Highlights
-
•
A real-time muscle ultrasound imaging aid which implemented a machine-learning model utilising texture analysis was developed.
-
•
The model’s training accuracy was 81 %, and generalization performance validation showed 78 % per image and 93 % per patient.
-
•
Real-time analysis and automatic evaluation can enhance diagnostic muscle ultrasound without interfering with workflow.
Keywords: Muscle ultrasound, Texture analysis, Real-time diagnosis, Machine learning, Neuromuscular disorders
Abstract
Objective
Many artificial intelligence approaches to muscle ultrasound image analysis have not been implemented on usable devices in clinical neuromuscular medicine practice, owing to high computational demands and lack of standardised testing protocols. This study evaluated the feasibility of using real-time texture analysis to differentiate between various pathological conditions.
Methods
We analysed 17,021 cross-sectional ultrasound images of the biceps brachii of 75 participants, including 25 each with neurogenic disorders, myogenic disorders, and healthy controls. The size and location of the regions of interest were randomly selected to minimise bias. A random forest classifier utilising texture features such as Dissimilarity and Homogeneity was developed and deployed on a mobile PC, enabling real-time analysis.
Results
The classifier distinguished patients with an accuracy of 81 %. Echogenicity and Contrast from the Co-Occurrence Matrix were significant predictive features. Validation on 15 patients achieved accuracies of 78 %/93 % per image/patient over 15-second videos, respectively. The use of a mobile PC facilitated real-time estimation of the underlying pathology during ultrasound examination, without influencing procedures.
Conclusions
Real-time automatic texture analysis is feasible as an adjunct for the diagnosis of neuromuscular disorders.
Significance
Artificial intelligence using texture analysis with a light computational load supports the semi-quantitative evaluation of neuromuscular ultrasound.
1. Introduction
Recent advances in muscular ultrasound technology have significantly enhanced the detection and visualisation of the distribution and properties of the muscles affected in neuromuscular diseases, reaching a precision comparable to that of magnetic resonance imaging (MRI) (Pillen et al., 2008, Pillen et al., 2016, Van Alfen et al., 2018). Muscle ultrasonography is particularly valued for its practicality as a convenient bedside assessment tool capable of identifying a wide range of neuromuscular conditions (Walker, 2004, Hobson-Webb and Simmons, 2019, Albayda and Van Alfen, 2020). Modern clinical ultrasound equipment can visualise muscle fibres as small as 100 µm in diameter, and fascicles wrapped in endomysium could be clearly identified (Hodges et al., 2003). This non-invasive tool provides several key features regarding the underlying muscle pathological conditions, such as muscle echogenicity changes, atrophy, degeneration, particularly fatty degeneration, unusual intramuscular structures, fascial thickening, and involuntary muscle contractions (Misawa et al., 2011, Pillen and Van Alfen, 2011).
Despite these advantages, the qualitative analysis of muscle ultrasound remains highly dependent on the expertise of the observer, and its quantitative analysis is significantly influenced by the characteristics of the equipment and the associated high computational costs (Pillen et al., 2006, Pillen et al., 2007, Wijntjes et al., 2022). As such, the diagnostic process relies heavily on visual inspection, and lacks quantifiability (Wijntjes and Van Alfen, 2021, Fukushima et al., 2022).
Many artificial intelligence (AI) approaches to muscle ultrasound image analysis have limitations in real-time clinical applications in neuromuscular medicine because of their computational demands and non-standardized testing methods (Burlina et al., 2017, Marzola et al., 2021). However, recent research has explored the use of artificial intelligence (AI) and machine learning techniques to objectively quantify offline muscle ultrasound images. Texture analysis methods, such as the grey-level co-occurrence matrix (GLCM) and grey-level run-length matrix (GLRLM), have demonstrated the potential to distinguish between normal, neurogenic, and myogenic conditions based on image texture features (Sogawa et al., 2017, Nagawa et al., 2021, Paris and Mourtzakis, 2021, De Jong et al., 2023).
Texture analysis quantifies the variation in the surface intensity or patterns in an image, providing valuable information about the underlying structure. The GLCM analyses the frequency of pixel pairs with specific values in a specified spatial relationship. The GLRLM assesses the lengths of consecutive runs of pixels with the same value. These methods capture detailed textural features that are not easily discernible by the human eye, enabling the differentiation between various tissue types and pathological conditions.
The objective of this study was to evaluate the feasibility of real-time muscle ultrasound quantification using texture analysis to differentiate between pathological conditions. First, we developed a machine-learning model to estimate background pathologies based on image patterns, after which we constructed an externally available environment that could provide real-time estimation results during the examinations. This study addressed the important issue of ensuring objectivity in muscle ultrasound analysis, with the aim of improving the diagnostic accuracy and accessibility of muscle ultrasound.
