Abstract
Cell state control is crucial for normal tissue development and cancer cell mimicry of stem/progenitor states, contributing to tumor heterogeneity, therapy resistance, and progression. Here, we demonstrate that the cell surface glycoprotein Mcam maintains the tumorigenic luminal progenitor (LP)-like epithelial cell state, leading to Basal-like mammary cancers. In the Py230 mouse mammary carcinoma model, Mcam knockdown (KD) destabilized the LP state by deregulating the Ck2/Stat3 axis, causing a switch to alveolar and basal states, loss of an estrogen-sensing subpopulation, and resistance to tamoxifen—an effect reversed by Ck2 and Stat3 inhibitors. In vivo, Mcam KD blocked generation of Basal-like tumors and Sox10+Krt14+ cells. In human tumors, MCAM loss was largely exclusive of the Basal-like subtype, linked instead to proliferative Luminal subtypes, including often endocrine-resistant Luminal B cancers. This study has implications for developing therapies targeting MCAM, CK2, and STAT3 and their likely effective contexts.
Subject terms: Breast cancer, Cell signalling
Introduction
Breast cancers exhibit cellular and subtype heterogeneity that complicate treatment and contribute to disease progression. Whereas estrogen- and/or progesterone-hormone receptor expressing (HR+) and HER2 amplified (HER2+) cancers are treated with molecular therapies targeting their molecular drivers, triple negative breast cancers (TNBC) lack these molecular targets (reviewed in Testa et al.1). Transcriptional profiling has identified intrinsic breast cancer subtypes that partially overlap with these clinical subtype designations and with cellular compartments of the normal breast2. For instance, Luminal A and B subtypes are typically HR+ (i.e. ER/PR+), the ‘HER2-enriched’ intrinsic subtype comprises most HER2 amplified tumors, and the ‘Basal-like’ designation encompasses most TNBCs3.
The origins of this heterogeneity remain controversial, and the molecular mediators are incompletely understood. However, it is generally held that tumor subtype reflects particular combinations of driver mutations and intrinsic characteristics of the cell of origin1,4,5. As such, experiments in mice targeting drivers to different mammary gland cell types give rise to different subtypes of breast cancer6,7. Nevertheless, cell plasticity has emerged as a significant contributor to intra- and inter-tumoral heterogeneity, likely playing a crucial role in therapy resistance, dormancy, and metastasis in breast and other cancers8–10. Examples include epithelial-to-mesenchymal transition (EMT)—a classic plasticity mechanism implicated in tumor cell migration, invasion, and metastasis7,11—and cell lineage switching within the epithelial compartment, a well-documented mechanism of therapy evasion and progression in diverse cancers12–14. Thus, for example, hormone-dependent cancers can sometimes transition into hormone-independent phenotypes that evade conventional hormone-blocking therapy. In mammary carcinoma, this conversion has been associated with EMT or alternatively a state transition to a hormone insensitive, alveolar progenitor program classically driven by Elf5, and Stat3/515–17. Identifying and understanding the molecular regulators of epithelial lineage plasticity in breast and other cancers could reveal strategies to maintain therapy sensitivity or uncover new therapeutic vulnerabilities. In this regard, we chose to examine the membrane-integral, cell-surface glycoprotein MCAM (CD146; MUC18; S-Endo-1; and Gicerin) as a regulator of epithelial plasticity in breast cancer based on several prior observations. Our prior studies showed that Mcam is broadly expressed in multipotent fetal mammary stem cells18,19. Mcam is also an established marker of pericytes noted for their high intrinsic cellular plasticity, although lower-level expression in a variety of other cells and tissues, including luminal progenitor (LP) compartments, has been reported19–21. MCAM is upregulated in a variety of cancers and has different activities depending on the target cancer/cell type20,21. In breast cancers, elevated expression of MCAM has been associated with the Basal-like subtype and poor prognosis20,22. Experimental overexpression (OE) of MCAM in MCF7 and SKBR-3 cells (HR+ and HER2+ cell lines, respectively) drives EMT and enhances tumorigenesis in xenograft settings23,24. However, MCAM has also been reported to be tumor suppressive and recent work has challenged the notion that it is widely oncogenic25,26.
Mechanistically, MCAM has been implicated in a variety of biological processes relevant to mammary development, cell identity, and cancer. MCAM can mediate cell-cell interactions between heterotypic cells during extravasation of immune and tumor cells and promotes homotypic cell interactions through cognate binding partners that are yet to be identified20. MCAM binds S100A8/9, Galectin-1/3 and Laminin-α4 as extracellular ligands and regulates Integrin β1 and CD44 levels on the cell surface27–29. Inside the cell, MCAM regulates integrin-mediated signaling through PIP2 and FAK activation, and ERM binding, and cooperates in trafficking of a discrete internalized organellar structure associated with Wnt-mediated planar cell polarity30–32. The MCAM cytoplasmic tail possesses putative binding/phosphorylation sites for Src family kinases, PKA, PKC and CK2 that may orchestrate coupling and sequestering of broad-spectrum kinases to and from their targets33. To date, it is unknown how these signals are integrated into cell fate choices, particularly in luminal breast cancer cells where MCAM has thus far only been studied under conditions of acquired or experimental OE20,23,31,33.
Despite notable differences in tissue architecture and the distribution of certain progenitor markers between mouse and human mammary epithelia34–36, key similarities make the mouse mammary gland an informative model for understanding normal and neoplastic mammary cell states37. In particular, conserved features of the LP-like state and its potential contribution to aggressive Basal-like mammary carcinomas has been demonstrated38–40. Thus, in the present study, we used a LP/stem cell-like mammary tumor cell line derived from the MMTV-PyMT mouse model (Py230) that exhibits lineage and tumor subtype plasticity7, and compared findings with a number of additional cell lines and human archival tumor data. We found that Mcam is a potent regulator of cell state in mouse mammary carcinoma, therapy resistance and tumorigenicity, and that changes in human MCAM track with molecular subtype and disease aggressiveness. Mcam’s effects are mediated by its governance of the master regulatory kinase Ck2. Mcam KD altered Ck2 substrate utilization, including its activation of Stat3 and inactivation of Pten. Inhibition of Ck2/Stat3 reversed these signaling and transcriptional state changes, and the associated insensitivity to tamoxifen—a common characteristic of human Luminal B breast cancers, which we note often harbor MCAM loss. These results may inform the development of combination breast cancer therapies that incorporate MCAM and/or CK2 and STAT inhibitors with existing molecularly targeted therapies and point to opportunities as well as potential challenges in clinical management of breast cancer.
Results
Mcam maintains epithelial phenotypes in mammary cancer cells
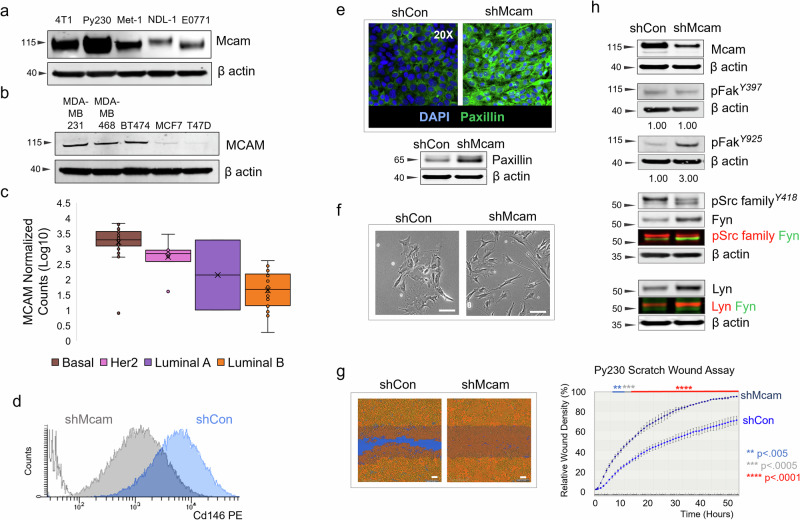
Although experimental OE of MCAM has been reported to drive EMT and basal phenotypes in breast cancer cells21,24,41, we found that diverse murine/human breast cancer cell lines and patient derived organoid models expressed MCAM including luminal and LP-like models (Fig. 1a–c)7,42. MCAM/Mcam expression has previously been reported in Basal-like models and only sporadically in more luminal lines22,43. To test the requirements for Mcam in controlling luminal gene expression patterns, cell state plasticity and mammary tumorigenicity, we focused our initial attention on the murine Py230 cell line, given its experimental tractability, reported LP-like phenotype, robust Mcam expression, and reported subtype plasticity7,42. We knocked Mcam down by transient transfection with a panel of Mcam-directed siRNAs (Supplementary Fig. 1a–c) or by stable transduction with lentiviral vectors (LV) bearing an Mcam-directed shRNA construct (Fig. 1d–h, Supplementary Fig. 1d–g). Although transfection efficiency and KD were modest in transient transfections, a subset of cells in siMcam-transfected cultures exhibited qualitative differences in adhesion and cellular morphology with an elongated spindeloid-like phenotype (Supplementary Fig. 1a–c). Although basal/mesenchymal phenotypes were unexpected in KD cells, we obtained corroborating results using sorted stable LV-transduced Mcam KD Py230 cells compared to cells bearing scrambled control vectors (Fig. 1e–h). Three independently derived stable Mcam KD and control clone pairs (clones A–C) were generated and showed the same cellular and molecular phenotypes observed in transient KD cells including elongated cellular morphology and augmented Paxillin levels (Fig. 1d–h, Supplementary Fig. 1d, e). Throughout our work changes observed in Mcam KD clones were largely consistent, though some variance in population distributions was noted, potentially owing to a loss of cell state control in the relatively plastic Py230 cell line.
