Abstract
Hepatitis B virus (HBV), the causative agent of B-type hepatitis in humans, is a hepatotropic DNA-containing virus that replicates via reverse transcription. Because of its narrow host range, there is as yet no practical small-animal system for HBV infection. The hosts of the few related animal viruses, including woodchuck hepatitis B virus and duck hepatitis B virus, are either difficult to keep or only distantly related to humans. Some evidence suggests that tree shrews (tupaias) may be susceptible to infection with human HBV, albeit with low efficiency. Infection efficiency depends on interactions of the virus with factors on the surface and inside the host cell. To bypass restrictions during the initial entry phase, we used recombinant replication-defective adenovirus vectors, either with or without a green fluorescent protein marker gene, to deliver complete HBV genomes into primary tupaia hepatocytes. Here we show that these cells, like the human hepatoma cell lines HepG2 and Huh7, are efficiently transduced by the vectors and produce all HBV gene products required to generate the secretory antigens HBsAg and HBeAg, replication-competent nucleocapsids, and enveloped virions. We further demonstrate that covalently closed circular HBV DNA is formed. Therefore, primary tupaia hepatocytes support all steps of HBV replication following deposition of the genome in the nucleus, including the intracellular amplification cycle. These data provide a rational basis for in vivo experiments aimed at developing tupaias into a useful experimental animal system for HBV infection.
Hepatitis B virus (HBV), an enveloped DNA-containing virus that replicates through an RNA intermediate (38), is the type member of the family Hepadnaviridae and the causative agent of B-type hepatitis in humans. Chronic infections are widespread and associated with a greatly increased risk for the development of liver cirrhosis and, eventually, primary liver carcinoma (5, 35). Several fundamental aspects of the HBV replication cycle have been elucidated, mainly by genetic techniques using transfection of cloned HBV DNA into suitable human hepatoma cell lines, such as Huh7 and HepG2 (27, 28). These cells support virus production, but neither they nor other cell lines can be infected. A related and similarly central problem is the lack of a feasible small-animal model of human HBV infection.
All hepadnaviruses have very narrow host ranges. Efficient infection by HBV is well documented for only humans and chimpanzees and, in cell culture, for primary hepatocytes from these hosts. The current generic animal models, the woodchuck-woodchuck hepatitis B virus (WHV) and the pekin duck-duck hepatitis B virus systems (28), are useful in many respects but suffer from two different limitations. Woodchucks are difficult to keep, and their use is restricted to a few appropriately equipped facilities; even then, most studies have used outbred, wild-caught animals (33). Ducks, by contrast, are convenient experimental animals (19) but very distantly related to humans.
An ideal experimental system would consist of HBV, or a very closely related virus, plus a suseptible host that is as easily kept and as well characterized as the mouse. Some mouse-based surrogate systems are available; however, although HBV-transgenic mice (1, 9) produce substantial amounts of virus (14), they are not infectable; mice carrying heterologous liver cells (18, 29, 32) require immunosuppression to prevent rejection of the xenotransplant. Because the hepadnavirus host range is controlled on several levels, many barriers would have to be surmounted in order to establish a productive infection in a nonauthentic host. Certainly important is the interaction with cell surface molecules that mediate virus entry (Fig. 1A). The large viral envelope protein is essential for these early steps (7) and, for duck hepatitis B virus, a cellular membrane-associated carboxypeptidase seems crucially involved in, but not sufficient for, infection (6, 21, 42). Virtually nothing is known about the receptors for the mammalian hepadnaviruses. Intracellularly, hepadnavirus replication depends on host factors that can interact with diverse cis elements on the viral genome. These are not only tissue specific but also species specific, as shown, e.g., by the inactivity of WHV enhancer I in human hepatoma cells (12). Further, virus-host interactions may also contribute to species specificity. An example is the apparent lack of covalently closed circular DNA (cccDNA) formation in HBV-transgenic mice. cccDNA is a central intracellular intermediate in the HBV replication cycle. It is initially formed after nuclear deposition, by the nucleocapsid (20), of the partially double-stranded DNA genome present in extracellular virions. During natural infection, new capsids deliver progeny genomes to the nucleus such that a pool of some 10 to 100 copies of cccDNA is established (41, 47). Hence, mice apparently lack factors required for both HBV uptake and intracellular amplification. The chances for transspecies transmission are therefore likely to be higher for a more closely related virus-host pair.
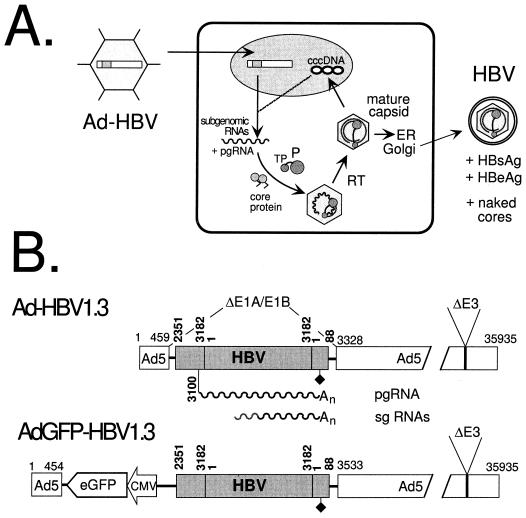
FIG. 1.
Ad vector-mediated HBV genome delivery. (A) Schematic outline of the transduction system. The replication-defective Ad-HBV vector infects an appropriate cell and deposits its recombinant genome in the nucleus. The authentic HBV regulatory elements on the transduced HBV genome control the transcription of subgenomic and genomic HBV RNAs, which serve as mRNAs. pgRNA is used to translate core and P protein and then is packaged into newly forming immature nucleocapsids. Using its terminal protein (TP) domain as a primer, P protein reverse transcribes (RT) the pgRNA, yielding mature DNA-containing nucleocapsids. These can either bud into the secretory pathway and be exported as enveloped virions or redirect the DNA genomes to the nucleus for conversion into cccDNA. In addition to virions, HBsAg and HBeAg, and possibly naked cores, are expected to be released from the transduced cells. ER, endoplasmic reticulum. (B) Replication-defective Ad-HBV vectors used in this study. The bars symbolize the Ad vector genomes (the horizontal numbers are nucleotide positions in Ad5) in which a 1.3× HBV genome (shaded; the vertical numbers are HBV nucleotide positions according to reference 30) replaces the E1A-E1B region; E3 is also deleted. HBV subgenomic (sg) and genomic transcripts (wavy lines) are controlled by the authentic elements, including the polyadenylation signal (diamond). pgRNA initiates at position 3100. Ad-GFP-HBV1.3, in addition, contains a CMV-driven enhanced GFP expression cassette. An, poly(A) tail.
One potential nonauthentic host for human HBV is the Asian tree shrew, or tupaia. Tupaias, classified in their own order (Scandentia), are the closest relatives of the primate lineage. Breeding colonies of the rat-size animals have been established at numerous places. Some evidence for their susceptibility to infection with human HBV has been reported (43, 48). Though the in vivo data were not really conclusive, results obtained with primary tupaia hepatocytes (PTHs) suggest that infection is possible (J. Köck, F. von Weizsäcker, and H. E. Blum, personal communication). However, the very low efficiency made it difficult to directly prove the net formation of complete progeny virions. We therefore reasoned that replacing the authentic entry process with a more effective HBV genome delivery procedure should allow us to investigate the ability of PTHs to support all intracellular steps of the hepadnavirus replication cycle. If so, this would provide a rational basis for in vivo experiments aimed at artificially initiating HBV infection in this species.
None of the established methods for delivery of cloned HBV DNA is ideal for in vivo applications. Naked DNA (45) and liposomes (39) reach only a limited number of hepatocytes. Recombinant baculoviruses are effective in cultured cells (10, 11), but the current vectors are subject to complement inactivation in vivo. By contrast, replication-defective adenovirus (Ad) vectors are widely used in gene therapy studies (2). They infect various cells from different hosts, including resting cells; they are relatively easily prepared at high titers by growth on cell lines complementing their replication defect, such as the human embryonal kidney line 293; and, with few exceptions (40), they appear to be safe.
We therefore generated replication-defective Ad vectors that carry complete HBV genomes under the control of the authentic regulatory elements (Fig. 1A). These Ad-HBV vectors (Fig. 1B) were used to transduce PTHs, and the generation of viral gene products and virions was compared to that observed in the human hepatoma cell lines Huh7 and HepG2. Our data indicate that (i) Ad vectors can efficiently infect primary hepatocytes from tupaias, (ii) transduction yields levels of hepadnavirus structural proteins that are as high as or higher than those obtained in human hepatoma cells, (iii) these gene products assemble into replication-competent hepadnavirus capsids that are released as complete virions, and (iv) primary tupaia hepatocytes support the formation of viral cccDNA.