2. Methods
2.1. Patients and ultrasound examination procedures
This study included patients who visited our clinic to undergo muscular ultrasound (US) for clinically suspected neuromuscular disorders between August 2021 and July 2023. Patients with concurrent diseases, poor image quality, or focal neurological conditions (e.g. carpal tunnel syndrome) that did not meet the respective disease diagnostic criteria were excluded. As such, only patients with neurogenic or myogenic disorders who met the diagnostic criteria were included in the study. Neurogenic disorders are diagnosed on the basis of comprehensive clinical evaluations, electrodiagnostic assessments (nerve conduction studies and needle electromyography), blood tests, and genetic testing (Hobson-Webb and Simmons, 2019, Masrori and Van Damme, 2020, Koczwara et al., 2022). Myopathy was diagnosed through clinical evaluations, creatine kinase blood level testing, electromyography, and muscle biopsies, following the recommended diagnostic guidelines (Emery, 2002, Koczwara et al., 2022). The healthy control group comprised individuals with normal neurological examination results, no history of diabetes or hormonal disorders, no use of neurotoxic drugs, no exposure to industrial materials, and no family history of neurological diseases.
Muscular ultrasound examinations on the biceps brachii muscle were conducted by two experienced physicians (YN and SM) using a LOGIQe US device (GE Healthcare, Buckinghamshire, England), with a fixed linear transducer operating at a frequency of 12 MHz (Walker, 2004). Subjects were positioned supine, with the probe placed at the centre of the upper arm. Ultrasound images were captured while the subjects performed controlled flexion and extension arm movements and were stored as 15-second films in an Audio Video Interleaved Format. The study included 75 patients evenly divided into healthy control, neurogenic, and myogenic groups, each consisting of 25 patients. The healthy control group had an average age of 38.16 ± 11.93 years. The neurogenic group, averaging 57.31 ± 19.88 years, included cases with amyotrophic lateral sclerosis (7 cases), progressive muscular atrophy (7 cases), spinal muscular atrophy (4 cases), spinal and bulbar muscular atrophy (3 cases), and cervical spondylotic amyotrophy (4 cases). The myogenic group, with an average age of 57.80 ± 16.42 years, comprised cases with Duchenne muscular dystrophy (7 cases), Becker muscular dystrophy (8 cases), myotonic dystrophy (4 cases), facioscapulohumeral muscular dystrophy (3 cases), and mitochondrial diseases (3 cases). The biceps brachii muscle strength was assessed using the Medical Research Council (MRC) scale. The neurogenic group had a mean MRC score of 4.4 ± 0.6 (range: 3–5), while the myogenic group had a mean MRC score of 3.1 ± 1.2 (range: 2–5). The study protocol was approved by our Institutional Review Board (approval no. B220238). The requirement for written informed consent was waived because of the study’s retrospective anonymised design, and consent was obtained through an opt-out form. This form included an outline of the research, purpose and methods, and use of materials, and included a point of contact to allow patients to refuse to participate at any time. The authors declare no conflicts of interest.
2.2. Texture analysis and machine learning of ultrasound images
The ultrasound video images for each case were randomly divided into 60 training sets and 15 test sets. A comprehensive region of interest (ROI) was established on one side of the biceps brachii muscle, and the ROI size was randomly altered in each frame (Supplementary Video 1). Initially, 38,700 frames were extracted from the training dataset. Frames with an ROI smaller than 3,000 pixels were excluded. This diagnostic system was constructed using open-source software, including scikit-image and scikit-learn, which are freely available Open CV Library, 2024. The use of open-source materials allows other researchers and medical institutions to easily access, replicate, and improve the system, thereby enhancing the transparency and reproducibility of research. The program includes an innovative identification module capable of detecting and quantifying motion within images using a background subtraction method that calculates the absolute difference between the background image generated by the preceding moving averaged image and the current image (Open CV. accumulateWeighted, 2024, Gijsbertse et al., 2018). The motion ratio was determined by dividing the number of pixels that shifted within the ROI by the total number of pixels within the ROI. Additionally, a motion-detection function identified frames showing voluntary muscle contractions, involuntary muscle contractions, and blurring caused by the examiner's hand movements. Specifically, images with a motion ratio greater than 0.5 were excluded from the analysis. The final number of frames used for the training was 17,021. The test set was then calculated in the same manner, and the final number of frames used for validation was 3,555.
For texture analysis and machine learning, we used the OpenCV, scikit-image, and scikit-learn libraries in Python 3.9. In our study, we computed the echo values of the ultrasound images as histograms using the OpenCV library, and calculated the histogram features, including the mean and standard deviation. Texture features were extracted using a grey-level co-occurrence matrix (GLCM) from the scikit-image library, which is a statistical tool that quantifies the frequency of occurrence of pixel pairs in a specified direction and distance within an image (Molinari et al., 2015). We configured the GLCM to analyse the pixel pairs in the 0° direction at a two-pixel distance. Each GLCM element quantifies the frequency of two-pixel values appearing at a set offset, enabling the derivation of texture features such as Contrast, Dissimilarity, Homogeneity, Angular Second Moment, and Correlation. These features help elucidate the frequency of adjacent pixel values, thereby enhancing texture understanding and aiding in differentiating between regions within the image.