Fig. 1. Cellular and molecular signaling changes following Mcam KD in Py230 cells.
a Western blot of mouse breast cancer cell lines. b Western blot of human breast cancer cell lines. c Normalized MCAM counts in PDMs, stratified across breast cancer subtypes by PAM50 analysis. Whiskers mark the 5th and 95th percentile; boxes demarcate 25th, 50th and 75th percentile. d Surface Mcam expression (FACS) in control and Mcam KD Py230 cells. e Representative immunofluorescent Paxillin staining and western blot in Mcam KD Py230 cells and scrambled controls. f Phase contrast image of low density Mcam KD Py230 cells showing a more elongated spindeloid appearance than controls. Scale bar = 100 μm. g IncuCyte scratch wound assay indicates Mcam KD increases migration of Py230-A cells. Scale bar = 100 μm. Two-way ANOVA with Sidak’s multiple comparisons test between shCon and shMcam. ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent SD. h Western blot analysis from the same blot as panel (e) with the same β-actin of Mcam and altered focal adhesion related signaling in Mcam KD and control Py230-A cells.
Previous studies have placed Mcam downstream of Syk and Lck kinases44. However, Py230 shMcam cells also exhibited alterations in focal adhesion signaling including elevated activation of focal adhesion kinase and alterations in Src family kinases downstream of Mcam, including Fyn and Lyn (Fig. 1h)30. These changes were correlated with enhanced migration in scratch wound assays but no significant alteration to proliferation or survival relative to scrambled shRNA controls (Fig. 1g, Supplementary Fig. 1f). However, Mcam/MCAM KD did alter growth kinetics of Py230 in 3D organoid culture as well as the 3D growth of the patient derived organoid model, HCI-011 (Supplementary Fig. 1g)45. These data indicate that in contrast to studies where MCAM OE drives mesenchymal phenotypes in epithelial cells, endogenous MCAM expression can sustain distinct mammary epithelial phenotypes in breast cancer cells.
Mcam stabilizes a luminal progenitor cell state in Py230 cells
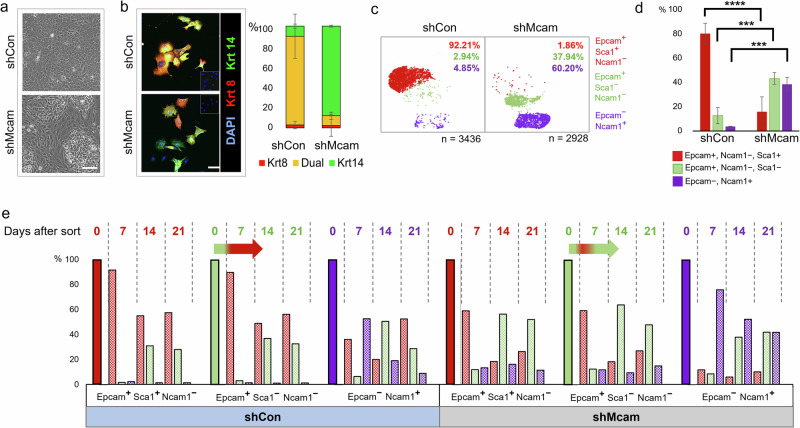
Changes in cell morphology and morphological heterogeneity were also evident in confluent Mcam KD Py230 cultures relative to controls, suggesting alterations in cellular differentiation states or their relative distributions (Fig. 2a). Examination of the lineage related keratins, Krt8 and Krt14, upon Mcam KD revealed a marked skewing of the Krt8/14 co-expression that typifies the uncommitted stem-progenitor state of the parental and control cell line (Fig. 2b). To further characterize Mcam-associated cell state changes, we sequenced transcriptomes from several thousand individual control and Mcam KD Py230 cells (Fig. 2c, Supplementary Fig. 2a, b). UMAP graphical representation of the data revealed three major transcriptional cell states with differential representation between control and Mcam KD cells (Fig. 2c, Supplementary Fig. 2a, b). Surveying the levels of previously described markers of distinct mammary cell states46, we observed that transcript levels for Epcam and Sca1 delineate the major transcriptional states identified in the UMAPs as Epcam+Sca1High, Epcam+Sca1Low and Epcam-Sca1High. We further determined that the Epcam-cells are Ncam1+ whereas Epcam+ cells are Ncam- (Fig. 2c). Although capture of the Epcam-Ncam+ state in single cell sequencing was variable, flow cytometric analysis (FACS) of surface protein expression generally mirrored scRNA-seq data and confirmed population skewing upon Mcam KD (Fig. 2d, Supplementary Fig. 2c, d). FACS and scRNA-seq of additional independent clones demonstrated consistent loss of the Epcam+Sca1High cell state in Mcam KD cells, although as noted above some variance in the final population frequencies was observed (Supplementary Fig. 2b–d). Upon Mcam OE with a silent mutation-bearing shRNA-resistant vector in our Py230 Mcam KD cells, we restored predominance of the Epcam+Sca1High profile while Epcam+Sca1low and Epcam-Ncam+ cell populations were restored to near baseline proportions (Supplementary Fig. 2e–h). We also determined that all three populations were able to interconvert and re-establish parental distributions within 3 weeks of cell sorting by FACS, although kinetics for redistribution from the Epcam-Ncam+ of each genotype were slower than other populations (Fig. 2e). Critically, while sorted Epcam+Sca1Low cells of both genotypes rapidly (within one week) generated Epcam+Sca1High cells, the Epcam+Sca1High cell type was depleted in Mcam KD cells within two weeks (Fig. 2e, arrows). Thus, Mcam KD alters cell state distributions in Py230 by destabilizing the Epcam+Sca1High phenotype.
Fig. 2. Cell-state change following Mcam KD in Py230 cells.
a Phase contrast image of confluent Mcam KD and control Py230-A cultures. Scale bar = 100 μm. b Altered expression of lineage associated Krt8 (red) and Krt14 (green) in Py230-A Mcam KD cultures. Scale bar = 100 μm. Error bars represent SEM. n = 2. c scRNA-seq profiles from Py230-A control and Mcam KD cells reveal three major subpopulations that are identifiable by expression of previously described mammary lineage markers46. d FACS analysis in triplicates confirms proportional skewing among cells expressing these three marker proteins that mirrored subpopulations identified by scRNA-seq (n = 3 independent clones per genotype). Error bars represent SD. Two-way ANOVA with Tukey multiple comparison test: ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. e Interconversion of Py230-A subpopulations upon FACS separation and culture for 0, 7, 14 or 21 days. Horizontal arrows show immediate conversion of Epcam+ Ncam1low Sca1low cells to Epcam+Ncam1low Sca1high within seven days followed by reversion into Sca1low predominance in Mcam KD cells by day 14.
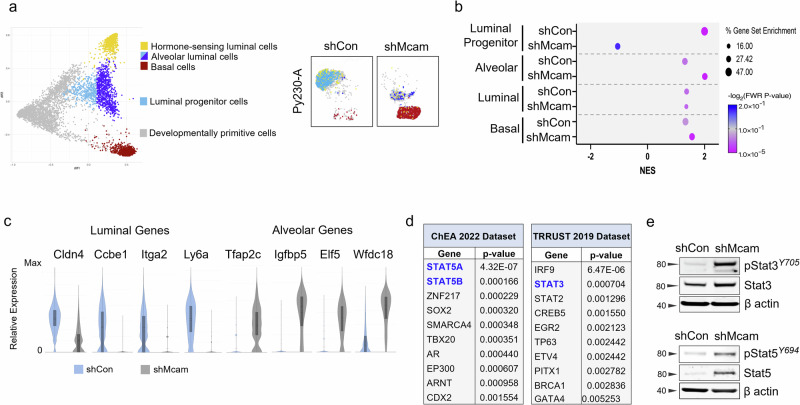
Epcam and Sca1 have previously been proposed to distinguish alveolar progenitors (AP; EpcamhighSca1low) from hormone sensing luminal progenitors (HSP/LP; EpcamhighSca1high)46. We took a comparative transcriptomics approach to determine whether Py230 transcriptomes mimic these progenitor states more broadly. We determined that Py230 cells shared expression similarity with previously published expression profiles for HSP/LP transcriptional states, while the majority of Mcam KD Py230 cells resembled AP and basal cell states of the normal gland (Fig. 3a–c, Supplementary Fig. 3a–d)18,47. Additionally, although we noted upregulation of genes previously ascribed to neural crest fates in Mcam KD Py230 cells in vivo, we determined that several of these genes are also expressed in normal mammary alveolar and basal cell types and do not necessarily indicate loss of mammary epithelial specification (Fig. 3c, Supplementary Fig. 3b)18,47,48.
Fig. 3. A luminal progenitor to alveolar/basal lineage switch accompanies Mcam loss in Py230 cells.
a The top and bottom five differentially expressed genes between the adult mammary cell types identified in the diffusion map from Giraddi et al. (left)18, identify different cell states in Py230-A control and Mcam KD cells on the same scRNAseq coordinate map from Fig. 2c (right). b Gene set enrichment analysis (GSEA) showing enrichment of shCon and shMcam for LP and alveolar cell states, respectively compared to normal mouse mammary cell populations from Giraddi et al.18. c scRNA-seq analysis for relative expression distribution of luminal and alveolar cell state markers in Py230 control (blue) and Mcam KD (grey) cells. d Differentially expressed genes collectively overrepresented in shMcam-A compared to shCon-A Py230 cells analyzed for enrichment using the ChEA and TRRUST datasets from the Enrichr database. The top 10 enriched transcription factors expected to regulate shMcam upregulated genes in each analysis are shown. e Upregulated phosphorylation and expression of Stat3 and Stat5 in shMcam Py230-A cells relative to controls.
