Abstract
Abscisic acid (ABA) is the primary preventing factor of seed germination, which is crucial to plant survival and propagation. ABA-induced seed germination inhibition is mainly mediated by the dimeric PYR/PYL/RCAR (PYLs) family members. However, little is known about the relevance between dimeric stability of PYLs and seed germination. Here, we reveal that stabilization of PYL dimer can relieve ABA-induced inhibition of seed germination using chemical genetic approaches. Di-nitrobensulfamide (DBSA), a computationally designed chemical probe, yields around ten-fold improvement in receptor affinity relative to ABA. DBSA reverses ABA-induced inhibition of seed germination mainly through dimeric receptors and recovers the expression of ABA-responsive genes. DBSA maintains PYR1 in dimeric state during protein oligomeric state experiment. X-ray crystallography shows that DBSA targets a pocket in PYL dimer interface and may stabilize PYL dimer by forming hydrogen networks. Our results illustrate the potential of PYL dimer stabilization in preventing ABA-induced seed germination inhibition.
Subject terms: Plant hormones, Abiotic, Small molecules
The authors reveal that stabilization of PYL dimer can relieve ABA-induced inhibition of seed germination using a computationally designed compound DBSA. Crystal structure and other experiments show that DBSA selectively binds to the dimeric PYL.
Introduction
The success of seed germination determines the propagation of most higher plant species1. Moreover, seed germination is a critical and yield-defining stage in the growth of crops2. In some main crops, such as rice, wheat and corn, an improvement in the seed germination rate might increase the yield by 20%-50% or even higher3. Well-germinated seeds accelerate crop development, shortening the period of crop maturity and reducing the time to biomass accumulation and yield in agricultural production4. But, seeds often overreact to adverse conditions and fail to germinate and grow when conditions become favorable5. Therefore, overcoming adverse conditions to increase the seed germination rate is a major challenge in improving crop yield and ensuring food security6,7.
Among the phytohormones in plants, abscisic acid (ABA) is the primary preventer of seed germination8,9. ABA is perceived by the soluble Pyrabactin Resistance 1/PYR1-Like/Regulatory Component of ABA Receptor (PYR/PYL/RCAR, simplified as PYLs) family of receptors, which consists of dimers (PYR1, PYL1-PYL3) and monomers (PYL4-PYL13)10,11. Upon ABA binding, a gate-latch-lock conformational change occurs in PYLs12, forming an interface for PYL binding to and inhibition of group A type 2 C protein phosphatases (PP2Cs), therefore blocking the interactions between the PP2Cs and downstream effector proteins13,14. The inhibition of PP2Cs leads to the activation of SNF1-related kinase 2 s (SnRK2s), and further elicits the function of ABA in seed germination inhibition15,16. Vaidya et al. revealed that blocking the interactions between PP2Cs and PYLs could relieve inhibition of seed germination in Arabidopsis, barley and tomato using a chemical molecule antabactin17. In summary, the generation of a ABA-PYL-PP2C complex plays a key role in the activation of ABA signaling and the inhibition of seed germination in plants18,19. Therefore, PYLs have become an important target in the investigations of ABA signaling and the regulation of seed germination inhibition20,21.
Dimeric PYLs play vital roles in the ABA-induced inhibition of seed germination. Classical genetic approaches have been used to examine the function of different PYLs members on seed germination. Park et al. revealed that the simultaneous disruption of pyr1, pyl1, and pyl4 genes leads to the abolishment of ABA-induced inhibition of seed germination9,13. Nishimura et al. revealed that pyr1;pyl1;pyl2;pyl4 quadruple mutant lines exhibit strong ABA insensitivity in seed germination10,22. Nevertheless, mutational analysis of ABA receptor function is always limited by the issue of gene redundancy23,24. Chemical genetics is a powerful tool to investigate the function of PYLs because its ability to bypass the gene redundancy. Okamoto et al. designed an ABA functional analogue, quinabactin, which inhibits seed germination in vitro, and revealed that the effects of quinabactin in vegetative tissues are primarily mediated by the dimeric ABA receptors PYR1 and PYL125. Vaidya et al. further discovered a more potent ABA receptor agonist, opabactin, which exhibits ~10-fold higher activity in inhibiting seed germination than ABA26. The dimeric PYR1, PYL1, and PYL2 are identified as the main targets of opabactin in regulating seed germination. These studies showed that dimeric PYLs are crucial for ABA signaling in the regulation of seed germination. However, little is known about the relationship between the dimer stability of PYLs and seed germination inhibition mediated by ABA.
In this study, we revealed that stabilizing PYL dimers is a powerful way to relieve ABA-induced inhibition of seed germination using a chemical genetic approach. A binding pocket located at the PYL dimer interface was discovered, and a computational fragment virtual screening was performed to design the chemical probe di-nitrobensulfamide (DBSA). DBSA exhibits around ten-fold higher binding affinity to PYR1 (Kd = 2.34 μM) compared with ABA (Kd = 21.95 μM). In vitro and in vivo experiments suggested that DBSA can effectively release ABA-induced seed germination inhibition through PYL dimers, and repress the expression level of ABA-responsive genes. X-ray crystal structures confirmed that DBSA binds to the pocket in the PYL1 dimer interface. The protein oligomeric state experiment suggested that the presence of DBSA can stabilize PYR1 in a dimeric state, while ABA led to the dissociation of PYR1 dimer by using size exclusion chromatography and multi-angle laser light detection (SEC-MALLS). Molecular dynamics (MD) simulations indicated that DBSA stabilizes the PYL1 dimer by forming hydrogen bond networks. The results reveal that stabilizing the PYL dimer can prevent the ABA-induced inhibition of seed germination. These findings lay the foundation for the chemical control of ABA signaling in seed germination.
Results
Identification of a binding pocket in the PYL dimer interface
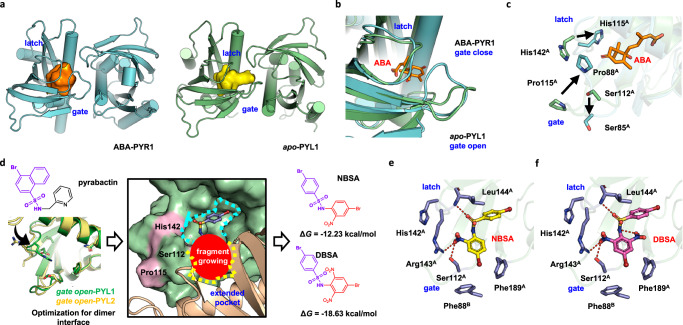
To design a chemical probe that could stabilize PYL dimers, a possible binding cavity on the dimer interface was identified first. As the conformational changes of PYL dimers caused by ABA was essential for the activation of ABA signaling pathway, the comparison of dimeric ABA-bound PYL and dimeric apo-PYL might give the important guidance for stabilizer design. It was generally thought that one compound is sufficient to stabilize PYL dimer, but single ABA-bound PYL1 dimer or dimeric apo-PYR1 structures were lacking for the comparison of surface conformation change. Therefore, the binding pockets in single ABA-bound PYR1 dimer and apo-PYL1 dimer were used as they shared highly conserved ligand binding pockets among dimeric PYL members27. We found the binding pocket in ABA-bound PYR1 is located deeply inside and far away from the dimer interface (Fig. 1a). However, the binding pocket extends to the dimer interface in apo-PYL1. Although two binding pockets are partly overlapped, the volume of decreases from 565.65 Å3 to 475.53 Å3 after ABA binding. This difference is mainly attributed to the conformational changes of two loops, which are commonly defined as gate loop and latch loop, near the dimer interface12,28. Once ABA binds to dimeric PYLs, the gate loop undergoes a conformational change from an open state to a closed state and moves away from the dimer interface (Fig. 1b). The extended binding pocket only exists in apo-PYL dimers which show more stable dimer interactions than ABA-bound PYLs. Therefore, targeting this extended binding pocket with a chemical probe might block the conformational changes of gate loop and latch loop to stabilize the PYL dimer.
