Abstract
Human inborn errors of immunity include rare disorders entailing functional and quantitative antibody deficiencies due to impaired B cells called the common variable immunodeficiency (CVID) phenotype. Patients with CVID face delayed diagnoses and treatments for 5 to 15 years after symptom onset because the disorders are rare (prevalence of ~1/25,000), and there is extensive heterogeneity in CVID phenotypes, ranging from infections to autoimmunity to inflammatory conditions, overlapping with other more common disorders. The prolonged diagnostic odyssey drives excessive system-wide costs before diagnosis. Because there is no single causal mechanism, there are no genetic tests to definitively diagnose CVID. Here, we present PheNet, a machine learning algorithm that identifies patients with CVID from their electronic health records (EHRs). PheNet learns phenotypic patterns from verified CVID cases and uses this knowledge to rank patients by likelihood of having CVID. PheNet could have diagnosed more than half of our patients with CVID 1 or more years earlier than they had been diagnosed. When applied to a large EHR dataset, followed by blinded chart review of the top 100 patients ranked by PheNet, we found that 74% were highly probable to have CVID. We externally validated PheNet using >6 million records from disparate medical systems in California and Tennessee. As artificial intelligence and machine learning make their way into health care, we show that algorithms such as PheNet can offer clinical benefits by expediting the diagnosis of rare diseases.
INTRODUCTION
Human inborn errors of immunity (IEIs), also referred to as primary immunodeficiencies, are rare, often monogenic diseases that confer susceptibility to infection, autoimmunity, and auto-inflammation. There are now more than 500 distinct IEIs, and dozens more are discovered each year because of the availability of whole exome or genome sequencing (1). One of the most common IEIs is the common variable immunodeficiency (CVID) phenotype, a heterogeneous group of disorders characterized by a state of functional or quantitative antibody deficiency and impaired B cell responses. Most patients with CVID have recurrent sinopulmonary infections, but they can have autoimmunity (for example, autoimmune hemolytic anemia) or immune dysregulation (for example, enteritis and granulomata). The prevalence of CVID is around 1 in 25,000 individuals worldwide (2).
Despite advances in genome sequencing technologies and the increased capacity of diagnosis for IEIs, the spectrum of genetic etiologies of the CVID phenotype is not fully understood. At least 68 genes have been implicated in the disease (3), but, for most individuals, no specific genetic cause can yet be identified (3, 4). More recently, it has been proposed that the genetic basis of CVID can be described by a polygenic architecture, where disease risk is conferred by cumulative effects across the genome (5, 6) or by a combination of genetic, epigenetic, and environmental effects on B cells (4). Because there is no single causal genetic mechanism, there is no genetic test available for providing definitive diagnoses. Furthermore, the broad genetic variability and epigenetic alterations that contribute to B cell dysfunction lead to heterogeneous CVID presentations, making it even more difficult to diagnose. Because the immune system is intertwined with nearly all organs and tissues, the clinical presentation of rare immune phenotypes such as CVID intersects with many medical specialties. For example, these patients are often followed in otorhinolaryngology clinics for sinus infections and pulmonology clinics for pneumonia. This causes a scattering of patients across multiple clinical subspecialties and leads to substantial delays in diagnosis and treatment. This consequential delay is one of the major challenges in initiating clinical care for patients with CVID, averaging 5 years in children (7) to 15 years in adults (2, 8). This protracted delay in treatment increases both morbidity and mortality (8–10). There is a critical and unmet need to reduce the diagnostic delay for CVID and promptly provide these patients with treatments such as immunoglobulin replacement and immune-modulators.
The recent availability of large-scale electronic health records (EHRs) has enabled the computational assessment of patients’ phenotypic characteristics solely on the basis of their medical records (11–15), enabling the systematic and scalable review for millions of individuals. A fundamental difficulty in this approach is having a priori knowledge about how the patterns of CVID are represented solely through EHRs. We refer to these patterns describing the manifestations of CVID as the EHR signature of the disease. Because there is not a single clinical presentation for CVID, constructing an EHR signature for the CVID phenotype is not straightforward. Here, we set out to develop a computational algorithm, PheNet, that gleans EHR signatures from the records of patients with CVID and computes a numerical score that prioritizes patients most likely to have CVID. To test whether we could shorten the diagnostic odyssey of patients with CVID, we examined their PheNet scores, looking back in time in the EHR data. To test PheNet’s ability to find previously undiagnosed patients with CVID, we applied PheNet to a large EHR dataset at UCLA (University of California Los Angeles) and performed blinded chart reviews of the top-ranked individuals. To demonstrate PheNet’s ability to learn locally and act broadly, we applied PheNet to >6 million records across five disparate health systems in California and Tennessee. Those with high scores would be candidates for referral to an immunology specialist.
Artificial intelligence and machine learning are rapidly entering into the medical realm. We show that approaches such as PheNet can learn from the EHR both to expedite the diagnosis of patients with CVID and to identify phenotypic patterns of rare diseases.
RESULTS
Summary and description of the “ground-truth” UCLA CVID cohort
Central to our approach was training and validating our model using a dataset of individuals with a known CVID diagnosis. To find these individuals, we searched the UCLA EHR for International Classification of Diseases (ICD-10) billing code D80.* (codes that broadly include immunodeficiencies with predominantly antibody defects), which produced approximately 3200 individuals. To consider only the data of those who correctly received an immunodeficiency diagnosis, medical records were then manually reviewed by a clinical immunologist. We defined individuals as having the CVID phenotype per standard definitions (2, 16): documented low serum immunoglobulin G (IgG) concentrations, absent or defective antibody responses to vaccine-or infection-associated antigens, age more than 2 years, and having no other explanation for low immunoglobulin concentrations (such as losses).
This procedure narrowed the list to 197 individuals. For 186 of these individuals, we found documented low IgG laboratory values (hypogammaglobulinemia) before immunoglobulin replacement therapy (IgRT). In the other 11, hypogammaglobulinemia was documented only in the written notes. Regardless, all our “ground-truth” individuals were diagnosed and documented by immunology-certified physicians as having CVID (fig. S1), but a strict, modern definition would regard these patients as having either CVID, “unclassified antibody deficiencies,” or combined immunodeficiency. For example, one patient formally diagnosed with CVID was later discovered to have nuclear factor κB subunit 1 deficiency, which affects both T and B cells and is thus a combined immunodeficiency. Table S1 offers clinical details of our case cohort.
Some of our patients with CVID showed both low and normal IgG concentrations in our EHR data because they were treated with IgRT, which would raise IgG values to the normal range, including a few who started IgRT before coming to UCLA. For model training and validation, we proceeded with n = 197 case individuals. Model weights were not changed if we used the more limited set of 186 individuals. We constructed a matched control cohort from the EHR of 1106 individuals on the basis of sex, self-identified race/ethnicity, age (closest within a 5-year window), and the number of days recorded in the EHR (closest within a 180-day window) (Fig. 1).
Fig. 1. Overview of PheNet model training and application within a discovery cohort.
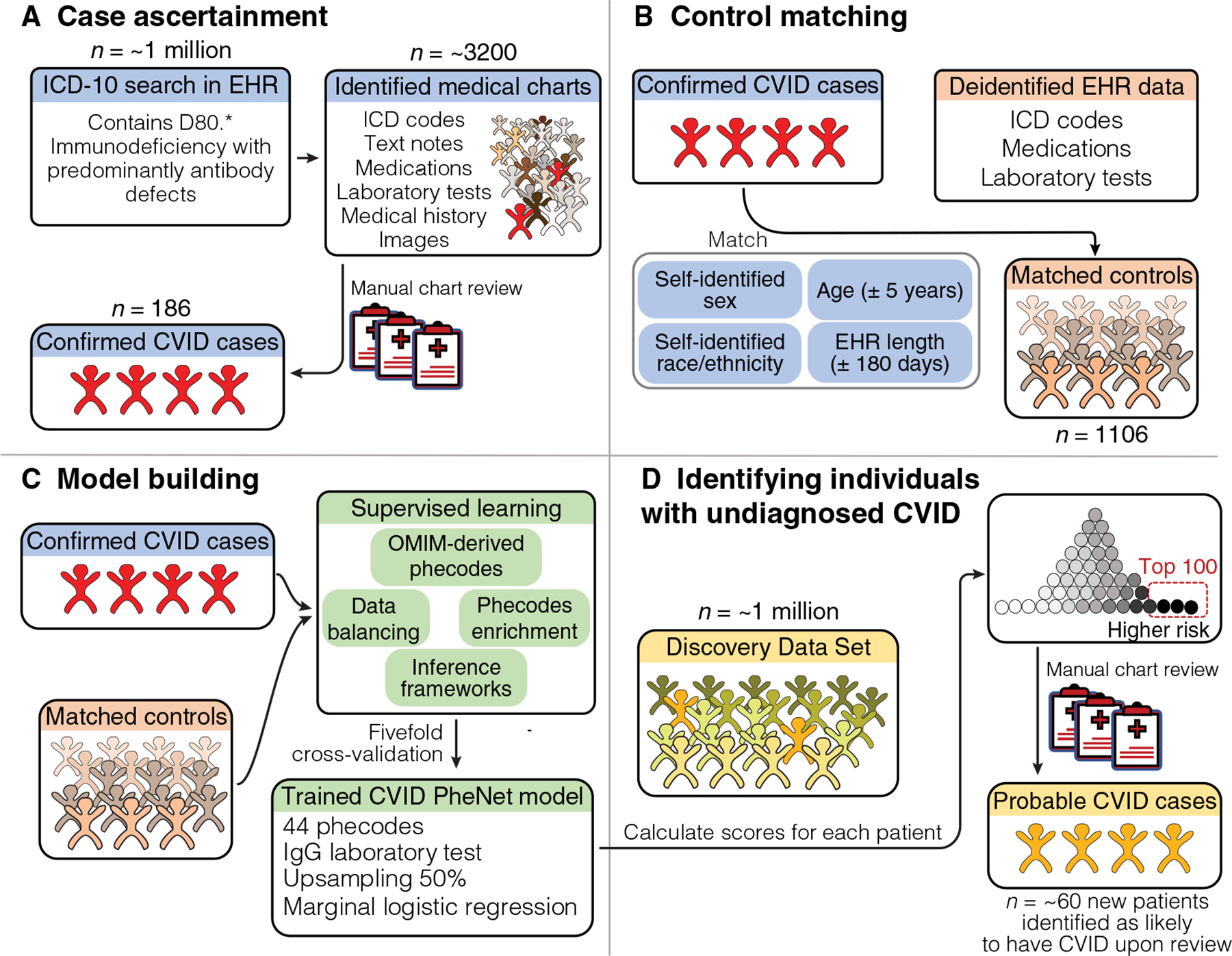
We present a visual summary of case/control cohort construction, PheNet model training, and application within a discovery dataset at UCLA Health. (A) The workflow for constructing a case cohort of clinically diagnosed patients with CVID from medical charts. (B) Criteria used to create a matched control cohort from the EHR (n = 1106). (C) Construction of the prediction model, including feature selection from phecodes, inclusion of laboratory values, a variety of inference frameworks, and data balancing techniques. (D) Example of how the PheNet model can be applied within a discovery cohort to identify patients with a high likelihood of CVID, who could then be further assessed by manual chart review to confirm diagnosis.