MATERIALS AND METHODS
Cells and cell lines.
The human hepatoma cell lines Huh7 and HepG2 were grown under standard conditions as previously described (26). Cell densities at confluency were around 1.2 × 107 cells per 10-cm-diameter dish. For preparation of primary tupaia hepatocytes, animals from an in-house breeding colony were anesthesized, and the livers were surgically exposed and perfused using a two-step protocol as previously described (43). Cells were seeded on collagen I-coated plates (Becton Dickinson) at a density of 4 to 5 × 106/10-cm-diameter dish and maintained in BioCoat hepatocyte defined medium (Becton Dickinson). Viability was usually around 70 to 90%. The cells could routinely be maintained for at least 2 to 3 weeks.
Bacteria and plasmids.
Plasmids were amplified in Escherichia coli Top10 cells (Invitrogen), and homologous recombinations were performed in strain BJ5183 (8, 15). Shuttle plasmid pTG9530-HBV1.3 was generated by inserting, through several steps, a 1.3× HBV genome (subtype ayw), starting at nucleotide position 2351 (with position 1 corresponding to the first nucleotide of the core gene [30]), extending through a unit-length genome, and ending at position 88 after the polyadenylation signal, into the unique SalI restriction site in plasmid pTG9530 (8). The shuttle plasmid pAdTrack-HBV1.3 was similarly constructed by insertion of the same 1.3× HBV genome between the SalI and XhoI sites in plasmid pAdTrack (15). The shuttle plasmids and the corresponding Ad backbone plasmids pTG4656 and pAdEasy1, containing Ad5 genomes with E1A-E1B B plus E3 deletions, were kindly provided by M. Lusky (Transgene, Molsheim, France) and T. C. He (Howard Hughes Medical Institute, Baltimore, Md.).
Generation of recombinant Ads.
Recombination between the shuttle and the Ad backbone plasmids was performed essentially as described previously (8, 15). For the pTG system, recombinants were preselected by colony hybridization using an HBV-specific probe. Subsequently, the desired recombinants were identified by restriction digestion, usually after retransformation of the DNA minipreparations into Top10 cells to increase plasmid yields. Plasmid DNAs from one well-characterized clone each were used to excise the Ad vector part using the flanking PacI restriction sites. Linear DNAs were transfected into 293 cells using FuGENE 6 Reagent (Roche Diagnostics) according to the manufacturer's recommendations. After 8 to 10 days, vector particles were released from the cells by three freeze-thaw cycles, and the lysates were used to infect 293 cells (13). After reamplification, the resulting Ad-HBV1.3 and AdGFP-HBV1.3 viruses were concentrated by cesium chloride centrifugation. CsCl was removed by passage through a NAP-25 gel filtration column (Amersham-Pharmacia). The purified viruses were kept in storage buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 1 mg of bovine serum albumin/ml, 40% glycerol) at −20°C as liquid stocks. Physical titers were determined via absorbance at 260 nm, and infectious titers were determined by plaque assays on 293 cells (13). The yields from 40 10-cm-diameter dishes were usually between 3 × 1011 and 11 × 1011 PFU per ml, with physical titers being about 20-fold higher.
Infection of cultured cells.
Cells were counted after trypsin digestion using a hemocytometer. Virus stock aliquots containing the appropriate number of PFU to obtain the desired multiplicity of infecton (MOI) were mixed with 2 ml of Biocoat medium per 10-cm-diameter dish (PTHs) or with Dulbecco's modified Eagle medium (Huh7 and HepG2) and added to the cells. After 1.5 h at 37°C with occasional rocking, the medium was added to 10 ml. Media were changed every 2 days. Conditioned media were stored at 4°C. Cells were harvested at the desired times, washed with TBS buffer (25 mM Tris-Cl [pH 7.4] 137 mM NaCl, 2.7 mM KCl) and stored at −80°C. The total protein concentrations in cell lysates were determined using the Bradford assay (Bio-Rad).
Secretory HBV antigens.
HBsAg levels were determined using the HBsAg Monoclonal II enzyme-linked immunosorbent assay (ELISA) kit (Dade-Behring). Total amounts of HBsAg were calculated by comparison with serial dilutions of a standard HBsAg-positive human serum (a gift of W. Gerlich, University of Giessen). HBeAg was assayed with the IMx HBe 2 kit (Abbott) and compared to positive and negative control sera provided by the manufacturer.
Intracellular replication-competent core particles.
To release intracellular cores, cells were resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 100 μg of, RNase A/ml, 0.7% NP-40) and incubated at 37°C for 15 min. Nuclei and cellular debris were removed by centrifugation. To the supernatants, Mg2+ acetate (final concentration, 10 mM) and DNase I (final concentration, 500 μg/ml) were added, and the mixture was incubated at 37°C for 45 min to digest nonencapsidated DNA. For native agarose gel electrophoresis, about 1% of the lysate was loaded on a 1% agarose gel. The gel was blotted on a positively charged nylon membrane (Roche Diagnostics) by capillary transfer in TNE buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA). After being soaked in 0.2 N NaOH–150 mM NaCl and neutralized in 0.2 M Tris-HCl; (pH 7.5)–1.5 M NaCl (5 min each), the membrane was fixed by UV-cross-linking (1.5 J/cm2) and briefly rinsed with water. HBV-specific nucleic acids were detected with a digoxigeuin (DIG)-labeled probe obtained by random priming (DIG labeling and detection kit; Roche Diagnostics) on a 3.2-kb EcoRI fragment containing a complete linear HBV genome. Core protein was detected using the polyclonal rabbit anti-HBcAg antiserum H800 (4) and a peroxidase-conjugated secondary antibody with a chemiluminescent substrate (ECL-Plus; Amersham-Pharmacia).
Extracellular HBV DNA.
For enzymatic enrichment of HBV DNA contained in enveloped particles, a procedure similar to a previously reported method was used (44). In brief, particles from 5 to 20 ml of culture medium were precipitated by 10% polyethylene glycol (PEG 8000) for 1 h on ice. After centrifugation, the pellets were resuspended in 800 μl of 50 mM Tris (pH 8.0) and, after the addition of Mg2+ acetate to a final volume of 5 mM, incubated with 750 μg of pronase/ml at 37°C for 1 h and then with 500 μg of DNase I/ml. After 1 h, the samples were adjusted to 15 mM EDTA, 0.5% sodium dodecyl sulfate (SDS) and 500 μg of proteinase K/ml and incubated at 45°C for 1 h. After phenol extraction, the liberated nucleic acids were precipitated with ethanol and resuspended in Tris-EDTA (TE) buffer. For density gradient purification, the PEG pellets were resuspended in 1.5 ml of TNE buffer and loaded on CsCl step gradients (five steps, each of 2 ml, from 1.1 to 1.5 g/ml). After centrifugation at 32,000 rpm for 35 h at 4°C in a Kontron TST 41.14 rotor, 22 500-μl fractions were collected. Densities were determined via the refractive index, and HBsAg concentrations were determined by ELISA. For DNA extraction, appropriate gradient fractions were diluted with 2 volumes of 50 mM Tris-Cl (pH 8.0), Mg2+ acetate was added to 5 mM concentration, and the samples were incubated with DNase I (500 μg/ml) and RNase A (200 μg/ml), followed by SDS-proteinase K treatment and ethanol precipitation as described above. For Southern blotting, the same DIG-labeled HBV probe described above was used.
Western blot analysis of HBV envelope proteins.
Proteins from 5 μl of HBsAg-positive CsC1 fractions were separated by SDS-polyacrylamide gel electrophoresis (12.5% polyacrylamide, 0.1% SDS) and blotted to polyvinylidene difluoride membranes (Bio-Rad). Individual envelope proteins were detected with the following antibodies: human monoclonal antibody 4/7B (31), recognizing an epitope around amino acids 178 to 186 of S protein (a gift of R. A. Heijtink, Organon Teknika, Oss, The Netherlands); mouse monoclonal antibody S26 (25), specific for an epitope around the motif QDPR in the pre-S2 region (provided by V. Bichko, Scriptgen, Waltham, Mass.); and mouse monoclonal antibody MA18/07 (16, 37), directed against an epitope close to the N terminus of the pre-S1 domain (a gift of W. Gerlich). Detection was performed with peroxidase-conjugated secondary antibodies and the ECL-Plus system.
Nuclear cccDNA detection.