Fig. 1. Computational design of DBSA targeting a binding pocket at the PYL dimer interface.
a An extended binding pocket was discovered in the gate-open apo-PYL1 dimer interface (PDB code: 3KAY). This binding pocket partly overlaps with the ABA-binding pocket in the gate-closed PYR1 dimer (PDB code: 3K3K). b The binding pockets differ due to conformational changes in the gate loop and latch loop. The gate loop and latch loop transitions to closed state from open state after ABA binding. c Ser112 at gate loop as well as Pro115 and His142 at latch loop of PYL1 along with their homologous residues in PYR1 (Ser85, Pro88, and His115, respectively) exhibit conformational changes during conformation transition. d Fragment screening was performed to discover chemical probes targeting PYL1 dimer interface. Structural optimization and molecular docking were performed on pyrabactin to suit gate open and latch open PYL1 (PDB code: 3KAY) and PYL2 (PDB code: 3NR4), and find a conformation towards dimer interface. NBSA exhibited the lowest binding free energy. Based on structural optimization, DBSA showed the largest improvement of binding free energy in structural optimization. e NBSA binds to the pocket at the dimer interface and forms several hydrogen bonds with the PYL1 dimer. f DBSA forms an intramolecular hydrogen bond that stabilizes its conformation and forms a series of hydrogen bonds with residues at the PYL1 dimer interface. The hydrogen bonds are shown by red dotted lines.
To design a chemical probe that hinders the conformation transition, the structural basis of these conformational changes was further analyzed. Among residues located in the gate loop and latch loop, Ser112, Pro115 and His143 of PYL1 undergo obvious conformational changes upon ABA binding compared with their homologous residues Ser85, Pro88 and His115 in PYR1 (Fig. 1c). The side chains of proline in the gate loop and histidine in the latch loop flip away from the protein surface towards the protein cavity and interact with ABA, which is related to the dissociation of PYL dimer and provides an interaction interface for the binding of downstream PP2Cs29,30. Moreover, the side chain of serine in the gate loop turns outward and interacts with PP2Cs, which hinders the substrate binding of PP2Cs and inhibits the activity of PP2Cs29. We hypothesized that hindering the conformational transition of these residues would maintain the dimeric conformation of PYLs and block the dissociation of PYL dimer. Therefore, we aimed to design a chemical probe to block the conformational changes of key residues Ser112, Pro115 and His143 in the extended binding pocket to stabilize the PYL1 dimer.
Computationally designed ligand to stabilize PYL dimer
Based on the extended binding pocket in the apo-PYL1 dimer interface, chemical probes that may stabilize the PYL1 dimer were designed by computational virtual screening (Fig. 1d). Pyrabactin could act as ABA receptor agonist for PYR1 and PYL1 or antagonist for PYL230,31. We analyzed the binding mode of pyrabactin as antagonist, and found steric clash was occurred to PYR1 and PYL1 (Supplementary Fig. 1)32. To avoid steric clash and find a proper conformation for dimer stabilizer design, the pyridine group should be removed, and molecular docking was performed on the rest of molecule. It could be noticed that conformation towards dimer interface was easy to cause steric clash with gate open and latch open PYL1. Therefore, molecule was finally optimized into 4-bromobenzenesulfonamide group. More importantly, molecular dynamics simulations indicated that 4-bromobenzenesulfonamide group was binding tightly to gate-open and latch-open PYL1 by forming hydrogen bonds with Arg143. Through computational-based optimization, 4-bromobenzenesulfonamide group was selected as a starting structure for fragments growing to discover dimer stabilizer. Subsequently, fragments from PADFrag, a library of high-frequency fragments from approved drugs and pesticides, were linked to 4-bromobenzenesulfonamide using ACFIS 2.0 web server33,34. Newly generated chemical probes were sorted according to their binding free energy with PYL dimer. Nitrobensulfamide (NBSA, Supplementary Note 1) exhibited the lowest binding free energy (ΔG = −12.23 kcal/mol, Supplementary Table 1). The predicted binding mode of NBSA revealed that it might form hydrogen bonds with Ser112, Arg143 and Leu144 (Fig. 1e). A series of structural modifications of NBSA were made using AILDE web server (Supplementary Data 1)35. We found that DBSA (Supplementary Note 1), which contains an additional nitro group, exhibited the largest improvement in the binding free energy (ΔG = −18.63 kcal/mol). The additional intramolecular hydrogen bonds of DBSA were predicted to enhance the conformation of DBSA (Fig. 1f). Moreover, DBSA was predicted to interact with Ser112, and its binding at the dimer interface pocket was hypothesized to hinder the Pro115 ring flip and the conformational change of His143. Thus, DBSA might bind to the pocket at the dimer interface, and thus hinder the conformational changes of the gate loop and latch loop by interacting with the key residue Ser112 to stabilize the PYL dimer.
DBSA is mainly an antagonist of the dimeric ABA receptors
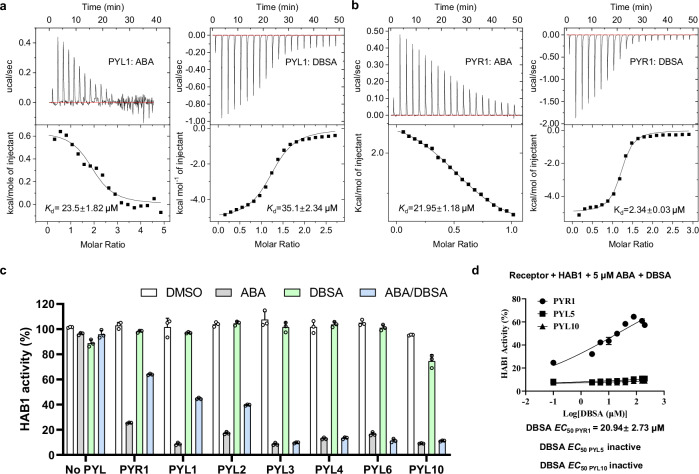
To verify the reliability of our computational design, the binding affinity of DBSA and ABA for dimeric PYLs were first evaluated by isothermal titration calorimetry (ITC). The binding affinity of NBSA could not be determined due to its poor solubility. It is noticed that ABA bind to PYL1 with an apparent dissociation constant (Kd) of 23.5 μM (Fig. 2a), which was comparable to the literature (~50 μM), and this finding validates the reliability of our data36. DBSA possessed an affinity for PYL1 (Kd = 35.1 μM) equivalent to that of ABA. In addition, we found that ABA showed a Kd value of 21.95 μM for PYR1 (Fig. 2b). Surprisingly, DBSA exhibited a much stronger binding affinity for PYR1 (Kd = 2.34 μM), which was an order of magnitude lower than that of ABA for PYR1. But for monomeric PYLs (PYL5, PYL6 and PYL10), DBSA showed no obvious binding affinity (Supplementary Fig. 2). In addition, a ΔH change is observed after dropping ABA into a cell containing PYR1 and DBSA, which indicated that the binding of DBSA to PYL prevents ABA from binding to PYL (Supplementary Fig. 3). The significant binding affinity improvement of DBSA for PYR1 dimer (around ten-fold) than that of ABA supports the feasibility of our computational design.
