphoid tissues and provide the first experimental evidence that a variety of cultured B-cell populations can support productive infection by BLV.
MATERIALS AND METHODS
Animals and tissues.
The sheep used for the isolation of B cells were Suffolk lambs obtained from the Department of Animal and Poultry Science, University of Saskatchewan, Saskatoon, Canada. Ileal Peyer's patch tissue was collected from 6- to 8-week-old lambs within 15 min after a lethal intravenous injection of Euthanyl (MTC Pharmaceuticals Ltd.). Tissue was collected and handled as described previously (12). Blood was collected from 6- to 12-month-old lambs using the procedure described previously (8). Sheep that were inoculated intradermally with cultured cells were maintained in isolation at the Centre d'Etude et de Recherches Vétérinaires et Agrochimiques (Brussels, Belgium). Animals were injected at 6 months of age with 107 cells collected from the different B-cell cultures, and blood and sera were then collected from each sheep at weekly intervals for the next year. Anti-p24 serum antibody titers were determined with a competitive enzyme-linked immunosorbent assay as previously described (33). For detection of BLV provirus, 10-ml samples of total blood were frozen and used for PCR amplification as previously described (40).
Cells and media.
OVK (ovine kidney) cells, an uninfected cell line, and FLK, a BLV-infected fetal lamb kidney cell line (44), were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) (Gibco), 1 mM sodium pyruvate, 2 mM glutamine, nonessential amino acids, and kanamycin (100 μg/ml). YR2 is a latently infected, Tax-deficient ovine B-cell line that was established from leukemic B cells isolated from a BLV-infected sheep (23, 41). YR2LTaxSN is a BLV-producing B-cell line resulting from retrovirus vector-mediated tax gene transfer into native YR2 cells (40). M267 is a clonal lymphoma-derived B-cell line isolated from a BLV-infected sheep. The B-cell lines were maintained in Optimem medium (Gibco) supplemented with 10% FBS and additives as described for adherent cell lines. J558L cells expressing murine CD154 were maintained and used for coculture with sheep B cells as described previously (14). Griebel and Ferrari (14) confirmed that there was a specific interaction between murine CD154 and CD40 expressed on sheep B cells. Clone 2 (sIgM+) and clone 4 (sIgG1+) B-cell clones were established from the jejunal Peyer's patch of a lamb by limiting-dilution culture following long-term coculture with CD154 and recombinant human interleukin-2 (IL-2), IL-4, IL-7, and IL-15 (15). The culture medium for cloned Peyer's patch B cells, freshly isolated iPfB cells, and blood lymphocytes was AIM-V (Gibco-BRL) supplemented with 2% FBS and 2 × 10−5 M β-mercaptoethanol (Sigma). Blood lymphocytes were cocultured with CD154 and recombinant human IL-2 in a manner similar to that described for iPfB cells (14). Recombinant human IL-2, IL-4, IL-7 and IL-15 were purchased from Peprotech EC (London, U.K.), and each cytokine was used at a final concentration of 10 ng/ml. All cell cultures were incubated at 37°C in a 5% CO2 humidified atmosphere. Viable cells were identified by trypan blue dye exclusion or propidium iodide exclusion (2.5 μg/ml), and cell number was counted with a hemacytometer or a Coulter particle counter. Proliferation assays with 2 × 105 cells/well were conducted in flat-bottomed 96-well tissue culture plates (Falcon) in a final volume of 200 μl. During the last 4 h of a 72-h incubation period, the cultures were pulsed with 0.4 μCi of [3H]thymidine (Amersham). [3H]thymidine incorporation was determined using standard methods for cell harvesting and liquid scintillation counting.
BLV infection of ovine B-cell cultures.
BLV-producing FLK cells were seeded at 106 cells in 100-mm culture dishes with 10 ml of medium and cultured for 48 h. Cloned FLK proviruses were shown to be infectious in vitro and in vivo (37). Expression of viral capsid protein is a marker for late-stage expression, when full-length and singly spliced transcripts are translated into the structural proteins that are assembled into virions (34). Flow cytometric analysis of intracellular p24 confirmed that the majority of FLK cells were producing viral proteins, and a competitive enzyme-linked immunosorbent assay (33) determined that supernatants collected from 48-h-cultured FLK cells contained 850 ng of p24 per ml (data not shown). For all in vitro infection assays, 10 × 106 B cells/well were incubated in a six-well plate with 1 ml of cell-free culture supernatant from either FLK or OVK cells in a final culture volume of 5 ml. After a 96-h infection period, the B cells were collected, washed with phosphate-buffered saline, and transferred to new cultures with fresh medium and the appropriate costimulation.
Southern blot analysis.
High-molecular-weight cellular DNA was prepared by 0.05% sodium dodecyl sulfate (SDS) and pronase (0.2 mg/ml) disruption of cells, followed by extraction with phenol-chloroform and ethanol precipitation. Genomic DNA was analyzed by Southern blot as previously described (40). The nylon-bound digested DNA was hybridized with a 32P-labeled 8.3-kb SacI full-length BLV probe (4 × 108 to 5 × 108 cpm/ml) (41). The amount of hybridized sequence was quantified by PhosphoImager analysis (Molecular Dynamics) or conventional autoradiography.
Primers for PCR.
The sequences of the BLV primers used in PCR experiments were as follows; nucleotide positions according to Sagata (38) are shown in parentheses, and 5′ long terminal repeat (LTR) positions are given for primers located in the LTR region: Tax1 sense (7321 to 7340), 5′-GATGCCTGGTGCCCCCTCTG-3′; Tax2 antisense (7604 to 7623), 5′-ACCGTCGCTAGAGGCCGAGG-3′; EnvA sense (4766 to 4788): 5′-TCCTGGCTACTAACCCCCCCGT-3′; EnvB antisense (5756 to 5777), 5′-TCCAGTGAGCCCCACTGACAGG-3′; LTRA sense (1 to 22), 5′-TGTATGAAAGATCATGCAGGCC-3′; LTRC antisense (577 to 599), 5′-GCCGCCGAGGGGGTGGGTCCAGA-3′; U5 sense (470 to 490), 5′-TTCTCGCGGCCCGCGCTCTCT-3′; and U3 antisense (415 to 434), 5′-GCCAGACGCCCTTGGAGCGC-3′.