We developed a model using the scikit-learn library to differentiate various neuromuscular disorders using diverse data analysis algorithms. To ensure that data from the same patient were not used in both the training and testing phases, we classified the features extracted from each frame individually per patient. After feature selection, the model was trained using a random forest classifier (RFC) (Scikit-learn. random forest classifier). RFC, which integrates multiple decision trees, bases its decisions on a majority vote among the trees. To ensure the generalisability of the RFC model, we applied Group K-Fold cross-validation (Scikit-learn. Group K-Fold cross-validation), dividing the dataset into five groups. This method prevents information leakage and ensures no overlap between the training and validation sets, with each group being used once as the validation set and the remainder used for training. RFC was executed 300 times per fold, after which the results were averaged. This cross-validation process was repeated five times to confirm the reliability of the results. This strategy allowed our model to accurately assess its generalisability across different patient groups, thereby enhancing the reliability of the predictions for new patient data.
2.3. Generalisation performance verification
To validate the generalisability of the model, we further conducted additional evaluations using 15 patients and healthy subjects who were not included in the training dataset. Two approaches are used for this evaluation. The first method involved a muscle imaging approach, in which ROIs of varying sizes were extracted from each frame, and the predicted labels for each frame were compared with the correct labels. The second involved a per-patient approach, in which the entire 15-second video for each patient was considered. The prediction rate was defined as the number of most likely predicted labels per frame divided by the total number of frames evaluated. The accuracy rate was denoted as the percentage of frames that displayed the correct labels. Using these approaches, the generalisation performance of the model was assessed using an independent dataset separate from the training data. This study adhered to the Checklist for Artificial Intelligence in medical imaging (CLAIM), ensuring rigorous standards for AI deployment in clinical settings, and enhancing the reliability and reproducibility of our results (Mongan et al., 2020).
2.4. Application implementation for real-time texture analysis
To develop an application that could display underlying pathological assumptions based on real-time texture analysis of current ultrasound images, we used the OpenCV and Scikit-Image libraries. This program portrays the echogenicity through a grayscale histogram consisting of 255 shades in the lower-right corner of the screen, and further allows users to select an ROI within muscle ultrasound videos. Random ROIs were extracted from the selected ROIs, and the program computed the mean and standard deviation of the echogenicity. This conducts texture analysis using GLCM and detects motion.
The computed texture features included mean echogenicity; standard deviation of histogram features; and texture descriptors such as Contrast, Dissimilarity, Homogeneity, Angular Second Moment (ASM), and Correlation from the GLCM.
The accumulated assumptions of the texture analysis results were calculated using a pretrained machine-learning model. The application was implemented on a mobile tablet (Surface Pro 8; Microsoft Corporation, WA, USA) running on the Windows operating system and connected to the external output of an ultrasound examination device.
3. Results
3.1. Machine learning analysis
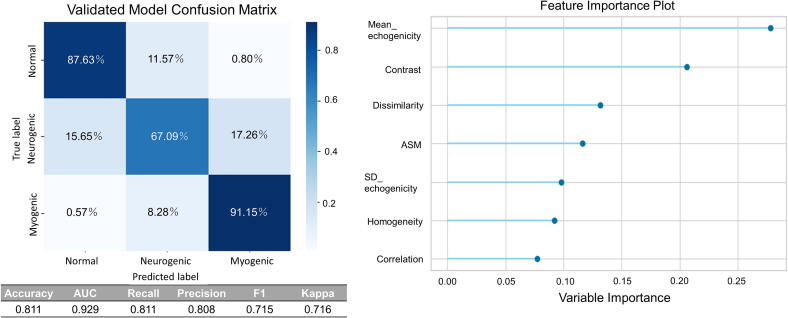
Fig. 1 illustrates the results of the machine learning analyses, focusing on seven key features. Using the holdout method for evaluation, the model achieved an accuracy of 81.11 % in classifying the three distinct groups: normal, neurogenic, and myogenic. Notably, the mean echogenicity derived from the histograms and contrast from the co-occurrence matrix were identified as the most significant features in determining the classifications.
Fig. 1.
Performance Evaluation of the random forest Classifier using Ultrasound Data. This figure overviews the comprehensive two-part analysis of the ultrasound data processed using a random forest classifier. The left panel displays the confusion matrix for the validated model, showing the accuracy of diagnoses based on the regions of interest (ROI). The evaluation employed the holdout method with matrix entries representing the percentages of correct and incorrect classifications for Normal, Neurogenic, and Myogenic conditions. The right panel shows a Feature Importance Plot that ranks the factors most significantly influencing the decision-making process of the model. Notably, Mean echogenicity and Contrast were highlighted as the top features that significantly affected the model's predictions.