Episomal DNAs, including HBV cccDNA, were prepared by a modification (J. Summers, personal communication) of a previously described protocol (49). In brief, cells were resuspended in lysis buffer containing 0.2% NP-40, mixed with an equal volume of 0.15 N NaOH containing 6% SDS, incubated at 37°C for 20 min, and neutralized by adding 3 M K+ acetate (pH 5.5) to a final concentration of 0.6 M. After 30 min on ice, cellular debris and chromosomal DNA were removed by centrifugation at 20,000 × g for 15 min at 4°C. After phenol extraction, soluble nucleic acids in the supernatant were precipitated by adding 0.7 volumes of isopropanol. The pellets were resuspended in 500 μl of restriction buffer 4 (New England Biolabs) containing 40 U of HpaI/ml, 400 μg of RNase A/ml, and 300 U of Plasmid-Safe DNase (Epicentre Technologies)/ml and incubated at 37°C for 4 to 6 h. Subsequently, a second NaOH treatment was performed by adding 0.2 N NaOH to 0.05 N final concentration. After 10 min at 37°C, the reaction was neutralized by adding 3 M K+ acetate to 0.6 M final concentration, the samples were extracted with phenol, and nucleic acids were ethanol precipitated. Southern blot analysis was performed as described above.
RESULTS
Design and generation of recombinant Ad vectors carrying complete HBV genomes.
In order to test whether all HBV control elements are functional in PTHs, we used 1.3× overlength HBV genomes (subtype ayw) for generating the recombinant vectors. They imitate the circular HBV cccDNA as a transcriptional template by having duplicated terminal regions (Fig. 1B). The borders were essentially the same as those of an overlength genome used to generate HBV-transgenic mice that produce complete virions (14). The high liver specificity of most of the authentic hepadnavirus control elements significantly adds to the biosafety of the Ad-HBV vectors.
The previous tedious methods for generating recombinant Ad vectors have been superseded by procedures in which homologous recombination between a small, manipulatable shuttle plasmid and a large Ad backbone plasmid is performed entirely in a suitable E. coli strain. The resulting plasmid derivative of the recombinant Ad vector genome is characterized by conventional molecular biology methods, and transfection of the excised vector genome into suitable eukaryotic cells gives rise to a clonal population of Ad vector particles.
The vectors used in this study lack the E1A and E1B regions essential for initiating Ad replication and, in addition, the E3 region involved in subverting the host's immune response (46). Their replication is therefore restricted to a complementing cell line, such as 293. Two different systems were used. The first was that reported by Chartier et al. (8). The 1.3× HBV DNA was cloned into the shuttle plasmid pTG9530 and recombined with the Ad backbone plasmid pTG4656 (Fig. 1). The second was the AdEasy system (15), which utilizes a more advanced antibiotic selection scheme and, in addition, provides an enhanced green fluorescent protein (GFP) cassette in the Ad part. This allows for direct monitoring of virus production and infection efficiency by fluorescence microscopy. The yields of desired plasmid recombinants were variable with the first system, sometimes requiring colony hybridization to identify several potential positives within about 100 colonies. The second system was more robust, yielding between one and four positives among 10 plasmids analyzed. One well-defined clone obtained by each method was used to generate Ad-HBV1.3 and its counterpart, Ad-GFP-HBV1.3 (Fig. 1B). After amplification and concentration on CsCl gradients, both yielded infectious titers on 293 cells in the range of 3 × 1011 to 11 × 1011 PFU per ml.
Infection and Ad-HBV-mediated production of secretory HBV antigens in primary tupaia hepatocytes and Huh7 and HepG2 cells.
As a first test for successful transduction as well as potential cytotoxicity, PTHs and Huh7 and HepG2 cells were incubated with Ad-HBV1.3 and Ad-GFP-HBV1.3 at increasing MOIs. All of the cells were similarly susceptible to infection, since incubation at an MOI of 1 to 5 with the GFP vector induced GFP expression in the majority of cells (not shown). At higher doses, increasingly strong cytopathic effects (CPE) became macroscopically visible within the first few days postinfection (p.i.), eventually leading to the killing of most cells. The dose inducing this fast CPE differed substantially among the different cells and was also influenced by the vector backbone. Ad-HBV1.3 was tolerated by PTHs at higher doses (an MOI of 625) than those tolerated by Huh7 (MOI, 125), and HepG2 (MOI, 15) cells. Ad-GFP-HBV1.3 appeared to be toxic to PTHs above an MOI of 25, while no significant difference in the toxicity of Ad-HBV1.3 was observed with Huh7 and HepG2 cells. A similarly high relative toxicity for PTHs but not the hepatoma cell lines was also seen with a control vector containing only a cytomegalovirus (CMV) promoter-controlled GFP expression cassette (Ad-GFP) and a vector carrying a CMV-driven WHV genome but not with a 1.3× WHV construct (data not shown). This suggests that the CMV promoter, in an unknown fashion, contributes to the increased toxicity of the corresponding Ad vectors in PTHs.
The cultures were kept for up to 20 (PTH) or 30 (hepatoma lines) days, and aliquots from the culture supernatants, collected every 2 days, were analyzed by ELISA for HBsAg and HBeAg. For HBsAg, absolute concentrations were estimated by comparison with an HBsAg-positive standard serum. All of the cells supported the production of both secretory antigens (shown for PTHs and Huh7 cells [Fig. 2]). For the first few days, HBsAg increased in a dose-dependent manner, except at doses inducing fast CPE; this was particularly evident with Ad-GFP-HBV1.3 in PTHs, where the lowest dose (MOI, 5) gave the highest HBsAg titers (Fig. 2, middle-left graph). Infection of PTHs from the same preparation with Ad-HBV1.3 at an MOI of 25, by contrast, was again well tolerated and gave very similar values. With doses not inducing fast CPE, HBsAg expression peaked between days 4 and 10 p.i. and then declined. The slope was steeper at relatively high MOIs that had yielded higher antigen levels in the beginning. An accompanying decrease in cell numbers may reflect the fact that prolonged infection with the Ad vector, while not immediately killing the cells, still exerts a slow-acting CPE. However, the remaining cells were difficult to count because they grew in patchy islets; in addition, the metabolic activity and viability of the primary cells is known to decrease after a few days in culture.
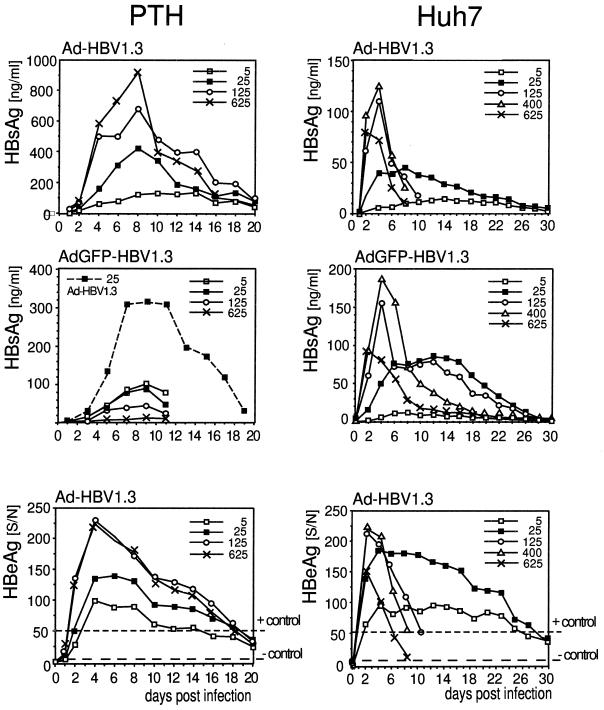
FIG. 2.
Dose dependence and time course of secretory HBV antigen production in transduced PTHs and Huh7 cells. Culture supernatants from the indicated cells infected with various MOIs of Ad-HBV1.3 and Ad-GFP-HBV1.3 were collected every 2 days. Aliquots were analyzed by ELISA for HBsAg (upper two rows) and for HBeAg (bottom row). HBeAg concentrations are indicated by S/N ratios (mean sample absorbance minus the background absorbance [S] divided by the background absorbance [N]), with the values obtained for positive and negative control sera indicated by the upper and lower dashed lines. Ad-GFP-HBV1.3 was much more toxic to PTHs than Ad-HBV1.3; hence, above an MOI of 5, the HBsAg values decreased rather than increased (middle-left panel). By contrast, a parallel infection of the same cell batch with Ad-HBV1.3 (dashed line) gave results similar to those obtained before.
As for the absolute amounts of HBsAg, PTHs gave clearly higher maximum levels (up to 900 ng/ml/2 days) than the human hepatoma cell lines (up to 200 ng/ml for Huh7 cells and up to 300 ng/ml for HepG2 cells), although the number of PTHs per dish was two- to threefold lower. To account for a potentially larger size and higher protein synthesis capacity of the primary cells, we compared the total protein concentrations in lysates from PTHs and Huh7 cells. PTHs consistently gave values between 7 and 8 mg/ml of lysate from a 10-cm-diameter dish, while the proliferating Huh7 cells yielded from 9 to 14 mg/ml. Hence, although the difference was less pronounced, the maximum amounts of HBsAg remained higher in PTHs than in Huh7 cells when normalized to total protein rather than cell numbers.
