Abstract
The virion host shutoff protein (vhs) of herpes simplex virus (HSV) triggers global shutoff of host protein synthesis and accelerated mRNA turnover during virus infection and induces endoribonucleolytic cleavage of exogenous RNA substrates when it is produced in a rabbit reticulocyte (RRL) in vitro translation system. Although vhs induces RNA turnover in the absence of other HSV gene products, it is not yet known whether cellular factors are required for its activity. As one approach to addressing this question, we expressed vhs in the budding yeast Saccharomyces cerevisiae. Expression of vhs inhibited colony formation, and the severity of this effect varied with the carbon source. The biological relevance of this effect was assessed by examining the activity of five mutant forms of vhs bearing previously characterized in-frame linker insertions. The results indicated a complete concordance between the growth inhibition phenotype in yeast and mammalian host cell shutoff. Despite these results, expression of vhs did not trigger global mRNA turnover in vivo, and cell extracts of yeast expressing vhs displayed little if any vhs-dependent endoribonuclease activity. However, activity was readily detected when such extracts were mixed with RRL. These data suggest that the vhs-dependent endoribonuclease requires one or more mammalian macromolecular factors for efficient activity.
Herpes simplex virus (HSV) is a large enveloped DNA virus that replicates in the nuclei of infected mammalian cells. During lytic infection, more than 80 genes are expressed in the order of immediately early, early, and late through execution of a complex genetic regulatory program. Several of the viral regulatory proteins are contained in the tegument of the HSV virion. One of the best characterized of these is the virion host shutoff protein (vhs) encoded by HSV gene UL41. vhs triggers early shutoff of cellular protein synthesis, disruption of polysomes and rapid degradation of preexisting mRNAs (9, 10, 12, 13, 24, 25, 31, 32, 35, 46). Three lines of evidence indicate that the vhs protein is both necessary and sufficient for early host shutoff. First, several mutations that lead to a vhs-deficient phenotype have been mapped to the UL41 locus, and targeted disruption of the UL41 gene eliminates early shutoff (11, 36, 41, 43). Second, viral recombinants in which the UL41 gene of HSV type 1 (HSV-1) has been replaced by the corresponding gene from HSV-2 display the more robust shutoff phenotype characteristic of HSV-2 (14). Third, vhs blocks reporter gene expression when it is produced as the only viral protein in transiently transfected mammalian cells (21, 33).
In addition to triggering degradation of cellular mRNAs, vhs also significantly destabilizes HSV mRNAs belonging to all three temporal classes. This effect is believed to sharpen the transitions between the successive phases of viral protein synthesis by tightly coupling changes in the rate of mRNA synthesis to altered mRNA levels (12, 24, 25, 31, 32, 35, 46). Although vhs significantly destabilizes viral mRNAs, the vhs activity delivered by the infecting virion is partially dampened by a newly synthesized viral protein, allowing viral mRNAs to accumulate after host mRNAs have been degraded (12). Two lines of evidence suggest that the virion transactivator VP16 serves this negative regulatory role. First, vhs binds directly to VP16 (42). Second, VP16 null mutants undergo vhs-induced termination of viral protein synthesis at intermediate times post infection, and this effect is inhibited by VP16 supplied in trans (26).
Although the mechanism of action of vhs has yet to be precisely defined, the currently available evidence strongly suggests that vhs is either itself an RNase or else a subunit of an RNase that also includes one or more cellular subunits. Extracts of HSV-infected cells and partially purified virions contain a vhs-dependent RNase activity (22, 23, 44, 48) that is inhibited by anti-vhs antibodies (48). In addition, vhs induces endoribonucleolytic cleavage of a variety of reporter mRNAs when it is expressed as the only HSV protein in a rabbit reticulocyte lysate (RRL) expression system (7, 8, 48). Moreover, vhs displays weak but significant amino acid sequence similarity to the FEN-1 family of nucleases that are involved in DNA replication and repair in eukaryotes and archaebacteria (5), and recent studies have shown that human FEN-1 cleaves both RNA and DNA substrates (45). Although the foregoing data indicate that the vhs protein is a required component of the vhs-dependent endoribonuclease, they leave open the possibility that the enzyme also contains one or more cellular subunits or requires cellular factors for its activity.
As one approach to testing a possible requirement for cellular factors, we studied the effects of expressing vhs in a heterologous eukaryotic system, the budding yeast Saccharomyces cerevisiae. S. cerevisiae has been successfully utilized for the expression of other HSV proteins, including glycoprotein B (30), DNA polymerase (17), and thymidine kinase (29, 47, 49). We report here that the expression of vhs inhibits colony formation and that this effect displays the same mutational sensitivity spectrum as host shutoff in mammalian cells. However, expression of vhs did not trigger global mRNA turnover in vivo, and cell extracts of yeast expressing vhs displayed little if any vhs-dependent endoribonuclease activity. Activity was restored by adding RRL to the extracts, indicating that the vhs-dependent RNase requires one or more mammalian factors.
MATERIALS AND METHODS
Plasmids.
Two different yeast expression vectors were used to express wild-type and mutant forms of vhs in yeast: pYGAL and pYEX-BX. pYGAL bears the galactose-inducible GAL10 promoter, and pYEX-BX contains the copper-inducible CUP1 promoter. pYGAL was generated from pJAY99 by inserting a 375-bp BglII-HindIII fragment from pPGK (generously donated by John Glover, McMaster University) into the SphI-HindIII sites of pJAY99 (after making the BglII and SphI sites flush with T4 DNA polymerase). This fragment of pPGK contains the 3′ untranslated region of the yeast 3-phosphoglycerate kinase gene which bears a yeast polyadenylation signal and transcription termination sequence (19). pJAY99 was derived by Jacques Archambault in the Friesen laboratory (University of Toronto) by cloning the GAL10 promoter region (16) into the EcoRI-SmaI sites of pFL39. pFL39 is a pUC19-based low-copy-number plasmid bearing the TRP1 gene, an autonomously replicating sequence, and the centromere of yeast chromosome VI (CEN6) (3).
A yeast expression vector bearing the vhs open reading frame (ORF) under the control of the GAL10 promoter was constructed in two steps. First, a plasmid (pvhsRI) (42) containing the vhs ORF and 0.3 kb of the 3′ flanking sequences with an engineered NcoI site at the vhs initiation codon was modified by inserting a StuI linker immediately upstream of the NcoI site, generating pvhs Stu. Second, the StuI-HincII fragment of pvhs Stu bearing the vhs ORF and 3′ flanking sequences was subcloned between the SmaI-HincII sites of pYGAL, generating pYGAL vhs. The vhs1 point mutation and several vhs linker insertion mutations were transferred into pYGAL vhs by replacing the 1.7-kb NcoI-PstI fragment of pYGAL vhs with the corresponding 1.7-kb NcoI-KpnI fragment from pCMV vhs1, R27, pN138-HA, pSc243, pS344-HA, and pM384 (after making the KpnI and PstI sites flush with T4 DNA polymerase) (21), generating the plasmids pYGAL vhs1, pYGAL R27, pYGAL N138-HA, pYGAL Sc243, pYGAL S344-HA, and pYGAL M384, respectively.
pYEX-BX (Clontech) contains the yeast CUP1 promoter (pCUP1), the leu2-d gene (a LEU2 gene with a truncated, but partially functional, promoter), the 2μ origin of DNA replication, and the URA3 gene. The NcoI-HindIII fragment of pYGAL vhs bearing the vhs ORF and 3′ flanking sequences was cloned between the BamHI-SalI sites of pYEX-BX (after making all four ends flush with the Klenow fragment of DNA polymerase I), generating pYEX-BX vhs. pYEX-BX vhs1 was generated in the same way, using the NcoI-HindIII fragment of pYGAL vhs1. A pYEX-BX vector bearing a doubly tagged vhs ORF (pYEX-BX2.1vhs) was generated by inserting the NcoI-EcoRI fragment from pSP62.1vhs (7) into the BamHI-EcoRI sites of pYEX-BX (after repairing the NcoI and BamHI ends with the Klenow fragment of DNA polymerase I). pYEX-BX2.1vhs1 was generated in the same way, using the NcoI-EcoRI fragment of pSP62.1vhs1.
The vhs in vitro translation vector (pSP6vhs) and in vitro transcription vectors (pCITE-1 and pSPSR19N) encoding substrate RNAs (pCITE-1 and SRPα RNA, respectively) have been described previously (7, 8).
Bacterial strains and growth media.