3.2. Generalization performance validation
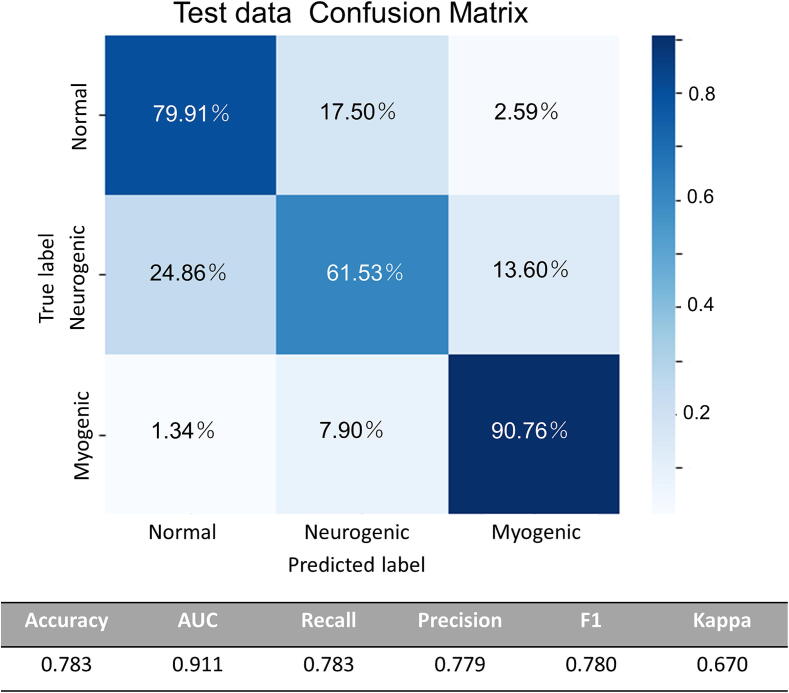
The results of the validation of the generalisation performance of the model are presented in detail. Fig. 2 illustrates the outcomes of the muscle image approach, including a confusion matrix. This method involved comparing the predicted labels with the correct labels for each frame of the muscle ultrasound data. Using this approach, the model achieved an overall accuracy of 78.3 %. Specifically, it correctly classified 79.91 % normal cases, 61.53 % neurogenic cases, and 90.76 % myogenic cases. However, it misclassified 17.50 % of the normal cases as neurogenic, 24.86 % of neurogenic cases as normal, and 7.90 % of myogenic cases as neurogenic, indicating areas for potential improvement.
Fig. 2.
Generalization Performance on Test Data Using Muscle Image Approach The efficacy of the muscle image approach on the test data illustrated in Fig. 3, which presents a confusion matrix that compares the predicted labels with the actual labels per frame, yielding an accuracy of 78.3%.
In contrast, the per-patient approach analysed the entire 15-second video for each patient, considering the highest prediction rate among the diagnostic categories: 'Normal', 'Neurogenic', and 'Myogenic'. This approach successfully distinguished 14 of the 15 patients, with an accuracy greatly exceeding 50 %. A detailed analysis revealed that the model accurately diagnosed all five normal cases, 4 of the five neurogenic cases, and five myogenic cases. The overall accuracy of this approach was 93.3 %, which was significantly higher than that of the frame-by-frame analysis. This high accuracy indicates the strong capability of the model to generalise across different datasets, and suggests that the per-patient approach may be more robust for clinical applications.
The confusion matrix in Fig. 2 and detailed case-by-case results in Table 1 collectively demonstrate the model's performance and potential clinical applicability. The muscle image approach provides a granular frame-by-frame analysis, whereas the per-patient approach offers a more holistic assessment, both of which provide valuable insights into the model's diagnostic capabilities.
Table 1.
Generalization Performance on Test Data Using the Patient Approach. This table compiles the outcomes of applying the quantitative analysis tool to muscle ultrasonography data. Each row corresponds to a unique clinical case, detailing the actual condition of the patient ('True label'), alongside the diagnosis inferred by the predictive model ('Predicted label'), which is selected based on the highest prediction rate. The diagnostic categories were as follows: The Accuracy rate column quantifies the precision of the model predictions on a case-by-case basis. In addition, the prediction rate was delineated for each diagnostic category, reflecting the probabilistic assessment of each model condition.