The HBeAg profiles, in general, paralleled those obtained for HBsAg, except that the maximum levels obtainable were not higher in PTHs than in Huh7 cells.
These data showed that the HBV S promoter and the core promoter-enhancer II responsible for transcription of the precore mRNA directing HBeAg synthesis are active in PTHs. The relatively higher production of HBsAg than of HBeAg in PTHs may reflect differential promoter activities but could also be influenced by the efficiencies of antigen secretion. The data further suggested that the initial dose-dependent increase in antigen production was due to an increased number of Ad vectors within one cell, which eventually reached a cytotoxic level. Because the number of viruses infecting one cell follows a Poisson distribution, it is plausible that the long-term-surviving cells at high MOIs were those that had received relatively few copies. Similar bell-shaped curves, with an initial dose-dependent signal increase and a subsequent decline due to increased cell death, were seen in all later titration experiments. Notably, during none of these experiments was any spread of the replication-defective vector observed on either type of cell either by plaque formation with Ad-HBV1.3 or by an increase of GFP-positive cells with AdGFP-HBV1.3. Hence, the presence of the entire HBV genome, including the HBx gene, did not complement the replication defect of the Ad vectors with E1A and E1B deleted.
Generation of intracellular replication-competent nucleocapsids.
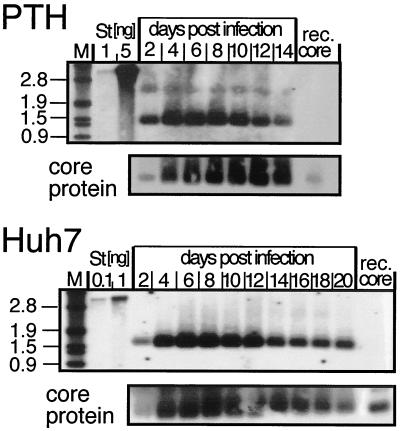
The formation of genome-containing nucleocapsids indicates the synthesis of functional pregenomic RNA (pgRNA), core protein, and P protein (Fig. 1A). We therefore compared intracellular nucleocapsid generation in PTHs and Huh7 cells transduced with both Ad vectors. In native agarose gel electrophoresis (4), intact cores migrate as distinct bands. After transfer to a membrane, core protein can be detected by antibodies and packaged nucleic acid can be detected by hybridization with an appropriate probe; while RNA may be partially degraded, DNA is stable under these conditions. When analyzed accordingly, intracellular nucleocapsids were detected in both PTHs and Huh7 cells. The dose dependence paralleled that of HBsAg and HBeAg production, with a similar cell- and vector-typical decline at high MOIs because of CPE. A time course for Ad-HBV1.3-transduced PTHs and Huh7 cells is shown in Fig. 3. The HBV DNA signals increased sharply from day 2 to day 4 and then slowly declined. Detection of core protein at the same positions indicated that the DNA was encapsidated. A semi quantitative determination of the DNA signals by densitometric scanning showed comparable intensities for the samples from PTHs, corresponding to 1/100 of the lysate from three dishes (total cell count, 1.3 × 107), and Huh7 cells, corresponding to 1/100 of the lysate from one dish (1.6 × 107 cells). Hence, nucleocapsid yields per dish, in contrast to HBsAg and in accord with HBeAg production, were not higher in PTHs than in Huh7 cells but almost identical on a per-cell basis. Therefore, it was evident that the tupaia cells support generation of functional HBV pgRNA and that the cell factors involved in nucleocapsid assembly (17) interact productively with the HBV gene products.
FIG. 3.
Time course of replication-competent-nucleocapsid formation in transduced PTHs and Huh7 cells. Nucleocapsids from cytoplasmic lysates of PTHs and Huh7 cells, infected at MOIs of 125 and 50, respectively, with Ad-HBV1.3, were prepared at the indicated times, subjected to gel electrophoresis in native agarose gels, and blotted on nylon membranes. DNA and core protein were detected by chemiluminescence, using a DIG-labeled HBV DNA probe (upper gels) and a core-specific antiserum (lower gels). M, DIG-labeled DNA marker, with fragments sizes in kilobases indicated on the left; St, linear 3.2-kb HBV genome; rec. core, 5 ng of particles from recombinant core protein amino acids 1 to 149.
Formation of extracellular enveloped virions in PTHs.
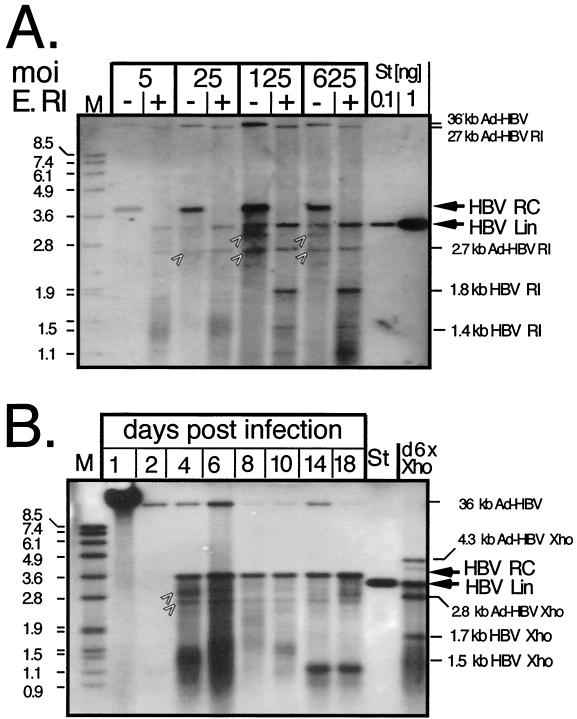
Next, we analyzed whether PTHs support HBV virion production. The first assay exploited protection by the lipid envelope of DNA in virions from the concerted action of pronase and DNase, whereas naked DNA, and DNA in nonenveloped particles, is digested. Virion DNA is subsequently isolated via proteinase K-SDS treatment (22). Extracellular particles from cells infected with increasing MOIs of Ad-HBV1.3 were concentrated by PEG precipitation. One part of each sample was digested with EcoRI to linearize the relaxed circular (RC) DNA, the major nucleic acid form expected in enveloped HBV virions. Southern blotting (Fig. 4A) showed a prominent, specifically hybridzing band slightly above the 3.6-kb marker whose intensity increased with vector dosage; the decrease in the signal from an MOI of 125 to an MOI of 625 correlated, as before, with increased CPE. Weaker bands in the undigested samples corresponded to 3.2-kb linear HBV DNA and a faster-migrating smear of immature HBV DNAs; the exact identities of the bands at about 2.6 and 3.0 kb are not clear. In the top region of the gel, small and slightly variable amounts of residual Ad vector DNA were visible. Upon EcoRI digestion, the HBV RC-DNA band disappeared while the intensity of the 3.2-kb linear band increased as expected. The mobilities of the bands appearing at about the 1.9- and 1.4-kb positions are consistent with their being EcoRI cleavage products of linear HBV DNA. The Ad-HBV1.3 vector should produce two HBV-containing fragments of 2.6 and 27 kb, which were also visible. Similar results were obtained with Huh7 cells (not shown). These data suggested that PTHs are competent for HBV virion formation.
FIG. 4.
Enzymatic assay for enveloped HBV virion formation in PTHs. (A) Dose dependence. Particles in supernatants from PTHs transduced with the indicated MOIs of Ad-HBV1.3 and collected on days 9 and 10 p.i. were PEG precipitated. Naked DNA and DNA in nonenveloped particles was digested by pronase plus DNase. DNA from enveloped particles was subsequently released by proteinase K-SDS treatment. Aliquots were loaded directly (lanes −) or after digestion with EcoRI (E. RI) (lanes +) and detected with a DIG-labeled HBV probe. M, DIG-labeled DNA marker with fragment sizes in kilobases; St, linear 3.2-kb HBV genome. The positions of HBV RC-DNA and linear (Lin) DNA and of the assignable HBV and Ad-HBV1.3 EcoRI fragments are indicated on the right. The arrows denote bands in the undigested samples whose exact natures are not known. (B) Time course. Supernatants from PTHs transduced with Ad-HBV1.3 at an MOI of 100 were collected every 2 days as indicated, except for the day 18 sample, which was collected over 4 days. Extracellular DNA was analyzed as for panel A. An aliquot from the day 6 sample was digested with XhoI (lane d6X Xho), which linearizes the circular HBV genome and generates a 1.7- plus a 1.5-kb fragment from linear molecules. Ad-HBV1.3 produces two HBV-containing fragments of 4.3 and 2.7 kb. St, 50 pg of linear 3.2-kb HBV genome.