To identify underlying regulatory programs contributing to MCAM mediated cell state control, we examined enriched transcription factor target categories among genes that were differentially expressed between control and Mcam KD Py230 cells using EnrichR49. In addition to Elf5, a classic alveolar lineage specifier (Fig. 3c, Supplementary Fig. 3c)50, genes upregulated in Mcam KD cells were significantly enriched for target genes of the STAT family of transcription factors, including alveolar markers Tfap2c, Igfbp5, and Wfdc18 (Fig. 3c, d, Supplementary Fig. 3e, f)16,17,51–53. We therefore examined Stat3 and Stat5a as potential mediators of Py230 cell state change consequent to Mcam KD. Activated Stat3 and Stat5A were upregulated in Mcam KD Py230, providing a potential mechanistic basis for the alveolar cell state switch adopted by Mcam KD cells and for the upregulation of Stat target genes described above (Fig. 3e, Supplementary Fig. 3g–i). While increased Stat5a could be explained by its overexpression, total Stat3 protein levels were only modestly upregulated though it was strongly overactivated in Mcam KD cells relative to MCAM expressing controls (Fig. 3e, Supplementary Fig. 3g, h). Altogether, this data suggests that Mcam maintains the HSP/LP-like cell state in Py230 cells through control of STAT activity and target gene activation, while the loss of Mcam allows a Stat3-driven alveolar/basal lineage switch.
Mcam governs Ck2 substrate utilization attenuating Stat3 activity in Py230 cells
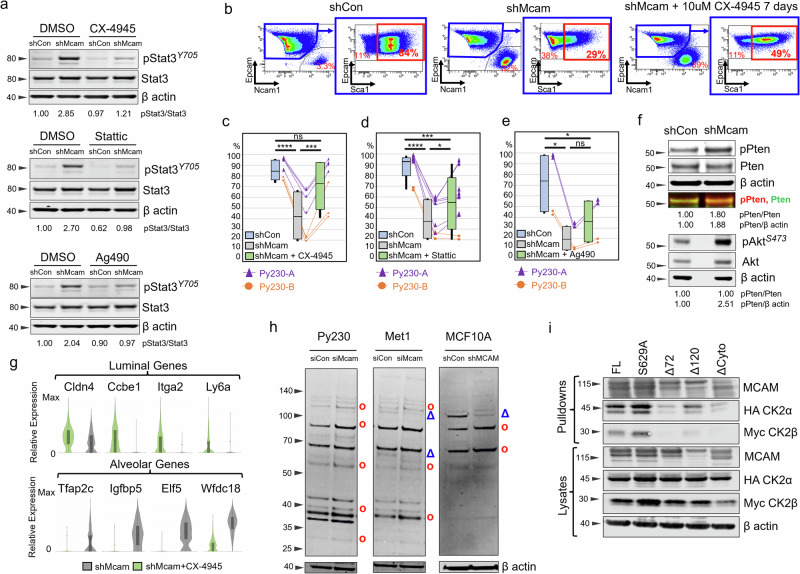
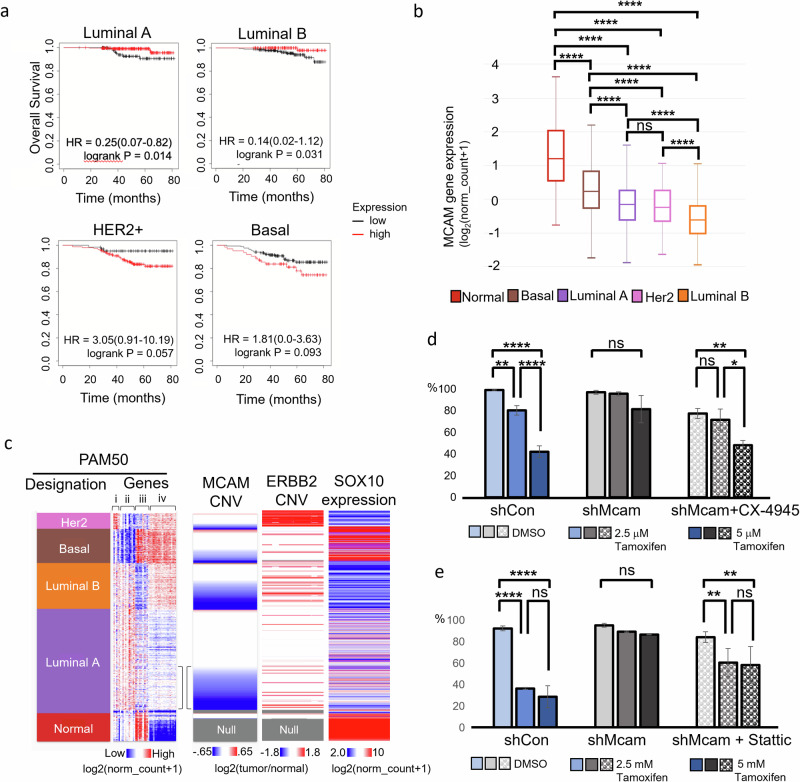
To determine whether hyper-activation of Stat3 was necessary for the HSP/LP → AP switch, we treated Py230 cells with Stattic, a selective STAT3 inhibitor. Additionally, we looked at the effects of inhibitors of upstream regulators of STAT signaling including Ag490, an inhibitor of JAK2 and EGFR, and the CK2 inhibitors CX-4935 and GO289, (Fig. 4a–e, g, Supplementary Fig. 4a–d)51,54,55. Levels of phosphorylated Stat3 (pStat3) in Py230 Mcam KD cells were decreased to near baseline levels with Stattic or by inhibition of either upstream kinase (Fig. 4a). Cell state skewing associated with Mcam KD was dramatically reversed by CX-4945, and to a lesser degree by Stattic, whereas effects of Ag490 were not significant with respect to restoration of a predominant Epcam+Sca1High LP-like cell state (Fig. 4b–e, Supplementary Fig. 4a, b, e–h). Cytotoxicity was negligible for all three reagents (Supplementary Fig. 4f–h and data not shown). The role of Ck2 in Mcam KD phenotypes was intriguing as CK2 has not previously been implicated as a functional downstream target of MCAM, although CK2 has previously been implicated in breast tumorigenesis and cell state control of mammary cells56,57. In previous studies, reduction of CK2 levels in the colorectal cancer cell line, LoVo, led to reduced E-cadherin and other signs of EMT58. However, in our study, Ck2 inhibition did not significantly alter EMT features and Ck2 levels were unaffected by Mcam KD (Supplementary Fig. 4d, h, i). Rather, we noted Mcam KD was associated with augmented Ck2 activity toward Stat3 and phosphorylation of the PI3K negative regulator Pten at Ck2 specific sites (Ser380/Thr382/Thr383) that promote its degradation and consequent Akt hyperactivation (Fig. 4f, Supplementary Fig. 4j)58,59. Ck2 inhibition reversed these effects and restored expression of luminal markers while depleting alveolar markers (Fig. 4g, Supplementary Fig. 4d). These data indicate that Mcam controls cell state in Py230 cells in a manner dependent on Ck2 and Stat3.
Fig. 4. Overactivity of Ck2 toward Stat3 in Py230 shMcam cells drives the alveolar progenitor phenotype.
a Upregulated Stat3 expression and phosphorylation in Py230-A cells and its inhibition by the kinase inhibitors, CX-4945 10 μM, Ag490 50 μM, Stattic 1 μM for 30 min in WB analysis. b Representative FACS analysis of shCon and shMcam Py230-A cells stained for Epcam, Ncam1, and Sca1 with gating (blue outline) of Epcamhigh Ncam1low cells to examine Sca1 levels. Representative plots of shMcam cells treated with 10 μM CX-4945 for 7 days (right two panels) show restoration of the Epcamhigh Ncam1low Sca1high population. c–e Compiled FACS analysis showing the rescue of Epcamhigh Ncam1low Sca1high subpopulation frequencies in Py230-A and Py230-B cells treated with CX-4945 10 μM for 7 Days (c), with Stattic 1 μM for 7 days (d), and with Ag490 30 μM for 7 days (e). Whiskers mark the 5th and 95th percentile; boxes demarcate 25th, 50th, and 75th percentile. One-way ANOVA (Geisser-Greenhouse correction) with Tukey multiple comparison test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. f Western blot analysis in Py230-A cells showing higher basal levels of pPten (Ser380/Thr382/383) and pAKT in Mcam KD. g Gene expression in Py230-A cells treated with CX-4945 10 μM for 7 Days (scRNA-seq). h Western blot analysis in select cell lines showing differential representation of phosphorylated CK2 substrates in siCon and siMcam transfected cell lines (Py230 and Met1) in culture for 48 h post transfection or with shCon and shMCAM transduction (MCF10A). red circle = increased in MCAM KD, blue triangle = decreased in MCAM KD. i Co-immunoprecipitation western blot analysis of a transiently co-transfected Flag-tagged Mcam variants and HA-Ck2a/Myc-Ck2b expression constructs in HEK293A cells. FL full length Mcam, S629A = point mutant in the CK2 phosphorylation site of Mcam, Δ72 = deletion of the distal 72 amino acids of the Mcam tail, Δ120 = deletion of the membrane proximal 120 amino acids of the Mcam tail, ΔCyto = complete deletion of the cytoplasmic tail of Mcam.
Using an antibody that detects phosphorylated Ck2 (pCK2) sites across diverse proteins, we observed altered Ck2 phosphorylation target patterns as a function of Mcam/MCAM status in a variety of lines, including Py230, another PyMT derived line, Met1, and the immortalized human non-transformed epithelial cell line MCF10A (Fig. 4h, Supplementary Fig. 4k, l). CX-4945 treatment confirmed Ck2 specificity of the majority of detected proteins (Supplementary Fig. 4m). Further, Flag ‘pull downs’ of Flag-tagged Mcam rescue constructs co-precipitated tagged CK2α/β proteins when co-expressed in HEK293A cells, indicating Mcam regulation of Ck2 may be direct and dependent on the terminal 72 amino acids of MCAM’s cytoplasmic tail (Fig. 4i). Mutation of a putative Ck2 phosphorylation site target residue (S629A) did not compromise binding (Fig. 4i)33. Thus, Mcam control of Ck2 activity likely involves both binding between Mcam and Ck2 and coupling of Ck2 to select regulatory co-factors and substrates.