Fig. 2. DBSA shows low ABA receptor binding affinity and is a potent antagonist of multiple ABA receptors.
a The Kd values of ABA and DBSA to PYL1 using ITC. b The Kd values of ABA and DBSA to PYR1 using ITC. c Antagonistic effect of DBSA on HAB1 activity through a phosphatase assay. PYLs and HAB1 were present at a molar ratio of 1:1 (0.4 μM: 0.4 μM) for PYR1/PYL1/PYL2/PYL3 and 2:1 (0.8 μM: 0.4 μM) for PYL5/PYL6/PYL10. Various PYL-HAB1 combinations were incubated with the indicated chemicals (5 μM ABA, 50 μM DBSA or 5 μM ABA and 50 μM DBSA). n = 3 biologically replicates. d Antagonistic effect of various concentrations of DBSA on HAB1 activity through different PYL members. EC50 values were obtained by nonlinear fits of dose-response data. DBSA were tested at 0 μM to 200 μM, and the concentration of ABA was 5 μM. The concentration of PYR1, PYL5 and PYL10 were 0.4 μM, 0.8 μM and 0.8 μM, respectively, while HAB1 proteins were used at the molar ratio of 0.4 μM. n = 3 biologically replicates. For c, d, the data are presented as the mean ± SD. For (c), the line within the box marks the median and the whiskers represent the minimum and maximum values. Source data are provided as a Source Data file.
To further evaluate whether DBSA interferes with the interaction between PYLs PP2Cs, the PP2C activity of Arabidopsis thaliana HAB1 was detected using phosphatase assay37. Treatment with 5 μM ABA inhibited the activity of HAB1 by 70% or more with seven PYLs (PYR1, PYL1, PYL2, PYL3, PYL5, PYL6 and PYL10) because PP2Cs act as co-receptor of PYL to increase the binding affinity (Fig. 2e). In contrast, more than 90% HAB1 activity was maintained after incubation with 50 μM DBSA and all tested PYLs. Importantly, DBSA alleviated the ABA-induced inhibition dimeric PYLs to (PYR1, PYL1, PYL2) HAB1, but could not antagonize such inhibition for PYL3 (a cis-homodimer) and monomeric PYL5, PYL6 and PYL10. In addition, DBSA showed the highest antagonistic effect to PYR1 with an EC50 value of 20.94 μM, but it could not antagonize ABA for PYL5 and PYL10 even at high concentration (Fig. 2f). Another thing interesting thing is that high concentration of DBSA could inhibit HAB1 activity directly, which might limit the antagonistic effect of DBSA (Supplementary Fig. 4). Together, these data indicated that DBSA are effectively in alleviating the inhibition of HAB1 induced by ABA for PYL dimers and is thus a dimeric ABA receptor antagonist.
DBSA relieves ABA-induced inhibition of seed germination
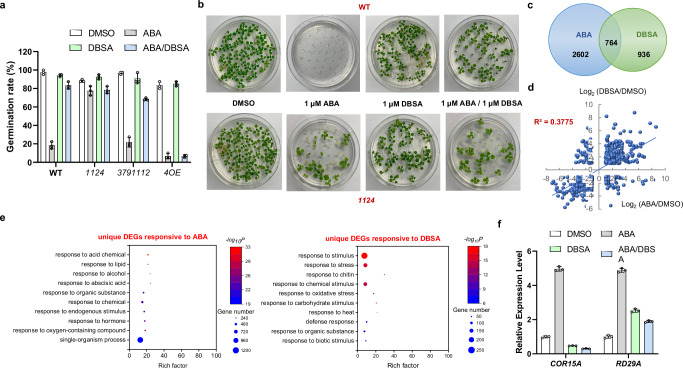
To validate the function of DBSA in seed germination regulation in vivo, germination assays were performed. Wild type (WT), PYL dimer quadruple deletion mutant (pyr1/pyl1/pyl2/pyl4, 1124), PYL monomer multiple deletion mutant (pyl3/pyl7/pyl9/pyl11/pyl12, 3791112), PYL4 overexpression (4OE) seeds were used to verify the relationship between DBSA and PYL dimers in ABA signaling. As shown in Fig. 3a, the germination rates of WT, 3791112 and 4OE seeds were lower than 30% after treatment with ABA (1 µM), and 4OE seeds exhibited ABA more sensitive phenotype. But 1124 seeds showed ABA insensitive phenotype as literature13. In contrast, DBSA (1 µM) showed almost no influence to seed germination (seed germination rate >90%). More importantly, the ABA-induced seed germination inhibition was partly abolished by the treatment with DBSA (1 µM) in WT and 3791112 seeds, which resulted in a seed germination rate higher than 70%. But it could not antagonize ABA induced seed germination inhibition in 4OE seeds. In addition, DBSA showed similar effects of antagonizing ABA to seedling growth of WT and 1124 mutants as seed germination (Fig. 3b). These results indicated that DBSA might reverse such inhibition mainly via PYL dimers.
Fig. 3. DBSA relieves ABA-induced inhibition of seed germination and seedling growth, and does not induce ABA-responsive gene expression in Arabidopsis thaliana.
a Germination rate of seeds exposed to ABA (1 µM), DBSA (1 µM) or ABA and DBSA (1 µM:1 µM). DMSO (0.05%) was used as a control. PYL dimer quadruple deletion mutant (pyr1/pyl1/pyl2/pyl4, 1124), PYL monomer multiple deletion mutant (pyl3/pyl7/pyl9/pyl11/pyl12, 3791112), PYL4 overexpression (4OE) seeds are considered germinated when green cotyledons expand (n = 3 biologically replicates). b Seeding growth of WT and 1124 seeds under the treatment of ABA (1 µM), DBSA (1 µM) or ABA and DBSA (1 µM:1 µM). n = 3 biologically replicates. c DBSA and ABA treatment caused different gene expression patterns. d The transcript levels of the common DBSA- and ABA-responsive genes were poorly correlated. The scatter plot shows the log2-transformed expression levels of DBSA-responsive DEGs (y-axis) and ABA-responsive DEGs (x-axis) relative to the DMSO control. e The representative GO terms and pathways enriched in ABA-specific response and DBSA-specific response DEGs based on functional enrichment analysis (p < 0.01). f Induction of abiotic stress marker genes in Arabidopsis thaliana (Col-0) after ABA or DBSA treatment as determined by qRT-PCR. Ten-day-old seedlings were treated with 10 μM ABA or 100 μM DBSA or 10 μM ABA and 100 μM DBSA for 6 h before RNA extraction, and 0.05% DMSO was used as control (n = 3 biologically replicates). For a and f, the data are presented as the mean ± SD, the line within the box marks the median and the whiskers represent the minimum and maximum values. For f, P values are indicated by two-tailed Student’s t test. Source data are provided as a Source Data file.
DBSA does not induce ABA-responsive gene expression
To characterize the effect of DBSA and ABA on Arabidopsis thaliana gene expression, we used RNA sequencing (RNA-seq) to profile the transcriptomes of ten-day-old plants. It was noticed that 3366 and 1700 differentially expressed genes (DEGs) were identified as induced by ABA and DBSA, respectively, while 2602 genes were specifically responsive to ABA and 936 genes were specifically responsive to DBSA (Fig. 3c and Supplementary Data 2-3). In addition, the expression profiles obtained with DBSA and ABA were weakly correlated at the response in transcript level (R2 = 0.3775 with a cut-off false discovery rate <0.05, Fig. 3d). The statistical data of DEGs indicated that DBSA may regulate the Arabidopsis thaliana in a different way compared with ABA.
A gene ontology (GO) analysis was performed on the DEGs response to different chemicals. It can be found that none of the top ten enriched unique DEG terms in response to ABA or DBSA were the same (Fig. 3e and Supplementary Data 4-6). Importantly, ABA activating process genes were detected after the treatment of ABA, but not for the treatment of DBSA (Fig. 3e and Supplementary Fig. 5). The expression of 28 genes involved in seed germination was induced by ABA (GO:0009845, Supplementary Data 5), but none of the genes was found to respond to DBSA treatment (Supplementary Data 6). The RNA-seq result suggested that DBSA may be not able to activate the expression level of ABA-induced genes.