The experiment was repeated in a time course fashion with particles from supernatants collected from days 1 to 18 p.i. The day 1 sample (Fig. 4B), containing the Ad-HBV1.3 inoculum, showed a strong signal of the Ad vector, which at this high concentration was only partially digested by pronase. Most but not all vector particles were removed during subsequent medium changes, as indicated by the continued presence of small amounts of this band in later samples. From day 4 on, new bands corresponding to HBV RC-DNA appeared, with a peak at day 6. The day 18 signal is slightly more intense because medium was collected for 4 rather than 2 days. Digestion of the day 6 sample with XhoI generated a band comigrating with the 3.2-kb linear HBV DNA marker and a 1.7-kb band probably derived from digestion of linear HBV DNA, plus the expected HBV-containing XhoI fragments from the Ad vector. In addition, smaller, less distinct bands probably representing replicative intermediates, such as single-stranded DNA, were observed throughout (see below). These data indicated a continuous release of HBV virions from the PTHs for almost 3 weeks.
The presence of immature replicative HBV intermediates with slightly varying mobilities and intensities at different time points might have indicated varying degrees of maturation; however, a more likely explanation, supported by the small but detectable amounts of Ad vector DNA, was that complete enzymatic removal of DNA in nonenveloped particles was not easily controlled. To obtain independent physical proof for the presence of intact virions, extracellular particles were subjected to CsCl density gradient centrifugation, which separates enveloped virions from nonenveloped cores and Ad vector particles according to their buoyant densities (around 1.24 and 1.35 g/ml, respectively). Because no protease treatment was included, naked cores and the remaining vector particles were left intact.
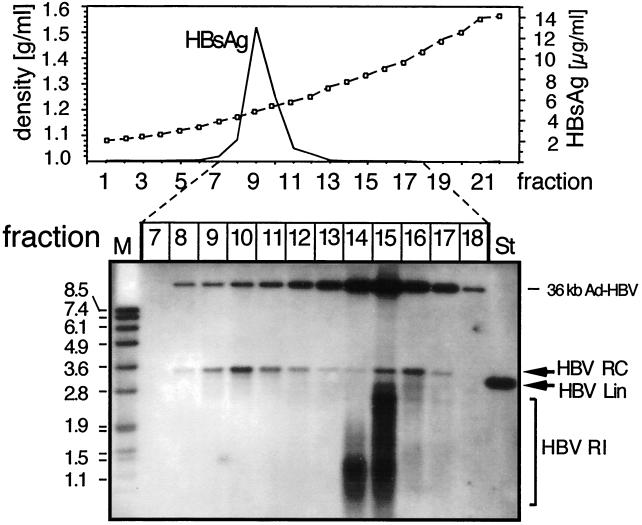
The density and HBsAg profiles, and the distribution of HBV-specific DNAs in the gradient, are shown in Fig. 5. HBsAg was detectable in fractions 7 to 11, with a strong peak in fraction 9 (density, 1.20 g/ml). Nearly pure RC-DNA was present in fractions 9 to 12 (density range, 1.20 to 1.25 g/ml), peaking in fraction 10 and therefore consistent with the presence of mature enveloped HB virions. A second peak occurred in fractions 14 to 16 (density range, 1.31 to 1.37 g/ml), which was accompanied, especially in the lower density fractions, by faster-migrating nucleic acids. The presence of core protein (not shown) indicated that they derived from naked cores at various stages of maturation that were partially separated according to their increasing DNA contents. Fraction 15 also contained the bulk of the remaining Ad vector DNA at the expected density. These data confirmed that PTHs are proficient in the production of complete enveloped HBV virions carrying largely mature DNA genomes.
FIG. 5.
Physical separation of enveloped particles by density gradient centrifugation. Extracellular particles in the day 7 and day 8 supernatants of PTHs transduced with Ad-HBV1.3 at an MOI of 100 were concentrated by PEG precipitation and centrifuged through a CsCl gradient. Because no protease treatment was included, naked cores and Ad-HBV particles were left intact. The density and HBsAg profiles are shown in the graph. Subsequently, DNAs contained in fractions 7 to 18, covering a density range from 1.16 to 1.42 g/ml, were extracted and analyzed by Southern blotting using a DIG-labeled HBV probe as described in the legend to Fig. 4. HBV RI, replicative intermediates; St, 50 pg of a 3.2-kb linear HBV genome; M, DIG-labeled DNA marker with fragment sizes in kilobases.
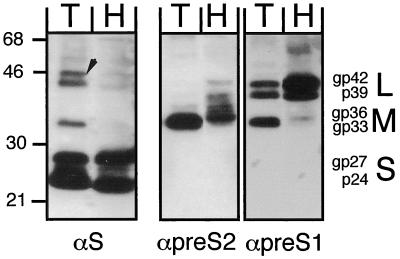
HBsAg peak fractions from a similar gradient were also used to directly test, by Western blotting, whether all three of the HBV envelope proteins, i.e., S, M, and L, were formed in the tupaia cells. For comparison, the corresponding fraction from an identical gradient loaded with particles from Ad-HBV1.3-transduced Huh7 cells was analyzed in parallel (Fig. 6). The human monoclonal anti-S antibody 4/7B (31) detected similar amounts of nonglycosylated (p24) and glycosylated (gp27) S proteins in both samples. Of the additional slower-migrating bands in the PTH sample, the 33-kDa band comigrated exactly with the M protein band specifically detected in the pre-S2 blot with the pre-S2-specific antibody S26 (37) and therefore may represent M protein. By contrast, the two larger bands, at first glance resembling the L proteins p39 and glycosylated gp42 in the pre-S1-specific blot developed with monoclonal antibody 18/07 (16), are probably nonspecific. The largest one runs more slowly than gp42, and the second, though comigrating with gp42, should also be present in the Huh7 lane, which, according the pre-S1 blot, contained even more of both L proteins than the PTH lane. The M protein patterns were slightly different in samples from human and tupaia cells, possibly reflecting some differences in the glycan moieties of the mono- and diglycosylated forms (gp33 and gp36), while no obvious differences were observed for the L proteins except for a higher ratio of M to L protein in PTHs versus Huh7 cells. Together, these data showed that tupaia hepatocytes support the production of all structural HBV proteins and their assembly into enveloped virions.
FIG. 6.
Comparison of HBV envelope proteins produced in PTHs and Huh7 cells. Secreted particles from Ad-HBV1.3-transduced PTHs (lanes T; MOI, 125; 9 ml of medium from days 7 and 8) and Huh7 cells (lanes H; MOI, 400; 20 ml of medium from days 3 and 4) were separated on similar CsCl gradients as shown in Fig. 5, and the proteins from the HBsAg peak fractions were analyzed by Western blotting with monoclonal antibodies specific for S (4/7B), pre-S2 (S26), and pre-S1 (MA18/07); the αpre-S2 and αpre-S1 blots were obtained sequentially without intermediate stripping. Detection was performed using a chemiluminescent substrate. The positions of protein size markers (in kilodaltons) are indicated on the left, and the various forms of the S, M, and L proteins are given on the right. The arrow marks a doublet of bands in the T lane of the αS blot that superficially resemble L proteins but are probably nonspecific.
Primary tupaia hepatocytes support formation of HBV cccDNA.
Next, we analyzed the transduced PTHs, in parallel with transduced Huh7 cells, for the presence of cccDNA. The procedure aimed at removing, as far as possible, other DNAs that might give rise to bands that could be mistaken for cccDNA, e.g., particle-borne replicative intermediates or fragments from the Ad-HBV1.3 vector. To this end, the soluble nucleic acids obtained after twofold SDS-NaOH treatment were incubated with Plasmid-safe DNase (10), an ATP-dependent nuclease that digests linear DNA much more effectively than circular DNA, in particular covalently closed circular molecules, and in addition with HpaI, a restriction enzyme that cuts the Ad-HBV vector but not HBV DNA.
