Abstract
Insulin resistance (IR) was found to be a critical element in the pathogenesis of Parkinson’s disease (PD), facilitating abnormal α-synuclein (α-Syn) aggregation in neurons and thus promoting PD development. However, how IR contributes to abnormal α-Syn aggregation remains ill-defined. Here, we analyzed six PD postmortem brain transcriptome datasets to reveal module genes implicated in IR-mediated α-Syn aggregation. In addition, we induced IR in cultured dopaminergic (DA) neurons overexpressing α-Syn to identify IR-modulated differentially expressed genes (DEGs). Integrated analysis of data from PD patients and cultured neurons revealed 226 genes involved in α-Syn aggregation under IR conditions, of which 53 exhibited differential expression between PD patients and controls. Subsequently, we conducted an integrated analysis of the 53 IR-modulated genes employing transcriptome data from PD patients with different Braak stages and DA neuron subclasses with varying α-Syn aggregation scores. Protein tyrosine phosphatase receptor type O (PTPRO) was identified to be closely associated with PD progression and α-Syn aggregation. Experimental validation in a cultured PD cell model confirmed that both mRNA and protein of PTPRO were reduced under IR conditions, and the downregulation of PTPRO significantly facilitated α-Syn aggregation and cell death. Collectively, our findings identified PTPRO as a key regulator in IR-mediated α-Syn aggregation and uncovered its prospective utility as a therapeutic target in PD patients with IR.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05436-4.
Keywords: Parkinson’s disease, Insulin resistance, α-Synuclein aggregation, PTPRO, Dopaminergic neurons
Introduction
Insulin resistance (IR), characterized by impaired cellular response to insulin, is a typical feature of type 2 diabetes mellitus (T2DM) [1]. The occurrence of IR is not limited to specific organs and can be observed in various tissues, including the brain, contributing to the pathogenesis of multiple diseases [2]. Given the brain is an insulin-sensitive organ, understanding insulin signaling in the brain has recently received increased attention [3]. An emerging body of evidence reveals the presence of IR in the brains of individuals with neurodegenerative diseases, such as Parkinson’s disease (PD) and Alzheimer’s disease (AD), even in the absence of concurrent T2DM [4, 5]. Dysregulation of insulin signaling is considered a crucial factor in the pathogenesis and development of these neurodegenerative disorders [6–8].
PD stands out as the most common motoric neurodegenerative disease, and its prevalence is on the rise [9, 10]. Peripheral IR is demonstrated in 60% of PD patients, accompanied by a rapid and worsening progression of PD [11, 12]. Notably, brain IR can occur prior to peripheral IR in PD, evidenced by a reduction in insulin receptor substrate-1 (IRS-1) expression in neurons [13–15]. Phosphorylation at serine residue 312 of IRS-1 (IRS-1pS312) promotes the proteasomal degradation of IRS-1, leading to impaired functional insulin signaling [16]. Increased IRS-1pS312 has been detected in the substantia nigra of PD patients [17], correlating with a higher severity of tremor [18]. These findings underscore the importance of brain IR in the progression of PD, highlighting the essential need to comprehend the molecular mechanisms underlying IR.
Misfolded and aggregated α-synuclein (α-Syn), which forms the Lewy bodies in midbrain dopaminergic (DA) neurons, is a prominent feature of PD [19]. The aggregation of α-Syn is implicated in the loss of DA neurons, another key pathological feature of PD [20]. α-Syn aggregation disrupts the function of multiple organelles, such as mitochondria, endoplasmic reticulum (ER), Golgi, lysosomes, and autophagosomes, ultimately leading to the degeneration of DA neurons [21]. Previous research has revealed that IR can facilitate the aggregation of α-Syn. In mice with T2DM, the α-Syn accumulation can be observed in the midbrain and substantia nigra [22]. Additionally, elevated expression and phosphorylation of α-Syn occur in IR cell models, contributing to its aggregation [23]. These findings uncover the significant role of IR in α-Syn aggregation, thereby impacting the progression of PD, although the underlying mechanism remains incompletely understood.
In this study, we conducted a comprehensive transcriptome analysis focused on the modulators involved in IR-mediated α-Syn aggregation, utilizing both single-nucleus RNA-Seq (snRNA-Seq) and bulk transcriptome data. Our findings uncovered that PTPRO serves as an IR-mediated modulator influencing α-Syn aggregation in DA neurons, thus contributing to the progression of PD. Notably, PTPRO expression was significantly reduced in the PD cell model under IR conditions, and its downregulation promoted α-Syn aggregation in the PD cell model. Intervention targeting PTPRO might be a promising therapeutic strategy against PD with IR conditions.
Materials and methods
Data collection and processing
The transcriptome sequence data from the human substantia nigra (SN) of PD patients and controls were acquired from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. The following six independent datasets were used, GSE20141, GSE20163, GSE20164, GSE20292, GSE24378 and GSE7621, and integrated into a combined dataset with batch correction using the "ComBat" algorithm. Additionally, we enrolled the GSE49036 dataset, which includes RNA sequencing data and corresponding Braak stage information, as well as the GSE178265 dataset, which contains single-nucleus transcriptomics data. Moreover, the GSE8397, GSE20186, and GSE26927 datasets were incorporated to validate the expression of PTPRO in PD. The IR-related gene set and the α-Syn aggregation-related gene set were obtained from KEGG (https://www.kegg.jp/kegg/) and GeneCards (https://www.genecards.org/), respectively (Supplementary Table 1).
Gene set variation analysis (GSVA)
GSVA, a nonparametric and unsupervised approach, was adopted to interrogate the variations in concerning molecular features among groups based on transcriptomic data. The gene set enrichment scores for individual samples were calculated by “GSVA” R package.
Consensus clustering analysis
Consensus clustering, an unsupervised method of clustering, was utilized to classify the PD samples into distinct subtypes that are associated with IR based on the IR-related genes using “ConsensusClusterPlus” R package.
Weighted correlation network analysis (WGCNA)
Scale-free co-expression networks were inferred using the "WGCNA" R package. Gene modules that exhibited high correlation with IR-related PD subtypes that had high α-Syn aggregation were identified within the combined dataset. Furthermore, gene modules that were highly associated with Braak stage were identified in the GSE49036 dataset.
Functional enrichment analysis
The “clusterProfiler” R package was utilized to operate Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis.
RNA sequencing
Total RNA was extracted from differentiated Lund Human Mesencephalic (LUHMES) cells in the negative control (siCon) and IRS-1 siRNA (siIRS-1) groups using TRIzol reagent (Invitrogen, USA), with each group consisting of three replicates. The integrity and concentration of RNA were measured by Agilent 2100 bioanalyzer (Agilent Technologies, USA). After library preparation and quality control, RNA samples were sequenced on the Illumina Novaseq platform.
Differential expression analysis
Differentially expressed genes (DEGs) between the PD and control groups were identified using the “limma” R package with the cut-off criteria of p < 0.05. DEGs between α-Syn overexpressing DA neurons with IR and control cells were analyzed using the “DESeq2” R package with significance criterion of |log2FC|≥ 0.6 and p < 0.05.
Single-nucleus RNA sequencing analysis
The single nuclei transcriptome of postmortem human substantia nigra pars compacta (SNpc) of both PD patients and matched controls were acquired from the GEO database (GSE178265). This dataset included both NR4A2-positive and -negative nuclei profiles and the NR4A2-positive profiles were enrolled in our study. Cells were clustered using R package “Seurat” and annotated based on marker genes reported previously. The α-Syn aggregation score was calculated using the GSVA method. Based on this score, the DA neurons were categorized into four subsets (agg1, agg2, agg3, agg4). The “FindAIIMarkers” function of “Seurat” R package was used to determine the DEGs of each cluster.