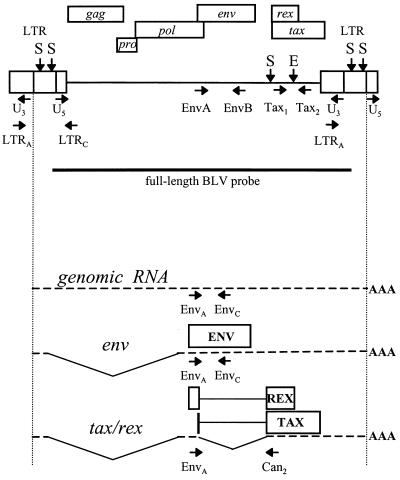
For the detection of BLV-specific mRNA, three different primers were used; the numbers indicating their positions in the genome correspond to the sequence of BLV-FLK (38). To detect full-length gag-pol (8.4 kb) and singly spliced env (4.4 kb) BLV mRNAs, a primer upstream of the second splice donor, EnvA (described above) was used with primer EnvC (4921 to 4942) (5′-CCTAGGGACAGGGAGCATCTCC-3′), located downstream of the second splice donor site but upstream of the second splice acceptor site, into the second intron. EnvA was used as a 5′ primer with Can2 (7314 to 7333) (5′-GGCACCAGGCATCGATGGTG-3′) as the complementary primer for the detection of the 2.1-kb doubly spliced tax/rex mRNA. Can2 is located downstream of the second splice junction.
DNA PCR.
Genomic DNA was amplified in 100-μl reaction mixtures containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.2 mM deoxynucleoside triphosphates, 0.5 μM each primer, and 1 U of Taq polymerase (Roche). The amplification sequence consisted of a 5-min step at 94°C, 36 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C, and a 10-min step at 72°C.
RNA PCR.
For reverse transcription (RT)-PCR experiments, total RNA was extracted using Tripure reagent according to the manufacturer's protocol (Roche) and treated for 1 h at 37°C with 40 U of DNase I (Promega). Then 1 μg of RNA was reverse transcribed and amplified using the Titan RT-PCR system according to the standard protocol supplied by the manufacturer (Roche).
Flow cytometry.
The phenotype of cells in all B-cell cultures was analyzed prior to collecting cell pellets to analyze proviral integration and viral expression. B cells were identified with monoclonal antibodies specific for the following surface antigens: IgM (PIg45A clone), IgG1 (BIg715A clone), and major histocompatibility complex (MHC) class II (TH14B clone) (VMRD Inc., Pullman, Wash.). CD72 (DU2-104 clone) was a generous gift from Wayne Hein, Wallaceville Animal Research Center, Upper Hut, New Zealand. T cells were identified with monoclonal antibodies specific for CD5 (clone ST1a), CD4 (clone 17D-13), CD8 (clone E95), and γδTCR (clone 86D). Cell labeling and analysis were done as described previously (16). For the detection of intracellular BLV p24, the cells were permeabilized with Permeafix (Ortho Diagnostics), incubated with anti-p24 monoclonal antibodies (clones 4H6, 4′F3, 7G6, 3B1, 2′C1, 4′G9, 2B1, and 5F7; provided by D. Portetelle, Facultés universitaires Sciences agronomiques Gembloux, Belgium), and monoclonal antibody binding was detected with fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Sigma), as previously described (40).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism 2.01 software (Graphpad Software, Inc., San Diego, Calif.). Differences among groups were determined using a one-way analysis of variance and Tukey's multiple comparison test if the analysis of variance revealed a significant difference (P < 0.05) among groups. Data are presented as the mean and 1 standard deviation (SD) of values from three experiments.
RESULTS
BLV infection of iFpB cells.
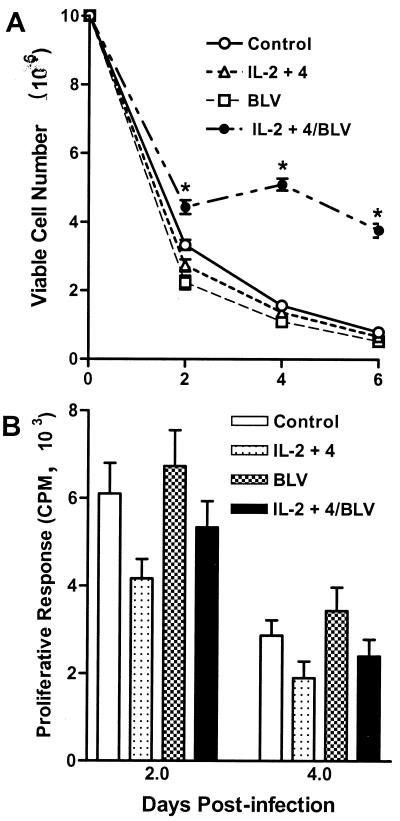
To determine whether BLV could infect ovine B cells in vitro, we first used a pure B-cell population isolated from lymphoid follicles of the ileal Peyer's patch (iPfB cells) of sheep (12). IPfB cells are a relatively homogeneous population of immature, sIgM+ B cells (18) with 35 to 40% of freshly isolated cells in S phase (17). The permissiveness of the host cell to many and perhaps all oncoretroviruses is cell cycle dependent, and infection of growth-arrested cells appears to be unique to lentiviruses in the retrovirus family (25). Therefore it was deemed most appropriate to select a B-cell target population with a high mitotic index. In the first experiment, iPfB cells were incubated with FLK (BLV-positive) or OVK (BLV-negative) culture supernatants added to culture medium alone or medium supplemented with recombinant human IL-2 and IL-4. Viable-cell numbers were determined, and cell pellets were collected for analysis of provirus integration and viral gene expression at 24 and 96 h postinfection (p.i.). These cultures are hereafter referred to as short-term iPfB-cell cultures. In the absence of exogenous cytokines, viable iPfB-cell number decreased rapidly, with most B cells dead within 6 days after exposure to either OVK or FLK culture supernatant. In contrast, with the addition of exogenous human IL-2 and IL-4, FLK-exposed iPfB-cell cultures displayed a significantly (P < 0.05) increased number of viable cells relative to OVK-exposed cultures (Fig. 1A). The level of [3H]thymidine incorporation, however, was not significantly different for viable cells collected from FLK- and OVK-exposed iPfB cultures (Fig. 1B). These observations suggested that BLV infection and costimulation with exogenous IL-2 and IL-4 might enhance iPfB-cell survival.
FIG. 1.
BLV infection alters the survival of iPfB cells in short-term cultures. iPfB cells were cultured with OVK (control) or FLK (BLV) culture supernatant in medium alone or medium supplemented with 10 ng of recombinant human IL-2 and IL-4 (IL-2+4) per ml. For each experiment, triplicate cultures were established with 10 × 106 cells/well, and cells from a single well were collected and analyzed at 2, 4, and 6 days p.i. (A) Viable-cell number was determined by counting total cell number with a Coulter particle counter and then determining the percent viable cells (propidium iodide exclusion) with a flow cytometer (FACScan). Incubation with BLV significantly (P < 0.05) increased viable-cell number in the presence (IL-2+4/BLV) but not in the absence (BLV) of recombinant human IL-2 and -4. (B) Sufficient viable cells were recovered from all treatment groups to assay spontaneous lymphocyte proliferation on days 2 and 4 p.i. A total of 2 × 105 viable cells were plated in triplicate cultures, and cells were pulsed for 4 h with 0.4 μCi of [3H]thymidine per well. The level of spontaneous proliferation was not significantly different among treatment groups at the two time points assayed. Data presented are the mean + one SD of values from three experiments. ∗, P < 0.05.