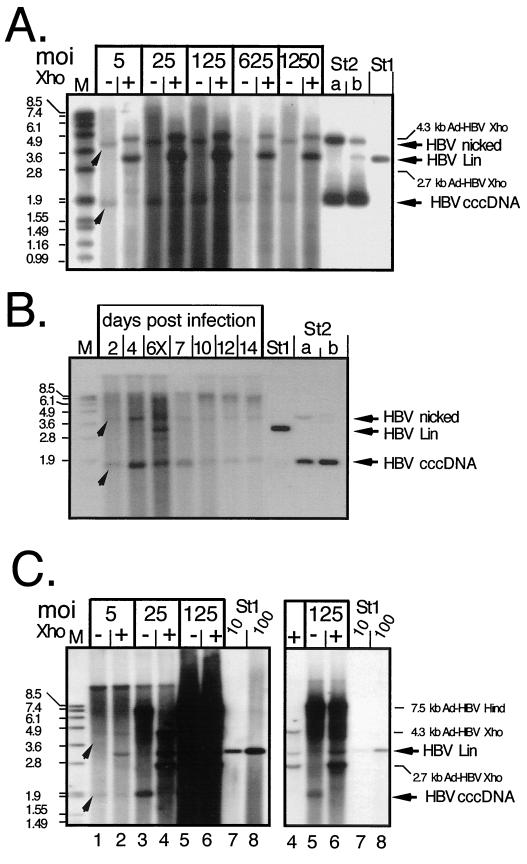
A representative Southern blot, obtained with PTHs infected with Ad-HBV1.3 at MOIs of 5 to 1,250, is shown in Fig. 7. All directly loaded samples showed two specifically hybridizing bands; the one at about the 1.9-kb position comigrated with the cccDNA form of a 3.2-kb pUC plasmid containing about 500 nucleotides of HBV sequence (lanes St2); the second, between the 3.6 and 4.8-kb markers, comigrated with the open circular form of this plasmid. Incubation with XhoI, which cuts once inside HBV cccDNA, generated a new band comigrating with linear 3.2-kb HBV DNA (lane St1). The weaker new bands after XhoI digestion, at 4.3 and 2.7 kb, correspond to the only two XhoI fragments of the Ad-HBV1.3 vector that contain HBV sequences. Considering the extraction procedure, it was unlikely that the band migrating at the nicked plasmid position corresponded to particle-borne RC-DNA; rather, it represented cccDNA that was nicked during the harsh extraction procedure. This was corroborated by mixing an aliquot of the plasmid DNA preparation (St2b) with noninfected cells and taking it through the entire procedure (St2a), which also increased the nicked DNA form. As observed before, the initial dose-dependent increase in yield leveled off at an MOI of about 125 and then declined because of CPE. Qualitatively similar results were obtained with transduced Huh7 cells (see below).
FIG. 7.
cccDNA generation in Ad-HBV-transduced PTHs and Huh7 cells. (A) Dose dependence in PTHs. Episomal DNAs were isolated at day 7 p.i. from PTHs transduced with Ad-HBV1.3 at the indicated MOIs and further treated to remove most noncircular molecules. One-half of each sample was directly loaded (lanes −), and the other half was loaded after incubation with XhoI (lanes +). HBV-specific DNAs were detected with a DIG-labeled probe. St1, 50 pg of linear 3.2-kb HBV DNA; St2, 3.2-kb pUC plasmid containing about 0.5 kb of HBV sequence. St2b contained 400 pg of directly loaded plasmid, and St2a contained 800 pg of plasmid that had been mixed with noninfected cell lysate and taken through the entire extraction procedure. The arrowheads indicate the positions of HBV cccDNA and its nicked form. As we found out later, the apparent signal increase after XhoI linearization was due to a specific cccDNA detection problem with the batch of agarose used in these experiments. (B) Time course in PTHs. PTHs were infected with Ad-HBV1.3 at an MOI of 100, and episomal DNAs were isolated at the indicated times. The sample from day 6 was partially digested with XhoI (lane 6X) to visualize the position of linear DNA. St1 and St2, reference DNAs as described for panel A. (C) Dose dependence in Huh7 cells. Episomal DNAs from cells infected at the indicated MOIs with Ad-HBV1.3 were analyzed as described above, except that during preparation HindIII rather than HpaI was used to cut DNAs not corresponding to HBV cccDNA. HindIII digestion of Ad-HBV1.3 generates a single HBV-containing fragment of 7.5 kb whose further cleavage by XhoI gives the same fragments as in panel A. The righthand gel shows a fivefold-shorter exposure of lanes 4 to 8, to resolve individual bands in the samples at an MOI of 125. St1, reference DNA (10 and 100 pg) as in panel A.
Somewhat surprisingly, the intensities of the XhoI-linearized bands appeared stronger than the sum of the cccDNA plus nicked forms in the nondigested lanes. This observation was reproduced in many experiments, and further controls using plasmid DNAs confirmed a selective underrepresentation of the cccDNA signals, especially at low concentrations. Various changes in the Southern blotting protocol had only minor effects on cccDNA detectability. Eventually, we found that a different batch of agarose yielded improved results (see below). Because linear DNA was not affected, a rough estimate of the minimal number of cccDNA molecules produced could be derived by densitometrically comparing the intensities of the linear 3.2-kb XhoI product bands with that of the 50 pg of the 3.2-kb HBV DNA marker (lane St1) on the same gel. Accordingly, the sample at an MOI of 5 contained about twice as much DNA; for a 3.2-kb double-stranded DNA, 100 pg correspond to about 3 × 107 molecules. At an average of about 5 × 106 PTHs, this translates into about six copies per cell. The stronger signals obtained with the samples at MOIs of 25 and 125 indicate that this number can be further increased.
Next, we monitored the time course of cccDNA generation in PTHs infected with Ad-HBV1.3 an MOI of 100, this time using a different batch of agarose that allowed better cccDNA detection (Fig. 7B). A linear HBV genome (St1) and the same HBV-containing plasmid used before (St2) taken through the extraction (lane a) or directly loaded (lane b) served as controls. Two bands corresponding to cccDNA and its nicked form were weakly visible in the day 2 sample, increased at day 4, and then declined. The day 6 sample was partially digested with XhoI (lane 6X) to visualize the position of linear DNA; the overall signal intensity was similar to that at day 4. As mentioned before, the number of viable cells started decreasing around that time, certainly contributing to the decreased cccDNA signals at later timer points. Together, these experiments proved that PTHs are able to support the intracellular HBV amplification cycle.
As a control, we also analyzed cccDNA formation in Ad-HBV1.3-transduced Huh7 cells. In this dose dependence experiment (Fig. 7C), HindIII rather than HpaI was used during the DNA preparations; this enzyme does not cut the HBV genome but produces a single HBV-containing 7.5-kb Ad-HBV fragment which is further cleaved by XhoI into the same 4.3-and 2.7-kb fragments described above. In this experiment, the Ad-HBV vector DNA was only partially removed, leading to a relatively high background at the higher doses. Nonetheless, an HBV-specific band migrating at about the 1.9-kb marker position was clearly detectable; it disappeared upon digestion with XhoI, while a new band became visible at the 3.2-kb position, indicative of cccDNA formation in the hepatoma cells. Although the high background made accurate determinations difficult, densitometric scanning of various exposures allowed for at least an approximate estimate of cccDNA concentrations. Based on the 10- and 100-pg standards, these ranged from slightly less than 5 pg with vector at an Mol of 5 over about 50 pg at an Mol of 25 to about 150 pg at an Mol of 125. Because one-half of the DNA preparation from 1.7 × 107 cells was loaded per lane, these values correspond numerically to 0.2, 1, and 3 copies of cccDNA per Huh7 cell. While the accuracy of these estimates is limited by the many steps involved in sample preparation, they show that PTHs are certainly not less efficient in cccDNA formation than the human hepatoma cells.
DISCUSSION
Tupaias would offer several advantages over the established animal models if they could be efficiently infected with HBV or a very closely related virus. At present, however, the evidence for their in vivo susceptibility to HBV is, at best, indirect (48). In vitro infection of cultured primary tupaia liver cells with serum HBV is possible but inefficient (43). In its present form, it does not appear suited for detailed studies of infection itself or of antivirals, and the in vitro data cast doubt on the chances of establishing an in vivo infection by this route.
The Ad-HBV vector-based delivery system described here proved well suited to efficiently transduce complete HBV genomes into PTHs. As desired, the greatly increased amounts of viral gene products allowed us to directly test whether tupaia hepatocytes support all downstream events of the HBV replication cycle. This was indeed the case, as shown by the generation of complete HBV virions and nuclear HBV cccDNA in amounts comparable to or higher than those obtained in human hepatoma cell lines. These data suggest the system will also be useful for in vivo production of HBV in tupaias or other potential hosts. Furthermore, the formation of HBV cccDNA in PTHs demonstrates that the complete genomic replication cycle from DNA to RNA and back is carried out. The error-prone transcription and reverse-transcription steps should allow for the relatively rapid generation of variant genomes and hence increase the chances for adapting HBV to tupaias.
The Ad-HBV vector system.
