Abstract
As a powerful paradigm, artificial intelligence (AI) is rapidly impacting every aspect of our day-to-day life and scientific research through interdisciplinary transformations. Living human organoids (LOs) have a great potential for in vitro reshaping many aspects of in vivo true human organs, including organ development, disease occurrence, and drug responses. To date, AI has driven the revolutionary advances of human organoids in life science, precision medicine and pharmaceutical science in an unprecedented way. Herein, we provide a forward-looking review, the frontiers of LOs, covering the engineered construction strategies and multidisciplinary technologies for developing LOs, highlighting the cutting-edge achievements and the prospective applications of AI in LOs, particularly in biological study, disease occurrence, disease diagnosis and prediction and drug screening in preclinical assay. Moreover, we shed light on the new research trends harnessing the power of AI for LO research in the context of multidisciplinary technologies. The aim of this paper is to motivate researchers to explore organ function throughout the human life cycle, narrow the gap between in vitro microphysiological models and the real human body, accurately predict human-related responses to external stimuli (cues and drugs), accelerate the preclinical-to-clinical transformation, and ultimately enhance the health and well-being of patients.
Keywords: Artificial intelligence, Human living organoids, Organoid intelligence, In vitro miniature organ models, Multidisciplinary techniques, Preclinical-to-clinical transformation
Graphical abstract
Highlights
-
•
Construction strategies and evaluation techniques for human living organoid (LO) research are reviewed.
-
•
Artificial intelligence (AI)-enabled LOs in biomedicine are highlighted.
-
•
Challenges and perspectives to advance the development of AI-supported LO systems.
1. Introduction
Since the end of 2022, the advent of an AI chatbot (ChatGPT) developed by OpenAI has caused a buzz around the world due to its human-like responses to text input. This technology is promoting researchers to reorient the development direction of all walks of life and driving their scales to brand-new heights. Artificial Intelligence (AI), also known as machine intelligence, is a simulation model that provides targeted services for specific scenarios by highly reproducing complex human behaviors such as feeling, memory, generalizing, reasoning, creating, deciding, coping, and so on [1]. The emergence of big data has promoted the training of AI algorithms on large data sets, thus improving the performance of tasks. AI can be trained on large amounts of data to learn from experience, decouple and apply causality, make predictions, optimize strategies, classify objects, adapt to new situations, and perform complex tasks [2,3]. It contains machine learning, natural language processing, language synthesis, computer vision, and robotics, etc [4]. It is worth noteworthy that machine learning, as a cutting-edge technology in the field of AI, has been widely applied to biological research, disease discover and drug screening in preclinical assay and clinical applications [[5], [6], [7], [8]]. For instance, Hamamoto's team was able to accurately predict integrated survival subtypes by training the reverse-phase protein array and distinguish between high-risk and low-risk patients [8]. Gulshan et al. created an algorithm by applying deep learning for automated detection of diabetic retinopathy and diabetic macular edema in retinal fundus photographs [7]. In the face of the 2019 pandemic, deep learning could effectively distinguish normal versus abnormal chest radiographs [6].
Advances in stem cells, microfabrication and biomaterials have given rise to novel in vitro miniature organ models—living human organoids (LOs) [9]. In general, LOs refer to 3D living cell culture models originating from human primary tissues or stem cells, which present abundant cell types, genotypes, histological features and functions similar to human real organs, especially human tissues-derived organoids [9,10]. Currently, various LOs (e.g., brain, retina, lung, intestine, tumor, and placenta, etc) have been established to explore the effects of matrix materials (e.g., types, stiffness and degradability) and signaling factors (e.g., BMP and WNT) on stem cells regulation and organoid development [[11], [12], [13], [14], [15], [16], [17], [18], [19]]. For instance, Lutolf's team showed that high matrix stiffness significantly enhanced intestinal stem cell proliferation through a yes-associated protein-dependent mechanism, whereas a soft matrix and laminin-based adhesion were required for stem cell differentiation and organoid generation [15]. Hofbauer et al. found that mesodermal WNT-BMP signaling directed cavity morphogenesis via HAND1 during the development of hiPS-derived cardioids [20]. Kotton and colleagues found that periodic regulation of Wnt signaling can induce rapid differentiation of human stem cells into proximal lung organoids via NKX2-1+ progenitor cell intermediates. Furthermore, NKX2-1+ progenitor cells had high levels of Wnt activation, but essentially responded to the decrease in Wnt signaling by rapidly entering the proximal airway spectrum at the expense of fate on the far side [18]. Emerging LOs, on the other hand, have been used in human pathophysiology studies such as developmental research, disease modeling, drug screening, tissue regeneration and personalized medicine [5,19,[21], [22], [23], [24], [25]]. Particularly, LOs can adapt local in vivo environment after transplantation, restore the expression of region-specific markers, assume different regional identities, finally realize cell-based therapy [[26], [27], [28], [29], [30], [31], [32], [33], [34]]. For example, Sampaziotis et al. transplanted cholangiocyte organoids into a living human liver with extracorporeal thermostatic perfusion to repair the biliary tree and improve bile characteristics [30]. Of particular note, the FDA (US Food and Drug Administration) Modernization Act 2.0, approved in December 2022, eliminates the age-old requirement for animal testing of all drugs and will no longer require animal testing before entering clinical trials. This release is undoubtedly a high endorsement of organoid technology, and it has also set off a wave of research on organoids in academia and enterprises. Recently, organoid technology ushered in an important milestone in the field of preclinical research and drug development: For the first time, the US FDA approved a drug (Sanofi sutimlimab) to apply for a new indication to enter clinical trials based solely on the preclinical efficacy data obtained in the organoid chip research, combined with existing safety data [35]. This is the first time the FDA has included organ-on-a-chip and microphysiological systems in the Act as a separate evaluation system for non-clinical trials of drugs. This breakthrough provides a new direction for the search and development of rare disease drugs, and also provides a new idea for drug research and development enterprises to apply for drug clinical trials and new indications. Therefore, LOs provide valuable and effective models for drug screening and discovery to identify candidate treatments to tackle rare and emerging diseases [36,37]. Chen and coworkers determined the effectiveness of FDA-approved drugs such as imatinib, mycophenolic acid and quinacrine dihydrochloride) in inhibiting the entry of SARS-CoV-2 into lung and colonic Los [38]. Overall, the rapid development of LOs in the biomedical field has broken the limitations of animal models and traditional cell cultures to a certain extent, bringing a new dawn to the biomedical research and pharmaceutical fields.
Despite the great potential of LOs, the establishment of organoid systems is highly human-dependent, and therefore time-consuming and laborious in the process of optimizing strategies, data analysis, and efficacy verification. From the perspective of construction, maturity, heterogeneity, variability and standardization remain the major factors restricting the development of LOs. These factors determine the fidelity, repeatability and rationality of in vitro organoid models, affect the accuracy and reliability of evaluation results, as well as lead to poor comparability of different literature data. In terms of evaluation methods, multiple biosensors, imaging devices and omics analysis tools (e.g., genomics, single-cell omics and metabolomics) have become key assessment methods for LOs. However, due to the heterogeneity and multi-focus of LOs, it is difficult to quickly and accurately obtain the hierarchy and spatial features of organoids and dynamic behavior characteristics between organoids and microenvironments through image analysis. The functional analysis from multiple omics tests reveals a large amount of data, which poses a great challenge to analyze and decouple the complex network relationships. Therefore, a new paradigm for LO research is urgently needed to break the limitations of traditional systems, quickly optimize the establishment methods, efficiently parse data, precisely improve functional verification, and then modify the research strategy.
Therefore, we can't help but think about how AI technology can accelerate the development of LOs in fundamental research and clinical therapy in biomedicine. In the context of technological innovation, researchers at Johns Hopkins University in the United States have creatively conceived a technology similar to human organs—“organoid intelligence (OI)”—brain organoids as hardware to develop biocomputing [39]. In 2023, this team envisioned computers running on human brain organoids and announced related research plans [39]. In the rapid development of the information age, harnessing the power of AI for LO research, combined with gene editing, imaging analysis, and multi-omics sequencing, could aid in organoid system improvement, clinical decision-making and clinical trial performance analysis. AI can predict the differentiation pathway of stem cells, infer the production and development of LOs, and ameliorate the LO culture environment, attempting to build more sophisticated and authentic in vitro physiological models in a highly human-relevant manner [[40], [41], [42], [43]]. With the help of AI, researchers can efficiently and accurately analyze the structure and function of LOs through image-based training and sequencing [44,45]. Additionally, AI can be used to analyze big data from LO experiments, allowing for in-depth parsing of the underlying mechanisms of biological behavior, forecasting disease occurrence and mimicking pharmacological action [4,[46], [47], [48]]. For instance, a machine-learning framework could identify translational biomarkers and predict drug responses to cancer patients by combining network-based analyses and pharmacogenomic data based on preclinical bladder LO models [48]. Overall, the rise of AI reduces tedious human labor, enhances the reliability and validity of models, provides more comprehensive biological information. The integration of multidisciplinary technology has also driven a deep and comprehensive understanding of LOs. Undoubtedly, OI is a breakthrough product of the integration and collision of emerging HO and AI technologies and has epoch-making significance.
Here, we comprehensively summarize how AI aids in driving the revolutionary advances of LOs in life science, precision medicine and pharmaceutical science in an unprecedented way. Firstly, various construction strategies such as the hanging-drop method, droplet microfluidic technology, microfluidic organ chip, 3D bioprinting are introduced in detail, as well as their respective features and applications are also included. Secondly, we shed light on the state-of-the-art multidisciplinary techniques (e.g., image devices, biosensors, and multi-omics) for monitoring and analyzing in vitro LO models. Thirdly, we highlighted the cutting-edge achievements and the prospective applications of AI in LOs, particularly in biological study (e.g., AI-guided system optimization and environmental control) and preclinical applications (e.g., disease occurrence, disease diagnosis and prediction and drug screening). More importantly, we shed light on the new research trends harnessing the power of AI for LO research in the context of multidisciplinary technologies. Moreover, current challenges of the integration of AI and LOs, as well as potential solutions and future development direction were discussed. Taken together, AI-driven LOs hold the promise of closing the gap between in vitro microphysiological models and the real human body, accurately predicting relevant human responses to external stimuli (cues and drugs), accelerating preclinical to clinical transformation, and ultimately improving patient health and well-being.
2. CURRENT construction strategues of in vitro LOs
Living human organoids (LOs)—prevailing in vitro 3D human organotypic models—refer to the self-assembly of cellular clusters originating from human pluripotent stem cells (hPSCs), adult stem cells and tumor tissues. These 3D constructs replicate some traits of in vivo counterparts such as hereditary characters, complex spatial morphology, cell-cell/matrix interactions, and organ-specific function. Currently, LOs have been widely applied in biomedicine (e.g., tissue/organ development, disease modeling, drug testing and precision medicine). In recent years, multiple engineering assembly strategies have developed to establish in vitro LOs (Fig. 1), aiming to improve the correlation of the tissue/organ behavior between real human body and in vitro experiments. The core of these approaches is to reproduce the ecological niche of the native cells, such as pivotal biochemical and biophysical signaling from the bloodstream or neighboring cells, the formation of intercellular junctions, as well as interactions and functional transduction with the ECM. These organoids display a variety of properties that should enhance their potential for biological computing. While efforts to abstract biological principles never stop, the creation of artificial LO systems is gaining increasing interest. On the other hand, researchers in tissue engineering are also actively carrying out research.
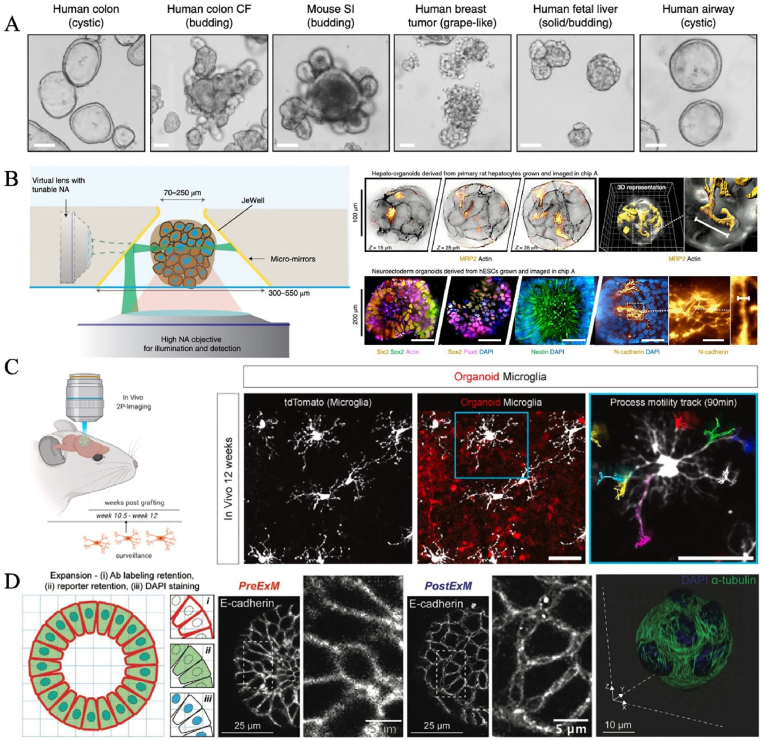
Fig. 1.
Schematic of engineering strategies (e.g., droplet microfluidic, organ chip, 3D bioprinting, etc.) for the construction of in vitro human organoids in various biomedical applications.
2.1. HANGING-DROP method
Historically, Harrison (1907) invented a hanging-drop technique for culturing nerve fibers [49]. Later, this method was used to produce spheroids and organoids. They took advantage of the surface tension to keep the overhanging drop in place, and gravity caused the cells at the bottom of the droplet to rapidly aggregate. The technique is easy to use and versatile, and can obtain a large number of uniform spheres in a relatively simple manner with an efficiency of about 100 % (one spheroid/drop). In addition, tightly packed spheroids are easier to generate than loose cell aggregates, but the morphology of spheroids depends on the cell type. Currently, various organoids can be formed by this method, including inner-ear organoids, proximal tubule organoids, mammary organoids, intestinal organoids, etc [[50], [51], [52], [53], [54]]. For instance, Parigoris et al. constructed tubuloids based on hanging drop possessed reversed polarity, in which the basolateral side faces inward, and the apical side is outward-facing. These organoids exhibited time- and dose-dependent responses to proteinuric conditions [51]. In addition, the use of commercial hanging drop array platforms (e.g., 96- and 384-well plates) makes this method more broadly accessible than previously reported hanging drop systems and enables compatible with direct live microscopy imaging [55,56]. Another concern with this technique is that the culture media in the droplet evaporates, resulting in increased osmotic pressure, which limits the time for growth and analysis [57]. To overcome the limitation, Bashir's group modified previous designs to cover the modular array of droplets encapsulated cells with mineral oil, effectively preventing culture media from evaporation and allowed for measuring response to drugs over time with real-time confocal microscopy [58].
2.2. Droplet microfluidic
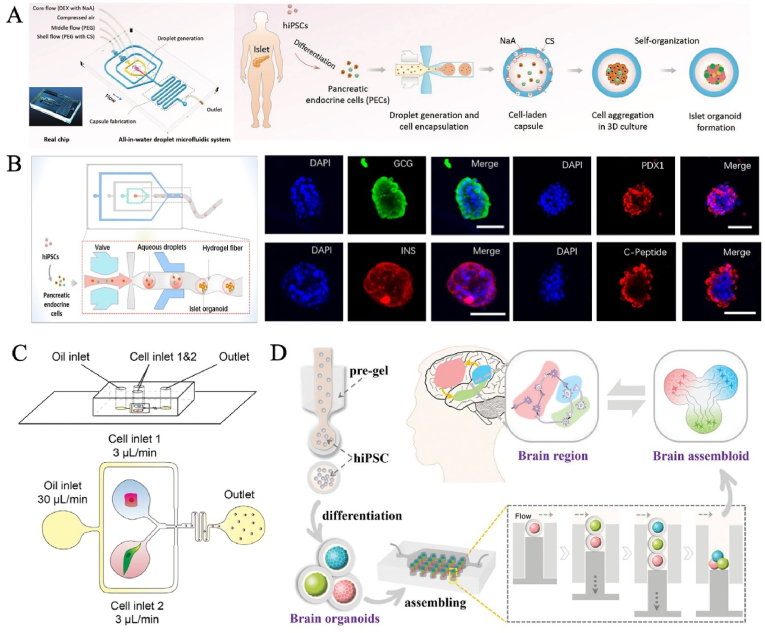
Droplet microfluidics has been utilized as a versatile tool in molecule detection, single-cell analysis, multifunctional materials fabrication, tissue engineering, drug screening, etc [59]. It is a scientific technology to generate and manipulate nano-micron discrete droplets in microchannel structures by using the surface tension and shear force at the interface of dispersed phase and continuous phase, which has been widely used organoid carriers [60,61]. In general, stem cells or progenitors are resuspended into the medium as a dispersed phase, and then cut to form the liquid drops by a continuous phase, and finally cell microcarriers are generated. Over time, cells self-organize, proliferate and sequentially differentiate into given LOs. The outstanding features are high throughput (up to thousands of Hertz), uniformity (size and shape), controllability (flow and component) and compartmentalization (independent bioreactor). Therefore, the construction of LOs using this technique requires consideration of the design of microfluidic channels, the size of the required spheroids, the type of gel used and the cross-linking method. The emergence of droplet microfluidics allows the production of organoids to break through the significant variability and low throughput of themselves [60]. Due to its small volume, droplet allows high-density cell encapsulation and increases cell paracrine signaling. Meanwhile, it precisely defines the heterogeneous cell composition in each small droplet and ensures inter-organoid consistency, intra-organoid complexity and inter-batch reproducibility [62]. At present, this strategy used microfluidic droplets as structural templates and was used in functional organ fabrication, drug testing, organ transplantation in organoid research [61,[63], [64], [65]] (Fig. 2A). The human pancreatic islet organoids in droplet-filled hydrogel fibers created by Wang et al. contained four classical pancreatic-lineage cell types (α-,β-, γ, and δ-cells) that showed sensitive insulin secretion function [66] (Fig. 2B). Compared to the conventional organoid models, microfluidic droplet encapsulation-guided tumor organoid growth and organization promoted parental tumor phenotype recapitulation with high purity and high rates [62]. Certainly, different systems can also be designed according to needs to achieve cell-cell interactions [67,68]. For instance, Dowbaj et al. coencapsulated isolated liver portal mesenchyme and ductal cell organoids into microgels, which recapitulate aspects of their cellular interactions in vitro [69] (Fig. 2C). Zhao's group developed human brain assembloids composed of cortical, hippocampal, and thalamic organoids from human induced pluripotent stem cells (hiPSCs) by coding and fusing microcarriers on droplet microfluidics [70] (Fig. 2D). The produced assembloids showed active neural migration and interaction and good cell viability. This novel droplet-engineered organoid technology is expected to be a latest tool aimed at advancing the manipulation of neural circuits to recapitulate human-level brain function, including learning plasticity and memory. So far, the technology is still in its infancy. The engineering concept is not limited to the brain, but extends to reshaping the complex biomimetic multi-organoid systems (e.g., brain-spinal-muscle organoid axis) in vitro and exploring the physiopathologic interactions.
Fig. 2.
Representative organoid systems based on droplet microfluidics. (A) Droplet microfluidic system for the formation of islet organoids. Reproduced from Liu et al. Copyright 2020, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (B) The human pancreatic islet organoids in droplet-filled hydrogel fibers were created. Copyright 2021, ACS Publications. (C) Isolated liver portal mesenchyme and ductal cell organoids were coencapsulated into microgels based on droplet microfluidic. Copyright 2023, Cell Press. (D) Human brain assembloids were composed of cortical, hippocampal, and thalamic organoids by coding microcapsules based on droplet microfluidic. Copyright 2023, Wiley-VCH GmbH [61,69,70].