DNA from short-term iPfB-cell cultures (24 and 96 h p.i.) were analyzed by PCR amplification with primer pairs specific for BLV proviral sequences (Tax1/Tax2, EnvA/EnvB, and LTRA/LTRC) and Southern blot hybridization using a 32P-labeled full-length BLV probe. A diagram of the BLV genome and relevant primers used for PCR analysis is presented in Fig. 2. BLV-specific 283-bp tax, 990-bp env, and 570-bp LTR amplification products were detected in FLK-exposed iPfB cells at both 24 and 96 h p.i., confirming the presence of BLV proviral sequences in these cultures (Fig. 3A). DNA prepared from 96-h OVK-exposed iPfB was negative, as expected, and the BLV-infected M267 B-cell line was used as a positive control.
FIG. 2.
Diagram of the BLV provirus and major transcripts. The locations of the two LTRs and the gag, pro, pol, env, tax, and rex genes are represented. Vertical arrows indicate restriction sites: S, SacI; E, EcoRI. The position and direction of the PCR primers are indicated on the provirus map. A horizontal bar indicates the region that was used as a probe. Only the genomic, env, and tax/rex transcripts are represented below. Alternatively spliced RNAs are not shown. The translation products of the spliced transcripts and the positions of the RT-PCR primers are indicated.
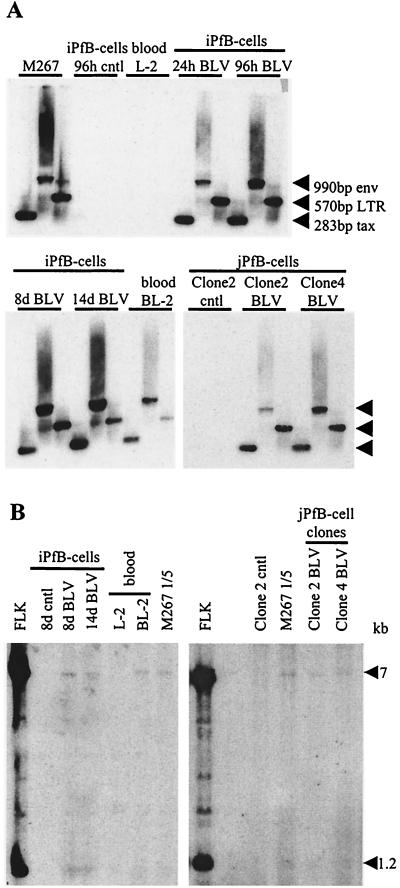
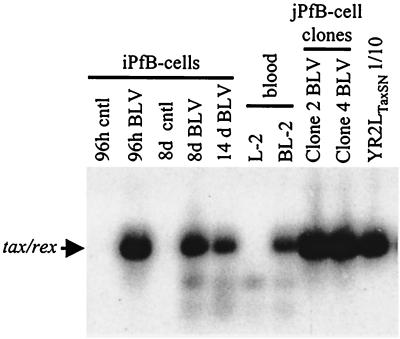
FIG. 3.
Detection of proviral sequences in FLK-exposed B-cell cultures derived from ovine lymphoid tissues. B cells were exposed to culture supernatant of BLV-infected FLK cells or mock infected by incubation with OVK supernatant. Cells were then collected for DNA isolation after the indicated period of time. The following BLV-infected samples were analyzed: short-term iPfB cells cultured for 24 and 96 h (24h BLV and 96h BLV, respectively); cytokine/CD154 iPfB cells cultured for 8 and 14 days (8d BLV and 14d BLV, respectively); blood-derived B cells cultured for 60 days (BL-2); and jejunal Peyer's patch B-cell (jPfB) clones 2 and 4 cultured for 48 days. M267 is a BLV-infected control cell line. Control samples included the following OVK-exposed cultures: short term iPfB cells cultured for 96 h (iPfB-cells 96h cntl); cytokine/CD154 iPfB cells cultured for 8 days (iPfB-cells 8d cntl); blood-derived B cells cultured for 60 days (L-2); and jPfB clone 2 cells cultured for 48 days (jPfB-cell Clone2 cntl). (A) DNA was amplified with the tax, env, and LTR primers and analyzed by Southern blot hybridization with a 32P-labeled full-length BLV probe. The amplified products of tax, env, and LTR were 283, 990, and 570 bp, respectively. (B) Evaluation of provirus loads by Southern blot analysis following SacI digestion. The FLK cell line (four provirus copies per cell) and fivefold dilutions of the BLV-infected M267 cells (one provirus copy per cell) were included for comparison.
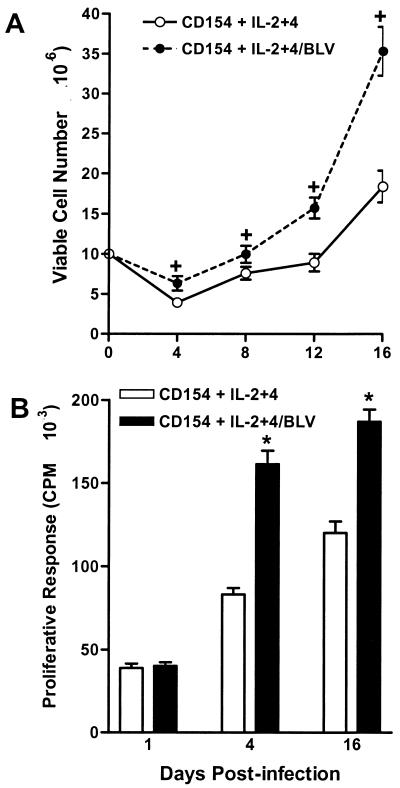
The rapid cell death in short-term iPfB-cell cultures limited the value of these cultures for studying BLV infection. However, these cultures did provide evidence that B cells could be infected in vitro. Therefore, we investigated further the possibility of stable proviral integration and sustained viral gene expression in primary B-cell cultures by infecting iPfB cells costimulated with murine CD154 and exogenous recombinant human γ-chain-common cytokines. These culture conditions had previously been shown to specifically block iPfB-cell death and induce a sustained B-cell proliferative response (13). The iPfB-cell cultures were maintained by transferring the cells every 3 to 4 days p.i. to fresh medium and restimulating with CD154 and cytokines. These iPfB-cell cultures are referred to as cytokine/CD154 iPfB-cell cultures. CD154 and exogenous cytokines supported the growth of iPfB cells, but in addition, there was a significant increase (P < 0.01) in the number of viable cells present in the FLK-exposed cultures (Fig. 4A). [3H]thymidine incorporation assays indicated that BLV infection increased the number of viable CD154-stimulated iPfB cells through a significant (P < 0.05) increase in cell proliferation (Fig. 4B). Thus, the effect of BLV infection on B-cell growth or survival appeared to be modulated by exogenous cytokines and other cosignals (Fig. 1 and 4).