Seventy-two percent of our 186 individuals with “ground-truth” antibody deficiency self-identified with a sex of female, and 28% identified as male. Other demographics are shown in Table 1. Previously reported cohorts had a similar demographic profile that also showed a majority female proportion (3, 17). We found that the average age of individuals when first diagnosed with CVID was 55 years (SD, 19.5 years), which is consistent with other work showing that most individuals are diagnosed with CVID after age 40 years (9) (fig. S2A). Of all patients with CVID, 22% had “uncomplicated CVID,” meaning a history of only infections (21 of 186) or infections along with just asthma (20 of 186) but no autoimmune or severe inflammatory phenotypes. This proportion of individuals with uncomplicated CVID is on par with other cohorts (18, 19). Most of our individuals had many encounters in the EHR (Table 1 and fig. S2B), but there were 6 individuals of the 197 CVID cases who had fewer than 10 encounters in our EHR. This could reflect individuals who came to UCLA only to receive a formal diagnosis or those who only came for a second opinion but largely had their care at a different center.
Table 1. Demographics of cohorts of CVID cases and controls.
Numbers in parentheses are SDs. All our individuals had self-identified sex of either male or female; intersex or other options were not assessed.
| Cases | Controls | |
|---|---|---|
| Sample size | 186 | 1106 |
| Age (years) | ||
| Mean | 55 (19.5) | 55 (20.1) |
| Median | 58.5 | 57 |
| Sex (%) | ||
| Male | 28 | 30 |
| Female | 72 | 70 |
| Mean number of unique iCD codes | 102 (92.2) | 54 (55.5) |
| Mean medical record length (years) | 14 (9.7) | 14 (8.4) |
Constructing a CVID risk score model from EHR-derived phenotypes
We used the curated set of cases to learn the EHR signature for CVID as follows. From ICD codes of our cases, we derived phecodes (20, 21) capturing a limited and clinically meaningful set of phenotypes from the EHR. To prevent overfitting, we selected only a subset of phecodes (of the possible ~1800 codes) that best captured the phenotypic patterns of CVID. We first selected phecodes matching the clinical description of CVID listed in the Online Mendelian Inheritance in Man (OMIM) (22) database, resulting in 34 phecodes. Then, leveraging the annotated data specifically for this study, we included additional phecodes. We identified any phecodes that had significantly higher frequency in the cases as compared with the controls (Bonferroni adjustment for multiple testing, P < 1 × 10−5). Then, using cross-validation, we included those phecodes that would improve the model. To prevent bias, we excluded the actual phecode for CVID itself (phecode 279.11) from the set of features. We varied the number of additional phecodes to control the trade-off between adding more information to the model and overfitting due to the increased data dimensionality. We found that 10 additional phecodes (44 total) added performance while preventing overfitting (fig. S3A).
We next compared a variety of prediction methods to learn a function that best mapped the feature set to each individual’s CVID status. We evaluated methods that varied in model complexity, including linear methods such as marginal logistic regression of each feature, penalized joint models like ridge regression, as well as non-linear methods such as random forest regression (fig. S3B). We found that marginal regression and ridge regression achieved similarly high performance [area under the curve for the receiver operating curve (AUC-ROC) and area under the curve for the precision recall curve (PR) for marginal regression: 0.95 and 0.83, respectively, versus AUC-ROC and AUC-PR for ridge regression: 0.96 and 0.88]. We opted to use marginal regression to maintain straightforward inter-pretability of the regression coefficients.
In addition to the set of phecodes, we included the laboratory test for IgG concentrations. Because we were interested in identifying individuals with abnormally low IgG values, we discretized laboratory measurements as a categorical variable where patients’ results were either normal, low, or no IgG test was recorded in their medical record. We assessed the performance of the model when including the IgG laboratory test result and found that the inclusion of this single feature substantially increased performance (AUC-ROC and AUC-PR for including IgG: 0.95 and 0.83 versus AUC-ROC and AUC-PR for excluding IgG: 0.89 and 0.73) (fig. S3C).
To account for case imbalances associated with predicting rare diseases, we performed random upsampling and downsampling of the cases to achieve a more balanced dataset (fig. S3,D and E). Comparing various upsampled ratios, we found that a ratio of 0.50 provided optimal performance. These results show the optimization process for PheNet.
Our final prediction model included the 34 phecodes selected from OMIM, the 10 additional phecodes learned from the case cohort, the IgG laboratory test, and an upsampling ratio of 0.50. Using fivefold cross-validation, we showed that the average PheNet scores for individuals with CVID had a significantly higher risk score than the matched controls (Cochran-Armitage test, P < 2.2 × 10−16) (fig. S4A). Note that the PheNet risk score assesses whether the patient likely already has CVID but has simply not yet been diagnosed. PheNet scores did not vary by age, but the performance of PheNet was better for older individuals than younger ones (fig. S4,B and C). These data show that PheNet scores enrich for individuals with CVID.
PheNet is more accurate than existing phenotype risk scores for predicting CVID
We compared PheNet against the PheRS method, a phenotype risk score methodology for detecting rare, Mendelian diseases (11). Because PheRS is an unsupervised method, we were able to train a PheRS model for CVID prediction within the UCLA EHRs. We found that PheNet performed 17% better than PheRS when comparing AUC-ROC and 42% better when comparing AUC-PR (Fig. 2,A and B).
Fig. 2. PheNet is more accurate than existing phenotype risk scores for predicting CVID.
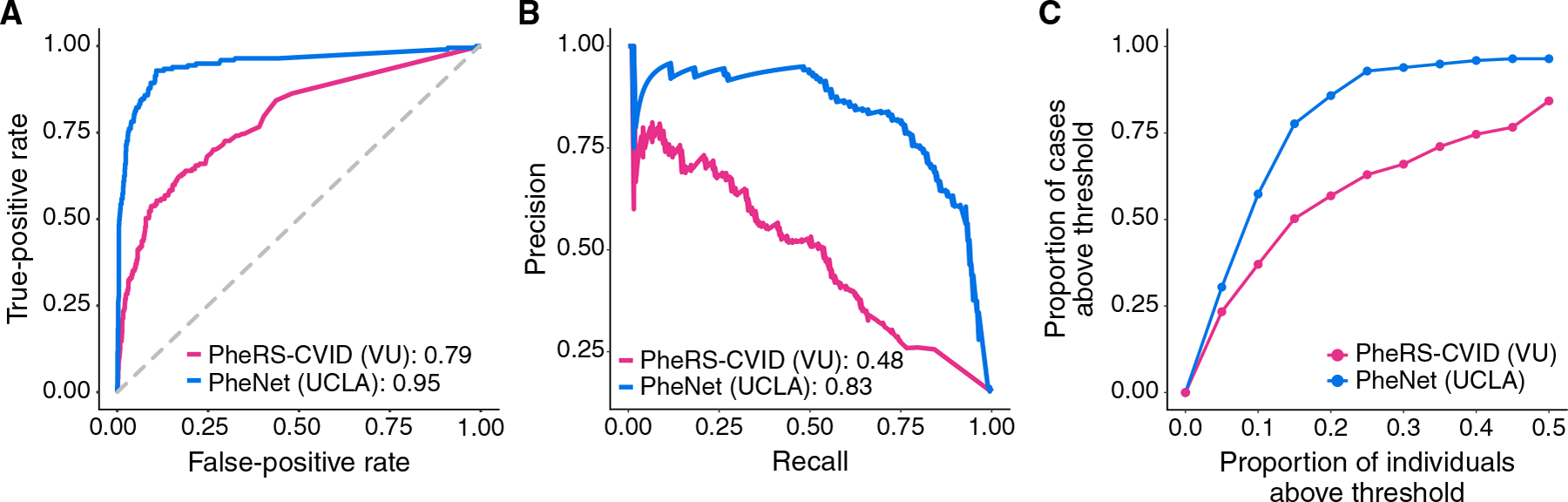
Performance metrics comparing the performance of PheNet and PheRS-CVID within UCLA Health population case and control cohorts. The PheNet and PheRS-CVID models were trained using weights trained from EHR data. Receiver operating characteristic (A) and precision-recall (B) curves across the different prediction models are shown. AUC is provided in the legend. (C) Individuals with a PheNet score of >0.90 and the proportion of CVID cases captured within the varying percentiles of PheNet and PheRS scores.
In practice, individuals with high-risk scores would be candidates for follow-up. Setting a threshold score of 0.90, we found that 57% of cases could be detected within the top 10% of individuals ranked by PheNet score (Fig. 2C); the other cases were all detected but at lower scores. In contrast, PheRS captured only 37% of cases at the same threshold. These results show that PheNet outperforms PheRS.
