Summary
Kissing bugs are known to produce anticoagulant venom that facilitates blood-feeding. However, it is unknown how this saliva evolved and if the venom produced by the entomophagous ancestors of kissing bugs would have helped or hindered the trophic shift. In this study, we show that venoms produced by extant predatory assassin bugs have strong anticoagulant properties mediated chiefly by proteolytic degradation of fibrinogen, and additionally contain anticoagulant disulfide-rich peptides. However, venom produced by predatory species also has pain-inducing and membrane-permeabilizing activities that would be maladaptive for blood-feeding, and which venom of the blood-feeding species lack. This study demonstrates that venom produced by the predatory ancestors of kissing bugs was exapted for the trophic switch to blood-feeding by virtue of its anticoagulant properties. Further adaptation to blood-feeding occurred by downregulation of venom toxins with proteolytic, cytolytic, and pain-inducing activities, and upregulation and neofunctionalization of toxins with anticoagulant activity independent of proteolysis.
Subject areas: Pharmacology, Entomology, Biochemistry, Evolutionary biology
Graphical abstract
Highlights
-
•
Extant assassin bugs and ancestors of kissing bugs had venom rich in proteases
-
•
Assassin bug venom causes pain and degrades fibrinogen to inhibit coagulation
-
•
Kissing bug venom is anticoagulant by other mechanisms and does not cause pain
-
•
Assassin bug coagulotoxicity was an exaptation favoring evolution of blood-feeding
Pharmacology; Entomology; Biochemistry; Evolutionary biology
Introduction
The family Reduviidae (Insecta: Hemiptera: Heteroptera) comprises 25 subfamilies, 24 of which are known as assassin bugs and retain the ancestral trophic strategy of predation on other arthropods. The remaining subfamily, Triatominae (kissing bugs), underwent a trophic shift to obligate hematophagy (blood feeding) approximately 30 mya.1 Kissing bugs have received considerable attention as vectors of Chagas disease, a trypanosomal infection that results in >7,000 deaths annually and significantly diminished quality of life for the >7 million people with this infection.2
Kissing bug salivary glands secrete saliva or venom (terms used interchangeably hereafter since an injected bioactive saliva fits the common definition of venom3). This saliva is adapted to facilitate blood-feeding, having manifold effects on host hemostatic systems by diverse mechanisms such as releasing nitrous oxide; the binding or degradation biogenic amines, ADP, thromboxane A2 and prostaglandins; and through inhibition of thrombin, the tenase complex, clotting factor XII, and platelet aggregation.4,5 The toxins that underlie these effects include apyrase, 5′ nucleosidase, and a radiation of the lipocalin family containing multiple instances of neofunctionalization. Apart from anticoagulant activity and vasodilation, kissing bug salivas are reported to be painless or analgesic, with unsuspecting victims often unable to sense a bite.4,5
By contrast, predatory assassin bugs produce venoms that are adapted to facilitate predation, and consequently they are potently and rapidly insecticidal.6,7,8,9 Assassin bugs also use their venom for defense against predators, and envenomation of mammals typically induces a strong pain response and dermonecrosis.4 The composition of assassin bug venom is markedly different from that of kissing bugs, being rich in proteases, pore-forming toxins, and peptides that both quickly paralyze prey and liquefy their internal organs.6,10 However, to date only assassin bugs in subfamilies Harpactorinae6,10,11,12 and Reduviinae7,13,14 have been examined using transcriptomics and proteomics, whereas bugs that are phylogenetically closer to the Triatominae have been neglected. Nevertheless, the available data suggest assassin bug venom composition has been relatively stable over evolutionary time, as many features of assassin bug venom are shared with distantly related heteropterans such as giant fish-killing water bugs (Belostomatidae) and other heteropterans that form part of the same predatory lineage.15,16 The drastically different composition of triatomine venom therefore suggests a rapid evolution of venom composition associated with their trophic shift to hematophagy (blood feeding).
However, other aspects of this trophic shift remain enigmatic. The first assassin bugs to attempt blood-feeding would have encountered multiple obstacles. Strong pain induced by their venom would be expected to increase the likelihood of their removal by hosts. Furthermore, without toxins adapted to circumvent the hemostatic system, the host cardiovascular system would act rapidly to prevent blood loss by vasoconstriction and clotting. One previous finding that highlights the importance of host defenses is that kissing bugs with ablated salivary glands are unable to obtain blood meals.17 According to one hypothesis, these evolutionary barriers were reduced in the microhabitat that is the nests of birds and mammals, where helpless juvenile hosts were unable to defend themselves against reduviids.1,18 While this would have circumvented removal by the host after activation of pain and itch responses, it remains unknown how the other defenses of the hemostatic system were overcome. We hypothesize that the venoms of ancestral predatory reduviids were also able to inhibit coagulation, constituting a possible exaptation for a trophic shift to hematophagy.
To test this hypothesis, we examined the venom composition of two assassin bugs, Ectomocoris sp. (Peiratinae) and Oncocephalus sp. (Stenopodainae) by proteomics and transcriptomics. Due to their phylogenetic positions, the composition of these bugs’ venom provides insights into the probable composition of venom produced by the predatory ancestors of kissing bugs. We tested the ability of venoms produced by the predatory reduviids Havinthus rufovarius and Pristhesancus plagipennis to disrupt clotting of human plasma and fibrinogen, compared to the blood-feeding reduviid Triatoma pallidipennis. We report that these predatory reduviid venoms show strong anticoagulant properties, in excess even of the documented anticoagulant properties of blood-feeding triatomines. These data suggest the obstacles to blood-feeding by the first triatomines may have been reduced by venom that was simultaneously an adaptation to predation and an exaptation for blood-feeding. We also examined the ability of predatory and blood-feeding reduviid venoms to activate mammalian pain-sensing neurons. Consistent with their ecology, we show that P. plagipennis venom has pain-inducing and cytolytic activity, while the venom of the blood-feeding T. pallidipennis does not.
Results
Predatory ancestors of kissing bugs possessed venom similar to extant predatory reduviids
To investigate the likely venom composition of the predatory ancestors of kissing bugs, we examined the venom composition of two assassin bugs from subfamilies not previously investigated, Peiratinae and Stenopodainae. Both of these subfamilies belong to the “higher Reduviidae” clade as used by Hwang et al.1 (Figure 1), which also contains Triatominae and the speciose subfamilies Reduviinae and Harpactorinae to which assassin bugs with previously characterized venom belong. Within higher Reduviidae, Peiratinae is the earliest diverging subfamily, while Stenopodainae is closely related to Triatominae.1
Figure 1.
Evolution of reduviid venoms across higher Reduviidae
Pie charts show the number of protein precursors detected by LC-MS/MS of venom for Ectomocoris sp. (Peiratinae) and Oncocephalus sp. (Stenopodainae) (this study), T. pallidipennis (Triatominae),19Platymeris rhadamanthus (Reduviinae),7 and Pr. plagipennis (Harpactorinae).6,10 The phylogenetic tree in the lower panel is from Hwang and Weirauch.1 Photos by Ian McMillan (Ectomocoris sp.), Graeme V. Cocks (Oncocephalus sp.), Elizabeth Arellano (T. pallidipennis), Eric Isselée (Pl. rhadamanthus), and Jiayi Jin (Pr. plagipennis).
Venom was extracted from Ectomocoris sp. (Peiratinae) and Oncocephalus sp. (Stenopodainae) and analyzed by comparing mass spectra obtained by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with species-specific amino acid sequence databases obtained from venom gland RNA sequencing (RNA-seq) using protocols established for other species of reduviids.20,21 These experiments revealed 109 and 153 peptide and protein primary structures present in the venom of Ectomocoris sp. and Oncocephalus sp., respectively. Both the peiratine and stenopodaine bug possessed venom that is highly similar in composition to venom produced by the predatory harpactorine and reduviine assassin bugs whose venom composition has been previously reported (Figure 1, Data S1).6,7,10 All electrostimulated venoms from predatory species featured high numbers of proteases (42–56% of primary structures detected), the majority of which (39–42%) were accounted for by S1 proteases. Proteases accounted for 60.0% and 39.7% of spectral counts originating from Ectomocoris sp. and Oncocephalus sp. venoms, respectively, indicating proteases are not just numerous but highly abundant in both venoms, similar to previously published data for other predatory reduviids (Data S1).6,7,10 The other protein classes present revealed high levels of conservation in venom composition across higher Reduviidae, with protein families such as redulysins, cystatins, CUB domain proteins, and numerous protein families known only from heteropteran venoms, represented with minimal variation.
In comparison, the reported composition of venom from the kissing bug Triatoma pallidipennis19 is markedly different, being dominated by 51 (65% of proteins detected) different members of the triabin/lipocalin family. Only a single protease (M2 family) was detected, and the putatively cytolytic redulysin family is absent. Notably, despite the composition of T. pallidipennis venom being so different from predatory reduviids, more than 80% of proteins detected in T. pallidipennis venom have close sequence homologs (BLASTp E-value <0.001 in a search of all known reduviid venom proteins against all) among venom proteins of the four predatory species shown in Figure 1. These include at least three classes (triabins/lipocalins, 5′ nucleosidases, and inositol phosphate phosphatases) that are reported to facilitate blood-feeding by kissing bugs5 that are present as trace components of venoms produced by predatory species. These data strongly suggest the predatory ancestors of kissing bugs possessed venom highly similar to that produced by extant predatory assassin bugs and contained numerous proteases. Adaptation to blood-feeding by kissing bugs was achieved by the massive expansion of a small number of venom protein families, and downregulation and loss of most others.
Reduviid venoms display diverse coagulotoxic effects
We hypothesized that, due to their protease content, venoms of predatory reduviids have anticoagulant activity. In this study we compared venom activity of the kissing bug T. pallidipennis to that of the relatively common harpactorine species P. plagipennis and H. rufovarius. We were unable to obtain enough individual Oncocephalus sp. or Ectomocoris sp. bugs to collect sufficient venom for activity assays, however our proteotranscriptomic analyses of these venoms indicated that they were very similar in composition, and therefore likely share similar activity, to those of P. plagipennis and H. rufovarius.