2.3. Organ CHIP
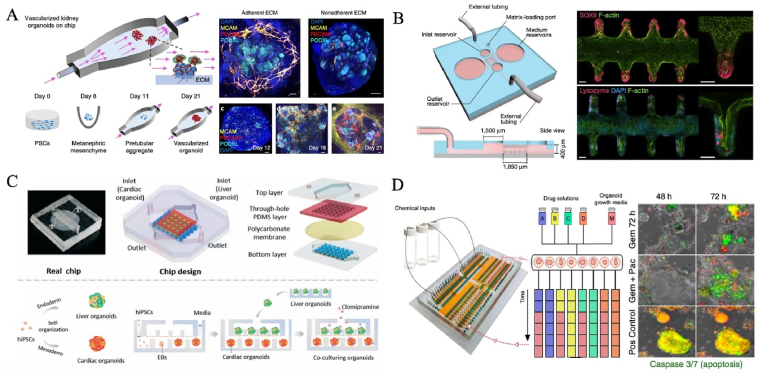
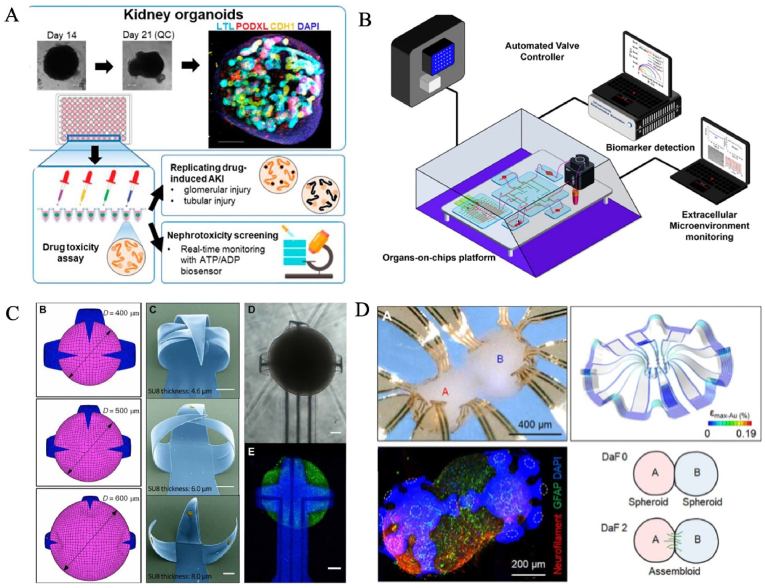
Organ chip is a transformative biomedical technology that has been born in recent years, and was listed as one of the “Top ten emerging technologies” by the World Economic Forum in Davos in 2016, and is also the core key technology to be overcome in the biomedical field. The organ chip is a miniaturized cell culture platform that comprises microchannels inhabited by living cells, evolving from microfabrication technologies and bioengineering strategies [71]. The outstanding advantage of organ chips is the controllable regulation and high integration of cell niche, so it is considered as a kind of in vitro microphysiological system with strong comprehensive ability. It can in vitro partially replicate organ-specific structures (e.g., villi, gas-liquid interface, blood vessel, and neural networks, etc) and realize key functions (e.g., blood-brain barrier, rhythmic respiration, peristalsis, and angiogenesis, etc) [[72], [73], [74]]. At present, this technology has been successfully applied to recreate every system of the human body, including nervous, motor, digestive, respiratory, immune, urinary, reproductive and circulatory [[75], [76], [77], [78], [79], [80], [81]]. Notably, state-of-the-art organ chip endows organoids with in vivo-like ecological niches (e.g., biochemical and biomechanical signals) in a controllable manner, termed organoids-on-chips (OOCs) technology [82]. Cutting-edge OOCs facilitate the near-physiological biological function of organoids, as well as advance their applications in the biomedical field by harnessing the power of microfluidic chips to improve the high fidelity of Los [[82], [83], [84], [85], [86], [87]]. Homan et al. found that flow enhanced the vascularization and tubular morphological maturation (e.g., podocyte, endogenous pool, vascular network, perfusable lumen, cellular polarity and adult gene expression) of kidney organoids in vitro, displaying the formation of capillary loops abutting foot processes in the mammalian embryonic kidney, and mirror early stages of glomerular development in vivo [83] (Fig. 3A). The individual and collective behavior of cells can also be affected by geometric constraints, which lead to different dynamics of tissue evolution and influence organ development processes [88]. In another case, Lutolf's team made full use of microstructure of the microfluidic chip, successfully induced intestinal stem cells to organize tube-shaped epithelia with an perfused mini-gut lumen and a similar spatial arrangement of crypt- and villus-like domains to that in vivo [85] (Fig. 3B). Organ chips can provide a screening and research model closer to the real physiological and pathological conditions of human body for researching pharmacokinetics, as well as assessing the safety and efficacy of drugs and vaccines. Qin's group used a liver-heart organoids-on-chip to simulate in vivo-like organ interaction such as function promotion during organ development and hepatic metabolism-dependent cardiotoxicity [89] (Fig. 3C). With the deepening of research on organoids-on-chip technology, a more superior alternative model can be constructed by using its unique properties, which provides a practical solution to many problems faced by disease modeling and drug evaluation [90].
Fig. 3.
Organ chip technology for the generation of human organoids. (A) An organ chip for enhancing the vascularization and tubular morphological maturation of kidney organoids. Copyright 2019, Spring Nature. (B) On-chip mini-intestines were realized by adopting scaffold-guided organoid morphogenesis. Copyright 2020, Spring Nature. (C) Liver-cardiac organoids on a chip, resembling human liver-heart axis for studying hepatic metabolism-dependent cardiotoxicity. Copyright 2021, The Royal Society of Chemistry. (D) An automated multiplexer control system for combinatorial and dynamic drug screening on human tissue-derived tumor organoids. Copyright 2020, Spring Nature [83,85,89,94].
Organ chips can also enhance the monitoring and analysis of LOs by integrating a variety of technologies, such as real-time imaging systems, automated sampling systems and biosensors [[91], [92], [93]]. Notably, Schuster et al. established an automated multiplexer control system for individual, combinatorial, sequential and dynamic drug testing and screening on tumor organoids from human tissues in a microfluidic system [94] (Fig. 3D). In the platform, similar personalized characteristics can be reproduced in vivo, that is, the individualized response of different patients-based organoids to drug treatment regimens [94]. Interestingly, temporarily revised treatment regimens could be more efficient than constant-dose monotherapy or combination therapy. Inspired by this work, it is noted that this strategy will need to be modified and intensified. In other words, the whole process from early detection of the disease, effective treatment to full recovery is underway.
2.4. 3D bioprinting
Bioprinting is a rapid prototyping technique, also known as additive manufacturing, invented by Charles Hull Started 30 years ago [95]. It is a prospective bioengineering technique to precisely position the living cells, biomaterials and signal factors for the construction of objects layer-by-layer on a computer-supported basis [95]. An important type of 3D printing is to autonomous self-assembly and construction of organ/tissue blocks that resemble the corresponding precise architecture in time and space [96]. Notably, the lungs, with their multilayered dendrite structure and highly branched network of blood vessels, are vital tissues responsible for air exchange, which is difficult to recreate in vitro. Grigoryan et al. printed a space-filled mathematical topology to enable the creation of a bionic alveolar model with entangled vascular systems, showing the oxygenation and flow of human red blood cells during tidal ventilation and proximal airway dilation [97].
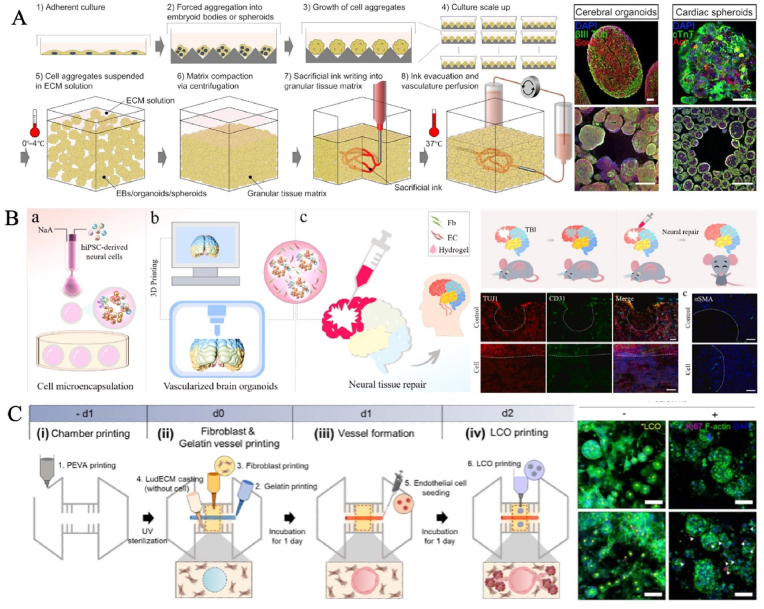
3D bioprinting allows for the precise design of physiologically relevant organoids, including shape, structure, mechanical properties, cellular arrangement, and biological cues to mimic natural tissues [98]. The balance between complex structural patterns during the printing process and the self-organizing properties of organoids supported by a network that runs through the entire structure is crucial. A critical feature of 3D bioprinting is bioinks. Optimizing specialized bioinks to guide stem cell proliferation and differentiation, while increasing the resolution of phenotypic analysis as per defined geometries and patterns, will further expand the range of possible bioprinted organoid models [[99], [100], [101]]. Indeed, 3D bioprinting can combine living cells, biomaterials, and bioactive factors to create 3D cellular structures that replicate the in vivo organ architecture [102]. Most importantly, in order to ensure cell survival, the automated rapid prototyping technology allows complex tissues to be assembled in a very short time, thereby expanding the production of organoids and tissue constructs on a large scale. For instance, Lewis's team rapidly patterned patient-specific-induced pluripotent stem cell-derived functional tissues with high cell density, maturation, and desired functionality. As an exemplar, they created a cardiac tissue derived from cardiac organoids that exhibited bulk perfusable vascularization and mature function such as fusion and beating synchronously over 7 days [103] (Fig. 4A). This strategy opened a new avenue to rapidly assemble organ-specific tissues with embedded vascular systems at therapeutic scales. Besides, personalized bioprinting of complex organs/tissues based on patients' physiological data can be carried out according to the specific needs of patients. So, another potential direction of 3D bioprinting with high cellular density, bigger sizes and higher throughput is building delivery and transplantation systems of organoids in regenerative medicine. 3D bioprinting allows to generate of tissue-level cell densities, complexity, and automation in fabrication, which can fabricate functional and vascularized whole organs [95,104]. For instance, Zhu et al. formed brain organoids with complex vascular networks by integrating brain organoids, endothelial cells and fibroblastin hydrogel via 3D printing [105] (Fig. 4B). In vivo animal studies have shown functional neural connectivity and nerve regeneration from graft to host, suggesting organ-level tissue repair. Therefore, it can resolve the huge challenges faced by clinical transplantation therapy such as insufficient supply of healthy donor tissues and immune rejection of the recipients, and it is expected to repair or replace dysfunctional tissues. Based on 3D bioprinting technology, Jang's team successfully built a vascularized lung cancer model that integrates patient-derived lung cancer organoids, lung fibroblasts, and perfusable blood vessels to more accurately evaluate drugs for the underlying disease [106] (Fig. 4C). Undoubtedly, this disruptive technology will play a crucial role in comprehending disease mechanisms, and developing novel regenerative therapies over the next decade.
Fig. 4.
The generation of organoids by 3D bioprinting. (A) As an exemplar, they created a cardiac tissue derived from cardiac organoids, exhibiting bulk perfusable vascularization and mature function. Copyright 2019, American Association for the Advancement of Science. (B) The construction of vascularized human brain organoids by 3D printing in neural tissue repair. Copyright 2021, Elsevier B.V. (C) Vascularized lung cancer organoid models for precise drug evaluation. Copyright 2023, IOP Publishing Ltd [103,105,106].
2.5. Electrospinning
Electrospinning is a process that uses electrical forces to fabricate fibrous 2D or 3D fine scaffolds ranging from 2 nm to several micrometers in scale from polymer solutions [107,108]. Over the past decade, the emergence of electrospinning-based cell culture platforms with various geometries has shown immense capabilities in replicating bionic ECM and topological morphology for guiding cell alignment, migration, differentiation and regeneration [[109], [110], [111], [112]]. In general, electrospun scaffolds are valuable for heart regeneration because they provide an environment capable of providing synchronous beating of heart muscle cells, promoting the anisotropic structure and the systolic function of the heart tissue in vivo [113]. Ritzau-Reid et al. used melt electrospinning writing to make tuneable geometric scaffolds that could guide lumenogenesis and the formation of interconnected and spatially discrete brain organoids derived from embryoids [114]. This methodological development has opened up new opportunities for constructing physiological in vitro organotypic models. However, electrospinning techniques have often been criticized for denaturing biomolecules due to conformational changes in organic solvent environments [114]. This shortcoming can be overcome by selecting the chemistry of the polymer skeleton to effectively enable and regulate the degradation profile [115].
3. Developing multidisciplinary techniques for assessing LOs
In general, developmental biology has inspired organoid engineering strategies to build in vivo-like organoid physiology in vitro. Verifying the success of LO model, i.e., evaluating various biochemical and physical parameters in OOC devices, is crucial. Accurate assessment of the LO function is also crucial, affecting the reliability of the established models. In the following part, we will introduce the developing multidisciplinary techniques for assessing LOs in detail. In addition, we have summarized the construction techniques of organoids and the multidisciplinary evaluation technologies in Table 1.
Table 1.
The summary of AI-enabled LOs in biomedicine.
| Organoid types | Cell sources | Extracellular matrix | Construction methods | Assessment techniques | Key results |
|---|---|---|---|---|---|
| Brain [70,105,114,[116], [117], [118]] | hiPSCs [70,105,114,118]; hESCs [114,116,118] |
Matrigel [105,114,117,118]; NaA [70,105]; Laminin [116,117]; CMC [70]; Fibronectin [105]; |
Droplet microfluidic [70,105]; 3D bioprinting [105]; Hanging drop [[116], [117], [118]]; Electrospinning [114] |
SEM [70,114,117]; HIM [116]; FM [70]; CM [162]; CLSM [105,[116], [117], [118]]; Calcium imaging [70,105]; Glutamate biosensor [116]; scRNA-seq [168]; |
|
| Pancreas [61,66,94] | Pancreatic ductal adenocarcinoma tissue [94]; hiPSCs [61,66] | Matrigel [94]; NaA [61,66]; PEG [66] |
Organ chip [94]; Droplet microfluidic [61,66] |
FM [61,66,94]; SEM [66]; Time-lapse imaging [94] |
|
| Kidney [51,83,119,120] | hiPSCs [51,119,120]; Primary renal proximal tubule epithelium [83] |
Geltrex [83,119,120]; Matrigel [51] |
Organ chip [83]; Hanging drop [51,119,120] |
CLSM [83,119,120]; FM [51,83,119]; SEM [83]; TEM [83]; ATP/ADP biosensor [119]; RNA-sequencing [51,119,120]; scRNA-seq [120]; Proteomics [120] |
|
| Intestine [85,[121], [122], [123]] | Intestinal stem cells [85,122,123]; Colorectal tissue [121] |
Collagen I [85]; Matrigel [85,121,122]; PEG [122]; BME [123] |
Organ chip [85]; Hanging drop [[121], [122], [123]] |
CLSM [85,121,122]; SEM [85]; FM [121]; TEM [85,123]; Calcium sensor [123]; scRNA-seq [85,123]; RNA-sequencing [122]; Whole exome-seq [121] |
|
| Lung [106,124] | Lung tissue [106]; Lung carcinoma cell lines [124] |
LudECM hydrogel, Matrigel [106]; Collagen I, Methylcellulose [124] |
3D bioprinting [106]; Hanging drop [124] |
CLSM, SEM, FM [106]; Electrical impedance sensor [124]; Proteomics [106] |
Modeling the complex microenvironment of lung cancer and lung fibrosis, as well as dynamic monitoring of drug effects |
| Retina [125,126] | hiPSCs [125,126]; hESCs [126] |
Matrigel [151] | Hanging drop [125,126] | FLIM, IM, OCT [125]; SEM, CLSM, FM [126]; HSpec, Tomography [126]; Electrochemical biosensor, Impedance sensors, oxygen sensors [151] |
|
| Liver [69,127] | Primary hepatocytes [127]; Portal mesenchymal cells; Ductal epithelium [69] | GleMA [127]; Matrigel [69] |
3D bioprinting [127]; Droplet microfluidic [69] |
CLSM [69]; Electrochemical biosensor [127] |
|
| Breast [128] | Normal and cancer tissue | BME | Hanging drop | Multispectral 3D imaging; scRNA-seq | Revealing dynamic behaviour and killing mechanisms of engineered T cells in patient-derived cancerous organoids |
| Heart [97] | hiPSCs | Geltrex | Electrospinning | CLSM | Randomly oriented microfibre scaffolds promote survival and angiogenesis of cardiomyocytes and endothelial cells |
| Liver-heart axis [89,129] | Primary hepatocytes [129]; hiPSCs [89,129] | GelMA [129]; Matrigel [89] |
Organ chip | CLSM [89]; Electrochemical sensor, pH sensor, Oxygen sensor [129] |
Continuous in situ monitoring of organoid behaviors and assessing drug safety |
| Liver-islet axis [90] | hiPSCs | Matrigel | Organ chip | CLSM; RNA sequencing |
Recapitulating human liver-islet axis in normal and type 2 Diabetes |
| Brain, heart, kidney [103] | hiPSCs | Collagen I; Matrigel |
3D bioprinting | FM; CLSM; Calcium imaging |
Biomanufacturing of organ-specific tissues with vascularized networks |
| Lung, colon, kidney, liver, breast [130] | Mammary glands; Organoid biobank |
BME; Matrigel |
Hanging drop | CLSM; LSFM; FM; Super-resolution confocal; Multiphoton and light-sheet microscopy |
Capturing and quantifying 3D structure and cellular composition of organoid |
| Breast, intestine [131] | Mammary glands; Colorectal cancer tissue |
Matrigel | Hanging drop | Deep learning; CNNS; FIB-SEM |
Quantitative analysis of subcellular structures |
| Intestine, liver, brain [132] | hESCs; Primary hepatocytes |
Matrigel | Organ chip | LSFM; CLSM; Deep learning; Time-lapse imaging; Multi-scale imaging |
Multiscale phenotypic quantification of organoids |
Human Induced Pluripotent Stem Cells (hiPSCs); Human embryonic stem cells (hESCs); Fluorescence microscope(FM); Confocal laser scanning microscope (CLSM); Confocal microscopy (CM); Scanning electron microscopy (SEM); Transmission electron microscopy (TEM); Inverted microscope (IM); Focused ion beam-scanning electron microscopy (FIB-SEM); Light-sheet fluorescence microscopy (LSFM); Optical coherence tomography (OCT); Hyperspectral imaging (HSpec); Fluorescence lifetime imaging microscopy (FLIM); Scanning helium ion microscopy (HIM); Single-cell sequencing (scRNA-seq); Sodium alginate (NaA); Basement membrane extract (BME); Poly(ethylene glycol) (PEG); Gelatin methacryloyl (GelMA); Sodium carboxymethyl cellulose (CMC); Lung-derived decellularized extracellular matrix (LudECM hydrogel); *N.A. indicates that the specified feature has not been measured or reported.
3.1. Live imaging techniques
High-quality images are necessary for reliable analysis of 3D LO systems. At present, multiple microscopy such as optical, electron, fluorescence, multiphoton and laser scanning confocal microscopy are the most common detection ways [130]. Optical imaging is ideal for evaluating LO behavior because of its subcellular resolution, depth of penetration throughout the LOs, and functional endpoints [133]. Mahamid's group realized correlative imaging from the light microscopy millimeter scale of entire organoids to the nanometer-scale volume electron microscopy that captures subcellular ultrastructure [131]. Fluorescence microscopy and laser scanning confocal microscopy have great advantages for visualization of 3D multi-scaled LOs, characterization of the cell types of LOs and phenotypic similarity between in vitro LOs and their original tissues (Fig. 5A). demonstrated organoids [130] Multiphoton microscopy is characterized by optical sectioning and deep imaging, which greatly promote the deep understanding of intact living organoids.
Fig. 5.
Representative achievements of live imaging technique in studying organoids. (A) The visualization of 3D multi-scaled organoids by high-resolution 3D imaging. Copyright 2019, Springer Nature. (B) A 3D single-objective light-sheet imaging device that integrated microarray LOs culture module, beam steering module, laser control and acquisition software. Copyright 2022, Springer Nature. (C) Two-photon imaging revealed that human brain organoid-resident microglia actively. Copyright 2023, Elsevier Inc. (D) 3D imaging of colorectal cancer organoids identifies responses to Tankyrase inhibitors. Copyright 2022, Wiley-VCH GmbH [121,130,132,134].
Unlike histological analyses that reconstruct organ developmental processes by fixing multiple specimens at different times, live imaging techniques provide a more direct way to dynamically characterize organ behavior online. They provide insights into LOs in terms of structure and function during growth in vitro, including morphological parameters (e.g., size, number, etc.), multi-scale phenotypic analysis, as well as metabolic changes. For instance, Viasnoff and colleagues developed a 3D single-objective light-sheet imaging device that integrated a microarray LO culture module, beam steering module, laser control and acquisition software in a standard commercial inverted microscope [132] (Fig. 5B). It could perform 3D live imaging up to 300 LOs/hour at subcellular resolution, enhancing user-friendliness with maximum automated manipulations. In this work, the authors monitored the development of neuroectoderm LOs by using stem cells expressing fluorescent proteins and capturing one z stack of 70 optical planes/15 min over 8 days. Based on the local cell arrangements, it was easy to find that Sox2+/Pax6+ rosette structures began to appear in LOs after 4–5 days. In addition, high-resolution imaging systems enabled rare event detection in LOs such as the localization of rare cellular clusters composed of a few cells expressing specific markers [132]. Browne et al. detected increased glycolytic activity and accumulation of retinol and retinoic acid in human stem-cell-derived retinal organoids, using fluorescence lifetime imaging microscopy and hyperspectral imaging, respectively, which coincided with the occurrence of photoreceptors [125]. In vivo, two-photon imaging displayed that human brain organoid-resident microglia actively participated in the monitoring of the human brain environment and reacted to local and systemic inflammatory perturbation [134] (Fig. 5C).
Another interesting domain of 3D imaging is conducting patient-specific drug testing, which can allow the clinic to move from watching and waiting to predict treatment decisions to making informed treatment plans based on how patient-specific LOs respond to treatment [133]. For instance, using image-based assays, Badder et al. demonstrated that morphometric analyses can capture subtle alterations in colorectal cancer organoid responses to Wnt inhibitors that are consistent with activity against a cancer stem cell subpopulation [121] (Fig. 5D). 3D imaging analysis based on patient-derived tumor organoid platforms could aid in quantify apoptotic and tumor-stroma regulation profiles and measure responses to standard-of-care regimens in individualized cancer treatment [133,135]. Overall, 3D imaging can reserved as a valuable phenotypic readout to quantitatively asses drug-intervened effects in relevant preclinical models.