To characterize the role of DBSA in regulating some important genes in ABA pathway, we detected the expression levels of COR15A and RD29A in seedlings using quantitative real-time PCR (qRT-PCR, Fig. 3f). COR15A expression was markedly activated by ABA treatment, but was reduced by DBSA alone or in combination with ABA. RD29A expression was very strongly induced by ABA but not DBSA alone or in combination with ABA. These results indicated that DBSA may repress the ABA-induced gene expression and act as an ABA antagonist.
DBSA binds to the pocket of PYL1 dimer interface
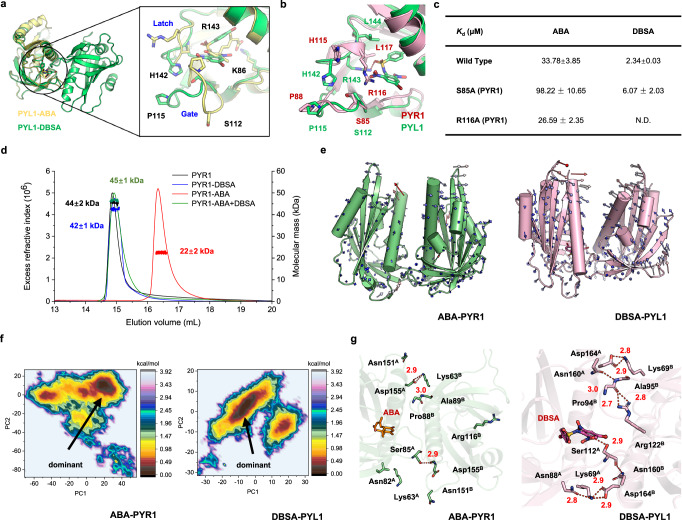
X-ray crystallography was performed to verify the binding mode between DBSA and PYL1 dimer. The structure of dimeric PYL1-DBSA complex was obtained at a resolution of 2.29 Å (PDB code 9J6I, Supplementary Table 2). Consistent with our hypothesis, DBSA is located at the binding pocket of dimer interface with a gate open and latch open conformation compared with ABA bind to a gate close and latch close conformation (Fig. 4a and Supplementary Fig. 6). In addition, the binding mode of DBSA was similar to our predicted binding mode (Supplementary Fig. 7): the root-mean square deviation (RMSD) values were 1.06 Å (protein backbone atoms), 1.48 Å (protein heavy atoms) and 0.86 Å (ligand heavy atoms), respectively, which indicated that our computational model was reliable. For the detailed binding mode of DBSA, the gate loop of PYL1-DBSA complex was in its open state, and the key residues Ser112, Pro115 and His142 were in conformations similar to those in the apo-PYL1 structure. Furthermore, as predicted, hydrogen bond was observed between DBSA and the side chain of Arg143, but the hydrogen bond between DBSA and Ser112 was not found (Fig. 4b). For the binding mode of DBSA to PYR1, the structural alignment predicted that DBSA might share a similar binding mode compared to PYL1, and mutants were built for homological residue S85 (S112 in PYL1) and R116 (R143 in PYL1). The Kd value coincides well with our prediction, DBSA showed no binding to R116A and a weaker binding affinity to S85A (Fig. 4c). The crystal structure indicated that DBSA targeted the binding pocket at the dimeric PYL interface and hindered ABA-induced conformational changes in key loops.
Fig. 4. DBSA targets PYL1 dimer interface and stabilizes the PYL1 dimer via hydrogen bond networks.
a Crystal structure of PYL1 with DBSA (PDB code 9J6I, green) and ABA (PDB code 3JRS, yellow). ABA induced a gate-close and latch-close conformation, which DBSA bind to a DBSA induced a gate-open and latch- open conformation. b PYL1 was maintained in the latch-open and gate-open state in the crystal structure (shown in green stick model). DBSA forms hydrogen bonds with the side chain of Arg143. DBSA was predicted to bind to latch-open and gate-open PYR1 and interact with R116 (shown in pink stick model). c Alanine mutation result showed that DBSA may form strong interaction with R116 of PYR1. d The aggregation states of PYR1 dimer induced by ABA, DBSA or ABA/DBSA co-treatment were detected by SEC-MALLS. e The motion trends of ABA-PYR1 and DBSA-PYL1 complexes. f The dominant conformations were determined using FEL analysis. g The hydrogen networks mediated by ABA of PYR1 or DBSA of PYL1 in the dimer interfaces.
To further verify the effect of DBSA on PYR1 dimer stabilization, the protein oligomeric state was detected using SEC-MALLS approach. The elution volume and detected molecular weight of apo-PYR1 was 14.74–15.45 mL and 44 ± 2 kDa (Fig. 4d), respectively, which is similar to literature and indicated that it was in the dimeric state (theoretical mass of 43 kDa)36. After the treatment of ABA, the elution volume was changed to 16.02–17.05 mL, and the molecular mass was 22 ± 2 kDa, which is existing in PYR1 monomer state. While for DBSA treatment and DBSA/ABA co-treatment, the elution volumes were 14.82–15.47 mL and 14.66–15.51 ml, respectively. The molecular weight of 42 ± 1 kDa for DBSA-PYR1 and 45 ± 1 kDa for DBSA/ABA-PYR1 indicated that proteins were in dimeric states. These results suggested that ABA led to a dimer dissociation, and DBSA reached our goal to stabilize the PYL dimer.
DBSA antagonizes ABA signaling pathway by stabilizing PYL dimers via hydrogen bond networks
To understand the mechanism that DBSA antagonize the ABA signaling pathway, MD simulations and binding free energy calculation were carried on PYR1-ABA, apo-PYL1 and PYL1-DBSA complexes. After a 500-ns MD simulation, DBSA remains in a stable state with PYL1 dimer (Supplementary Fig. 8) and maintains hydrogen bonds with Arg143. The binding free energy between two monomers of ABA-bound PYR1, apo-PYL1, and DBSA-bound PYL1 proteins were −62.19 kcal/mol, −71.19 kcal/mol and −91.18 kcal/mol, respectively (Supplementary Table 3). Therefore, DBSA exhibits stabilization effect to the PYL dimer as expected.
To further determine the mechanism that DBSA stabilize the PYR1 dimer, the detailed interactions were analyzed. The residue correlation analysis revealed that ABA-PYL1 and DBSA-PYL1 may exhibit more active conformation changes (Supplementary Fig. 9). Further, the motion trend analysis showed that after ABA binding, PYR1 monomers moved far away from each other (Fig. 4e). However, PYL1 monomers moved towards each other upon DBSA binding, which suggested that DBSA might strengthen the interaction between PYL1 monomers compared with apo-PYL1 (Fig. 4e and Supplementary Fig. 10). The dominant conformation during MD simulations were determined using free energy landscape analysis (Fig. 4f and Supplementary Fig. 11). Only three stable (hydrogen bond occupancy > 50%) hydrogen bonds were noted at the ABA-PYL1 dimer interface (Fig. 4g and Supplementary Table 4), and the apo-PYL1 had a higher number of hydrogen bonds (four hydrogen bonds, Supplementary Fig. 12). Surprisingly, ten hydrogen bonds were observed at the dimer interface of DBSA-PYL1. During the simulation, Lys69A, Asn88A, Ser112A, Asnl60B and Asp164B formed a hydrogen bond network near DBSA. At the other end of the interface, Lys69B, Pro94B, Ala95B, Arg122B, Asnl60A and Asp164A formed a hydrogen bond network. The result of MD simulations suggested that DBSA stabilized PYL dimers via hydrogen bond networks.