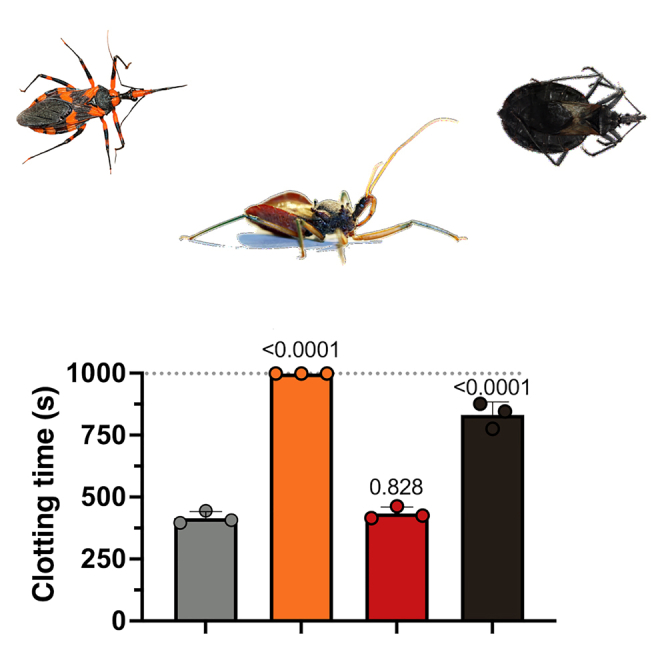
To test the coagulotoxicity of reduviid venoms, we added venom (20 μg/mL) to human plasma and measured spontaneous clotting time on a STA-R Max coagulation analyzer up to a maximum of 999 s. Control plasma spontaneously clotted in 415 ± 25 s (mean ± SD), while venom from the blood-feeding T. pallidipennis caused a delay in clotting time to 832 ± 51 s (p = 0.0012, unpaired t test) (Figure 2A), illustrating anti-coagulation effects.
Figure 2.
Coagulotoxicity of reduviid venoms
(A) Effect of venoms (20 μg/mL) on spontaneous clotting of human recalcified plasma after 120 s of incubation at 37°C. Note that venoms that require long incubation times to exert anticoagulant effects through fibrinogen destruction would show no activity on this assay. Data are mean ± SD of n = 3; dotted gray line indicates maximal assay time of 999 s (reached by H. rufovarius). Numbers above the bars indicate p values of t tests against control.
(B–D) Activity of venom (20 μg/mL) on clotting of fibrinogen (4 mg/mL; “Fib. destruction”) and plasma (all other tests). Error bars show SD. ND, not determined; Fib., fibrinogen; Inh., inhibition; Prothromb., prothrombinase. X-fold shift of zero indicates no change in clotting time. H. rufovarius photo by Jeff Gruber.
By comparison, the predatory reduviid venoms tested showed variation in their inhibition of spontaneous plasma clotting. H. rufovarius venom completely abolished clotting up to the maximum assay time of 999 s (p = 0.0006, unpaired t test), whereas P. plagipennis venom had no significant effect compared to control (435 ± 21 s; p = 0.41, unpaired t test).
To investigate the mechanism(s) by which reduviid venoms exert coagulotoxic effects, we used STA-R Max assays to test activity on specific clotting factors, including destruction of fibrinogen, and inhibition of FVIIa, FIXa, FXa, FXIa, thrombin, and the prothrombinase complex (Figures 2B–2D; see STAR methods). H. rufovarius venom (20 μg/mL) had no effect, compared to negative control, on inhibition of thrombin (p > 0.999, unpaired t test) and prothrombinase (p = 0.094, unpaired t test), with small but statistically significant effects observed on FIXa (p = 0.021), FXIa (p = 0.004), FVIIa (p = 0.0017), and FXa (p = 0.002, unpaired t tests). By contrast, in the fibrinogen destruction assay, H. rufovarius venom caused a 182-fold increase in clotting time compared to the control (p < 0.0001, unpaired t test) (Figure 2B). P. plagipennis venom (20 μg/mL) also produced strong anticoagulant effects due to fibrinogen destruction (177-fold) in this assay (Figure 2C).
Concentration- and incubation time-dependent anticoagulatory effects of predatory reduviid venoms
To further investigate the anticoagulant properties of predatory reduviid venoms, we measured how their ability to prevent fibrinogen clotting depends on venom concentration and incubation time with fibrinogen, prior to addition of thrombin (Figure 3). The anticoagulant effects of predatory reduviid venoms on fibrinogen were concentration-dependent. With a fixed incubation time of 1,800 s, negative controls and low concentrations of venom <2 μg/mL formed clots quickly (in <15 s), while venom concentrations >10 μg/mL completely inhibited clotting within the maximum assay time of 999 s (Figures 3A and 3B). The EC50 values for clot inhibition H. rufovarius and P. plagipennis were 4.9 and 5.0 μg/mL, respectively (Figures 3A and 3B).
Figure 3.
Concentration-response and incubation-time response curves for anticoagulant effects of predatory reduviid venoms on fibrinogen clotting
(A) and (C) show data for H. rufovarius and panels B and D show data for P. plagipennis venoms. (A) and (B) show clotting times for human fibrinogen (4 mg/mL) incubated with H. rufovarius and P. plagipennis venoms (0.05–20 μg/mL), respectively, for 1,800 s at 37°C before addition of thrombin. The gray dotted line shows maximal assay time of 999 s. A lack of clot observed for the highest venom concentrations and longest incubation times indicate complete fibrinogen destruction (inability to clot in presence of thrombin) by the venom. The dashed line corresponds to the negative control (i.e., buffer in place of venom). Data points are n = 3 ± SD error bars. (C) and (D) show dependence of incubation time by venom (20 μg/mL) activity on fibrinogen (4 mg/mL) before addition of thrombin. A four-variable nonlinear regression was fitted to the data.
With a fixed venom concentration of 20 μg/mL venom, shorter incubation times before addition of thrombin negated the ability of each venom to prevent clotting (Figures 3C and 3D). The effective incubation time to inhibit 50% of the anticoagulant effect (ET50 value) was slightly shorter for H. rufovarius (405 s) compared to that for P. plagipennis (520 s). These data show that venoms of both H. rufovarius and P. plagipennis possess similar fibrinogen-destroying abilities, despite their different observed effects on whole plasma clotting (Figure 2A).
Fibrinogen destruction depends on multiple classes of venom proteases
To investigate which molecules in predatory reduviid venoms are responsible for destroying fibrinogen, we tested the ability of protease inhibitors to prevent fibrinogen destruction (by venoms that exhibited fibrinogen destruction) after an 1,800 s incubation with venom (Figure 4). For H. rufovarius venom, we found that the metalloprotease inhibitor prinomastat (0.2 mM) had no effect (ANOVA p > 0.999), but the serine protease inhibitor AEBSF (2 mM) prevented the majority of the anticoagulant effect of venom (ANOVA p < 0.0001), rescuing clotting time from the assay maximum of 999 s to 66.5 ± 0.3 s, which is closer, but still significantly different, to the control value of 5.0 ± 0.3 s (t test p < 0.0001).
Figure 4.
Pharmacological inhibition of anticoagulant effects of predatory reduviid venoms
Venom (20 μg/mL) from either (A) Hr, H. rufovarius or (B) Pp, P. plagipennis was applied to the fibrinogen destruction assay in the presence of different protease inhibitors at concentrations of 0.2–2 mM. Error bars show SD. The dotted gray line indicates maximal assay time of 999 s. Numbers above the bars indicate p values of one-way ANOVAs with Dunnett’s multiple comparisons tests compared to venom without inhibitors (Hr or Pp).
By contrast, destruction of fibrinogen by P. plagipennis was not inhibited by addition of the metalloprotease inhibitor prinomastat, the cysteine protease inhibitor E-64, or either of the serine protease inhibitors AEBSF and PMSF. Addition of a commercial cocktail that inhibits both serine and cysteine proteases (Roche cOmplete) did however reduce clotting time (14.0 ± 0.0 s) to values closer to, albeit still significantly different from, the control (t test p = 0.0008). These data suggest that serine proteases are primarily responsible for fibrinogen destruction by H. rufovarius venom, whereas both serine and cysteine proteases in P. plagipennis venom are capable of degrading fibrinogen under these assay conditions.
Reduviid venoms reduce clot strength
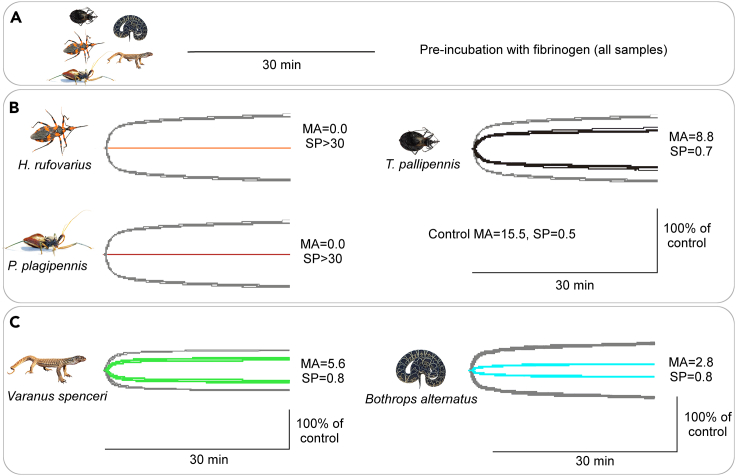
To investigate the effect of reduviid venoms on clot strength, we performed thromboelastography in the presence of fibrinogen and thrombin (Figure 5). In this assay, fibrinogen is incubated with venom, or buffer (negative control), for an initial 30 min; if no clot is observed in that time, thrombin is added to the same cup to induce clot formation, and clot strength is monitored over a subsequent 30 min period. During incubation with fibrinogen during the initial reading period, H. rufovarius and P. plagipennis venoms and the negative control had no effect (Figure 5A). In controls (with no venom), strong clots were formed during the secondary reading period in the presence of thrombin (gray lines in Figures 5B and 5C). By comparison, both H. rufovarius and P. plagipennis venom (20 μg/mL) completely prevented the ability of fibrinogen to form a clot in the presence of thrombin (Figure 5B), showing apparently stronger anticoagulant activity in this assay compared to venom of the blood-feeding T. pallidipennis, as well as reptile venoms reported to have anticoagulant activities such as those produced by Spencer’s monitor Varanus spenceri22 or the crossed pit viper Bothrops alternatus23 (Figure 5C).