Plasmids were maintained and amplified in two Escherichia coli strains. Plasmids derived from pYGAL were maintained and amplified in strain DH5α (F− endA1 hsdR17 [rk− mk+] supE44 thi-1 λ recA gyrA96 relA1 Δ[argF-laczya] U169 θ80 lacZΔM15) (18), while plasmids derived from pYEX-BX were maintained and amplified in strain HB101 (F− Δ[gpt-proA]62 leuB6 supE44 ara-14 galK2 lacY1 Δ[mcrC-mrr] rpsL20 [Strr] xyl-5 mtl-1 recA13) (28). Both strains were cultured at 37°C in Luria-Bertani medium (LB; 1.0% [wt/vol] Bacto Tryptone, 0.5% [wt/vol] yeast extract, 1.0% [wt/vol] NaCl) in a shaker incubator set at 250 rpm. Derivatives transformed with recombinant plasmids were isolated on LB agar plates (LB with 1.5% [wt/vol] agar) containing 100 μg of ampicillin per ml.
Yeast strains.
Two different yeast strains were used: YPH500 (MATα ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 lec2-Δ1) (40), generously supplied by John Glover (McMaster University), and W303-1A (MATa SUC3 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) (27), generously supplied by Ivan Sadowski (University of British Columbia). YPH500 was used as the host for plasmids derived from the pYGAL vector, while W303-1A was used as the host for plasmids derived from pYEX-BX.
Yeast media and growth conditions.
YPH500 and W303-1A were cultured and maintained in YEPD (2% Bacto Peptone, 1% yeast extract, 2% dextrose). Yeast strains transformed with plasmids were cultured in YNBD or YNBG (0.67% yeast nitrogen base without amino acids, containing 2% dextrose or 2% galactose, respectively) supplemented with the appropriate nutrients for plasmid selection (uracil, l-lycine, adenine, l-histidine, and l-leucine in the case of pYGAL derivatives and strain YPH500; adenine, l-histidine, and tryptophan in the case of pYEX-BX derivatives and strain W303-1A). All yeast strains were cultured at 30°C in a shaker incubator set at 200 rpm.
Yeast transformation.
YPH500 was transformed using either of the following methods. The first is a modification of a protocol described elsewhere (1). Briefly, yeast cells cultured to log phase in YEPD were washed three times with double-distilled water (ddH2O) and once with ice-cold 1.0 M sorbitol (Sigma). The final cell pellet was resuspended into ice-cold 1.0 M sorbitol and placed on ice until use. Then, 1 μg of transforming plasmid DNA was added to 20 μl of the yeast cell suspension. The cells were electroporated at 250 V with 4 KΩ resistance in a Cell-Porator (Gibco-BRL). The electroporated cells were immediately removed and suspended in 100 μl of ice-cold 1.0 M sorbitol, and the entire cell suspension was spread onto selective YNBD plates. Alternatively, cells were transformed using a modification of the lithium acetate (LiAc) procedure (6). Briefly, 1.0 ml of log-phase yeast cells in YEPD were pelleted by centrifugation. Then, 1 μg of transforming plasmid DNA was mixed with the cell pellet. A total of 500 μl of PLATE medium (40% [wt/vol] PEG 4000; 100 mM LiAc; 10 mM Tris, pH 7.5; 1 mM EDTA) was added to the mixture, and the cells were resuspended by pipetting up and down several times. The cell suspension was incubated at room temperature without mixing. After incubation for at least 1 day, 50 μl of the mixture was taken from the bottom of the tube and mixed with 50 μl of ddH2O, and the entire cell suspension was spread onto selective YNBD plates.
W303-1A was transformed using a slightly different LiAc method (6), with the following modification. Briefly, log-phase cells were collected by centrifugation, washed with 1 ml of TE-LiAc, which was made fresh from 10× filter-sterilized stocks (10× TE [0.1 M Tris-HCl, 0.01 M EDTA; pH 7.5], 10× LiAc [1 M LiAc pH 7.5 adjusted with diluted acetic acid]) and resuspended at 2 × 109 cells/ml in 1× TE-LiAc. Then, 50 μl of the cells were mixed with 1 μg of transforming DNA, 50 μg of single-stranded salmon sperm carrier DNA, and 300 μl of 40% PEG 4000 solution (40% PEG 4000–1× TE–1× LiAc, which was made fresh from 50% PEG 4000, 10× TE, and 10× LiAc). The mixture was mixed thoroughly and then incubated at 30°C for 30 min with agitation. After incubation, the mixture was heat shocked for 15 min at 42°C. Cells were collected and resuspended in 1 ml of 1× TE and then plated onto selective YNBD plates.
Preparation of cell extracts for Western blot analysis.
Yeast cultures were grown to an optical density at 600 nm (OD600) of 2 to 3, and then split into two portions. One culture was induced with 0.5 mM CuSO4 for 5 h, while the other was left untreated. Cells were collected, washed once with ice-cold ddH2O, and then lysed by boiling them 10 min in 2 volumes of 2× sample buffer (125 mM Tris, pH 6.8; 600 mM 2-mercaptoethanol; 6% sodium dodecyl sulfate, 20% glycerol; 0.005% bromophenol blue). The lysate was clarified by centrifugation for 10 min and stored at −70°C until use.
Extracts of HSV-1-infected Vero cells were prepared as described elsewhere (21), with the following modification. Briefly, Vero cells in 35-mm-diameter dishes were infected with the mutant virus HSV-1 Pvhs N138-HA at 10 PFU per cell. After infection for 12 h, the cells were lysed by boiling for 10 min in 2 volumes of 2× sample buffer. The lysate was stored at −70°C until use. HSV-1 Pvhs N138-HA encodes a mutant version of vhs bearing the hemagglutinin (HA) tag following residue 138 (21).
Western blot analysis.
Western blot analysis was conducted as described elsewhere (21). Briefly, samples were separated by electrophoresis through a sodium dodecyl sulfate–9% polyacrylamide gel and then transferred to a nitrocellulose filter. The protein was detected with a 1/500 dilution of rabbit antiserum against the HA epitope (Boehringer Mannheim) and a 1/500 dilution of sheep anti-rabbit immunoglobulin conjugated with horseradish peroxidase (Boehringer Mannheim). Bound secondary antibody was visualized by using Renaissance Chemiluminescence Reagent (NEN) according to the manufacturer's protocol.
Effects of vhs on mRNA stability in vivo.
Cultures of W303 pYEX-BX and W303 pYEX-BXvhs were grown to an OD600 of 0.6 in YNBD, and a 10-ml aliquot was withdrawn for RNA extraction (t = 0). The remainder of the culture was induced with the addition of 0.5 mM CuSO4 for 30 min. Another aliquot was then removed for RNA extraction, and transcription was inhibited in the remainder of the culture using 1,10-phenanthroline (Sigma) at a concentration of 100 μg/ml (34). Subsequent aliquots were removed every 30 min over a 3-h time course.
Yeast cells were harvested by centrifugation, and RNA was extracted using the RNEasy Mini Protocol for the isolation of total RNA from yeast (Qiagen). The standard enzymatic lysis version of the protocol was used, which initially generates spheroplasts using zymolyase (Seikagaku Corp.). RNA was quantitated, and 10 μg was resolved on a 1.2% agarose formaldehyde gel. RNA was blotted onto a Genescreen membrane (NEN), and the blot was hybridized to a radiolabeled probe for the S. cerevisiae PDA1 gene (encoding the E1α subunit of pyruvate dehydrogenase). Hybridization was performed in ExpressHyb (Clontech) according to the manufacturer's protocol. Visualization and quantitation of the products were performed using a STORM PhosphorImager and ImageQuant software (Molecular Dynamics).
Preparation of yeast extracts for vhs activity assay.
Frozen cells were prepared as described elsewhere (39). Briefly, cells grown in YNBG to an OD600 of ∼3 were induced with 0.15 mM CuSO4 for 5 h and then harvested by centrifugation at 4,000 rpm at 4°C for 4 min in a Beckman J-Lite rotor. The wet weight of the cells was measured, and then the cells were washed sequentially in ice-cold ddH2O, 1.3 volumes of extraction buffer (100 mM HEPES-KOH, pH 7.9; 245 mM KCl; 5 mM EGTA; 1 mM EDTA; and freshly made 2.5 mM dithiothreitol [DTT]), and 1.3 volumes of extraction buffer supplemented with protease inhibitors (0.2 mM phenylmethylsulfonyl fluoride, 10 mM benzamidine hydrochloride, and 3.5 μg of pepstatin A, 5 μg of leupeptin, and 10 μg of aprotinin per ml). The cell pellet was loaded into a syringe, squeezed into liquid nitrogen, and then stored at −70°C.