MCAM loss hallmarks aggressive luminal tumors
Considering Mcam’s activity in HSP/LP/AP cell state transitions in Py230 cells and its expression in some cells with more luminal features, we re-examined MCAM’s relationship to tumor aggressiveness and subtype using The Cancer Genome Atlas (TCGA) database. While we confirmed that elevated MCAM levels correlated with poorer prognosis in Basal-like and HER2-enriched subtypes, the inverse was true in a subset of Luminal breast cancers when examining patients treated with chemotherapy (Fig. 5a). Chemotherapy treated Luminal cancers tend to reflect more aggressive tumors than those treated with endocrine therapy as an adjuvant monotherapy60. Examining MCAM expression across the various breast cancer subtypes, we found that MCAM was generally reduced across all breast cancer subtypes relative to tumor adjacent normal tissue samples, yet more highly expressed in Basal-like, and Her2-enriched tumors than in Luminal A/B tumors as previously reported (Fig. 5b, Supplementary Fig. 5 a)22. It should be noted that MCAM expression levels in TCGA may also reflect different levels of stromal involvement since stromal cells can express high levels of MCAM.
Fig. 5. MCAM loss marks aggressive luminal tumors.
a Kaplan-Meier analysis of TCGA data breast cancer data, stratifying patients receiving chemotherapy between high and low MCAM expression. b MCAM expression across breast cancer subtypes and normal tumor adjacent tissue in TCGA data. One-Way ANOVA with Tukey Kramer Multiple Comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; n = 1247. Error bars represent SD. c TCGA breast cancer data organized by PAM50 designation and MCAM copy number variation (CNV) with expression of PAM50 gene set, ERBB2 copy number variation, and SOX10 expression. The PAM50 gene set can be broken down roughly into groups of genes related to: ERBB2 expression (i), luminal genes including hormone receptor expression (ii), basal genes (iii), and proliferation genes (iv). Brackets highlight ‘transitional’ Luminal A with increased proliferation and MCAM CNV loss. d, e IncuCyte proliferation assays of Py230-A control and Mcam KD cells with tamoxifen treatment shows that pretreatment with CX-4945 10 μM for 7 days (d), and pretreatment with Stattic 1 μM for 7 days (e) rescue the tamoxifen sensitivity in Mcam KD cells. One-Way ANOVA with Tukey Kramer Multiple Comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent SD.
The Luminal B intrinsic subtype is distinguishable from Luminal A tumors by its poorer prognosis, more frequent endocrine resistance, and greater proliferation1,4. However, underlying mechanisms that give rise to these distinctions remain unclear. We noted that reduced MCAM gene copy number was surprisingly frequent in Luminal B breast cancer and was also evident in approximately one third of Luminal A tumors (Fig. 5c, Supplementary Fig. 5a). In the Luminal A subtype MCAM loss was associated with a trend toward increased expression of proliferation associated PAM50 genes and therefore a trend toward a more Luminal B like phenotype (Fig. 5c, Supplementary Fig. 5b)5,61,62. This likely reflects concomitant amplification of Cyclin D1 at 11q, in many cases62. Interestingly, among patient derived models (PDMs) that formed estrogen independent outgrowths, two out of four instances showed concomitant reduction in MCAM, one of which (HCI-040) showed evidence of the same 11q perturbations observed in TCGA (Supplementary Fig. 5b)45. We also noted a correlation between MCAM copy number reduction and ‘triple positive’ (TPBC) status in archival TCGA data (Fig. 5c). Clinically, despite their amplified ERBB2, such tumors are usually treated as ER+ luminal tumors (Fig. 1c, Supplementary Fig. 5d).
Frontline treatments for luminal, ER+ breast cancers include a variety of estrogen blocking therapies such as the selective estrogen receptor modulator, tamoxifen. When grown in media containing estrogens, Py230 cells express estrogen receptor and become estrogen sensitive (Supplementary Fig. 5e, f)7. In light of this, our observation that Stat3 inhibition reverts Mcam KD Py230 cells to an HSP/LP-related transcriptional state, and prior reports that inhibition of Stat3 can increase the sensitivity to tamoxifen treatment63, we tested the sensitivity of control and Mcam KD Py230 cells to tamoxifen using an in vitro proliferation assay (Fig. 5d, e). We found control cells are more sensitive to tamoxifen than Mcam KD cells, and that sensitivity can be restored in Mcam KD Py230 cells by treatment with inhibitors of Stat3 or Ck2 (Fig. 5d, e). However, Py230 cells did not show sensitivity to the selective estrogen receptor degrader, Fulvestrant, in pilot studies (data not shown). Together, these data further indicate that Mcam maintains access to a hormone-sensing cell state through modulation of Ck2 and downstream Stat3 signaling.
MCAM promotes tumorigenicity through cell state control
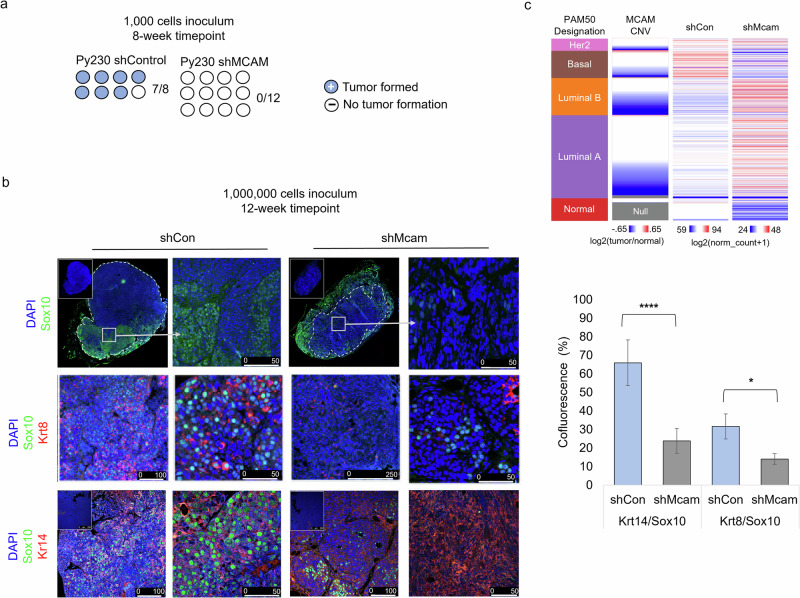
Finally, we examined whether cell state and signaling changes observed in Mcam KD cells in vitro alter tumorigenic potential of the cells or whether they alter the neural crest-associated, Basal-like, Sox10-positive phenotype previously associated with Py230 cells in vivo (Fig. 5c)42. Surprisingly, given the apparent increase in basal/mesenchymal traits observed when Mcam is KD in these cells in 2D culture, they were less tumorigenic than control cells in the graft setting when low cell numbers (1000) were injected (Fig. 6a). When 1,000,000 cells were orthotopically transplanted into the mammary fat pads, we saw formation of both control and Mcam KD-derived tumors, but the tumors that form were fundamentally different in terms of size, histopathological appearance, and expression of select cell type markers (Fig. 6b). We found that Py230 cells exhibit expression of Sox10 in vivo though they lack such expression in 2D culture, consistent with a prior report (Fig. 6b)42. Sox10 expression in vivo marked large invasive peripheral regions of Py230 tumors consistent with the previously proposed neural crest like differentiation state. We determined cells in such regions co-express Krt14 suggesting they are likely basal-like or LP-like regions (Figs. 5c, 6b). However, these large uniform Sox10+Krt14+ positive regions were absent in Mcam KD tumors and the many fewer Sox10+ cells that were found in Mcam KD tumors were usually either Krt8/14 negative or were found in organized Krt8+ epithelial structures resembling the Sox10+, PR- luminal cells previously described in the normal mouse mammary epithelium (Fig. 6b)48. Gene expression profiling of control and Mcam KD tumors confirmed cell state skewing by Mcam KD that was consistent with a block to the LP-related basal-like phenotype (Fig. 6c). Overall, this data suggests that Mcam is required for the generation of Basal-like tumors that arise from a LP-like precursor, and that loss of MCAM expression in human patients predisposes to more aggressive Luminal tumors.
Fig. 6. Mcam promotes tumorigenic phenotypes and tumor cell plasticity.
a Reduced tumor seeding capacity of Mcam KD Py230 cells (1000 cell orthotopic injection) after 8 weeks of growth. shControl n = 8, shMcam n = 12. b Altered cell type distribution in tumors in vivo generated from orthotopic injection of 10e6 Py230 control or Mcam KD cells (left) n = 4 per condition. IHC analysis shows differential expression and distribution of cell type markers between control and Mcam KD tumors. Sox10+ stromal tumor margins are demarcated from tumor parenchyma (dashed lines). Regions of interest in low magnification images marked by white squares. Quantification of marker co-fluorescence utilizing representative tumors from (b) and additional 10e6 Py230 control or Mcam KD cell-derived tumors (right). Two-way t-test unequal variances test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent SD. c TCGA breast cancer data organized by PAM50 designation, MCAM copy number loss, and relative expression of genes with ten greatest rank changes up or down between control and shMcam Py230 tumor grafts (bulk RNASeq). Two-way ANOVA p < 0.0001.
Discussion
Lineage plasticity is critical for proper mammary gland development and maintenance; however, in the context of cancer, it can drive increased tumor heterogeneity and treatment failure8,9,12,13,64. Here we demonstrate a functional role for Mcam in governing epithelial-related LP/alveolar/basal-like cell state switches in mammary carcinoma cells via Ck2 and Stat3. This may represent a key molecular mechanism for understanding and ultimately targeting developmental plasticity in the generation of breast cancer intra- and inter-tumoral heterogeneity and related therapy resistance.
Although prior studies have shown that experimental overexpression of MCAM in Luminal breast cancer cells can promote EMT41, our study shows an increase in EMT-related cellular features (e.g., cell elongation and increased migration) when Mcam is knocked down in the LP-related Py230 mouse mammary carcinoma line. However, these features appear to reflect changes within the epithelial program hierarchy rather than EMT per se since the cells retain transcriptional similarity to various defined cell types of the normal mammary epithelium and do not exhibit coherent changes in classic EMT genes following reduction of Mcam or modulation of its downstream signaling (Supplementary Figure 4d). This is consistent with other studies distinguishing MCAM effects from classic EMT26,65.