Figure 5.
Reduviid venoms reduce strength of clots
(A) Lack of clot formation prior to addition of thrombin.
(B) Overlaid thromboelastography traces (n = 3) of reduviid venoms (20 μg/mL) (colored traces), compared to the control (gray traces). “MA” = maximum amplitude (mm), where larger indicates stronger clot; “SP” = split point (min), where larger values indicate a slower forming clot.
(C) Comparison data showing clot-weakening effect of reptile venoms. Left, Spencer’s monitor Varanus spenceri; data reproduced,22 photo by Daniel Bromley. Right, crossed pit viper Bothrops alternatus; data reproduced,23 photo by Dr Kristof Zyskowski.
Peptides in venoms of predatory reduviids have anticoagulant activity
We demonstrated that venoms of the predatory reduviids H. rufovarius and P. plagipennis have anticoagulant activity due to the proteases they contain, but we reasoned that because kissing bugs utilize other classes of proteins beyond proteases as anticoagulants, there are likely to be additional molecules in predatory reduviid venoms with anticoagulant activity. To test this hypothesis, we examined the ability of three assassin bug peptides with domains similar to serine protease inhibitors to inhibit the clotting of human plasma (Figure 6). To do so, P. plagipennis venom Kazal domain peptide 1 (PpK1) was produced recombinantly, whereas Pr11a1 and Pr11a2, two pacifastin family peptides from the venom of Platymeris rhadamanthus, were produced by solid-phase peptide synthesis. At a concentration of 10 μM, each of these three peptides showed significant anticoagulant activity on human plasma (Figure 6B). Thus, although these predatory venoms exhibit anticoagulation via their proteolytic activity, they also contain minor components with additional anticoagulant effects, likely through inhibition of protease activity. These data illustrate that kissing bug anticoagulant toxins may have evolved from assassin bug venom toxins that possess weaker anticoagulant effects but nevertheless may have been selected for during the trophic shift to blood-feeding. For example, rhodniin, which is a picomolar-affinity inhibitor of thrombin from kissing bug venom,24 may have evolved from an assassin bug Kazal domain venom peptide similar to PpK1 (Figure 6C).
Figure 6.
Anticoagulant effect of predatory reduviid venom peptides
(A) The upper alignment shows the primary structures of the P. plagipennis venom peptide PpK1 and the C-terminal domain of rhodniin (Rhodniin_C), a thrombin inhibitor produced by the kissing bug Rhodnius prolixus. The lower alignment shows the pacifastin-domain peptides Pr11a1 and Pr11a2 aligned to locust Schistocerca piceifrons pacifastin (NCBI Protein: XP_047097550.1). Stars underneath alignments show similarity in ClustalW format, with cysteines (highlighted gray), cationic residues (blue), anionic residues (red), hydrophobic residues (pink), and proline (brown).
(B) Changes in clotting time induced by venom peptides (10 μM). Error bars show SD. Numbers above the bars show significance compared to control (n = 3, unpaired t test).
(C) AlphaFold 2 predicted structure of assassin-bug PpK1 compared to the crystal structure of kissing-bug rhodniin_C (PDB: 1TBR).
Venoms of predatory, but not blood-feeding reduviids, cause pain
To investigate the pain-inducing capacity of predatory and blood-feeding reduviids, we tested the effects of the venoms on mammalian dorsal root ganglion (DRG) cells, which are primarily pain-sensing neurons. In this assay, [Ca2+]i is monitored using a pre-loaded calcium-sensitive dye and a fluorescence microscope. Venom (100 μg/mL) from the predatory P. plagipennis caused a strong increase in dye fluorescence in all cells, with subsequent dye leakage into the extracellular medium (Figure 7A). After application of P. plagipennis venom, 20 mM KCl produced no response, indicating that transmembrane ionic gradients have been destroyed. This result is consistent with previous results for predatory reduviids suggesting direct membrane disruption as a primary mechanism of action7 and is consistent with pain-causing activity when injected into mammals—the typical consequence of natural defensive envenomations by predatory assassin bugs.4 By contrast, application of venom of the blood-feeding T. pallidipennis (100 μg/mL) to DRGs produced no change in [Ca2+]i nor other observable changes. Subsequent addition of 20 mM KCl produced a strong [Ca2+]i increase, indicating intact transmembrane ionic gradients.
Figure 7.
Venoms of predatory, but not blood-feeding, reduviids permeabilize mammalian neurons
(A) Fluorescence micrographs showing calcium responses following application of reduviid venoms (100 μg/mL) to DRG cells.
(B) Time course of [Ca2+]i response to venom (660 μg/mL, upper panel) and concentration-response relationship (lower panel). Data are mean ± SEM of n = 3 replicates.
(C) Time course of membrane permeabilization in response to venom (660 μg/mL, upper panel) and concentration-response relationship (lower panel). Data are mean ± SEM of n = 3 replicates.
To quantify the cytolytic activity of reduviid venoms, we measured [Ca2+]i and nucleic acid exposure in the human neuroblastoma cell line SH-SY5Y using a fluorescent imaging plate reader (FLIPR) duplex assay that measures [Ca2+]i, concurrently with membrane permeability indicated by access of propidium iodide to intracellular nucleic acids (Figures 7B and 7C).25,26 P. plagipennis venom induced sustained propidium iodide responses and transient [Ca2+]i signals, consistent with membrane disruption, DNA and RNA exposure, and subsequent dye leakage into the extracellular space that contains a Ca2+-sensitive dye quencher, terminating the signal. The EC50 values for this effect were (mean ± SEM) 70 ± 3 μg/mL and 258 ± 10 μg/mL for [Ca2+]i increases and nucleic acid exposure, respectively. By contrast, venom of the blood-feeding T. pallidipennis caused no effect at concentrations up to 660 μg/mL. These results are consistent with the venoms of predatory reduviids, but not blood-feeding reduviids, causing pain in mammals via disruption of the membrane of pain-sensing neurons.4
Because the venom of the blood-feeding Triatoma infestans has been reported to inhibit NaV channels,27 which could confer analgesic properties, we tested the ability of T. pallidipennis venom to modulate NaV channels in SHSY5Y cells using a fluorescence-imaging assay, and NaV1.7 channels expressed in HEK293 cells using electrophysiology. However, we found no significant modulation of NaV channels in the presence of T. pallidipennis venom (Figure S1).
Discussion
In this study we investigated the ability of venoms from predatory and blood-feeding reduviids to modulate mammalian blood clotting and activate pain-sensing neurons. We found that venom from both predatory and blood-feeding reduviids exhibit anticoagulant activities, albeit through different modes of action, whereas only venom from predatory species activated mammalian neurons consistent with the capacity to cause pain.
Limitations of the study
A more detailed picture of venom evolution in Reduviidae would examine venom produced by the early-diverging phymatine complex as well as venoms of non-reduviid cimicomorphs, prey specialists such as the arachnophagous Emesinae and the myrmecophagous Holoptilinae, and some of the many groups that employ hunting specializations, such as the use of plant resins to catch prey.1 Within Triatominae, examination of saliva produced by additional species from multiple lineages (especially those that switched to blood-feeding independently, if the subfamily is shown to be polyphyletic) and including generalists and specialists on different host taxa and species associated especially with nests and burrows will be informative. The venoms of predatory reduviids such as Zelurus spp. and Opisthacidius spp. that are most closely related to Triatominae and share some behaviors such as habitation of bird nests by Opisthacidius spp.1 may also provide more information about the evolution of triatomine saliva.
We searched for, but did not find, any evidence supporting the previously proposed hypothesis that kissing bug venom has analgesic activity due to its ability to inhibit NaV1.7 channels.27 However, these data do not rule out the possibility that kissing bug venom could induce analgesia by a different mechanism or under different assay conditions. For example, kissing bug venom could modulate other ion channels or receptors involved in nociception, such as NaV1.1, NaV1.8, or transient receptor potential (TRP) channels.28,29,30
Evolution of venom in higher Reduviidae
We found that assassin bugs from the earliest-diverging subfamily of higher Reduviidae (Peiratinae), as well as a subfamily closely related to Triatominae (Stenopodainae) have venom that is highly similar in composition to that produced by previously examined reduviids from Harpactorinae and Reduviinae. This finding suggests that venom composition has been largely stable due to purifying selection among the higher Reduviidae. This is consistent with a switch to predation from herbivory that gave rise to the common ancestor of Heteroptera 250–300 million years ago, and which was accompanied by the adaptation of saliva into toxic venom.15 Predation has likely remained as a conserved trophic strategy from the last common ancestor of Heteroptera through to extant Reduviidae,15 which is likely to favor purifying rather than diversifying selection31 in contrast to heteropteran groups that have switched to trophic strategies such as blood-feeding or back to phytophagy. Nevertheless, this near homogeneity of venom composition in Reduviidae is perhaps surprising considering that reduviid predators have evolved numerous instances of prey specialization and specialized hunting strategies that might be expected to co-evolve with venom. Possibly, further studies focusing on species with more specialized hunting strategies, or different kinds of venom bioactivities, will uncover more nuanced venom adaptations. Alternatively, it is possible that the protease-rich venoms of predatory reduviids are simply well-suited to myriad different hunting strategies. These data are consistent with other examples where venoms are surprisingly similar despite great differences in biology, for example between solitary and eusocial bees.32
In any case, the near homogeneity of venom from different predatory reduviid subfamilies examined, and in particular the high number of S1 proteases present, suggests that the predatory ancestors of Triatominae would likely have had a similar venom composition to extant predatory higher reduviids when they first fed on blood. For this reason, we consider that comparing the anticoagulant activities of venoms produced by extant predatory and blood-feeding assassin bugs provides insights into venom exaptation and adaptation during the evolution of blood-feeding in Triatominae.