Extracts were prepared by the method of Schultz et al. (38). Briefly, ∼3 g of frozen cells were used to prepare the extract in an unmodified home coffee mill. The mill was chilled by covering the blades with dry ice and running it until the dry ice was a powder. Frozen cells were added and processed in the cold room for 5 min. The powder was transferred to a cold beaker, and 1.3 volumes of extraction buffer with protease inhibitors was added. The powder was thawed, mixed, and then centrifuged at 100,000 × g for 2 h. The supernatant (minus the lipid pellicle) was collected by tube puncture and dialyzed overnight against 50 volumes of vhs assay buffer (1.6 mM Tris-acetate, 80 mM potassium acetate, 2.0 mM magnesium acetate, 0.1 mM DTT, 0.25 mM ATP, and 20 U of RNase inhibitor [Sigma]; adjust to pH 7.8 with acetic acid). The extracts were stored at −70°C. Protein concentration in the yeast extract was determined by the method of Bradford (4).
In vitro transcription and RNA labeling.
vhs and vhs1 mRNAs destined for in vitro translation were produced according to a procedure that has been described elsewhere (7). Uncapped, internally labeled reporter RNAs were generated in a similar way, except that the cap primer was omitted and the reaction period was shortened to 30 min. The pCITE-1 reporter RNA transcribed from pCITE-1 was generated using T7 RNA polymerase and Eco47III-linearized plasmid DNA as a template to yield a runoff transcript of ca. 2.3 kb (8). The SRPα reporter mRNA was generated using SP6 RNA polymerase and EcoRV-linearized pSPSR19N as a template to yield a runoff transcript of 2.4 kb (7).
In vitro translation.
In vitro translation of vhs using RRL has been described previously (7, 8).
vhs activity assay.
Reporter mRNAs were added to RRL controls (blank RRL and RRL containing the pretranslated vhs), yeast extracts, or yeast extracts mixed with blank RRL. The amount of yeast extract used for each reaction was based on the total protein concentration, and ∼100 μg of total protein was used in each reaction. All reactions were adjusted to the same final volume using the vhs assay buffer. Where indicated, an equal volume of blank RRL was added to the yeast extracts. Reactions were incubated at 30°C. At various times after addition of the reporter RNA, aliquots were obtained and RNA was recovered using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Briefly, samples were added to a mixture of RNase-free H2O and 100 mM EDTA to bring the final EDTA concentration to 10 mM and the final volume of the mixture to 100 μl. To this mixture, 350 μl of buffer RLT (with 10 μl of β-mercaptoethanol/ml of RLT; Qiagen) and 250 μl of ethanol (95%) were added. The mixture was loaded onto an RNeasy Mini Spin column and centrifuged for 15 s at ≥10,000 rpm. The column was washed twice with wash buffer RPE (Qiagen), and then the RNA was eluted with RNase-free water. The eluted RNA was precipitated with 95% ethanol and a 1/10 volume of 3 M sodium acetate. The RNA pellet was washed with 70% ethanol, dried, and then resuspended in RNase-free water.
Agarose gel electrophoresis and Northern blot analysis.
The details of electrophoresis and Northern blot analysis have been described elsewhere (7).
Markers.
RNA markers were generated as previously described (7). Briefly, pSPSR19N DNA was linearized with EcoRV, PvuII, SmaI, and NruI. These linearized DNA templates were used in in vitro transcription to produce runoff transcripts of 2,422, 1,628, 800, and 429 nucleotides (nt), respectively.
RESULTS
vhs inhibits colony formation in S. cerevisiae.
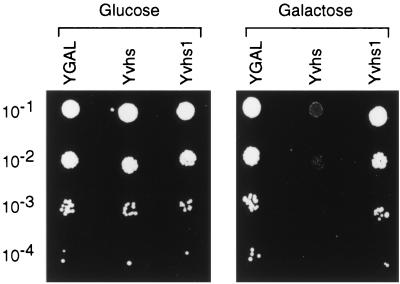
As reviewed in the introduction, previous data strongly suggest that vhs is an integral component of the vhs-dependent endoribonuclease but leave open the possibility that one or more mammalian subunits or cofactors are required for its activity. As one approach to testing the possible involvement of cellular cofactors, we explored the consequences of expressing vhs in the budding yeast S. cerevisiae. Strain YPH500 was transformed with a plasmid bearing the vhs ORF under the control of the galactose-inducible GAL10 promoter (Yvhs); as controls, we also derived strains bearing the empty expression vector (YGAL) and a vector specifying the inactive vhs1 mutant form of vhs (Yvhs1). Strains were grown to saturation in selective glucose medium, and then serial dilutions were spotted onto minimal plates containing either galactose or glucose as the carbon source (Fig. 1). As expected, the control YGAL strain harboring the empty expression vector formed equivalent numbers of colonies on glucose and galactose plates. In contrast, the Yvhs strain displayed a large reduction in the number of visible colonies when cells were plated in the presence of galactose; only a few small colonies were observed at the 10−2 dilution, and no colonies were observed at dilutions of >10−2 (Fig. 1, see also Fig. 2). The vector encoding the vhs1 mutant form of vhs had little if any effect (Yvhs1). These results indicate that induction of vhs expression from the galactose-inducible GAL10 promoter strongly inhibits colony formation and that the vhs1 point mutation eliminates this phenotype.
FIG. 1.
Expression of vhs inhibits yeast colony formation. Strains YGAL, Yvhs, and Yvhs1 were grown to saturation in YNBD, diluted 10−1 to 10−4 in sterile ddH2O, and 2 μl of each dilution was spotted onto YNBD (glucose) and YNBG (galactose) plates. The plates were incubated at 30°C for 3 and 5 days, respectively.
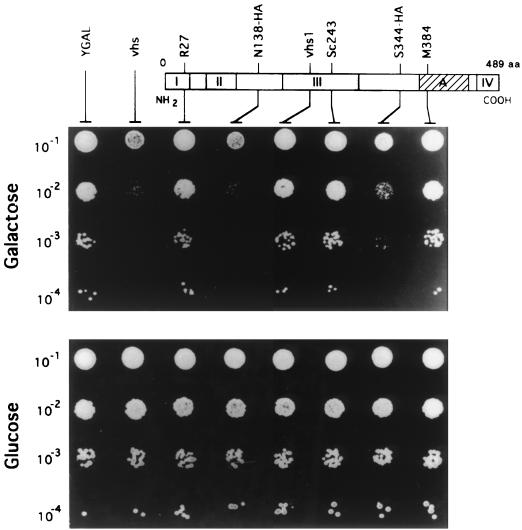
FIG. 2.
The growth inhibition phenotype requires the same regions of the vhs polypeptide as mammalian shutoff. Strains YGAL, Yvhs (vhs), YR27 (R27), YN138-HA (N138-HA), YSc243 (Sc243), YS344-HA (S344-HA), and YM384 (M384) were grown to saturation in YNBD, diluted 10−1 to 10−4 in sterile ddH2O, and 2 μl of each dilution was spotted onto YNBD (glucose) and YNBG (galactose) plates. The plates were incubated at 30°C for 3 and 5 days, respectively. The upper portion of the figure displays a linear representation of the 489-residue vhs polypeptide, with the positions of in-frame linker insertion mutations indicated. Regions conserved between the vhs proteins of alphaherpesvirus (conserved regions I, II, III, IV, and A [2, 21]) are presented as shaded and hatched boxes.
The vhs induced phenotype displays the same mutational sensitivity spectrum as host shutoff in mammalian cells.
We compared the mutational sensitivity spectrum of the growth inhibition phenotype to that of mammalian host shutoff, as an additional test of the biological relevance of the foregoing results. Previous mutational studies have indicated that two regions of the vhs polypeptide tolerate in-frame insertions, while a minimum of three regions of the protein are essential for its function (21). These results are exemplified by the phenotypes of five representative mutants: R27, Sc243, and M384 bear in-frame insertions that disrupt highly conserved regions of the vhs polypeptide and are inactive in mammalian cells; in contrast, N138-HA and S344-HA alter regions of vhs that are deleted from the vhs homologues of some alphaherpesviruses and retain full activity (21). We cloned these five mutations into the pYGAL vector and determined their effects on yeast cell growth using the dilution patch test (Fig. 2). The results revealed a complete concordance between the yeast and mammalian assay systems: expression of vhs, N138-HA, and S344-HA strongly inhibited colony formation, while the empty vector, R27, Sc243, and M384 had no effect (Fig. 2). These data provide a strong indication that the growth inhibition phenotype in yeast requires the same regions of the vhs polypeptide as does the shutoff phenotype in mammalian cells and are consistent with the hypothesis that this effect reflects a biologically relevant activity of vhs.
Carbon source-dependent inhibition of growth.