To further investigate the potential bias due to EHRs from different institutions, we compared the accuracy of PheRS using a model that was trained in the Vanderbilt EHR. Because the PheRS method is unsupervised, we could do a systematic performance comparison when using models trained at UCLA and Vanderbilt (VU). We found that the pretrained feature weight for the phecode 561.1 (diarrhea) was not available in the Vanderbilt EHR; thus, we excluded this feature from both models for this set of analyses. We found that the PheRS AUC-ROC and AUC-PR were almost exactly the same at the two institutions (PheRS-UCLA, 0.79 and 0.48; and PheRS-VU, 0.79 and 0.49). This shows that the EHR signature for CVID is similar between the institutions and not likely a major source of bias (fig. S5).
We performed additional analyses to assess whether the performance of PheNet was biased because scores were computed for individuals on the basis of EHR information obtained both before and after their diagnoses. To test whether a more temporally restricted set of EHR data could still have appropriate predictive power, we created a “censored” testing dataset that limited the information to only that present in an individual’s medical record before their “ICD-based diagnosis” of CVID. Because we did not have access to the exact date of patients’ formal diagnoses (because of a date shift in the EHR not present in the manually reviewed medical records), we estimated the date on the basis of the occurrences of the ICD-10 code for CVID (D83.9) within the EHR. The cohort of patients with CVID was formally identified through manual chart review, and this ICD-based procedure was only used to identify the approximate date of diagnosis within the EHR. The training dataset was still trained on all data points up to the present day regardless of the diagnosis date because this did not affect test performance. Using this more restricted test set, we found a modest reduction in performance, but we were still able to capture a large percentage of patients with CVID. Specifically, we found 46% of cases compared with 57% of cases within the top 10% of patients ranked by PheNet (fig. S6). When comparing AUC-ROC and AUC-PR, we saw a 17.7 and 51.7% decrease in performance, respectively. This drop in performance could be because some patients did not have substantial medical history at UCLA preceding their diagnosis, which would limit the phenotypes in our EHR and thus prediction power. When limiting our assessment to only those patients with at least 1 year of UCLA EHR data before their diagnosis (n = 58), we found only an 8.1 and 44.6% drop in performance for AUC-ROC and AUC-PR, respectively. Thus, given an adequate medical history, there is limited performance bias when using all EHR information up to the present.
PheNet provides an early diagnosis
We next sought to quantify the utility of PheNet as a predictive tool for identifying whether patients could be diagnosed earlier by conducting an analysis using the UCLA EHR data. The dataset comprised all individuals at UCLA with at least one encounter and at least one ICD code, for a total of ~880,000 individuals, including our previously established case cohort. Using fivefold cross-validation, we divided the data into 80% training and 20% testing and ran PheNet on each fold of the data. To mirror how PheNet would be used in practice, we limited the testing data to only features that appeared in the EHR before an individual’s ICD-based diagnosis. For different scoring thresholds, we captured patients with CVID at various times both before and after their diagnosis (fig. S7). In practice, the score threshold could be chosen according to specific goals and the resources available. For example, one would recommend using a high stringency score threshold, thus capturing fewer individuals, if patients were to be followed up individually, which is a resource-expensive undertaking.
To ensure that individuals had an adequate amount of medical history before their diagnosis of CVID, we restricted the analysis to individuals with at least 1 year of EHR data before their ICD-based diagnosis (n = 58). We set a threshold PheNet score of 0.9 for this analysis and found that PheNet identified 64% of individuals with CVID before the date of their ICD-based diagnosis (Fig. 3). PheNet could identify these individuals as likely having CVID ~244 days before their ICD diagnosis (median) (SD, 374). For example, the individual shown in Fig. 4 reached the top percentile of the PheNet score distribution 41 days before their record reflected any immunodeficiency diagnoses. This patient had accumulated seven phecodes that influenced their PheNet score in the years before diagnosis. Then, the patient’s record revealed measurement of a modestly low IgG concentration, which further increased their risk score. This example demonstrates the advantage of aggregating information from both phenotypes and laboratory tests to identify individuals as high risk. These results show that PheNet has substantial utility for not only identifying undiagnosed individuals with CVID but also providing earlier diagnosis than they might have in a usual clinical scenario.
Fig. 3. PheNet can identify patients with CVID before their original diagnosis dates.
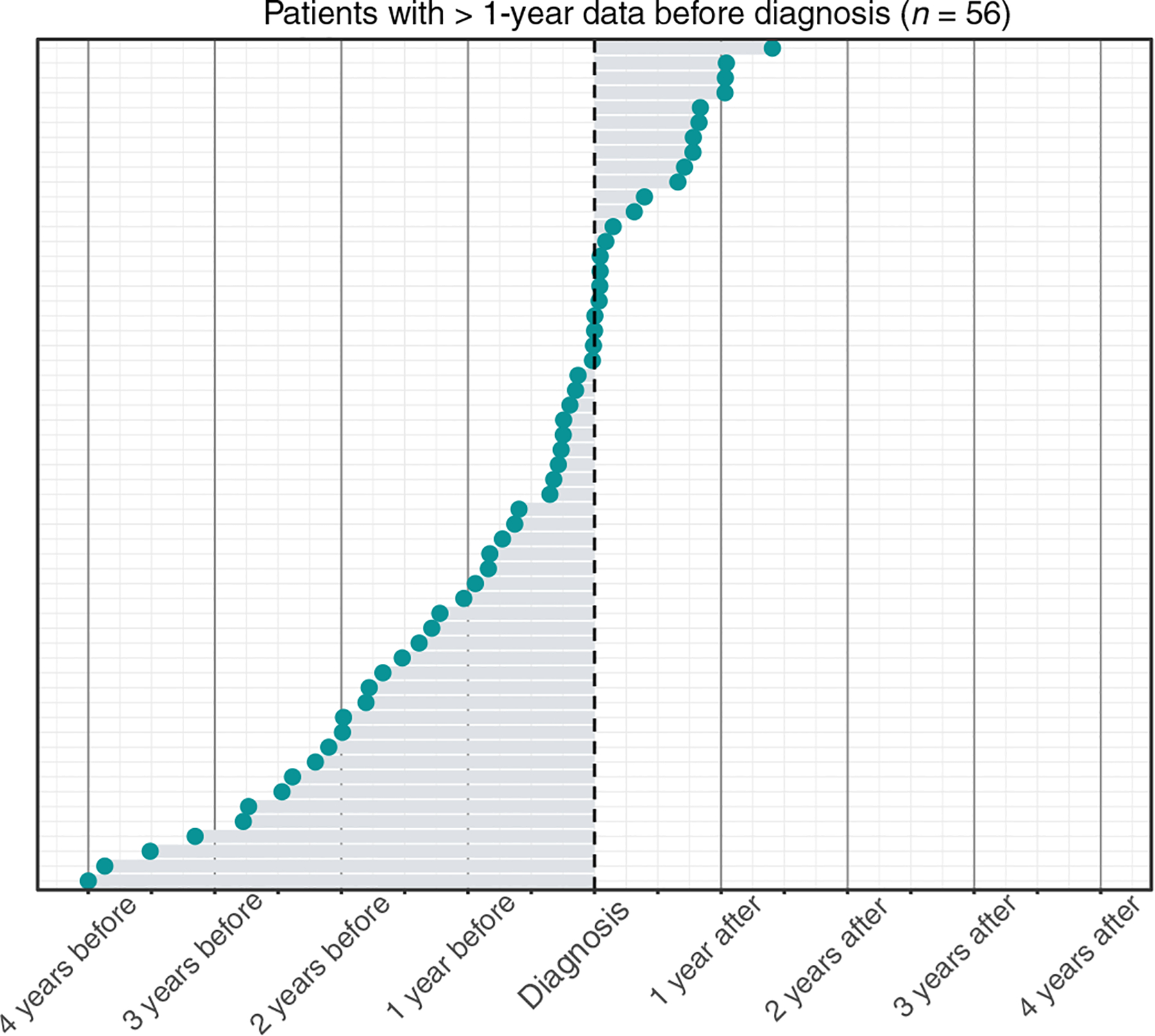
Distribution of the time between individuals’ ICD-based diagnoses for CVID and the time point at which individuals’ risk score > 0.90 (denoted at the blue circles). Only individuals with at least 1 year of EHR data before their ICD-based diagnosis were included. Two of the 58 individuals were excluded from the graph because their PheNet scores were not >0.9 at any point in time. ICD-based diagnoses were determined as the time point when individuals first accumulated at least two CVID ICD codes (D83.9) within a year.
Fig. 4. Early CVID diagnosis by PheNet: Sample patient’s timeline.