Cell culture
As described previously [24], proliferating LUHMES cells were maintained in proliferation medium consisted of Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 Ham (DMEM/F-12; Sigma-Aldrich, USA) with 1 × N-2 supplement (STEMCELL, CA) and 40 ng/mL recombinant human FGF-basic (Biolegend, CA). Cells were cultured in a humidified incubator containing 5% CO2 at 37 °C. To start differentiation, the medium was replaced with differentiation medium containing DMEM/F-12 supplemented with 1% N-2 supplement, 1 µg/ml tetracycline (Sigma-Aldrich, USA), 0.5 µg/ml N6,2′-O-Dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt (Dibutyryl cyclic-AMP; Sigma-Aldrich, USA), and 2 ng/mL recombinant human GDNF (PeproTech, USA). Cell culture flasks and dishes (Thermo Fisher Scientific, USA) were pre-coated with 0.1 mg/mL poly-l-ornithine solution (PLO; Sigma-Aldrich, USA) for cell proliferation and differentiation. For cell differentiation, an additional coating with 5 µg/ml bovine fibronectin (Sigma-Aldrich, USA) was applied. High insulin-induced IR in differentiated dopaminergic neurons was achieved by administration of insulin at 3 μM for 24 h, as previously described [25].
Lentivirus transfection and RNA interference
To achieve the overexpression of α-Syn in differentiated LUHMES cells, adenoviruses serotype 5 (AV5)-α-Syn purchased from VectorBuilder (China) was added to the cell culture medium at a multiplicity of infection (MOI) of 50. After 24 h, the medium containing the virus was replaced with fresh differentiation medium. Transient knockdown of IRS-1 and PTPRO was achieved by using small interfering RNAs (siRNAs) from GenePharma (Shanghai, China). Cells were transfected utilizing Lipofectamine 3000 reagent (Invitrogen, USA) according to the manufacturer’s protocols.
Quantitative real-time PCR (qRT‒PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, USA). cDNA was prepared utilizing reverse transcription kit (Toyobo, China) according to the manufacturer’s instructions. qRT‒PCR was performed on Mx-3000P Quantitative PCR System with SYBR Green Real-time PCR System Mix (Toyobo, China). The comparative CT method (2−ΔΔCT) was used for relative gene expression analysis. β-actin was used as the internal control. qRT-PCR was performed on three biological replicates, each consisting of three technical replicates. The sequences of primers used were as follows: PTPRO-F: 5’-ATGACTTCAGCCGTGTGAGA-3’, PTPRO-R: 5’-TGTTGCAGGACCATCTTCCA-3’; β-actin-F: 5′-GACAGGATGCAGAAGGAGAT-3′, β-actin-R: 5′-TGATCCACATCT-GCTGGAAGGT-3′.
Western blotting
Protein was extracted from differentiated LUHMES cells. Western blotting was performed as described previously [26]. The primary antibodies used were: PTPRO (Proteintech Group, 67,000–1-Ig), IRS-1 (Cell Signaling Technology, 3407), α-Syn (Thermo Fisher Scientific, 701,085), and β-actin (Cell Signaling Technology, 14,074).
Identification of potential therapeutic small molecule compounds
The Connectivity Map (CMap) database (https://clue.io/) offers predictive capabilities for molecularly targeted drugs base on differential gene expression profiles. In this study, 150 upregulated and 150 downregulated DEGs between the high-PTPRO expression group and the low-PTPRO expression group were submitted to the CMap database. Compounds with negative score were screened as potential small molecule drugs that may counteract PTPRO downregulation. The 3D protein structures of the compounds were obtained from the PubChem accessible chemical database (https://pubchem.ncbi.nlm.nih.gov/).
Statistical analysis
Statistical analyses were performed by R 4.2.0. Wilcoxon test and Student’s t test were conducted to compare differences between two groups, dependently. One-way ANOVA was performed to compare differences in more than two groups. P-value < 0.05 was considered statistically significant.
Results
IR is positively associated with α-Syn aggregation in PD patients’ transcriptome profile
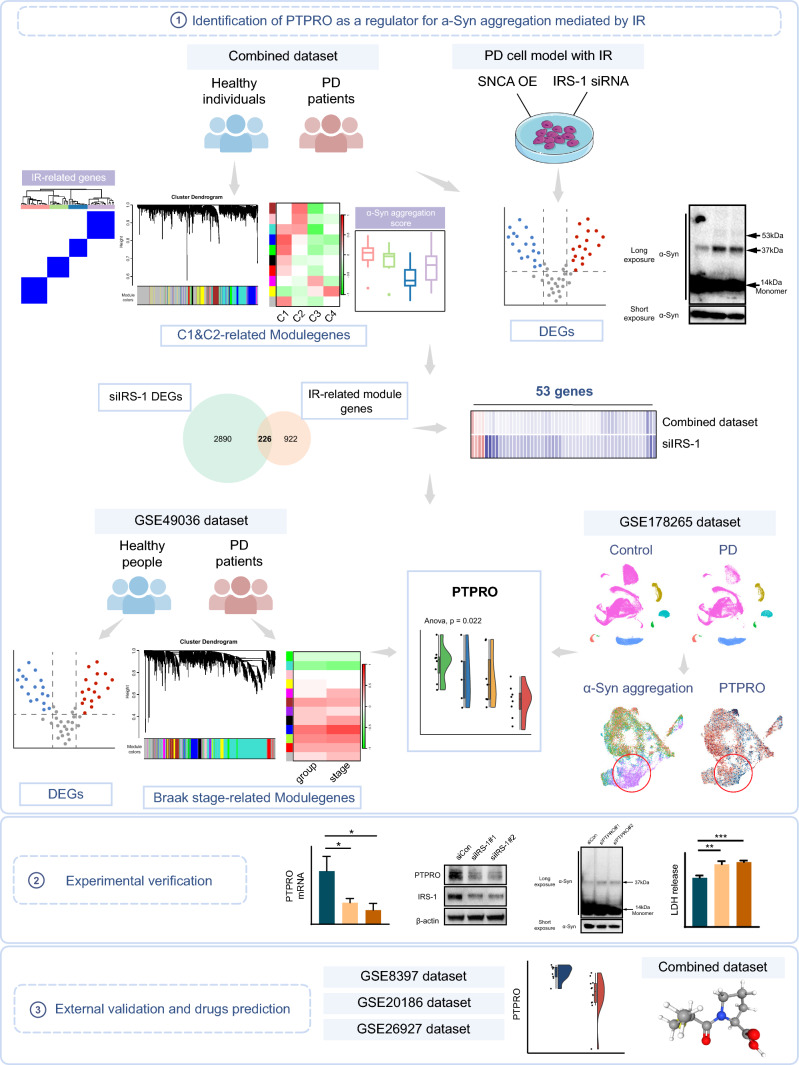
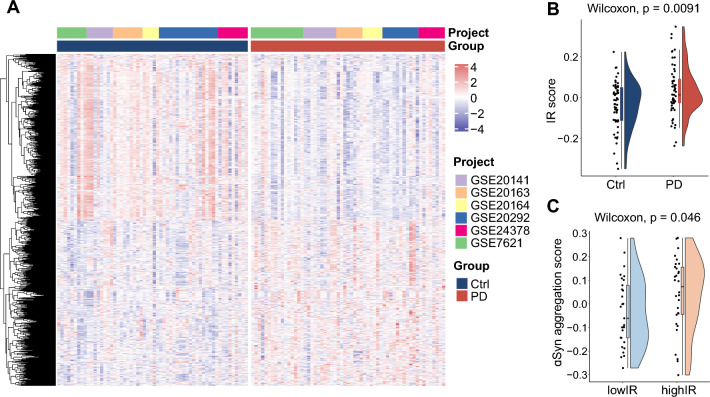
The overall research workflow systematically describes our study (Fig. 1). Previous research has presented evidence that IR contributes to α-Syn aggregation in neuronal cells [23]. To comprehensively investigate the effect of IR on α-Syn aggregation at the transcriptome level, six independent datasets containing transcriptome data from human substantia nigra (SN) of PD patients were enrolled in our research. The datasets were integrated and batch corrected using the “ComBat” algorithm from the "sva" R package to obtain a combined dataset, which contains 59 PD patients and 58 control samples (Fig. 2A). The IR scores, assessed in all samples using gene set variation analysis (GSVA) based on IR-related genes, were significantly higher in PD patients than those observed in control samples (Fig. 2B). The PD samples were then stratified into high IR group and low IR group based on their IR scores. The α-Syn aggregation scores in PD patients were calculated by GSVA based on α-Syn aggregation-related genes. The results indicated that the group with high IR scores exhibited a greater α-Syn aggregation score (Fig. 2C), suggesting the modulation of α-Syn aggregation by IR.