| Case | True label | Prediction label | Accuracy rate | Prediction rate |
||
|---|---|---|---|---|---|---|
| Normal | Neurogenic | Myogenic | ||||
| 1 | Normal | Normal | 0.963 | 0.963 | 0.036 | 0 |
| 2 | Normal | Normal | 0.96 | 0.96 | 0.035 | 0.003 |
| 3 | Normal | Normal | 0.842 | 0.842 | 0.157 | 0 |
| 4 | Normal | Normal | 0.867 | 0.867 | 0.132 | 0.001 |
| 5 | Normal | Normal | 0.885 | 0.885 | 0.092 | 0.022 |
| 6 | Neurogenic | Neurogenic | 0.589 | 0.098 | 0.589 | 0.312 |
| 7 | Neurogenic | Neurogenic | 0.853 | 0.123 | 0.853 | 0.022 |
| 8 | Neurogenic | Normal | 0.159 | 0.84 | 0.159 | 0 |
| 9 | Neurogenic | Neurogenic | 0.708 | 0.004 | 0.708 | 0.286 |
| 10 | Neurogenic | Neurogenic | 0.795 | 0.198 | 0.795 | 0.005 |
| 11 | Myogenic | Myogenic | 0.906 | 0 | 0.093 | 0.906 |
| 12 | Myogenic | Myogenic | 0.982 | 0 | 0.017 | 0.982 |
| 13 | Myogenic | Myogenic | 0.813 | 0.004 | 0.182 | 0.813 |
| 14 | Myogenic | Myogenic | 0.98 | 0 | 0.019 | 0.98 |
| 15 | Myogenic | Myogenic | 0.923 | 0.01 | 0.065 | 0.923 |
3.3. Real-time texture analysis during ultrasound examination
The results of our real-time AI diagnostic tool were demonstrated on a tablet PC with data transmitted from the ultrasound system via a capture board. This tool enables the real-time estimation of muscle pathology using ultrasound without interfering with standard examination procedures. The system interface allows the examiner to adjust the size of the ROI and the sensitivity of the analysis, ensuring precise and customisable assessments.
The echogenicity of the muscle tissue, highest predictive classification, and its probability were dynamically displayed on the screen. The probabilities of each classification were visualised through comprehensive graphs, aiding in understanding the likelihood of different muscle conditions such as 'Normal', 'Neurogenic', and 'Myogenic'. The prediction results were displayed in the upper left corner of the screen only when the ROI size exceeded 3,000 pixels and the motion ratio was less than 0.5. This real-time feedback allowed for the immediate identification of pathological conditions, thereby enhancing the examiner’s evaluation.
The system tracked and represented temporal changes in the echogenicity and motion ratio within the ROI using blue and green lines, respectively. These visual indicators allow continuous monitoring of the muscle condition over time, which is crucial for detecting dynamic changes in muscle pathology. A histogram of echogenicity is also provided, offering a detailed real-time analysis of muscle tissue characteristics.
These capabilities are shown in Fig. 3 and Supplementary Video 2, and have been demonstrated in clinical settings. The use of this tool during standard ultrasound examination procedures provides semi-quantitative information to the examiner, without disrupting the workflow.
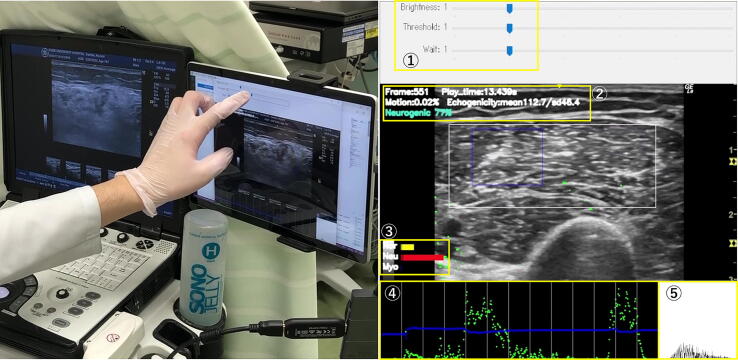
Fig. 3.
Operation of the Real-Time Automatic Muscular Ultrasound Diagnostic Tool. This image shows the examiner using the automatic diagnostic tool for muscle ultrasound analysis. The system allows adjustments in the size of the Region of Interest (ROI) and sensitivity of the analysis via a user interface (①). The echogenicity, as well as the highest predictive classification and its probability, are dynamically displayed on the screen (②). The probability of each classification was visualised using graphs (③). Additionally, temporal changes in echogenicity and motion ratio within the ROI were tracked and are represented by blue and green lines, respectively (④). A histogram of the echogenicity is also provided in real-time feedback (⑤).
4. Discussion
This study effectively demonstrated the feasibility and efficacy of employing real-time texture analysis using RFC to diagnose neuromuscular disorders by muscle ultrasound. Our method significantly reduced the computational demands associated with artificial intelligence techniques, making it practical for clinical use. Validation of this model in additional patients with video assessments has underscored its practical utility in clinical settings. The ability to perform texture analysis in real-time without disrupting clinical workflows represents a significant advancement over traditional diagnostic methods. By directly integrating quantitative analysis into the clinical examination process, our approach further enhances diagnostic precision and markedly accelerates decision making, thereby improving patient outcomes and clinical efficiency.