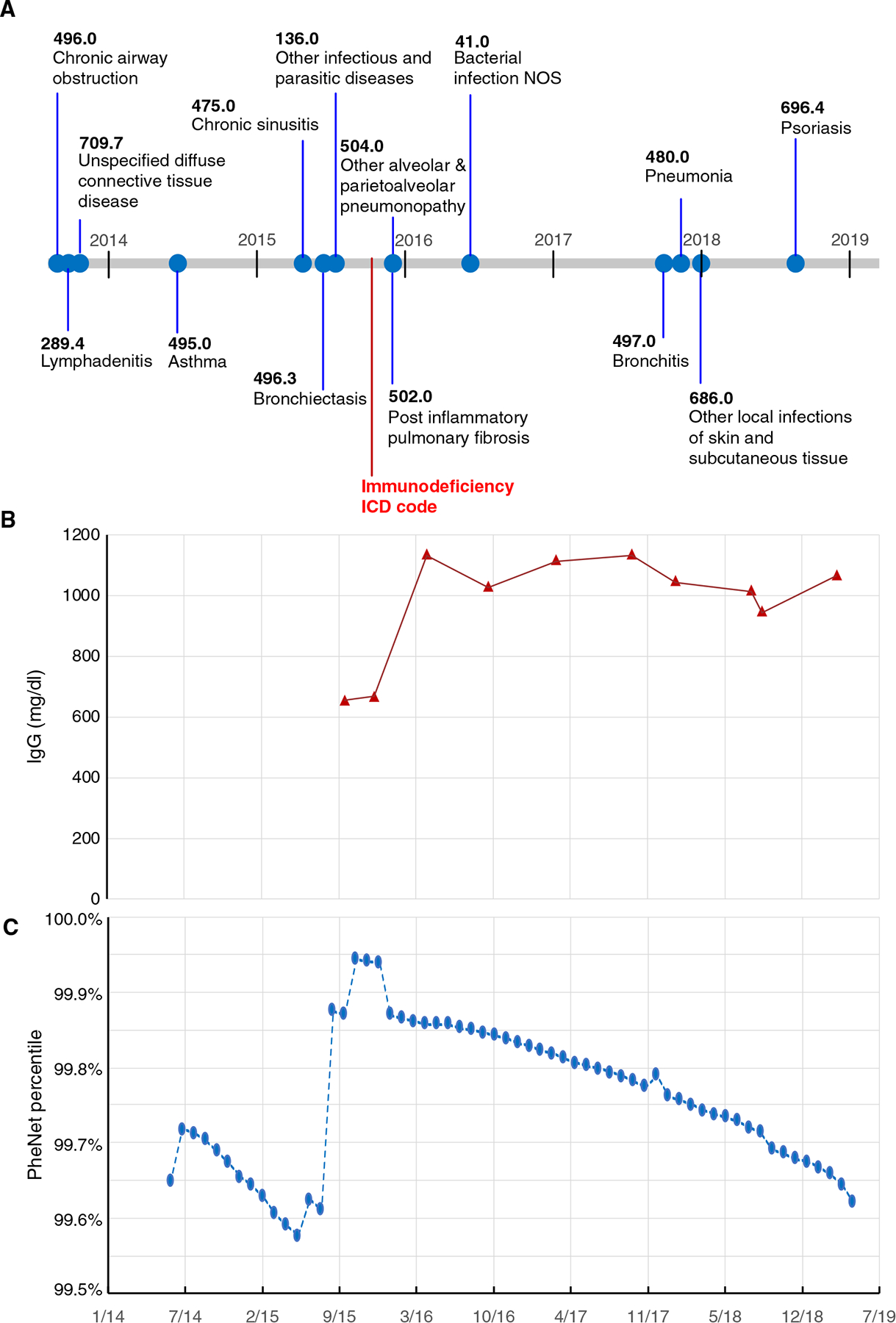
(A) All CVID-relevant phecodes on a sample patient’s record. The point when the patient received their first immunodeficiency billing code is denoted by the red star. (B) The patient’s IgG laboratory results over time; the first two points were low, and then the patient was started on immunoglobulin replacement therapy. (C) The percentile of the patient’s risk score computed over time. Specifically, we show that the patient reached the 99th percentile of the PheNet score distribution 41 days before their medical record showed evidence of immunodeficiency. Note that the patient’s timeline has been date-shifted. NOS, not otherwise specified.
Identifying undiagnosed individuals with CVID
To validate the utility of PheNet for identifying undiagnosed patients with CVID, we conducted an analysis using the EHR data from more than 880,000 individuals at UCLA as the discovery cohort. We removed from consideration all individuals who were deceased or who had phecodes corresponding to solid organ transplants, cystic fibrosis, or infection with the human immunodeficiency virus, resulting in the removal of 42,346 individuals. Individuals with these disorders may exhibit similar clinical profiles as those with CVID, but their phenotypes are likely due to their immunocompromised conditions, not a primary immune disease. We then selected the top 100 individuals identified by PheNet and a control group of 100 randomly selected individuals from the patient population. On average, the group of top 100 individuals had an average of 15.5 years of medical history, and the randomly selected group had 7.1 years (table S2).
We scrambled these two sets of patients and performed a clinical chart review for these individuals. Medical records were directly examined by a clinical immunologist who was blinded to the groups and not informed that they were validating a risk score algorithm for CVID. The clinician had access to each individual’s full medical record including notes, images, and scanned documents, which were not available to the PheNet algorithm. Each individual was ranked according to an ordinal scale from 1 to 5 quantifying the likelihood of having CVID where 1 was defined as “near certainty not CVID” and 5 was “definitive as CVID,” meaning that the patient met the diagnosis criteria. From the list of top 100 ranked individuals, 74% of individuals were assigned a score of 3, 4, or 5, indicating that they were highly probable as having CVID (Fig. 5). Eight percent of individuals were assigned a score of 5, meaning that they were very likely diagnosable with CVID in having low immunoglobulin concentrations and poor humoral responses to vaccine antigens or having a prior outside physician diagnosis of CVID. In contrast, the individuals who were randomly chosen exclusively had scores of 1, 2, or 3, and 90% of individuals had a score of 1 or 2, indicating that they likely would not have CVID. These results validate that our approach is useful to identify undiagnosed patients with CVID and overcome the major challenge of initiating care in a timely manner.
Fig. 5. PheNet identifies undiagnosed individuals with CVID.
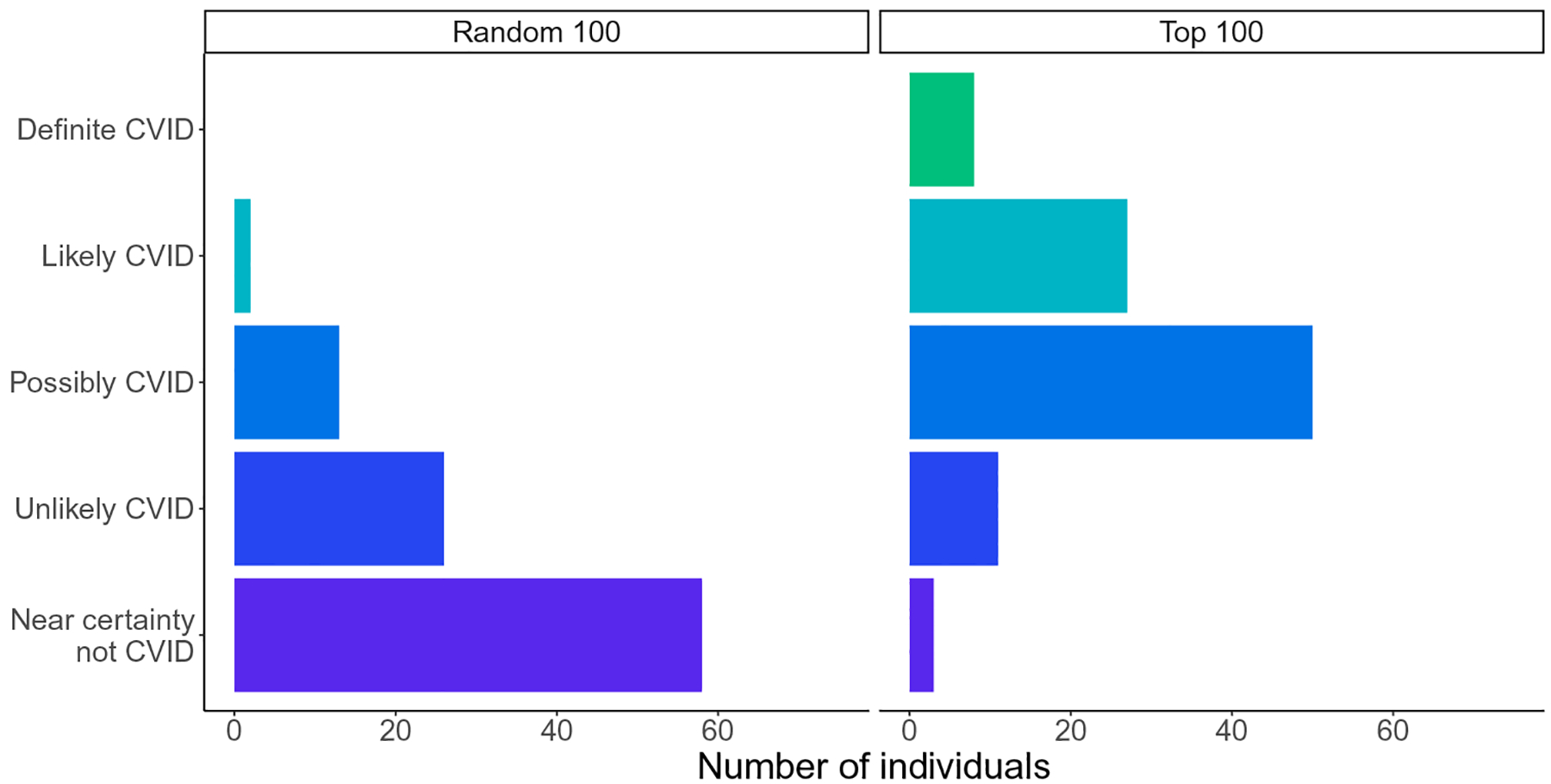
CVID clinical validation scores for the top n = 100 individuals with the highest PheNet score and n = 100 randomly sampled individuals. Each individual was ranked according to an ordinal scale from 1 to 5 quantifying the likeliness of having CVID where 1 was defined as near certainty not CVID and 5 was definitive as CVID.
In addition to prediction performance, it is also important to understand the symptoms that contributed to each individual’s increased risk status. In practice, it would not be sufficient to only identify individuals to refer to an immunology clinic, but it is also necessary to explain exactly which factors contributed to their identification. Examining the regression coefficients from the model in the form of odds ratios, we identified the phenotypes that were most predictive (table S3). Some of the most predictive features (for example, thrombocytopenia) were not provided from the OMIM clinical description but were obtained from the set of enriched phecodes identified from our case cohort, further emphasizing the benefit of including a well-curated case cohort in the prediction model. The signs and symptoms that contributed to each of the top 100 individuals’ risk scores are shown in Fig. 6. Overall, there were wide variations in the symptoms of each individual, demonstrating the utility of methods that aggregate both symptoms and laboratory results to identify patients at risk. There was no single feature present in all 100 individuals with the highest PheNet scores, underscoring the lack of any single clinical manifestation as being pathognomonic of CVID. We also observed that most individuals had a mixture of both autoimmune and infection-related phecodes, further demonstrating the heterogeneity of the CVID phenotype. These patterns were consistent with those observed in our case cohort (fig. S8). In contrast, most randomly selected individuals did not have major symptoms matching the CVID patterns estimated by PheNet, and the signs and symptoms present within this group were those that were among the most common in the general population such as upper respiratory infections and asthma (fig. S9).