Although Live imaging techniques have been widely applied in organoid research, there are still some limitations. Firstly, organoids are difficult to image because of the limited penetration depth of high-resolution microscopes and depth-dependent light attenuation, which can limit the understanding of signal transduction pathways and characterization of intimate cell-extracellular matrix (ECM) interactions. Super-resolution imaging permitted to visualization of the organoids-extracellular matrix (ECM) interactions by identifying the localization of ECM proteins secreted by cells. For example, the Anseth team from the University of Colorado Boulder developed a photo transfer by allyl sulfide exchange-expansion microscopy (PhASE-ExM). It combined two technologies: 1) propoly(ethylene glycol) hydrogels suitable for intestinal organoid growth and organoid crypt formation by reversible addition-fragmentation chain transfer reaction mediated by a radical cleavage of allyl sulfide, and 2) photo-expansion microscopy relied on swellable hydrogels generated with thiol-acrylate mixed-mode photopolymerization. COLIV accumulated at hinge regions of differentiated organoids alongside regions of thick ITGB1, with discontinuous COLIV and minimal ITGB1 at crypt base regions [122]. Secondly, most of imaging techniques-based analysis remain semi-automatic. After the data is generated by image acquisition methods, it is necessary to manually analyze the image with the help of image identification tools (e.g., ImageJ, Matlab languages, etc) [136]. Thirdly, the amount of generated data is becoming challenging to analyze manually. Organoids are large structures with high phenotypic complexity and are imaged on a wide range of platforms, from simple benchtop stereoscopes to high-content confocal-based imaging systems. The large volumes of images, resulting from hundreds of organoids cultured at once, are becoming increasingly difficult to inspect and interpret.
3.2. Biosensors
LOs may experience physiological signal alternation from their development and aging or responses to external stimuli such as toxins, drugs, and microbial invasion. These signals are usually vital in maintaining functions such as gastrointestinal including pH regulation, electrolyte balance, and barrier functions [137,138]. So, targeting specific signals, biosensor technology can help us learn more about the status of organoids such as development, metabolism, and microenvironment, providing continuous data on organ-specific responses in human physiopathology [139]. Today, a variety of biosensors (electrochemical, mechanical and optical biosensors) have been used to enable in-situ and real-time monitoring of pivotal cues.
Currently, 2D multi-electrode arrays have been used to enable a noninvasive and long-term stable bioelectronic interface with LOs in vitro for monitoring of electrophysiological properties of LOs. LOs are typically cultured above a microelectrode array, which detects action potentials, mechanical signals, and biochemical activities at the bottom of the LOs, thereby transmitting these signals to an external amplification system for continuous signal recording [126]. Rapid firing rates and network bursting events could be observed via a multi-electrode array platform, providing important insights into the differentiation, development, and electrophysiological maturation of brain organoids [140]. Biosensors can be used to monitor changes in biochemical substances during the organoid culture for assessing cellular metabolism and response to external stimuli [116,119,124,141]. For instance, Nasr et al. exploited functionalized borosilicate glass capillaries as an electrochemical sensors to monitor glutamate release in cerebral organoids produced by human embryonic stem cells (hESC) that different regions of the brain [116]. Susa et al. employed reporter kidney organoids generated with an ATP/ADP biosensor for drug nephrotoxicity assessment [119] (Fig. 6A). Wu et al. simultaneously test the drug anticarcinogen inhibition and side effects of on lung-heart-liver cancer spheroids platform can be quantitatively evaluated by a connected multiwell interdigitated electrode array [124]. In summary, biosensors based on 3D tumor spheroids have high predictive value for drug drug screening and discovery, and are expected to be well used in pharmacy and clinical medicine. Biosensors can detect specific biomarkers in complex biological environments (e.g., medium), which often contain excessive amounts of nonspecific proteins and interfering substance, but trace amounts of biomarkers of interest. For instance, Khademhosseini's team integrated a label-free electrochemical biosensor on a microfluidic chip for long-term continual measuring of cell-secreted soluble biomarkers (e.g., albumin and glutathione-S-transferase-alpha GST-α) with an LOD of 0.023 and 0.01 ng/mL from primary hepatocyte organoids under drug exposure by fully automated manner [127]. Notably, the biosensor has been designed with regenerative process capability, capable of long-term monitoring at up to 25 regenerates without any significant loss of sensor sensitivity [127]. A similar sensing method, using a more complex biological system, was proposed by Zhang et al., who combined liver and cardiac organoids to detect cardiac biomarker secretion following the addition of acetaminophen (paracetamol) and doxorubicin [129].
Fig. 6.
Representative achievements of biosensors in studying organoids. (A) Kidney organoids were generated with an ATP/ADP biosensor for drug nephrotoxicity assessment. Copyright 2023, Frontiersin.org. (B) Modular physical, biochemical and optical sensors on an organoid chip, sensing microenvironment factors and the organ's dynamic response to pharmaceutical compounds over time. Copyright 2017, PNAS. (C) Multielectrode arrays-brain organoids interface to allow electrophysiological output recording. Copyright 2022, American Association for the Advancement of Science. (D) Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assemblies. Copyright 2021, American Association for the Advancement of Science [117,119,129,145].
In addition, other biosensors such as bioelectrochemical, mechanobiological, chemical and optical sensors have been coupled to develop accurate, physiologically relevant organoid models to study organ behaviors, disease occurrence, organ dysfunction and drug intervention [129,139,142]. Notably, Khademhosseini's team assembled modular physical, biochemical and optical sensors on an organoid chip that could sense microenvironment factors in real-time as well as the organ's dynamic response pharmaceutical compounds over time [129] (Fig. 6B). Mechanobiology biosensors will demonstrate powerful measurements of forces to better understand the mechanobiology of cell-cell/matrix interplay, and how mechanical forces affect the biological behavior of Los [142,143].
Although recording physiological signals via 2D microelectrode arrays is an often-used means of assessing organ function, network communication, and response to chemicals and biologicals, the accessibility to 3D LOs is poor due to the planar structure limitations (e.g., limited recording contact area) of 2D microelectrode arrays [139]. Recently, 3D flexible bioelectronics with shape-matched multiple metal electrodes covering the entire scalp, allowing sampling of electrical signals from all over its 3D geometries, which have demonstrated their promising applications in the latest electrophysiological measurements of LOs [144]. Notably, Gracias's group developed multielectrode arrays (MEA)-brain organoids interface to allow electrophysiological output recording [117] (Fig. 6C). Guided by mechanics simulations, the self-folding polymer shells covered with patterned multielectrode nanostructures and probes could capture electrophysiological recording across the entire organoid surface for up to 4 weeks and the responds to glutamate stimulation in an ultra-high-resolution 3D spatiotemporal manner [117]. The groundbreaking interfaces can be integrated with the aforementioned microfluidic chips, supporting the scalability and durability of the platform as well as biochemical signals through spatial patterns and gradients. Together, they create a robust biosystem to gain an iterative, in-depth understanding of organoid behavior and responses to a range of highly modifiable environmental and input stimuli, which in turn will allow us to explore their ability to generalize the molecular mechanisms of learning and memory formation, and ultimately to realize their computational potential for stimulation. Rogers and coworkers designed a 3D multifunctional neural interface with optical, electrochemical, and thermal sensing modules to a cortical spheroid, capturing multimodal stimulation and recordings such as 3D spatiotemporal mapping of spontaneous neural activity [145] (Fig. 6D). Of note, a deformable framework that completely covers the entire 4 Π solid angle allows for mechanical deformation to accommodate changes in organoid size during neural growth [145].
3.3. Multi-omics
With the progress of scientific research, it has been found that the simple study of a certain direction (genome, proteome, transcriptome, etc.) cannot explain all biomedical problems. Scientists have proposed to reflect the function and metabolism of human organs/tissues through the study of the interaction between genes, proteins and their molecules from a holistic perspective. It provides new avenues for exploring organ development, studying pathology and discovering potential therapeutic strategies. At present, proteomics, transcriptomics, genomics and metabolomics have been widely applied in the functional analysis of LOs.
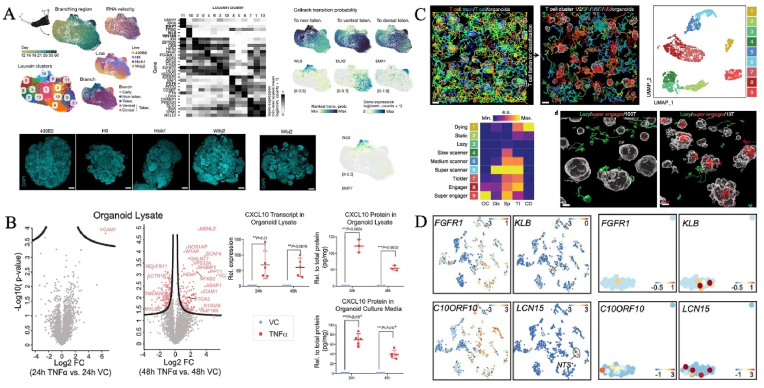
Often-used omics tools are classical bulk sequencing and single-cell profiling, which can reveal cellular and molecular changes. Bulk RNA analysis and qPCR provide simple transcriptomic readouts with limited resolution, while single-cell RNA sequencing (scRNA-seq) offers the possibility of identifying cell diversity, identities, differentiation trajectories, gene regulatory networks, phenotypic landscape and even transcriptional biases in LOs under different regimens [[146], [147], [148], [149]]. Single-cell technologies and LO systems can be leveraged to reconstruct human developmental events. Treutlein and partners revealed distinct regulome and effectors of dorsoventral telencephalon specification in the brain organoid from a single-cell multiome view [118]. Notably, two distinct GLI3 regulome are critical to telencephalic fate decisions: one regulated the dorsoventral mode with HES4/5 as a direct GLI3 target, and the other controlled ganglionic eminence diversification in late development [118] (Fig. 7A). Apart from transcriptional profiling, organoid genomic atlas at single-cell resolution have also been incorporated to uncover human-specific traits such as epigenetic landscape [150,151]. By comparing cell-type transcriptome maps of in vitro human retinal organoids and in vivo retinal tissues developed in vitro, it is possible to map disease-related genes to specific cell types [152]. In addition, the combination of large-scale proteomic and transcriptional profiling contributes to a fuller understanding of the disease process. For instance, Lassé et al. uncovered the cellular origins of proteins using organoid proteome-transcriptome integration. Proteome alterations in cytokine stressor (TNFα)-induced organoids help stratify diseased human kidney tissue [120] (Fig. 7B). Large-scale transcriptome mapping is also a promising technique for characterizing behavioral phenotypic heterogeneity in cellular immunotherapies as well as potentially deciphering and optimizing personalized solid tumor-targeted cell therapies [128] (Fig. 7C). Dekkers et al. established a 3D imaging-transcriptomic system to explore the dynamic interactions of T cells and patient-derived tumor organoids [128]. Of note, type I interferon could initiate resistant organoids for engineered T cell-mediated killing. Besides, secretomics can reveal the landscape of secreted products such as hormone levels secreted by human intestinal endocrine cell organoids [123] (Fig. 7D). With the integration of multiple disciplines, a variety of techniques (e.g., Raman microspectroscopy and nuclear magnetic resonance) are widely used to investigate enzymatic activity and drug efficacy [[153], [154], [155]].
Fig. 7.
Representative achievements of multi-omics in studying organoids. (A) Distinct regulomes and effectors of dorsoventral telencephalon specification from the perspective of single cell multiomics. Copyright 2022, Springer Nature. (B) Proteome alterations in cytokine stressor (TNFα)-induced organoids help stratify diseased human kidney tissue. Copyright 2023, Springer Nature. (C) An integrated organoid omics map extends deciphering and optimizing personalized solid tumor-targeted cell therapies. Copyright 2023, Springer Nature. (D) High-resolution mRNA and Secretome Atlas for revealing hormone levels secreted by human intestinal endocrine cell organoids. Copyright 2020, CellPress [118,120,123,128].
4. Overview of AI and AI-enabled LOs
4.1. Challenges of LO research
Undoubtedly, the development of LOs has propelled researchers to gain a deeper understanding of health, disease mechanisms, and drug efficacy. However, LOs still face certain limitations in their construction and assessment technologies. In terms of the in vitro construction of LOs, LOs rely heavily on animal-derived matrix gels such as Matrigel, which have poor reproducibility, high batch-to-batch variability, and potential immunogenicity problems [156]. Although various biomaterials have been tried to replace animal-derived substrates, such as polyethylene glycol (PEG), polylactic-glycolic acid (PLGA), polyacrylamide (PAm), etc. [157], the development of biomaterials with appropriate properties remains challenging due to the complex composition of the native matrix and its dynamic changes with organ development and disease progression. Therefore, it is extremely difficult to develop high-fidelity biomaterials relying solely on traditional experimental methods of trial and error. Similarly, optimization strategies for each component of the LO culture process (such as cell type, growth factors, cell response behavior to external stimuli, etc) are mostly derived from labor experiments. In the emerging field of intelligence, this traditional optimization model is very undesirable, and it is inevitable to be replaced.
Comprehensive and accurate assessment of organoid function is also extremely important. Yet there is much room for improvement. On the one hand, when processing a large amount of data generated by organoids, it is difficult to obtain high-quality data, and insufficient sample size and completeness may also affect the reliability of data. On the other hand, data analysis lacks standardized protocols and real-time monitoring technologies, resulting in large organoid variability, and manual processing and subjective interpretation increase uncertainty. In addition, the lack of automation in the current organoid culture and data analysis process limits the efficiency and accuracy of high-throughput screening, and affects the practical application of organoid models in clinical transformation.
4.2. Snapshot of AI
4.2.1. Definition and development history
AI is a sub-discipline of computer science. Technically, the term AI is named as a model proposed to figure out a specific problem or offer a particular service. As early as 1950, Alan Turing proposed the Turing Test as a standard to determine whether a machine has intelligence [158]. The term AI was first introduced by John McCarthy during the Dartmouth Conference in 1956, which is widely recognized as the inception of AI as an academic discipline [159]. This pivotal event marked the beginning of the study of machines simulating human cognitive processes, thus laying the foundation for the field of artificial intelligence as we understand it today. The 1950s–1970s were the early stages of AI development, with the emergence of symbolism, early reasoning systems, early neural networks, and expert systems [160,161]. Fueled by the booming World Wide Web, the 1980s to 2000 ushered in a second wave of development that marked advances in machine learning, speech recognition, and neural networks for pattern recognition [158]. The popularity of the internet and the emergence of big data have provided new data resources and application scenarios for AI. Until now, deep learning, machine learning, convolutional neural networks, and generative adversarial networks have achieved significant advancements and impacts. Although it is difficult to accurately trace the specific time point when artificial intelligence began to be applied to organoid research, it can be determined that with the breakthrough of artificial intelligence technology in the early 21st century, and the important progress of organoid technology in 2009, the application of artificial intelligence in organoid research began to become more extensive and in-depth. The authors have shown very detailed details [161], so I won't go into too much detail here.
4.2.2. Types of machine learning
In the past decade or so, AI has developed rapidly. With the help of AI technology, computers can efficiently process and analyze large amounts of biomedical data. Machine learning is a core area of AI and the basis for most AI applications today. It enables the computer system to automatically learn and summarize the rules and knowledge contained in the data, and train AI models for efficient prediction and analysis, so as to replace the manual completion of complex tasks or forward-looking predictions, and assist in the promotion of clinical diagnosis and treatment and scientific experiments.
Currently, there are three main types of machine learning, including supervised learning, unsupervised learning, and reinforcement learning, which can be targeted to solve different problems. Specifically, supervised learning entails the training of algorithms by analyzing labeled training datasets, intending to enable the prediction of outputs for previously unseen data [161]. Commonly utilized algorithms in this domain include linear regression, logistic regression, decision trees, random forests, support vector machines (SVM), neural networks, and convolutional neural networks (CNNs). In contrast to supervised learning, unsupervised learning employs algorithms to process unlabeled data to uncover underlying structures and patterns within the dataset [[162], [163], [164], [165], [166], [167]]. Clustering and association rule mining are prevalent tasks in unsupervised learning. Reinforcement learning focuses on developing optimal behavioral strategies through the interaction of algorithms with the environment [168]. The algorithms learn to perform actions based on feedback from the environment, in the form of rewards or penalties, to maximize long-term cumulative rewards. Common algorithms in the field of reinforcement learning include Q-learning, Deep q-network (DQN), State-action-reward-state-action (SARSA), Monte carlo tree search (MCTS), etc.
In general, machine learning endows AI systems with the capacity for learning and adaptation, enabling them to extract knowledge from historical data, confront unforeseen challenges, manage extensive datasets, and continuously improve their efficacy, performing complex tasks that were previously unattainable. As the core impetus of the current AI wave, machine learning is anticipated to maintain an important role in the ongoing advancements.
4.2.3. Models of machine learning
Common machine learning models and algorithms exhibit different performance and applicability on different problems and data sets. The selection of a particular model often depends on the specific needs of the task, the characteristics of the data, and the expected performance of the model. For example, a CNN has excellent performance in image data processing and can automatically extract image features, so it is mainly used for efficient recognition of medical images and videos, histological analysis, and diagnosis of diseases [167,169]. Recurrent neural network (RNN) is well-suited for time series prediction, performing natural language processing tasks in healthcare systems (e.g., the generation of health reports), however, the model may struggle to capture long-distance dependencies [166]. Decision tree (DT) is tailored for dealing with classification and regression problems, and their biggest advantages are clear logic, intuition, and strong interpretability [170]. However, they are easy to overfit and sensitive to noisy data constraints. Therefore, pruning and other strategies may be needed to optimize model performance in practical applications. SVM can effectively process high-dimensional data and is compatible with image recognition and disease classification based on gene expression data [163]. Unfortunately, SVM may encounter computational efficiency problems when dealing with large data sets, and kernel function and parameter selection have significant effects on model performance, requiring careful adjustment. Overall, different models can be adapted to diverse tasks in the field of medicine, covering many aspects from disease diagnosis, patient risk stratification, treatment effect prediction, and medical risk assessment. In addition, researchers can also take full advantage of the differences between different models for the same task, to build a more powerful model, which is a hybrid model. For example, medical images can be analyzed and features extracted based on CNN, and then diseases can be classified according to the features of medical images using SVM to achieve automatic diagnosis.
4.2.4. Advantages and limitations
In today's society, AI is becoming essential because it can solve complex problems in a highly efficient way. The advantages of AI in the biomedical field include its ability to process and analyze large amounts of complex data, the ability to quickly identify patterns and associations, improve the accuracy and efficiency of disease diagnosis, and innovative applications in drug discovery and genomics research [171]. However, its limitations lie in its reliance on high-quality, accurately labeled data, possible data bias and interpretative difficulties, and ethical and legal issues in clinical practice, including patient privacy protection and algorithmic transparency [172]. In addition, there are still challenges in the ability of AI systems to generalize, cope with unknown situations, and collaborate with human experts [171]. We will describe in detail how to overcome these limitations in our conclusions and future outlook.
4.3. Overview of AI-enabled LOs in biomedicine
The combination of organoids and AI is a new field of organoid research, which has great application potential in biomedical applications. Firstly, AI can help optimize every component of the LO culture process (e.g., cell types, growth factors, matrix gels, and cell behaviors in response to external stimuli, etc.). Secondly, the advent of AI, a new paradigm, has accelerated data extraction and analysis of LOs, predicted morphogenetic dynamics from images, and identified and interpreted complex biological patterns and processes, enabling spatial control of multicellular models and better LO design. AI can effectively process the large amount of omics data generated in organoid research, including genomics, metabolomics, single-cell omics, etc., thereby improving the efficiency and accuracy of data analysis. Thirdly, By simulating the microstructure and function of human organs, artificial intelligence LO can provide accurate in vitro models for disease modeling, disease prediction, drug screening, and toxicity testing, to reduce the cost of drug research and development and accelerate the process of new drugs to market. In personalized medicine, patient-derived LOs are used for disease model reconstruction and treatment plan customization to improve the pertinence and effectiveness of treatment. This interdisciplinary integration opens new avenues for disease mechanism research, precision medicine, and regenerative medicine, although there are still challenges in technology integration, data interpretation, and ethical regulations.
5. Advances in AI-enabled LOs in biomedicine
AI originates in computer science and aims to highly replicate complex human behaviors such as feeling, memory, summarizing, reasoning, creating, deciding, coping, etc. Today, AI has drove LO research in the biomedical field. It is most exciting to see what the holds of AI are for these rapidly growing LOs.
5.1. AI-guided optimization of LO culture system
Machine learning is a subset of AI that uses algorithms to learn from data iteration to decision-making and prediction in an automated manner [173]. The learning potential of in vitro LOs can be optimized by manipulating intrinsic cell paradigms (e.g., gene editing, complex organ composition and vasculature) and external environmental stimuli (e.g., electrical, chemical and optogenetic cues). That is, AI will input complex biological signals to LOs, in turn, LOs will perceive them and output results to AI through implantable biosensors, high-resolution imaging and real-time measurement technologies. These outputs can be used as biofeedback to facilitate organoid learning or directly for computation purposes, realizing biological computing solutions based on AI, machine learning, living organoid systems and big data management. Specifically, AI can help deal with every component of LO (cell types, growth factors, matrix gels, cell behaviors in response to external stimuli, etc.).