Although Mcam KD decreased overall aggressiveness in terms of tumorigenic seeding and growth of grafted cells in mice—a result that is at least superficially consistent with prior studies correlating higher MCAM expression to tumor aggressiveness20,22,24— the cell state changes within the mammary epithelial hierarchy that are governed by Mcam may also contribute to tumor heterogeneity and therapy evasion such as we demonstrated for the selective estrogen antagonist, Tamoxifen. Fulvestrant-insensitive breast cancer cells have previously been reported to display different degrees of response to estrogen and tamoxifen stimulation, and it may be key to determine if the lack of Fulvestrant effects we observed are related to reported roles for CK2 in governing ER stability66,67.
Consistent with recent studies identifying MCAM as a marker of KRT14/KRT19 double-positive LP cells of the normal human breast, that demonstrate shared expression profiles with Basal-like breast cancers40,68, we observed that the LP-like state maintained by Mcam in Py230 cells is functionally required for the formation of Py230 tumors with Basal-like expression profiles (e.g., broad positivity for Sox10 and Krt14). In this regard, it is also noteworthy that recent studies have suggested that the Basal-like subtype arises from LPs and a process of ‘involution mimicry’, related to a key developmental context involving LP and alveolar fate switches50,68.
The striking emergence of Basal-like phenotypes in vivo from the LP phenotype of Py230 cells observed in vitro was also noted by Dravis et al. 42 This observation contrasts somewhat with original reports of the multi-subtype potential of the line7, and with prior analyses (including our own) demonstrating co-clustering of most PyMT tumor expression profiles with Luminal human tumors69,70. However, Dravis et al. also showed that the tumors, while having Basal-like features, retained luminal cell lineage commitment programs. Indeed, as in Bao et al. we also find Esr1 expression in subsets of Py230 cells7,48. The apparent block to both hormone-sensing (HSP)-like and Sox10-positive neural crest-like/Basal-like breast cancer cell states that we observe with Mcam KD suggests that the Mcam-regulated LP state may be a gate-keeper to multiple epithelial cell states and may contribute to propagation of various tumor subtypes.
In human archival data breast cancer data sets, MCAM is most highly expressed in Basal-like tumors, though it is also expressed in other subtypes65. However, its expression in these data sets may be attributable to both parenchymal and stromal cells. Additionally, the different effects in these distinct compartments have been proposed as a basis for MCAM’s conflicted literature as a tumor suppressor vs tumor promoter, though tumor promoting activities within the epithelium have primarily been attributed to promotion of EMT71. In contrast, the CNVs we analyzed in these data sets are predicted to reflect changes in the genetically less stable tumor clones that are under adaptive/selective pressures during tumorigenesis and progression. Although mutual exclusivity of Mcam loss with the generation of the aggressive Sox10+Krt14+ Basal-like phenotypes is consistent between our studies in the Py230 model and archival human data, MCAM loss in human patients did not specifically correlate with reduced aggressiveness as it did in Py230. In human data, although MCAM loss was predominantly associated with Luminal subtypes, it was more prevalent among more proliferative (by gene signature) Luminal A tumors and among the more proliferative, more aggressive and more frequently hormone therapy resistant Luminal B subtype61. The recurrent genetic aberration at 11q, which typifies most of the TCGA breast cancers with MCAM copy number reduction in our study and has been associated with tamoxifen resistance previously62, usually involves concomitant loss of several additional 11q genes that may contribute to cell phenotype in human patients (e.g., PGR, ATM, etc.), as well as amplification of Cyclin D1, a likely contributor to their elevated proliferation profile2,4,62. Thus, it is reasonable to assume that several additional genetic alterations may cooperate with MCAM loss in the context of Luminal B tumors in humans. In contrast to the majority of human specimens showing MCAM loss, isolated experimental modulation of Mcam independent of these other changes suggests a potential role for MCAM in therapeutic sensitivity through cell state change but did not lead to an overall similarity with Luminal B tumors (Fig. 6c). Although, MCAM expression among HR+ breast cancer patient-derived models (PDMs) was enriched among those that (like Py230) co-express ERBB2 and classify as Luminal B (Supplementary Fig. 5d)72.
We report MCAM expression across diverse human and mouse mammary carcinoma cell lines, including those associated with luminal cell states, and the surprising finding that Mcam can alter Ck2 substrate utilization in diverse ways depending on the ‘ground state’ of the cells. Tumor promotion versus suppression as a function of MCAM levels could therefore also be ‘ground state’ dependent. Although Stat3 appears to be a critical substrate in the LP to alveolar/basal switches, we observe in the Py230 model that Ck2 has numerous substrates that have shown to be involved in diverse processes in the cell including but not limited to: hormone receptor stability transcription factor activation, adhesion, cell energetics, and chromatin remodeling73. Thus, understanding the precise levels of MCAM needed to properly scaffold CK2 with various downstream effectors of cell state determination could be key to designing combination therapies that effectively manage shifting therapeutic vulnerabilities deriving from the plastic cell state change potential of mammary LPs. For instance, newer CK2 or STAT3 inhibitors may be of interest in combination with hormone receptor-targeted therapies for some cancers56,63. STATs and associated factors are not only strongly implicated in alveolar cell state control in the normal gland but also have well documented roles in breast and other cancers17,63,72. Indeed, a recent study demonstrated STAT3 inhibition can increase the tamoxifen sensitivity of tamoxifen-resistant breast cancer cells63. Alternatively, if MCAM expression permits access to invasive neural crest-like states in vivo, it may be more effective to inhibit MCAM and seek out new vulnerabilities of the resultant hormone-insensitive cell state. Altogether, our work indicates that MCAM plays a critical role in mammary carcinoma cell state determination via CK2 and STAT3 control, with implications in both breast cancer intra- and inter-tumoral heterogeneity and therapy.
Methods
Cell lines and cell culture
Met-1 and NDL-1 cell lines were provided by Dr. Alexander Borowsky74,75. Authenticated 4T1 (RRID:
CVCL_0125), E0771 (RRID: CVCL_GR23), MCF10A (RRID: CVCL_0598), MDA-MB-231 (RRID: CVCL_0062) and MDA-MB-468 (RRID: CVCL_0419) cell lines were obtained from ATCC. 4T1, E0771, NDL-1, Met-1, MDA-MB-231, and MDA-MB-468 cells were cultured in Dulbecco’s modified Eagle’s media (DMEM) with ciprofloxacin 10 μg/ml and 10% fetal calf serum. MCF10A cells were cultured in Dulbecco’s modified Eagle’s medium/F12 with 5% horse serum, 10 μg/ml insulin, 20 ng/ml epidermal growth factor, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone, and 10 μg/ml ciprofloxacin. Py230 cell line was kindly provided by L. Ellies7. Identity of gifted lines is authenticated herein through molecular assays (e.g., scRNASeq,) that match profiles reported for these lines. Passage numbers were tracked and minimized, and experiments were repeated 2–8 times including repetition with early passage frozen aliquots. Py230 cells were cultured in “Py230 Media” composed of F12-Kaighn’s Modified Media with 5% fetal calf serum, ciprofloxacin 10 μg/ml, amphotericin B 2.5 μg/ml, and Mito+ serum extender (Corning #355006). Py230 organoids were cultured in Py230 media with 4% Matrigel. HCI-011 organoids were kindly provided by Dr. Alana Welm and cultured according to their published protocol45. They were grown in PDxO base medium (Advanced DMEM/F12 with 5% FBS, 10 mM HEPES, 1× Glutamax, 1 μg ml–1 hydrocortisone, 50 μg ml–1 gentamicin and 10 ng ml–1 hEGF) with 10 μM Y-27632, 100 ng ml–1 FGF2 and 1 mM NAC. All cells were grown in sterile humidified tissue culture incubators at 37 °C with 5% CO2 and ambient ( ~ 17–18%) O2. Cells were transduced with lentiviral concentrates in the presence of 7 μg/ml polybrene for up to 16 h with transduction efficiency (not shown) suggesting MOI ⟪1. CX-4945/Silmitasertib (Selleck Chem #S2248), (E/Z)-GO289 (MedChem Express, #HY-115519), Ag490 (Selleck Chem #S1143), Stattic/S7947 (S7947, Sigma Aldrich #19983-44-9), and tamoxifen citrate (Selleck Chem #S1972) were resuspended in DMSO according to manufacturers’ recommendations and diluted to final concentrations in media immediately prior to being added to cells.
Lentiviral vectors
Viral vector plasmids were obtained from OriGene Technologies, Inc. (Rockville, MD), including the mouse Mcam shRNA lentiviral plasmid (Cat # TL514377), a scrambled shControl, and a custom shRNA-resistant (via silent-mutation) mouse Mcam construct subcloned into a lentiviral gene expression vector (pLenti-C-mCFP-P2A-BSD), (# PS100107). Completed vectors were sequenced confirmed.
siRNA construct
siRNA constructs were obtained from OriGene Technologies, Inc. (Rockville, MD). This includes the three unique 27 mer mouse Mcam siRNA duplexes (SR418656A-C) and a Trilencer-27 Universal Scrambled Negative Control siRNA Duplex (SR30005). Cells were transfected utilizing Lipofectamine 3000 Transfection Reagent (Invitrogen, #L3000008) according to manufacturer’s instructions.
Western blot
Cell were lysed in RIPA buffer (1X PBS (137 mM NaCl; 2.7 mM KCl; 4.3 mM Na2PO4; 1.47 mM KH2PO4) with 1% Nonidet P-40 Substitute (Sigma #74385), 0.5% Sodium deoxycholate, 0.1% SDS) with the Halt Protease & Phosphatase inhibitor cocktail (Thermo Fisher Scientific, #78440), quantified with the RC-DC Protein Quantification Assay (BioRad) and equivalent concentrations (10–16 ug) loaded per sample on precast NuPAGE 4–12% Bis-Tris gradient gels (Invitrogen). Separated proteins were transferred to nitrocellulose and probed overnight with primary antibodies under agitation. Secondary antibodies were incubated with blots for 1 h. Intervening washes used TBST pH7.5 (10 mM Tris, 15 mM NaCl, 0.05%Tween 20), Blots were imaged on an Odyssey CLx Imager (LI-COR).