Traditionally, muscle ultrasound images or videos have been used to visually identify whether muscles are normal or pathological; however, unlike needle electromyography, they cannot distinguish between neurogenic and myogenic origins. Visual evaluations can detect obvious abnormalities, such as changes in brightness, atrophy, tumours, or abnormal movements; however, they require substantial expertise and achieve only approximately 70 % efficacy in differentiating between healthy and abnormal muscle tissues (Pillen et al., 2006). Electromyography findings are often need to complement these visual evaluations to ensure an accurate assessment.
The grayscale histogram provides data on the distribution of image brightness, but is insufficient for assessing textural characteristics such as patterns and granularity. Thus, computing textural features using methods such as GLCM and GLRLM is essential (Sogawa et al., 2017, Nagawa et al., 2021, Paris and Mourtzakis, 2021). However, these calculations require specialised software and result in high computational costs (Molinari et al., 2015), emphasising the need for more efficient methods that preserve discriminative power.
In neuromuscular disorders, the underlying pathological changes and textural differences in ultrasound images vary significantly under different conditions (Zaidman and Van Alfen, 2016; Filosto et al., 2022). Because of the lack of a universally optimal method for distinguishing background pathologies, we employed ensemble learning, which integrates multiple methods (Sogawa et al., 2017, Tannemaat et al., 2023). A major advantage of our tool is its ability to display the results from machine learning algorithms in real time. Currently, muscle ultrasonography primarily serves as a supplementary tool to electromyography for the diagnosis of neuromuscular diseases. Unless these AI-driven assessments become more reliable and the evaluation process becomes quicker, traditional diagnostic techniques that depend on invasive procedures, such as electromyography and muscle biopsy, will likely remain prevalent (Pillen et al., 2008, Van Alfen et al., 2018, Hobson-Webb and Simmons, 2019). The benefits of quantitative muscle ultrasound analysis over electromyography include a wider ROI and more consistent evaluations, without sampling bias. Moreover, it is non-invasive and enables analysis both during muscle contractions and at rest. Nevertheless, it is uncertain to what extent the image variation is due to the pathology itself versus other factors, such as muscle atrophy due to malnutrition or differences in body size. Furthermore, the variability in imaging techniques presents additional challenges. AI analysis using extensive training data and enhanced analytical methods may mitigate these variabilities (Verdú-Díaz et al., 2020, Paris and Mourtzakis, 2021). Our study is the first to demonstrate that even with limited training data, integrating multiple analysis methods can achieve rapid, high-performance classification, proving invaluable for clinical applications.
This study had several limitations. First, the cohort used for machine learning was relatively small, and the training dataset primarily consisted of neuromuscular disorders with confirmed diagnoses such as motor neuron diseases and muscular dystrophy, excluding acute and subacute neuromuscular conditions. Although the overall accuracy was reported to be 78 %, the discrimination between normal and myogenic cases was comparatively high, whereas it remained low for neurogenic conditions. This disparity likely stems from textural variations associated with different stages of the disease (Nodera et al., 2016, Nodera et al., 2018). To enhance the classification accuracy, adopting a patient-specific approach that averages decisions across individual frames may be beneficial. Further improvements will require an increase in the number of cases and the inclusion of texture differences related to the disease stages in the training dataset. Furthermore, whether real-time AI classification for neuromuscular ultrasound has a diagnostic value should be confirmed in another study using this device.
In addition, our study's training and test data were acquired using the same ultrasound machine with identical settings. The sensitivity of ultrasound devices can vary significantly based on the equipment and settings, meaning our model may not be universally applicable across all devices. However, our methodology allows for creating a customized learning model specific to the equipment used, enabling real-time quantitative muscle ultrasound tailored to specific devices. As our next objective, we aim to investigate how texture features vary across different presets from various manufacturers and how these variations impact learning outcomes. This approach aims to develop a standardized model that can be utilized across different ultrasound devices, ensuring consistent performance.
This study demonstrates the potential of real-time quantitative analysis of muscle ultrasound as a non-invasive diagnostic method for neuromuscular disorders. This enables a faster and more accurate diagnostic process in a clinical setting. Moving forward, our aim is for this technology to be widely adopted in clinical practice and become a new standard in the management of neuromuscular diseases.
5. Conclusions
This study represents a pioneering effort in applying real-time analysis and automatic evaluation to enhance the utility of ultrasound in clinical settings. Real-time semi-quantitative analysis of muscle ultrasound facilitates the objective and detailed assessment of muscle conditions, thus advancing the clinical application of ultrasound in the management of neuromuscular diseases.
Funding
This work was supported by JSPS KAKENHI Grant number 24K14297.