FIG. 4.
BLV infection alters the growth and proliferation of iPfB cells costimulated with CD154 and recombinant human IL-2 and -4. iPfB cells were cocultured with γ-irradiated J558L cells, expressing a membrane form of murine CD154, at a ratio of 1:10 (J558L cell to iPfB cell), and the medium was supplemented with 10 ng of recombinant human IL-2 and -4 (CD154 + IL-2+4) per ml. Cultures were established with 10 × 106 iPfB cells exposed to culture supernatant from either OVK (CD154 + IL-2+4) or FLK (CD154 + IL-2+4/BLV) cells. A total of 4 × 106 viable iPfB cells were transferred every 4 days to fresh medium supplemented with CD154 plus IL-2 and IL-4. (A) Viable-cell number was determined at each cell passage by counting total cell number with a Coulter particle counter and then determining the percent viable cells (propidium iodide exclusion) with a flow cytometer (FACScan). Incubation with BLV significantly (P < 0.001) increased viable-cell number throughout the 16-day culture period. (B) iPfB-cell proliferation was assayed at each passage by culturing 2 × 105 viable cells in triplicate cultures for 4 h. During this time, cells were pulsed with 0.4 μCi of [3H]thymidine per well. Spontaneous proliferation was significantly (P < 0.05) increased following exposure to BLV. Data presented are the mean + 1 SD of values from three experiments. ∗, P < 0.05, +, P < 0.01.
PCR analysis of the cell pellets collected at 8 and 14 days p.i. revealed the presence of provirus in BLV-infected cytokine/CD154 iPfB cultures (Fig. 3A). The provirus load was then evaluated by Southern blot analysis of genomic DNA restriction digests and quantification by PhosphorImager analysis. SacI cleaves the BLV provirus twice in each LTR for all known variants. An additional internal restriction site is present in variants derived from American strains, to which the provirus in the FLK cell line belongs (Fig. 2) (38). Two bands of low intensity at 7 and 1.2 kb were generated with DNA isolated from the cytokine/CD154 FLK-exposed iPfB cells collected at days 8 and 14 p.i. (Fig. 3B). A comparison of the intensity of bands on Southern blots was made between these FLK-exposed cytokine/CD154 iPfB cells, FLK-cellular DNA (four proviral copies per cell), and a fivefold dilution of DNA isolated from M267 cells, which harbor a single proviral copy. From this comparison, we concluded that there was approximately 1 provirus copy for every 5 cells in the FLK-exposed iPfB cells.
There are multiple provirus integration sites in BLV-infected B cells prior to the generation of a leukemic B-cell clone. EcoRI cleaves at one location in the BLV genome (Fig. 2), and hybridization with a full-length BLV probe generates both a 5′ and a 3′ proviral DNA-genomic DNA flanking sequence fragment for each integrated provirus copy. EcoRI-digested DNA from FLK-exposed cytokine/CD154 iPfB-cell cultures did not generate discrete bands, as would be expected if the provirus integration site was identical in each cell (data not shown). This observation was consistent with provirus integration at multiple sites in the genome of FLK-exposed cytokine/CD154 iPfB cells. Collectively, our data indicate that the immature, rapidly dividing, sIgM+ B cells isolated from sheep ileal Peyer's patches were permissive for BLV infection.
BLV transcription is initiated when infected leukocytes are isolated from the blood of aleukemic animals and cultured ex vivo (20, 34). In contrast, viral transcription is absent in transformed B-cell lines derived from leukemic sheep (41–43). Thus, it was of interest to determine whether viral RNA was expressed following BLV infection of iPfB cells. Analysis of viral gene transcription first focused on the tax/rex transcripts. Transcriptional activity was evaluated by RT-PCR using the doubly spliced tax/rex RNA splice-specific primers EnvA and Can2 (Fig. 2) and Southern blot analysis with a full-length BLV probe (Fig. 5). tax/rex transcripts were detected in both the short-term and the cytokine/CD154 FLK-exposed iPfB-cell cultures as well as in YR2LTaxSN cells, which constitutively express the virus. These data confirmed that the doubly spliced BLV transcripts were synthesized in the BLV-infected iPfB cells. Furthermore, since it has been shown that the presence of singly spliced BLV env transcripts parallels transcription of full-length BLV gag-pol RNA during both early and late expression (34), we also used the EnvA and EnvC set of primers (Fig. 2) and found that both mRNA species were present (data not shown). The use of EnvC as a complementary primer eliminated the possibility of amplifying doubly spliced mRNA, as this primer is located downstream of the second splice donor but upstream of the second splice acceptor site, in the second intron of the tax/rex transcript. Thus, it was evident that viral transcription was occurring following infection by the FLK supernatant virus.
FIG. 5.
Amplification of tax/rex BLV transcripts in FLK-exposed B-cell cultures. DNase I-treated total RNA preparations were amplified by RT-PCR with BLV primers EnvA and Can2 and analyzed by Southern blot hybridization with a 32P-labeled full-length BLV probe. Samples analyzed included FLK-exposed 96-h culture of short-term iPfB cells (iPfB 96h BLV); 8- and 14-day cultures of cytokine/CD154 iPfB-cells (iPfB 8d BLV and iPfB 14d BLV); blood-derived B cells cultured for 60 days (BL-2); and 48-day cultures of jPfB clone 2 and clone 4 cells (jPfB Clone2 BLV and jPfB Clone4 BLV). Control samples analyzed included OVK-exposed short-term iPfB cells (iPfB 96h cntl); cytokine/CD154 iPfB cells (iPfB 8d cntl); and blood-derived B cells (L-2). A 10-fold dilution of YR2LTaxSN was used as a positive control. Amplified products were 193 bp in length.
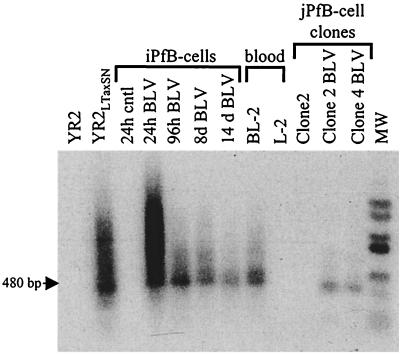
Circular extrachromosomal DNA is always present during the acute phase of retroviral infection in vitro and in vivo and for this reason is also considered a marker of active viral replication (3). Extrachromosomal BLV proviral forms have been detected during the early phase of infection (24), and in vitro studies have demonstrated that an accumulation of unintegrated circular BLV DNA resulted from a process of reinfection rather than intracellular reverse transcription of newly synthesized BLV RNA (36). Therefore, we screened FLK-exposed B-cell cultures for viral replication by a qualitative PCR method with the U3/U5 primer pair (Fig. 2) that has been shown to selectively amplify circular BLV DNA having two linked LTRs (47). Amplification products of 480 bp were generated with DNA isolated from the short-term and the cytokine/CD154 FLK-exposed iPfB-cell cultures and from YR2LTaxSN cells, which are productively infected with BLV (Fig. 6). As expected, the latently infected YR2 cell line was negative. The presence of circular forms supports the conclusion that viral integration and transcription were also associated with viral replication in both the short-term and cytokine/CD154 iPfB-cell cultures.