DBSA exhibits a mode of action that is different from the known PYL ligands
To understand the mechanistic difference between DBSA and existing PYLs chemical modulators, we analyzed the binding modes of these molecules with PYLs. DBSA binds to the pocket at PYL1 dimer interface in the latch-open and gate-open conformation (Fig. 5a). A gate-closed conformation is observed for PYR1-AS6, and AS6 antagonize ABA signaling pathway by occupying the position of 3’-tunnel to block the interface of PYL-PP2C interaction38. Antabactin is the most potent ABA-receptor antagonist yet, and it also binds to the gate-closed/latch-closed PYL10 and occupying the position of conserved Trp lock of PP2Cs to destroy the interface of PYL-PP2C interaction. Pyrabactin selectively activates PYL1 and PYR1, and binds to a gate-closed PYR113. Although it binds to PYL2 in a gate-open state, a trimer was found in a lattice, and did not have effect to plant phenotype. Quinabactin (AM1) exhibits agonist activity towards more ABA receptors, and adopt a gate-closed conformation with PYL225. Opabactin and analogs 3CB is a pan-ABA receptor agonist which bind PYL10 in a gate-closed conformation26. Therefore, by targeting the pocket in gate-open PYL dimer interface, DBSA exhibits a different binding mode compared with existing chemical probes.
Fig. 5. Mode of action of DBSA compared with those of reported ABA receptor agonists and antagonists.
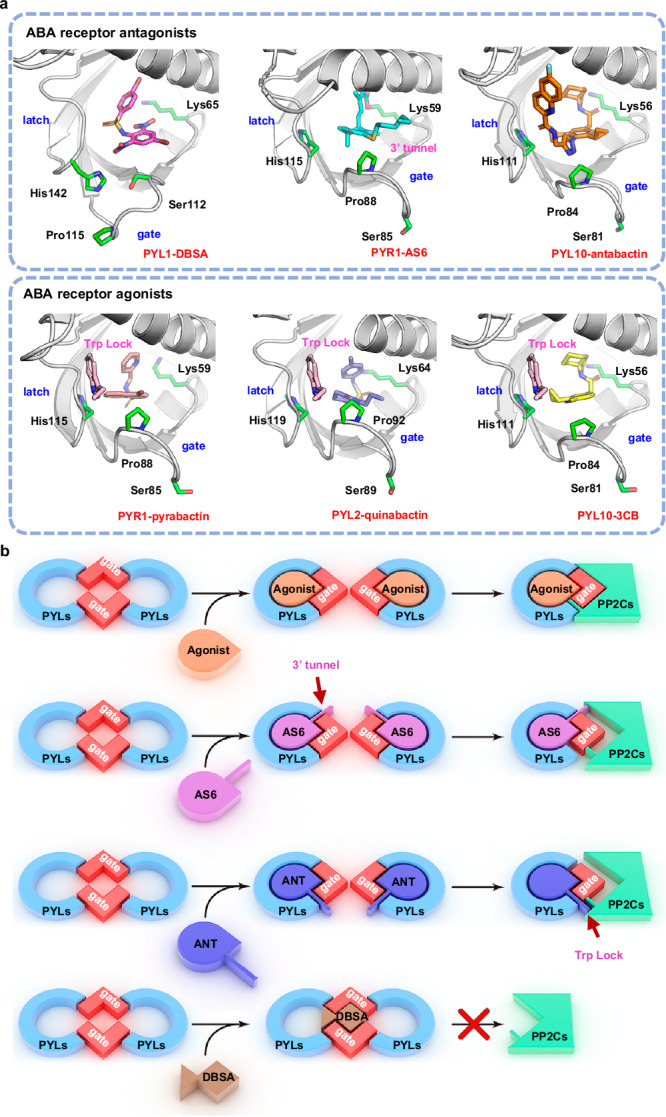
a Binding mode of different ligands with PYLs. PYL1-DBSA (PDB code 9J6I), PYR1-AS6 (PDB code 3WG8), PYL10-antabactin (PDB code 7MLD), PYR1-pyrabactin (PDB code 5UR4), PYL2-quinabactin (PDB code 4LA7) and PYL10-3CB (analog of opabactin, PDB code 6NWC). PYR1-pyrabactin, PYL2-quribaction, PYL10−3CB, PYL10-antabactin and PYL5-AS6 are in latch-closed and gate-closed conformations, whereas PYL1-DBSA is in latch-open and gate-open conformation. For the latch-closed and gate-closed agonist binding conformations, a conserved Trp lock of PP2Cs were insert into the PYL pockets. b Mode of action of ABA receptor agonists, ABA-mimic receptor antagonists AS6, antabactin and DBSA. ABA receptor agonists cause gate-closed conformations and PYL dimer dissociation, which inhibits PP2Cs. ABA-mimic receptor antagonists AS6 and antabactin also cause gate-closed conformations and PYL dimer dissociation, but AS6 obstructs the interaction between PP2C and PYLs by occupying the 3’ tunnel, and antabactin blocks the conserved Trp lock of HAB1 to PYL. DBSA stabilizes the PYL dimer, which results in PP2C activation.
The differences in binding mode suggests a distinct molecular mechanism through which DBSA regulates PYL-PP2C interaction. ABA and ABA receptor agonists induce conformation changes in PYLs from latch-open and gate-open to latch-closed and gate-closed state, which results in dimer dissociation and then monomer PYLs repress the activity of PP2Cs (Fig. 5b)39. Most of antagonists also mediate such conformation transition and dissociation of dimeric PYLs. These antagonists hinder the binding of PYL monomers with PP2Cs by occupying the interaction interface via steric clash group. But PP2Cs are usually only partly inhibited, and these antagonists often exhibit modest in vivo activities. DBSA functions as an ABA receptor antagonist with a mode of action that stabilizes the PYL dimer. DBSA targets the binding pocket at the dimer interface and interacts with the PYL dimer, which prevents PYL dimer dissociation. The PYL dimer is unable to bind PP2Cs; thus, PP2C activity is not inhibited and ABA signaling is not activated. In summary, the known chemical probes cause dimer dissociation and regulates PYL-PP2C interaction, whereas DBSA stabilizes PYL dimer and blocks the interaction between PYL and PP2C.
Discussion
In this study, using the chemical genetic approach, including chemical design, chemical synthesis, molecular modeling, biochemistry, structural biology and botany, we revealed that stabilizing PYL dimer can relieve the inhibition of seed germination induced by ABA. A binding pocket was found in the dimer interface of PYL1, and computational fragment-based virtual screening was performed to discover PYL dimer stabilizers, NBSA and DBSA. DBSA exhibited an approximately 10-fold improvement in receptor affinity (PYR1 Kd = 2.34 μM) compared with ABA (PYR1 Kd = 21.95 μM). The activity of HAB1 in the presence of PYLs and ABA was recovered after treatment with DBSA. Transcriptome profiling showed that DBSA does not induce the expression of ABA-responsive genes. In vivo experiments revealed that DBSA strongly counteracts the ABA-induced inhibition of seed germination via PYL dimers. Furthermore, X-ray crystallography confirmed that DBSA binds to the pocket in PYL1 dimer interface with a gate-open state. And the alanine mutation result indicated that DBSA shared similar binding mode in PYR1, and Arg116 played an important role to the binding affinity. In the SEC-MALLS experiment, DBSA maintains the dimeric state of PYR1, while ABA led to the dissociation of PYR1 dimer. MD simulations revealed that that DBSA may stabilize PYL dimer via hydrogen bond networks with residues in the dimer interface. Together, these results demonstrate that the stabilization of PYL dimers can relieve the seed germination inhibition induced by ABA.