Both predatory and blood-feeding reduviid venoms are anticoagulant
Venom produced by the predatory species H. rufovarius and P. plagipennis prevented fibrinogen clotting through direct proteolytic destruction of fibrinogen (Figures 2 and 5). This is the same mechanism by which venoms of varanid lizards have been shown to be anticoagulant,22 and is consistent with the high content of proteolytic enzymes that has been previously reported in venoms of predatory reduviids.10 Notably, the predatory reduviid venoms have substantially higher potency than many reptile venoms reported to have anticoagulant effects.22,23
Given that P. plagipennis venom had strong anticoagulant effects in the fibrinogen destruction assay (Figure 3) and in thromboelastography (Figure 5), it was initially surprising that it showed no observable effect on spontaneous clotting of recalcified plasma (Figure 2A), which contains 1.5–4.5 mg/mL fibrinogen.33,34 This can, however, be explained by the relationship between the degree of fibrinogen destruction and the ability to form a clot within a given time frame. The steep slopes we observed in concentration- and time-response curves for inhibition of fibrinogen clotting by venom (Figure 3) suggest there is a critical threshold of fibrinogen destruction, below which clotting remains feasible, and above which clotting is impossible, similar to previously reported results on multiple snake venoms.35,36 That is, H. rufovarius venom required a shorter effective time of incubation (405 s) compared to P. plagipennis venom (520 s) to destroy sufficient fibrinogen to inhibit clotting. Under the conditions of our assay, spontaneous clotting occurred in 415 s, which is in between these values. Thus, the two harpactorine venoms produced markedly different results on this assay despite possessing very similar bioactivity.
Regarding which molecules are responsible for this destruction of fibrinogen by H. rufovarius or P. plagipennis venom, we found that metalloproteinases were not responsible but that serine proteases within H. rufovarius venom were largely responsible for the anticoagulant activity, as indicated by AEBSF (a serine protease inhibitor) inducing a 14-fold reduction in the anticoagulant activity. Cysteine proteases were also found to be partially responsible, albeit to a lesser degree, as indicated by E-64 (a cysteine protease inhibitor) reducing venom activity by 0.41-fold. In contrast, destruction of fibrinogen by P. plagipennis was more resistant to inhibition by the protease inhibitors tested. Neither AEBSF or PMSF (two different serine protease inhibitors), nor E-64 (cysteine protease inhibitor), were capable of rescuing fibrinogen from destruction by P. plagipennis venom. Only inhibition of multiple classes of proteases (cysteine and serine proteases) by addition of an inhibitor cocktail (Roche “cOmplete”) was sufficient to reduce clotting time to near control values, albeit still significantly different than the control. These data are consistent with a study showing that P. plagipennis venom contains both S1 (trypsin-like) and C1 (cathepsin B-like) proteases,10 and it suggests that multiple enzyme classes contribute to the fibrinogen-destroying anticoagulant activity of this venom. Our results demonstrate that proteases are the most important anticoagulants in the venoms of predatory reduviids, but we also showed that peptides that are minor venom components can have anticoagulant properties. Although we did not determine the mechanism by which these peptides act, we hypothesize they inhibit one or more serine proteases in the coagulation cascade.
We found that venom of the kissing bug T. pallidipennis also inhibits spontaneous clotting of recalcified plasma (Figure 2A) but this is not due to fibrinogen destruction (Figure 2D). T. pallidipennis venom also weakened clots in the thromboelastography assay (Figure 5A, lower trace). These results are consistent with the well-established anticoagulant activities of triatomine venoms reviewed by Ribeiro et al.5 and the vastly different protein composition of venoms produced by predatory reduviids, which are rich in proteases and pore-forming proteins, compared to those of blood-feeding reduviids that are rich in lipocalin family proteins.10,15 T. pallidipennis venom has been reported to contain triabin, an inhibitor that prevents thrombin from converting fibrinogen to fibrin.37 While such an activity may be responsible for the thromboelastography results, we found no evidence for thrombin or FXa inhibition in specific coagulometer assays (Figure 2D). Since T. pallidipennis venom is reported to produce anticoagulant activity via mechanisms that our assays do not measure, such as inhibition of collagen-induced platelet aggregation by pallidipin,38 the full coagulotoxic capacity of T. pallidipennis venom may not be observed in our experiments.
Venoms of predatory reduviids, but not blood-feeding reduviids, cause pain
Besides anticoagulant activity, hematophagous parasites may produce analgesic compounds that limit their detection by the host. The venoms of kissing bugs have been reported to have an analgesic effect39 that could possibly be mediated by their reported inhibition of mammalian NaV channels.27 Conversely, venoms of predatory reduviids are reported to cause rapid pain responses when injected into vertebrates,9 which would be expected to lead to rapid detection and possibly removal by the host.
We found that venoms of predatory and blood-feeding reduviids showed marked differences in their ability to permeabilize mammalian peripheral sensory neurons or a neuroblastoma cell line (Figure 6). While the cytolytic activity of predatory reduviid venoms is likely to underlie pain experienced after envenomation,4 triatomine venom from blood-feeding reduviids lacks these activities.
Adaptation and exaptation in reduviid venom evolution
The venom of predatory assassin bugs is used to induce paralysis, death, and extra-oral digestion of arthropod prey, as well as deterring predators by causing pain or death.4 Although P. plagipennis (and probably all predatory assassin bugs) is capable of injecting two distinct, complex venoms,6 we analyzed venom harvested by electrostimulation, which is known to originate in the larger posterior main gland, and to have potent insecticidal, cytolytic, and proteolytic activity.6,7,10 Predatory assassin bugs do not feed on vertebrate blood and, thus, the anticoagulant activity of their venoms is not an adaptation. The most likely explanation for the potent fibrinogen-destroying activity of predatory reduviid venoms is broad-spectrum proteolytic activity that is adapted to induce death of prey and initiate extra-oral digestion. The assays performed in this study do not closely recapitulate actual blood-feeding events by Reduviidae (e.g., incubation times of 1,800 s are longer than the duration of most blood meals; (25)), and actual venom protein concentrations are likely to be three to four orders of magnitude higher than the concentrations tested here. Nevertheless, our results clearly indicate a strong potential for disruption of hemostatic systems by predatory reduviid venoms. We propose that this anticoagulant activity of predatory reduviid venom due to its high proteolytic activity is best understood as an adaptation for predation and an exaptation for the development of hematophagy.
If predatory reduviid venoms possessed exaptations that lowered the evolutionary barrier to blood-feeding, they were nevertheless lacking the sophisticated adaptations displayed by the saliva of extant Triatominae. Predatory reduviid venoms also cause rapid and strong pain responses, likely due to the cytolytic and proteolytic activities that are adaptative for both predation and defense. For adoption of blood-feeding, pain-causing activities would have constituted a drawback, possibly restricting triatomines to feeding on defenseless bird and mammal juveniles, as has been previously hypothesized.1,18 Nevertheless, hematophagy limited to young juveniles would have exposed the early Triatominae to different selection pressures compared to their predatory ancestors. New selection pressures on triatomine venom composition likely resulted in the loss of most proteolytic and pore-forming toxins; the loss of pain-inducing toxins; and increased expression, duplication, and neofunctionalization of toxins with anticoagulant activity, such as lipocalins. Lipocalin family proteins confer many anticoagulant properties of triatomine bug venom and are present in the venoms of predatory bugs, albeit only as minor components.10 While production of a large protein such as a lipocalin in active form is beyond the scope of this study, we found that some disulfide-rich peptides, which are also minor components of venoms produced by predatory reduviids, also possess anticoagulant properties. This demonstrates that predatory reduviid venoms contain multiple anticoagulant molecules that would have been exposed to the changing selection pressures during the switch to hematophagy, eventually resulting in the adaptations present of triatomine saliva today. The absence of pain-inducing activity in triatomine venoms is likely an adaptation for hematophagy, and one that may have allowed the transition from obligate feeding on juvenile vertebrates in nests to the broader range of vertebrate hosts parasitized by kissing bugs today.40
In conclusion, the venoms of predatory reduviids are capable of strong anticoagulant activity, and this is likely due to the presence of proteases that destroy fibrinogen. Thus, venoms of both blood-feeding and predatory reduviids have anticoagulant properties, albeit conferred by different toxins that act on distinct molecular targets. While venoms of predatory bugs induce strong neuronal permeabilization, venoms of blood-feeding bugs do not. We propose that the anticoagulant properties of predatory reduviids nevertheless constitute an exaptation that would have lowered the evolutionary barrier to the development of blood-feeding by kissing bugs, whereas pain-inducing properties are likely to have been attenuated over time to exploit blood-feeding from able adult animals rather than nestlings. This study provides insights into adaptations and exaptations during the trophic switch to hematophagy by the Triatominae.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andy Walker (a.walker@imb.uq.edu.au).
Materials availability
Materials generated in this study, including expression plasmid for peptide PpK1, and recombinant or synthetic peptides can be requested from the lead contact.
Data and code availability
-
•
Data: RNA-Seq data in FASTQ format are available from the USA National Center for Biotechnology Information (NCBI)’s Sequence Read Archive: BioProject ID PRJNA1046250. The amino acid sequences of venom toxins and the contigs encoding them are available from GenBank (NCBI Nucleotide: PP510767–PP510872, PP516297–PP516299, PP517412–PP517564). The CO1 barcoding genes of the Ectomocoris sp. and Oncocephalus sp. used in this study are available from GenBank (NCBI Nucleotide: PP506657, PP506658), respectively. Mass spectral data of venom is available in the ProteomeXchange PRotein IDEntification (PRIDE: PXD050854).