FIG. 6.
Detection of circular nonintegrated provirus forms in FLK-exposed B-cell cultures. Circular extrachromosomal DNA with two linked LTRs was amplified with the U3/U5 primer pair, and amplicons were hybridized with a 32P-labeled full-length BLV probe. Samples analyzed included FLK-exposed 24- and 96-h-cultured short-term iPfB cells (iPfB 24h BLV and iPfB 96h BLV); 8- and 14-day cultures of cytokine/CD154 iPfB cells (iPfB 8d BLV and iPfB 14d BLV); blood-derived B cells cultured for 60 days (BL-2); and 48-day cultures of jPfB clone 2 and clone 4 B cells (jPfB Clone2 BLV and jPfB Clone4 BLV). Control samples analyzed included the following OVK-exposed B cells: iPfB cells (24h cntl); blood-derived B cells (L-2); and jPfB clone 2 cells (Clone2 cntl). DNA from productively infected YR2LTaxSN cells and the YR2 cell line, with a silent provirus, were included as positive and negative controls, respectively. The amplified products were 480 bp in length. MW, molecular size markers.
Collectively, our observations indicate that freshly isolated iPfB cells are permissive for BLV infection by virus particles present in the supernatant of FLK cells. Furthermore, provirus expression is sustained within the B-cell cultures despite the low percentage of infected cells and the high levels of cell proliferation and cell death that occur in this culture system. Therefore, features of a patent viral infection were observed in these cultures.
BLV infection of B cells from blood.
BLV infection in sheep most frequently results in B-cell leukemia. Therefore, we investigated whether it was possible to also infect B cells present in blood. Freshly isolated blood mononuclear cells were incubated with either BLV-negative OVK or BLV-positive FLK culture supernatant for 96 h. These cultures are hereafter referred to as L-2 and BL-2 cultures, respectively. To support the specific growth of blood B cells, it was necessary to cocultivate blood lymphocytes with murine CD154 and recombinant human IL-2. Blood mononuclear cells were passaged and restimulated every 3 to 4 days p.i. Following a 3-week culture period, flow cytometric analysis revealed that over 97% of viable cells were B cells (CD72+, MHC class II+, and either sIgM+ or sIgG1+), and there were no detectable T cells (CD4+, CD8+, or γδTCR+) (data not shown).
DNA from L-2 and BL-2 cells cultured for 60 days p.i. were analyzed for the presence of proviral sequences and viral expression as described for the iPfB-cell cultures. The BL-2 but not the L-2 cells were positive in PCR amplification with BLV-specific primers (Fig. 3A), and Southern blot analysis indicated a low provirus load of approximately 1 provirus copy per 5 BL-2 cells (Fig. 3B). Furthermore, BLV provirus was actively transcribed in BL-2 cells, with doubly spliced BLV tax/rex mRNA (Fig. 5), and both mRNAs for the structural proteins were detected by RT-PCR (data not shown). Finally, BL-2 cells were examined for viral replication activity, and as expected, PCR analysis revealed the presence of circular nonintegrated forms (Fig. 6).
In conclusion, we demonstrated that murine CD154-activated blood B cells were permissive for BLV infection. Coculture of B cells with murine CD154 and recombinant human IL-2 supported the growth of infected B cells in which there was replication and transcription of proviral DNA. Furthermore, a low level of BLV infection persisted for at least 60 days under these culture conditions. Thus, it is evident that BLV can infect not only the immature, rapidly dividing B cells present in the ileal Peyer's patch but also activated B cells isolated from blood.
BLV infection of jejunal Peyer's patch B-cell clones.
The experiments with B cells cultured from ileal Peyer's patch and blood indicated that B cells were permissive for BLV infection. However, the initial cultures from blood contain numerous monocytes (8), and macrophages and mesenchymal cells are present in cell suspensions prepared from ileal Peyer's patch lymphoid follicles (12). Therefore, it was possible that the initial BLV infection in these cultures involved cells other than B lymphocytes. To conclusively prove that BLV infection and viral expression can involve only B cells, we used two cloned B-cell populations derived from the jejunal Peyer's patch of a lamb (15). Furthermore, we chose an sIgM+ (clone 2) and an sIgG1+ (clone 4) B-cell clone to determine if the state of B-cell differentiation restricted BLV infection.
Clone 2 and clone 4 cells were exposed for 96 h to culture supernatant from either FLK or OVK cells. The presence of proviral DNA, nonintegrated circular forms, and BLV transcripts was evaluated by PCR and RT-PCR in cell pellets collected at days 10, 20, and 48 p.i. These analyses revealed that the provirus was present and transcribed in both the clone 2 (sIgM+) and the clone 4 (sIgG+) FLK-exposed B cells. Southern blot analysis of SacI DNA digests suggested that approximately 1 of every 10 cells had integrated provirus. The results from samples collected at day 48 p.i. are shown in Fig. 3A, 3B, 5, and 6. There was no significant difference in viable B-cell number when the growth of BLV-infected cells was compared to that of uninfected (OVK supernatant) B-cell clones (data not shown). Thus, BLV infection in approximately 10% of B cells did not have a significant impact on the growth of these cloned B-cell populations.
In conclusion, by using cloned B cells, we clearly demonstrated that BLV can directly infect ovine B cells. Furthermore, we determined that BLV replication can occur in a pure B-cell population and that viral transcription is T-cell independent. Also, by using clone 2 (sIgM+) and clone 4 (sIgG+) B cells, we demonstrated that in vitro BLV infection is not limited to sIgM+ B cells.
Production of infectious virus by infected B cells.