Immunoprecipitation
HEK293A cells were transfected with ps14-Mock76, an rTA expression construct to later induce CK2 expression, utilizing Lipofectamine 3000 (Invitrogen; L3000008). 16 h later cells were sorted for GFP+ cells and plated. After 32 h cells were transfected with an equimolar mix of CK2α/β plasmid (Addgene; #27093) and the relevant MCAM tail variant. Following another 16-hour incubation cells were treated with DOX. After 24 h cells were washed with PBS + EDTA, collected, resuspended in IP Lysis buffer (Pierce; 87787) with the Halt Protease & Phosphatase inhibitor cocktail (Thermo Fisher Scientific, #78440) and allowed to rotate at 4 °C for 1 h. 10ug of rabbit anti-FLAG antibody (Invitrogen; #740001) was added and allowed to incubate overnight at 4 °C. The following day, magnetic Protein A/G beads (Pierce; 88802) were added according to the manufacturer’s instructions and samples were rotated at 4 °C for 1 h prior to magnetic separation, washing the A/G beads, and sample preparation for western blot analysis. Westerns were probed for HA-Tag (Invitrogen; #26183) and Myc (Invitrogen; #MA1-980) for CK2 α and β respectively.
Immunohistochemistry
Freshly dissected tissues were fixed overnight at 4 °C in 10% neutral buffered formalin (Sigma; HT501128). Fixed tissues were transferred to 70% ethanol for storage at 4 °C. Processing for histology followed standard protocols with paraffin embedding, sectioning at 5 μm thickness, baking for 1 h at 55 °C, and deparaffinizing with CitriSolv (Decon Labs, 89426-268) and rehydration through graded alcohol/water washes. Antigen retrieval was achieved by boiling samples 15 min in citrate buffer (10 mM citric acid, 0.05% Tween 20, pH 6.0) prior to overnight primary antibody staining. Slides were mounted in Fluormount-G (Electron Microscopy Sciences). Slides were imaged on a Leica SP8 White light Laser Confocal microscope. Quantification was performed using the QuPath image analysis software (https://qupath.readthedocs.io/en/0.4/docs/intro/citing. html) or utilizing the Leica LasX colocalization analysis (version 3.5.7.23225; https://www.leica-microsystems.com/products/microscope-software/details/product/leica-las-x-ls/) with 30% thresholding as appropriate.
Antibodies
A complete list of antibodies used is available in Table 1.
Table 1.
Commercial antibodies used in this study
| Antibody | Clone | Manufacturer | Catalogue # | Assay | Dilution | RRID |
|---|---|---|---|---|---|---|
| anti-Akt | 40D4 | Cell Signaling Technology | 2920 | Western Blot | 1:2000 | AB_1147620 |
| anti-phospho-Akt (Ser473) | D9E | Cell Signaling Technology | 4060 | Western Blot | 1:2000 | AB_2315049 |
| anti-β-actin | W16197A | Biolegend | 664802 | Western Blot | 1:2000 | AB_2721349 |
| anti-phospho-Stat3 (Tyr705) | D3A7 | Cell Signaling B1:H36 Technology | 9145 | Western Blot | 1:1000 | AB_2491009 |
| anti-Stat3 | 124H6 | Cell Signaling Technology | 9139 | Western Blot | 1:1000 | AB_331757 |
| anti-Fyn | E3 | Santa Cruz Biotechnology | SC-365913 | Western Blot | 1:500 | AB_10842309 |
| anti-Lyn | C13F9 | Cell Signaling Technology | 2796 T | Western Blot | 1:1000 | AB_2138391 |
| anti-phospho-src-famly (Tyr418) | EP503Y | Abcam | Ab40660 | Western Blot | 1:1000 | AB_776106 |
| anti-Mcam | 281 | Novus Biologicals | NBP2-90721 | Western Blot | 1:1000 | unavailable |
| anti-MCAM | EPR3208 | Abcam | Ab75769 | Western Blot | 1:1000 | AB_2143375 |
| anti-phospho-Pten (Ser380/Thr382/383) | 44A7 | Cell Signaling Technology | 9549 | Western Blot | 1:1000 | AB_659891 |
| anti-Pten | A2B1 | Santa Cruz Biotechnology | SC-7974 | Western Blot | 1–500 | AB_628187 |
| anti-phospho-Fak (Ty925) | polyclonal | Cell Signaling Technology | 3284 | Western Blot | 1:1000 | AB_10831810 |
| anti-phospho-Fak (Tyr397 | D20B1 | Cell Signaling Technology | 8556 | Western Blot | 1:1000 | AB_10891442 |
| anti-Fak | polyclonal | Cell Signaling Technology | 3285 | Western Blot | 1:1000 | AB_2269034 |
| anti-phospho-Stat5 (Tyr694) | D47E7 | Cell Signaling Technology | 4322 | Western Blot | 1:400 | AB_10544692 |
| anti-Stat5 | ST5-8F7 | Thermo Fisher Scientific | 33–5900 | Western Blot | 1:200 | AB_2533129 |
| anti-Rabbit-IgG-AF680 | polyclonal | Thermo Fisher Scientific | A-21076 | Western Blot | 1:1000 | AB_2535736 |
| anti-Mouse-IgG-AF790 | polyclonal | Thermo Fisher Scientific | A11375 | Western Blot | 1:1000 | AB_2534146 |
| anti-Rat-IgG-H&LDyLight-800 | polyclonal | Thermo Fisher Scientific | SA5-10032 | Western Blot | 1:10000 | AB_2556612 |
| anti-Paxillin | Y113 | Abcam | Ab32084 | Western Blot | 1:5000 | AB_779033 |
| HA-Tag | 2-2.2.14 | Invitrogen | 26183 | Western Blot | 1:1000 | AB_10978021 |
| c-Myc | 9E10 | Invitrogen | MA1-980 | Western Blot | 1:1000 | AB_558470 |
| phospho-CK2 substrate | MultiMab | Cell Signaling Technology | 8738S | Western Blot | 1:1000 | AB_2797653 |
| anti-Sox10 | SD204-04 | Thermo Fisher Scientific | MA5-32398 | Immunocytochemistry | 1:50 | AB_2809676 |
| anti-Krt14 | LL002 | Thermo Fisher Scientific | MA5-11599 | Immunocyto-chemistry | 1:50 | AB_10982092 |
| anti-Krt8 | TROMA-1 | DSHB U of Iowa | Ab-531826 | Immunocyto-chemistry | 1:50 | AB_531826 |
| anti-Rabbit-IgG-AF568 | polyclonal | Thermo Fisher Scientific | A-11011 | Immunocyto-chemistry | 1:200 | AB_143157 |
| anti-Mouse-IgG-AF647 | polyclonal | Thermo Fisher Scientific | A28181 | Immunocyto-chemistry | 1:200 | AB_2536165 |
| anti-Rat-IgG-AF647 | polyclonal | Thermo Fisher Scientific | A-21247 | Immunocyto-chemistry | 1:200 | AB_141778 |
| anti-Mcam | ME9F1 | Biolegend | 313605 | FACS | 1:100 | AB_345299 |
| anti-CD326 | G8.8 | Thermo Fisher Scientific | 25-5791-80 | FACS | 1:160 | AB_1724047 |
| anti-Sca1 | D7 | Thermo Fisher Scientific | 17-5981-81 | FACS | 1:300 | AB_469486 |
| anti-Ncam1 | EPR21827 | Abcam | Ab220360 | FACS | 1:500 | AB_2927664 |
| rabbit anti-FLAG | polyclonal | Invitrogen | 740001 | Immuno-precipitation | 1:50 | AB_2610628 |
scRNA sequencing
All protocols used to generate scRNA-seq data on 10x Genomics Chromium Controller platform including library prep, instrument and sequencing setting can be found on: https://www.10xgenomics.com/support/single-cell-gene-expression/documentation. The Chromium Next GEM Single Cell 3’ Kit v3.1 (PN-1000268) was used to barcode individual cells with 16 nt 10× Barcode, to tag cell specific transcript molecules with 12 nt Unique Molecular Identifier (UMI) and to capture poly (A) mRNA with 30 nt ploy(dT) sequence according to the manufactures. The following protocol based on 10× Genomics user guide (CG000315) was performed by High-Throughput Genomics Shared Resource at Huntsman Cancer Institute, University of Utah. Briefly, Py230 single cell suspension was isolated by trypsinization and resuspended in phosphate buffered saline with 0.04% bovine serum albumin. The cell suspension was filtered through 40-micron cell strainer. Viability and cell count were assessed on Countess 2 (Invitrogen, Carlsbad, CA). Equilibrium to targeted cell recovery of 6000 cells along with 10× Gel Beads and reverse transcription reagents were loaded to Chromium Single Cell Chip G (PN-1000120) to form Gel-Bead-In Emulsions (GEMs), the nano-droplets. Within individual GEMs, barcoded cDNA generated from captured mRNA was synthesized by reverse transcription at the setting of 53 °C for 45 min followed by 85 °C for 5 min. Subsequent fragmentation, end repair and A-tailing, adaptor ligation and sample indexing with dual index (PN-1000215) were performed in bulk according to the user guide. The resulting barcoded libraries were qualified using Agilent D1000 ScreenTape on Agilent Technology 2200 TapeStation system and quantified by quantification PCR using KAPA Biosystems Library Quantification Kit for Illumine Platforms (KK4842). Multiple libraries were then normalized, pooled and sequenced on NovaSeq 6000 with 150 × 150 paired end mode.