Fig. 6. EHR signatures of individuals at high risk for CVID.

Each row shows a clinical feature from the PheNet model, and each column is a patient’s EHR profile. The top 100 individuals identified by PheNet are shown where the lowest to highest ranked individuals are displayed from left to right. Boxes are colored according to phenotype category (autoimmune/inflammation, infection, and other). The PheNet scores ranged from 18.3 to 30.9. By contrast, the PheNet scores for random comparators (shown in fig. S9) ranged from −0.8 to 4.3.
Validation and replication of PheNet at five other medical centers
For general applicability, we next tested whether PheNet could be applied to additional databases. We validated the generalizability of PheNet using de-identified clinical data collected from the University of California medical centers including University of California Los Angeles; University of California San Francisco (UCSF); University of California Davis (UCD); University of California San Diego (UCSD but not Rady Children’s Hospital); and University of California Irvine (UCI). We also ran the algorithm using the de-identified EHR at Vanderbilt University Medical Center. In aggregate, these comprised more than 5.6 million patient records (first row of Table 2). We scored and ranked each individual using the PheNet weights calculated solely from UCLA training data as above; that is, no training was performed on the UC-wide or the Vanderbilt data. To assess the utility of the scores, we asked whether PheNet could identify patients who had at least one encounter with a diagnosis code of CVID (ICD code “D83.9”) (second row of Table 2). When ranking patients by PheNet scores, we found a notable enrichment of patients with a diagnosis code of CVID in the top-ranked patients. For example, among the top 5000 patients ranked in the UCSF EHR by PheNet, 320 patients carried a CVID ICD diagnosis code (49% of the total patients at UCSF bearing the code). By comparison, a random ranking of patients in the UCSF EHR found only ~2.6 patients with a CVID ICD code among the top 5000 patients. Our results demonstrate an enrichment of CVID cases among those with high PheNet scores. For example, in the top 100 ranked individuals, the average enrichment for CVID diagnosis across all six institutions was ~434-fold over random.
Table 2. External validation of PheNet identifying patients with CVID ICD codes in five other medical systems outside UCLA.
The first row shows the number of individuals in each EHR from six medical systems (UCLA, the four other University of California medical centers, and Vanderbilt University Medical Center). Second row shows the number of patients bearing a CVID ICD-10 code (D83.9) in each EHR. The following rows show the number of individuals with CVID ICD codes identified among the top x PheNet-ranked individuals. Results for individuals ranked in the UCLA EHR are also shown in the first column for reference. In paratheses, we show the fold enrichment of patients within each PheNet ranking bin as compared with randomized ranking. We also show the proportion of individuals with the CVID ICD code that were captured in each ranking bin.
| UCLA | UCSF | UCD | UCSD | UCI | Vanderbilt | |
|---|---|---|---|---|---|---|
| Total patients | 1,647,930 | 1,226,707 | 796,662 | 739,153 | 589,924 | 1,342,173 |
| Total patients with cVID IcD code | 853 | 647 | 230 | 528 | 641 | 852 |
| PheNet top 100 | 29 | 23 | 16 | 30 | 28 | 24 |
| (560x, 3%) | (436x, 4%) | (554x, 7%) | (420x, 6%) | (258x, 4%) | (378x, 3%) | |
| PheNet top 1000 | 153 | 130 | 82 | 147 | 171 | 136 |
| (296x, 18%) | (246x, 20%) | (284x, 36%) | (206x, 28%) | (157x, 27%) | (214x, 16%) | |
| PheNet top 5000 | 373 | 320 | 143 | 315 | 412 | 315 |
| (144x, 44%) | (121x, 49%) | (99x, 62%) | (88x, 60%) | (76x, 64%) | (99x, 37%) | |
| PheNet top 10,000 | 479 | 425 | 156 | 378 | 496 | 421 |
| (93x, 56%) | (81x, 66%) | (54x, 68%) | (53x, 72%) | (46x, 77%) | (66x, 49%) |
These findings showcase the power of our approach to prioritize patients suspected for CVID for follow-up analyses. These results show that the training data derived from cases in the UCLA EHR have high applicability across multiple, disparate health systems.
As much as PheNet could enrich the identification of those individuals with a CVID ICD code, we sought to understand whether the individuals who were highly ranked bore CVID-type phenotypes. To ascertain that our approach was capturing individuals who resembled CVID, we examined the phenotypes of the top 100 ranked individuals identified in each of the five UC hospitals (graphically depicted in fig. S10). For each individual, we asked what proportion of infection, autoimmune/inflammation, or other phenotypes were present in their EHR. We found that each individual had abundant diagnoses that fit the EHR-signatures of CVID (Fig. 7). For example, each individual in the top 100 ranked at UCSF carried an average of ~35% of the 14 assessed infection phenotypes and ~ 40% of the 17 assessed autoimmune/inflammation phenotypes. The proportions of phenotypes are comparable across institutions, although the training data only came from UCLA’s EHR. Together, these results confirm that PheNet identifies individuals with CVID phenotypes and maintains robust interoperability across multiple databases.
Fig. 7. Burden of CVID phenotypes among the top 100 ranked individuals at each institution.
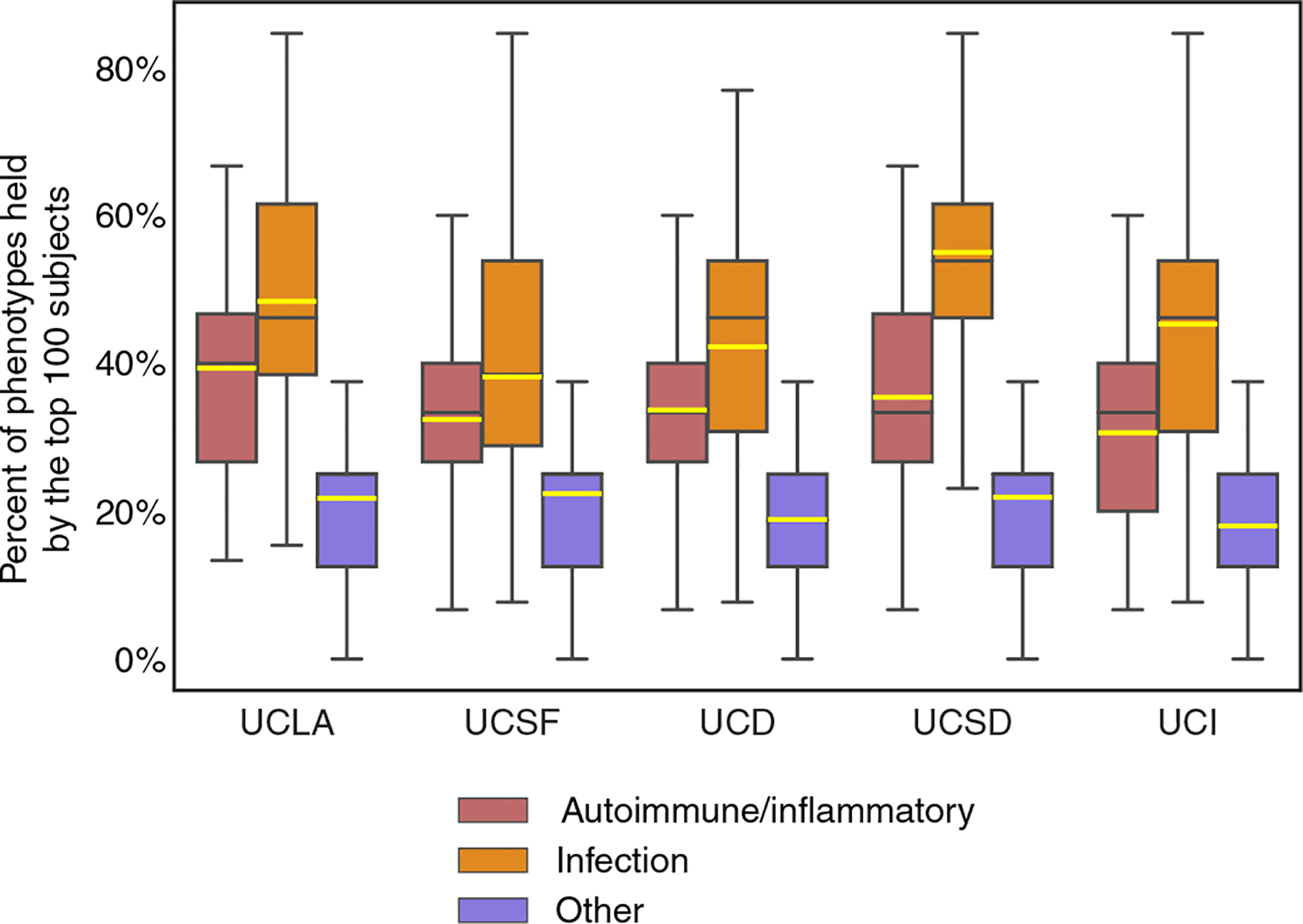
To externally validate and demonstrate broad applicability in databases outside UCLA, we examined the clinical phenotypes for each individual of the top 100 ranked at each institution for features of autoimmunity, infection, and other (the phenotypes shown in Fig. 6) from their electronic records. Averages (yellow line), 25th to 75th percentiles (box), median (black line), and range (whiskers) are shown. For example, each UCSF individual ranked in the top 100 by PheNet has in their medical record on average 35% of the 14 assessed infection phenotypes and 40% of the 17 assessed autoimmunity/inflammation phenotypes.