Declaration of Generative AI and AI-assisted technologies in the writing process
During preparation for this study, the authors used Chat GPT to edit the manuscript. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Conflict of interest
None of the authors declared any conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cnp.2024.08.003.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Albayda J., Van Alfen N. Diagnostic value of muscle ultrasound for myopathies and myositis. Curr. Rheumatol. Rep. 2020;22(11):82. doi: 10.1007/s11926-020-00947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlina P., Billings S., Joshi N., Albayda J. Automated diagnosis of myositis from muscle ultrasound: exploring the use of machine learning and deep learning methods. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0184059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong L., Greco A., Nikolaev A., Weijers G., Van Engelen B.G.M., De Korte C.L., Fütterer J.J. Three-dimensional quantitative muscle ultrasound in patients with facioscapulohumeral dystrophy and myotonic dystrophy. Muscle Nerve. 2023;68(4):432–438. doi: 10.1002/mus.27943. [DOI] [PubMed] [Google Scholar]
- Emery A.E.H. The muscular dystrophies. Lancet. 2002;359(9307):687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- Filosto M., Pichiecchio A., Diaz-Manera J., Santini F. Imaging of neuromuscular diseases. Front. Media SA. 2022 doi: 10.3389/978-2-88976-628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K., Takamatsu N., Yamamoto Y., Yamazaki H., Yoshida T., Osaki Y., Haji S., Fujita K., Sugie K., Izumi Y. Early diagnosis of amyotrophic lateral sclerosis based on fasciculations in muscle ultrasonography: a machine learning approach. Clin. Neurophysiol. 2022;140:136–144. doi: 10.1016/j.clinph.2022.06.005. [DOI] [PubMed] [Google Scholar]
- Gijsbertse K., Bakker M., Sprengers A., Wijntjes J., Lassche S., Verdonschot N., de Korte C.L., van Alfen N. Computer-aided detection of fasciculations and other movements in muscle with ultrasound: development and clinical application. Clin. Neurophysiol. 2018;129(11):2567–2576. doi: 10.1016/j.clinph.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Hobson-Webb L.D., Simmons Z. Ultrasound in the diagnosis and monitoring of amyotrophic lateral sclerosis: a review. Muscle Nerve. 2019;60(2):114–123. doi: 10.1002/mus.26487. [DOI] [PubMed] [Google Scholar]
- Hodges P.W., Pengel L.H.M., Herbert R.D., Gandevia S.C. Measurement of muscle contraction with ultrasound imaging. Muscle Nerve. 2003;27(6):682–692. doi: 10.1002/mus.10375. [DOI] [PubMed] [Google Scholar]
- Koczwara K.E., Lake N.J., Desimone A.M., Lek M. Neuromuscular disorders: finding the missing genetic diagnoses. Trends Genet. 2022;38(9):956–971. doi: 10.1016/j.tig.2022.07.001. [DOI] [PubMed] [Google Scholar]
- Marzola F., Van Alfen N., Doorduin J., Meiburger K.M. Deep learning segmentation of transverse musculoskeletal ultrasound images for neuromuscular disease assessment. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104623. [DOI] [PubMed] [Google Scholar]
- Masrori P., Van Damme P. Amyotrophic lateral sclerosis: a clinical review. Eur. J. Neurol. 2020;27(10):1918–1929. doi: 10.1111/ene.14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa S., Noto Y., Shibuya K., Isose S., Sekiguchi Y., Nasu S., Kuwabara S. Ultrasonographic detection of fasciculations markedly increases diagnostic sensitivity of ALS. Neurology. 2011;77(16):1532–1537. doi: 10.1212/WNL.0b013e318233b36a. [DOI] [PubMed] [Google Scholar]
- Molinari F., Caresio C., Acharya U.R., Mookiah M.R.K., Minetto M.A. Advances in quantitative muscle ultrasonography using texture analysis of ultrasound images. Ultrasound Med. Biol. 2015;41(9):2520–2532. doi: 10.1016/j.ultrasmedbio.2015.04.021. [DOI] [PubMed] [Google Scholar]
- Mongan J., Moy L., Kahn C.E. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol. Artif. Intell. 2020;2(2) doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa K., Suzuki M., Yamamoto Y., Inoue K., Kozawa E., Mimura T., Nakamura K., Nagata M., Niitsu M. Texture analysis of muscle MRI: machine learning-based classifications in idiopathic inflammatory myopathies. Sci. Rep. 2021;11(1):9821. doi: 10.1038/s41598-021-89311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodera H., Takamatsu N., Matsui N., Mori A., Terasawa Y., Shimatani Y., Osaki Y., Maruyama K., Izumi Y., Kaji R. Intramuscular dissociation of echogenicity in the triceps surae characterizes sporadic inclusion body myositis. Eur. J. Neurol. 2016;23(3):588–596. doi: 10.1111/ene.12899. [DOI] [PubMed] [Google Scholar]
- Nodera H., Sogawa K., Takamatsu N., Mori A., Yamazaki H., Izumi Y., Kaji R. Age-dependent texture features in skeletal muscle ultrasonography. J. Med. Invest. 2018;65(3):274–279. doi: 10.2152/jmi.65.274. [DOI] [PubMed] [Google Scholar]
- Open CV Library. [Cited 2024-05-18]. Available from: https://opencv.org/.