Bulk RNA-sequencing
RNA was collected from snap frozen biological replicates of 10e6 Py230 shControl/shMcam cell-derived tumors that were collected 12-weeks after inoculation. RNA was isolated via QIAzol-chloroform extraction followed by column-based purification and On-Column DNase Digestion (Qiagen #79254). The aqueous phase was brought to a final concentration of 50% ethanol, and RNA was purified using the RNeasy Lipid Tissue Mini Kit according to the manufacturer’s instructions (Qiagen #74804). Library preparation was performed using the Illumina TruSeq Stranded mRNA Library Prep with UDI (Illumina; poly(A) selection). Sequencing was performed using the NovaSeq 6000 (50 × 50 bp paired-end sequencing; 25 million reads per sample).
Bioinformatic analysis
All downstream analysis of sequencing data was completed using Loupe Browser (6.0.0) (RRID:SCR_018555), Enrichr (RRID:SCR_001575), Appyters (RRID:SCR_021245), GraphPad Prism (RRID:SCR_002798) and RStudio (4.1.2) (RRID:SCR_000432). For Enrichr analysis, we first compared single cell RNAseq data from Py230 shCon and shMcam populations globally to identify differentially expressed genes (DEG) in Loupe Browser. The DEG lists were then analyzed for categorical enrichment using ChEA and TRRUST datasets in Enrichr analysis73. DEG analysis for cell subtypes utilized the default clustering method of Seurat with the Louvain algorithm to iteratively group cells together77. We then used the Seurat default differential expressed gene (DEG) test [FindMarkers()] (non-parametric Wilcoxon rank sum test). The top 100 DEGs were then used for downstream comparative analysis. From the Py230 dataset, DEG lists were generated by comparing the shCon luminal to shMcam luminal clusters and similarly for the alveolar clusters identified through marker gene comparisons. Results for specific genes were visualized in Loupe Browser generated UMAP projections. Reanalysis of primary mouse mammary data from Giraddi et al. used the published diffusion map coordinates and the markers Krt14, Krt8 and Wfdc18, to designate 4 adult groups/cell-types, basal, luminal differentiated, luminal progenitor, and alveolar18. We used GSEA software (RRID:SCR_005724), and Molecular Signature Database (MSigDB; RRID:SCR_016863) to compare gene sets with 100,000 permutations in GSEA Pre-ranked. GSEA data was plotted with GraphPad Prism (RRID:SCR_002798). Kaplan Meier plots were generated using TCGA data visualized utilizing KMplot and the auto-select cutoff values for Basal (3.45), Luminal A (3.35), Luminal B (3.29) and Her2 (2.97)78. Heatmaps and MCAM CNV analysis utilized the UCSC Xena Browser61 (RRID:SCR_018938).
Tumor studies
10-week-old nude female mice were from Charles River Laboratories (Wilmington, MA). 1000 or 1,000,000 MMTV-PyMT Py230 cells were resuspended in 20 μL complete Matrigel (Corning) and orthotopically injected into the #4 mammary glands of anesthetized (by inhalation of isoflurane mixed with medical air) mice. Tumor volume was measured at least once a week. No animals originally part of this study were excluded. At endpoint, mice were euthanized according to AVMA guidelines and tissues were harvested for processing. Mouse euthanasia was performed by exposure to carbon dioxide (CO2), followed by cervical dislocation. The maximum mammary tumor size permitted is 2 cm diameter, which was not exceeded in these studies. All animal studies were performed ethically in accordance with protocols approved by IACUC oversite committee at the University of Utah. All mice were housed in an accredited animal facility at HCI and maintained in a controlled environment under specific pathogen-free conditions.
Statistics
Replicates and experimental repetition are indicated in figures and figure legends. Error bars are representative of the standard deviation (SD) or standard error of the mean (SEM) as denoted in figure legends. For statistical analysis, One-Way ANOVA with Tukey Kramer Multiple Comparisons, Repeated Measures One-Way ANOVA (Geisser-Greenhouse correction) w/ Tukey multiple comparison tests, and Two-Way ANOVA with Sidak’s multiple comparisons were used. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Gene mapping and differential gene expression analysis utilized established default methods embedded in the Cell Ranger, CLoupe browser and Seurat computational packages.
Supplementary information
Updated supplementary figures and legends
Acknowledgements
We thank C. Trejo for technical assistance and M. Kruithof-de Julio, T. Oliver, and D. Salomon for valuable discussions. We also acknowledge generous support from the Huntsman Cancer Foundation, and from the Halt Cancer at X foundation. BLG was also supported in part by the AAUW American Fellowship. We thank the University of Utah/HCI Cancer Center Support Grant (P30 CA42014), and staff in the Flow Cytometry, HSC Cell Imaging, Preclinical Research Resource, Biostatistics, High-Throughput Genomics and Bioinformatics shared core resources. Research reported in this publication utilized the Preclinical Research Resource, Flow Cytometry Core, Biorepository and Molecular Pathology, and High-Throughput Genomics and Bioinformatics shared resources at Huntsman Cancer Institute at the University of Utah and was supported by the National Cancer Institute of the National Institutes of Health CCSG support grant under Award Number P30CA042014. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was supported by the Huntsman Cancer Foundation, Halt Cancer at X Foundation.
Author contributions
Concept: B.T.S. Experimental Design: O.B., B.L.G., B.T.S. Execution: O.B., B.L.G., D.W.F., B.H., B.T.S. Analysis: B.L.G., E.M.M., D.A.-T. Interpretation: all authors. Writing: O.B., B.L.G., B.T.S.
Data availability
Datasets generated during and/or analyzed during the current study are either publicly available (TCGA; https://www.cancer.gov/tcga), are deposited at Gene Expression Omnibus (GEO) under GSE233093 or are within the article and its supplementary data files.
Code availability
All codes and R-packages used in the study are publicly available and have been disclosed in Methods or are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ozlen Balcioglu, Brooke L. Gates.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-024-00687-7.
References
- 1.Testa, U., Castelli, G. & Pelosi, E. Breast cancer: a molecularly heterogenous disease needing subtype-specific treatments. Med. Sci.8, 18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA98, 10869–10874 (2001). 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho, N. Molecular subtypes and imaging phenotypes of breast cancer. Ultrasonography35, 281–288 (2016). 10.14366/usg.16030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ades, F. et al. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J. Clin. Oncol.32, 2794–2803 (2014). 10.1200/JCO.2013.54.1870 [DOI] [PubMed] [Google Scholar]
- 5.Kensler, K. H. et al. PAM50 molecular intrinsic subtypes in the nurses’ Health Study Cohorts. Cancer Epidemiol. Biomark. Prev.28, 798–806 (2019). 10.1158/1055-9965.EPI-18-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith, B. A. et al. Targeting the PyMT oncogene to diverse mammary cell populations enhances tumor heterogeneity and generates rare breast cancer subtypes. Genes Cancer3, 550–563 (2012). 10.1177/1947601913475359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao, L., Cardiff, R. D., Steinbach, P., Messer, K. S. & Ellies, L. G. Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast Cancer Res.17, 137 (2015). 10.1186/s13058-015-0615-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl, G. M. & Spike, B. T. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer3, 14 (2017). 10.1038/s41523-017-0012-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong, D., Hughes, C. J. & Ford, H. L. Cellular plasticity in breast cancer progression and therapy. Front. Mol. Biosci.7, 72 (2020). 10.3389/fmolb.2020.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu, M. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science339, 580–584 (2013). 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Y. & Weinberg, R. A. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front. Med.12, 361–373 (2018). 10.1007/s11684-018-0656-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zewdu, R. et al. An NKX2-1/ERK/WNT feedback loop modulates gastric identity and response to targeted therapy in lung adenocarcinoma. Elife10, e66788 (2021). 10.7554/eLife.66788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orstad, G. et al. FoxA1 and FoxA2 control growth and cellular identity in NKX2-1-positive lung adenocarcinoma. Dev. Cell57, 1866–1882 e1810 (2022). 10.1016/j.devcel.2022.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, X. et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature461, 495–500 (2009). 10.1038/nature08361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti, R. et al. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells30, 1496–1508 (2012). 10.1002/stem.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haricharan, S. & Li, Y. STAT signaling in mammary gland differentiation, cell survival and tumorigenesis. Mol. Cell Endocrinol.382, 560–569 (2014). 10.1016/j.mce.2013.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caffarel, M. M. et al. Constitutive activation of JAK2 in mammary epithelium elevates Stat5 signalling, promotes alveologenesis and resistance to cell death, and contributes to tumourigenesis. Cell Death Differ.19, 511–522 (2012). 10.1038/cdd.2011.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraddi, R. R. et al. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep.24, 1653–1666.e1657 (2018). 10.1016/j.celrep.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spike, B. T. et al. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell10, 183–197 (2012). 10.1016/j.stem.2011.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, Z. et al. CD146, from a melanoma cell adhesion molecule to a signaling receptor. Signal Transduct. Target Ther.5, 148 (2020). 10.1038/s41392-020-00259-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imbert, A. M. et al. CD146 expression in human breast cancer cell lines induces phenotypic and functional changes observed in Epithelial to Mesenchymal Transition. PLoS One7, e43752 (2012). 10.1371/journal.pone.0043752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, Y. K. et al. MCAM/CD146 promotes tamoxifen resistance in breast cancer cells through induction of epithelial-mesenchymal transition, decreased ERalpha expression and AKT activation. Cancer Lett.386, 65–76 (2017). 10.1016/j.canlet.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 23.Huang, C. Y. & Wu, G. J. METCAM/MUC18 promoted tumorigenesis of human breast cancer SK-BR-3 cells in a dosage-specific manner. Taiwan J. Obstet. Gynecol.55, 202–212 (2016). 10.1016/j.tjog.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 24.Zabouo, G. et al. CD146 expression is associated with a poor prognosis in human breast tumors and with enhanced motility in breast cancer cell lines. Breast Cancer Res. 11, R1 (2009). 10.1186/bcr2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma, A., Somasundaram, I. & Chabaud, M. B. CD146 as a prognostic marker in breast cancer: a meta-analysis. J. Cancer Res Ther.20, 193–198 (2024). 10.4103/jcrt.jcrt_738_22 [DOI] [PubMed] [Google Scholar]