The unique conformational change mechanism of PYL dimers induced by ABA draws a lot of attentions. At first, scientists found that ABA bound to latch-closed and gate-closed conformation of PYLs and interacted with PP2Cs12. Therefore, the transition of latch loop and gate loop and the dimer dissociation were considered to occur at the same time. But the subsequent studies revealed that PYLs are always in the equilibrium state of latch-closed and latch-open in the absence of ABA, and the latch-closed state is more preferred to the binding of ABA40. The latch-closed PYL along with ABA lead to the conformation change of gate loop, and such change is a dynamic process. The existence of PP2Cs can stabilize PYLs in gate-closed and latch-closed conformation. Therefore, PP2Cs are the co-receptors to ABA binding and lead to an improved binding affinity of ABA to PYL dimer. Almost all of current ABA receptor modulators adopted such mechanism. But PYL monomers are usually in the latch-closed and gate-closed state, and exhibit ABA-independent PP2C inhibitory activity. So, the function difference between PYL monomers and dimers remains unclear. To utilize the co-receptor factor of PP2Cs to improve activity, ABA receptor agonist and antagonist need to precisely bind to the similar but functionally different conformation, which increase the difficulty of molecular design. We designed a PYL high affinity ligand DBSA via computational approach, which could antagonize ABA signaling pathway by stabilizing dimeric PYL receptors. The high similarity between experiment and predicted model indicated our computational methods were reliable, which could be used for the discovery of more ligands with unique functions. The stabilization of PYL dimers might become a promising approach to explore the biofunction difference between PYL dimers and monomers.
A chemical probe DBSA was designed to reveal the relationship between PYL dimer stabilization and seed germination. But our work also raises several questions that need to be addressed in the future. First, DBSA antagonize the interaction between PYL monomers and PP2Cs, but the influence of monomers to seed germination is not fully evaluated. Second, the function of DBSA in other ABA-dependent processes such as stomatal regulation needs to be studied in the future. Third, the activity of DBSA in alleviating ABA-induced inhibition of seed germination in crop species needs to be studied. In addition, high concentration of DBSA might inhibit the seed germination due to its HAB1 directly inhibition (Supplementary Fig. 13).
Our findings are significant for understanding the regulation and chemical intervention of ABA signaling. ABA is one of the most important plant hormones and exerts a series of effects on plant growth and development in addition to its roles in water relations, seed germination, and gene regulation. This study revealed that ABA signaling can be regulated by the stability of PYL dimer. Compared with the traditional antagonism mechanism of blocking the PYL-PP2C interaction, stabilizing PYL dimers could bypass the redundancy between PYL family members, which in theory may result in a markedly improved antagonistic effect. The different mode of action of DBSA in relieving the ABA-induced inhibition of seed germination might open a venue for the regulation and chemical intervention of ABA signaling and seed germination.
Methods
Molecular docking
The molecular docking study was performed using AutoDock Vina 1.1.241. The receptor proteins were retrieved from the RCSB Protein Data Bank42. The protein structures were prepared by adding hydrogen atoms, repairing side chains and removing water molecules. Then protein was then subjected to an energy minimization. The ligand was then docked into the binding pocket, and twenty poses were exported for further analysis.
Computational fragment-based drug discovery
The 4-bromobenzenesulfonamide in complex at a gate-open state PYL1 was selected for fragment growing using ACFIS 2.0. The protein-ligand complex was first subjected to energy minimization, and then the conformation ensemble was then produced for flexible fragment growing. High-frequency fragments in FDA drugs from PADFrag were added to the starting point to generate new ligands. The Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method was employed to evaluate the binding free energy of generated ligands using Amber 16 package43.
Ligand directing evolution
The ligand directing evolution of NBSA was performed using AILDE web server. The complex of PYL1 and NBSA was selected as the starting structure. After energy minimization and a short-term MD simulation was performed using Amber 16 package. Each hydrogen atom of NBSA was randomly replaced by ten substituents with different properties. Energy minimization was performed, and the binding free energy of generated ligand with PYL1 then calculated using the MM-PBSA method using Amber 16 software package.
Protein expression and purification
The full-length PYR1, PYL1, PYL6 and PYL10 were cloned into pET15b vector, while PYL2, PYL3 and PYL5 were cloned into pET28a vector, and HAB1 was cloned into pET15s vector to generate N‐terminal His6‐tagged recombinant proteins. The plasmid was transformed into E. coli BL21 (DE3). One litre of lysogeny broth medium supplemented with 100 mg ml−1 ampicillin was inoculated with a transformed bacterial preculture and shaken at 37 °C until the cell density reached an OD600 of 1.0–1.2. Protein expression was induced with 0.2 mM isopropyl-β-D-thiogalactoside at 20 °C for 12–16 h. The cells were collected by centrifugation, homogenized in buffer A (25 mM Tris, pH 8.0, 150 mM NaCl), and lysed by a lysozyme (Sigma-Aldrich). Cell debris was removed by centrifugation at 14,000 g and 4 °C for 1 h, and the supernatant was loaded onto a column equipped with Ni2+ affinity resin (Ni-NTA, Qiagen), washed with buffer B (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 10 mM imidazole), and eluted with buffer C (25 mM Tris-HCl, pH 8.0, and 250 mM imidazole). The protein was then separated by cation exchange chromatography (Source 15Q, GE Healthcare) using a linear NaCl gradient in buffer A. The N-terminal His tag was removed by dRICE. The purified protein was subjected to gel filtration chromatography (Superdex-200 Increase 10/300 GL, GE Healthcare) in a buffer containing 25 mM Tris, pH 8.0, 150 mM NaCl. The buffer used for HAB1 added extra 2 mM MgCl2. The peak fractions were pooled for crystallization immediately or stored at −80 °C.
ITC of PYR1 and PYL1 with ABA or DBSA
ITC experiments were performed using an iTC200 microcalorimeter (MicroCal) in ITC buffer (25 mM HEPES, 100 mM NaCl, pH 7.5) at 25 °C. ABA or DBSA were dissolved in ITC buffer, adjusted to pH 7.5 and used directly in titration experiments. Both protein and ligand solutions were degassed extensively and their concentrations were determined precisely using a UV/Vis spectrophotometer. In the Isothermal titration calorimetry (ITC) assay, the final concentrations of PYR1 and PYL1 were in the range of 50 to 100 μM and ABA or DBSA were in the range of 500 to 1000 μM, respectively. Each titration consisted of 19 injections, and the Kd value was fitted using a one-site binding model by MicroCal ITC200 analysis software Origin 7.0 (Malvern).
Phosphatase activity assay
The Ser/Thr phosphatase assay system (Promega) was used to measure the phosphatase activity, and the protocol of phosphatase activity assay was performed using a previously described protocol44. The reaction was performed in a 50 μl reaction volume with PYR/PYL-HAB1 at a molar ratio of 1:1 (0.4 μM: 0.4 μM) for PYR1, PYL1, PYL2 and PYL3 or and 2:1 (0.8 μM: 0.4 μM) for PYL5, PYL6 and PYL10, while 5 μM ABA, 50 μM DBSA or 5 μM ABA and 50 μM DBSA were added if needed.
Seed germination, seedling growth, and chemical treatments
WT Col-0 and mutant (Arabidopsis thaliana) seeds (pyr1/pyl1/pyl2/pyl4, pyl3/pyl7/pyl9/pyl11/pyl12 and pyl4 overexpression) were stratified for 4 days before they were sown on half-strength MS solid medium containing 1% sucrose and the indicated compounds, with 40 seeds per 6 cm plate and three plates for each chemical. The plates were kept in a growth chamber at 22 °C under long-day conditions. Seeds were evaluated daily and were considered germinated when the green cotyledons appeared. Chemicals used for treatment groups included 0.05% DMSO as a mount control, 1 µM ABA, 1 µM DBSA, and 1 µM ABA/1 µM DBSA, respectively45. For the seedling experiment, 1 µM ABA, 1 µM DBSA, 1 µM ABA / 1 µM DBSA were used for WT and mutant (pyl0;1;2;4) seedlings, respectively. Three replicates of each treatment were established. The seeding establishment were observed after 14-days growing.