- Open CV. accumulateWeighted. [Cited 2024-05-18]. Available from: https://docs.opencv.org/3.4/d7/df3/group__imgproc__motion.html#ga4f9552b541187f61f6818e8d2d826bc7.
- Paris M.T., Mourtzakis M. Muscle composition analysis of ultrasound images: a narrative review of texture analysis. Ultrasound Med. Biol. 2021;47(4):880–895. doi: 10.1016/j.ultrasmedbio.2020.12.012. [DOI] [PubMed] [Google Scholar]
- Pillen S., Verrips A., Van Alfen N., Arts I.M.P., Sie L.T.L., Zwarts M.J. Quantitative skeletal muscle ultrasound: diagnostic value in childhood neuromuscular disease. Neuromuscul. Disord. 2007;17(7):509–516. doi: 10.1016/j.nmd.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Pillen S., Arts I.M.P., Zwarts M.J. Muscle ultrasound in neuromuscular disorders. Muscle Nerve. 2008;37(6):679–693. doi: 10.1002/mus.21015. [DOI] [PubMed] [Google Scholar]
- Pillen S., Boon A., Van Alfen N. In: Handbook of Clinical Neurology, Neuroimaging Part II. Masdeu J.C., González R.G., editors. Elsevier; 2016. Chapter 42. Muscle ultrasound; pp. 843–853. [DOI] [PubMed] [Google Scholar]
- Pillen S., Van Alfen N. Skeletal muscle ultrasound. Neurol. Res. 2011;33(10):1016–1024. doi: 10.1179/1743132811Y.0000000010. [DOI] [PubMed] [Google Scholar]
- Pillen S., Van Keimpema M., Nievelstein R.A.J., Verrips A., Van Kruijsbergen-Raijmann W., Zwarts M.J. Skeletal muscle ultrasonography: visual versus quantitative evaluation. Ultrasound Med. Biol. 2006;32(9):1315–1321. doi: 10.1016/j.ultrasmedbio.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Scikit-learn. Group K-Fold cross-validation. [Cited 2024-05-18]. Available from: https://scikit-learn.org/stable/modules/cross_validation.html#group-k-fold.
- Scikit-learn. Random forest classifier. [Cited 2024-05-18]. Available from: https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html.
- Sogawa K., Nodera H., Takamatsu N., Mori A., Yamazaki H., Shimatani Y., Izumi Y., Kaji R. Neurogenic and myogenic diseases: quantitative texture analysis of muscle US data for differentiation. Radiology. 2017;283(2):492–498. doi: 10.1148/radiol.2016160826. [DOI] [PubMed] [Google Scholar]
- Tannemaat M.R., Kefalas M., Geraedts V.J., Remijn-Nelissen L., Verschuuren A.J.M., Koch M., Kononova A.V., Wang H., Bäck T.H.W. Distinguishing normal, neuropathic and myopathic EMG with an automated machine learning approach. Clin. Neurophysiol. 2023;146:49–54. doi: 10.1016/j.clinph.2022.11.019. [DOI] [PubMed] [Google Scholar]
- Van Alfen N., Gijsbertse K., De Korte C.L. How useful is muscle ultrasound in the diagnostic workup of neuromuscular diseases? Curr. Opin. Neurol. 2018;31(5):568–574. doi: 10.1097/WCO.0000000000000589. [DOI] [PubMed] [Google Scholar]
- Verdú-Díaz J, Alonso-Pérez J, Nuñez-Peralta C, Tasca G, Vissing J, Straub V, Fernández-Torrón R, Llauger J, Illa I, Díaz-Manera J., 2020. Accuracy of a machine learning muscle MRI-based tool for the diagnosis of muscular dystrophies. Neurology 94(10), e1094–102–e1102. 10.1212/WNL.0000000000009068. [DOI] [PubMed]
- Walker F.O. Neuromuscular ultrasound. Neurol. Clin. 2004;22(3):563–590. doi: 10.1016/j.ncl.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Wijntjes J., Van Alfen N. Muscle ultrasound: present state and future opportunities. Muscle Nerve. 2021;63(4):455–466. doi: 10.1002/mus.27081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijntjes J., Van der Hoeven J., Saris C.G.J., Doorduin J., Van Alfen N. Visual versus quantitative analysis of muscle ultrasound in neuromuscular disease. Muscle Nerve. 2022;66(3):253–261. doi: 10.1002/mus.27669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman C.M., Van Alfen N. Ultrasound in the assessment of myopathic disorders. J. Clin. Neurophysiol. 2016;33(2):103–111. doi: 10.1097/WNP.0000000000000245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.