- 26.Shih, L. M., Hsu, M. Y., Palazzo, J. P. & Herlyn, M. The cell-cell adhesion receptor Mel-CAM acts as a tumor suppressor in breast carcinoma. Am. J. Pathol.151, 745–751 (1997). [PMC free article] [PubMed] [Google Scholar]
- 27.Ruma, I. M. et al. MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-kappaB and ROS formation upon ligand binding. Clin. Exp. Metastasis33, 609–627 (2016). 10.1007/s10585-016-9801-2 [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa, T. et al. Laminins 411 and 421 differentially promote tumor cell migration via alpha6beta1 integrin and MCAM (CD146). Matrix Biol.38, 69–83 (2014). 10.1016/j.matbio.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 29.Ouhtit, A., Abdraboh, M. E., Hollenbach, A. D., Zayed, H. & Raj, M. H. G. CD146, a novel target of CD44-signaling, suppresses breast tumor cell invasion. Cell Commun. Signal15, 45 (2017). 10.1186/s12964-017-0200-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb, D. J. et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol.6, 154–161 (2004). 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- 31.Ye, Z. et al. Wnt5a uses CD146 as a receptor to regulate cell motility and convergent extension. Nat. Commun.4, 2803 (2013). 10.1038/ncomms3803 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Q., Zhang, S., Chen, J. & Xie, Z. The interplay between integrins and immune cells as a regulator in cancer immunology. Int. J. Mol. Sci.24, 6170 (2023). 10.3390/ijms24076170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, G.-J. METCAM/MUC18 expression and cancer metastasis. Curr. Genom.6, 333–349 (2005). 10.2174/1389202054750211 [DOI] [Google Scholar]
- 34.Dontu, G. & Ince, T. A. Of mice and women: a comparative tissue biology perspective of breast stem cells and differentiation. J. Mammary Gland Biol. Neoplasia20, 51–62 (2015). 10.1007/s10911-015-9341-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray, G. K. et al. A human breast atlas integrating single-cell proteomics and transcriptomics. Dev. Cell57, 1400–1420.e1407 (2022). 10.1016/j.devcel.2022.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenbluth, J. M. et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun.11, 1711 (2020). 10.1038/s41467-020-15548-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfefferle, A. D., Spike, B. T., Wahl, G. M. & Perou, C. M. Luminal progenitor and fetal mammary stem cell expression features predict breast tumor response to neoadjuvant chemotherapy. Breast Cancer Res. Treat.149, 425–437 (2015). 10.1007/s10549-014-3262-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, E. et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res.12, R21 (2010). 10.1186/bcr2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim, E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med.15, 907–913 (2009). 10.1038/nm.2000 [DOI] [PubMed] [Google Scholar]
- 40.Isberg, O. G. et al. A CD146 FACS protocol enriches for Luminal Keratin 14/19 double positive human breast progenitors. Sci. Rep.9, 14843 (2019). 10.1038/s41598-019-50903-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng, Q. et al. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc. Natl Acad. Sci. USA109, 1127–1132 (2012). 10.1073/pnas.1111053108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dravis, C. et al. Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell34, 466–482.e466 (2018). 10.1016/j.ccell.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng, G. F., Cai, S. X. & Wu, G. J. Up-regulation of METCAM/MUC18 promotes motility, invasion, and tumorigenesis of human breast cancer cells. BMC Cancer11, 113 (2011). 10.1186/1471-2407-11-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakraborty, G., Rangaswami, H., Jain, S. & Kundu, G. C. Hypoxia regulates cross-talk between Syk and Lck leading to breast cancer progression and angiogenesis. J. Biol. Chem.281, 11322–11331 (2006). 10.1074/jbc.M512546200 [DOI] [PubMed] [Google Scholar]
- 45.Guillen, K. P. et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat. Cancer3, 232–250 (2022). 10.1038/s43018-022-00337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shehata, M. et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res.14, R134 (2012). 10.1186/bcr3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bach, K. et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun.8, 2128 (2017). 10.1038/s41467-017-02001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dravis, C. et al. Sox10 regulates stem/progenitor and mesenchymal cell states in mammary epithelial cells. Cell Rep.12, 2035–2048 (2015). 10.1016/j.celrep.2015.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protoc.1, e90 (2021). 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valdes-Mora, F. et al. Single-cell transcriptomics reveals involution mimicry during the specification of the basal breast cancer subtype. Cell Rep.35, 108945 (2021). 10.1016/j.celrep.2021.108945 [DOI] [PubMed] [Google Scholar]
- 51.De Vos, J., Jourdan, M., Tarte, K., Jasmin, C. & Klein, B. JAK2 tyrosine kinase inhibitor tyrphostin AG490 downregulates the mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription (STAT) pathways and induces apoptosis in myeloma cells. Br. J. Haematol.109, 823–828 (2000). 10.1046/j.1365-2141.2000.02127.x [DOI] [PubMed] [Google Scholar]
- 52.Yeo, S. K. et al. Single-cell RNA-sequencing reveals distinct patterns of cell state heterogeneity in mouse models of breast cancer. Elife9, e58810 (2020). 10.7554/eLife.58810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tai, C. I., Schulze, E. N. & Ying, Q. L. Stat3 signaling regulates embryonic stem cell fate in a dose-dependent manner. Biol. Open3, 958–965 (2014). 10.1242/bio.20149514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schust, J., Sperl, B., Hollis, A., Mayer, T. U. & Berg, T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol.13, 1235–1242 (2006). 10.1016/j.chembiol.2006.09.018 [DOI] [PubMed] [Google Scholar]
- 55.Zheng, Y. et al. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathway. Blood118, 156–166 (2011). 10.1182/blood-2010-01-266320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Filhol, O., Deshiere, A. & Cochet, C. Protein Kinase CK2, 363–382 (2013). 10.1002/9781118482490.ch14 [DOI] [Google Scholar]
- 57.Gray, G. K., McFarland, B. C., Rowse, A. L., Gibson, S. A. & Benveniste, E. N. Therapeutic CK2 inhibition attenuates diverse prosurvival signaling cascades and decreases cell viability in human breast cancer cells. Oncotarget5, 6484–6496 (2014). 10.18632/oncotarget.2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou, J. et al. Protein kinase CK2alpha is overexpressed in colorectal cancer and modulates cell proliferation and invasion via regulating EMT-related genes. J. Transl. Med9, 97 (2011). 10.1186/1479-5876-9-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller, S. J., Lou, D. Y., Seldin, D. C., Lane, W. S. & Neel, B. G. Direct identification of PTEN phosphorylation sites. FEBS Lett.528, 145–153 (2002). 10.1016/S0014-5793(02)03274-X [DOI] [PubMed] [Google Scholar]
- 60.Dai, X. et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 5, 2929–2943 (2015). [PMC free article] [PubMed] [Google Scholar]
- 61.Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol.38, 675–678 (2020). 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lundgren, K., Holm, K., Nordenskjold, B., Borg, A. & Landberg, G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 10, R81 (2008). 10.1186/bcr2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon, S. Y. et al. Inhibition of STAT3 enhances sensitivity to tamoxifen in tamoxifen-resistant breast cancer cells. BMC Cancer21, 931 (2021). 10.1186/s12885-021-08641-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ireland, A. S. et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell38, 60–78.e12 (2020). 10.1016/j.ccell.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Kruijff, I. E. et al. The prevalence of CD146 expression in breast cancer subtypes and its relation to outcome. Cancers10, 134 (2018). 10.3390/cancers10050134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaminska, K. et al. Distinct mechanisms of resistance to fulvestrant treatment dictate level of ER independence and selective response to CDK inhibitors in metastatic breast cancer. Breast Cancer Res.23, 26 (2021). 10.1186/s13058-021-01402-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams, C. C. et al. Identification of four novel phosphorylation sites in estrogen receptor alpha: impact on receptor-dependent gene expression and phosphorylation by protein kinase CK2. BMC Biochem.10, 36 (2009). 10.1186/1471-2091-10-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohler, K. T. et al. Ductal keratin 15(+) luminal progenitors in normal breast exhibit a basal-like breast cancer transcriptomic signature. NPJ Breast Cancer8, 81 (2022). 10.1038/s41523-022-00444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfefferle, A. D. et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol.14, R125 (2013). 10.1186/gb-2013-14-11-r125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herschkowitz, J. I. et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol.8, R76 (2007). 10.1186/gb-2007-8-5-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mannion, A. J. et al. Pro- and anti-tumour activities of CD146/MCAM in breast cancer result from its heterogeneous expression and association with epithelial to mesenchymal transition. Front. Cell Dev. Biol.11, 1129015 (2023). 10.3389/fcell.2023.1129015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hughes, K. & Watson, C. J. The multifaceted role of STAT3 in mammary gland involution and breast cancer. Int. J. Mol. Sci.19, 1695 (2018). 10.3390/ijms19061695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44, W90–W97 (2016). 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borowsky, A. D. et al. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin. Exp. Metastasis22, 47–59 (2005). 10.1007/s10585-005-2908-5 [DOI] [PubMed] [Google Scholar]
- 75.Siegel, P. M., Ryan, E. D., Cardiff, R. D. & Muller, W. J. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. EMBO J.18, 2149–2164 (1999). 10.1093/emboj/18.8.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balcioglu, O. et al. CRIPTO antagonist ALK4(L75A)-Fc inhibits breast cancer cell plasticity and adaptation to stress. Breast Cancer Res.22, 125 (2020). 10.1186/s13058-020-01361-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e3529 (2021). 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanczky, A. & Gyorffy, B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med Internet Res.23, e27633 (2021). 10.2196/27633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Updated supplementary figures and legends
Data Availability Statement
Datasets generated during and/or analyzed during the current study are either publicly available (TCGA; https://www.cancer.gov/tcga), are deposited at Gene Expression Omnibus (GEO) under GSE233093 or are within the article and its supplementary data files.
All codes and R-packages used in the study are publicly available and have been disclosed in Methods or are available from the corresponding authors on reasonable request.