First, it is key to offer optimal culture conditions for the development of LOs, as this largely determines the structural and functional characteristics of organ models in vitro. Many elements, such as PH, growth factors, oxygen levels, temperature, matrix property, biochemical factors and mechanical parameters have profound effects on cell viability, proliferation and differentiation. Unfortunately, the optimization and development of traditional cell culture schemes are highly dependent on artificial trial and error, which to a large extent hinders the rapid development of science and technology. Leveraging big data analysis, machine learning algorithms open up new avenues for experimental design to build more efficient and higher quality LO models. Camacho-Gomez et al. developed a framework that combined agent-based modeling and deep learning to unravel cell coordination and reproduce tumor organoid morphogenesis by self-organizing in different modes according to certain culture conditions [174]. In the context of big data, AI can be used to predict how microenvironmental conditions affect cell behaviors (e.g., proliferation, differentiation and migration) and accelerate organoid development [161,174,175]. Generally, the optimal parameters were found by repeating the candidate proposal, execution, validation, and prediction. For example, Kanda et al. presented a robotic AI platform that used a batch Bayesian optimization algorithm to autonomously guide stem cells to effectively differentiate into retinal pigment epithelial cells [176]. The system performed cell cultures under 143 different conditions from 200 million possible parameter combinations over 111 days, with a differentiation rate of up to 88 % in terms of pigmentation scores compared to pre-optimized cultures [176]. Singaraju et al. identified key determinants of cell differentiation and cardiac organoid formation, such as cardiac growth factors, by training a lasso-regularized linear regression algorithm [177]. Zhu's group proposed a deep neural network model to prospectively recognize the differentiated cell types from neural stem into neurons [175]. Remarkably, neurotrophic factors, hormones and nanoparticles were taken into account, highlighting the great potential of the AI-enabled strategy in screening cell culture protocols [175,178].
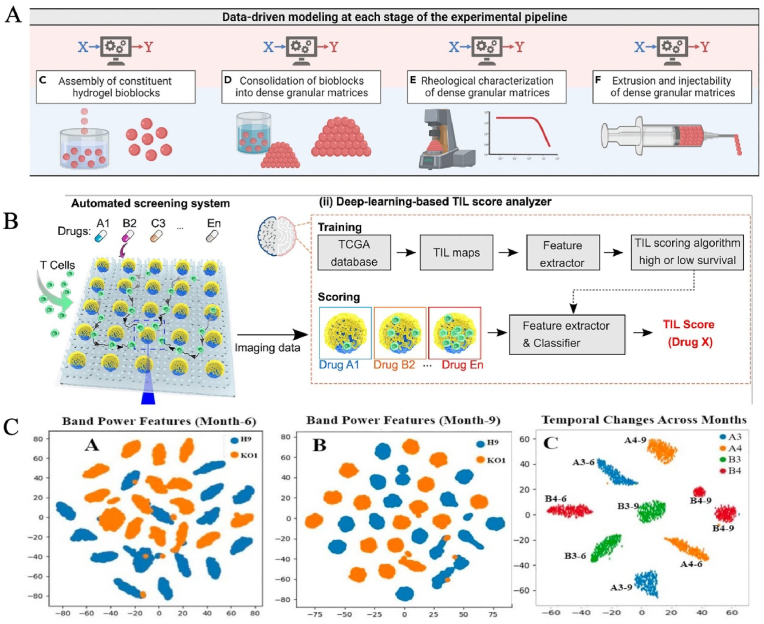
In addition, matrix gel is a pivotal part of the construction of in vitro LOs, which supplies physiologically relevant microenvironments. At present, Matrigel and decellularized extracellular matrices (dECM) are still the main sources of native matrix gels due to their good biocompatibility and bioactivity [179,180]. However, their heterogeneity, batch-to-batch variability and undefined protein components may result in the uncontrollability of the microenvironment and low reproducibility of Los [156]. Although a variety of biomaterials, such as synthetic and composite hydrogels, have been explored as alternatives to the animal-derived matrix, including polyethylene glycol (PEG), polylactic-glycolic acid (PLGA), polyacrylamide (PAm), etc. [157], there has been considerable difficulty in developing realistic biomaterials. On the one hand, the component of the native matrix is extremely complicated. On the other hand, with the development of organs and the occurrence of diseases, matrix properties (such as components, hardness, mechanics and biological cues) will also change dynamically. Therefore, it is extremely difficult to develop high-fidelity biomaterials with inherent complexity based only on traditional experimental methods. The advent of AI, a new paradigm, has provided new perspectives and feasible solutions for aiding the design and development of novel matrix gels. It can automatically analyze existing data sets, capture key information and identify feasible patterns, providing a powerful framework for predictive analysis and hypothesis generation. AI, as a powerful tool, is being applied to elucidate the relationships among structures, properties, processes and performances, discover the formation and degradation mechanisms of matrix gels, especially decoupling multidimensional communication between matrices/LO-LO. Verheyen et al. exploited modular machine learning to optimize and fabricate alginate bioblocks with tunable morphology, rheological and injectable characteristics [181] (Fig. 8A). Combined with nested and grouped cross-validation with multimetric evaluation, the data-driven models were utilized to evaluate predictability and modify design fundamentals to guide subsequent phases of development [181]. Li et al. calculated the physicochemical traits of more than 2000 polypeptides using quantitative structure-property relationships and applied machine learning algorithms to correlate their structure with self-assembly behavior [182]. Notably, the stiffness of AI-designed hydrogels is related to the diameter and crosslinking degree of the nanofibers. Combined with large-scale data computation, AI can screen the reaction conditions for novel material design in a high-throughput manner [183]. Ao et al. demonstrated an automated platform for screening immunocyte-solid tumor interplay, dynamically tracking T-cell infiltration and cytotoxicity within tumor spheroids (Fig. 8B). By recourse to a clinical data-driven deep learning and drug library, this system found an epigenetic modulator (lysine-specific histone demethylase 1 inhibitor) that, in combination with an immune checkpoint inhibitor such as anti-programmed cell death protein 1, effectively promoted T cell tumor invasion and improved immunotherapy in vivo [184].
Fig. 8.
AI-guided optimization of the LO culture system. (A) Optimization and fabrication of alginate bioblocks with tunable morphology, rheological and injectable characteristics by machine learning. Copyright 2023, Cell press. (B) Deep learning for cancer immunotherapy screening. Copyright 2022, PNAS. (C) Characterizing stem cell-derived organoids from electrical signals with machine learning. Copyright 2019, IEEE [181,184,186].
In vivo, the human organ can perceive and respond to external stimuli [185]. In vitro LO models should also be constructed to mimic or reproduce key functional properties of human organs as much as possible. External stimuli such as eelectricity, light, machinery and drugs are also an important elements in regulating the behaviors, development and maturity of LOs. The rapid development of AI technology has shown great potential to optimize these stimuli and monitor physiological parameters in LO research. For example, Huang's team characterized human embryonic stem cells (hESCs)-derived brain organoid behaviors from electrical signals with machine learning [186] (Fig. 8C). Machine learning and deep learning could characterize changes over time in local field potential (LFP) signals specific to chromodomain helicase DNA binding protein 2 (CHD2) mutated brain organoids. In combination with support vector machine method and convolutional neural network, LFP signatures could be applied successfully to distinguish CHD2-mutant LOs from controlled LOs [186]. So et al. integrated piezoresistive sensor with a deep learning-aided data process to advance in the precise evaluation of various stimuli (strength, hardness and shape) to human skin with high accuracy (96.9 %) [187]. Overall, solving an intricate and high-dimensional optimization issue remains a challenge. AI-guided optimization of the LO culture systems, such as the target experimental scheme, microenvrionment parameters and functional verification, is reaching LO research to a new milestone. The construction of miniaturized LO models with high fidelity of in vivo-like organs is helpful to comprehend the pathophysiology of the human body at the organ scale.
5.2. AI-accelerated the data extraction and analysis of LOs
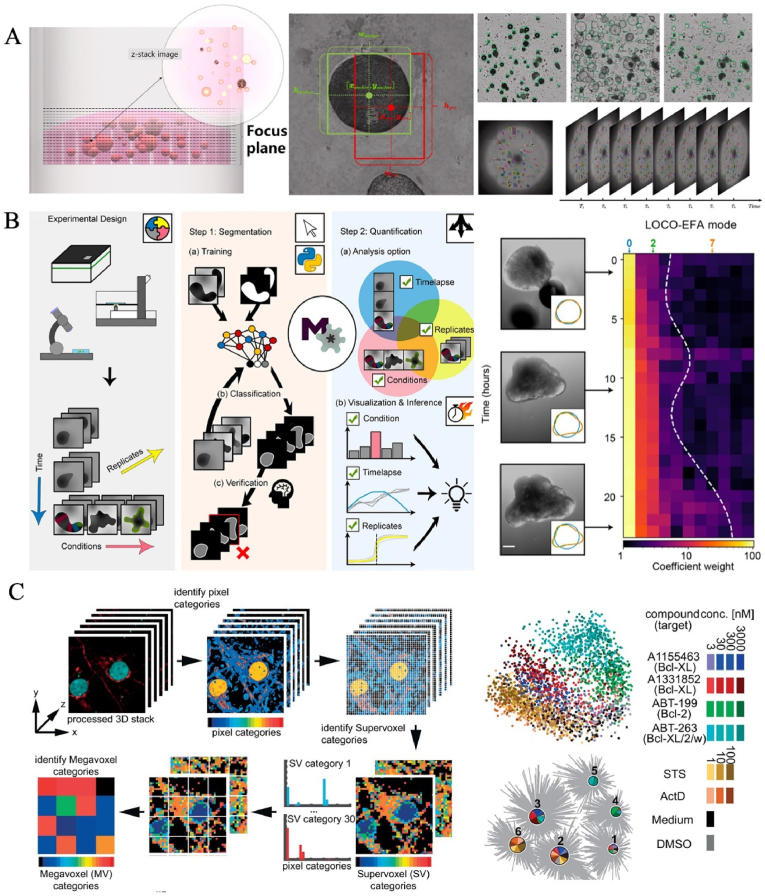
Accurate and efficient analysis of LO images is essential for the functional evaluation of LOs. Up to now, rapid and effective extraction of multi-dimensional characteristics of LOs from batch images remains a major challenge in the biomedical field. In general, tracking organoid behaviors at the single cell level is beneficial for elucidating cellular spatiotemporal dynamics, as well as understanding developmental processes and homeostasis [188]. LO image acquisition is generally on a single focal plane, and it is difficult to restore the 3D scale of LOs in terms of morphology (size and shape) and function (biomarker distribution and regional signal). Even 3D confocal microscopy can not produce large amounts of image data and can not adequately capture the complexity of organoid structures. Although endowed fluorescent signals for cells via gene modification is a common method to promote image segmentation and tracking, this process may alter the cell dynamics of the original tissue [161]. Because manual information acquisition is time-consuming, laborious, complex and lower accuracy, automated cell-tracking algorithms based on computer programs is the inevitable trend of development in LO research. The AI systems not only accelerate the exploration of experimental search space, but also effectively strengthen the data extraction and analysis of multi-dimensional LOs. Firstly, AI can evaluate the characteristic changes and visualize spatial patterns of LOs, including cell division, movement, growth, development and death according to segment images without the need for fluorescence or transgenic labeling [161,189]. Luo's team is the first to establish the first dataset of high-throughput organoid images for time organoid detection and tracking and proposed a new deep neural network that dynamically captures and tracks trajectories of organoids over time during culture, including irregular movement, morphological changes and growth kinetics [44] (Fig. 9A). AI can detect the infrastructure of organoids frame-by-frame, extract features, pair adjacent frames by identifying captured sequential images, and ultimately realize the detection and tracking of living organoids with the aid of the proposed dataset. It is also a problem to quantitatively and accurately analyze complex structures within LOs without compromising the quality of phenotypic measurements. AI, especially machine learning algorithms can be trained on large data sets of LO images, learning to recognize and quantify LO characteristics. Data-driven feature learning has the ability to classify, cluster and visualize 3D images, as well as rapid quantitative phenotyping analysis of LOs [190,191] (Fig. 9B). For instance, Mergenthaler et al. investigated the phenotypic responses of dense cortical neurons to pharmacological inhibition of anti-apoptotic proteins and the effects of oncogene on morphological alterations in human mammary acinar organoids [190] (Fig. 9C). In addition, the machine learning algorithm contributes to monitoring stem cell-derived islet organoid grafts labeled with particles and provides quantitative information about their presence in vivo [192].
Fig. 9.
AI accelerated the data extraction and analysis of LOs. (A) A new deep neural network was developed to dynamically capture and track trajectories of organoids over time during culture based on the dataset of high-throughput organoid images. Copyright 2021, Elsevier Ltd. (B) Data-driven feature learning has the ability to classify, cluster and visualize 3D images, as well as rapidly quantitative phenotyping analysis of LOs. Copyright 2021, The Company of Biologists Ltd. (C) Data-driven cell segmentation-free machine learning recorded the phenotypic responses of dense cortical neurons to pharmacological inhibition of anti-apoptotic proteins. Copyright 2021, PLoS [44,190,191].
As we all know, diverse analytical approaches facilitate a comprehensive understanding of biological data, elucidating how gene regulation influences biodiversity and functionality. Omics sequencing technologies (e.g., genomics, transcriptomics, metabolomics, etc) are continually emerging, providing foundational tools for understanding complex biological events during organogenesis. However, omics data encompass a variety of unstructured, semi-structured, and heterogeneous features, with an immense volume that poses a significant challenge to regular analytical modes. Therefore, high-throughput multi-omics data analysis based on traditional methods is arduous and intricate due to the diversity of data patterns and inherent limitations [161]. Also, traditional methods not only consume a significant amount of researchers' time and energy but also struggle to keep pace with the rapidly expanding volumes of data and the emergence of complex datasets. Consequently, there is a need for more efficient novel approaches to facilitate the realization of analysis tasks. In recent years, the synergistic potential of AI and omics technologies has advanced our understanding of biological complexity, such as the relationship between phenotypes and omics. Of note, a deep learning network, by integrating various tasks such as multi-modal fusion and cross-modal analysis, has the ability to analyze high-dimensional single-cell data (e.g., DNA, RNA accessibility, and proteins) with high accuracy in data integration and prediction [193]). Also, the proposed model is expected to directly assess the relationship between gene expression and other cell-specific patterns, aiding in the discovery of cell-specific regulatory dynamics. Another example is that an information barrier to single-cell RNA sequencing is the accurate labeling of data sets due to the complex, multi-dimensional, and ephemeral nature of cell states. Pioneering work by Wu's team introduced devCellPy, a high-precision (>90 %) machine learning model capable of automatically predicting cell types across complex multi-dimensional annotations, developmental datasets and species [194].
Consequently, AI can facilitate the analysis and mining of data across multi-scales and multi-modes, enhancing the signal-to-noise ratio, and thereby yielding LO data with superior spatiotemporal resolution. As biomedical research enters the era of big data, the adoption of efficient technological and analytical methods to support scientific conclusions and decision-making becomes increasingly critical. It must be acknowledged that the rapid advancement of AI is further bolstering biomedical research, creating a new interdisciplinary research paradigm of ‘AI for Science’. Despite the extensive integration of AI with omics technologies, which has yielded a plethora of innovative research findings, there are still some limitations and challenges. Notably, the prevailing algorithms frequently encounter difficulties in providing intelligible and discernible explanations in a biological context for the outputs of certain models. This may hinder the thorough elucidation of the underlying mechanisms governing the biological phenomena that occur throughout the LO developmental process. Therefore, there is an urgent need to develop new AI models and algorithms to drive the development of LOs.
5.3. Preclinical applications
AI-driven LO research is revolutionizing preclinical applications by providing near-physiological models for disease study and drug testing. It accelerates the understanding of complex biological processes and disease mechanisms and improves the efficiency of drug development. Herein, we summarize the preclinical applications of AI-enabled LO research (Table 2).
Table 2.
A summary of the preclinical applications of AI-enabled LO research.
| Organoid types | Cell resources | Disease types | AI tools | Study objectives |
|---|---|---|---|---|
| Brain [47] | hiPSCs | Parkinson | Machine learning; Random forest |
Neurotoxicity prediction and phenotypic characterisation monitoring |
| Pancreas [195] | Patient pancreas tissue | Ductal adenocarcinoma | Deep learning | Accurate identification and monitoring of pancreatic ductal adenocarcinoma carcinoid responses to chemotherapy and immunotherapy |
| Bladder [196,197] | Bladder cancer cell lines [196,197] | Bladder cancer | Deep learning [196,197]; Convolutional neural networks [197] |
Improving image segmentation accuracy and drug screening efficiency |
| Colorectum [46,198] | Adult stem cells [46]; Colorectal tissue [198] |
Colorectal cancer | Machine learning | |
| Islet [192] | hiPSCs | Type 1 diabetes | Machine learning | Achieving quantitative, long-term, and in vivo monitoring of human islet organoid transplantation |
| Liver [199] | N.A. | COVID-19 | Machine learning | Baricitinib has an anti-inflammatory effect and anti-COVID-19 viral activity |
| Lung, Pancreas [200] |
Lung and pancreatic tissues | Lung cancer; Pancreatic ductal adenocarcinoma cancer |
Deep learning; Convolutions neural networks |
Validating AI's advantages in partitioning cell arrest and cytotoxic drug responses |
| Colorectum; Bladder [48] |
Colorectal and bladder tissues | Colorectal cancer | Machine learning | Identifying biomarkers to accurately predict the response of cancer patients to 5-fluorouracil and cisplatin drugs |
Human Induced Pluripotent Stem Cells (hiPSCs); Corona Virus Disease 2019 (COVID-19); *N.A. indicates that the specified feature has not been measured or reported.
5.3.1. Disease modeling, diagnosis and prediction
In addition to biological research, the convergence of AI and LOs will show promise in disease modeling, diagnosis and risk prediction. Especially in patients with malignant tumors, which presents late with poor outcomes and high lifetime risk. Recently, Sander and colleagues successfully applied cutting-edge AI tools to predict the occurrence of pancreatic cancer 3 years before diagnosis based on disease codes, clinical datasets and disease trajectories of 9 million patients from two different healthcare systems: the Danish National Patient Registry (DNPR) and the United States Veterans Affairs (US-VA) [201]. Placido et al. trained a deep learning model in the cross-application of the Danish model with the US-VA dataset and showed enhanced performance (area under the receiver operating characteristic curve = 0.78) in the case of 3-year cancer occurrence. In a realistic surveillance program [201]. Placido et al. excluded diagnoses within 3 months prior to model training for pancreatic cancer diagnoses. Unexpectedly, among 0.1 % of highest-risk patients over 50 years of age who were predicted within 12 months of diagnosis, there was a large difference in relative risk between DNPR (58.7) and US-VA (33.1) [201]. This hinted that an absolute risk framework and/or retraining for a given clinical dataset might help improve the accuracy of risk predictions. Unfortunately, current research has mainly focused on clinical patient cases. Due to insufficient big data, and small treatment windows for advanced diseases, it is difficult to propose effective treatment plans for patients with specific diseases. LOs are excellent models and tools, which can remodel near-physiological human tissues in terms of structure and function [85,202]. Therefore, LOs can be served as in vivo substitution models for providing datasets, analyzing human pathology and highlighting the potential for AI applications. For instance, Feng et al. proposed a machine learning label transfer model that integrated hiPSC-derived heart organoids with the experimental and computational analysis, which allowed us to model heart development in normal and disease states. Notably, the model could characterize differentiated cells from hiPSC-derived heart organoids carrying an Ebstein's anomaly-associated genetic variant in NKX2-5, successfully establishing the congenital heart defects [203] (Fig. 10A). Schwamborn's team quantified the number of dopaminergic neurons and neuronal complexity in midbrain organoids derived from patients with Parkinson's disease, and used this data to establish a machine learning classifier to optimize data processing strategies. The cell atlas analysis and toxicity prediction in brain organoids under the neurotoxic compound exposure could also be obtained by machine learning [47] (Fig. 10B). Living organoid biobank is a unique biological system, which centralized stored various organoids for clinical therapy of diseases and academic research in life science. Notably, patients-derived tumor organoid living biobanks contain a mass of data (genetic backgrounds, mutant multiformity, and individualized properties) of specific diseases, and offer powerful biosystems for disease discovery, drug testing and personalized treatment in precision medicine [[204], [205], [206]]. Conventional in vitro models are frustrated to preserve the cellular fidelity and mutant diversity of parental tumors, as well as require a longer production time, which may delay treatment time. Currently, patient-derived tumor organoid biobanks have been established to capture tumoral and disease heterogeneity, as well as genotype-phenotype mapping [204,206,207]. Overall, the living organoid biobank, especially patients-derived tumor organoid living biobanks, will help scientists conduct disease research and treatment, and promote human health.
Fig. 10.
Preclinical applications of the convergence of AI and LOs in disease modeling, diagnosis and prediction, as well as drug testing and disease treatment. (A) Computational profiling of hiPSC-derived heart organoids reveals chamber defects associated with NKX2-5 deficiency. Copyright 2022, Springer Nature. (B) Machine learning-assisted neurotoxicity prediction in human midbrain organoids. Copyright 2020, Elsevier Ltd. (C) Machine learning for genetics-based classification and treatment response prediction in cancer of unknown primary. Copyright 2023, Springer Nature [47,203,214].