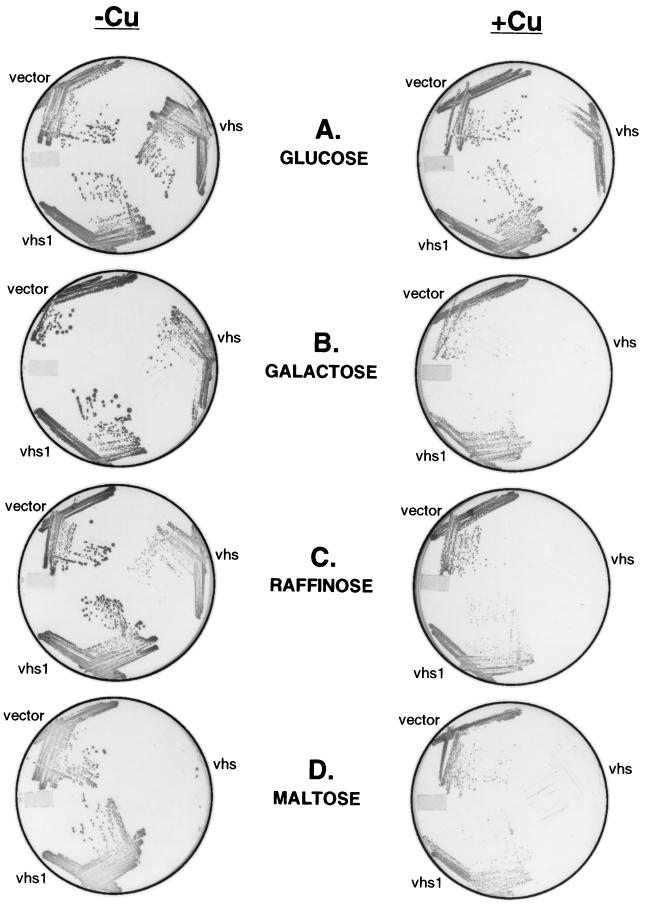
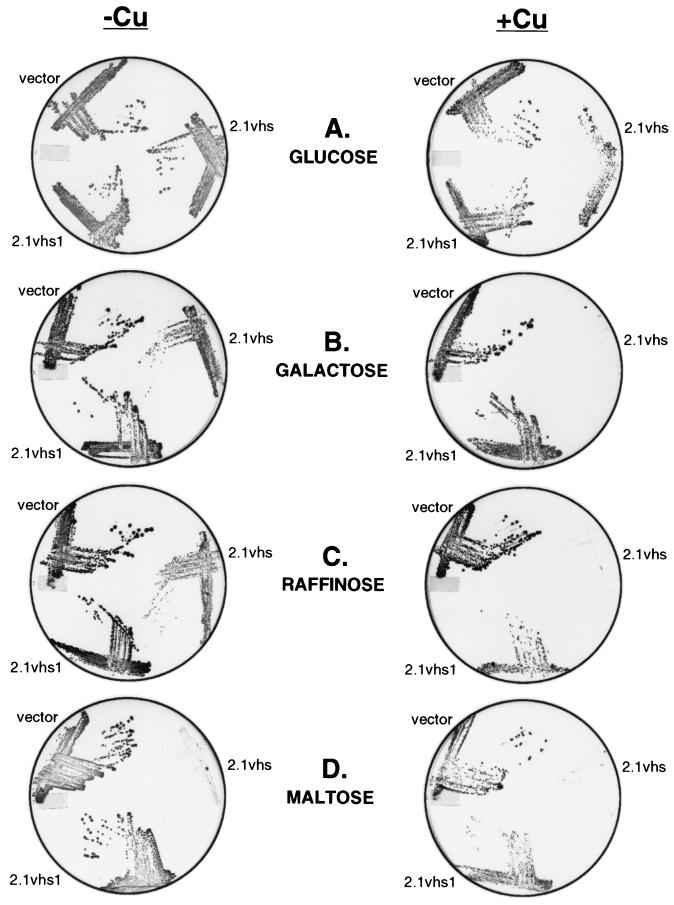
The pYGAL plasmid used in the preceeding experiments is a single-copy vector that directs accumulation of relatively low levels of vhs protein (data not shown). In addition, the GAL10 promoter is not well suited for eventual studies of the in vivo effects of vhs on protein synthesis and RNA turnover in yeast because the addition of the inducer (galactose) causes a large increase in cellular growth rate, protein synthesis, and mRNA levels. In order to increase the levels of vhs expression and reduce the global effects of the inducer on cell growth rates, we decided to use a multicopy plasmid containing the copper-inducible promoter derived from the yeast metallothionein (CUP1) gene (induction of the CUP1 promoter has been reported to have relatively little effect on cell growth or protein synthesis [15]). To this end, strain W303-1A was transformed with a plasmid bearing the vhs ORF under the control of the CUP1 promoter; the empty expression vector and a vhs1 mutant construct served as controls. Cells harboring each of these plasmids were grown to saturation in selective glucose medium; then, equal numbers of cells (based on the OD600) were spotted and streaked out on selective glucose plates containing or lacking the inducer (copper sulfate). Surprisingly, the vector encoding wild-type vhs did not prevent growth on glucose plates in the presence of copper sulfate, although fewer colonies were observed and the colony size was reduced (Fig. 3A).
FIG. 3.
Expression of vhs from the inducible CUP1 promoter inhibits colony formation. Strains W303 pYEX-BX (vector), W303 pYEX-BX vhs (vhs), and W303 pYEX-BX vhs1 (vhs1) were grown to saturation in YNBD, and then equal amounts of cells from each strain were spotted and streaked onto YNBD (glucose), YNBG (galactose), YNBR (raffinose), and YNBM (maltose) plates containing (+) or lacking (−) 0.5 mM copper sulfate (Cu). The plates were incubated at 30°C for 5 days.
Although this observation at first glance appears to conflict with the results described above (Fig. 1 and 2), the GAL10 constructs were assayed in the presence of galactose as the sole carbon source (in order to avoid catabolite repression of the GAL10 promoter), while glucose was used as the carbon source in the experiment shown in Fig. 3A. We therefore tested the effects of varying the carbon source on colony formation (Fig. 3). As shown in Fig. 3B and C, induction of vhs expression from the CUP1 promoter severely inhibited cell growth on solid medium when galactose or raffinose was used as the carbon source. Moreover, the vhs vector strongly inhibited cell growth in both the presence and the absence of copper sulfate when maltose was used as the carbon source (Fig. 3D). In all three cases, the vhs1 vector had no effect. Taken in combination, these data demonstrate that the severity of vhs-induced growth inhibition on solid medium varies markedly with the carbon source. Presumably, the failure of cells harboring the vhs vector to grow on maltose plates in the absence of inducer stems from low constitutive levels of vhs expression obtained with the CUP1 promoter (see Fig. 6).
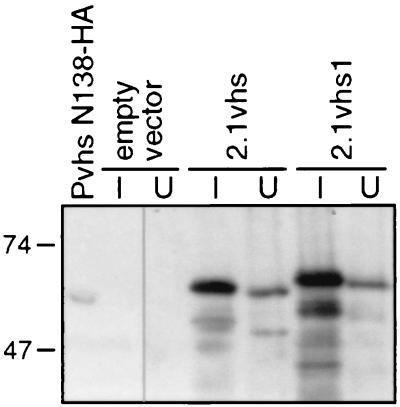
FIG. 6.
Levels of expression of 2.1vhs and 2.1vhs1 proteins. Strains W303 pYEX-BX (empty vector), W303 pYEX-BX 2.1vhs (2.1vhs), and W303 pYEX-BX 2.1vhs1 (2.1vhs1) were grown to saturation in YNBG, and the cultures were then split into two portions. One culture was induced with 0.5 mM CuSO4 for 5 h at 30°C (lanes I), while the other was left untreated (lanes U). Cell extracts were then analyzed for vhs expression by Western blot analysis using a monoclonal antibody directed against the HA epitope. A lysate of Vero cells infected for 12 h with HSV-1 Pvhs N138-HA (lane Pvhs N138-HA) at a multiplicity of infection (MOI) of 10 was included as a positive control.
Although the induction of vhs expression prevented the formation of visible colonies within 5 days on galactose plates, the expected number of colonies were observed when the plates were incubated for 10 days (data not shown). Thus, expression of vhs reduces the growth rate of yeast but is not lethal.