A number of advantages led to the widespread use of Ad vectors in gene therapy studies (2). In addition, by intraveneous injection, they can be targeted to the liver with reasonable specificity. We therefore chose this system as a vehicle for the transduction of complete HBV genomes. However, there are also potential disadvantages, including the originally tedious generation of recombinants and vector toxicity, at least at high doses (40). The new E. coli recombination systems (8, 15) have greatly facilitated recombinant Ad vector production, and the viruses are derived from easily characterizable plasmid clones. In our hands, and depending on the system, between 1 and 40% of the bacterial colonies contained the desired recombinant plasmids. It should therefore be possible to generate, within a reasonable time, vectors carrying HBV genomes with defined genetic alterations. As for toxicity, the Ad vectors are certainly more cytotoxic than the recently described baculovirus vectors (10). They induced dose-dependent CPE in all cells studied, although to different extents. We did not thoroughly quantitate the cytotoxicity of individual vector-cell combinations, but approximate values can be inferred from the dose dependence of HBsAg and HBeAg production (Fig. 2). The failure of an increased vector dosage to increase antigen production plausibly correlates with significant cytotoxicity, probably due to the increased gene dosage by multiple infections of one cell. Hence, the MOI giving the highest antigen level may be regarded as a guideline for the maximum useful dose. Notably, if only short-term expression is required, much higher doses, with correspondingly increased production of HBV gene products, may be employed.
Given that PTHs tolerated an MOI of at least 5 for Ad-GFP-HBV1.3 and much higher doses of Ad-HBV1.3 and assuming a liver contains on the order of 1011 hepatocytes, there are also few restrictions regarding the in vivo application of these vectors. The current data do not allow us to propose a specific mechanism for the increased toxicity of the GFP-encoding vector. However, the similarly high toxicity towards PTHs, but not hepatoma cells, observed with two other CMV-containing Ad vectors, including Ad-GFP without a hepadnavirus genome, suggests an involvement of this strong promoter. This does not rule out the possibility that cooperative effects between Ad and hepadnavirus sequences and/or gene products contribute to cytotoxicity.
The E1A-E1B deletion in the Ad vectors should require that complementing cell lines be used for vector particle formation. However, some safety concerns were raised by a report that a cell line derived from transfection with an HBx expression construct could partially compensate for an E1A defect (34). Because the X gene is present in our vectors, we carefully monitored many experiments with HepG2 and Huh7 cells, as well as with PTHs, for formation of plaques and, more sensitively, for an increase in GFP-positive cells after infection with Ad-GFP-HBV1.3. However, we never observed any spread of the infection, except in 293 cells. This was also true in long-term cultures, where even a poorly efficient complementation should have become detectable. Notably, others also have failed to detect a complementation by HBx when the Ad vector, as in our case, lacked both E1A and E1B (3, 24). While it is possible that the HBx protein was not expressed in sufficient amounts in our experiments, the data provide clear-cut evidence that the presence of an intact HBx gene per se is not sufficient to rescue the Ad vector defect.
Together, these data show that replication-defective Ad-HBV vectors provide an efficient and safe alternative for the transfer of complete HBV genomes into a variety of permanent cell lines and, importantly, primary cells, which are notoriously difficult to transfect. An added advantage over transfection is that, at low MOIs, only one or few HBV genomes rather than many copies per cell will be present.
Competence of primary tupaia hepatocytes for HBV virion formation.
As demonstrated, Ad-HBV-infected PTHs were as proficient as human hepatoma lines for the synthesis of all viral structural proteins, including P protein, and eventually virions. Regarding the envelope proteins, the only discernible qualitative difference between PTHs and Huh7 cells was a slightly modified glycosylation pattern for the M protein. Quantitatively, with close to 1 μg of HBsAg per ml of supernatant collected for 2 days, Ad-HBV1.3-transduced PTHs yielded three to five-times-higher maximum levels of antigen than Huh7 and HepG2 cells. This range is similar to that described for baculovectors (10) at MOIs of 100 to 400 (about 1 μg per 3 ml of supernatant collected for 1 day) and about 100 times more than the amount of HBsAg secreted by the permanent HBV expression line HepG2.2.15 (36). While these maximum numbers also reflect the higher tolerance of PTHs for high vector doses, increased HBsAg levels were also observed at low-to-medium MOIs. These differences were more pronounced when based on cell numbers but remained easily detectable when based on total protein content. They cannot, therefore, be explained solely by the larger size of the primary cells.
The levels of HBeAg, by contrast, were not significantly increased in PTHs over Huh7 cells. Possibly, the core promoter-enhancer II is not maximally activated in the tupaia cells. While a definite conclusion will require quantitative mRNA determinations, similar observations were made regarding nucleocapsid amounts. Semiquantitative estimates suggest that the yield of DNA-containing nucleocapsids per dish was about threefold lower in PTHs than in Huh7 cells but similar on a per-cell basis.
Irrespective of such quantitative considerations, PTHs clearly supported the formation of nucleocapsids containing HBV-specific nucleic acids and therefore of functional P protein. That the originally packaged pgRNA has authentic ends is demonstrated by the presence of HBV RC-DNA, whose synthesis is strongly affected by even minor changes in the 5′-terminal RNA regions (23). The generation of complete virions carrying mature RC-DNA genomes was made likely by an enzymatic procedure and was independently confirmed by density gradient separation of enveloped and nonenveloped particles. Hence, in contrast to WHV in human cell lines, there seems to be no species-specific impairment of HBV genome maturation in tupaia cells.
The supernatants also contained naked cores. Such nonenveloped cores are often found after transfection of hepatoma cells with efficient HBV expression constructs. Whether the same unknown release mechanism is operating in the Ad vector-transduced cells is unclear because vector-mediated CPE might also liberate intracellular cores.
HBV cccDNA formation in primary tupaia hepatocytes.
Because no cccDNA has been detected in HBV-transgenic mice, an important objective of this study was to directly prove the ability, or inability, of tupaia hepatocytes to support HBV cccDNA formation. A potential contaminant was the Ad-HBV vector, which is pertinent to the transduction system and, although it does not replicate, remained detectable for extended periods; another was particle-borne HBV DNA. RC-DNA in particular would yield an indistinguishable linearization product upon restriction digestion. We therefore used an extraction method that counterselected against non-cccDNA molecules, including RC-DNA, by double treatment with NaOH and, in addition, with Plasmid-Safe DNase (10). The final products were one band comigrating with a 3.2 kb-plasmid cccDNA standard at about the 1.9-kb position and a second band at the RC-DNA position. Both collapsed into a single 3.2-kb band upon digestion with XhoI. Because the single XhoI site in HBV lies outside the redundancy of the 1.3× HBV genome in the Ad-HBV vectors, they do not generate such a fragment. We therefore conclude that the fast-migrating band indeed corresponds to cccDNA while the second band represents a fraction of cccDNA molecules that were physically nicked during extraction. Consistently, a plasmid DNA preparation taken through the entire procedure also showed an increased proportion of nicked DNA.
Overall similar results were obtained with Ad-HBV1.3-transduced Huh7 cells. Whether the nominally somewhat lower cccDNA yields per cell in Huh7 cells versus PTHs are significant remains to be determined because the relatively high background made accurate measurements difficult. However, even if the results are interpreted conservatively, the PTHs performed at least as well as the human hepatoma cells. These results imply that hepatocytes from tupaias, in contrast to those from mice, contain all of the host factors required for the intracellular HBV amplification cycle. Alternatively, though less likely, there may be differences between HBV expression from an integrated transgene and that from an episomal Ad vector-transduced genome. This issue is being addressed by similar experiments with mouse and rat primary hepatocytes.
Together, our data established transduction of HBV genomes with replication-defective Ad vectors as an effective delivery method that may also find a use in applications currently requiring transgenic animals. With cultured cells, the Ad-HBV vectors are probably as useful as the recently introduced baculovirus transduction system, but in addition, they are directly suited for in vivo experiments. The ability of PTHs to produce HBV virions and to support intracellular cccDNA formation and their at least low-level susceptibility to HBV infection suggest that by using the Ad-HBV vectors it may be possible to artificially establish HBV infections in these animals. A pilot experiment indicates that a long-lasting and high-titer HBV antigenemia (with HBsAg levels above 10 μg/ml of serum) can indeed by induced by intravenous injection into newborn tupaias (S. Ren and M. Nassal, unpublished data). While HBV production should be achievable in other species as well, our data suggest that tupaias may be particularly suited to support a true HBV infection. If HBV expression can be maintained for sufficient periods, even a very low-level infection may ultimately enable the selection of HBV variants adapted to this experimentally practical animal system.