5.3.2. Drug testing and disease treatment
Since our understanding grew, with technology driving scale.in the rapid development of the information age, AI has Notably, harnessing the power of AI for living organoid research, combined with drug testing and multi-omics sequencing, could aid in clinical decision-making and clinical trial performance analysis. Of particular note, the FDA Modernization Act 2.0, approved in December 2022, eliminates the age-old requirement for animal testing of all drugs and will no longer require animal testing before entering clinical trials. This release is undoubtedly a high endorsement of organoid technology, and it has also set off a wave of research on organoids in academia and enterprises. It offers a unique opportunity for LOs to explore pathological traits and drug responses, discover innovative drugs, and give targeted therapies [196,197,208]. Combining large-scale AI-based omics analysis with diseased LO models allows for the identification of targets in preclinical and clinical trials and accurately assess the probability of a target passing a efficacy test. For example, by analyzing pharmacogenomic data from colorectal and bladder organoids, Kim and colleagues identified biomarkers associated with drug response that accurately predicted the patients' response to anti-cancer drugs [48]. In another case, network-based selection of PRKAB1 agonists were first-pass targets for intestinal barrier therapy that could effectively protect mucosal homeostasis and restore the leaky barrier in colon organoids [209]. In addition, AI can systematic pre-screen drug combinations at the pre-clinical stage to accelerate drug discovery. Julkunen et al. leveraged a comboFM framework to identify dosing combination effects of crizotinib and bortezomib in lymphoma cells to support lymphoma therapy [210]. Ghosh's team found that PRKAB1 agonists designed to up-regulate gene clusters on the healthy side of the network, combined with FDA-approved anti-inflammatory agents that inhibit pro-inflammatory gene expression on the diseased side, may show a synergistic therapeutic effect on inflammatory bowel disease (IBD) [209]. Notably, the inclusion of emerging organoid biobanks will help AI technology rapidly capture genotype-phenotype mapping of diseases in a short period, match with established disease databases, and develop alternative therapeutic options. Deben's team revealed clinically relevant response heterogeneity and invasive subclones in patients with pancreatic ductal adenocarcinoma at the organoid level [211]. Zhang's team used a tumor organoid biobank established in 144 liver cancer patients to perform a full-spectrum analysis of intra-tumor heterogeneity and a large-scale drug sensitivity screening [212]. Up to 75 % of drugs in clinical trials fail to make it to market due to safety and efficacy issues [213]. Imaging-Based Machine Learning Analysis of Patient-Derived Tumor Organoid Drug Response), indicating the urgent need for improved preclinical screening methodologies to advance the development of drugs.AI technology possesses the capability to construct sophisticated in silico drug screening paradigms, which facilitate the rapid triage of suboptimal compounds. These models are adept at engineering superior chemical entities, augmenting the enrichment of molecules with therapeutic potential, thereby attenuating the temporal and fiscal expenditures associated with the drug discovery process [196,197,200]. Using an AI-enabled approach, the researchers designed and synthesized a new compound, PKUF-01, which combined Lenvatinib and the c-Jun inhibitor veratramine, showing significant synergistic effects. Nowak-Sliwinska's group designed personalized treatment regimens for patients with colorectal cancer by combining patient-derived organoid efficacy tests and mathematical models based on outcomes [214] (Fig. 10C). The four low-dose combinations of rifenib, vemurafenib, palbocinb and lapatinib could inhibit cell viability by up to 88 %, signifcantly better than the clinical dose of FOLFOXIRI. AI models help predict human-related biological behaviors at the organoid level, quickly identifying potential pathways, proteins, mechanisms, and targets associated with disease [195,199]. It is worth considering that the establishment of organoid biobanks for different cancers, such as the culture system, is not standardized [215]. Overall, AI-enabled LO research is warranted to predict drug responses, provide a new therapeutic strategy, and guide clinical decision making. It is expected to transform the drug approval process, allowing biopharmaceutical companies to enter the drug development field directly through AI-enabled LO research.
Certainly, AI can also help doctors assist the consultation, assist analysis of images and electrical data, and make surgical plans, identify hard-to-detect traces, analyze clues, and more [198]. For example, Mao et al. improved the performance of medical inquiry by optimizing the algorithm model, and the accuracy rate of medical consultation reached 90.15 % [216]. An AIdecision support system with expert knowledge quality control algorithms can identify blood glucose patterns in the type 1 diabetes with the same accuracy as the board-certified endocrinologists [217]. Specifically, Jacobs's team generated 51,831 glucose observations to train the decision support engine performance in discerning high or low glycemic factors and to predict user-specific insulin adjustments from multiple potential recommendations [217]. After 12 weeks of continuous use, there were substantial improvements in blood glucose outcomes in virtual adults, adolescents and children. Importantly, the AI-based engine system can similarly identify insulin dosing problems in real-world human data, showing great application value in managing and improving glucose levels in type 1 diabetes. Vermeulen et al. achieved diagnostic turnaround times of less than 90 min in 25 surgeries by applying transfer-learned neural networks [218]. Notably, the diagnosis rate was as high as 72 %. Cancer Institute and the Massachusetts Institute of Technology (MIT) exploited a machine learning model called OncoNPC to predict the origin of cancer with an average accuracy of 80 percent, rising to 95 percent accuracy for tumors with high confidence predictions (about 65 percent of the total) [219]. The carcinoma of unknown primary focus (CUP) patients treated with the OncoNPC predictive approach had significantly longer survival than those treated with the inconsistent approach [219]. In a word, AI-assisted systems can assist decision-making and promote the implementation of personalized treatment, which is of great significance for doctors to formulate appropriate and accurate treatment strategies and avoid additional surgeries. For patients with pre-existing conditions or underlying risk factors, the next generation of technology will predict the onset of disease and intervene in a timely manner, ultimately achieving human health. We want to push the frontiers of artificial intelligence and organoid science to explore together how AI can better serve the study of human intelligence, inspire the development of artificial general intelligence, and provide innovative solutions to the challenges of organ function and disease.
6. Conclusion and future perspectives
Developing AI has displayed huge utilization potentiality in LO research, but AI-enabled LO models are still some limitations. In this part, we will present existing deficiencies, potential solutions, and future directions to advance the development of state-of-the-art in vivo LO systems.
6.1. Standardization
Undoubtedly, multiple self-assembly and engineering strategies have emerged to construct in vitro LOs, aiming to recapitulate the in vivo-like ecological niche of the human native organs. The road from LOs to general AI is still full of opportunities and challenges. In summarizing what has been achieved so far, some of the potential limits of AI-enabled LOs in the biomedical and biopharmaceutical fields have become apparent. We should understand how the human organ works and apply these principles to the development and innovation of general AI-enabled LOs. The standardization of LOs has always been a headache, which determines the normalization, conformity, recognition and extensibility of the field. In my opinion, several efforts can be made in the following aspects. Firstly, the current organoid systems still lack deficiencies such as coherent and mature blood systems, maturity functions, immune systems, complete life systems, dynamically changing microenvironments, as well as in vivo-like sensitivity and responsiveness to external stimulus (e.g., electrical, light, mechanical, and drug stimulation). These will limit the signal transmission (e.g., the mismatch between real input and actual input signals), identification and detection of AI. In particular, vascularized LOs show significant performance in connecting the circulatory system to support organ-organ communication and maintain systemic physiological function. To construct more bionic LOs, isolated organoids, microbiome, and immune cells should stem from human biopsies, blood and feces. A defined matrix should be developed to target the decellularized extracellular matrix in vivo. Besides, organ-organ interaction is important for organ development and maturity. For instance, the simple brain organoid model can not meet the needs of neuroscientific research to reveal the functional characteristics of the brain and to understand the mechanism of neuropsychiatric diseases [[220], [221], [222], [223]]. Also, the complex dynamic interplay between numerous cell types and regions ensure that the brain operates on well-defined neural circuits. It is exciting that the leading assembly technology developed in the last two years has brought new light to the complexity and functionality of LOs, especially in the development of whole brain organoids with network structures. Therefore, it is necessary to construct complex tissues with high fidelity. In recent decades, emerging organ chips have enabled to connection of different organs/tissues through precise fluid manipulation. Also, the mismatch between the size of the miniature LOs and the size of authentic human tissues is a problem. Herein, 3D bioprinting can remodel big-scale 3D tissues with an in vivo-like hierarchical structure to resolve this problem. In addition to improving their own systems of LOs, the compatibility of LOs and AI is also a point worth focusing on. That is, 3D and even all-round LO-AI interfaces should be flexible, and can sense the changes in size, hardness, biomarkers, and secretions over time during culture and make appropriate adjustments to monitor the development of LOs. Secondly, hierarchical classification of different diseases should be established to constrain the criteria of in vitro LO models. It is worth mentioning that the LO biobank opens up new ways to build physiological and pathological models relevant to humans in basic and clinical applications. Especially, as a rich resource, the establishment of tumor organoid biobanks contributes to obtaining mass data on specific diseases and further realizing personalized therapy. In my opinion, standardized research systems determine whether the subsequent function is widely recognized. Thirdly, to match the rapid development of clinical medicine and advance the preclinical to clinical transition, emerging tumor organoids-on-chip technology has been applied to in vitro organoid behavior monitor, disease modeling, and drug screening. The state-of-the-art chip devices are expected to be integrated into the AI-enabled LO systems to take advantage of their advantages. At the same time, multidisciplinary efforts are needed to establish standardization and guidelines in biology, including biomaterials and media formulations, sample collection, and operation specification. Various real-time readout systems integrated into organ-on-a-chip devices will enhance quality control processes and improve efficiency, standardization and repeatability.
6.2. Automation and individuation
Current technological development provides great opportunities to harness the power of AI for specific disease research and has great potential to give personalized treatment plans for patients. High-integrated systems involving AI, machine learning, living human organoids, microfluidics, genetic editing, biosensors, multi-omics, high-resolution imaging, soft materials, and more hold promise to advance AI-enabled LO models for biological research and personalized therapy. It must be done collaboratively by workers with different disciplinary backgrounds. In recent years, gene-editing systems like CRISPR/Cas9 have shown remarkable efficacy in identifying oncogenes, the study of drug-gene Interactions and the evaluation of drug-targeting treatment [223]. By accurately regulating the gene profiles of tumor organoids through gene editing, scientists could parse the correlation between gene mutations and anti-tumor drug sensitivity, gain insight into the mechanisms of development at the molecular level, and discover potential biomarkers and innovative drugs [224]. It is worth noting that automated microfluidic LOs-on-chips have advantages in high throughput and precise analysis of cells, but are not yet compatible with AI. The emerging systems may drive LO models to screen and reflect real-world patient treatment processes, and potentially facilitate treatment decisions for personalized treatment. Until these platforms are used more widely, an important next step is to evaluate how surveillance strategies designed for patients such as those at-risk groups can promote early disease detection in clinical trials and improve survival, while minimizing overdiagnosis and potential harm.
The essence of the algorithm is based on the analysis of massive data, constantly improve the accuracy of the models, help doctors and medical institutions enhance efficiency and lower the error rate. However, big data is constantly generated and endless, which means that the data obtained from a certain sector or a certain region is very limited, and the algorithm improved by this data is difficult to be universal, which also means that artificial intelligence is easier to have an advantage in the data with certainty, rather than real-time dynamic adjustment of the data. From a medical perspective, although evidence-based medicine is widely applied in the treatment process, the treatment of many complex diseases requires comprehensive data analysis, and patients are different, complex diseases need personalized treatment, algorithms can only provide auxiliary reference on data, and can not replace decision-making. User compliance is highly dependent on their living and eating habits, such a service essentially needs more personalization, that is, more human intervention, and the versatility of the algorithm is difficult to get the desired effect. This leaves digital therapy Mired in a paradox of its own creation, with the universality of algorithms struggling to cope with dynamic and uncertain data. In the future, researchers can enrich data sets with LOs, not just clinical samples, and make more attempts on other tasks, further pushing biomedical advances.
6.3. Regulatory situation
Research on organoids supported by artificial intelligence is making breakthroughs in biomedicine and life sciences. The so-called opportunities and challenges coexist. In particular, there are some regulatory and legal challenges and risks that we need to face and address, such as how to ensure its smart safety and control, how to ensure that it is ethical and responsible, and how to do economic restraint and regulation. Notably, patient-specific tumor organoid models are admirable in terms of ethical principles associated with reducing animal use, but their scale-up and clinical translation still face significant ethical issues. Due to rapid advances in technology, it is difficult to foresee the possible future use and storage of these models, which has led to concerns about obtaining informed consent from donors. The prowess of AI is underpinned by its capacity to aggregate and integrate a broad spectrum of disparate datasets, thereby facilitating tailored medical approaches and enabling ongoing learning processes. Nonetheless, this proficiency is contingent upon the utilization and governance of LO data from human tissues, which inherently intersects with issues on the privacy of patients [225]. Investigators are obliged to deliberate upon these concerns and establish a framework of guidelines that are designed to foster and uphold the principles of responsible research conduct [172]. For example, establishing cryptographic protocols or digital signatures can prevent data theft, tampering, or corruption. Differential privacy technology can also be used to process data in a privacy-preserving manner, ensuring that an attacker cannot identify specific information about any individual in the data set through the analysis results. In addition, ethical guidelines on commercialization are needed to ensure a fair distribution of benefits among donors/patients, researchers, commercial parties and other stakeholders involved in generating these models. The ethical concerns raised by LO research have partly focused on questions about creating entities that could potentially exhibit consciousness. Of course, AI-based organoid solutions are only for better service of humans for life sciences and effectively reducing the burden on public medical service. The computer will be built from human cells and is designed to mimic the complex processes in living organisms to enable efficient, intelligent computing. Overall, the construction of AI-enabled LO model systems and their research in biomedicine have just started, and still need to be further explored. AI-powered LOs have the potential to dramatically improve the way we approach medicine, and it's a heart-stirring field that deserves continued attention and investment.
6.4. Commercialization
A multitude of corporations globally, including Hubrecht Organoid Technology (HUB), Emulate, DaXiang Technology, and D1Med, have catalyzed the commercial progression of organoid technologies via diverse commercial strategies. This spectrum encompasses the provision of direct pharmaceutical services, technological licensing, product sales, and cooperative research endeavors. The 2019 signified a pivotal stage in the commercialization of organoids, witnessing the inception of six enterprises [226]. Organome and HUB are devoted to the establishment of organoid biobanks in a non-profit mode within the organoid realm, whereas their counterparts are focused on the manufacture, commercialization, or other organoid-centric services. Of note, SUN Biosciences and System1 Biosciences are pioneering the utilization of robotic automation for organoid fabrication. Recently, MIMETAS has declared a partnership with HUB to commercialize organoids integrated within microfluidic chips. In parallel with these organoid-focused companies, the collaborative venture between the US Type Culture Collection and the Human Cancer Model Initiative to cultivate patient-derived cancer organoids is also an important advance in the commercialization of organoids. For organoid companies, the road to commercialization needs further validation and exploration. Firstly, companies providing professional services need to establish a standardized service system and identify scalable application scenarios, so as to promote the commercialization process of the company. Secondly, the maturity of the technology platform largely determines the quality of organoids, which will become an important determinant of the competitive landscape of the industry. Thirdly, the inherent opacity of AI models, often referred to as their “black box” feature, can lead to challenges in model interpretability, thus limiting the widespread deployment of such models in organoid commercialization. Therefore, enhancing the interpretability of AI models helps to increase the confidence of scientific researchers and clinical practitioners in the prognostic insights generated by AI algorithms. Besides, computational resource requirements, ethical considerations, validation, and reproducibility are also factors to consider. Finally, advancing the commercialization of AI in organoids requires interdisciplinary research and collaboration among scientific researchers in the fields of biology, medicine and AI.
Overall, the research on AI-enabled LOs has just started, and there is a lot of room for development to improve the system. By integrating AI systems, researchers can in-depth understanding of human organ development, disease occurrence, and drug responses. The proposed AI systems have great potential for directing cell behavior and controlling cell niches (matrix gel and environment stimuli), enabling researchers to investigate the biological processes of LOs on a single-cell or cluster scale. Researchers can rapidly and precisely capture changes in phenotype, genotype and biological activity of LOs over time. This information can be used to train machine learning algorithms and further predict future development direction. Living imaging techniques, biosensors, and multi-omics advance the comprehensive assessment of LOs from multiple perspectives. Due to the difficulty of analyzing large amounts of data, AI is driving the evaluation of these technologies in an unprecedented way to match the rapid development of LOs in an automated and high-throughput mode.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable. This study did not involve any human participants, human data, or human tissue, nor did it involve any live animals. Therefore, no ethics approval was required, and consent to participate is not applicable.
CRediT authorship contribution statement
Hui Wang: Writing – original draft, Visualization, Funding acquisition, Conceptualization. Xiangyang Li: Validation, Data curation. Xiaoyan You: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Guoping Zhao: Writing – review & editing, Supervision, Resources, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This review was supported by the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Projects (TSBICIP-CXRC-008), Strategic Priority Research Program of the Chinese Academy of Sciences (XDC 0110300),Major Project of Haihe Laboratory of Synthetic Biology (E2M9560201), National Natural Science Foundation of China (32301210 & 31200035) and the China Postdoctoral Science Foundation (No. 2022M713330).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Xiaoyan You, Email: xiaoyanyou@haust.edu.cn.
Guoping Zhao, Email: gpzhao@sibs.ac.cn.