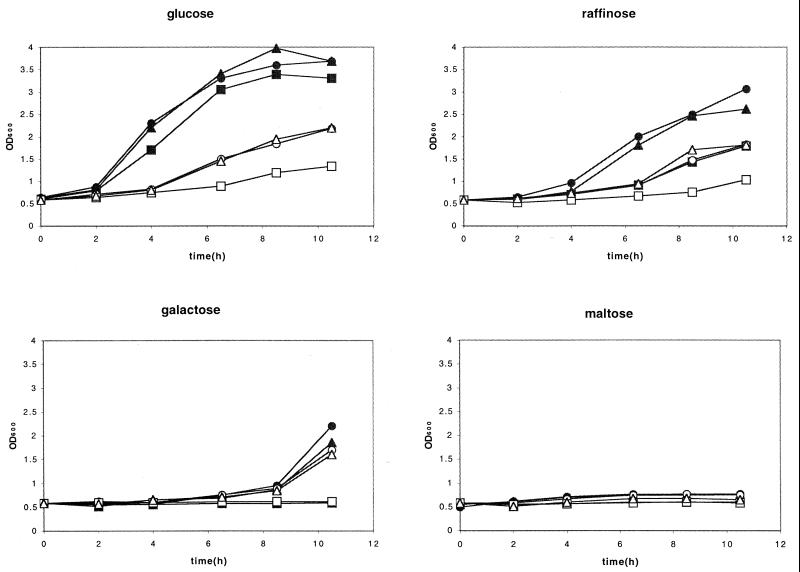
We also observed carbon source-dependent variation of the effect of vhs on growth in liquid medium (Fig. 4), but the pattern was not the same as that observed on solid medium (Fig. 3). In this experiment, cells were grown in YNBD and then washed and resuspended in medium containing glucose, raffinose, galactose, and maltose as the carbon source. The OD of the cultures was then monitored over time. Addition of the inducer significantly reduced the growth of cells harboring empty vector or the vhs1 expression plasmid in glucose or raffinose medium, and expression of wild-type vhs had a further inhibitory effect in the presence of these carbon sources. Thus, the marked difference in phenotype between glucose and raffinose that was observed on solid medium did not occur in liquid culture. In addition, cells harboring the vhs vector failed to grow in the presence of galactose in either the presence or absence of inducer over the time course of the experiment in liquid culture, and no growth was observed on maltose under any conditions. Taken in combination, these data indicate that the severity of the growth inhibition phenotype produced by vhs is highly dependent on culture conditions, varying both with carbon source and liquid versus solid medium.
FIG. 4.
Carbon source-dependent variation in the vhs-induced phenotype in liquid cultures. Strains W303 pYEX-BX (vector), W303 pYEX-BX vhs (vhs), and W303 pYEX-BX vhs1 (vhs1) growing in YNBD (glucose) were pelleted, washed in water, and resuspended in YNB containing the indicated carbon sources, in the presence or absence of 0.5 mM CuSO4. The OD600 of the cultures was then monitored over time. Closed symbols, not induced; open symbols, induced. Circles, empty vector; squares, vhs expression vector; triangles, vhs1 expression vector.
The foregoing data demonstrate that vhs strongly inhibits the growth of yeast under certain conditions and that this effect is eliminated by mutations that inactivate shutoff function in mammalian cells. The failure of certain mutant forms of vhs to inhibit the growth of yeast might stem from the loss of one or more functions of the vhs protein. Alternatively, the mutations might reduce the accumulation of the mutant protein. In order to distinguish between these possibilities in the case of the vhs1 mutant protein, we examined the levels of accumulation of epitope-tagged protein by Western blot analysis. The 2.1 version of vhs (7) bears eight tandem histidine residues inserted after residue 138 and an influenza virus HA epitope following residue 344 and retains full activity in the RRL in vitro system. We placed wild-type and vhs1 mutant versions of the 2.1vhs ORF under the control of the CUP1 promoter and then tested them for effects on cell growth and accumulation of vhs protein. As shown in Fig. 5, the 2.1 versions of wild-type and mutant vhs produced the same effects on cell growth on solid medium as the corresponding untagged proteins and were therefore suitable for the experiment. Cells harboring the 2.1vhs and 2.1vhs1 expression vectors were grown to saturation in selective galactose medium, induced with copper sulfate, and then harvested 5 h later. Extracts were then examined by Western blot analysis using an anti-HA monoclonal antibody (Fig. 6). As a control, we also examined an extract of Vero cells infected with HSV-1 Pvhs N138-HA (21), which encodes a mutant version of vhs bearing the HA tag following residue 138. The 2.1vhs and 2.1vhs1 vectors gave rise to the expected band of ca. 58 kDa, while cells containing empty vector showed no signal. Significant signals were observed for both wild-type and vhs1 mutant protein in the uninduced culture, but in both cases the level was markedly higher after induction. The vhs1 mutant protein accumulated to somewhat higher levels than the wild-type vhs after induction, demonstrating that the vhs1 mutation does not prevent protein accumulation. This observation in turn suggests that the failure of the vhs1 mutant protein to inhibit the growth of yeast stems from the loss of one or more functions of vhs.
FIG. 5.
Expression of epitope-tagged vhs inhibits colony formation. Strains W303 pYEX-BX (vector), W303 pYEX-BX 2.1vhs (2.1vhs), and W303 pYEX-BX 2.1vhs1 (2.1vhs1) were grown to saturation in YNBD, and then equal amounts of cells from each strain were spotted and streaked onto YNBD (glucose), YNBG (galactose), YNBR (raffinose), and YNBM (maltose) plates containing (+) or lacking (−) 0.5 mM copper sulfate (Cu). The plates were incubated at 30°C for 5 days.
Expression of vhs does not trigger global mRNA turnover in yeast.
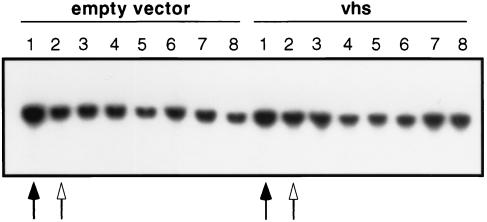
Vhs triggers global mRNA turnover in HSV-infected mammalian cells. To determine if this is also the case in yeast, we asked if the induction of vhs expression alters the in vivo stability of yeast PDA1 mRNA, encoding the E1α subunit of pyruvate dehydrogenase (Fig. 7). Cultures harboring the vhs expression vector and empty vector (growing in YNBD) were treated with 0.5 mM CuSO4 to induce vhs expression for 30 min, and then new transcription was inhibited by adding 1,10-phenanthroline to a final concentration of 100 μg/ml (34). The 30-min induction period was sufficient for full induction of vhs expression (data not shown). Aliquots of the culture were then withdrawn at 30-min intervals, and the levels of PDA1 mRNA were determined by Northern blot hybridization. PDA1 RNA levels dropped approximately threefold during the induction period in both cultures; however, RNA levels remained essentially constant after imposition of the transcriptional blockade. Thus, perhaps surprisingly, the induction of vhs expression did not detectably alter the stability of PDA1 RNA. We also obtained entirely analogous results when the Northern blots were probed for ACT1 mRNA encoding actin (data not shown).
FIG. 7.
Effect of vhs expression on the in vivo stability of PDA1 mRNA. Strains W303 pYEX-BX (empty vector) and W303 pYEX-BX vhs (vhs) were grown to an OD600 of 0.6 in YNBD, and a 10-ml aliquot was withdrawn for RNA extraction (solid arrow). The remainder of the culture was induced with the addition of 0.5 mM CuSO4 for 30 min. Another aliquot was then removed for RNA extraction (open arrow), and transcription was inhibited in the remainder of the culture with 100 μg of phenanthroline per ml. Aliquots were then removed every 30 min over a 3-h time course. Total RNA was analyzed for PDA1 mRNA levels by Northern blot hybridization. Lane 1, RNA extracted just before induction with CuSO4; lane 2, RNA extracted just before addition of phenanthroline; lanes 3, 4, 5, 6, 7, and 8, RNA extracted at 30, 60, 90, 120, 180, and 210 min after addition of phenanthroline, respectively.
Extracts of yeast expressing vhs do not display vhs-dependent endoribonuclease activity.
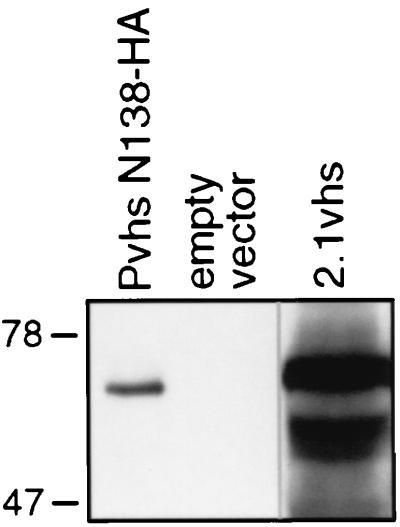
We next sought to determine whether cell extracts prepared from yeast expressing vhs display endoribonuclease activity comparable to that previously observed in RRL containing pretranslated vhs (7, 8). Strains harboring the 2.1vhs expression plasmid and empty vector were induced with copper sulfate, and whole-cell extracts were prepared as described in Materials and Methods. Western blot analysis confirmed that the extract prepared from 2.1vhs-expressing cells contained readily detectable amounts of full-length vhs protein (Fig. 8).