RNA-Seq and transcriptome analysis
Ten-day-old Arabidopsis WT (Col) seed seedlings were mock-treated or exposed to 50 μM ABA or DBSA for 3 h (biological duplicates), prior to total RNA extraction using the RNeasy Plant Mini Kit (QIAGEN). Multiplex RNA library construction and Illumina sequencing were carried out at the Annoroad Gene Technology Co. Ltd. (Beijing, China) according to the previous description with minor modifications46,47. Approximately 20 million reads of 50 base pair (bp) paired-ended sequences were generated per sample. The raw reads were mapped onto the Arabidopsis gene model based on HISAT48. Then, the number of reads mapped to each gene was then counted using HTSeq49. Finally, the differential expression was evaluated through DESeq50. Furthermore, the GO analysis of the DEGs was performed using agriGO51.
qRT-PCR analysis
Total RNA was reverse-transcribed using TransScript RT kit (Invitrogen) according to the manufacturer’s protocol. All quantitative RT-PCR assays were performed following a two-step protocol with the SYBR Premix Ex TaqTM II kit (TaKaRa) and a CFX96 Real-time system (BIORAD) according to the manufacturer’s instructions using the following primers: COR15A forward, 5’-GAGCCAAGCAGAGCAGCTTC-3’ and reverse, 5’-AGGATGTTGCCGTCACCTTTAG -3’. RD29A forward, 5’-GAAGACTGAAACTGGAATGGAC-3’ and reverse, 5’-GCTCCTGATTCACTACCAAAGC-3’. Each assay consisted of three biological replicates and was performed twice. ACT7 was used as an internal control in the quantitative RT-PCR assay.
Crystallization, data collection, and structure determination
Protein was concentrated to 10 mg ml−1 before crystallization trials. Crystallizations were performed using the sitting-drop vapour diffusion method at 18 °C by mixing equal volumes (1 μl) of protein with reservoir solution. The soaking method was applied to obtain the cocrystals of PYL1 and DBSA. A series of PYL1 truncations were prepared to optimize the cocrystals. After numerous trials, the diamond-like co-crystal of PYL1 (36-210)-DBSA in the well buffer containing 11.7% PEG 8000, 40 mM potassium phosphate dibasic, 19.5% glycerol and 3% ethanol was collected and cryoprotected by adding glycerol to a final concentration of 14%. All the diffraction data were collected at Shanghai Synchrotron Research Facility (SSRF) on beamlines BL17U or BL19U, integrated, and processed with the HKL2000 package. Further data processing was carried out using CCP4 suit. The structure of PYL1-DBSA complex was resolved by molecular replacement using the reported PYL1 (PDB code: 3KAY) as the search model with the program PHASER. All the structures were iteratively built with COOT 0.9.4 and refined with PHENIX program. The data collection and structure refinement statistics are summarized in Supplementary Table 2.
SEC-MALLS experiments
SEC was performed using a Superdex S200 column (GE Healthcare) equilibrated with 20 mM Tris pH 7.5, 150 mM NaCl and 1 mM β‐mercaptoethanol. For the experiments with chemicals, 1 mM ABA, 1 mM DBSA or 1 mM ABA and 1 mM DBSA were included in the equilibration buffer. Receptor proteins were injected at a concentration of 80 μM. All separations were performed at 20 °C with a flow rate of 0.5 ml/min. On‐line MALLS detection was performed with a DAWN‐EOS detector (Wyatt Technology Corp., Santa Barbara, CA) using a laser emitting at 690 nm. Data were analyzed and weight‐averaged molar masses were calculated using the ASTRA software (Wyatt Technology Corp.).
MD simulations
The MD simulation was performed using the AMBER 16 software package. The crystal structures of ABA-PYR1 (PDB code: 3K3K), apo-PYL1 (PDB code: 3KAY) and DBSA-PYL1 (PDB code: 9J6I) were utilized as the initial structures, and the protein and ligand were treated with ff14SB force field and general amber force field (gaff). The complex systems were first minimized by the steepest descent method and the conjugate gradient method. Subsequently, the systems were gradually heated from 0 K to 300 K in the NPT ensemble. The 500-ns MD simulation of each system was performed according to previous work. The snapshots extracted at every picosecond of the stable interval were used for structural and energetic analysis. The residue cross-correlation was analyzed based on the Nor_mod module of LARMD webserver52,53. The binding free energy was calculated using MMPBS.py module in Amber 16 software package.
Statistics and reproducibility
All statistical analyses were performed using GraphPad Prism 8.0 software. No statistical method was used to predetermine sample size. No data were excluded from the analysis. Samples were grown under the same conditions and randomly allocated in the growth chamber. Experimental plant material was collected randomly without any bias. Investigators were not blinded to allocation during the experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 32125033) to G.F. Hao, and the Postdoctoral Fellowship Program of CPSF (No. GZB20230198) to Z.Z.W. We thanks to Prof. Yang Zhao at Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences for providing PYL monomer multiple deletion mutant (pyl3/pyl7/pyl9/pyl11/pyl12) and Prof. Yuan Zheng at Henan University for providing pyl4 overexpression seeds.
Author contributions
G.-F.Y, G.-F.H. and J.-K.Z. conceived the research; Z.-Z.W, J.-F.Y and Z.-Y.H. designed and synthesized the chemicals; M.-J.C., J.D., R.-N.Y., Y.T., K.H.H and R.-J.P designed and performed most biochemical and physiological experiments; J.Y., L.L and P.Y performed the AlphaScreen assay and protein crystallization; M.-J.C. and M.-X.C analyzed the RNA-seq data; J.-H.L., Y.-Y.G. and Y.-N.L. performed the plant experiments; J.Y., L.L and P.Y analyzed the X-ray crystal structure data; Z.-Z.W performed the molecular simulation; Z.-Z.W and J.-F.Y drafted the manuscript. J.-K.Z., G.-F.H. and G.-F.Y revised and finalized the manuscript.
Peer review
Peer review information
Nature Communications thanks Armando Albert, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw RNAseq data generated in this study have been deposited in the sequence read archive (SRA) repository of NCBI under BioProject accessions: PRJNA1138801. The crystal structure of PYL1-DBSA complex can be accessed from the Protein Data Band under the accession codes 9J6I. All study data are included in the article and/or supporting information. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhi-Zheng Wang, Min-Jie Cao, Junjie Yan, Jin Dong.
Contributor Information
Jian-Kang Zhu, Email: zhujk@sustech.edu.cn.
Ge-Fei Hao, Email: gefei_hao@foxmail.com.
Guang-Fu Yang, Email: gfyang@mail.ccnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52426-y.