Fig. 1.
Schematic workflow of the study
Fig. 2.
IR score in PD patients is higher and is correlated with α-Syn aggregation score. A Transcriptome profile heatmap for PD and control nigrostriatal (SN) samples in the combined dataset. B The IR score was calculated utilizing the GSVA method for both PD and control samples. C The α-Syn aggregation score was evaluated using GSVA among PD patients with varying IR scores
Analysis of gene modules involved in IR-mediated α-Syn aggregation in PD
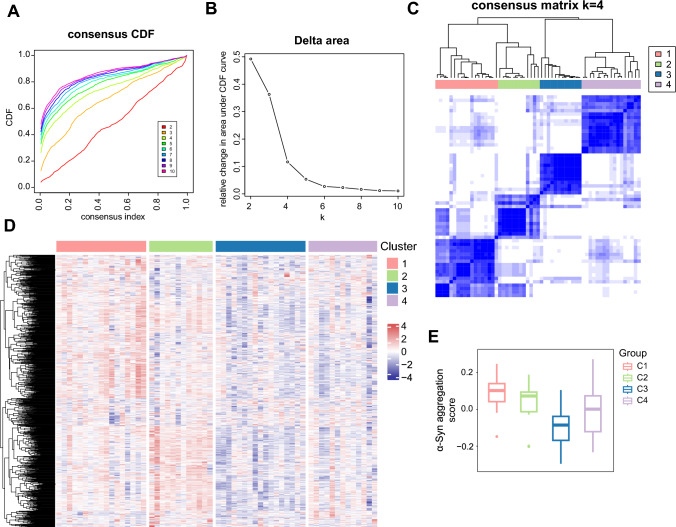
To investigate alterations in the transcriptome features influenced by IR in PD patients, consensus clustering was applied to categorize the gene expression profiles for 59 PD samples into distinct subclasses based on IR-related genes (Fig. 3A, B). According to the cumulative distribution function (CDF), four was determined as the optimal number of clusters. When k = 4, the consensus matrix heatmap demonstrated clear boundaries, indicating the stability of the cluster across multiple iterations (Fig. 3C). The heatmap unveiled the discrepancies in the expression profiles among the four clusters (Fig. 3D). Cluster 1 (C1) and cluster 2 (C2) exhibited higher scores of α-Syn aggregation calculated by GSVA (Fig. 3E). These findings indicate that the signature genes in C1 and C2 subtypes may be involved in the regulation of α-Syn aggregation through IR.
Fig. 3.
Four PD subtypes identified based on IR-related genes. A Consensus clustering cumulative distribution function (CDF) plot depicting consensus distributions for k = 2 to k = 10. B Delta area plot displaying the relative changes in the area under the CDF curve for each k. C Consensus clustering matrix of 59 PD samples when k = 4. D Heatmap demonstrating the differences among the four groups based on gene expression data. E The α-Syn aggregation scores of the four IR-related PD subtypes
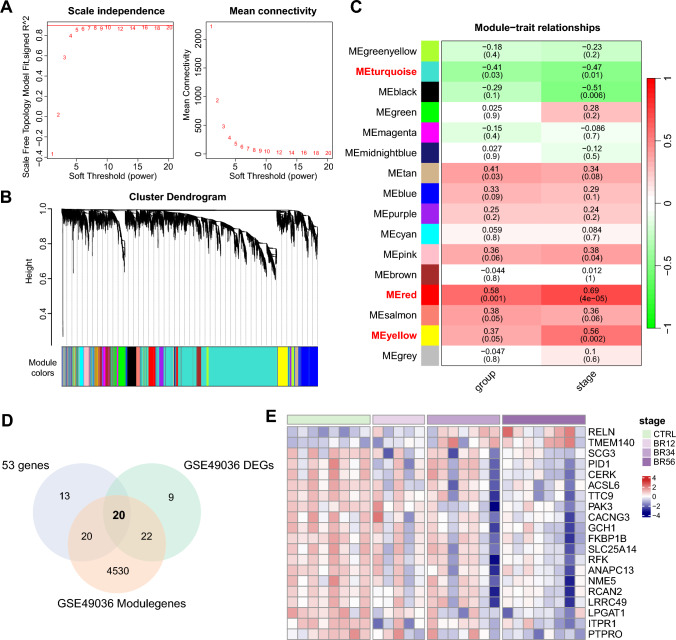
Subsequently, weighted gene co-expression network analysis (WGCNA) was employed to assess the signature genes within the C1 and C2 subtypes. The determination of the soft-threshold value, set at 9, was based on the scale-free topology model and mean connectivity (Fig. 4A). Following this, a hierarchical clustering algorithm yielded ten co-expression modules (Fig. 4B). The blue and green modules displayed a positive correlation with the C1 subtype, and the same for the brown and pink modules with the C2 subtype (Fig. 4C). Furthermore, we conducted GO and KEGG enrichment analyses with genes in the aforementioned four modules (Fig. 4D, E). GO enrichment analysis revealed that these signature genes were predominantly enriched in synaptic vesicle-related biological process, which can be disrupted by α-Syn aggregation [27], and protein folding chaperone function, which may regulate α-Syn conformation and aggregation [28]. Furthermore, KEGG pathway enrichment analysis demonstrated that these signature genes were principally enriched in protein degradation related pathways such as autophagy, ubiquitin mediated proteolysis, and protein processing in endoplasmic reticulum. Taken together, the gene modules that contribute to IR-induced aggregation of α-Syn were identified and analyzed in PD patients.
Fig. 4.