-
•
Code: No original code was generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Quantification and statistical analysis
Statistical tests were performed using Prism v.9.1.2 (GraphPad Software). Unpaired t-tests with Welch’s correction (i.e., no assumption of equal SDs) were used to compare the means of the controls with those of treatments (i.e., venom tests). One-way ANOVA with Dunnett’s multiple comparisons tests were used to test for significant effects of the means of inhibitors on venom activity.
Acknowledgments
The authors would like to thank Pam and Danny Cocks and Elizabeth Arellano for permission to use photos. We also thank Abhinandan Chowdhury for assistance with inhibitors. We gratefully acknowledge financial support from the Australian Research Council (Centre of Excellence grant CE200100012 to G.F.K.), and the Australian National Medical and Health Research Council (Ideas Grant APP1188959 to F.C.C., Investigator Grant APP2017461 to S.D.R., Principal Research Fellowship APP1136889 to G.F.K., and Investigator Grant APP2017086 to I.V.).
Author contributions
A.A.W. conceptualized the study. C.N.Z., F.C.C., S.D.R., G.C., R.S.M., M.J.H.-V., B.G.F., and A.A.W. designed experiments. C.N.Z., F.C.C., S.D.R., R.S.M., E.R.R., M.J.H.-V., and J.J. performed experiments. C.N.Z., F.C.C., S.D.R., R.S.M., J.J, and A.A.W. analyzed data; C.N.Z. and A.A.W. wrote the original draft. All authors edited the final manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21(ΔDE3) | Thermo Fisher Scientific | Cat# EC0114 |
| Biological samples | ||
| Oncocephalus sp. and Ectomocoris sp. insects | Private land in Brisbane, QLD, Australia | N/A |
| T. pallidipennis insects | Private houses in the county of Yautepec, Morelos, Mexico | N/A |
| Blood from uninfected CD-1 mice | Universidad Nacional Autónoma de México | N/A |
| Reduviid venom samples | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| PpK1, Pr11a1, and Pr11a2 peptides | This study | N/A |
| sequencing-grade trypsin | Sigma Aldrich | Cat# 7575 |
| plasma | Australian Red Cross | N/A |
| fibrinogen | Sigma Aldrich | Cat# F3879 |
| ‘cOmplete’ inhibitor | Roche | Cat# 11697498001 |
| prinomastat | Sigma Aldrich | Cat# 192329-42-3 |
| AEBSF | Sigma Aldrich | Cat# 101500 |
| recombinant tobacco etch virus protease | Gift of Dr. Natalie Saez | N/A |
| Fluo-4 a.m. calcium indicator | Thermo Fisher Scientific | Cat# F36206 |
| calcium-4 dye | Bio-Strategy | Cat# MDEVR8141 |
| propidium iodide | Sigma Aldrich | Cat# P4864 |
| veratridine | Sigma Aldrich | Cat# V5754 |
| tetrodotoxin | ABCAM Australia | Cat# ab120055 |
| Critical commercial assays | ||
| DNeasy kit | Qiagen | Cat# 74104 |
| Dynabeads mRNA Direct kit | Thermo Fisher Scientific | Cat# 61012 |
| Deposited data | ||
| RNA-Seq data in FASTQ format | This study | SRA: PRJNA1046250 |
| amino acid sequences of venom toxins | This study | GenBank: PP510767–PP510872, PP516297–PP516299, and PP517412–PP51756 |
| barcoding genes of the Ectomocoris sp. and Oncocephalus sp. | This study | GenBank: PP506657 and PP506658 |
| Mass spectral data | This study | PRIDE: PXD050854 |
| Experimental models: Cell lines | ||
| SH-SY5Y neuroblastoma cells | Gift of Prof. Irina Vetter | N/A |
| HEK293 cells stably expressing human NaV1.7 co-expressed with the β1 auxiliary subunit | SB Drug Discovery | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6 mice | Animal Resources Center | C57BL/6JArc |
| Recombinant DNA | ||
| pLICC expression vector encoding MBP:PpK1 fusion protein | GenScript | N/A |
| Software and algorithms | ||
| CLC Genomics Workbench | CLC Bio | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/ |
| Trinity v2.2.0 | Open software | https://github.com/trinityrnaseq/trinityrnaseq/releases |
| GetORF | Server | https://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html |
| ProteinPilot 4.0.8085 | SCIEX | https://sciex.com/products/software/proteinpilot-software |
| SignalP 4.1 | https://services.healthtech.dtu.dk/services/SignalP-4.1/ | |
| UniProt UniRef90 database | UniProt | https://www.uniprot.org/ |
| Pfam database | InterPro | https://www.ebi.ac.uk/interpro/download/Pfam/ |
| PRISM 10 | GraphPad | https://www.graphpad.com/ |
| Other | ||
| STA-R Max® coagulation analyser | Stago | https://www.stago.com.au/products-services/max-generation-analysers/sta-r-max3/ |
| Thromboelastograph® 5000 Haemostasis Analyser | Haemonetics® | Cat# 07-033 |
| poly-D-lysine-coated culture plate | Corning | Cat# 354651 |
| Ti-E deconvolution inverted microscope equipped with a Lumencor Spectra LED light source | Nikon | https://www.microscope.healthcare.nikon.com/en_AOM/products/inverted-microscopes/eclipse-ti2-series |
| 384-well black wall flat clear-bottom microplates | Corning | Cat# 3764 |
| FLIPR Penta System | Molecular Devices | https://www.moleculardevices.com/products/flipr-penta-high-throughput-cellular-screening-system |
| QPatch 16X | Sophion Bioscience | https://sophion.com/products/qpatch/ |
Experimental model and study participant details
Recombinant expression used BL21(ΔDE3) E. coli, which were maintained in Lysogeny Broth at 37°C with 180 rpm shaking.
Method details
Insects, venom collection and preparation
Ectomocoris sp., Oncocephalus sp., H. rufovarius, and P. plagipennis were collected from private land in Brisbane, Australia. Venom was harvested by inserting the rostrum into a pipette tip and applying a mild electric current (5–25 V) to the thorax. T. pallidipennis species were collected in private houses in the county of Yautepec, Morelos, Mexico. T. pallidipennis species were positively identified and analyzed for possible Trypanosoma infection. Non-infected insects were separated and bred in the laboratory. The triatomine colony was maintained at the Instituto de Biotecnología belonging to the Universidad Nacional Autónoma de México (UNAM). All insects were maintained at 28°C, 60% relative humidity and a light/dark photocycle of 12:12 h. They were fed with blood from uninfected CD-1 mice, 25–30 g. The experimental protocol was approved by the Ethics and Research Committee at Instituto de Biotecnología, UNAM, permit No. 271. Venom was harvested from unsexed non-infected adults starved for 1–3 weeks of the last feeding. To harvest venom, the rostrum was inserted into a capillary and the insect stimulated by manual touching. Venom was transferred to 2 mL freezing vials in dry ice and lyophilized for storage. Venom protein concentration was estimated from absorbance at 280 nm measured using a NanoDrop2000 spectrophotometer (Thermo Fisher, Sydney, NSW, Australia).
Transcriptomics and proteomics
Transcriptomic and proteomic experiments were carried out on Oncocephalus sp. and Ectomocoris sp. due to their informative phylogenetic positions. For RNA-Seq experiments, insects were anesthetized by CO2 for 10 min, and the venom gland complex (including posterior and anterior main glands and accessory glands) were removed and stored in >10-fold the tissue volume of RNAlater (Thermo Fisher) at −20°C until further analysis. Total RNA was extracted using a RNeasy kit (Qiagen, Hilden, Germany) and mRNA purified using a Dynabeads mRNA Direct kit (Thermo Fisher) according to the manufacturer’s instructions. Construction and sequencing of 150 bp insert paired-end TruSeq Stranded mRNA libraries was performed by the Institute for Molecular Bioscience Sequencing Facility at The University of Queensland, Australia. The multiplexed libraries were then sequenced as a part of a high-output run on an Illumina HiSeq 2500 instrument. One transcriptome was assembled using Trinity v2.2.0, and five transcriptomes were assembled using CLC Genomics Workbench (CLC Bio, Aarhus, Denmark) with k-mer values of 24, 34, 44, 54, and 64. Multiple assembly programs were used to reduce false negatives in the final dataset, relying on the ProtGroup algorithm (see below) to avoid false positive identification of duplicate sequences based on the same spectral data. A library of possible protein sequences was generated using GetORF (https://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) with open reading frame length >90 bp.
For liquid chromatography-tandem mass spectrometry (LC-MS/MS), reduced, alkylated and trypsinized samples were prepared by diluting the sample to be analyzed in reducing/alkylating solution (1% 2-iodoethanol, 0.25% triethylphosphine, 48.75% acetonitrile (ACN), 50 mM ammonium carbonate pH 11.0) and incubating the sample for 1 h at 37°C. Samples were then dried by vacuum centrifugation and reconstituted in digestion reagent (20 ng/μL sequencing-grade trypsin (#7575, Sigma Aldrich, St Louis, MO, USA) in 40 mM ammonium bicarbonate pH 8.0, 10% ACN) before quenching in an extraction reagent (50% ACN, 5% formic acid (FA)), vacuum centrifugation, and reconstitution in 1% FA. Reduced and alkylated (but not trypsinized) and native MS samples were generated by omitting the digestion step, or the reduction, alkylation, and digestion steps, respectively. Samples were analyzed using a 5600 Triple TOF mass spectrometer (SCIEX) equipped with a Turbo V ion source and coupled to a Nexera X2 LC system (Shimadzu, Kyoto, Japan) with a Zorbax 300SB-C18 column (#858750–902, Agilent, Santa Clara, CA, USA) incubated at 60°C. Peptides were eluted over 75 min using a gradient of 1–40% solvent B (90% ACN/0.1% FA) in solvent A (0.1% FA) at a flow rate of 0.2 mL/min. MS1 scans were collected between 350 and 2200 m/z, and precursor ions in the range m/z 350–1500 with a charge from +2 to +5 and signal >100 counts/s were selected for analysis, excluding isotopes within 2 Da. MS/MS scans were acquired with an accumulation time of 250 ms and cycle time of 4 s. The ‘Rolling collision energy’ option was selected in Analyst software, allowing collision energy to be varied dynamically based on m/z and z of the precursor ion. Up to 20 similar MS/MS spectra over m/z range 80–1500 were pooled from precursor ions differing by < 0.1 Da.