ACKNOWLEDGMENTS
We thank J. Summers for sharing protocols, R. Schneider for helpful comments on potential Ad vector complementation by HBx, and S. McNelly for excellent technical assistance. Several immunological reagents were kindly provided by V. Bichko, W. Gerlich, and R. A. Heijtink, and parental Ad vector plasmids were provided by M. Lusky and T. C. He. We also thank several members of the Regierungspräsidium Tübingen for continuous stimulating discussions.
S.R. is grateful for a fellowship from the University of Freiburg. This work was supported by grants from the Center for Clinical Research I (ZKF I-B7), the Bundesministerium für Bildung und Forschung (DLR 01KV98041), and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Akbar S K, Onji M. Hepatitis B virus (HBV) transgenic mice as an investigative tool to study immunopathology during HBV infection. Int J Exp Pathol. 1998;79:279–291. doi: 10.1046/j.1365-2613.1998.740406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr Opin Biotechnol. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 3.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg B S. Hepatitis B virus, the vaccine, and the control of primary cancer of the liver. Proc Natl Acad Sci USA. 1997;94:7121–7125. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breiner K M, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi resident protein, functions in the attachment and entry of avian hepatitis B viruses. J Virol. 1998;72:8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss V, Gerhardt E, Vieluf K, Wunderlich G. Functions of the large hepatitis B virus surface protein in viral particle morphogenesis. Intervirology. 1996;39:23–31. doi: 10.1159/000150471. [DOI] [PubMed] [Google Scholar]
- 8.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chisari F V. Hepatitis B virus transgenic mice: models of viral immunobiology and pathogenesis. Curr Top Microbiol Immunol. 1996;206:149–173. doi: 10.1007/978-3-642-85208-4_9. [DOI] [PubMed] [Google Scholar]
- 10.Delaney W E T, Isom H C. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology. 1998;28:1134–1146. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- 11.Delaney W E T, Miller T G, Isom H C. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (−)-β-2′,3′-dideoxy-3′-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob Agents Chemother. 1999;43:2017–2026. doi: 10.1128/aac.43.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Q, Summers J, Burch J B, Mason W S. Major differences between WHV and HBV in the regulation of transcription. Virology. 1997;229:25–35. doi: 10.1006/viro.1996.8422. [DOI] [PubMed] [Google Scholar]
- 13.Gall J. Use and application of adenovirus expression vectors. In: Spector D L, Goldman R D, Leinwand L A, editors. Cells—a laboratory manual. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 90.1–90.28. [Google Scholar]
- 14.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heermann K H, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multicomponent chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilan E, Burakova T, Dagan S, Nussbaum O, Lubin I, Eren R, Ben-Moshe O, Arazi J, Berr S, Neville L, Yuen L, Mansour T S, Gillard J, Eid A, Jurim O, Shouval D, Reisner Y, Galun E. The hepatitis B virus trimera mouse: a model for human HBV infection and evaluation of anti HBV therapeutic agents. Hepatology. 1999;29:553–562. doi: 10.1002/hep.510290228. [DOI] [PubMed] [Google Scholar]
- 19.Jilbert A R, Kotlarski I. Immune responses to duck hepatitis B virus infection. Dev Comp Immunol. 2000;24:285–302. doi: 10.1016/s0145-305x(99)00079-8. [DOI] [PubMed] [Google Scholar]
- 20.Kann M, Sodeik B, Vlachou A, Gerlich W H, Helenius A. Phosphorylation dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 22.Lenhoff R J, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loeb D D, Tian R, Gulya K J, Qualey A E. Changing the site of initiation of plus-strand DNA synthesis inhibits the subsequent template switch during replication of a hepadnavirus. J Virol. 1998;72:6565–6573. doi: 10.1128/jvi.72.8.6565-6573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucito R, Schneider R J. Hepatitis B virus X protein activates transcription factor NF-κ B without a requirement for protein kinase C. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meisel H, Sominskaya I, Pumpens P, Pushko P, Borisova G, Deepen R, Lu X, Spiller G H, Kruger D H, Grens E, et al. Fine mapping and functional characterization of two immuno-dominant regions from the preS2 sequence of hepatitis B virus. Intervirology. 1994;37:330–339. doi: 10.1159/000150397. [DOI] [PubMed] [Google Scholar]
- 26.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassal M. Hepatitis B virus replication: novel roles for virus-host interactions. Intervirology. 1999;42:100–116. doi: 10.1159/000024970. [DOI] [PubMed] [Google Scholar]
- 28.Nassal M. Macromolecular interactions in hepatitis B virus replication and particle formation. In: Cann A J, editor. Frontiers in molecular biology: DNA virus replication. Vol. 26. Oxford, United Kingdom: Oxford University Press; 2000. pp. 1–40. [Google Scholar]
- 29.Ohashi K, Marion P L, Nakai H, Meuse L, Cullen J M, Bordier B B, Schwall R, Greenberg H B, Glenn J S, Kay M A. Sustained survival of human hepatocytes in mice: a model for in vivo infection with human hepatitis B and hepatitis delta viruses. Nat Med. 2000;6:327–331. doi: 10.1038/73187. [DOI] [PubMed] [Google Scholar]
- 30.Pasek M, Goto T, Gilbert W, Zink B, Schaller H, MacKay P, Leadbetter G, Murray K. Hepatitis B virus genes and their expression in E. coli. Nature. 1979;282:575–579. doi: 10.1038/282575a0. [DOI] [PubMed] [Google Scholar]
- 31.Paulij W P, de Wit P L, Sunnen C M, van Roosmalen M H, Petersen-van Ettekoven A, Cooreman M P, Heijtink R A. Localization of a unique hepatitis B virus epitope sheds new light on the structure of hepatitis B virus surface antigen. J Gen Virol. 1999;80:2121–2126. doi: 10.1099/0022-1317-80-8-2121. [DOI] [PubMed] [Google Scholar]
- 32.Petersen J, Dandri M, Gupta S, Rogler C E. Liver repopulation with xenogenic hepatocytes in B and T cell deficient mice leads to chronic hepadnavirus infection and clonal growth of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1998;95:310–315. doi: 10.1073/pnas.95.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roggendorf M, Tolle T K. The woodchuck: an animal model for hepatitis B virus infection in man. Intervirology. 1995;38:100–112. doi: 10.1159/000150418. [DOI] [PubMed] [Google Scholar]
- 34.Schaack J, Maguire H F, Siddiqui A. Hepatitis B virus X protein partially substitutes for E1A transcriptional function during adenovirus infection. Virology. 1996;216:425–430. doi: 10.1006/viro.1996.0079. [DOI] [PubMed] [Google Scholar]
- 35.Seeger C, Mason W S. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sells M A, Chen M L, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sominskaya I, Pushko P, Dreilina D, Kozlovskaya T, Pumpen P. Determination of the minimal length of preS1 epitope recognized by a monoclonal antibody which inhibits attachment of hepatitis B virus to hepatocytes. Med Microbiol Immunol. 1992;181:215–226. doi: 10.1007/BF00215767. [DOI] [PubMed] [Google Scholar]
- 38.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi H, Fujimoto J, Hanada S, Isselbacher K J. Acute hepatitis in rats expressing human hepatitis B virus transgenes. Proc Natl Acad Sci USA. 1995;92:1470–1474. doi: 10.1073/pnas.92.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teichler Zallen D. US gene therapy in crisis. Trends Genet. 2000;16:272–275. doi: 10.1016/s0168-9525(00)02025-4. [DOI] [PubMed] [Google Scholar]
- 41.Tuttleman J S, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 42.Urban S, Schwarz C, Marx U C, Zentgraf H, Schaller H, Multhaup G. Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus-receptor interaction. EMBO J. 2000;19:1217–1227. doi: 10.1093/emboj/19.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter E, Keist R, Niederost B, Pult I, Blum H E. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology. 1996;24:1–5. doi: 10.1002/hep.510240101. [DOI] [PubMed] [Google Scholar]
- 44.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Will H, Cattaneo R, Darai G, Deinhardt F, Schellekens H, Schaller H. Infectious hepatitis B virus from cloned DNA of known nucleotide sequence. Proc Natl Acad Sci USA. 1985;82:891–895. doi: 10.1073/pnas.82.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wold W S M, Chinnadurai G. Adenovirus proteins that regulate apoptosis. In: Cann A J, editor. Frontiers in molecular biology: DNA virus replication. Vol. 26. Oxford, United Kingdom: Oxford University Press; 2000. pp. 200–232. [Google Scholar]
- 47.Wu T T, Coates L, Aldrich C E, Summers J, Mason W S. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 48.Yan R Q, Su J J, Huang D R, Gan Y C, Yang C, Huang G H. Human hepatitis B virus and hepatocellular carcinoma. I. Experimental infection of tree shrews with hepatitis B virus. J Cancer Res Clin Oncol. 1996;122:283–288. doi: 10.1007/BF01261404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]