Identification of modules in PD subtypes with high α-Syn aggregation score. A Soft thresholds determined based on the scale-free fit index and the mean connectivity. B Hierarchical cluster tree of co-expression modules. C Correlations between module genes and PD subtypes. D GO analysis of the high α-Syn aggregation PD subtypes-related module genes on biological processes, cellular components, and molecular functions. E KEGG pathways enriched in the module genes related to PD subtype with high α-Syn aggregation
Identification of IR-modulated genes implicated in α-Syn aggregation in DA neurons
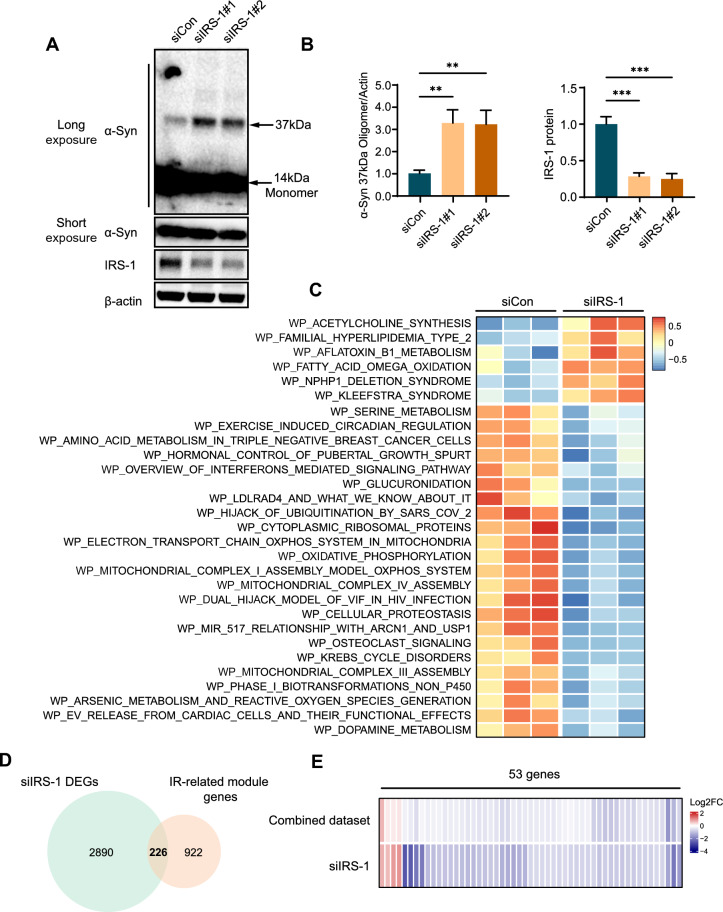
Considering that DA neurons are essential in the degeneration process of PD, we conducted further research using LUHMES cells, a type of neuronal cell known for their ability to differentiate into human DA neurons [24]. DA neurons differentiated from LUHMES cells overexpressing α-Syn were employed as the PD cell model. A previous study showed that heterozygous knockout of IRS-1 leads to IR in mice [29]. Our previous research has demonstrated that elevated IRS-1 expression can alleviate IR [25]. Thus, IRS-1 was silenced by siRNA to induce IR in the PD cell model. Enhanced aggregation of α-Syn was detected in the PD cell model under IR condition (Fig. 5A, B). Subsequently, bulk RNA sequencing was performed on the PD cell model induced with and without IR. The pathway enrichment scores of all the samples were quantified by GSVA. The result revealed significantly different enriched pathways between the PD cell model induced with and without IR (Fig. 5C). Pathways related to inflammation, oxidative phosphorylation, and cellular proteostasis were enriched, which are involved in α-Syn aggregation [30–33]. The DEGs between α-Syn overexpressing LUHMES cells with and without IR were identified using “DESeq2” R package. The identified DEGs were then intersected with module genes that contribute to IR-mediated aggregation of α-Syn in the combined dataset, yielding 226 IR-modulated genes associated with α-Syn aggregation in DA neurons (Fig. 5D). Among these, 53 genes exhibit differential expression between PD and control samples, suggesting their critical roles in IR-mediated PD pathogenesis (Fig. 5E).
Fig. 5.
Screening of IR-mediated genes promoting α-Syn aggregation in DA neurons. A α-Syn overexpressing DA neurons were transfected with siIRS-1 or siCon. IRS-1 and monomeric and oligomeric α-Syn were detected via WB. B Quantification of (A). ***P < 0.005, **P < 0.01. Data are shown as the means ± SEMs. C Heatmap of significantly differentially enriched pathways between the siCon and siIRS-1 groups. D Venn diagram of the intersection of DEGs between the PD cell model with and without IR and IR-related module genes. E Heatmap visualization of 53 overlapping genes obtained by intersecting IR-modulated genes and DEGs in the combined dataset between PD and control samples
PTPRO serves as a regulator for IR-mediated α-Syn aggregation and the progression of PD
To further assess the role of the 53 genes in regulating α-Syn aggregation and the progression of PD, the GSE49036 dataset containing 28 samples from PD patients with Braak stage information was enrolled. Genes associated with PD progression were identified using WGCNA. Soft threshold value was determined as 8, according to scale-free topology model and mean connectivity (Fig. 6A). Sixteen co-expression modules were generated by a hierarchical clustering algorithm (Fig. 6B). Positive correlations were observed between Braak stage and the red module as well as the yellow module, whereas a negative correlation was detected between Braak stage and the turquoise module (Fig. 6C). The Braak stage-related module genes were intersected with the DEGs identified between the PD and control samples in GSE49036 dataset, the results were then intersected with the 53 genes that may contribute to α-Syn aggregation under IR condition in the PD cell model to obtain 20 genes that associated with PD progression mediated by IR. (Fig. 6D). The expression of the twenty genes is indicated in the heatmap, reflecting an evident variation in gene expression pattern across different Braak stages (Fig. 6E).
Fig. 6.
Identification of twenty hub genes associated with PD progression by WGCNA. A Analysis of the scale-free fit index and the mean connectivity to determine the soft threshold. B Cluster dendrogram of co-expression modules in GSE49036. C Correlations between module genes and clinical traits. D Twenty overlapping genes were identified by intersecting the DEGs between PD and control samples with the Braak stage-related module genes, and by determining the intersection with the aforementioned 53 genes, as depicted in a Venn diagram. E Heatmap of the expression of the twenty genes in the different Braak stages
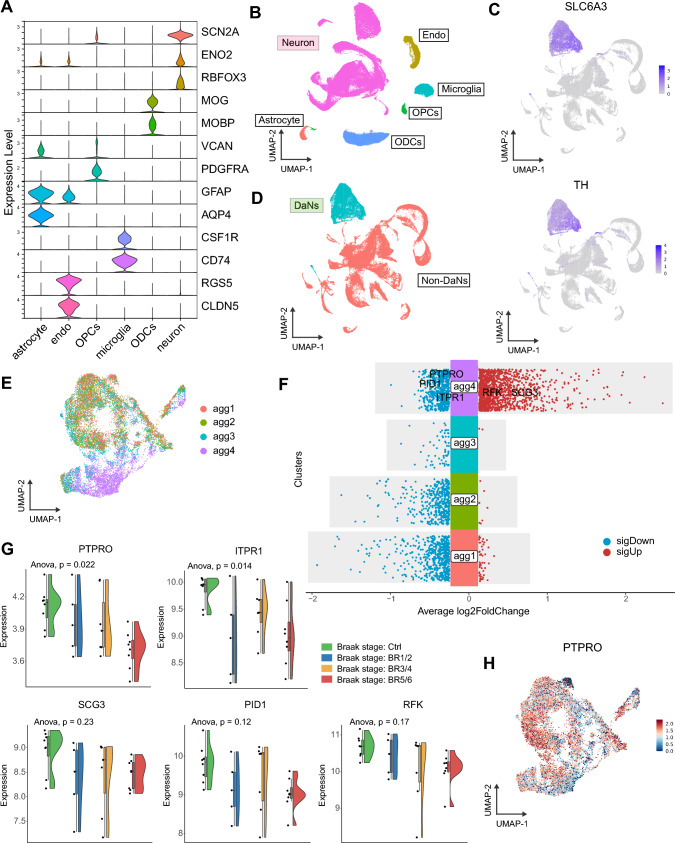
Next, we analyzed a snRNA-seq dataset with PD cohort (GSE178265), to gain in-depth insights into the role of the 20 hub genes on α-Syn aggregation in DA neurons. In the GSE178265 dataset, DA neuron nuclei were enriched using fluorescence-activated nuclei sorting (FANS) with NR4A2 as the specific marker. Then, the NR4A2-positive cells were clustered and annotated based on cell-specific marker genes reported in previous studies [34–38] (Fig. 7A). Six main cell types, including astrocytes, endothelial cells (endo), oligodendrocyte progenitor cells (OPCs), microglia, oligodendrocytes (ODCs) and neurons, were identified (Fig. 7B). DA neurons were further distinguished among neuron subtypes based on characteristic marker genes [36] (Fig. 7C, D). Subsequently, the DA neurons were divided into four subsets based on the α-Syn aggregation score, among which the agg4 subset exhibited the highest α-Syn aggregation score (Fig. 7E). We observed that 5 out of the 20 hub genes were present in the DEGs that identified between agg4 subset and other subsets, implying the importance of the 5 genes in α-Syn aggregation (Fig. 7F). Through evaluating the expression of these genes in groups across different Braak stages in the GSE49036 dataset, only PTPRO exhibited a negative correlation with Braak stages (Fig. 7G). PTPRO was significantly downregulated in the agg4 subset as compared to other subsets (Fig. 7H), exhibiting consistent alterations with the α-Syn aggregation score and highlighting its potential role in α-Syn aggregation.