The resulting spectral data in WIFF format were then compared with the amino acid sequence database using the Paragon 4.0.0.0 and ProtGroup algorithms implemented in ProteinPilot 4.0.8085 software (SCIEX, Framingham, MA, USA). The ProtGroup algorithm identifies the smallest number of sequences capable of explaining mass spectral data, thereby reducing false positive duplicated sequences in the final proteome. A mass tolerance of 50 mDa was used for both precursor and MS/MS ions. Low-quality identifications (<95% confidence at the protein level, or <2 tryptic fragments detected with >95% confidence) along with contaminant and decoy database identifications were removed. Label-free quantitation was performed using peptides with the highest spectral counts, unique amino acid sequence, and >90% confidence identification in the trypsinized whole venom sample. For each protein, the MS1 spectral counts from the three most abundant tryptic fragments were summed.
Each precursor sequence was annotated for a secretion signal sequence using SignalP 4.1.41 Homology to known sequences was annotated using BLAST searches against UniProt UniRef90 database; and HMMER searches42 against the Pfam database. Polypeptide precursors were grouped by BLAST searches against each other (threshold E < 0.001). The comparison of venom composition in Figure 1 shows the number of detected sequences in each delineated family. The final determined proteomes were submitted to NCBI.
Plasma and fibrinogen preparation
For coagulation studies, venom was frozen, lyophilized, and stored at −80°C. It was later reconstituted into a 50% glycerol working stock solution at 1 mg/mL, which was stored at −20°C in liquid form. Surplus plasma in 3.2% citrate from healthy human donors (18+ years old) was donated by the Australian Red Cross (Kelvin Grove, QLD, Australia) under Research Agreement #18-03QLD-09 and University of Queensland (UQ) Human Ethics Committee Approval #2016000256. Pooled plasma bags (∼280 mL each; blood type O+) were thawed at 37°C, prepared into 1.2 mL aliquots, flash frozen in liquid nitrogen, and immediately stored at −80°C until use. Human fibrinogen (Cat #F3879, Sigma Aldrich, St. Louis, MO, USA) was reconstituted in enzyme running buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.3) to a concentration of 4 mg/mL, vortexed until clear, separated into 1 mL aliquots, flash-frozen in liquid nitrogen, and immediately stored at −80°C until required. When required, plasma/fibrinogen aliquots were rapidly thawed at 37°C in a water bath and used immediately for experimentation.
Coagulotoxicity tests
Functional assays were conducted using T. pallidipennis, H. rufovarius, and P. plagipennis venoms, or lab-produced peptides, due to the greater amount of material available and the different feeding strategies used by these species. Coagulation tests were performed using an STA-R Max coagulation analyser (Stago, Asnières sur Seine, France). The time it took for the venom to cause plasma or fibrinogen to clot was measured automatically via a viscosity-based (mechanical) detection system, whereby opposing magnets oscillate a spherical pellet inside a 10 mm wide cuvette (250 μL total volume) until a clot forms, which stops the pellet. The maximum reading time of the machine was set to 999 s. Assay methods are detailed in Table S1. For coagulation factor inhibition tests, the final concentration of each factor in the assay was matched to the physiological concentration of that factor in plasma.34 Incubation times for assays with activated factors in the assay were limited to 120 s to preserve the activity of the sensitive factor, whereas fibrinogen remains stable for long periods at 37°C (see controls) and because some venoms (e.g., night adder, Causus spp.) require long incubation times for full activity to be observed.36 Changes in coagulation time were expressed as x-fold shift using the following formula, in which an x-fold shift of zero indicates no shift, whereas an x-fold shift of 1 indicates a 100% increase in clotting time:
Clot strength tests
The ability of venom to reduce the clot strength of human fibrinogen was measured using a Thromboelastograph 5000 Haemostasis Analyser (Haemonetics, Boston, MA, USA, Cat#07–033), as previously described,22 using ‘natural pins and cups’ (Lot#HMO3163, Haemonetics Australia, North Ryde, NSW, Australia). Method details are shown in Table S1. In brief, OK Buffer, calcium, phospholipid, venom (20 μg/mL final), and (lastly) fibrinogen were added to the cup and pipette-mixed before measurements commenced and proceeded for 30 min. Then, if no clot formed, thrombin was added to the cup and measurement continued for an additional 30 min.
Inhibitor tests
Inhibition of fibrinogenolytic venom activity was determined using a range of commercially available enzyme inhibitors that inhibit different toxin classes (Table S2). Inhibitors were made up according to manufacturer’s instructions and added in replacement of OK buffer in the fibrinogen-destruction assay at an incubating concentration of 10 mM (final 1 mM). The ‘cOmplete’ inhibitor came in tablet form and was made up into a 2× solution according to manufacturer’s instructions. The concentration of Prinomastat and AEBSF was chosen as per previous work, which at 0.2 mM (final) inhibited viper (Microvipera and Trimeresurus spp., respectively) venom at 20 μg/mL (the same concentration used in this study).43,44,45,46 PMSF was additionally tested on P. plagipennis venom only because AEBSF did not inhibit its venom activity, and these two serine protease inhibitors have previously acted differently on serine-protease containing snake venoms.44 Negative controls for inhibitor tests were performed by replacing venom in the assay with an equivalent blank. Where available, positive controls for the inhibitors were run using a venom previously described as being inhibited by the specific inhibitor used.
Preparation of reduviid venom peptides
Reduviid venom peptides were selected from the literature6,7 due to their primary structural homology to known protease inhibitors. P. plagipennis Kazal domain peptide 1 (PpK1) was produced via heterologous expression in the periplasm of Escherichia coli.47 Briefly, a gene encoding PpK1 was synthesized and subcloned into the pLICC vector containing an ampicillin (Amp) resistance marker to yield a gene encoding a His6:maltose binding protein:PpK1 fusion protein by GenScript (Piscataway, NJ, USA) as a commercial service. This plasmid was used to transform BL21(ΔDE3) E. coli. Cultures carrying the recombinant plasmid were then grown in 1 L cultures of LB-Amp media in 5 L baffled flasks at 24°C with 180 rpm shaking until they reached an optical density of 1.0. The incubation temperature and shaking were then changed to 16°C and 300 rpm, respectively, then toxin expression was induced with 1 mM isopropyl-β-D-thiogalactoside, and the cells were grown for 16 h. The cell pellet was harvested by centrifugation (6,000 rcf), resuspended in TN buffer (400 mM NaCl, 20 mM Tris-Cl pH 7.5), then cells were lysed using a constant pressure cell disrupter (28 kpsi/4°C). The fusion protein was purified by nickel-affinity chromatography using HisPur Ni-NTA beads (#88832, Thermo Fisher) and eluted with TN buffer containing 500 mM imidazole. This eluant was concentrated using an Amicon-30 centrifugal concentrator to 5 mL, mixed with 5 mL of 2× cleavage buffer (10 mL TN buffer, 1.8 mg reduced glutathione, 3.6 mg oxidized glutathione, 500 μL 1 mg/mL recombinant tobacco etch virus protease), and incubated at room temperature for 48 h. Trifluoroacetic acid (TFA) was added to 1% final concentration, the solution clarified by centrifugation, and then the peptide was purified via reversed-phase high-performance liquid chromatography (RP-HPLC) using a semipreparative C4 column (10 × 250 mm; #00G-4168-N0, Phenomenex, Torrance, CA, USA) and a gradient of solution B (90% acetonitrile (ACN), 0.043% TFA) in solution A (0.05% TFA). Fractions containing PpK1 were identified using matrix-assisted laser desorption/ionization mass spectrometry on an AB SCIEX 5800 instrument or by electrospray ionization using a LCMS2020 mass spectrometer (Shimadzu, Kyoto, Japan), lyophilized, and resuspended in water.
Platymeris rhadamanthus venom peptides Pr11a1 and Pr11a2 were synthesized using Fmoc-solid-phase peptide synthesis using a Liberty PRIME automated microwave synthesiser (CEM, Charlotte, NC, USA) at 0.1 mmol scale using preloaded Wang resins. Couplings were performed in dimethylformamide (DMF) using 5 equivalents of Fmoc-protected amino acid/0.25 M Oxyma Pure/2 M N,N′-diisopropylcarbodiimide, relative to resin substitution, for 1 min at 105°C. Fmoc was removed by treatment with 25% pyrrolidine/DMF (40 s at 100°C). Side-chain protecting groups used were Arg-Pbf, Lys-Boc, Ser/Tyr-tBu, Asn/His/Cys-Trt, and Glu-OtBu. Cleavage and simultaneous removal of side-chain protecting groups were carried out using a CEM Razor rapid peptide cleavage system for 30–60 min at 40°C–45°C in cleavage solution (92.5% TFA, 2.5% triisopropylsilane, 2.5% H2O, 2.5% anisole or 3,6-dioxa-1,8-octanedithiol). Cleavage solution was filtered, precipitated with ice-cold diethyl ether (6,000 × g, 5 min) three times, then resuspended in 0.1% TFA/50% ACN/H2O, filtered, and lyophilized. Crude peptides were purified using a preparatory Vydac C18 column and a linear gradient of 10–60% solvent B (0.043% TFA, 90% ACN) in solvent A (0.05% TFA in water) over 50 min. The expected mass was confirmed using a LCMS2020 mass spectrometer (Shimadzu). RP-HPLC fractions containing the synthetic venom peptide were lyophilized and resuspended in ultrapure water. Peptides were oxidatively folded in 100 mM Tris-HCl pH 8.0, 1 mM reduced glutathione, 1 mM oxidized glutathione for 24 h then quenched by addition of 0.5% TFA and purified using RP-HPLC.
Peptide concentration was estimated from A214 and A280 absorbance measured on a Nanodrop spectrophotometer using extinction coefficients calculated using programs available at https://web.expasy.org/protparam/and https://bestsel.elte.hu/extcoeff.php.