DISCUSSION
In this work, we used a machine learning approach to identify phenotypic patterns of the CVID phenotype, or EHR signatures, encoded in patients’ medical records and trained an algorithm to identify patients who likely have CVID but who have been, otherwise, “hiding” in the medical system. Because of the heterogeneity of clinical presentations for IEIs, patients with CVID can initially present to a wide range of clinical specialists who focus on the specific organ system involved (for example, the lung or liver) rather than directly to an immunologist for the underlying immune defect. This organ-based approach of our current health care system can result in tunnel vision and hinder a formal diagnosis in IEI, particularly for those patients who have multisystem manifestations that fluctuate over time. As a result, these patients face a diagnostic delay of 10 or more years. Each year of delay in the diagnosis of CVID results in an increase in infections, antibiotic use, emergency room visits, hospitalizations, and missed days of school and work totaling more than US $108,000 compared with the year after the diagnosis of CVID is made (in 2011 dollars, which is roughly $145,000 in 2023 dollars) (23). If the diagnostic delay for adults with CVID ranges between 10 and 15 years (8), then these results suggest that $1 million or more per patient with CVID is being misdirected by the current US health care system because of delays. The aggregate impact to the US health system of failing to diagnose CVID in a timely fashion could be hundreds of millions or billions of dollars. Beyond the economic impact, the nonquantifiable impacts on patients’ lives due to diagnostic delays are even more impactful. For example, previous studies have shown that undiagnosed patients suffer from anxiety and depression as they undergo costly tests and specialty visits (23).
The reduction of delays in diagnosis may enable patients to seek appropriate medical care to reduce morbidity and mortality. For example, early diagnosis may reduce the risk of irrevocable sequelae of invasive infections, such as bronchiectasis, encephalitis, or kidney failure. A number of efforts have attempted to codify a set of “warning signs” that offer guidance to frontline physicians such as primary care doctors. Most recognizable are the “10 warning signs” that have been widely disseminated by the Jeffrey Modell Foundation. Before EHRs, the statistical phenotyping necessary to assemble a full picture of heterogeneous IEIs like CVID was not possible, and so guidelines developed by expert committees led to gaps. These warning signs largely emphasized infections as a core feature of IEIs. Our results suggest that phenotypes of inflammation, autoimmunity, malignancy, and atopy should also be included. The majority of our patients showed a “complicated CVID phenotype,” defined as bearing both infections and autoimmunity/inflammation. Two analyses found that the original 10 warning signs were unable to identify many individuals with known IEIs because phenotypes aside from infections were missing (24, 25). When the warning signs were applied to adults versus children with known IEIs, adult patients were often missed (45% sensitivity for adults versus 64% for children) (26), suggesting the need to modify assessments on the basis of age. In other studies, the need for intravenous antibiotics, failure to thrive, or a relevant family history was found to be the only strong predictors of IEIs (27, 28). An algorithm developed by the Modell Foundation improved IEI diagnoses by using a summation of diagnostic codes (29), and another recent algorithm that summed weighted ICD codes from claims databases further improved diagnoses (30). Like most approaches based on databases of payor claims, this method did not include laboratory values. Retrospective gathering of features like we performed here has been useful in aggregating the phenotypic features of patients with IEIs into a score that can discern those with IEIs from those with secondary immunodeficiencies (31). Recent work used a Bayesian network model to score “risk” in a framework that categorized individuals into either high-, medium-, or low-risk categories of having any IEI (30). Their approach also classified each patient into a likely IEI categorization (for example, combined immunodeficiency and antibody deficiency). One limitation in Bayesian analyses is in the assessment of probabilities (and conditional probabilities) for rare events; this concern was partially alleviated by using a large cohort of children with known IEIs. However, as a result, that work suggested that 1% of all patients had medium-to-high risk of IEI, likely overestimating the true prevalence. That work highlights one of our limitations, too, that ascertaining a proper threshold for risk scores is fraught. A follow-up study from the same group followed a group of “high-risk” patients over 1 year after they were identified, subjected to manual chart review, and referred to immunology (32). This manual step reduced the number of referred patients to a reasonable number. Regardless, these efforts showcase both the potential and the unmet need for identifying previously undiagnosed patients in large health care systems and at least one potential advantage of using EHR data rather than claims data.
There are several inherent limitations to our study. The training data showed biases of older age (mean, 55 years) and female sex. In the future, we hope that two approaches of increasing the training data from additional institutions and employment of statistical approaches will reduce the impact of these biases. The prediction algorithm is derived primarily from ICD codes within the EHR. ICD codes represent an international standard, but the patterns of assigning specific ICD codes can vary among physicians and institutions (33, 34). We overcame this concern by using phecodes, a generalization of phenotypes derived from ICD codes and better suited for EHR research (35). However, even using phecodes requires a careful examination of their level of descriptive granularity. For example, one clinical description for CVID in OMIM includes hypothyroidism as a potential phenotypic feature. Accordingly, we used the phecode for “hypothyroidism” (phecode 244.2) in the prediction model. However, no individuals within the CVID cohort at UCLA actually had this phecode within their medical records. Upon further inspection, we found that UCLA patients instead bore the phecode “hypothyroidism NOS” (phecode 244.4). The lesson was that many phecodes under 244.* could equally apply and that small deviations in diagnosis-coding practices could affect algorithmic outputs. We ameliorated these deviations by not only using symptoms provided in OMIM but also by empirically learning important model features directly from the training data.
Another limitation of our work was the amount of longitudinal information available in the EHR. Patients move frequently and obtain care from a variety of settings (private practices, urgent care clinics, in addition to large health systems). Consequently, only a subset of a patient’s data is contained in a health system’s database. Because EHR vendors change with regular occurrence, many EHRs hold only a maximum of 5 to 15 years’ worth of data, which may not be enough to fully glean the necessary details of a patient’s health trajectory. We also did not consider the number of times a specific diagnosis appeared nor the order that the phecodes appeared on the medical record. Because many patients with CVID are characterized by recurrent infections, we believe that longitudinal information of multiple occurrences would increase the specificity of the model by disregarding individuals with single acute diagnoses. We also did not restrict the types of encounters when collecting the diagnosis codes (such as emergency department or outpatient clinic). Annotations of past and present ICD codes vary considerably across these settings. Instead, we wanted to use as much information as possible to increase the power of our model. However, limiting diagnoses that occur specifically during appointments or hospital visits (and not, say, phlebotomy encounters) could also increase specificity and better differentiate individuals with other immunocompromising conditions such as cancer. We hope to develop these extensions as future work.
Artificial intelligence approaches like PheNet can be used to expedite the referral of undiagnosed patients to immunologists. In the future, we will recruit patients identified by this algorithm to our immunology clinics. The impact of our work will benefit the rare disease community because there is an urgent need to identify patients early and efficiently.
MATERIALS AND METHODS
Study design
The objective of the research was to train a machine learning algorithm to recognize clinical features of patients with CVID from their EHR data so that undiagnosed patients could be identified. This study was not an interventional clinical trial. The study was approved by the UCLA Institutional Review Board (IRB). There was no direct contact with any human individuals, and consent was not required.
CVID case cohort
Central to our approach was training PheNet on a cohort of patients with a known diagnosis of CVID to serve as our “ground-truth” cohort. These patients with CVID were selected through the following process. First, we queried the UCLA EHR for all patients bearing an ICD code of D80.* (which indicates predominantly antibody deficiencies) under an IRB-approved and data compliance–approved protocol. This search revealed 3200 individuals. Most of these individuals, however, were obviously miscoded upon inspection and did not have an immunodeficiency. To consider only the EHR data of those who correctly received an immunodeficiency diagnosis, medical records were manually reviewed by a clinical immunologist. We defined individuals as having the CVID phenotype per standard definitions (2, 16): documented low serum IgG concentrations, absent or defective antibody responses to vaccine- or infection-associated antigens, age more than 2 years, and having no other explanation for low immunoglobulin concentrations (such as losses). This process helped eliminate patients who received an immunodeficiency ICD code only of the basis of acute occurrences of low IgG concentrations or for surreptitious access to immunoglobulin therapy.
This procedure narrowed the list to 197 individuals. For 186 of these individuals, we found documented low IgG concentrations (hypogammaglobulinemia) before IgRT within our EHR. For the other 11, hypogammaglobulinemia was documented only in the written notes. Regardless, each of these individuals was diagnosed and documented by immunology-certified physicians as having CVID. We note, however, that a modern definition (16) would regard these patients as having either CVID, unclassified antibody deficiencies, or combined immunodeficiency. For the purposes of training PheNet, we used n = 197 cases.
Control cohort
We constructed a case-matched control cohort using the following procedure. Of the possible ~880,000 patients in the UCLA Health EHR, we randomly selected individuals on the basis of the self-identified sex, self-identified race/ethnicity, age (closest within a 5-year window), and the number of days recorded in the EHR (closest within a 180-day window) that matched each individual in the case cohort. For age, we used the age listed on the individuals’ most recent encounter. The resulting procedure resulted in 1106 controls.
Data inclusion/exclusion criteria
When we sought to identify undiagnosed individuals across our EHR for a likelihood of having CVID, we, of course, excluded patients who already had ICD codes for CVID (“279.06,” D83.9, “Z94.2,” and “Z94.3”). We also excluded those who were deceased or with cancer diagnoses who developed secondary immunodeficiency because of immunosuppression, solid organ transplants, cystic fibrosis, or infection with the human immunodeficiency virus.
The experimental design entailed algorithmic chart review for training PheNet and then application of PheNet in the EHR system. Machine learning for PheNet was performed on the de-identified EHRs of patients at UCLA on the basis of our (de-identified) Discovery Data Repository. This data warehouse contains all UCLA Health patient information since the implementation of the EHR system in March 2013. The data include values such as laboratory tests, medications, billing codes, encounters, and other data. Lists of potential patients with CVID were converted over in bulk by the honest broker to our identified medical record system for review.