References
- 1.Wang H., Fu T., Du Y., Gao W., Huang K., Liu Z., Chandak P., Liu S., Van Katwyk P., Deac A., Anandkumar A., Bergen K., Gomes C.P., Ho S., Kohli P., Lasenby J., Leskovec J., Liu T.Y., Manrai A., Marks D., Ramsundar B., Song L., Sun J., Tang J., Velickovic P., Welling M., Zhang L., Coley C.W., Bengio Y., Zitnik M. Scientific discovery in the age of artificial intelligence. Nature. 2023;620:47–60. doi: 10.1038/s41586-023-06221-2. [DOI] [PubMed] [Google Scholar]
- 2.Jutel M., Zemelka-Wiacek M., Ordak M., Pfaar O., Eiwegger T., Rechenmacher M., Akdis C.A. The artificial intelligence (AI) revolution: how important for scientific work and its reliable sharing. Allergy. 2023;78:2085–2088. doi: 10.1111/all.15778. [DOI] [PubMed] [Google Scholar]
- 3.Ju H., Juan R., Gomez R., Nakamura K., Li G. Transferring policy of deep reinforcement learning from simulation to reality for robotics. Nat. Mach. Intell. 2022;4:1077–1087. [Google Scholar]
- 4.Renner H., Scholer H.R., Bruder J.M. Combining automated organoid workflows with artificial intelligence-based analyses: opportunities to build a new generation of interdisciplinary high-throughput screens for Parkinson's disease and beyond. Mov. Disord. 2021;36:2745–2762. doi: 10.1002/mds.28775. [DOI] [PubMed] [Google Scholar]
- 5.Swanson K., Wu E., Zhang A., Alizadeh A.A., Zou J. From patterns to patients: advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell. 2023;186:1772–1791. doi: 10.1016/j.cell.2023.01.035. [DOI] [PubMed] [Google Scholar]
- 6.Nabulsi Z., Sellergren A., Jamshy S., Lau C., Santos E., Kiraly A.P., Ye W., Yang J., Pilgrim R., Kazemzadeh S., Yu J., Kalidindi S.R., Etemadi M., Garcia-Vicente F., Melnick D., Corrado G.S., Peng L., Eswaran K., Tse D., Beladia N., Liu Y., Chen P.C., Shetty S. Deep learning for distinguishing normal versus abnormal chest radiographs and generalization to two unseen diseases tuberculosis and COVID-19. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-93967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P.C., Mega J.L., Webster D.R. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi S., Asada K., Takasawa K., Shimoyama R., Sakai A., Bolatkan A., Shinkai N., Kobayashi K., Komatsu M., Kaneko S., Sese J., Hamamoto R. Predicting deep learning based multi-omics parallel integration survival subtypes in lung cancer using reverse phase protein array data. Biomolecules. 2020;10:1460. doi: 10.3390/biom10101460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Z., Chen X., Dowbaj A.M., Sljukic A., Bratlie K., Lin L., Fong E.L.S., Balachander G.M., Chen Z., Soragni A., Huch M., Zeng Y.A., Wang Q., Yu H. Organoids. Nat Rev Methods Primers. 2022;2:94. doi: 10.1038/s43586-022-00174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie J., Hashino E. Organoid technologies meet genome engineering. EMBO Rep. 2017;18:367–376. doi: 10.15252/embr.201643732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shahbazi M.N., Siggia E.D., Zernicka-Goetz M. Self-organization of stem cells into embryos: a window on early mammalian development. Science. 2019;364:948–951. doi: 10.1126/science.aax0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri O., Cooper-White J., Janmey P.A., Mooney D.J., Shenoy V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature. 2020;584:535–546. doi: 10.1038/s41586-020-2612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prince E., Cruickshank J., Ba-Alawi W., Hodgson K., Haight J., Tobin C., Wakeman A., Avoulov A., Topolskaia V., Elliott M.J., McGuigan A.P., Berman H.K., Haibe-Kains B., Cescon D.W., Kumacheva E. Biomimetic hydrogel supports initiation and growth of patient-derived breast tumor organoids. Nat. Commun. 2022;13:1466. doi: 10.1038/s41467-022-28788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordonez-Moran P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 16.Shao Y., Taniguchi K., Gurdziel K., Townshend R.F., Xue X., Yong K.M.A., Sang J., Spence J.R., Gumucio D.L., Fu J. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater. 2017;16:419–425. doi: 10.1038/nmat4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alzamil L., Nikolakopoulou K., Turco M.Y. Organoid systems to study the human female reproductive tract and pregnancy. Cell Death Differ. 2021;28:35–51. doi: 10.1038/s41418-020-0565-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell. 2017;20:844–857 e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu J., Kalyani F.S., Liu L., Cheng T., Chen L. Tumor organoids: synergistic applications, current challenges, and future prospects in cancer therapy. Cancer Commun. 2021;41:1331–1353. doi: 10.1002/cac2.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofbauer P., Jahnel S.M., Papai N., Giesshammer M., Deyett A., Schmidt C., Penc M., Tavernini K., Grdseloff N., Meledeth C., Ginistrelli L.C., Ctortecka C., Salic S., Novatchkova M., Mendjan S. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299–3317 e22. doi: 10.1016/j.cell.2021.04.034. [DOI] [PubMed] [Google Scholar]
- 21.Kelley K.W., Pasca S.P. Human brain organogenesis: toward a cellular understanding of development and disease. Cell. 2022;185:42–61. doi: 10.1016/j.cell.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Corsini N.S., Knoblich J.A. Human organoids: new strategies and methods for analyzing human development and disease. Cell. 2022;185:2756–2769. doi: 10.1016/j.cell.2022.06.051. [DOI] [PubMed] [Google Scholar]
- 23.Tuveson D., Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 24.Kim W., Gwon Y., Park S., Kim H., Kim J. Therapeutic strategies of three-dimensional stem cell spheroids and organoids for tissue repair and regeneration. Bioact. Mater. 2023;19:50–74. doi: 10.1016/j.bioactmat.2022.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Zhang W., Maskey N., Yang F., Zheng Z., Li C., Wang R., Wu P., Mao S., Zhang J., Yan Y., Li W., Yao X. Urological cancer organoids, patients' avatars for precision medicine: past, present and future. Cell Biosci. 2022;12:132. doi: 10.1186/s13578-022-00866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu H., Gehart H., Artegiani B., C L.O.-I., Dekkers F., Basak O., van Es J., Chuva de Sousa Lopes S.M., Begthel H., Korving J., van den Born M., Zou C., Quirk C., Chiriboga L., Rice C.M., Ma S., Rios A., Peters P.J., de Jong Y.P., Clevers H. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606 e19. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Wimmer R.A., Leopoldi A., Aichinger M., Wick N., Hantusch B., Novatchkova M., Taubenschmid J., Hammerle M., Esk C., Bagley J.A., Lindenhofer D., Chen G., Boehm M., Agu C.A., Yang F., Fu B., Zuber J., Knoblich J.A., Kerjaschki D., Penninger J.M. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Wang X., Tan Z., Su Y., Liu J., Chang M., Yan F., Chen J., Chen T., Li C., Hu J., Wang Y. Human ESC-derived expandable hepatic organoids enable therapeutic liver repopulation and pathophysiological modeling of alcoholic liver injury. Cell Res. 2019;29:1009–1026. doi: 10.1038/s41422-019-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revah O., Gore F., Kelley K.W., Andersen J., Sakai N., Chen X., Li M.Y., Birey F., Yang X., Saw N.L., Baker S.W., Amin N.D., Kulkarni S., Mudipalli R., Cui B., Nishino S., Grant G.A., Knowles J.K., Shamloo M., Huguenard J.R., Deisseroth K., Pasca S.P. Maturation and circuit integration of transplanted human cortical organoids. Nature. 2022;610:319–326. doi: 10.1038/s41586-022-05277-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampaziotis F., Muraro D., Tysoe O.C., Sawiak S., Beach T.E., Godfrey E.M., Upponi S.S., Brevini T., Wesley B.T., Garcia-Bernardo J., Mahbubani K., Canu G., Gieseck R., Berntsen N.L., Mulcahy V.L., Crick K., Fear C., Robinson S., Swift L., Gambardella L., Bargehr J., Ortmann D., Brown S.E., Osnato A., Murphy M.P., Corbett G., Gelson W.T.H., Mells G.F., Humphreys P., Davies S.E., Amin I., Gibbs P., Sinha S., Teichmann S.A., Butler A.J., See T.C., Melum E., Watson C.J.E., Saeb-Parsy K., Vallier L. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 2021;371:839. doi: 10.1126/science.aaz6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J., Rabbani C.C., Gao H., Steinhart M.R., Woodruff B.M., Pflum Z.E., Kim A., Heller S., Liu Y., Shipchandler T.Z., Koehler K.R. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582:399–404. doi: 10.1038/s41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu M., Xiong L., Lyu Y., Zhang X., Shen J., Guan J., Chai P., Lin Z., Nie B., Li C., Xu J., Deng H. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res. 2021;31:259–271. doi: 10.1038/s41422-020-00453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto S., Kobayashi E., Fujii M., Ohta Y., Arai K., Matano M., Ishikawa K., Miyamoto K., Toshimitsu K., Takahashi S., Nanki K., Hakamata Y., Kanai T., Sato T. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature. 2021;592:99–104. doi: 10.1038/s41586-021-03247-2. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi R., Okubo T., Kudo Y., Ishikawa Y., Imaizumi T., Suzuki K., Shibata S., Katayama T., Park S.J., Young R.D., Quantock A.J., Nishida K. Generation of 3D lacrimal gland organoids from human pluripotent stem cells. Nature. 2022;605:126–131. doi: 10.1038/s41586-022-04613-4. [DOI] [PubMed] [Google Scholar]
- 35.Rumsey J.W., Lorance C., Jackson M., Sasserath T., McAleer C.W., Long C.J., Goswami A., Russo M.A., Raja S.M., Gable K.L., Emmett D., Hobson-Webb L.D., Chopra M., Howard J.F., Jr., Guptill J.T., Storek M.J., Alonso-Alonso M., Atassi N., Panicker S., Parry G., Hammond T., Hickman J.J. Classical complement pathway inhibition in a "Human-On-A-Chip" model of autoimmune demyelinating neuropathies. Adv. Ther. 2022;5 doi: 10.1002/adtp.202200030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin H., Sun Z., Bernards R. LICOB: a powerful organoid platform for drug discovery. Cell Res. 2024;34:11–12. doi: 10.1038/s41422-023-00878-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S., Shen J., Wang X., Sun X., Wu Y., Zhang M.R., Wang R., Hu K. Integration of organoids in peptide drug discovery: rise of the high‐throughput screening. View. 2023:4. [Google Scholar]
- 38.Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., Bram Y., Richardson C., Zhu J., Zhao Z., Redmond D., Houghton S., Nguyen D.T., Xu D., Wang X., Jessurun J., Borczuk A., Huang Y., Johnson J.L., Liu Y., Xiang J., Wang H., Cantley L.C., tenOever B.R., Ho D.D., Pan F.C., Evans T., Chen H.J., Schwartz R.E., Chen S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smirnova L., Caffo B.S., Gracias D.H., Huang Q., Morales Pantoja I.E., Tang B., Zack D.J., Berlinicke C.A., Boyd J.L., Harris T.D., Johnson E.C., Kagan B.J., Kahn J., Muotri A.R., Paulhamus B.L., Schwamborn J.C., Plotkin J., Szalay A.S., Vogelstein J.T., Worley P.F., Hartung T. Organoid intelligence (OI): the new frontier in biocomputing and intelligence-in-a-dish. Front. Sci. 2023;1 [Google Scholar]
- 40.Badai J., Bu Q., Zhang L. Review of artificial intelligence applications and algorithms for brain organoid research. Interdiscip Sci. 2020;12:383–394. doi: 10.1007/s12539-020-00386-4. [DOI] [PubMed] [Google Scholar]
- 41.Park K., Lee J.Y., Lee S.Y., Jeong I., Park S.Y., Kim J.W., Nam S.A., Kim H.W., Kim Y.K., Lee S. Deep learning predicts the differentiation of kidney organoids derived from human induced pluripotent stem cells. Kidney Res Clin Pract. 2023;42:75–85. doi: 10.23876/j.krcp.22.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kegeles E., Naumov A., Karpulevich E.A., Volchkov P., Baranov P. Convolutional neural networks can predict retinal differentiation in retinal organoids. Front. Cell. Neurosci. 2020;14:171. doi: 10.3389/fncel.2020.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunha A.B., Hou J., Schuelke C. Machine learning for stem cell differentiation and proliferation classification on electrical impedance spectroscopy. J Electr Bioimpedance. 2019;10:124–132. doi: 10.2478/joeb-2019-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bian X., Li G., Wang C., Liu W., Lin X., Chen Z., Cheung M., Luo X. A deep learning model for detection and tracking in high-throughput images of organoid. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104490. [DOI] [PubMed] [Google Scholar]
- 45.He C., Kalafut N.C., Sandoval S.O., Risgaard R., Sirois C.L., Yang C., Khullar S., Suzuki M., Huang X., Chang Q., Zhao X., Sousa A.M.M., Wang D. BOMA, a machine-learning framework for comparative gene expression analysis across brains and organoids. Cell Rep Methods. 2023;3 doi: 10.1016/j.crmeth.2023.100409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spiller E.R., Ung N., Kim S., Patsch K., Lau R., Strelez C., Doshi C., Choung S., Choi B., Juarez Rosales E.F., Lenz H.J., Matasci N., Mumenthaler S.M. Imaging-based machine learning analysis of patient-derived tumor organoid drug response. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.771173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monzel A.S., Hemmer K., Kaoma T., Smits L.M., Bolognin S., Lucarelli P., Rosety I., Zagare A., Antony P., Nickels S.L., Krueger R., Azuaje F., Schwamborn J.C. Machine learning-assisted neurotoxicity prediction in human midbrain organoids. Parkinsonism Relat. Disorders. 2020;75:105–109. doi: 10.1016/j.parkreldis.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Kong J., Lee H., Kim D., Han S.K., Ha D., Shin K., Kim S. Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat. Commun. 2020;11:5485. doi: 10.1038/s41467-020-19313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison R.G. Observations on the living developing nerve fiber. Exp. Biol. Med. 1906;4:140–143. [Google Scholar]
- 50.Chang S.Y., Carpena N.T., Mun S., Jung J.Y., Chung P.S., Shim H., Han K., Ahn J.C., Lee M.Y. Enhanced inner-ear organoid formation from mouse embryonic stem cells by photobiomodulation. Mol Ther Methods Clin Dev. 2020;17:556–567. doi: 10.1016/j.omtm.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parigoris E., Lee J.H., Liu A.Y., Zhao X., Takayama S. Extended longevity geometrically-inverted proximal tubule organoids. Biomaterials. 2022;290 doi: 10.1016/j.biomaterials.2022.121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee S., Chang J., Kang S.M., Parigoris E., Lee J.H., Huh Y.S., Takayama S. High-throughput formation and image-based analysis of basal-in mammary organoids in 384-well plates. Sci. Rep. 2022;12:317. doi: 10.1038/s41598-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panek M., Grabacka M., Pierzchalska M. The formation of intestinal organoids in a hanging drop culture. Cytotechnology. 2018;70:1085–1095. doi: 10.1007/s10616-018-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parigoris E., Lee S., Mertz D., Turner M., Liu A.Y., Sentosa J., Djomehri S., Chang H.C., Luker K., Luker G., Kleer C.G., Takayama S. Cancer cell invasion of mammary organoids with basal-in phenotype. Adv. Healthcare Mater. 2021;10 doi: 10.1002/adhm.202000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tung Y.C., Hsiao A.Y., Allen S.G., Torisawa Y.S., Ho M., Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136:473–478. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen A., Young J., Nielsen J.E., Joensen U.N., Toft B.G., Rajpert-De Meyts E., Loveland K.L. Hanging drop cultures of human testis and testis cancer samples: a model used to investigate activin treatment effects in a preserved niche. Br. J. Cancer. 2014;110:2604–2614. doi: 10.1038/bjc.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nath S., Devi G.R. Three-dimensional culture systems in cancer research: focus on tumor spheroid model. Pharmacol. Ther. 2016;163:94–108. doi: 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ganguli A., Mostafa A., Saavedra C., Kim Y., Le P., Faramarzi V., Feathers R.W., Berger J., Ramos-Cruz K.P., Adeniba O., Diaz G.J.P., Drnevich J., Wright C.L., Hernandez A.G., Lin W., Smith A.M., Kosari F., Vasmatzis G., Anastasiadis P.Z., Bashir R. Three-dimensional microscale hanging drop arrays with geometric control for drug screening and live tissue imaging. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abc1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suea-Ngam A., Howes P.D., Srisa-Art M., deMello A.J. Droplet microfluidics: from proof-of-concept to real-world utility? Chem. Commun. 2019;55:9895–9903. doi: 10.1039/c9cc04750f. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y., Liu M., Zhang Y., Liu H., Han L. Recent methods of droplet microfluidics and their applications in spheroids and organoids. Lab Chip. 2023;23:1080–1096. doi: 10.1039/d2lc00493c. [DOI] [PubMed] [Google Scholar]
- 61.Liu H., Wang Y., Wang H., Zhao M., Tao T., Zhang X., Qin J. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. 2020;7 doi: 10.1002/advs.201903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W., Jin H., Lou S., Yang H., Dai X., Ma S. Microfluidic droplet encapsulation-guided organoid growth promotes parental tumor phenotype recapitulation. Int. J. Cancer. 2024;154:145–154. doi: 10.1002/ijc.34706. [DOI] [PubMed] [Google Scholar]
- 63.Amirifar L., Besanjideh M., Nasiri R., Shamloo A., Nasrollahi F., de Barros N.R., Davoodi E., Erdem A., Mahmoodi M., Hosseini V., Montazerian H., Jahangiry J., Darabi M.A., Haghniaz R., Dokmeci M.R., Annabi N., Ahadian S., Khademhosseini A. Droplet-based microfluidics in biomedical applications. Biofabrication. 2022;14 doi: 10.1088/1758-5090/ac39a9. [DOI] [PubMed] [Google Scholar]
- 64.Tomasi R.F., Sart S., Champetier T., Baroud C.N. Individual control and quantification of 3D spheroids in a high-density microfluidic droplet array. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W., Li D., Jiang S., Galan E.A., Zhang Z., Huang L., Ma S. Microfluidic droplets as structural templates for Matrigel to enable 1-week large organoid modeling. Chem. Eng. Sci. 2021;238 [Google Scholar]
- 66.Wang H., Liu H., Zhang X., Wang Y., Zhao M., Chen W., Qin J. One-step generation of aqueous-droplet-filled hydrogel fibers as organoid carriers using an all-in-water microfluidic system. ACS Appl. Mater. Interfaces. 2021;13:3199–3208. doi: 10.1021/acsami.0c20434. [DOI] [PubMed] [Google Scholar]
- 67.Wang H., Liu H., Liu H., Su W., Chen W., Qin J. One‐step generation of core–shell Gelatin methacrylate (GelMA) microgels using a droplet microfluidic system. Advanced Materials Technologies. 2019;4 [Google Scholar]
- 68.Wang H., Liu H., He F., Chen W., Zhang X., Zhao M., Wang L., Qin J. Flexible generation of multi‐aqueous core hydrogel capsules using microfluidic aqueous two‐phase system. Advanced Materials Technologies. 2020;5 [Google Scholar]
- 69.Dowbaj A.M., Kohler T.N., Cordero-Espinoza L., Hollfelder F., Huch M. Generation of liver mesenchyme and ductal cell organoid co-culture using cell self-aggregation and droplet microfluidics. STAR Protoc. 2023;4 doi: 10.1016/j.xpro.2023.102333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Y., Zhang X., Sun L., Wang Y., Zhao Y. Engineering human brain assembloids by microfluidics. Adv. Mater. 2023;35 doi: 10.1002/adma.202210083. [DOI] [PubMed] [Google Scholar]
- 71.Sun W., Chen Y.-Q., Luo G.-A., Zhang M., Zhang H.-Y., Wang Y.-R., Hu P. Organs-on-chips and its applications. Chin. J. Anal. Chem. 2016;44:533–541. [Google Scholar]
- 72.Vunjak-Novakovic G., Ronaldson-Bouchard K., Radisic M. Organs-on-a-chip models for biological research. Cell. 2021;184:4597–4611. doi: 10.1016/j.cell.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan J., Li Z., Guo J., Liu S., Guo J. Organ-on-a-chip: a new tool for in vitro research. Biosens. Bioelectron. 2022;216 doi: 10.1016/j.bios.2022.114626. [DOI] [PubMed] [Google Scholar]
- 74.Wang H., You X.Y., Zhao G.P. Microbial volatile communication in human 3D intestinal organotypic models. Sci. Bull. 2023;68:1353–1358. doi: 10.1016/j.scib.2023.05.030. [DOI] [PubMed] [Google Scholar]
- 75.Fernández‐Costa J.M., Ortega M.A., Rodríguez‐Comas J., Lopez‐Muñoz G., Yeste J., Mangas‐Florencio L., Fernández‐González M., Martin‐Lasierra E., Tejedera‐Villafranca A., Ramon‐Azcon J. Training‐on‐a‐Chip: a multi‐organ device to study the effect of muscle exercise on insulin secretion in vitro. Advanced Materials Technologies. 2022;8 [Google Scholar]
- 76.Ramadan Q., Hazaymeh R., Zourob M. Immunity-on-a-Chip: integration of immune components into the scheme of organ-on-a-chip systems. Adv Biol. 2023 doi: 10.1002/adbi.202200312. [DOI] [PubMed] [Google Scholar]
- 77.Reshma S., Megha K.B., Amir S., Rukhiya S., Mohanan P.V. Blood brain barrier-on-a-chip to model neurological diseases. J. Drug Deliv. Sci. Technol. 2023;80 [Google Scholar]
- 78.Hou C., Gu Y., Yuan W., Zhang W., Xiu X., Lin J., Gao Y., Liu P., Chen X., Song L. Application of microfluidic chips in the simulation of the urinary system microenvironment. Mater Today Bio. 2023;19 doi: 10.1016/j.mtbio.2023.100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young R.E., Huh D.D. Organ-on-a-chip technology for the study of the female reproductive system. Adv. Drug Deliv. Rev. 2021;173:461–478. doi: 10.1016/j.addr.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen D.T., van Noort D., Jeong I.K., Park S. Endocrine system on chip for a diabetes treatment model. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa5cc9. [DOI] [PubMed] [Google Scholar]