Murine dorsal root ganglion cell assay
Dorsal root ganglia (DRG) from 5-week-old male C57BL/6 mice were dissociated and plated in DMEM (Invitrogen, Carlsbad, California, USA) containing 10% fetal bovine serum (FBS, Assaymatrix, Melbourne, VIC, Australia) and penicillin/streptomycin (Gibco, Carlsbad, California, USA) on a 96-well poly-D-lysine-coated culture plate (Corning, New York, NY, USA), then they were maintained overnight. Cells were loaded with Fluo-4 a.m. calcium indicator according to the manufacturer’s instructions (Thermo Fisher Scientific). After loading (30 min at 37°C then 30 min at room temperature), the dye-containing solution was replaced with assay solution (Hanks’ balanced salt solution, 20 mM HEPES). Fluorescence corresponding to the internal calcium concentration ([Ca2+]i) of 100–150 DRG cells was monitored in parallel using a Ti-E deconvolution inverted microscope (Nikon, Tokyo, Japan), equipped with a Lumencor Spectra LED light source. Images were acquired with a 20× objective at one frame per second (excitation 485 nm, emission 521 nm). Baseline fluorescence was monitored for 30 s. Assay solution was removed at 30 s and replaced with new assay solution (negative control for effect of buffer replacement). At 60 s, assay solution was removed and replaced with venom (50 μg/mL) in assay solution. Experiments involving the use of mouse tissue were approved by the UQ Animal Ethics Committee (TRI/IMB/093/17).
Human neuroblastoma assays
SH-SY5Y neuroblastoma cells were cultured in RPMI supplemented with 15% FBS, 2 mM glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were incubated at 37°C in a humidified 5% CO2 incubator and subcultured every 2–3 days in a 1:3 ratio using 0.05% trypsin/EDTA and D-PBS after 80–90% confluency. Cells were seeded in 384-well black wall flat clear-bottom microplates (Corning, NY, USA) at ∼40,000 cells/well and incubated at 37°C in a humidified 5% CO2 incubator for 48 h prior to assays.
Venoms from T. pallidipennis and P. plagipennis were evaluated for cytolytic activity using an FLIPR Penta System (Molecular Devices, San Jose, CA, USA).25,26 SH-SY5Y cells were loaded with 20 μL per well of dye solution prepared in physiological buffer containing (in mM) 140 NaCl, 11.5 glucose, 5.9 KCl, 1.4 MgCl2, 1.2 NaH2PO4, 5 NaHCO3, 1.8 CaCl2, 10 HEPES (pH 7.4), 0.05 propidium iodide (Sigma-Aldrich), and calcium-4 dye according to the manufacturer’s instructions (Molecular Devices). Cells were incubated for 30 min at 37°C in a humidified 5% CO2 incubator. Fluorescence responses were recorded using two reading modes: 470–495 nm excitation and 565–625 nm emission for propidium iodide fluorescence detection, and at 470–495 nm excitation and 515–575 nm emission for calcium 4 dye fluorescence detection. A reading interval of 10 s was used before venom addition to set the baseline, followed by readings after addition of serial diluted venoms to a total reading time of 10 min. Physiological solution was used as negative control and Triton X-100 at 0.025% prepared in physiological solution was used as positive control for cell lysis.
The ability of the T. pallidipennis venom to modulate the activity of voltage-gated sodium (NaV) channels was tested using SH-SY5Y cells loaded with 20 μL per well of dye solution prepared in physiological buffer and calcium-4 dye according to the manufacturer’s instructions. Cells were incubated for 30 min at 37°C in a humidified 5% CO2 incubator. Fluorescence responses were recorded using 470–495 nm excitation and 515–575 nm emission. A reading interval of 10 s was used before venoms dispensing to set the baseline, followed by readings after addition of serial diluted venom to a total reading time of 5 min, and then for an additional 5 min after addition of 50 μM veratridine. Physiological solution was used as negative control and 1 μM tetrodotoxin, a nonselective inhibitor of mammalian NaV channels, was used as positive control.
Fluorescence intensity kinetic reduction Area Under the Curve (AUC) or Maximum-Minimum (Max-Min) were used for the analysis. Curve fitting was performed in Prism 9.0 (GraphPad Software, San Diego, CA, USA) using nonlinear regression with log-inhibitor versus normalized response and variable Hill slope for EC50 determination.
Electrophysiology of NaV1.7
NaV channel currents were recorded from HEK293 cells stably expressing human NaV1.7 co-expressed with the β1 auxiliary subunit (SB Drug Discovery) using an automated whole-cell patch-clamp electrophysiology platform (QPatch 16X; Sophion Bioscience A/S, Ballerup, Denmark). The extracellular solution comprised (in mM): 1 CaCl2, 1 MgCl2, 5 HEPES, 3 KCl, 140 NaCl, 0.1 CdCl2, 20 TEA-Cl, pH 7.3 and 320 mOsm. The intracellular solution comprised (in mM): 140 CsF, 1/5 EGTA/CsOH, 10 HEPES,10 NaCl, pH 7.3 and 320 mOsm. Cells were maintained at a holding potential −80 mV and Na+ currents elicited by 20-ms voltage steps to 0 mV from a −120 mV conditioning pulse applied for 200 ms. To obtain concentration–responses, cells at holding potential were incubated for 2 min with increasing concentrations of T. pallidipennis venom.
Published: August 13, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110723.
Contributor Information
Christina N. Zdenek, Email: christinazdenek@gmail.com.
Andrew A. Walker, Email: a.walker@imb.uq.edu.au.
Supplemental information
Sheet 1, Ectomocoris sp. venom polypeptides detected from reduced, alkylated, and trypsinized electrostimulated venom. Sheet 2, Oncocephalus sp. venom polypeptides detected from reduced, alkylated, and trypsinized electrostimulated venom. Sheet 3, Peptides detected in native and reduced-alkylated electrostimulated venoms of Ectomocoris sp. and Oncocephalus sp.
References
- 1.Hwang W.S., Weirauch C. Evolutionary history of assassin bugs (Insecta: Hemiptera: Reduviidae): insights from divergence dating and ancestral state reconstruction. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organisation W.H. World Health Organisation; 2015. Investing to Overcome the Global Impact of Neglected Tropical Diseases. [Google Scholar]
- 3.Fry B.G., Roelants K., Champagne D.E., Scheib H., Tyndall J.D.A., King G.F., Nevalainen T.J., Norman J.A., Lewis R.J., Norton R.S., et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- 4.Walker A.A., Weirauch C., Fry B.G., King G.F. Venoms of heteropteran insects: A treasure trove of diverse pharmacological toolkits. Toxins. 2016;8:43. doi: 10.3390/toxins8020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribeiro J.M.C., Assumpção T.C., Francischetti I.M.B. An insight into the sialomes of bloodsucking Heteroptera. Psyche. 2012;2012 doi: 10.1155/2012/470436. [DOI] [Google Scholar]
- 6.Walker A.A., Mayhew M.L., Jin J., Herzig V., Undheim E.A.B., Sombke A., Fry B.G., Meritt D.J., King G.F. The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat. Commun. 2018;9:755. doi: 10.1038/s41467-018-03091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker A.A., Robinson S.D., Undheim E.A.B., Jin J., Han X., Fry B.G., Vetter I., King G.F. Missiles of mass disruption: Composition and glandular origin of venom used as a projectile defensive weapon by the assassin bug Platymeris rhadamanthus. Toxins. 2019;11:673. doi: 10.3390/toxins11110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wait L.C., Walker A.A., King G.F. Crouching tiger, hidden protein: Searching for insecticidal toxins in venom of the red tiger assassin bug (Havinthus rufovarius) Toxins. 2020;13:3. doi: 10.3390/toxins13010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards J.S. The action and compostion of the saliva of an assassin bug Platymeris rhadamanthus Gaerst. (Hemiptera, Reduviidae) J. Exp. Biol. 1961;38:61–77. doi: 10.1242/jeb.38.1.61. [DOI] [Google Scholar]
- 10.Walker A.A., Madio B., Jin J., Undheim E.A.B., Fry B.G., King G.F. Melt with this kiss: Paralyzing and liquefying venom of the assassin bug Pristhesancus plagipennis (Hemiptera: Reduviidae) Mol. Cell. Proteomics. 2017;16:552–566. doi: 10.1074/mcp.M116.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rügen N., Jenkins T.P., Wielsch N., Vogel H., Hempel B.-F., Süssmuth R.D., Ainsworth S., Cabezas-Cruz A., Vilcinskas A., Tonk M. Hexapod assassins' potion: Venom composition and bioactivity from the Eurasian assassin bug Rhynocoris iracundus. Biomedicines. 2021;9:819. doi: 10.3390/biomedicines9070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C., Li L., Wang Y., Wei S., Zhu J. Morphological, functional, compositional and transcriptional constraints shape the distinct venom profiles of the assassin bug Sycanus croceovittatus. Int. J. Biol. Macromol. 2023;250 doi: 10.1016/j.ijbiomac.2023.126162. [DOI] [PubMed] [Google Scholar]
- 13.Fischer M.L., Wielsch N., Heckel D.G., Vilcinskas A., Vogel H. Context-dependent venom deployment and protein composition in two assassin bugs. Ecol. Evol. 2020;10:9932–9947. doi: 10.1002/ece3.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao F., Tian L., Li X., Zhang Y., Wang T., Ma L., Song F., Cai W., Li H. Proteotranscriptomic analysis and toxicity assay suggest the functional distinction between venom gland chambers in twin-spotted assassin bug, Platymeris biguttatus. Biology. 2022;11:464. doi: 10.3390/biology11030464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker A.A., Hernández-Vargas M.J., Corzo G., Fry B.G., King G.F. Giant fish-killing water bug reveals ancient and dynamic venom evolution in Heteroptera. Cell. Mol. Life Sci. 2018;75:3215–3229. doi: 10.1007/s00018-018-2768-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer M.L., Yepes Vivas S.A., Wielsch N., Kirsch R., Vilcinskas A., Vogel H. You are what you eat—ecological niche and microhabitat influence venom activity and composition in aquatic bugs. Proc. Biol. Sci. 2023;290 doi: 10.1098/rspb.2022.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeiro J.M.C., Garcia E.S. The role of the salivary glands in feeding in Rhodnius prolixus. J. Exp. Biol. 1981;94:219–230. doi: 10.1242/jeb.94.1.219. [DOI] [Google Scholar]