Fig. 7.
SnRNA-seq revealed a significant correlation between PTPRO and α-Syn aggregation. A Violin plots of marker genes for the six cell subtypes. B UMAP plot of cell identity. C UMAP plots of specific markers (SLC6A3 and TH) for DA neurons. D UMAP plot of neuron subtype identity. E UMAP plot of four DA neuron subsets classified by the α-Syn aggregation score. F The DEGs of each cluster were identified by the FindAllMarkers function of the Seurat package. Five out of the twenty hub genes were indicated in the agg4 subset-specific DEGs. G The expression of PTPRO, ITPR1, SCG3, PID1, and PFK in groups across different Braak stages in GSE49036. H PTPRO expression in DA neurons displayed in the UMAP plot
Decreased PTPRO expression promotes α-Syn aggregation
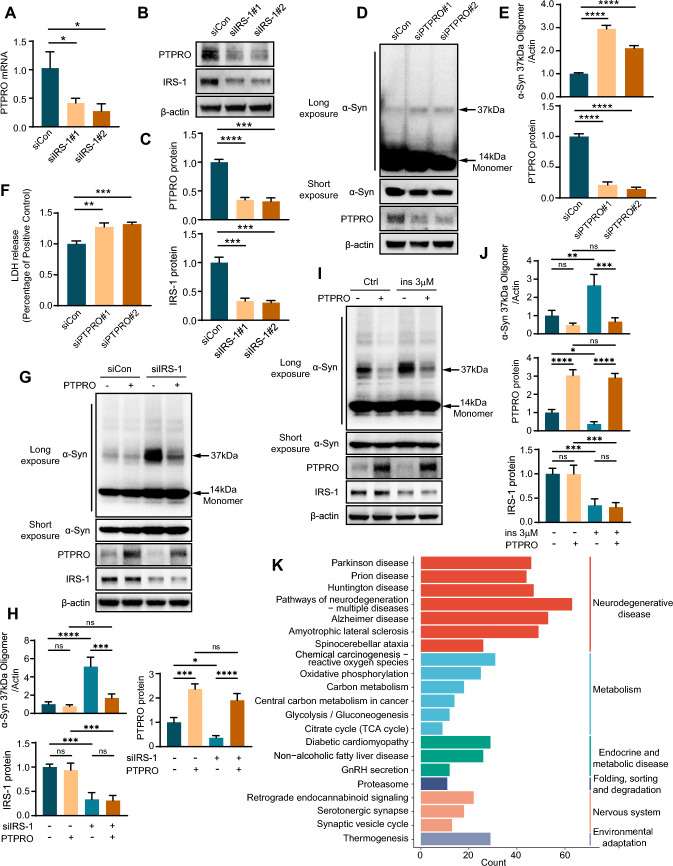
To validate the role of PTPRO in IR-mediated α-Syn aggregation, we scrutinized PTPRO expression in the PD cell model under IR condition. The decreased mRNA expression and protein level of PTPRO were observed in the PD cell model induced with IR (Fig. 8A–C). Furthermore, we explored the impact of PTPRO down-regulation on α-Syn aggregation. The decreased PTPRO markedly promoted α-Syn aggregation and cell death in the PD cell model (Fig. 8D–F). Conversely, overexpression of PTPRO significantly attenuated α-Syn aggregation in our PD cell model under IR conditions (Fig. 8G, H), further substantiating the critical role of PTPRO in IR-mediated α-Syn aggregation. Moreover, we employed an additional model of high insulin-induced IR. Consistent with our previous observations, induction of IR through high insulin treatment in the PD cell model resulted in increased α-Syn aggregation and concomitant decrease in PTPRO expression levels. Importantly, overexpression of PTPRO in this high insulin-induced IR model also led to a significant reduction in α-Syn aggregation (Fig. 8I, J), mirroring our previous findings. These results collectively underscore the importance of PTPRO in modulating IR-mediated α-Syn aggregation in PD cellular models. To better understand the function of PTPRO in α-Syn aggregation, the PD patients in the combined dataset were divided into two groups based on PTPRO expression. Following this, KEGG pathway enrichment analysis on DEGs between the two groups was performed. KEGG analysis revealed that the DEGs were abundant in pathways linked to neurodegenerative disease, metabolism and proteasome (Fig. 8K). These findings suggested that disordered metabolism and abnormal degradation of target proteins in DA neurons might be responsible for the downregulation of PTPRO which promotes α-Syn aggregation.
Fig. 8.
Reduced PTPRO expression detected under IR condition facilitates α-Syn aggregation in the PD cell model. siIRS-1 or siCon was transfected into α-Syn overexpressing LUHMES cells. The mRNA expression of PTPRO was analyzed using RT-qPCR (A), and the protein was determined using WB (B). C Quantification of (B). D α-Syn overexpressing LUHMES cells were transfected with PTPRO siRNA (siPTPRO) or negative control (siCon). WB was performed to detect PTPRO, as well as monomeric and oligomeric α-Syn. E Quantification of (D). F LDH assay was applied to assess cell death. G Lentivirus expressing PTPRO or control was introduced into LUHMES cells overexpressing a-Syn under control or IRS-1 knockdown conditions. WB assay was performed to assess the protein levels of monomeric and oligomeric α-Syn, PTPRO, and IRS-1. H Quantification of (G). I LUHMES cells overexpressing α-Syn were transduced with lentivirus expressing PTPRO or control, and treated with or without 3 μM insulin as indicated. Protein levels of monomeric and oligomeric α-Syn, PTPRO, and IRS-1 were assessed by WB assay. J Quantification of (I). K KEGG pathway analysis was conducted on DEGs between the high-PTPRO expression group and the low-PTPRO expression group in PD patients from the combined dataset. ****P < 0.0001, ***P < 0.005, **P < 0.01, *P < 0.05. The data are shown as the means ± SEMs
Validation of downregulated PTPRO expression in multiple PD datasets, and prediction of compound targeting for PTPRO
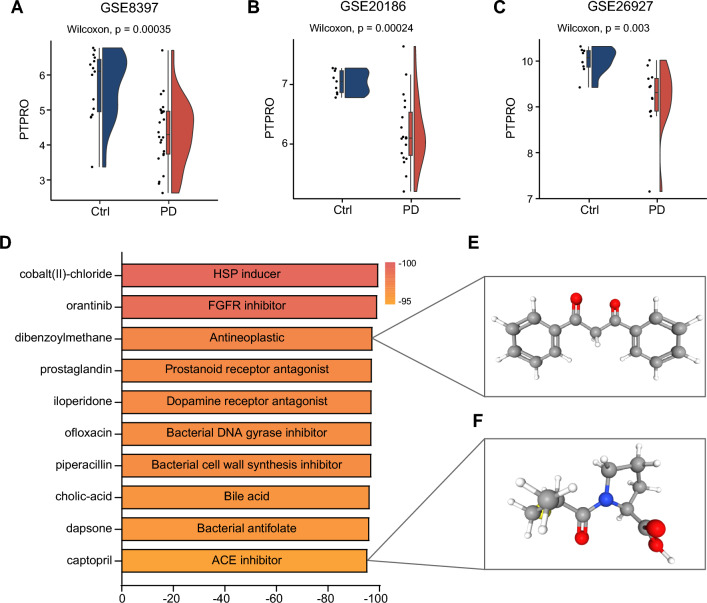
The downregulated expression of PTPRO was validated across three external PD datasets. A consistently significant downregulation of PTPRO expression was observed in PD patients (Fig. 9A–C), implying its potential significance in the progression of PD. Targeting PTPRO may therefore present as an effective strategy to mitigate the progression of PD facilitated by IR. To screen small molecule compounds capable of counteracting PTPRO dysregulation and potentially slowing PD progression, we utilized the CMap database to predict promising drugs based on the DEGs between the high-PTPRO expression group and the low-PTPRO expression group. Among the top 10 drugs exhibiting significantly negative enrichment scores, indicative of their ability to counteract gene expression changes induced by PTPRO downregulation, dibenzoylmethane and captopril emerged as promising candidates for PD treatment (Fig. 9D–F). Previous studies have demonstrated that dibenzoylmethane exhibit a neuroprotective effect in neurodegenerative mouse models [39, 40]. Similarly, captopril, an angiotensin-converting enzyme (ACE) inhibitor, has demonstrated neuroprotective effects in animal models of PD by suppressing the overproduction of reactive oxygen species [41, 42]. These findings suggest that these two compounds may impede the progression of PD associated with insulin resistance through counteracting PTPRO downregulation. Collectively, these results underscore the potential of PTPRO as a promising therapeutic target for personalized treatment approaches in PD patients with IR.