- 81.Huang Q., Yang T., Song Y., Sun W., Xu J., Cheng Y., Yin R., Zhu L., Zhang M., Ma L., Li H., Zhang H. A three-dimensional (3D) liver-kidney on a chip with a biomimicking circulating system for drug safety evaluation. Lab Chip. 2024;24:1715–1726. doi: 10.1039/d3lc00980g. [DOI] [PubMed] [Google Scholar]
- 82.Park S.E., Georgescu A., Huh D. Organoids-on-a-chip. Science. 2019;364(6444):960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Homan K.A., Gupta N., Kroll K.T., Kolesky D.B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M.T., Ferrante T., Bonventre J.V., Lewis J.A., Morizane R. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tao T., Wang Y., Chen W., Li Z., Su W., Guo Y., Deng P., Qin J. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip. 2019;19:948–958. doi: 10.1039/c8lc01298a. [DOI] [PubMed] [Google Scholar]
- 85.Nikolaev M., Mitrofanova O., Broguiere N., Geraldo S., Dutta D., Tabata Y., Elci B., Brandenberg N., Kolotuev I., Gjorevski N., Clevers H., Lutolf M.P. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 86.Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L., Fu J. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin B.J., Battula S., Zachos N., Kovbasnjuk O., Fawlke-Abel J., In J., Donowitz M., Verkman A.S. Microfluidics platform for measurement of volume changes in immobilized intestinal enteroids. Biomicrofluidics. 2014;8 doi: 10.1063/1.4870400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shang L., Ye F., Li M., Zhao Y. Spatial confinement toward creating artificial living systems. Chem. Soc. Rev. 2022;51:4075–4093. doi: 10.1039/d1cs01025e. [DOI] [PubMed] [Google Scholar]
- 89.Yin F., Zhang X., Wang L., Wang Y., Zhu Y., Li Z., Tao T., Chen W., Yu H., Qin J. HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs. Lab Chip. 2021;21:571–581. doi: 10.1039/d0lc00921k. [DOI] [PubMed] [Google Scholar]
- 90.Tao T., Deng P., Wang Y., Zhang X., Guo Y., Chen W., Qin J. Microengineered multi-organoid system from hiPSCs to recapitulate human liver-islet Axis in normal and type 2 diabetes. Adv. Sci. 2022;9 doi: 10.1002/advs.202103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim J., Kim J., Jin Y., Cho S.W. In situbiosensing technologies for an organ-on-a-chip. Biofabrication. 2023;15 doi: 10.1088/1758-5090/aceaae. [DOI] [PubMed] [Google Scholar]
- 92.Han J., Kang U., Moon E.-Y., Yoo H., Gweon B. Imaging technologies for microfluidic biochips. BioChip Journal. 2022;16:255–269. [Google Scholar]
- 93.Khan I., Prabhakar A., Delepine C., Tsang H., Pham V., Sur M. A low-cost 3D printed microfluidic bioreactor and imaging chamber for live-organoid imaging. Biomicrofluidics. 2021;15(2) doi: 10.1063/5.0041027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schuster B., Junkin M., Kashaf S.S., Romero-Calvo I., Kirby K., Matthews J., Weber C.R., Rzhetsky A., White K.P., Tay S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020;11:5271. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rawal P., Tripathi D.M., Ramakrishna S., Kaur S. Prospects for 3D bioprinting of organoids. Bio-Design and Manufacturing. 2021;4:627–640. [Google Scholar]
- 96.Juraski A.C., Sharma S., Sparanese S., da Silva V.A., Wong J., Laksman Z., Flannigan R., Rohani L., Willerth S.M. 3D bioprinting for organ and organoid models and disease modeling. Expet Opin. Drug Discov. 2023;18:1043–1059. doi: 10.1080/17460441.2023.2234280. [DOI] [PubMed] [Google Scholar]
- 97.Wanjare M., Kawamura M., Hu C., Alcazar C., Wang H., Woo Y.J., Huang N.F. Vascularization of engineered spatially patterned myocardial tissue derived from human pluripotent stem cells in vivo. Front. Bioeng. Biotechnol. 2019;7:208. doi: 10.3389/fbioe.2019.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y., Li G., Wang J., Zhou F., Ren X., Su J. Small joint organoids 3D bioprinting: construction strategy and application. Small. 2024;20 doi: 10.1002/smll.202302506. [DOI] [PubMed] [Google Scholar]
- 99.Layrolle P., Payoux P., Chavanas S. Message in a scaffold: natural biomaterials for three-dimensional (3D) bioprinting of human brain organoids. Biomolecules. 2022;13:25. doi: 10.3390/biom13010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chakraborty J., Chawla S., Ghosh S. Developmental biology-inspired tissue engineering by combining organoids and 3D bioprinting. Curr. Opin. Biotechnol. 2022;78 doi: 10.1016/j.copbio.2022.102832. [DOI] [PubMed] [Google Scholar]
- 101.Ji W., Hou B., Lin W., Wang L., Zheng W., Li W., Zheng J., Wen X., He P. 3D Bioprinting a human iPSC-derived MSC-loaded scaffold for repair of the uterine endometrium. Acta Biomater. 2020;116:268–284. doi: 10.1016/j.actbio.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 102.Teli P., Kale V., Vaidya A. Beyond animal models: revolutionizing neurodegenerative disease modeling using 3D in vitro organoids, microfluidic chips, and bioprinting. Cell Tissue Res. 2023;394:75–91. doi: 10.1007/s00441-023-03821-2. [DOI] [PubMed] [Google Scholar]
- 103.Skylar-Scott M.A., Uzel S.G.M., Nam L.L., Ahrens J.H., Truby R.L., Damaraju S., Lewis J.A. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen H., Wu Z., Gong Z., Xia Y., Li J., Du L., Zhang Y., Gao X., Fan Z., Hu H., Qian Q., Ding Z., Guo S. Acoustic bioprinting of patient-derived organoids for predicting cancer therapy responses. Adv. Healthcare Mater. 2022;11 doi: 10.1002/adhm.202102784. [DOI] [PubMed] [Google Scholar]
- 105.Zhu Y., Sun L., Fu X., Liu J., Liang Z., Tan H., Li W., Zhao Y. Engineering microcapsules to construct vascularized human brain organoids. Chem. Eng. J. 2021;424 [Google Scholar]
- 106.Choi Y.M., Lee H., Ann M., Song M., Rheey J., Jang J. 3D bioprinted vascularized lung cancer organoid models with underlying disease capable of more precise drug evaluation. Biofabrication. 2023;15 doi: 10.1088/1758-5090/acd95f. [DOI] [PubMed] [Google Scholar]
- 107.Kim D., Youn J., Lee J., Kim H., Kim D.S. Recent progress in fabrication of electrospun nanofiber membranes for developing physiological in vitro organ/tissue models. Macromol. Biosci. 2023;23 doi: 10.1002/mabi.202300244. [DOI] [PubMed] [Google Scholar]
- 108.Miranda C.C., Gomes M.R., Moco M., Cabral J.M.S., Ferreira F.C., Sanjuan-Alberte P. A concise review on electrospun scaffolds for kidney tissue engineering. Bioengineering. 2022;9:554. doi: 10.3390/bioengineering9100554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Joddar B., Natividad-Diaz S.L., Padilla A.E., Esparza A.A., Ramirez S.P., Chambers D.R., Ibaroudene H. Engineering approaches for cardiac organoid formation and their characterization. Transl. Res. 2022;250:46–67. doi: 10.1016/j.trsl.2022.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Beldjilali-Labro M., Jellali R., Brown A.D., Garcia Garcia A., Lerebours A., Guenin E., Bedoui F., Dufresne M., Stewart C., Grosset J.F., Legallais C. Multiscale-engineered muscle constructs: PEG hydrogel micro-patterning on an electrospun PCL mat functionalized with gold nanoparticles. Int. J. Mol. Sci. 2021;23:260. doi: 10.3390/ijms23010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dalton P.D., Woodfield T.B.F., Mironov V., Groll J. Advances in hybrid fabrication toward hierarchical tissue constructs. Adv. Sci. 2020;7 doi: 10.1002/advs.201902953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baklaushev V.P., Bogush V.G., Kalsin V.A., Sovetnikov N.N., Samoilova E.M., Revkova V.A., Sidoruk K.V., Konoplyannikov M.A., Timashev P.S., Kotova S.L., Yushkov K.B., Averyanov A.V., Troitskiy A.V., Ahlfors J.E. Tissue engineered neural constructs composed of neural precursor cells, recombinant spidroin and PRP for neural tissue regeneration. Sci. Rep. 2019;9:3161. doi: 10.1038/s41598-019-39341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kharaziha M., Nikkhah M., Shin S.R., Annabi N., Masoumi N., Gaharwar A.K., Camci-Unal G., Khademhosseini A. PGS:Gelatin nanofibrous scaffolds with tunable mechanical and structural properties for engineering cardiac tissues. Biomaterials. 2013;34:6355–6366. doi: 10.1016/j.biomaterials.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ritzau-Reid K.I., Callens S.J.P., Xie R., Cihova M., Reumann D., Grigsby C.L., Prados-Martin L., Wang R., Moore A.C., Armstrong J.P.K., Knoblich J.A., Stevens M.M. Microfibrous scaffolds guide stem cell lumenogenesis and brain organoid engineering. Adv. Mater. 2023;35 doi: 10.1002/adma.202300305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sahoo S., Ang L.T., Goh J.C., Toh S.L. Growth factor delivery through electrospun nanofibers in scaffolds for tissue engineering applications. J. Biomed. Mater. Res. 2010;93:1539–1550. doi: 10.1002/jbm.a.32645. [DOI] [PubMed] [Google Scholar]
- 116.Nasr B., Chatterton R., Yong J.H.M., Jamshidi P., D'Abaco G.M., Bjorksten A.R., Kavehei O., Chana G., Dottori M., Skafidas E. Self-Organized nanostructure modified microelectrode for sensitive electrochemical glutamate detection in stem cells-derived brain organoids. Biosensors. 2018;8:14. doi: 10.3390/bios8010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang Q., Tang B., Romero J.C., Yang Y., Elsayed S.K., Pahapale G., Lee T.J., Morales Pantoja I.E., Han F., Berlinicke C., Xiang T., Solazzo M., Hartung T., Qin Z., Caffo B.S., Smirnova L., Gracias D.H. Shell microelectrode arrays (MEAs) for brain organoids. Sci. Adv. 2022;8:eabq5031. doi: 10.1126/sciadv.abq5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fleck J.S., Jansen S.M.J., Wollny D., Zenk F., Seimiya M., Jain A., Okamoto R., Santel M., He Z., Camp J.G., Treutlein B. Inferring and perturbing cell fate regulomes in human brain organoids. Nature. 2023;621:365–372. doi: 10.1038/s41586-022-05279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Susa K., Kobayashi K., Galichon P., Matsumoto T., Tamura A., Hiratsuka K., Gupta N.R., Yazdi I.K., Bonventre J.V., Morizane R. ATP/ADP biosensor organoids for drug nephrotoxicity assessment. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1138504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lasse M., El Saghir J., Berthier C.C., Eddy S., Fischer M., Laufer S.D., Kylies D., Hutzfeldt A., Bonin L.L., Dumoulin B., Menon R., Vega-Warner V., Eichinger F., Alakwaa F., Fermin D., Billing A.M., Minakawa A., McCown P.J., Rose M.P., Godfrey B., Meister E., Wiech T., Noriega M., Chrysopoulou M., Brandts P., Ju W., Reinhard L., Hoxha E., Grahammer F., Lindenmeyer M.T., Huber T.B., Schluter H., Thiel S., Mariani L.H., Puelles V.G., Braun F., Kretzler M., Demir F., Harder J.L., Rinschen M.M. An integrated organoid omics map extends modeling potential of kidney disease. Nat. Commun. 2023;14:4903. doi: 10.1038/s41467-023-39740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Badder L.M., Hollins A.J., Herpers B., Yan K., Ewan K.B., Thomas M., Shone J.R., Badder D.A., Naven M., Ashelford K.E., Hargest R., Clarke A.R., Esdar C., Buchstaller H.P., Treherne J.M., Boj S., Ramezanpour B., Wienke D., Price L.S., Shaw P.H., Dale T.C. 3D imaging of colorectal cancer organoids identifies responses to Tankyrase inhibitors. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Blatchley M.R., Gunay K.A., Yavitt F.M., Hawat E.M., Dempsey P.J., Anseth K.S. In situ super-resolution imaging of organoids and extracellular matrix interactions via phototransfer by allyl sulfide exchange-expansion microscopy (PhASE-ExM) Adv. Mater. 2022;34 doi: 10.1002/adma.202109252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beumer J., Puschhof J., Bauza-Martinez J., Martinez-Silgado A., Elmentaite R., James K.R., Ross A., Hendriks D., Artegiani B., Busslinger G.A., Ponsioen B., Andersson-Rolf A., Saftien A., Boot C., Kretzschmar K., Geurts M.H., Bar-Ephraim Y.E., Pleguezuelos-Manzano C., Post Y., Begthel H., van der Linden F., Lopez-Iglesias C., van de Wetering W.J., van der Linden R., Peters P.J., Heck A.J.R., Goedhart J., Snippert H., Zilbauer M., Teichmann S.A., Wu W., Clevers H. High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell. 2020;181:1291–1306 e19. doi: 10.1016/j.cell.2020.04.036. [DOI] [PubMed] [Google Scholar]
- 124.Wu Q., Wei X., Pan Y., Zou Y., Hu N., Wang P. Bionic 3D spheroids biosensor chips for high-throughput and dynamic drug screening. Biomed. Microdevices. 2018;20:82. doi: 10.1007/s10544-018-0329-x. [DOI] [PubMed] [Google Scholar]
- 125.Browne A.W., Arnesano C., Harutyunyan N., Khuu T., Martinez J.C., Pollack H.A., Koos D.S., Lee T.C., Fraser S.E., Moats R.A., Aparicio J.G., Cobrinik D. Structural and functional characterization of human stem-cell-derived retinal organoids by live imaging. Invest. Ophthalmol. Vis. Sci. 2017;58:3311–3318. doi: 10.1167/iovs.16-20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tasnim K., Liu J. Emerging bioelectronics for brain organoid electrophysiology. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shin S.R., Kilic T., Zhang Y.S., Avci H., Hu N., Kim D., Branco C., Aleman J., Massa S., Silvestri A., Kang J., Desalvo A., Hussaini M.A., Chae S.K., Polini A., Bhise N., Hussain M.A., Lee H., Dokmeci M.R., Khademhosseini A. Label-free and regenerative electrochemical microfluidic biosensors for continual monitoring of cell secretomes. Adv. Sci. 2017;4 doi: 10.1002/advs.201600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dekkers J.F., Alieva M., Cleven A., Keramati F., Wezenaar A.K.L., van Vliet E.J., Puschhof J., Brazda P., Johanna I., Meringa A.D., Rebel H.G., Buchholz M.B., Barrera Roman M., Zeeman A.L., de Blank S., Fasci D., Geurts M.H., Cornel A.M., Driehuis E., Millen R., Straetemans T., Nicolasen M.J.T., Aarts-Riemens T., Ariese H.C.R., Johnson H.R., van Ineveld R.L., Karaiskaki F., Kopper O., Bar-Ephraim Y.E., Kretzschmar K., Eggermont A.M.M., Nierkens S., Wehrens E.J., Stunnenberg H.G., Clevers H., Kuball J., Sebestyen Z., Rios A.C. Uncovering the mode of action of engineered T cells in patient cancer organoids. Nat. Biotechnol. 2023;41:60–69. doi: 10.1038/s41587-022-01397-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Mousavi Shaegh S.A., Massa S., Riahi R., Chae S., Hu N., Avci H., Zhang W., Silvestri A., Sanati Nezhad A., Manbohi A., De Ferrari F., Polini A., Calzone G., Shaikh N., Alerasool P., Budina E., Kang J., Bhise N., Ribas J., Pourmand A., Skardal A., Shupe T., Bishop C.E., Dokmeci M.R., Atala A., Khademhosseini A. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dekkers J.F., Alieva M., Wellens L.M., Ariese H.C.R., Jamieson P.R., Vonk A.M., Amatngalim G.D., Hu H., Oost K.C., Snippert H.J.G., Beekman J.M., Wehrens E.J., Visvader J.E., Clevers H., Rios A.C. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019;14:1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- 131.D'Imprima E., Garcia Montero M., Gawrzak S., Ronchi P., Zagoriy I., Schwab Y., Jechlinger M., Mahamid J. Light and electron microscopy continuum-resolution imaging of 3D cell cultures. Dev. Cell. 2023;58:616–632 e6. doi: 10.1016/j.devcel.2023.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beghin A., Grenci G., Sahni G., Guo S., Rajendiran H., Delaire T., Mohamad Raffi S.B., Blanc D., de Mets R., Ong H.T., Galindo X., Monet A., Acharya V., Racine V., Levet F., Galland R., Sibarita J.B., Viasnoff V. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods. 2022;19:881–892. doi: 10.1038/s41592-022-01508-0. [DOI] [PubMed] [Google Scholar]
- 133.Walsh A.J., Cook R.S., Skala M.C. Functional optical imaging of primary human tumor organoids: development of a personalized drug screen. J. Nucl. Med. 2017;58:1367–1372. doi: 10.2967/jnumed.117.192534. [DOI] [PubMed] [Google Scholar]
- 134.Schafer S.T., Mansour A.A., Schlachetzki J.C.M., Pena M., Ghassemzadeh S., Mitchell L., Mar A., Quang D., Stumpf S., Ortiz I.S., Lana A.J., Baek C., Zaghal R., Glass C.K., Nimmerjahn A., Gage F.H. An in vivo neuroimmune organoid model to study human microglia phenotypes. Cell. 2023;186:2111–2126 e20. doi: 10.1016/j.cell.2023.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kang Y., Deng J., Ling J., Li X., Chiang Y.J., Koay E.J., Wang H., Burks J.K., Chiao P.J., Hurd M.W., Bhutani M.S., Lee J.H., Weston B.R., Maitra A., Ikoma N., Tzeng C.D., Lee J.E., DePinho R.A., Wolff R.A., Pant S., McAllister F., Katz M.H., Fleming J.B., Kim M.P. 3D imaging analysis on an organoid-based platform guides personalized treatment in pancreatic ductal adenocarcinoma. J. Clin. Invest. 2022;132 doi: 10.1172/JCI151604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bremond Martin C., Simon Chane C., Clouchoux C., Histace A. Recent trends and perspectives in cerebral organoids imaging and analysis. Front. Neurosci. 2021;15 doi: 10.3389/fnins.2021.629067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang J., Jiang Y., Ren Y., Liu Y., Wu X., Li Z., Ren J. Biomaterials and biosensors in intestinal organoid culture, a progress review. J. Biomed. Mater. Res. 2020;108:1501–1508. doi: 10.1002/jbm.a.36921. [DOI] [PubMed] [Google Scholar]
- 138.Kim G.A., Ginga N.J., Takayama S. Integration of sensors in gastrointestinal organoid culture for biological analysis. Cell Mol Gastroenterol Hepatol. 2018;6:123–131 e1. doi: 10.1016/j.jcmgh.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu J., Yin Y., Leng Y., Zhang J., Wang C., Chen Y., Li X., Wang X., Liu H., Liao Y., Jin Y., Zhang Y., Lu K., Wang K., Wang X., Wang L., Zheng F., Gu Z., Li Y., Fan Y. Emerging strategies of engineering retinal organoids and organoid-on-a-chip in modeling intraocular drug delivery: current progress and future perspectives. Adv. Drug Deliv. Rev. 2023;197 doi: 10.1016/j.addr.2023.114842. [DOI] [PubMed] [Google Scholar]
- 140.Fair S.R., Julian D., Hartlaub A.M., Pusuluri S.T., Malik G., Summerfied T.L., Zhao G., Hester A.B., Ackerman W.E.t., Hollingsworth E.W., Ali M., McElroy C.A., Buhimschi I.A., Imitola J., Maitre N.L., Bedrosian T.A., Hester M.E. Electrophysiological maturation of cerebral organoids correlates with dynamic morphological and cellular development. Stem Cell Rep. 2020;15:855–868. doi: 10.1016/j.stemcr.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zanotelli M.R., Goldblatt Z.E., Miller J.P., Bordeleau F., Li J., VanderBurgh J.A., Lampi M.C., King M.R., Reinhart-King C.A. Regulation of ATP utilization during metastatic cell migration by collagen architecture. Mol. Biol. Cell. 2018;29:1–9. doi: 10.1091/mbc.E17-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yousafzai M.S., Hammer J.A. Using biosensors to study organoids, spheroids and organs-on-a-chip: a mechanobiology perspective. Biosensors. 2023;13:905. doi: 10.3390/bios13100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu S., Kumari S., He H., Mishra P., Singh B.N., Singh D., Liu S., Srivastava P., Li C. Biosensors integrated 3D organoid/organ-on-a-chip system: a real-time biomechanical, biophysical, and biochemical monitoring and characterization. Biosens. Bioelectron. 2023;231 doi: 10.1016/j.bios.2023.115285. [DOI] [PubMed] [Google Scholar]
- 144.Kim M., Hwang J.C., Min S., Park Y.G., Kim S., Kim E., Seo H., Chung W.G., Lee J., Cho S.W., Park J.U. Multimodal characterization of cardiac organoids using integrations of pressure-sensitive transistor arrays with three-dimensional liquid metal electrodes. Nano Lett. 2022;22:7892–7901. doi: 10.1021/acs.nanolett.2c02790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park Y., Franz C.K., Ryu H., Luan H., Cotton K.Y., Kim J.U., Chung T.S., Zhao S., Vazquez-Guardado A., Yang D.S., Li K., Avila R., Phillips J.K., Quezada M.J., Jang H., Kwak S.S., Won S.M., Kwon K., Jeong H., Bandodkar A.J., Han M., Zhao H., Osher G.R., Wang H., Lee K., Zhang Y., Huang Y., Finan J.D., Rogers J.A. Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assembloids. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lukonin I., Serra D., Challet Meylan L., Volkmann K., Baaten J., Zhao R., Meeusen S., Colman K., Maurer F., Stadler M.B., Jenkins J., Liberali P. Phenotypic landscape of intestinal organoid regeneration. Nature. 2020;586:275–280. doi: 10.1038/s41586-020-2776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Atamian A., Cordon-Barris L., Quadrato G. Taming human brain organoids one cell at a time. Semin. Cell Dev. Biol. 2021;111:23–31. doi: 10.1016/j.semcdb.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zheng H., Feng Y., Tang J., Ma S. Interfacing brain organoids with precision medicine and machine learning. Cell Reports Physical Science. 2022;3 [Google Scholar]