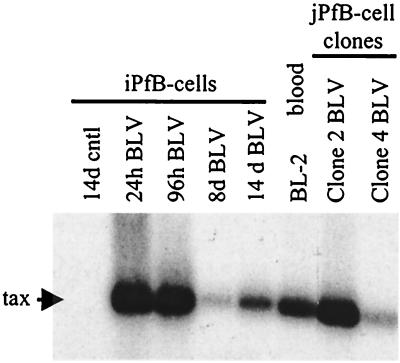
The above results analyzed viral replication and transcription but did not address the production of functional viral particles. To determine if the B cells infected in vitro with BLV were productively infected, we used an in vivo infection assay (40). This experimental approach is the most sensitive method for studying BLV infectivity (26, 40, 41, 48, 49). Furthermore, the in vivo ovine model is unique, because it allows us to address questions regarding retrovirus pathogenesis in a true biological context. Briefly, sheep were inoculated intradermally with 107 cells from each of the B-cell cultures (two animals per cell type for FLK-exposed B cells and one animal per cell type for control, OVK-exposed B cells). All animals injected with FLK-exposed B cells seroconverted within 2 to 3 weeks, whereas control animals remained seronegative. Similarly, PCR analysis with BLV-specific primers indicated that all sheep inoculated with FLK-exposed B cells had detectable provirus in blood by 3 weeks after injection (Fig. 7). Furthermore, blood samples were collected for over 1 year, and both antibody titers and BLV provirus loads increased gradually throughout this period (data not shown). These observations clearly demonstrated that the FLK-exposed B cells were productively infected by BLV. Since the initial in vitro infection was mediated though a cell-free viral supernatant procedure and not by a cocultivation strategy, there was no possibility of virus transmission by contaminating FLK cells. In conclusion, the in vivo experiments clearly demonstrated that B cells derived from a variety of ovine lymphoid tissues were productively infected in vitro by BLV.
FIG. 7.
PCR analysis of proviral sequences in blood samples from sheep inoculated with BLV-infected B cells. Blood was collected weekly following the injection of 107 BLV-infected or control B cells. Proviral sequences were amplified with BLV tax primers and analyzed by Southern blot hybridization with a 32P-labeled full-length BLV probe. BLV sequences were detected in blood samples collected 3 weeks p.i. from sheep injected with the following FLK-exposed B cell types (one animal per cell type): 24- and 96-h cultures of short-term iPfB cells (iPfB 24h BLV and iPfB 96h BLV); 8- and 14-day cultures of cytokine/CD154 iPfB cells (iPfB 8d BLV and iPfB 14d BLV); blood-derived B cells cultured for 60 days (BL-2); and 48-day cultures of jPfB clone 2 and clone 4 B cells (jPfB Clone2 BLV and jPfB Clone4 BLV). Control lane, sheep injected with OVK-exposed cytokine/CD154 14d iPfB cells (14d cntl).
DISCUSSION
The present investigation is the first demonstration of an in vitro culture system to examine B-cell infection by BLV. Integrated and extrachromosomal provirus forms were detected following BLV infection of ovine B cells isolated from a variety of lymphoid tissues. Furthermore, the presence of viral transcripts was demonstrated using RT-PCR, and in vivo infection experiments clearly demonstrated that infected B cells produced infectious virus. Finally, conclusive evidence of direct B-cell infection by BLV was obtained through the use of cloned B-cell populations. Collectively, these data confirmed that ovine B cells isolated from a variety of lymphoid tissues were productively infected in vitro with BLV.
The initial analysis of BLV-infected iPfB cells was limited by extensive apoptotic cell death and increased nuclease activity in these short-term cultures (30). The high mitotic index in these cultures, however, might have facilitated proviral integration. In fact, a high level of cell proliferation occurs in all ovine B-cell cultures costimulated with murine CD154 and exogenous γ-chain-common cytokines (12). This CD154 costimulation system was used to further investigate the permissiveness of B cells from blood and jejunal Peyer's patches to BLV infection. From these experiments, it was evident that BLV could infect not only the immature, rapidly dividing B cells present in ileal Peyer's patch but also the activated B cells isolated from blood and jejunal Peyer's patches.
PCR amplification revealed that all FLK-exposed B cells had detectable provirus, whatever the culture conditions and the length of the culture period. Evidence of provirus integration was provided by Southern blot experiments with restricted genomic DNA. Provirus load was consistently low, as expected following in vitro infection with viral supernatants. Despite the fact that this infection method is not as efficient as a cocultivation procedure, it was used to eliminate the possibility of detecting viral sequences from contaminating FLK producer cells. If it is assumed that each infected cell contained a single copy of provirus, then approximately 10 to 20% of cultured B cells had integrated provirus. This estimation of infected-cell frequency was based on integrated provirus forms, since much lower levels of circular, nonintegrated DNA are usually present during the course of a retroviral infection (3).
Circular extrachromosomal DNA is considered a marker of active viral replication and is always present during the acute phase of an in vitro or in vivo infection (reviewed in reference 3). We used a PCR method to specifically detect this form in the presence of both integrated and linear nonintegrated viral DNA (36). This PCR analysis detected circular extrachromosomal DNA in all BLV-infected B-cell cultures and provided evidence that viral replication was occurring in these cells. Although circular DNA is transcriptionally active, its integrated counterpart is a more efficient template for transcription and possesses a much longer half-life. Thus, the majority of viral mRNA necessary to produce viral proteins and, consequently, the majority of infectious particles are produced from integrated DNA. With the use of RT-PCR techniques, cultured B cells isolated from blood or jejunal or ileal Peyer's patches consistently produced genomic and subgenomic RNA following BLV infection. Thus, BLV infection and replication were not limited by any of the target B-cell populations that were evaluated.
The variety of culture systems used throughout the present investigation facilitated an analysis of the interaction between BLV and B cells. With long-term B-cell cultures, it was possible to analyze the persistence of BLV infection. Detection of BLV transcripts at 60 days p.i. confirmed that BLV infection could persist for a prolonged period in the B-cell cultures. Experiments with cloned sheep B cells provided conclusive proof of direct B-cell infection by BLV and clearly demonstrated that BLV infection was not restricted to sIgM+ B cells. The data from cloned B cells also confirmed that BLV replication could occur in a pure B-cell population. This observation supports the conclusion that BLV transcription is not T-cell dependent (20). The effect of BLV infection on B-cell survival and proliferation varied with culture conditions and the target B-cell population. This suggests that external stimuli and the state of B-cell activation may significantly influence the fate of a B cell following BLV infection. For example, the reduced level of cell death in iPfB-cell cultures costimulated with human IL-2 and -4 might be consistent with a previous report that BLV infection protects blood lymphocytes from ex vivo spontaneous apoptosis in sheep (4). Thus, the development of an in vitro model for BLV infection of B cells provides a novel approach to investigating the interaction between a retrovirus and B cells and to identifying factors that might influence this interaction.
Finally, the injection of BLV-infected B cells into naive sheep provided a sensitive method for detecting the production of functional viral particles. Inoculation with FLK-exposed B cells induced seroconversion, and provirus was first detected in blood samples at 3 weeks p.i. A gradual increase in virus load and antibody titers over a 1-year period confirmed that viral replication was occurring in the challenged animals. The 3-week delay in detecting provirus in blood and a gradual increase in virus load ruled out the possibility that inoculated, allogeneic B cells were the source of the provirus detected. Furthermore, it was previously shown that ovine B cells costimulated with murine CD154 and γ-chain-common cytokines die within 5 to 7 days after being deprived of these stimuli (13). The in vivo infection experiments with BLV-infected B-cell cultures proved that infection-competent progeny virus were produced following BLV infection of ovine B-cell cultures. Thus, the present investigation conclusively demonstrated that BLV can infect and replicate in both immature and mature ovine B cells. Ovine B-cell cultures permissive for BLV infection might provide a very useful model for investigating the leukemogenic process during B-cell transformation by a retrovirus.