To assess the generalizability of using PheNet scores for disparate EHR databases that did not participate in model training, we conducted validation on the UC Health Data Warehouse and Vanderbilt EHR. The clinical data of these EHRs contained ~6 million patients.
Mapping CVID clinical definition to phecodes
To represent features derived from the EHR in our model, we encoded features as phecodes using the ICD code to phecode mapping v1.2. These codings represent groupings of ICD codes developed to better represent phenotypic and clinical significance from the EHR and were originally used for phenome-wide association studies. To systematically select the set of phecodes describing CVID, we used the entries for CVID listed in the OMIM catalog (22), which provides clinical descriptions for thousands of rare diseases. Specifically, we selected the following OMIM IDs: 607594 (CVID1), 616576 (CVID12), 614700 (CVID8), 240500 (CVID2), 615577 (CVID10), 616873 (CVID13), 613495 (CVID5), 613494 (CVID4), 617765 (CVID14), 613493 (CVID3), 613496 (CVID6), 614699 (CVID7), and 615767 (CVID11). We then used a previously defined database annotating syndromes listed in OMIM with Human Phenotype Ontology (HPO) terms, a set of terms used to clinically describe human phenotypic abnormalities (36, 37). Using this database, we were able to systematically aggregate a list of HPO terms for CVID derived from the clinical descriptions within OMIM. We then used a previously defined mapping between HPO terms and phecodes (11) to translate the list of HPO terms into a list of phecodes that could be constructed using information directly from the EHR. Together, this process resulted in a total of 34 unique phecodes describing CVID.
Selecting model features derived from training cohorts
In addition to using features derived from OMIM (see the “Mapping CVID clinical definition to phecodes” section), we also included features learned specifically from the training cohort. Although features derived from OMIM may broadly categorize the disease, leveraging information specific to the training cohort can empirically add additional information not already encoded within OMIM. For example, there is variation in how institutions encode diagnoses within the EHR that may not be captured in OMIM clinical descriptions. In addition, OMIM descriptions are often derived from a limited number of cases because of the rare nature of the diseases.
To select cohort-specific features, we considered all phecodes present on the medical records of individuals in the case cohort. From a possible 1800 phecodes, we limited our selection to phecodes present in at least two CVID cases and excluded phecodes already selected from OMIM. We then selected the most highly enriched phecodes in the case cohort as compared with controls. We tested for the difference in proportions between the case and control groups for each phecode. Ranking phecodes by P value, we selected the top K phecodes. We explored multiple values of K phenotypes and set K = 10 to maximize performance and minimize overfitting (fig. S3A).
IgG laboratory tests
We included in the model measurements of IgG values. Low IgG concentrations (hypogammaglobulinemia) that lack another explanation (for example, losses in the stool due to lymphangiectasia) are required to make a diagnosis of CVID. Instead of using the raw measurement as a feature directly, we converted the values to a categorical scale where the laboratory value is encoded as “0” if the individual has never received an IgG test, “1” if the individual has had an IgG test ≥ 600 mg/dl (normal range), and “2” if the individual has had an IgG test <600 mg/dl (abnormal). If an individual had multiple recorded IgG tests, then we selected the lowest recorded value.
Model inference
For benchmarking experiments, we performed fivefold cross-validation within each experiment to quantify the accuracy of various inference frameworks. To address the imbalance of cases in our dataset, we created a more balanced training dataset using random upsampling with an upsampling ratio of 0.50 and downsampling controls to n = 10,000. We explored the trade-off of various upsample ratios and downsampling sample sizes (fig. S3,D and E). We estimated the weight of each feature using logistic regression (no penalty). We performed additional experiments to quantify performance using a variety of other inference methods, such as ridge regression, random forest, and the inverse-log frequency weighting scheme used by PheRS (fig. S3B). Hyperparameters used in the ridge regression and random forest models were selected using an additional fivefold cross-validation step within the training step.
Comparison with previous methods
We compared PheNet with the current state-of-the-art method PheRS (11) that also uses phecodes as features. PheRS selects phecodes that correspond to the OMIM clinical description of a given disease and then computes the log-inverse frequency of the phecode measured in the general patient population. This was then used as the feature weight in the algorithm, and the prediction score is a weighted sum of the weights and the presence of a given phecode, making this approach an unsupervised method that does not leverage any labeled case information. To compare methods, we used PheRS weights computed using the UCLA EHR from more than 880,000 patients and computed the risk scores for all patients using each method with fivefold cross-validation.
ICD-based diagnosis date
Although all individuals in the case cohort were verified to have CVID, we were not able to directly obtain the exact date of diagnosis from the manually reviewed records. For those individuals for whom we could not discern an exact date of diagnosis, we used a heuristic to estimate the date of diagnosis on the basis of occurrences of the ICD code for CVID (D83.9). We refer to this as the ICD-based diagnosis date to clarify that it does not constitute the precise date of a formal clinical diagnosis. However, there were eight individuals who did not have an ICD code for CVID, and, thus, we could not provide an estimated diagnosis date.
Assessing PheNet using retrospective EHR
We first encoded a patient’s most recent visit as time 0. We recorded the time of an encounter in a patient’s medical record as the number of days before their most recent visit. This provided us a common metric of time to use when performing analyses across all patients. We computed a patient’s PheNet score at 30-day intervals, spanning ~6 years (30 days × 12 months × 6 years). At each interval, we only considered features that were recorded up to and including that time point (and time before the given interval). To compute the score percentile for each patient with CVID, we used the scores of all other patients taken at time point 0 (the most recent visit) and then added the score of the single patient from the designated time point. Using this distribution, we computed the score percentile for the specific patient at that time point. This was then repeated for all patients with CVID across all time points. Because we only had EHR data from 2013, we did this to ensure that the overall distribution of scores at earlier time points was not skewed because there are many patients who do not have medical records in the electronic system at earlier time points. We then checked whether any patients reached the top of the score distribution at any time point before an individual’s ICD-based diagnosis (see ICD-based diagnosis date).
Clinical validation of individuals identified by PheNet
To validate our approach, we performed a clinical chart review for the top set of individuals prioritized by PheNet. First, we removed all individuals who were deceased or who had phecodes corresponding to solid organ transplants, cystic fibrosis, or human immunodeficiency virus, resulting in the removal of 42,346 individuals. These specific disorders could lead to immunodeficiency and have a similar profile to CVID, but the cause of their immunodeficiency is already explained. We then selected the top 100 individuals identified by PheNet and a control group of 100 randomly selected individuals from the patient population. For external validation in the UC Health data warehouse and Vanderbilt, we computed the PheNet score for all patients and counted those with ICD codes for CVID (279.06, D83.9, Z94.2, and Z94.3) for consistency.
Clinical charts were directly reviewed by a clinical immunologist who had access to each individual’s full medical record in a blinded review. The two lists were merged and scrambled, and the clinician was not aware of how the list of individuals was generated. Each individual was ranked according to an ordinal scale from 1 to 5 quantifying the likeliness of having CVID where 1 was defined as near certainty not CVID and 5 was definitive as CVID, meaning that the individual met the criteria of a physician diagnosis.
Statistical analysis
We built our marginal logistic regression, ridge regression, and random forest models using the sklearn package in Python. Statistical and computational analyses were also performed in Python. To find phecodes enriched in cases, we used the P value associated with the z score of the proportion of cases having each phecode compared with controls (these z scores were normally distributed). We used fivefold cross-validation to make the choice to include 10 phecodes in addition to the 34 OMIM phecodes, including IgG laboratory values as a feature, as well as to decide on marginal logistic regression as our model as compared with other models. We used sklearn to add upsampling and downsampling to the model and fivefold cross-validation to choose upsampling and downsampling rates. We used the Python package imblearn for all additional model-specific hyperparameter selection. To compare models, we used the metrics of AUC-ROC and AUC-PR, calculated using the sklearn python package, as well as positive predictive value. We used the Cochran-Armitage test to test for differences in score distributions in Fig. 5.
Supplementary Material
Acknowledgments:
We thank L. Dahm and the Center for Data-driven Insights and Innovation at UC Health (CDI2; www.ucop.edu/uc-health/functions/center-for-data-driven-insights-and-innovationscdi2.html) for analytical and technical support related to use of the UC Health Data Warehouse. We acknowledge the UCLA Institute for Precision Health for making data available for research.
Funding:
We acknowledge funding from the National Institutes of Health (NIH)/National Institutes of Allergy and Infectious Diseases (R01 AI153827 to M.J.B and B.P.) and DP5OD024579 (V.A.A.).
Competing interests:
L.A.B. is a consultant for Pharming and receives royalties from Nashville Biosciences. M.J.B. is a speaker for Grifols; consults for Pharming, Horizon, and Grifols; receives sponsored research funding from the NIH, the Bill and Melinda Gates Foundation, Pharming, and Chiesi; and serves on the scientific advisory board for ADMA Biologics and Alpine Immune Sciences.
Footnotes
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. PheNet code is available at DOI: 10.5281/zenodo.10947388.