Fig. 9.
Validation of downregulated PTPRO expression in multiple PD datasets, and prediction of compound targeting for PTPRO. PTPRO expression between PD and control samples in external datasets GSE8397 (A), GSE20186 (B), GSE26927 (C). D Candidate small-molecule compounds affecting downstream of PTPRO predicted by CMap. 3D structures of dibenzoylmethane (E) and captopril (F), retrieved from the PubChem database
Discussion
In this study, we provided a comprehensively analysis of the modulation of α-Syn aggregation by IR. Firstly, the module genes in PD patients regulated by IR were explored in PD subtypes exhibiting high α-Syn aggregation scores. Through the combination of analyses involving DEGs in the PD cell model modulated by IR, genes within IR-related modules from PD patients, and DEGs between PD and control samples, we successfully identified 53 IR-modulated genes in DA neurons associated with α-Syn aggregation. Subsequent investigations were focused on the functions of these 53 genes in PD progression and α-Syn aggregation. PTPRO emerged as a key regulator in IR-mediated α-Syn aggregation and PD progression. In the PD cell model, increased α-Syn aggregation and enhanced cell death were observed subsequent to PTPRO silencing. Moreover, PTPRO might serve as a promising therapeutic target for the individualized treatment in PD patients with IR.
Numerous identified risk factors contribute to the onset and progression of PD. One prominent risk factor for PD is the compromised insulin signaling in the brain [43], which is strongly correlated with the progression of PD, suggesting a crucial role of IR in the development of PD [44]. Previous research has exhibited an exacerbation of α-Syn accumulation within the midbrain and substantia nigra of mice with T2DM [22]. Another study observed that IR enhances the expression and phosphorylation of α-Syn, leading to its aggregation in an insulin-induced IR cell model [23]. Despite these findings, the precise mechanism through which IR promotes α-Syn aggregation remains to be elucidated.
In our research, the functional enrichment analysis of module genes revealed that pathways altered in IR-related PD subtypes with high α-Syn aggregation scores were notably enriched in biological processes related to synaptic vesicles and the chaperone function of protein folding. α-Syn is typically associated with synaptic vesicles in presynaptic terminals [45], and is believed to play a role in regulating neurotransmitter release and maintaining synaptic integrity [46–48]. Aggregated α-Syn was shown to disrupt normal synaptic function and axonal transport [27]. Protein folding chaperones, such as heat shock proteins (HSPs), are implicated in preventing the misfolding, oligomerization and aggregation of α-Syn [28, 49].
Moreover, our KEGG analysis highlighted additional enriched pathways associated with protein degradation, such as autophagy, ubiquitin-mediated proteolysis, and protein processing in the endoplasmic reticulum [50–52]. These pathways are involved in α-Syn degradation and clearance, emphasizing that IR may promote α-Syn aggregation in PD patients by compromising both protein folding and degradation mechanisms.
Furthermore, our investigation revealed that pathways related to inflammation, oxidative phosphorylation, and cellular proteostasis were enriched in DA neurons overexpressing α-Syn upon IR induction. Interferons mediate inflammation, and alterations in their signaling pathway may induce neuroinflammation, potentially contributing to the aggregation of α-synuclein [30, 31]. Oxidative stress resulting from dysregulated oxidative phosphorylation also fosters α-Syn aggregation [32]. IR may disrupt cellular proteostasis, a process maintained by regulated control of protein folding, post-translational modification, and protein degradation [53], resulting in abnormal α-Syn aggregation.
We identified 53 genes that potentially facilitate α-Syn aggregation mediated by IR in DA neurons. Among those, PTPRO was identified as a significant regulator in IR-mediated α-Syn aggregation and PD progression. PTPRO, a receptor-type protein tyrosine phosphatase, exhibits high expression in adult brain tissues [54], and is significantly associated with neurocognitive function [55]. Ptpro in the hippocampus shows a negative correlation with aging, and its deletion is associated with increased susceptibility to hippocampal neuronal death in mice treated with doxorubicin (DOX) [56]. Our analysis revealed that DEGs related to PTPRO are abundant in pathways linked to neurodegenerative diseases, metabolism, and the proteasome. While the precise mechanism through which PTPRO influences α-Syn aggregation remains to be fully elucidated, our pathway analysis suggests several potential mechanisms. PTPRO may be involved in regulating oxidative stress responses, as indicated by the enrichment of pathways related to reactive oxygen species, oxidative phosphorylation and glycolysis. Notably, oxidative stress related pathway was also identified in IR-related DEGs enriched pathways, contributing to the promotion of α-Syn aggregation, as previously discussed [32]. The enrichment of the proteasome pathway suggests PTPRO may regulate α-Syn aggregation by influencing proteasomal function. Impaired proteasome activity can contribute to the abnormal aggregation of α-Syn [57]. These potential mechanisms are not mutually exclusive and may interact in complex ways, creating a multifaceted impact on α-Syn aggregation. Further experimental studies are needed to elucidate the specific roles of PTPRO in these pathways and their relation to α-Syn aggregation in the context of IR and PD progression.
In summary, our research uncovered PTPRO as a crucial determinant in α-Syn aggregation mediated by IR. Nevertheless, it is important to note that the validation of PTPRO was confined to cultured DA neurons in the present study, and supporting evidence from mouse and human samples is still under investigation. Additionally, further exploration is needed to elucidate the molecular mechanism underlying the regulation of α-Syn aggregation by PTPRO and to evaluate the therapeutic potential of targeted small molecule drugs.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from the Jinan Science and Technology Bureau of Shandong Province (2021GXRC029), Fundamental Research Funds for the Central Universities (2022JC019), National Natural Science Foundation of China (Nos. 82273195, 82273286, 82201402), Youth Taishan Scholar Program of Shandong Province (tsqn202211316), Science and Technology Innovation Major Project, Ministry of Science and Technology of China (2021ZD0201600), Taishan Pandeng Scholar Program of Shandong Province (tspd20210322), Natural Science Foundation of Shandong Province of China (ZR2022MH313, ZR2023QH240, ZR2023LZL004), Shandong Province Youth Innovation Plan (2022KJ011), China Postdoctoral Science Foundation (2022TQ0196), Shandong Postdoctoral Science Foundation (SDCX-ZG-202302028).