ACKNOWLEDGMENTS
This work was supported by funds from the Fonds MEDIC, the Fonds National de la Recherche Scientifique, the Fondation BEKALES, the Fondation Rose et Jean Hoguet, NATO Collaborative Research grant 960219, the Alberta Agriculture Research Institute (AARI), and the Saskatchewan Health Services Utilization and Research Commission (HSURC).
REFERENCES
- 1.Birkebak T A, Palmer G H, Davis W C, Knowles D P, McElwain T F. Association of GP51 expression and persistent CD5+ B-lymphocyte expansion with lymphomagenesis in bovine leukemia virus infected sheep. Leukemia. 1994;8:1890–1899. [PubMed] [Google Scholar]
- 2.Burny A, Willems L, Callebaut I, Adam E, Cludts I, Dequiedt F, Droogmans L, Grimonpont C, Kerkhofs P, Mammerickx M, Portetelle D, Van den Broeke A, Kettmann R. Bovine leukemia virus: biology and mode of transformation. In: Minson A C, Neil J C, McRae M A, editors. Viruses and cancer. Cambridge, U.K: Cambridge University Press; 1994. pp. 213–234. [Google Scholar]
- 3.Cara A, Reitz M S J. New insight on the role of extrachromosomal retroviral DNA. Leukemia. 1997;11:1395–1399. doi: 10.1038/sj.leu.2400776. [DOI] [PubMed] [Google Scholar]
- 4.Dequiedt F, Hanon E, Kerkhofs P, Pastoret P P, Portetelle D, Burny A, Kettmann R, Willems L. Both wild-type and strongly attenuated bovine leukemia viruses protect peripheral blood mononuclear cells from apoptosis. J Virol. 1997;71:630–639. doi: 10.1128/jvi.71.1.630-639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987;61:2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derse D, Martarano L. Construction of a recombinant bovine leukemia virus vector for analysis of virus infectivity. J Virol. 1990;64:401–405. doi: 10.1128/jvi.64.1.401-405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domenech A, Llames L, Goyache J, Suarez G, Gomez-Lucia E. Macrophages infected with bovine leukaemia virus (BLV) induce humoral response in rabbits. Vet Immunol Immunopathol. 1997;58:309–320. doi: 10.1016/s0165-2427(97)00043-3. [DOI] [PubMed] [Google Scholar]
- 8.Dudler L, Griebel P, Hein W. Separation of mononuclear cells from blood. In: Lefkovits I, editor. Immunological methods manual. London, U.K: Academic Press; 1997. pp. 2075–2078. [Google Scholar]
- 9.Felber B K, Derse D, Athanassopoulos A, Campbell M, Pavlakis G N. Cross-activation of the Rex proteins of HTLV-I and BLV and of the Rev protein of HIV-1 and nonreciprocal interactions with their RNA responsive elements. New Biol. 1989;1:318–328. [PubMed] [Google Scholar]
- 10.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 11.Gallo R C, Nerurkar L S. Human retroviruses: their role in neoplasia and immunodeficiency. Ann NY Acad Sci. 1989;567:82–94. doi: 10.1111/j.1749-6632.1989.tb16461.x. [DOI] [PubMed] [Google Scholar]
- 12.Griebel P. Culture of ileal Peyer's patches B cells. In: Lefkovits I, editor. Immunological methods manual. London, U.K: Academic Press; 1997. pp. 2079–2086. [Google Scholar]
- 13.Griebel P, Beskorwayne T, Van den Broeke A, Ferrari G. CD40 signaling induces B cell responsiveness to multiple members of the gamma chain-common cytokine family. Int Immunol. 1999;11:1139–1147. doi: 10.1093/intimm/11.7.1139. [DOI] [PubMed] [Google Scholar]
- 14.Griebel P, Ferrari G. CD40 signalling in ileal Peyer's patch B cells: implications for T cell-dependent antigen selection. Int Immunol. 1995;7:369–379. doi: 10.1093/intimm/7.3.369. [DOI] [PubMed] [Google Scholar]
- 15.Griebel P J, Beskorwayne T, Godson D L, Popowych Y, Hein W. Cloning non-transformed sheep B cells. J Immunol Methods. 2000;237:19–28. doi: 10.1016/s0022-1759(99)00247-1. [DOI] [PubMed] [Google Scholar]
- 16.Griebel P J, Davis W C, Reynolds J D. An analysis of the growth and differentiation of B cells isolated from follicles of the ileal Peyer's patch of sheep. Immunology. 1992;75:601–607. [PMC free article] [PubMed] [Google Scholar]
- 17.Griebel P J, Ferrari G. Evidence for a stromal cell-dependent, self-renewing B cell population in lymphoid follicles of the ileal Peyer's patch of sheep. Eur J Immunol. 1994;24:401–409. doi: 10.1002/eji.1830240220. [DOI] [PubMed] [Google Scholar]
- 18.Griebel P J, Ghia P, Grawunder U, Ferrari G. A novel molecular complex expressed on immature B cells: a possible role in T cell-independent B cell development. Dev Immunol. 1996;5:67–78. doi: 10.1155/1996/21973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heeney J L, Valli P J, Jacobs R M, Valli V E. Evidence for bovine leukemia virus infection of peripheral blood monocytes and limited antigen expression in bovine lymphoid tissue. Lab Investig. 1992;66:608–617. [PubMed] [Google Scholar]
- 20.Jensen W A, Sheehy S E, Fox M H, Davis W C, Cockerell G L. In vitro expression of bovine leukemia virus in isolated B-lymphocytes of cattle and sheep. Vet Immunol Immunopathol. 1990;26:333–342. doi: 10.1016/0165-2427(90)90117-b. [DOI] [PubMed] [Google Scholar]
- 21.Kerkhofs P, Heremans H, Burny A, Kettmann R, Willems L. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J Virol. 1998;72:2554–2559. doi: 10.1128/jvi.72.3.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kettmann R, Burny A, Callebaut I, Droogmans L, Mammerickx M, Willems L, Portetelle D. Bovine leukemia virus. In: Levy J A, editor. The retroviridae. New York, N.Y: Plenum Press; 1994. pp. 39–81. [Google Scholar]
- 23.Kettmann R, Cleuter Y, Gregoire D, Burny A. Role of the 3′ long open reading frame region of bovine leukemia virus in the maintenance of cell transformation. J Virol. 1985;54:899–901. doi: 10.1128/jvi.54.3.899-901.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettmann R, Mammerickx M, Portetelle D, Gregoire D, Burny A. Experimental infection of sheep and goat with bovine leukemia virus: localization of proviral information on the target cells. Leukocyte Res. 1984;8:937–944. doi: 10.1016/0145-2126(84)90047-x. [DOI] [PubMed] [Google Scholar]
- 25.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammerickx M, Palm R, Portetelle D, Burny A. Experimental transmission of enzootic bovine leukosis to sheep: latency period of the tumoral disease. Leukemia. 1988;2:103–107. [PubMed] [Google Scholar]