REFERENCES AND NOTES
- 1.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, Klein C, Morio T, Oksenhendler E, Picard C, Puel A, Puck J, Seppänen MRJ, Somech R, Su HC, Sullivan KE, Torgerson TR, Meyts I, Human inborn errors of immunity: 2022 Update on the classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol 42, 1473–1507 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla FA, Barlan I, Chapel H, Costa-Carvalho BT, Cunningham-Rundles C, de la Morena MT, Espinosa-Rosales FJ, Hammarström L, Nonoyama S, Quinti I, Routes JM, Tang MLK, Warnatz K, International Consensus Document (ICON): Common variable immunodeficiency disorders. J. Allergy Clin. Immunol. Pract 4, 38–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abolhassani H, Hammarström L, Cunningham-Rundles C, Current genetic landscape in common variable immune deficiency. Blood 135, 656–667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez NJ, Posadas-Cantera S, Caballero-Oteyza A, Camacho-Ordonez N, Grimbacher B, There is no gene for CVID—Novel monogenetic causes for primary antibody deficiency. Curr. Opin. Immunol 72, 176–185 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Silva SL, Fonseca M, Pereira MLM, Silva SP, Barbosa RR, Serra-Caetano A, Blanco E, Rosmaninho P, Pérez-Andrés M, Sousa AB, Raposo AASF, Gama-Carvalho M, Victorino RMM, Hammarstrom L, Sousa AE, Monozygotic twins concordant for common variable immunodeficiency: Strikingly similar clinical and immune profile associated with a polygenic burden. Front. Immunol 10, 2503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, Kim CE, Hou C, Wang F, Chiavacci R, Kugathasan S, Sleasman JW, Baldassano R, Perez EE, Chapel H, Cunningham-Rundles C, Hakonarson H, Genome-wide association identifies diverse causes of common variable immunodeficiency. J. Allergy Clin. Immunol 127, 1360–1367. e6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urschel S, Kayikci L, Wintergerst U, Notheis G, Jansson A, Belohradsky BH, Common variable immunodeficiency disorders in children: Delayed diagnosis despite typical clinical presentation. J. Pediatr 154, 888–894 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Slade CA, Bosco JJ, Giang TB, Kruse E, Stirling RG, Cameron PU, Hore-Lacy F, Sutherland MF, Barnes SL, Holdsworth S, Ojaimi S, Unglik GA, De Luca J, Patel M, McComish J, Spriggs K, Tran Y, Auyeung P, Nicholls K, O’Hehir RE, Hodgkin PD, Douglass JA, Bryant VL, van Zelm MC, Delayed diagnosis and complications of predominantly antibody deficiencies in a cohort of Australian adults. Front. Immunol 9, 694–694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baloh C, Reddy A, Henson M, Prince K, Buckley R, Lugar P, 30-Year review of pediatric- and adult-onset CVID: Clinical correlates and prognostic indicators. J. Clin. Immunol 39, 678–687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graziano V, Pecoraro A, Mormile I, Quaremba G, Genovese A, Buccelli C, Paternoster M, Spadaro G, Delay in diagnosis affects the clinical outcome in a cohort of cvid patients with marked reduction of iga serum levels. Clin. Immunol 180, 1–4 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Bastarache L, Hughey JJ, Hebbring S, Marlo J, Zhao W, Ho WT, Van Driest SL, McGregor TL, Mosley JD, Wells QS, Temple M, Ramirez AH, Carroll R, Osterman T, Edwards T, Ruderfer D, Velez Edwards DR, Hamid R, Cogan J, Glazer A, Wei W, Feng Q, Brilliant M, Zhao ZJ, Cox NJ, Roden DM, Denny JC, Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science 359, 1233–1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morley TJ, Han L, Castro VM, Morra J, Perlis RH, Cox NJ, Bastarache L, Ruderfer DM, Phenotypic signatures in clinical data enable systematic identification of patients for genetic testing. Nat. Med 27, 1097–1104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banda JM, Sarraju A, Abbasi F, Parizo J, Pariani M, Ison H, Briskin E, Wand H, Dubois S, Jung K, Myers SA, Rader DJ, Leader JB, Murray MF, Myers KD, Wilemon K, Shah NH, Knowles JW, Finding missed cases of familial hypercholesterolemia in health systems using machine learning. NPJ Digit. Med 2, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M, Sundberg P, Yee H, Zhang K, Zhang Y, Flores G, Duggan GE, Irvine J, Le Q, Litsch K, Mossin A, Tansuwan J, Wang D, Wexler J, Wilson J, Ludwig D, Volchenboum SL, Chou K, Pearson M, Madabushi S, Shah NH, Butte AJ, Howell MD, Cui C, Corrado GS, Dean J, Scalable and accurate deep learning with electronic health records. NPJ Digit. Med 1, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S-I, Celik S, Logsdon BA, Lundberg SM, Martins TJ, Oehler VG, Estey EH, Miller CP, Chien S, Dai J, Saxena A, Blau CA, Becker PS, A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat. Commun 9, 42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidel MG, Kindle G, Gathmann B, Quinti I, Buckland M, Van Montfrans J, Scheible R, Rusch S, Gasteiger LM, Grimbacher B, Mahlaoui N, Ehl S, Abinun M, Albert M, Cohen SB, Bustamante J, Cant A, Casanova J-L, Chapel H, Basile GDS, De Vries E, Dokal I, Donadieu J, Durandy A, Edgar D, Espanol T, Etzioni A, Fischer A, Gaspar B, Gatti R, Gennery A, Grigoriadou S, Holland S, Janka G, Kanariou M, Klein C, Lachmann H, Lilic D, Manson A, Martinez N, Meyts I, Moes N, Moshous D, Neven B, Ochs H, Picard C, Renner E, Rieux-Laucat F, Seger R, Soresina A, Stoppa-Lyonnet D, Thon V, Thrasher A, Van De Veerdonk F, Villa A, Weemaes C, Warnatz K, Wolska B, Zhang S-Y, The European Society for Immunodeficiencies (ESID) registry working definitions for the clinical diagnosis of inborn errors of immunity. J. Allergy Clin. Immunol. Pract 7, 1763–1770 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Farmer JR, Ong M-S, Barmettler S, Yonker LM, Fuleihan R, Sullivan KE, Cunningham-Rundles C; Summary and descriptionThe USIDNET Consortium J. E. Walter, Common variable immunodeficiency non-infectious disease endotypes redefined using unbiased network clustering in large electronic datasets. Front. Immunol 8, 1740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrabba M, Salvi M, Baselli LA, Serafino S, Zarantonello M, Trombetta E, Pietrogrande MC, Fabio G, Dellepiane RM, Long-term follow-up in common variable immunodeficiency: The pediatric-onset and adult-onset landscape. Front. Pediatr 11, 1125994 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C, Morbidity and mortality in common variable immune deficiency over 4 decades. Blood 119, 1650–1657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastarache L, Using phecodes for research with the electronic health record: From PheWAS to PheRS. Annu. Rev. Biomed. Data Sci 4, 1–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC, PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–1210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKusick VA, Mendelian inheritance in man and its online version, OMIM. Am. J. Hum. Genet 80, 588–604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modell V, Gee B, Lewis DB, Orange JS, Roifman CM, Routes JM, Sorensen RU, Notarangelo LD, Modell F, Global study of primary immunodeficiency diseases (PI)-diagnosis, treatment, and economic impact: An updated report from the Jeffrey Modell Foundation. Immunol. Res 51, 61–70 (2011). [DOI] [PubMed] [Google Scholar]
- 24.MacGinnitie A, Aloi F, Mishra S, Clinical characteristics of pediatric patients evaluated for primary immunodeficiency. Pediatr. Allergy Immunol 22, 671–675 (2011). [DOI] [PubMed] [Google Scholar]
- 25.O’Sullivan MD, Cant AJ, The 10 warning signs. Curr. Opin. Allergy Clin. Immunol 12, 588–594 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Bjelac JA, Yonkof JR, Fernandez J, Differing performance of the warning signs for immunodeficiency in the diagnosis of pediatric versus adult patients in a two-center tertiary referral population. J. Clin. Immunol 39, 90–98 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Reda SM, El-Ghoneimy DH, Afifi HM, Clinical predictors of primary immunodeficiency diseases in children. Allergy Asthma Immunol. Res 5, 88–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subbarayan A, Colarusso G, Hughes SM, Gennery AR, Slatter M, Cant AJ, Arkwright PD, Clinical features that identify children with primary immunodeficiency diseases. Pediatrics 127, 810–816 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Modell V, Quinn J, Ginsberg G, Gladue R, Orange J, Modell F, Modeling strategy to identify patients with primary immunodeficiency utilizing risk management and outcome measurement. Immunol. Res 65, 713–720 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Rider NL, Miao D, Dodds M, Modell V, Modell F, Quinn J, Schwarzwald H, Orange JS, Calculation of a primary immunodeficiency “risk vital sign” via population-wide analysis of claims data to aid in clinical decision support. Front. Pediatr 7, 70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toms K, Gkrania-Klotsas E, Kumararatne D, Analysis of scoring systems for primary immunodeficiency diagnosis in adult immunology clinics. Clin. Exp. Immunol 203, 47–54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rider NL, Coffey M, Kurian A, Quinn J, Orange JS, Modell V, Modell F, A validated artificial intelligence-based pipeline for population-wide primary immunodeficiency screening. J. Allergy Clin. Immunol 151, 272–279 (2022). [DOI] [PubMed] [Google Scholar]
- 33.O’Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM, Measuring diagnoses: ICD code accuracy. Health Serv. Res 40, 1620–1639 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horsky J, Drucker EA, Ramelson HZ, Accuracy and completeness of clinical coding using ICD-10 for ambulatory visits. AMIA Annu. Symp. Proc 2017, 912–920 (2017). [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T, Zhao J, Carroll R, Bastarache L, Denny JC, Theodoratou E, Wei W-Q, Mapping ICD-10 and ICD-10-CM codes to phecodes: Workflow development and initial evaluation. JMIR Med. Inform 7, e14325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GCM, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jähn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park S-M, Riggs ER, Scott RH, Sisodiya S, Van Vooren S, Wapner RJ, Wilkie AOM, Wright CF, Silfhout ATV-V, de Leeuw N, de Vries BBA, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN, The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 42, D966–D974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groza T, Köhler S, Moldenhauer D, Vasilevsky N, Baynam G, Zemojtel T, Schriml LM, Kibbe WA, Schofield PN, Beck T, Vasant D, Brookes AJ, Zankl A, Washington NL, Mungall CJ, Lewis SE, Haendel MA, Parkinson H, Robinson PN, The Human Phenotype Ontology: Semantic unification of common and rare disease. Am. J. Hum. Genet 97, 111–124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. PheNet code is available at DOI: 10.5281/zenodo.10947388.