FIG. 8.
Western blot analysis of yeast extracts. Strains W303 pYEX-BX (empty vector), and W303 pYEX-BX 2.1vhs (2.1vhs) were grown to an OD600 of 2 to 3 in YNBG and then induced with 0.15 mM CuSO4 for 5 h at 30°C. Whole-cell extracts were prepared, and the vhs protein in the extracts was detected by Western blot analysis using a monoclonal antibody directed against the HA epitope. A lysate of Vero cells infected for 12 h with HSV-1 Pvhs N138-HA (lane Pvhs N138-HA) at an MOI of 10 was included as a positive control.
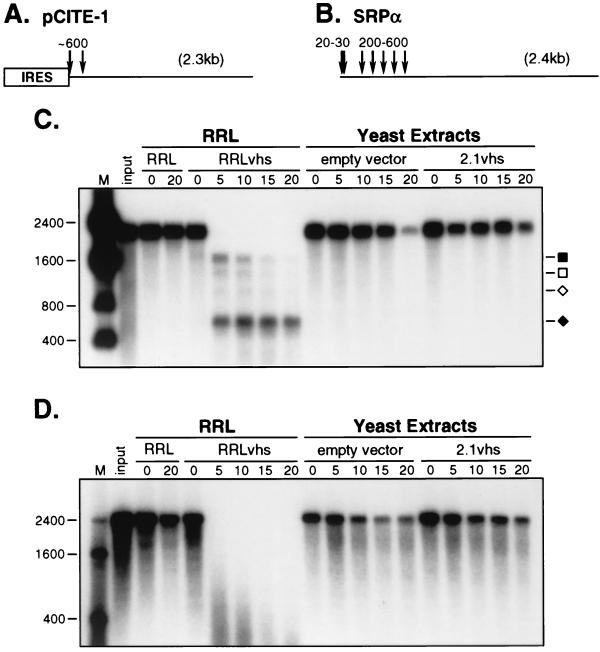
Two RNA substrates were used for the in vitro assays for vhs-dependent RNase activity: pCITE-1 RNA, which bears the internal ribosome entry site (IRES) of encephalomyocarditis virus (EMCV) at its 5′ end (Fig. 9A), and signal recognition particle α mRNA (SRPα RNA, Fig. 9B). Internally labeled substrate RNA was added to the extracts, and samples withdrawn at various times were then analyzed by agarose-formaldehyde gel electrophoresis (Fig. 9C and D). As a control, the RNA substrates were also incubated in RRL containing pretranslated vhs (RRLvhs) and blank RRL (RRL). Previous studies using the RRL assay system have shown that the vhs-dependent endoribonuclease preferentially cleaves pCITE-1 RNA immediately 3′ of the IRES, generating 5′ and 3′ products of ca. 600 and 1,800 nt, respectively (8). The 600-nt fragment is stable throughout the course of the reaction, while the 1,800-nt product is subject to further decay. We observed a similar pattern when pCITE-1 RNA was added to RRLvhs, with the exception that additional products of 1,500 and 1,000 nt were also observed (Fig. 9C). In marked contrast, extracts of yeast expressing 2.1vhs were devoid of detectable vhs-dependent RNase activity: the pCITE-1 RNA was as stable as in the extract of cells harboring empty expression vector. Moreover, no discrete vhs-induced degradation intermediates were observed. Similar results were obtained with SRPα RNA (Fig. 9D). In this case, the RRL system does not produce stable degradation products (7); rather, the RNA is initially cleaved at a cluster of sites located over the 5′ quadrant of the RNA, and the 5′ and 3′ products of these initial cleavages are subjected to rapid further decay to low-molecular-weight species (7). SRPα RNA was as stable in extracts of yeast expressing vhs as in control extracts.
FIG. 9.
Lack of endoribonuclease activity in extracts of yeast expressing 2.1vhs. Internally labeled pCITE-1 and SRPα RNAs were added to control RRL (RRL), RRL containing the pretranslated vhs (RRLvhs), and extracts of yeast containing (2.1vhs) and lacking (empty vector) 2.1vhs protein. RNA was extracted at the indicated time points, resolved on a 1% agarose–1.8% formaldehyde gel, and transferred to a GeneScreen Plus membrane, and the RNA signal was detected by autoradiography (panels C and D). (A and B) Diagrams of the pCITE-1 and SRPα RNA substrates, indicating the positions of the initial vhs-induced cleavage events in the RRL system. The EMCV IRES present on pCITE-1 is indicated. (C and D) Analysis of endoribonuclease activity on pCITE-1 and SRPα RNAs, respectively. The solid square and diamond indicate the previously described 5′ and 3′ degradation products of pCITE-1, respectively, while the open square and diamond indicate the additional 1,500- and 1,000-nt products, respectively, described in the text. The sizes of RNA markers (M) are indicated in nucleotides at the left.
Taken in combination, these data demonstrate that extracts of yeast expressing 2.1vhs display little if any vhs-dependent endoribonuclease activity. Similar results were also obtained with extracts of yeast cells expressing unmodified vhs (data not shown). Inasmuch as the yeast extracts analyzed contain far more vhs protein than the RRL reactions (data not shown), these results suggested that vhs protein produced in yeast has little or no endoribonuclease activity.
Reconstitution of endoribonuclease activity by a mammalian factor(s).
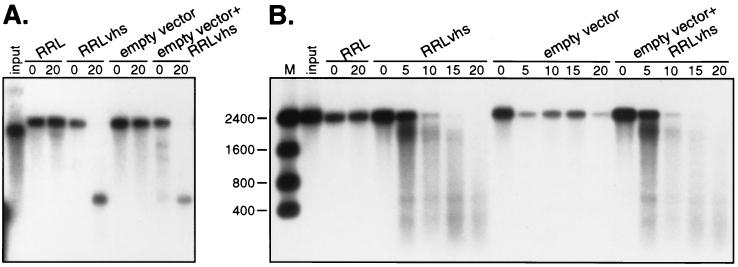
The failure to detect vhs-dependent RNase activity in extracts of yeast containing the 2.1vhs protein could be due to the presence of an inhibitor of the enzyme in the yeast extract or else reflect the absence of a required mammalian cofactor. We evaluated these possibilities in a series of mixing experiments. To test for the presence of an inhibitor, we mixed extract prepared from yeast harboring empty expression vector with an equal volume of RRL containing pretranslated vhs and then assayed for activity on pCITE-1 and SRPα RNA (Fig. 10A and B, respectively). The yeast extract had no significant effect on the vhs-dependent endoribonuclease produced in RRL, arguing that the inactivity of yeast extracts containing vhs does not stem from the presence of an inhibitor.
FIG. 10.
Yeast extract does not contain an inhibitor of the vhs-dependent RNase. Internally labeled pCITE-1 and SRPα RNAs were added to control RRL (RRL), RRL containing pretranslated vhs (RRLvhs), yeast extract from cells harboring empty vhs expression vector (empty vector), and yeast extract mixed with RRLvhs (empty vector+RRLvhs). Samples were incubated and processed as described in the legend to Fig. 9. Panels A and B show an analysis of the activity of pCITE-1 and SRPα RNAs, respectively. The sizes of RNA markers (M) are indicated in nucleotides at the left.
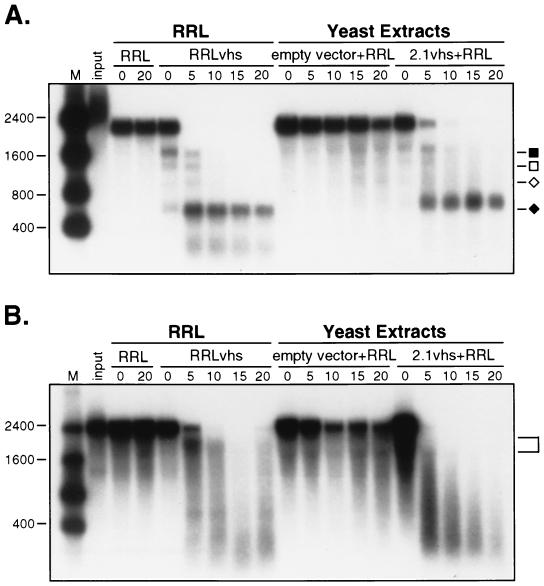
We next asked if RRL contains one or more factors capable of stimulating the activity of 2.1vhs present in yeast extracts. Blank RRL was added to extracts of control and 2.1vhs expressing yeast, and the resulting mixtures were tested for activity on pCITE-1 and SRPα RNAs (Fig. 11A and B, respectively). The extracts used in this experiment are the same as those used in the experiment depicted in Fig. 9, and these two experiments were conducted in parallel using the same reagents. Strikingly, the addition of blank RRL clearly reconstituted activity on both substrates. Moreover, the pattern of degradation intermediates observed in the reconstituted reaction was similar to that in the RRL reaction; in particular, pCITE-1 RNA gave rise to the 5′ and 3′ products characteristic of cleavage immediately 3′ to the EMCV IRES at early times, and the 5′ product was stable throughout the course of the reaction. These results indicate that RRL contains one or more factors that greatly stimulate the in vitro activity of vhs produced in yeast.