- 27.Meirom R, Moss S, Brenner J. Bovine leukemia virus-gp51 antigen expression is associated with CD5 and IgM markers on infected lymphocytes. Vet Immunol Immunopathol. 1997;59:113–119. doi: 10.1016/s0165-2427(97)00056-1. [DOI] [PubMed] [Google Scholar]
- 28.Milan D, Nicolas J F. Activator-dependent and activator-independent defective recombinant retroviruses from bovine leukemia virus. J Virol. 1991;65:1938–1945. doi: 10.1128/jvi.65.4.1938-1945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirsky M L, Olmstead C A, Da Y, Lewin H A. The prevalence of proviral bovine leukemia virus in peripheral blood mononuclear cells at two subclinical stages of infection. J Virol. 1996;70:2178–2183. doi: 10.1128/jvi.70.4.2178-2183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motyka B, Reynolds J D. Apoptosis is associated with the extensive B cell death in the sheep ileal Peyer's patch and the chicken bursa of Fabricius: a possible role in B cell selection. Eur J Immunol. 1991;21:1951–1958. doi: 10.1002/eji.1830210825. [DOI] [PubMed] [Google Scholar]
- 31.Murakami K, Aida Y, Kageyama R, Numakunai S, Ohshima K, Okada K, Ikawa Y. Immunopathologic study and characterization of the phenotype of transformed cells in sheep with bovine leukemia virus-induced lymphosarcoma. Am J Vet Res. 1994;55:72–80. [PubMed] [Google Scholar]
- 32.Murakami K, Okada K, Ikawa Y, Aida Y. Bovine leukemia virus induces CD5− B cell lymphoma in sheep despite temporarily increasing CD5+ B cells in asymptomatic stage. Virology. 1994;202:458–465. doi: 10.1006/viro.1994.1362. [DOI] [PubMed] [Google Scholar]
- 33.Portetelle D, Mammerickx M, Burny A. Use of two monoclonal antibodies in an ELISA test for the detection of antibodies to bovine leukaemia virus envelope protein gp51. J Virol Methods. 1989;23:211–222. doi: 10.1016/0166-0934(89)90135-3. [DOI] [PubMed] [Google Scholar]
- 34.Powers M A, Radke K. Activation of bovine leukemia virus transcription in lymphocytes from infected sheep: rapid transition through early to late gene expression. J Virol. 1992;66:4769–4777. doi: 10.1128/jvi.66.8.4769-4777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pozzatti R, Vogel J, Jay G. The human T-lymphotropic virus type I tax gene can cooperate with the ras oncogene to induce neoplastic transformation of cells. Mol Cell Biol. 1990;10:413–417. doi: 10.1128/mcb.10.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyes R A, Cockerell G L. Unintegrated bovine leukemia virus DNA: association with viral expression and disease. J Virol. 1996;70:4961–4965. doi: 10.1128/jvi.70.8.4961-4965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rovnak J, Boyd A L, Casey J W, Gonda M A, Jensen W A, Cockerell G L. Pathogenicity of molecularly cloned bovine leukemia virus. J Virol. 1993;67:7096–7105. doi: 10.1128/jvi.67.12.7096-7105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz I, Bensaid A, Polack B, Perrin B, Berthelemy M, Levy D. In vivo leukocyte tropism of bovine leukemia virus in sheep and cattle. J Virol. 1994;68:4589–4596. doi: 10.1128/jvi.68.7.4589-4596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van den Broeke A, Bagnis C, Ciesiolka M, Cleuter Y, Gelderblom H, Kerkhofs P, Griebel P, Mannoni P, Burny A. In vivo rescue of a silent tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type tax gene. J Virol. 1999;73:1054–1065. doi: 10.1128/jvi.73.2.1054-1065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van den Broeke A, Cleuter Y, Chen G, Portetelle D, Mammerickx M, Zagury D, Fouchard M, Coulombel L, Kettmann R, Burny A. Even transcriptionally competent proviruses are silent in bovine leukemia virus-induced sheep tumor cells. Proc Natl Acad Sci USA. 1988;85:9263–9267. doi: 10.1073/pnas.85.23.9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van den Broeke A, Cleuter Y, Droogmans L, Burny A, Kettmann R. Isolation and culture of B lymphoblastoid cell lines from bovine leukemia virus-induced tumors. In: Lefkovits J, editor. Immunological methods manual. London, U.K: Academic Press; 1997. pp. 2127–2132. [Google Scholar]
- 43.Van den Broeke A, Cleuter Y, Portetelle D, Mammerickx M, Kettmann R, Burny A. Viral expression in bovine leukemia virus-induced tumor cells. Dev Biol Stand. 1990;72:77–80. [PubMed] [Google Scholar]
- 44.Van Der Maaten M J, Miller J M. Replication of bovine leukemia virus in monolayer cell cultures. Bibl Haematol. 1976;43:360–362. doi: 10.1159/000399166. [DOI] [PubMed] [Google Scholar]
- 45.Vernau W, Jacobs R M, Valli V E, Heeney J L. The immunophenotypic characterization of bovine lymphomas. Vet Pathol. 1997;34:222–225. doi: 10.1177/030098589703400307. [DOI] [PubMed] [Google Scholar]
- 46.Willems L, Heremans H, Chen G, Portetelle D, Billiau A, Burny A, Kettmann R. Cooperation between bovine leukaemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 1990;9:1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willems L, Kerkhofs P, Burny A, Mammerickx M, Kettmann R. Lack of LTR and ENV genetic variation during bovine leukemia virus-induced leukemogenesis. Virology. 1995;206:769–772. doi: 10.1016/s0042-6822(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 48.Willems L, Kerkhofs P, Dequiedt F, Portetelle D, Mammerickx M, Burny A, Kettmann R. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc Natl Acad Sci USA. 1994;91:11532–11536. doi: 10.1073/pnas.91.24.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willems L, Kettmann R, Dequiedt F, Portetelle D, Voneche V, Cornil I, Kerkhofs P, Burny A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willems L, Portetelle D, Kerkhofs P, Chen G, Burny A, Mammerickx M, Kettmann R. In vivo transfection of bovine leukemia provirus into sheep. Virology. 1992;189:775–777. doi: 10.1016/0042-6822(92)90604-n. [DOI] [PubMed] [Google Scholar]
- 51.Yip M T, Chen I S. Modes of transformation by the human T-cell leukemia viruses. Mol Biol Med. 1990;7:33–44. [PubMed] [Google Scholar]