- 18.Schofield C.J. Biosystematics and evolution of the Triatominae. Cad. Saúde Pública. 2000;16:S89–S92. doi: 10.1590/s0102-311x2000000800010. [DOI] [PubMed] [Google Scholar]
- 19.Hernández-Vargas M.J., Gil J., Lozano L., Pedraza-Escalona M., Ortiz E., Encarnación-Guevara S., Alagón A., Corzo G. Proteomic and transcriptomic analysis of saliva components from the hematophagous reduviid Triatoma pallidipennis. J. Proteomics. 2017;162:30–39. doi: 10.1016/j.jprot.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 20.Walker A.A., Robinson S.D., Hamilton B.F., Undheim E.A.B., King G.F. Deadly proteomes: A practical guide to proteotranscriptomics of animal venoms. Proteomics. 2020;20 doi: 10.1002/pmic.201900324. [DOI] [PubMed] [Google Scholar]
- 21.Walker A.A., Rosenthal M., Undheim E.E.A., King G.F. Harvesting venom toxins from assassin bugs and other heteropteran insects. J. Vis. Exp. 2018;134 doi: 10.3791/57729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobson J.S., Zdenek C.N., Hay C., Violette A., Fourmy R., Cochran C., Fry B.G. Varanid lizard venoms disrupt the clotting ability of human fibrinogen through destructive cleavage. Toxins. 2019;11:255. doi: 10.3390/toxins11050255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourke L.A., Zdenek C.N., Tanaka-Azevedo A.M., Silveira G.P.M., Sant’Anna S.S., Grego K.F., Rodrigues C.F.B., Fry B.G. Clinical and evolutionary implications of dynamic coagulotoxicity divergences in Bothrops (lancehead pit viper) venoms. Toxins. 2022;14:297. doi: 10.3390/toxins14050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedrich T., Kröger B., Bialojan S., Lemaire H.G., Höffken H.W., Reuschenbach P., Otte M., Dodt J. A Kazal-type inhibitor with thrombin specificity from Rhodnius prolixus. J. Biol. Chem. 1993;268:16216–16222. doi: 10.1016/S0021-9258(19)85408-X. [DOI] [PubMed] [Google Scholar]
- 25.Kramer S., Kotapati C., Cao Y., Fry B.G., Palpant N.J., King G.F., Cardoso F.C. High-content fluorescence bioassay investigates pore formation, ion channel modulation and cell membrane lysis induced by venoms. Toxicon: X. 2024;21:100184. doi: 10.1016/j.toxcx.2024.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardoso F.C. High-throughput bioassays applied to phylogenetic and envenomation studies of spider and snake venoms. Venom Week. 2022;VIII:18–21. [Google Scholar]
- 27.Dan A., Pereira M.H., Pesquero J.L., Diotaiuti L., Beirão P.S. Action of the saliva of Triatoma infestans (Heteroptera: Reduviidae) on sodium channels. J. Med. Entomol. 1999;36:875–879. doi: 10.1093/jmedent/36.6.875. [DOI] [PubMed] [Google Scholar]
- 28.Osteen J.D., Herzig V., Gilchrist J., Emrick J.J., Zhang C., Wang X., Castro J., Garcia-Caraballo S., Grundy L., Rychkov G.Y., et al. Subtype-selective spider toxins implicate NaV1.1 voltage-gated sodium channels in mechanical pain. Nature. 2016;534:494–499. doi: 10.1038/nature17976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai J., Gold M.S., Kim C.-S., Bian D., Ossipov M.H., Hunter J.C., Porreca F. Inhibition of neuropathic pain by decreased expression of the tetrodotoxin-resistant sodium channel, NaV1.8. Pain. 2002;95:143–152. doi: 10.1016/s0304-3959(01)00391-8. [DOI] [PubMed] [Google Scholar]
- 30.Julius D. TRP Channels and Pain. Annu. Rev. Cell Dev. Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 31.Sunagar K., Moran Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi N., Szanto T.G., He J., Schroeder C.I., Walker A.A., Deuis J.R., Vetter I., Panyi G., King G.F., Robinson S.D. Venom composition and pain-causing toxins of the Australian great carpenter bee Xylocopa aruana. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-26867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe G.D.O., Rumley A., Mackie I.J. Plasma fibrinogen. Ann. Clin. Biochem. 2004;41:430–440. doi: 10.1258/0004563042466884. [DOI] [PubMed] [Google Scholar]
- 34.Palta S., Saroa R., Palta A. Overview of the coagulation system. Indian J. Anaesth. 2014;58:515–523. doi: 10.4103/0019-5049.144643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zdenek C.N., Youngman N.J., Hay C., Dobson J., Dunstan N., Allen L., Milanovic L., Fry B.G. Anticoagulant activity of black snake (Elapidae: Pseudechis) venoms: Mechanisms, potency, and antivenom efficacy. Toxicol. Lett. 2020;330:176–184. doi: 10.1016/j.toxlet.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Coimbra F.C.P., Dobson J., Zdenek C.N., op den Brouw B., Hamilton B., Debono J., Masci P., Frank N., Ge L., Kwok H.F., Fry B.G. Does size matter? Venom proteomic and functional comparison between night adder species (Viperidae: Causus) with short and long venom glands. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2018;211:7–14. doi: 10.1016/j.cbpc.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Noeske-Jungblut C., Haendler B., Donner P., Alagon A., Possani L., Schleuning W.D. Triabin, a highly potent exosite inhibitor of Thrombin. J. Biol. Chem. 1995;270:28629–28634. doi: 10.1074/jbc.270.48.28629. [DOI] [PubMed] [Google Scholar]
- 38.Haendler B., Becker A., Noeske-Jungblut C., Krätzschmar J., Donner P., Schleuning W.D. Expression, purification and characterisation of recombinant pallidipin, a novel platelet aggregation inhibitor from the haematophageous triatomine bug Triatoma pallidipennis. Blood Coagul. Fibrinolysis. 1996;7:183–186. doi: 10.1097/00001721-199603000-00018. [DOI] [PubMed] [Google Scholar]
- 39.Lavoipierre M.M., Dickerson G., Gordon R.M. Studies on the methods of feeding of blood-sucking arthropods. I. The manner in which triatomine bugs obtain their blood-meal, as observed in the tissues of the living rodent, with some remarks on the effects of the bite on human volunteers. Ann. Trop. Med. Parasitol. 1959;53:235–250. doi: 10.1080/00034983.1959.11685921. [DOI] [PubMed] [Google Scholar]
- 40.Georgieva A.Y., Gordon E.R.L., Weirauch C. Sylvatic host associations of Triatominae and implications for Chagas disease reservoirs: a review and new host records based on archival specimens. PeerJ. 2017;5:e3826. doi: 10.7717/peerj.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. http://www.nature.com/nmeth/journal/v8/n10/abs/nmeth.1701.html#supplementary-information [DOI] [PubMed] [Google Scholar]
- 42.Eddy S.R. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury A., Zdenek C.N., Dobson J.S., Bourke L.A., Soria R., Fry B.G. Clinical implications of differential procoagulant toxicity of the palearctic viperid genus Macrovipera, and the relative neutralization efficacy of antivenoms and enzyme inhibitors. Toxicol. Lett. 2021;340:77–88. doi: 10.1016/j.toxlet.2020.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Debono J., Bos M.H.A., Frank N., Fry B. Clinical implications of differential antivenom efficacy in neutralising coagulotoxicity produced by venoms from species within the arboreal viperid snake genus Trimeresurus. Toxicol. Lett. 2019;316:35–48. doi: 10.1016/j.toxlet.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues C.F.B., Zdenek C.N., Bourke L.A., Seneci L., Chowdhury A., Freitas-de-Sousa L.A., de Alcantara Menezes F., Moura-da-Silva A.M., Tanaka-Azevedo A.M., Fry B.G. Clinical implications of ontogenetic differences in the coagulotoxic activity of Bothrops jararacussu venoms. Toxicol. Lett. 2021;348:59–72. doi: 10.1016/j.toxlet.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Gold A.M. Sulfonyl fluorides as inhibitors of esterases. III. Identification of serine as the site of sulfonylation in phenylmethanesulfonyl α-chymotrypsin. Biochemistry. 1965;4:897–901. doi: 10.1021/bi00881a016. [DOI] [PubMed] [Google Scholar]
- 47.Saez N.J., Cristofori-Armstrong B., Anangi R., King G.F. A strategy for production of correctly folded disulfide-rich peptides in the periplasm of E. coli. Methods Mol. Biol. 2017;1586:155–180. doi: 10.1007/978-1-4939-6887-9_10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1, Ectomocoris sp. venom polypeptides detected from reduced, alkylated, and trypsinized electrostimulated venom. Sheet 2, Oncocephalus sp. venom polypeptides detected from reduced, alkylated, and trypsinized electrostimulated venom. Sheet 3, Peptides detected in native and reduced-alkylated electrostimulated venoms of Ectomocoris sp. and Oncocephalus sp.
Data Availability Statement
-
•
Data: RNA-Seq data in FASTQ format are available from the USA National Center for Biotechnology Information (NCBI)’s Sequence Read Archive: BioProject ID PRJNA1046250. The amino acid sequences of venom toxins and the contigs encoding them are available from GenBank (NCBI Nucleotide: PP510767–PP510872, PP516297–PP516299, PP517412–PP517564). The CO1 barcoding genes of the Ectomocoris sp. and Oncocephalus sp. used in this study are available from GenBank (NCBI Nucleotide: PP506657, PP506658), respectively. Mass spectral data of venom is available in the ProteomeXchange PRotein IDEntification (PRIDE: PXD050854).
-
•
Code: No original code was generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.