FIG. 11.
A mammalian cofactor(s) is required for reconstituting the endoribonuclease activity of the vhs protein produced in yeast. Internally labeled pCITE-1 and SRPα RNAs were added to control RRL (RRL), RRL containing pretranslated vhs (RRLvhs), yeast extract from cells harboring empty vhs expression vector mixed with blank RRL (empty vector+RRL), and yeast extract containing 2.1vhs mixed with blank RRL (2.1vhs+RRL). The yeast extract containing 2.1vhs used in this experiment is exactly the same as in Fig. 9, and these two experiments were done at the same time on the same day. Samples were incubated and processed as described in the legend to Fig. 9. Panels A and B show an analysis of the activity of pCITE-1 and SRPα RNAs, respectively. The sizes of RNA markers (M) are indicated in nucleotides at the left. Symbols in panel A are as described in the legend to Fig. 9. The bracket in panel B indicates the ca. 1,800-nt early degradation intermediate.
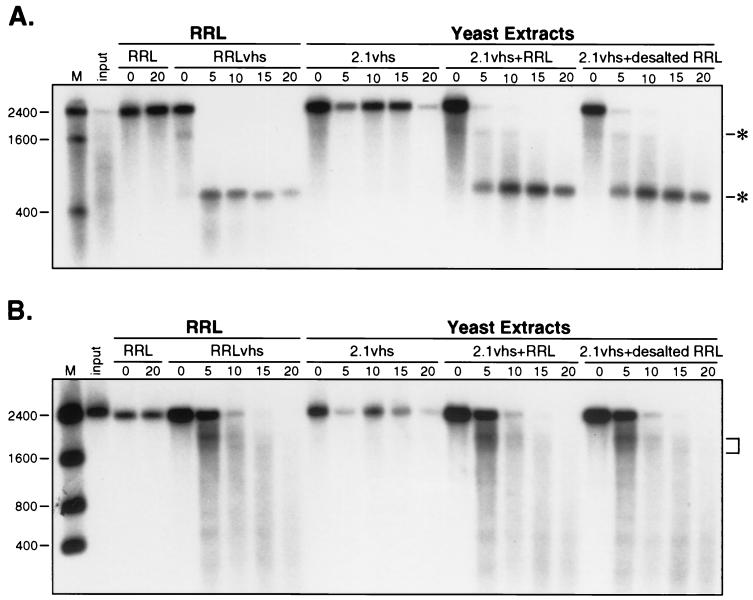
As a first step in characterizing the nature of the required cofactor, we desalted blank RRL by passage over Sephadex G-25 and assayed the excluded fraction for its ability to reconstitute activity on extracts of yeast expressing 2.1vhs (Fig. 12). The results clearly demonstrated that activity was recovered in the excluded fraction, indicating that the required factor is a macromolecule (i.e., ≥5 kDa).
FIG. 12.
The mammalian cofactor is a macromolecule. Internally labeled pCITE-1 and SRPα RNAs were added to control RRL (RRL), RRL containing pretranslated vhs (RRLvhs), yeast extract containing 2.1vhs (2.1vhs), yeast extract containing 2.1vhs mixed with blank RRL (2.1vhs+RRL), and yeast extract containing 2.1vhs mixed with desalted blank RRL (2.1vhs+desalted RRL). Samples were incubated and processed as described in the legend to Fig. 9. Panels A and B show an analysis of the activity of pCITE-1 and SRPα RNAs, respectively. The sizes of RNA markers (M) are indicated in nucleotides at the left. The asterisks in panel A indicate the 5′ and 3′ early degradation products. The bracket in panel B indicates the ca. 1,800-nt early degradation intermediate.
DISCUSSION
The experiments described here have led to two seemingly contradictory sets of findings. First, expression of vhs in the budding yeast S. cerevisiae strongly inhibits cell growth under certain culture conditions. This effect displays the same mutational sensitivity spectrum as host shutoff in mammalian cells, arguing that growth inhibition stems from one or more biologically relevant functions of vhs. Inasmuch as vhs appears to trigger shutoff in mammalian cells through its associated RNase activity, the simplest interpretation of these results is that vhs inhibits the growth of yeast by degrading one or more key cellular RNAs. Second, notwithstanding the foregoing, expression of vhs does not trigger global mRNA turnover in vivo, and vhs protein produced in yeast does not display significant endoribonuclease activity in crude extracts. However, vhs-dependent endoribonuclease activity is restored by adding RRL to the extract. These data argue that the vhs-dependent endoribonuclease requires one or more mammalian factors for activity.
How can one reconcile these seemingly discrepant sets of observations? One possibility is that growth inhibition in yeast results from a fortuitous event that is entirely unrelated to mammalian host shutoff. For example, vhs might interact by chance with a yeast protein and interfere with its normal activity. According to this scenario, the concordance of growth inhibition in yeasts with host shutoff in HSV-infected cells could simply be a consequence of impaired folding of the mutant proteins that lack activity in both systems. However, all of the mutant forms of vhs examined in the present study accumulate in infected cells to the same levels as the wild-type protein and are packaged into the tegument of HSV virions (21, 36). Both of these observations seem incompatible with gross alterations in protein folding. In addition, the vhs1 point mutation (Thr-214→Ile) maps to one of the regions of strongest homology to FEN-1 nucleases (5). These considerations suggest that the mutations abolish vhs function in yeast by inactivating the enzymatic activity of the vhs protein or by abolishing an interaction between vhs and a biologically relevant yeast factor (for example, the yeast homologue of a required mammalian cofactor).
If, as argued above, growth inhibition in yeast reflects the mammalian host shutoff function of vhs, then why do cell extracts of yeast expressing vhs lack detectable RNase activity? The simplest explanation is that our failure to detect RNase activity stems from technical limitations of the in vitro assay system. For example, the levels of RNase activity may be too low to be readily detected in cell extracts but nonetheless sufficient for growth inhibition in vivo, or the putative yeast homologue of the required mammalian cofactor may be inactive or labile under our in vitro conditions. However, this explanation cannot easily account for our inability to detect any effect of vhs on the in vivo stability of PDA1 or ACT1 mRNA. These latter observations seem more compatible with the hypothesis that vhs-induced growth inhibition does not involve global effects on mRNA stability. A second possibility is that the vhs-dependent endoribonuclease produced in yeast associates with a yeast cofactor that targets it in a sequence-specific fashion to a small subset of yeast RNAs, leading to growth inhibition. According to this hypothesis, the functional RNase complex produced in yeast lacks activity on the pCITE-1 and SRPα RNA substrates used in the in vitro assay (and on PDA1 and ACT1 mRNA in vivo) because it lacks an appropriate RNA targeting subunit. Another possibility is that vhs binds the yeast homologue of a mammalian cofactor and inhibits its normal function (leading to growth inhibition), without forming a functional RNase. Distinguishing between these and other alternative explanations will likely require identification of the mammalian stimulatory factor detected in this study.
The observation that vhs inhibits the growth of yeast when cells are grown on solid medium with galactose, raffinose, and (especially) maltose as the sole carbon source but has less effect in glucose is intriguing. However, understanding the molecular basis of this phenomenon will require determining precisely how vhs inhibits cell growth. We have found that the severity of the growth inhibition phenotype in liquid cultures does not correlate with the levels of vhs protein that accumulate under the various growth conditions (data not shown), suggesting that another mechanism is involved. Perhaps the effect stems from the selective action of vhs on some of the mRNAs or proteins that are required to metabolize these alternative carbon sources. Alternatively, it is conceivable that vhs activity is directly or indirectly altered by one of the signaling pathways that respond to changes in carbon source (reviewed in reference 37).