References
- 1.Rajjou, L. et al. Seed Germination and Vigor. Annu. Rev. Plant Biol.63, 507–533 (2012). 10.1146/annurev-arplant-042811-105550 [DOI] [PubMed] [Google Scholar]
- 2.Purugganan, M. D. & Jackson, S. A. Advancing crop genomics from lab to field. Nat. Genet.53, 595–601 (2021). 10.1038/s41588-021-00866-3 [DOI] [PubMed] [Google Scholar]
- 3.Tester, M. & Langridge, P. Breeding Technologies to Increase Crop Production in a Changing World. Science327, 818 (2010). 10.1126/science.1183700 [DOI] [PubMed] [Google Scholar]
- 4.Challinor, A. J., Koehler, A. K., Ramirez-Villegas, J., Whitfield, S. & Das, B. Current warming will reduce yields unless maize breeding and seed systems adapt immediately. Nat. Clim. Change6, 954–958 (2016). 10.1038/nclimate3061 [DOI] [Google Scholar]
- 5.Miransari, M. & Smith, D. L. Plant hormones and seed germination. Environ. Exp. Bot.99, 110–121 (2014). 10.1016/j.envexpbot.2013.11.005 [DOI] [Google Scholar]
- 6.Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell167, 313–324 (2016). 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rifna, E. J., Ratish Ramanan, K. & Mahendran, R. Emerging technology applications for improving seed germination. Trends Food Sci. Technol.86, 95–108 (2019). 10.1016/j.tifs.2019.02.029 [DOI] [Google Scholar]
- 8.Lin, Q. et al. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun.6, 7981 (2015). 10.1038/ncomms8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, Z. et al. Counteraction of ABA-Mediated Inhibition of Seed Germination and Seedling Establishment by ABA Signaling Terminator in <em>Arabidopsis</em>. Mol. Plant13, 1284–1297 (2020). 10.1016/j.molp.2020.06.011 [DOI] [PubMed] [Google Scholar]
- 10.Nishimura, N. et al. Structural Mechanism of Abscisic Acid Binding and Signaling by Dimeric PYR1. Science326, 1373 (2009). 10.1126/science.1181829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park, S.-Y. et al. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature520, 545–548 (2015). 10.1038/nature14123 [DOI] [PubMed] [Google Scholar]
- 12.Melcher, K. et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature462, 602–608 (2009). 10.1038/nature08613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, S.-Y. et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science324, 1068 (2009). 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, Y. et al. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science324, 1064 (2009). 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- 15.Soon, F.-F. et al. Molecular Mimicry Regulates ABA Signaling by SnRK2 Kinases and PP2C Phosphatases. Science335, 85 (2012). 10.1126/science.1215106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii, H. et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature462, 660–664 (2009). 10.1038/nature08599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaidya, A. S. et al. Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo. Proc. Natl Acad. Sci.118, e2108281118 (2021). 10.1073/pnas.2108281118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Née, G. et al. Delay of germination1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun.8, 72 (2017). 10.1038/s41467-017-00113-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, C.-Z. et al. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J.80, 654–668 (2014). 10.1111/tpj.12670 [DOI] [PubMed] [Google Scholar]
- 20.Carrera-Castaño, G., Calleja-Cabrera, J., Pernas, M., Gómez, L. & Oñate-Sánchez, L. An Updated Overview on the Regulation of Seed Germination. Plants9, 703 (2020). 10.3390/plants9060703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima, K. & Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep.32, 959–970 (2013). 10.1007/s00299-013-1418-1 [DOI] [PubMed] [Google Scholar]
- 22.Nishimura, N. et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J.61, 290–299 (2010). 10.1111/j.1365-313X.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCourt, P. Genetic analysis of hormone signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol.50, 219–243 (1999). 10.1146/annurev.arplant.50.1.219 [DOI] [PubMed] [Google Scholar]
- 24.Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R. & Abrams, S. R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol.61, 651–679 (2010). 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- 25.Okamoto, M. et al. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl Acad. Sci.110, 12132 (2013). 10.1073/pnas.1305919110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaidya, A. S. et al. Dynamic control of plant water use using designed ABA receptor agonists. Science366, eaaw8848 (2019). 10.1126/science.aaw8848 [DOI] [PubMed] [Google Scholar]
- 27.Durrant, J. D., Votapka, L., Sørensen, J. & Amaro, R. E. POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics. J. Chem. Theory Comput.10, 5047–5056 (2014). 10.1021/ct500381c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santiago, J. et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature462, 665–668 (2009). 10.1038/nature08591 [DOI] [PubMed] [Google Scholar]
- 29.Yin, P. et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol.16, 1230–1236 (2009). 10.1038/nsmb.1730 [DOI] [PubMed] [Google Scholar]
- 30.Peterson, F. C. et al. Structural basis for selective activation of ABA receptors. Nat. Struct. Mol. Biol.17, 1109–1113 (2010). 10.1038/nsmb.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, J.-F. et al. Molecular Determinants Elucidate the Selectivity in Abscisic Acid Receptor and HAB1 Protein Interactions. Front. Chem.8, 10.3389/fchem.2020.00425 (2020). [DOI] [PMC free article] [PubMed]
- 32.Yuan, X. et al. Single Amino Acid Alteration between Valine and Isoleucine Determines the Distinct Pyrabactin Selectivity by PYL1 and PYL2 *. J. Biol. Chem.285, 28953–28958 (2010). 10.1074/jbc.M110.160192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, X.-X., Wang, Z.-Z., Wang, F., Hao, G.-F. & Yang, G.-F. ACFIS 2.0: an improved web-server for fragment-based drug discovery via a dynamic screening strategy. Nucleic Acids Res.51, W25–W32 (2023). 10.1093/nar/gkad348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, J.-F. et al. PADFrag: A Database Built for the Exploration of Bioactive Fragment Space for Drug Discovery. J. Chem. Inf. Modeling58, 1725–1730 (2018). 10.1021/acs.jcim.8b00285 [DOI] [PubMed] [Google Scholar]
- 35.Wu, F. et al. Auto In Silico Ligand Directing Evolution to Facilitate the Rapid and Efficient Discovery of Drug Lead. iScience23, 10.1016/j.isci.2020.101179 (2020). [DOI] [PMC free article] [PubMed]
- 36.Dupeux, F. et al. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J.30, 4171–4184 (2011). 10.1038/emboj.2011.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Z. et al. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun.6, 8138 (2015). 10.1038/ncomms9138 [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi, J. et al. Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions. Nat. Chem. Biol.10, 477–482 (2014). 10.1038/nchembio.1524 [DOI] [PubMed] [Google Scholar]
- 39.Lozano-Juste, J., García-Maquilón, I., Ruiz-Partida, R. & Rodriguez, P. L. Drug Discovery for Thirsty Crops. Trends Plant Sci.25, 844–846 (2020). 10.1016/j.tplants.2020.07.001 [DOI] [PubMed] [Google Scholar]
- 40.Moreno-Alvero, M. et al. Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor. Mol. Plant10, 1250–1253 (2017). 10.1016/j.molp.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 41.Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Computational Chem.31, 455–461 (2010). 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose, P. W. et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res.45, D271–D281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, B. R. III et al. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput.8, 3314–3321 (2012). 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- 44.Hao, Q. et al. The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins. Mol. Cell42, 662–672 (2011). 10.1016/j.molcel.2011.05.011 [DOI] [PubMed] [Google Scholar]
- 45.Cao, M.-J. et al. Combining chemical and genetic approaches to increase drought resistance in plants. Nat. Commun.8, 1183 (2017). 10.1038/s41467-017-01239-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen, M.-X. et al. Comprehensive transcriptome and proteome analyses reveal a novel sodium chloride responsive gene network in maize seed tissues during germination. Plant, Cell Environ.44, 88–101 (2021). 10.1111/pce.13849 [DOI] [PubMed] [Google Scholar]
- 47.Chen, M.-X. et al. Full-Length Transcript-Based Proteogenomics of Rice Improves Its Genome and Proteome Annotation1. Plant Physiol.182, 1510–1526 (2020). 10.1104/pp.19.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc.11, 1650–1667 (2016). 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders, S. & Huber, W. Differential expression analysis for sequence count data. Nature Precedings, 10.1038/npre.2010.4282.1 (2010). [DOI] [PMC free article] [PubMed]
- 51.Du, Z., Zhou, X., Ling, Y., Zhang, Z. & Su, Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res.38, W64–W70 (2010). 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, J.-F., Wang, F., Chen, Y.-Z., Hao, G.-F. & Yang, G.-F. LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinforma.21, 2206–2218 (2020). 10.1093/bib/bbz141 [DOI] [PubMed] [Google Scholar]
- 53.Gao, Y.-Y., Yang, W.-C., Ashby, C. R. & Hao, G.-F. Mapping cryptic binding sites of drug targets to overcome drug resistance. Drug Resistance Updates67, 100934 (2023). 10.1016/j.drup.2023.100934 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw RNAseq data generated in this study have been deposited in the sequence read archive (SRA) repository of NCBI under BioProject accessions: PRJNA1138801. The crystal structure of PYL1-DBSA complex can be accessed from the Protein Data Band under the accession codes 9J6I. All study data are included in the article and/or supporting information. Source data are provided with this paper.