- 149.Kadur Lakshminarasimha Murthy P., Sontake V., Tata A., Kobayashi Y., Macadlo L., Okuda K., Conchola A.S., Nakano S., Gregory S., Miller L.A., Spence J.R., Engelhardt J.F., Boucher R.C., Rock J.R., Randell S.H., Tata P.R. Human distal lung maps and lineage hierarchies reveal a bipotent progenitor. Nature. 2022;604:111–119. doi: 10.1038/s41586-022-04541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchis-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D., Qian Z., Heide M., Huttner W.B., Khaitovich P., Paabo S., Treutlein B., Camp J.G. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 151.Uzquiano A., Kedaigle A.J., Pigoni M., Paulsen B., Adiconis X., Kim K., Faits T., Nagaraja S., Anton-Bolanos N., Gerhardinger C., Tucewicz A., Murray E., Jin X., Buenrostro J., Chen F., Velasco S., Regev A., Levin J.Z., Arlotta P. Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell. 2022;185:3770–3788 e27. doi: 10.1016/j.cell.2022.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T.M., Krol J., Szikra T., Cuttat R., Waldt A., Papasaikas P., Diggelmann R., Patino-Alvarez C.P., Galliker P., Spirig S.E., Pavlinic D., Gerber-Hollbach N., Schuierer S., Srdanovic A., Balogh M., Panero R., Kusnyerik A., Szabo A., Stadler M.B., Orgul S., Picelli S., Hasler P.W., Hierlemann A., Scholl H.P.N., Roma G., Nigsch F., Roska B. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020;182:1623–1640 e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sapir G., Steinberg D.J., Aqeilan R.I., Katz-Brull R. Real-time non-invasive and direct determination of lactate dehydrogenase activity in cerebral organoids-A new method to characterize the metabolism of brain organoids? Pharmaceuticals. 2021;14:878. doi: 10.3390/ph14090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Becker L., Fischer F., Fleck J.L., Harland N., Herkommer A., Stenzl A., Aicher W.K., Schenke-Layland K., Marzi J. Data-driven identification of biomarkers for in situ monitoring of drug treatment in bladder cancer organoids. Int. J. Mol. Sci. 2022;23:6956. doi: 10.3390/ijms23136956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choo N., Ramm S., Luu J., Winter J.M., Selth L.A., Dwyer A.R., Frydenberg M., Grummet J., Sandhu S., Hickey T.E., Tilley W.D., Taylor R.A., Risbridger G.P., Lawrence M.G., Simpson K.J. High-throughput imaging assay for drug screening of 3D prostate cancer organoids. SLAS Discov. 2021;26:1107–1124. doi: 10.1177/24725552211020668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Czerwinski M., Spence J.R. Hacking the matrix. Cell Stem Cell. 2017;20(1):9–10. doi: 10.1016/j.stem.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu H., Wang Y., Cui K., Guo Y., Zhang X., Qin J. Advances in hydrogels in organoids and organs-on-a-chip. Adv. Mater. 2019;31 doi: 10.1002/adma.201902042. [DOI] [PubMed] [Google Scholar]
- 158.Haenlein M., Kaplan A. A brief history of artificial intelligence: on the past, present, and future of artificial intelligence. Calif. Manag. Rev. 2019;61:5–14. [Google Scholar]
- 159.Miller G.A. The cognitive revolution: a historical perspective. Trends Cognit. Sci. 2003;7(3):141–144. doi: 10.1016/s1364-6613(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 160.Kulkarni P.S., N A.S. History and growth of artificial intelligence. Interantional Journal of Scientific Research in Engineering and Management. 2023;7:1–11. [Google Scholar]
- 161.Bai L., Wu Y., Li G., Zhang W., Zhang H., Su J. AI-enabled organoids: construction, analysis, and application. Bioact. Mater. 2024;31:525–548. doi: 10.1016/j.bioactmat.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodríguez J.J., Kuncheva L.I. Rotation forest:: a new classifier ensemble method. IEEE Trans. Pattern Anal. Mach. Intell. 2006;28:1619–1630. doi: 10.1109/TPAMI.2006.211. [DOI] [PubMed] [Google Scholar]
- 163.Hussain S.F. A novel robust kernel for classifying high-dimensional data using Support Vector Machines. Expert Syst. Appl. 2019;131:116–131. [Google Scholar]
- 164.Dreiseitl S., Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J. Biomed. Inf. 2002;35:352–359. doi: 10.1016/s1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 165.Ren W., Zhang J., Pan J., Liu S., Ren J., Du J., Cao X., Yang M.-H. Deblurring dynamic scenes via spatially varying recurrent neural networks. IEEE Trans. Pattern Anal. Mach. Intell. 2022;44:3974–3987. doi: 10.1109/TPAMI.2021.3061604. [DOI] [PubMed] [Google Scholar]
- 166.Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat. Med. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 167.Wang J., Hu X. Convolutional neural networks with gated recurrent connections. IEEE Trans. Pattern Anal. Mach. Intell. 2022;44:3421–3435. doi: 10.1109/TPAMI.2021.3054614. [DOI] [PubMed] [Google Scholar]
- 168.Alkhayrat M., Aljnidi M., Aljoumaa K. A comparative dimensionality reduction study in telecom customer segmentation using deep learning and PCA. Journal of Big Data. 2020;7 [Google Scholar]
- 169.Alghodhaifi H., Alghodhaifi A., Alghodhaifi M. Predicting invasive ductal carcinoma in breast histology images using convolutional neural network. Proc Naecon Ieee Nat. 2019:374–378. [Google Scholar]
- 170.Zou J., Huss M., Abid A., Mohammadi P., Torkamani A., Telenti A. A primer on deep learning in genomics. Nat. Genet. 2018;51:12–18. doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Maramraju S., Kowalczewski A., Kaza A., Liu X., Singaraju J.P., Albert M.V., Ma Z., Yang H. AI‐organoid integrated systems for biomedical studies and applications. Bioengineering & Translational Medicine. 2024;9 doi: 10.1002/btm2.10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhou J., Huang C., Gao X. Patient privacy in AI-driven omics methods. Trends Genet. 2024;40:383–386. doi: 10.1016/j.tig.2024.03.004. [DOI] [PubMed] [Google Scholar]
- 173.Antonopoulos I., Robu V., Couraud B., Kirli D., Norbu S., Kiprakis A., Flynn D., Elizondo-Gonzalez S., Wattam S. Artificial intelligence and machine learning approaches to energy demand-side response: a systematic review. Renew. Sustain. Energy Rev. 2020;130 [Google Scholar]
- 174.Camacho-Gomez D., Sorzabal-Bellido I., Ortiz-de-Solorzano C., Garcia-Aznar J.M., Gomez-Benito M.J. A hybrid physics-based and data-driven framework for cellular biological systems: application to the morphogenesis of organoids. iScience. 2023;26 doi: 10.1016/j.isci.2023.107164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhu Y., Huang R., Wu Z., Song S., Cheng L., Zhu R. Deep learning-based predictive identification of neural stem cell differentiation. Nat. Commun. 2021;12:2614. doi: 10.1038/s41467-021-22758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kanda G.N., Tsuzuki T., Terada M., Sakai N., Motozawa N., Masuda T., Nishida M., Watanabe C.T., Higashi T., Horiguchi S.A., Kudo T., Kamei M., Sunagawa G.A., Matsukuma K., Sakurada T., Ozawa Y., Takahashi M., Takahashi K., Natsume T. Robotic search for optimal cell culture in regenerative medicine. Elife. 2022;11 doi: 10.7554/eLife.77007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Singaraju J.P., Kadiresan A., Bhoi R.K., Gomez A.H., Ma Z., Yang H. Organalysis: multifunctional image preprocessing and analysis software for cardiac organoid studies. Tissue Eng. C Methods. 2023;29:572–582. doi: 10.1089/ten.tec.2023.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kagan B.J., Kitchen A.C., Tran N.T., Habibollahi F., Khajehnejad M., Parker B.J., Bhat A., Rollo B., Razi A., Friston K.J. In vitro neurons learn and exhibit sentience when embodied in a simulated game-world. Neuron. 2022;110:3952–3969 e8. doi: 10.1016/j.neuron.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wlodarczyk-Biegun M.K., Del Campo A. 3D bioprinting of structural proteins. Biomaterials. 2017;134:180–201. doi: 10.1016/j.biomaterials.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 180.Liaw C.Y., Ji S., Guvendiren M. Engineering 3D hydrogels for personalized in vitro human tissue models. Adv. Healthcare Mater. 2018;7 doi: 10.1002/adhm.201701165. [DOI] [PubMed] [Google Scholar]
- 181.Verheyen C.A., Uzel S.G.M., Kurum A., Roche E.T., Lewis J.A. Integrated data-driven modeling and experimental optimization of granular hydrogel matrices. Matter. 2023;6:1015–1036. [Google Scholar]
- 182.Li F., Han J., Cao T., Lam W., Fan B., Tang W., Chen S., Fok K.L., Li L. Design of self-assembly dipeptide hydrogels and machine learning via their chemical features. Proc. Natl. Acad. Sci. U. S. A. 2019;116:11259–11264. doi: 10.1073/pnas.1903376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li X., Yang X., Liu L., Zhou P., Zhou J., Shi X., Wang Y. A microarray platform designed for high-throughput screening the reaction conditions for the synthesis of micro/nanosized biomedical materials. Bioact. Mater. 2020;5:286–296. doi: 10.1016/j.bioactmat.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ao Z., Cai H., Wu Z., Hu L., Nunez A., Zhou Z., Liu H., Bondesson M., Lu X., Lu X., Dao M., Guo F. Microfluidics guided by deep learning for cancer immunotherapy screening. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2214569119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cho J., Lee H., Rah W., Chang H.J., Yoon Y.-s. From engineered heart tissue to cardiac organoid. Theranostics. 2022;12:2758–2772. doi: 10.7150/thno.67661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hasib M.M., Lybrand Z., Estevez V.N., Hsieh J., Huang Y.F. 2019 Ieee Embs International Conference on Biomedical & Health Informatics (Bhi) 2019. Charactering hESCs organoids from electrical signals with machine learning. [Google Scholar]
- 187.So C., Kim J.U., Luan H., Park S.U., Kim H., Han S., Kim D., Shin C., Kim T.-i., Lee W.H., Park Y., Heo K., Baac H.W., Ko J.H., Won S.M. Epidermal piezoresistive structure with deep learning-assisted data translation. npj Flexible Electronics. 2022;6:70. [Google Scholar]
- 188.Ulman V., Maska M., Magnusson K.E.G., Ronneberger O., Haubold C., Harder N., Matula P., Matula P., Svoboda D., Radojevic M., Smal I., Rohr K., Jalden J., Blau H.M., Dzyubachyk O., Lelieveldt B., Xiao P., Li Y., Cho S.Y., Dufour A.C., Olivo-Marin J.C., Reyes-Aldasoro C.C., Solis-Lemus J.A., Bensch R., Brox T., Stegmaier J., Mikut R., Wolf S., Hamprecht F.A., Esteves T., Quelhas P., Demirel O., Malmstrom L., Jug F., Tomancak P., Meijering E., Munoz-Barrutia A., Kozubek M., Ortiz-de-Solorzano C. An objective comparison of cell-tracking algorithms. Nat. Methods. 2017;14:1141–1152. doi: 10.1038/nmeth.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kok R.N.U., Hebert L., Huelsz-Prince G., Goos Y.J., Zheng X., Bozek K., Stephens G.J., Tans S.J., van Zon J.S. OrganoidTracker: efficient cell tracking using machine learning and manual error correction. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mergenthaler P., Hariharan S., Pemberton J.M., Lourenco C., Penn L.Z., Andrews D.W. Rapid 3D phenotypic analysis of neurons and organoids using data-driven cell segmentation-free machine learning. PLoS Comput. Biol. 2021;17 doi: 10.1371/journal.pcbi.1008630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gritti N., Lim J.L., Anlas K., Pandya M., Aalderink G., Martinez-Ara G., Trivedi V. MOrgAna: accessible quantitative analysis of organoids with machine learning. Development. 2021;148(18) doi: 10.1242/dev.199611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sun A., Hayat H., Liu S., Tull E., Bishop J.O., Dwan B.F., Gudi M., Talebloo N., Dizon J.R., Li W., Gaudet J., Alessio A., Aguirre A., Wang P. 3D in vivo magnetic particle imaging of human stem cell-derived islet organoid transplantation using a machine learning algorithm. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.704483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tang X., Zhang J., He Y., Zhang X., Lin Z., Partarrieu S., Hanna E.B., Ren Z., Shen H., Yang Y., Wang X., Li N., Ding J., Liu J. Explainable multi-task learning for multi-modality biological data analysis. Nat. Commun. 2023;14:2546. doi: 10.1038/s41467-023-37477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Galdos F.X., Xu S., Goodyer W.R., Duan L., Huang Y.V., Lee S., Zhu H., Lee C., Wei N., Lee D., Wu S.M. devCellPy is a machine learning-enabled pipeline for automated annotation of complex multilayered single-cell transcriptomic data. Nat. Commun. 2022;13:5271. doi: 10.1038/s41467-022-33045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ferreira N., Kulkarni A., Agorku D., Midelashvili T., Hardt O., Legler T.J., Ströbel P., Conradi L.C., Alves F., Ramos-Gomes F., Markus M.A. OrganoIDNet: a deep learning tool for identification of therapeutic effects in PDAC organoid-PBMC co-cultures from time-resolved imaging data. Cell. Oncol. 2024 doi: 10.1007/s13402-024-00958-2. [DOI] [PubMed] [Google Scholar]
- 196.Wang X., Wu C., Zhang S., Yu P., Li L., Guo C., Li R. A novel deep learning segmentation model for organoid-based drug screening. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1080273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang S., Li L., Yu P., Wu C., Wang X., Liu M., Deng S., Guo C., Tan R. A deep learning model for drug screening and evaluation in bladder cancer organoids. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1064548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Okamoto T., Natsume Y., Doi M., Nosato H., Iwaki T., Yamanaka H., Yamamoto M., Kawachi H., Noda T., Nagayama S., Sakanashi H., Yao R. Integration of human inspection and artificial intelligence-based morphological typing of patient-derived organoids reveals interpatient heterogeneity of colorectal cancer. Cancer Sci. 2022;113(8):2693–2703. doi: 10.1111/cas.15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schultz M.B., Vera D., Sinclair D.A. Can artificial intelligence identify effective COVID‐19 therapies? EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.202012817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Deben C., De La Hoz E.C., Compte M.L., Van Schil P., Hendriks J.M.H., Lauwers P., Yogeswaran S.K., Lardon F., Pauwels P., Van Laere S., Bogaerts A., Smits E., Vanlanduit S., Lin A. OrBITS: label-free and time-lapse monitoring of patient derived organoids for advanced drug screening. Cell. Oncol. 2022;46:299–314. doi: 10.1007/s13402-022-00750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Placido D., Yuan B., Hjaltelin J.X., Zheng C., Haue A.D., Chmura P.J., Yuan C., Kim J., Umeton R., Antell G., Chowdhury A., Franz A., Brais L., Andrews E., Marks D.S., Regev A., Ayandeh S., Brophy M.T., Do N.V., Kraft P., Wolpin B.M., Rosenthal M.H., Fillmore N.R., Brunak S., Sander C. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 2023;29:1113–1122. doi: 10.1038/s41591-023-02332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kim E., Choi S., Kang B., Kong J., Kim Y., Yoon W.H., Lee H.R., Kim S., Kim H.M., Lee H., Yang C., Lee Y.J., Kang M., Roh T.Y., Jung S., Kim S., Ku J.H., Shin K. Creation of bladder assembloids mimicking tissue regeneration and cancer. Nature. 2020;588:664–669. doi: 10.1038/s41586-020-3034-x. [DOI] [PubMed] [Google Scholar]
- 203.Feng W., Schriever H., Jiang S., Bais A., Wu H., Kostka D., Li G. Computational profiling of hiPSC-derived heart organoids reveals chamber defects associated with NKX2-5 deficiency. Commun. Biol. 2022;5:399. doi: 10.1038/s42003-022-03346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kawasaki K., Toshimitsu K., Matano M., Fujita M., Fujii M., Togasaki K., Ebisudani T., Shimokawa M., Takano A., Takahashi S., Ohta Y., Nanki K., Igarashi R., Ishimaru K., Ishida H., Sukawa Y., Sugimoto S., Saito Y., Maejima K., Sasagawa S., Lee H., Kim H.G., Ha K., Hamamoto J., Fukunaga K., Maekawa A., Tanabe M., Ishihara S., Hamamoto Y., Yasuda H., Sekine S., Kudo A., Kitagawa Y., Kanai T., Nakagawa H., Sato T. An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell. 2020;183:1420–1435 e21. doi: 10.1016/j.cell.2020.10.023. [DOI] [PubMed] [Google Scholar]
- 205.Ramos Zapatero M., Tong A., Opzoomer J.W., O'Sullivan R., Cardoso Rodriguez F., Sufi J., Vlckova P., Nattress C., Qin X., Claus J., Hochhauser D., Krishnaswamy S., Tape C.J. Trellis tree-based analysis reveals stromal regulation of patient-derived organoid drug responses. Cell. 2023;186:5606–5619 e24. doi: 10.1016/j.cell.2023.11.005. [DOI] [PubMed] [Google Scholar]
- 206.Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S., Liu D.A., Qian X., Petrov D., Lucas T., Chen H.I., Dorsey J.F., Christian K.M., Binder Z.A., Nasrallah M., Brem S., O'Rourke D.M., Ming G.L., Song H. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204 e22. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H., Korving J., van Boxtel R., Duarte A.A., Lelieveld D., van Hoeck A., Ernst R.F., Blokzijl F., Nijman I.J., Hoogstraat M., van de Ven M., Egan D.A., Zinzalla V., Moll J., Boj S.F., Voest E.E., Wessels L., van Diest P.J., Rottenberg S., Vries R.G.J., Cuppen E., Clevers H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386 e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 208.Fillioux L., Gontran E., Cartry J., Mathieu J.R.R., Bedja S., Boilève A., Cournède P.H., Jaulin F., Christodoulidis S., Vakalopoulou M. Spatio-temporal analysis of patient-derived organoid videos using deep learning for the prediction of drug efficacy. Ieee Int Conf Comp V. 2023:3932–3941. [Google Scholar]
- 209.Sahoo D., Swanson L., Sayed I.M., Katkar G.D., Ibeawuchi S.R., Mittal Y., Pranadinata R.F., Tindle C., Fuller M., Stec D.L., Chang J.T., Sandborn W.J., Das S., Ghosh P. Artificial intelligence guided discovery of a barrier-protective therapy in inflammatory bowel disease. Nat. Commun. 2021;12:4246. doi: 10.1038/s41467-021-24470-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Julkunen H., Cichonska A., Gautam P., Szedmak S., Douat J., Pahikkala T., Aittokallio T., Rousu J. Leveraging multi-way interactions for systematic prediction of pre-clinical drug combination effects. Nat. Commun. 2020;11:6136. doi: 10.1038/s41467-020-19950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Le Compte M., De La Hoz E.C., Peeters S., Fortes F.R., Hermans C., Domen A., Smits E., Lardon F., Vandamme T., Lin A., Vanlanduit S., Roeyen G., Van Laere S., Prenen H., Peeters M., Deben C. Single-organoid analysis reveals clinically relevant treatment-resistant and invasive subclones in pancreatic cancer. npj Precis. Oncol. 2023;7:128. doi: 10.1038/s41698-023-00480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yang H., Cheng J., Zhuang H., Xu H., Wang Y., Zhang T., Yang Y., Qian H., Lu Y., Han F., Cao L., Yang N., Liu R., Yang X., Zhang J., Wu J., Zhang N. Pharmacogenomic profiling of intra-tumor heterogeneity using a large organoid biobank of liver cancer. Cancer Cell. 2024;42:535–551 e8. doi: 10.1016/j.ccell.2024.03.004. [DOI] [PubMed] [Google Scholar]
- 213.Tavana B., Chen A. Determination of drugs in clinical trials: current status and outlook. Sensors. 2022;22:1592. doi: 10.3390/s22041592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ramzy G.M., Norkin M., Koessler T., Voirol L., Tihy M., Hany D., McKee T., Ris F., Buchs N., Docquier M., Toso C., Rubbia-Brandt L., Bakalli G., Guerrier S., Huelsken J., Nowak-Sliwinska P. Platform combining statistical modeling and patient-derived organoids to facilitate personalized treatment of colorectal carcinoma. J. Exp. Clin. Cancer Res. 2023;42:79. doi: 10.1186/s13046-023-02650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Foo M.A., You M., Chan S.L., Sethi G., Bonney G.K., Yong W.P., Chow E.K., Fong E.L.S., Wang L., Goh B.C. Clinical translation of patient-derived tumour organoids- bottlenecks and strategies. Biomark. Res. 2022;10:10. doi: 10.1186/s40364-022-00356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mao Y., Zhang L. Optimization of the medical service consultation system based on the artificial intelligence of the internet of things. IEEE Access. 2021;9:98261–98274. [Google Scholar]
- 217.Tyler N.S., Mosquera-Lopez C.M., Wilson L.M., Dodier R.H., Branigan D.L., Gabo V.B., Guillot F.H., Hilts W.W., El Youssef J., Castle J.R., Jacobs P.G. An artificial intelligence decision support system for the management of type 1 diabetes. Nat. Metab. 2020;2:612–619. doi: 10.1038/s42255-020-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vermeulen C., Pages-Gallego M., Kester L., Kranendonk M.E.G., Wesseling P., Verburg N., de Witt Hamer P., Kooi E.J., Dankmeijer L., van der Lugt J., van Baarsen K., Hoving E.W., Tops B.B.J., de Ridder J. Ultra-fast deep-learned CNS tumour classification during surgery. Nature. 2023;622:842–849. doi: 10.1038/s41586-023-06615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Moon I., LoPiccolo J., Baca S.C., Sholl L.M., Kehl K.L., Hassett M.J., Liu D., Schrag D., Gusev A. Machine learning for genetics-based classification and treatment response prediction in cancer of unknown primary. Nat. Med. 2023;29:2057–2067. doi: 10.1038/s41591-023-02482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Makrygianni E.A., Chrousos G.P. From brain organoids to networking assembloids: implications for neuroendocrinology and stress medicine. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.621970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Pasca S.P. Assembling human brain organoids. Science. 2019;363:126–127. doi: 10.1126/science.aau5729. [DOI] [PubMed] [Google Scholar]
- 222.Ozaki H., Suga H., Arima H. Hypothalamic-pituitary organoid generation through the recapitulation of organogenesis. Dev. Growth Differ. 2021;63:154–165. doi: 10.1111/dgd.12719. [DOI] [PubMed] [Google Scholar]
- 223.Song T., Kong B., Liu R., Luo Y., Wang Y., Zhao Y. Bioengineering approaches for the pancreatic tumor organoids research and application. Adv. Healthcare Mater. 2024;13 doi: 10.1002/adhm.202300984. [DOI] [PubMed] [Google Scholar]
- 224.Fujii M., Clevers H., Sato T. Modeling human digestive diseases with CRISPR-cas9-modified organoids. Gastroenterology. 2019;156:562–576. doi: 10.1053/j.gastro.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 225.Alonso A., Siracuse J.J. Protecting patient safety and privacy in the era of artificial intelligence. Semin. Vasc. Surg. 2023;36:426–429. doi: 10.1053/j.semvascsurg.2023.06.002. [DOI] [PubMed] [Google Scholar]
- 226.Choudhury D., Ashok A., Naing M.W. Commercialization of organoids. Trends Mol. Med. 2020;26:245–249. doi: 10.1016/j.molmed.2019.12.002. [DOI] [PubMed] [Google Scholar]