Our finding that desalted RRL reconstitutes vhs-dependent RNase activity in yeast extracts containing vhs provides strong, albeit indirect, evidence that the RNase requires one or more mammalian macromolecules for activity. Although it is formally possible that the expression of vhs induces the synthesis of a yeast RNase that requires one or more components present in RRL for activity, the observation that the reconstituted nuclease activity produces the same degradation intermediates on IRES-bearing substrates as the vhs-dependent RNase produced in RRL makes this possibility seem unlikely. A simpler interpretation is that vhs forms an integral part of the nuclease detected in our experiments, as previously shown for the vhs-dependent nuclease present in extracts of HSV virions (48). Assuming that this is so, we can think of at least three distinct possible mechanisms of action for the required mammalian factor(s). (i) It might be an enzyme needed for a required posttranslational modification of vhs. It is interesting to note that vhs produced during HSV infection is phosphorylated (43), and the inactivating vhs1 mutation alters the pattern of phospho-isoforms that accumulate (36). These observations suggest that phosphorylation may be functionally important. (ii) It might be a required regulatory or catalytic subunit of the RNase. There is no direct evidence that vhs itself has nuclease activity, and so it is possible under this scenario that the cellular factor is an RNase that is activated by vhs rather than vice versa. (iii) It might serve as a targeting subunit that selectively delivers the nuclease to mRNAs as opposed to other cytoplasmic transcripts. In this latter context we note that the vhs-dependent nuclease initially degrades the 5′ end of at least some mRNAs in vivo (22) and in vitro (7), and picornavirus IRES elements strongly target vhs-dependent cleavage events to adjacent RNA sequences (8). IRES elements provide an alternative, cap-independent mode for translation initiation in eukaryotes and function by recruiting translational initiation factors to the RNA (20). We have previously argued that these observations are consistent with the possibility that vhs selectively targets mRNAs by interacting with one or more components of the translational initiation apparatus that act upstream of loading of the 40S ribosomal subunit (8). Possibilities ii and iii are not mutually exclusive, and it is possible that RRL provides more than one required factor.
Our finding that the vhs-dependent endoribonuclease requires one or more mammalian factors for activity seems at first glance inconsistent with the previous conclusion of Zelus et al. (48) that the nuclease is active in the absence of cellular cofactors. These authors based their conclusion on the observation that partially purified HSV virions contain a vhs-dependent RNase and the assumption that their virion preparations lack cellular proteins. However, virions purified by the protocol used in their study likely contain at least some contaminating cellular components, and it is in any case possible that the required cellular factor is packaged along with vhs into the virus particle. Additionally or alternatively, if the required mammalian factor acts by inducing a posttranslational modification of vhs, it likely becomes dispensable after vhs is modified.
The simple in vitro assay described here will allow rapid purification and identification of the mammalian factor(s) that is required for vhs action. We expect that this will greatly enhance our understanding of the mode and regulation of vhs activity.
ACKNOWLEDGMENTS
We thank Carol Lavery, Joanne Duncan, and Rob Maranchuk for superb technical assistance; Kim Ellison for advice, discussions, and a critical review of the manuscript; John Glover and Rick Rachubinski for help with the pYGAL vector and yeast expression systems; and Mike Schultz and Troy Harkness for advice on preparing yeast extracts.
This work was supported by a grant from the National Cancer Institute of Canada, an establishment grant from the Alberta Heritage Foundation for Medical Research, and more recently by a grant from the Canadian Institutes of Health Research. J.R.S. was a Terry Fox Senior Scientist of the National Cancer Institute of Canada.
REFERENCES
- 1.Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 2.Berthomme H, Jacquemont B, Epstein A. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology. 1993;193:1028–1032. doi: 10.1006/viro.1993.1221. [DOI] [PubMed] [Google Scholar]
- 3.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Doherty A J, Serpell L C, Ponting C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elble R. A simple and efficient procedure for transformation of yeasts. Biol Technol. 1992;13:13–15. [PubMed] [Google Scholar]
- 7.Elgadi M M, Hayes C E, Smiley J R. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgadi M M, Smiley J R. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenwick M L, Clark J. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J Gen Virol. 1982;61:121–125. doi: 10.1099/0022-1317-61-1-121. [DOI] [PubMed] [Google Scholar]
- 10.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 11.Fenwick M L, Everett R D. Inactivation of the shutoff gene (UL41) of herpes simplex virus type 1 and 2. J Gen Virol. 1990;71:2961–2967. doi: 10.1099/0022-1317-71-12-2961. [DOI] [PubMed] [Google Scholar]
- 12.Fenwick M L, Owen S A. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J Gen Virol. 1988;69:2869–2877. doi: 10.1099/0022-1317-69-11-2869. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick M L, Walker M J. Suppression of synthesis of cellular macromolecules by herpes simplex virus. J Gen Virol. 1978;41:37–51. doi: 10.1099/0022-1317-41-1-37. [DOI] [PubMed] [Google Scholar]
- 14.Fenwick M L, Everett R D. Transfer of UL41, the gene controlling virion-associated host cell shutoff, between different strains of herpes simplex virus. J Gen Virol. 1990;71:411–418. doi: 10.1099/0022-1317-71-2-411. [DOI] [PubMed] [Google Scholar]
- 15.Fürst P, Hu S, Hackett R, Hamer D. Copper activates methallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988;55:705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- 16.Guarente L, Yocum R R, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA. 1982;79:7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haffey M L, Stevens J T, Terry B J, Dorsky D L, Crumpacker C S, Wietstock S M, Ruyechan W T, Field A K. Expression of herpes simplex virus type 1 DNA polymerase in Saccharomyces cerevisiae and detection of virus-specific enzyme activity in cell-free lysates. J Virol. 1988;62:4493–4498. doi: 10.1128/jvi.62.12.4493-4498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Hitzman R A, Hagie F E, Hayflick J S, Chen C Y, Seeburg P H, Derynck R. The primary structure of the Saccharomyces cerevisiae gene for 3-phosphoglycerate kinase. Nucleic Acids Res. 1982;10:7791–7808. doi: 10.1093/nar/10.23.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 21.Jones F E, Smibert C A, Smiley J R. Mutational analysis of the HSV virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J Virol. 1995;67:4863–4871. doi: 10.1128/jvi.69.8.4863-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karr B M, Read G S. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology. 1999;264:195–204. doi: 10.1006/viro.1999.9986. [DOI] [PubMed] [Google Scholar]
- 23.Krikorian C R, Read G S. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 27.Livingstone-Zatchej M, Meier A, Suter B, Thoma F. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 1997;25:3795–3800. doi: 10.1093/nar/25.19.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.McNeil J B, Friesen J D. Expression of the herpes simplex virus thymidine kinase gene in Saccharomyces cerevisiae. Mol Gen Genet. 1981;184:386–393. doi: 10.1007/BF00352510. [DOI] [PubMed] [Google Scholar]
- 30.Nozaki C, Makizumi K, Kino Y, Nakatake H, Eto T, Mizuno K, Hamada F, Ohtomo N. Expression of herpes simplex virus glycoprotein B gene in yeast. Virus Res. 1985;4:107–113. doi: 10.1016/0168-1702(85)90024-3. [DOI] [PubMed] [Google Scholar]
- 31.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate early (alpha) mRNA. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pak A S, Everly D N, Knight K, Read G S. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 34.Parker R, Herrick D, Peltz S W, Jacobson A. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:415–423. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- 35.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate-early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheffler I E, de la Cruz B J, Prieto S. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Int J Biochem Cell Biol. 1998;30:1175–1193. doi: 10.1016/s1357-2725(98)00086-7. [DOI] [PubMed] [Google Scholar]
- 38.Schultz M C, Hockman D J, Harkness T A A, Garinther W I, Altheim B A. Chromatin assembly in a yeast whole-cell extract. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schultz M C, Choe S Y, Reeder R H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smibert C A, Smiley J R. Differential regulation of endogenous and transduced β-globin genes during infection of erythroid cells with a herpes simplex virus type 1 recombinant. J Virol. 1990;64:3882–3894. doi: 10.1128/jvi.64.8.3882-3894.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smibert C A, Popova B, Xiao P, Capone J P, Smiley J R. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 44.Sorenson C M, Hart P A, Ross J. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 1991;19:4459–4465. doi: 10.1093/nar/19.16.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens A. Endonucleolytic cleavage of RNA at 5′ endogenous stem structures by human flap endonuclease 1. Biochem Biophys Res Commun. 1998;251:501–508. doi: 10.1006/bbrc.1998.9499. [DOI] [PubMed] [Google Scholar]
- 46.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zealey G R, Goodey A R, Piggott J R, Watson M E, Cafferkey R C, Doel S M, Carter B L A, Wheals A E. Amplification of plasmid copy number by thymidine kinase expression in Saccharomyces cerevisiae. Mol Gen Genet. 1988;211:155–159. doi: 10.1007/BF00338407. [DOI] [PubMed] [Google Scholar]
- 48.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X-L, Ward C, Weissbach A. Control of herpes simplex virus thymidine kinase gene expression in Saccharomyces cerevisiae by a yeast promoter sequence. Mol Gen Genet. 1984;194:31–41. doi: 10.1007/BF00383493. [DOI] [PubMed] [Google Scholar]