Author contributions
ST, QT, and GL designed this work. ST, HC, RZ, QZ, and ZG performed the statistical analyses. ST performed all experiment. ST and HC wrote the original manuscript. PW, HX, QT, and GL revised the manuscript. All authors read and approved the final manuscript.
Funding
Several funding sources that supported this study were provided in the Acknowledgments.
Availability of data and materials
The publicly available datasets referred in this study are downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Transcriptome data from IR-induced PD cell model reported in this study are available from the corresponding author upon reasonable request. R code for this study is available from the authors upon reasonable request.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qilin Tang, Email: Qilin.Tang@sdu.edu.cn.
Gang Li, Email: dr.ligang@sdu.edu.cn.
References
- 1.Reaven GM (2005) The insulin resistance syndrome: definition and dietary approaches to treatment. Annu Rev Nutr 25:391–406 10.1146/annurev.nutr.24.012003.132155 [DOI] [PubMed] [Google Scholar]
- 2.DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ et al (2015) Type 2 diabetes mellitus. Nat Rev Dis Primers 1:15019 10.1038/nrdp.2015.19 [DOI] [PubMed] [Google Scholar]
- 3.Milstein JL, Ferris HA (2021) The brain as an insulin-sensitive metabolic organ. Mol Metab 52:101234 10.1016/j.molmet.2021.101234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mieczkowski J, Kocyk M, Nauman P, Gabrusiewicz K, Sielska M, Przanowski P et al (2015) Down-regulation of IKKβ expression in glioma-infiltrating microglia/macrophages is associated with defective inflammatory/immune gene responses in glioblastoma. Oncotarget 6:33077–33090 10.18632/oncotarget.5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craft S, Watson GS (2004) Insulin and neurodegenerative disease: shared and specific mechanisms. Lancet Neurol 3:169–178 10.1016/S1474-4422(04)00681-7 [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Pozo VA, Tamayo-Trujillo R, Cadena-Ullauri S, Frias-Toral E, Guevara-Ramírez P, Paz-Cruz E et al (2023) The Molecular mechanisms of the relationship between insulin resistance and Parkinson’s disease pathogenesis. Nutrients 15:3585 10.3390/nu15163585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mielke JG, Taghibiglou C, Liu L, Zhang Y, Jia Z, Adeli K et al (2005) A biochemical and functional characterization of diet-induced brain insulin resistance. J Neurochem 93:1568–1578 10.1111/j.1471-4159.2005.03155.x [DOI] [PubMed] [Google Scholar]
- 8.Dineley KT, Jahrling JB, Denner L (2014) Insulin resistance in Alzheimer’s disease. Neurobiol Dis 72 Pt A:92–103 10.1016/j.nbd.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J et al (2017) Parkinson disease. Nat Rev Dis Primers 3:17013 10.1038/nrdp.2017.13 [DOI] [PubMed] [Google Scholar]
- 10.Ou Z, Pan J, Tang S, Duan D, Yu D, Nong H et al (2021) Global trends in the incidence, prevalence, and years lived with disability of Parkinson’s disease in 204 countries/territories from 1990 to 2019. Front Public Health 9:776847 10.3389/fpubh.2021.776847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Valbuena I, Amat-Villegas I, Valenti-Azcarate R, Carmona-Abellan MDM, Marcilla I, Tuñon M-T et al (2018) Interaction of amyloidogenic proteins in pancreatic β cells from subjects with synucleinopathies. Acta Neuropathol 135:877–886 10.1007/s00401-018-1832-0 [DOI] [PubMed] [Google Scholar]
- 12.Cheong JLY, de Pablo-Fernandez E, Foltynie T, Noyce AJ (2020) The association between type 2 diabetes mellitus and Parkinson’s disease. J Parkinsons Dis 10:775–789 10.3233/JPD-191900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talbot K, Wang H-Y, Kazi H, Han L-Y, Bakshi KP, Stucky A et al (2012) Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J Clin Invest 122:1316–1338 10.1172/JCI59903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moloney AM, Griffin RJ, Timmons S, O’Connor R, Ravid R, O’Neill C (2010) Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol Aging 31:224–243 10.1016/j.neurobiolaging.2008.04.002 [DOI] [PubMed] [Google Scholar]
- 15.Kakoty V, Kc S, Kumari S, Yang C-H, Dubey SK, Sahebkar A et al (2023) Brain insulin resistance linked Alzheimer’s and Parkinson’s disease pathology: an undying implication of epigenetic and autophagy modulation. Inflammopharmacology 31:699–716 10.1007/s10787-023-01187-z [DOI] [PubMed] [Google Scholar]
- 16.Greene MW, Sakaue H, Wang L, Alessi DR, Roth RA (2003) Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by Serine 312 phosphorylation. J Biol Chem 278:8199–8211 10.1074/jbc.M209153200 [DOI] [PubMed] [Google Scholar]
- 17.Bassil F, Delamarre A, Canron M-H, Dutheil N, Vital A, Négrier-Leibreich M-L et al (2022) Impaired brain insulin signalling in Parkinson’s disease. Neuropathol Appl Neurobiol 48:e12760 10.1111/nan.12760 [DOI] [PubMed] [Google Scholar]
- 18.Chou S-Y, Chan L, Chung C-C, Chiu J-Y, Hsieh Y-C, Hong C-T (2020) Altered insulin receptor substrate 1 phosphorylation in blood neuron-derived extracellular vesicles from patients with Parkinson’s disease. Front Cell Dev Biol 8:564641 10.3389/fcell.2020.564641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M (1997) Alpha-synuclein in Lewy bodies. Nature 388:839–840 10.1038/42166 [DOI] [PubMed] [Google Scholar]
- 20.Wegrzynowicz M, Bar-On D, Calo’ L, Anichtchik O, Iovino M, Xia J et al (2019) Depopulation of dense α-synuclein aggregates is associated with rescue of dopamine neuron dysfunction and death in a new Parkinson’s disease model. Acta Neuropathol 138:575–595 10.1007/s00401-019-02023-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong YC, Krainc D (2017) α-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat Med 23:1–13 10.1038/nm.4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Zhai Y-Q, Xu L-L, Qiao C, Sun X-L, Ding J-H et al (2014) Metabolic inflammation exacerbates dopaminergic neuronal degeneration in response to acute MPTP challenge in type 2 diabetes mice. Exp Neurol 251:22–29 10.1016/j.expneurol.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 23.Hong C-T, Chen K-Y, Wang W, Chiu J-Y, Wu D, Chao T-Y et al (2020) Insulin resistance promotes Parkinson’s disease through aberrant expression of α-synuclein, mitochondrial dysfunction, and deregulation of the polo-like kinase 2 signaling. Cells 9:E740 10.3390/cells9030740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Q, Gao P, Arzberger T, Höllerhage M, Herms J, Höglinger G et al (2021) Alpha-synuclein defects autophagy by impairing SNAP29-mediated autophagosome-lysosome fusion. Cell Death Dis 12:854 10.1038/s41419-021-04138-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian S, Tan S, Jia W, Zhao J, Sun X (2021) Activation of Wnt/β-catenin signaling restores insulin sensitivity in insulin resistant neurons through transcriptional regulation of IRS-1. J Neurochem 157:467–478 10.1111/jnc.15277 [DOI] [PubMed] [Google Scholar]
- 26.Wang P, Zhao J, Sun X (2021) DYRK1A phosphorylates MEF2D and decreases its transcriptional activity. J Cell Mol Med 25:6082–6093 10.1111/jcmm.16505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundblad M, Decressac M, Mattsson B, Björklund A (2012) Impaired neurotransmission caused by overexpression of α-synuclein in nigral dopamine neurons. Proc Natl Acad Sci U S A 109:3213–3219 10.1073/pnas.1200575109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auluck PK, Chan HYE, Trojanowski JQ, Lee VMY, Bonini NM (2002) Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295:865–868 10.1126/science.1067389 [DOI] [PubMed] [Google Scholar]
- 29.Shirakami A, Toyonaga T, Tsuruzoe K, Shirotani T, Matsumoto K, Yoshizato K et al (2002) Heterozygous knockout of the IRS-1 gene in mice enhances obesity-linked insulin resistance: a possible model for the development of type 2 diabetes. J Endocrinol 174:309–319 10.1677/joe.0.1740309 [DOI] [PubMed] [Google Scholar]
- 30.Picca A, Guerra F, Calvani R, Romano R, Coelho-Júnior HJ, Bucci C et al (2021) Mitochondrial dysfunction, protein misfolding and neuroinflammation in Parkinson’s disease: roads to biomarker discovery. Biomolecules 11:1508 10.3390/biom11101508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindestam Arlehamn CS, Dhanwani R, Pham J, Kuan R, Frazier A, Rezende Dutra J et al (2020) α-Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson’s disease. Nat Commun 11:1875 10.1038/s41467-020-15626-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Won SJ, Fong R, Butler N, Sanchez J, Zhang Y, Wong C et al (2022) Neuronal oxidative stress promotes α-synuclein aggregation in vivo. Antioxidants (Basel) 11:2466 10.3390/antiox11122466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciechanover A, Kwon YT (2015) Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 47:e147 10.1038/emm.2014.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamath T, Abdulraouf A, Burris SJ, Langlieb J, Gazestani V, Nadaf NM et al (2022) Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat Neurosci 25:588–595 10.1038/s41593-022-01061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smajić S, Prada-Medina CA, Landoulsi Z, Ghelfi J, Delcambre S, Dietrich C et al (2022) Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 145:964–978 10.1093/brain/awab446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R et al (2020) A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun 11:4183 10.1038/s41467-020-17876-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR et al (2020) Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat Med 26:131–142 10.1038/s41591-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marzi SJ, Leung SK, Ribarska T, Hannon E, Smith AR, Pishva E et al (2018) A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat Neurosci 21:1618–1627 10.1038/s41593-018-0253-7 [DOI] [PubMed] [Google Scholar]
- 39.Halliday M, Radford H, Zents KAM, Molloy C, Moreno JA, Verity NC et al (2017) Repurposed drugs targeting eIF2α-P-mediated translational repression prevent neurodegeneration in mice. Brain 140:1768–1783 10.1093/brain/awx074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano K, Kitao Y, Tabata Y, Miura H, Sato K, Takuma K et al (2007) A dibenzoylmethane derivative protects dopaminergic neurons against both oxidative stress and endoplasmic reticulum stress. Am J Physiol Cell Physiol 293:C1884-1894 10.1152/ajpcell.00305.2007 [DOI] [PubMed] [Google Scholar]
- 41.Perez-Lloret S, Otero-Losada M, Toblli JE, Capani F (2017) Renin-angiotensin system as a potential target for new therapeutic approaches in Parkinson’s disease. Expert Opin Investig Drugs 26:1163–1173 10.1080/13543784.2017.1371133 [DOI] [PubMed] [Google Scholar]
- 42.Sonsalla PK, Coleman C, Wong L-Y, Harris SL, Richardson JR, Gadad BS et al (2013) The angiotensin converting enzyme inhibitor captopril protects nigrostriatal dopamine neurons in animal models of parkinsonism. Exp Neurol 250:376–383 10.1016/j.expneurol.2013.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Pablo-Fernandez E, Goldacre R, Pakpoor J, Noyce AJ, Warner TT (2018) Association between diabetes and subsequent Parkinson disease: a record-linkage cohort study. Neurology 91:e139–e142 10.1212/WNL.0000000000005771 [DOI] [PubMed] [Google Scholar]
- 44.Athauda D, Foltynie T (2016) Insulin resistance and Parkinson’s disease: a new target for disease modification? Prog Neurobiol 145–146:98–120 10.1016/j.pneurobio.2016.10.001 [DOI] [PubMed] [Google Scholar]
- 45.Sharma M, Burré J (2023) α-Synuclein in synaptic function and dysfunction. Trends Neurosci 46:153–166 10.1016/j.tins.2022.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK et al (2010) Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65:66–79 10.1016/j.neuron.2009.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC (2010) Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329:1663–1667 10.1126/science.1195227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lashuel HA, Overk CR, Oueslati A, Masliah E (2013) The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat Rev Neurosci 14:38–48 10.1038/nrn3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu S, Tan J, Qin L, Lv L, Yan W, Zhang H et al (2021) Molecular chaperones and Parkinson’s disease. Neurobiol Dis 160:105527 10.1016/j.nbd.2021.105527 [DOI] [PubMed] [Google Scholar]
- 50.Xilouri M, Brekk OR, Stefanis L (2016) Autophagy and alpha-synuclein: relevance to Parkinson’s disease and related synucleopathies. Mov Disord 31:178–192 10.1002/mds.26477 [DOI] [PubMed] [Google Scholar]
- 51.Rott R, Szargel R, Shani V, Hamza H, Savyon M, Abd Elghani F et al (2017) SUMOylation and ubiquitination reciprocally regulate α-synuclein degradation and pathological aggregation. Proc Natl Acad Sci U S A 114:13176–13181 10.1073/pnas.1704351114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Costa CA, Manaa WE, Duplan E, Checler F (2020) The endoplasmic reticulum stress/unfolded protein response and their contributions to Parkinson’s disease physiopathology. Cells 9:2495 10.3390/cells9112495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurtishi A, Rosen B, Patil KS, Alves GW, Møller SG (2019) Cellular proteostasis in neurodegeneration. Mol Neurobiol 56:3676–3689 10.1007/s12035-018-1334-z [DOI] [PubMed] [Google Scholar]
- 54.Bodden K, Bixby JL (1996) CRYP-2: a receptor-type tyrosine phosphatase selectively expressed by developing vertebrate neurons. J Neurobiol 31:309–324 [DOI] [PubMed] [Google Scholar]
- 55.LeBlanc M, Kulle B, Sundet K, Agartz I, Melle I, Djurovic S et al (2012) Genome-wide study identifies PTPRO and WDR72 and FOXQ1-SUMO1P1 interaction associated with neurocognitive function. J Psychiatr Res 46:271–278 10.1016/j.jpsychires.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 56.Yao Z, Dong H, Zhu J, Du L, Luo Y, Liu Q et al (2023) Age-related decline in hippocampal tyrosine phosphatase PTPRO is a mechanistic factor in chemotherapy-related cognitive impairment. JCI Insight 8:e166306 10.1172/jci.insight.166306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bi M, Du X, Jiao Q, Chen X, Jiang H (2021) Expanding the role of proteasome homeostasis in Parkinson’s disease: beyond protein breakdown. Cell Death Dis 12:154 10.1038/s41419-021-03441-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The publicly available datasets referred in this study are downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Transcriptome data from IR-induced PD cell model reported in this study are available from the corresponding author upon reasonable request. R code for this study is available from the authors upon reasonable request.