Abstract
Bovine herpesvirus 4 (BoHV-4) is a gammaherpesvirus of cattle. The complete long unique coding region (LUR) of BoHV-4 strain 66-p-347 was determined by a shotgun approach. Together with the previously published noncoding terminal repeats, the entire genome sequence of BoHV-4 is now available. The LUR consists of 108,873 bp with an overall G+C content of 41.4%. At least 79 open reading frames (ORFs) are present in this coding region, 17 of them unique to BoHV-4. In contrast to herpesvirus saimiri and human herpesvirus 8, BoHV-4 has a reduced set of ORFs homologous to cellular genes. Gene arrangement as well as phylogenetic analysis confirmed that BoHV-4 is a member of the genus Rhadinovirus. In addition, an origin of replication (ori) in the genome of BoHV-4 was identified by DpnI assays. A minimum of 1.69 kbp located between ORFs 69 and 71 was sufficient to act as a cis signal for replication.
Bovine herpesvirus 4 (BoHV-4) was isolated from cattle with various clinical symptoms, e.g., skin lesions, genital and respiratory diseases, and malignant catarrhal fever, and also from apparently healthy animals (reviewed in reference 28), although its pathogenic role remains unclear. BoHV-4 replicates in animal cell lines like Georgia bovine kidney cells, baby hamster kidney cells, Crandell feline kidney cells, and equine dermal cells as well as in human cell lines like human embryonic lung cells and HeLa cells (16, 39).
Due to its behavior in cell culture as well as its morphogenesis, BoHV-4 was originally classified as a betaherpesvirus (36). The formation of high-density inclusion bodies and giant cells after infection of tissue culture cells, which is well-known for cytomegaloviruses, supported this initial classification (36, 38).
Studies on the genome structure and determination of first gene sequences revealed that BoHV-4 is a member of the Gammaherpesvirinae: (i) the virus has a B-type genome structure according to Roizman's classification of herpesvirus genomes (34) with a long unique genome region (LUR) flanked by polyrepetitive DNA (prDNA) elements, (ii) the genome contains a thymidine kinase gene, (iii) genes sequenced so far show striking homology to those of the gammaherpesviruses Kaposi's sarcoma-associated herpesvirus (or human herpesvirus 8 [HHV-8]) and herpesvirus saimiri (HVS), and (iv) the gene arrangement is collinear with those of other gammaherpesviruses (7, 8, 17, 22, 26, 27).
The 2,267-nucleotide sequence of a single prDNA element of BoHV-4 strain 66-p-347 was published previously (6). Sequence analysis of this prDNA element showed similarities to the pac-1 and pac-2 consensus sequences of the cleavage and packaging site which is essential for the replication of the viral genome of HSV (14). The total number of prDNA elements was estimated to be 15 on average, with a random distribution at both BoHV-4 termini (17). Analysis of the predicted open reading frames (ORFs) of the prDNA indicated no homology to sequences for known proteins. It was shown that one prDNA element is sufficient for cleavage and packaging and therefore plays a key role in the replication of BoHV-4 (6).
Partial sequence information of the BoHV-4 LUR, e.g., for the glycoprotein B (gB) gene (22), the region coding for the putative terminase (7), two immediate-early transcripts (44, 45), and two late transcripts (3, 4) and the LUR sequences outside the conserved gene blocks (27), was published previously. However, these sequence data have three limitations: (i) they were obtained from different strains, (ii) the available sequence files describing the genome regions outside the conserved gene blocks contain ambiguous base pairs, and (iii) individual or virus-specific ORFs may be found within the conserved gene blocks.
In this article the complete DNA sequence of the LUR of BoHV-4 strain 66-p-347 is presented. We describe its coding capacity and discuss the common and unique features of the BoHV-4 genome in comparison to those of other gammaherpesviruses.
It has been shown that BoHV-4 replication is restricted to the S phase of the cell cycle (42). In this study, we mapped an origin of replication (ori). Together with the recently published cis elements necessary for cleavage and packaging of the BoHV-4 genome as well as the gene map presented here, sufficient information is now available for constructing recombinant BoHV-4 genomes as vectors for several applications.
MATERIALS AND METHODS
Virus propagation, cloning, and sequence determination.
BoHV-4 isolate 66-p-347 was propagated on Madin-Darby bovine kidney cells. DNA was prepared as described earlier (17). In brief, virions were harvested from the supernatant of infected cells by centrifugation through a sucrose cushion. Viral DNA was purified by CsCl equilibrium gradient centrifugation and randomly shotgun cloned into M13mp18. For this purpose, 20 μg of viral DNA was sonicated and the resulting fragments were purified by agarose gel electrophoresis. The fraction of 800 to 1,200 bp was eluted from the gel, and the DNA fragments were blunted with T4 DNA polymerase. These fragments were subcloned into a SmaI-digested M13mp18 sequencing vector. The M13 subclones were sequenced with a combination of dye terminator and dye primer chemistries (PE Biosystems). Data were collected using ABI 377 automated sequencers (PE Biosystems). A total of 2,500 M13 clones were sequenced to cover the complete viral genome sixfold. Sequence data were further processed by the program REAP (Bernd Drescher, Institut für Molekulare Biotechnologie, Jena, Germany) and assembled by the program GAP4 (Roger Staden, Medical Research Council, Cambridge, United Kingdom).
Nucleotide and protein sequence analysis.
Potential coding regions were identified with the programs GRAIL2, GENSCAN, XPOUND, and GENEMARK (5, 10, 37, 40) using matrices for human and gammaherpesvirus DNA and MacVector (version 6.01; Oxford Molecular Group). For nucleotide and protein sequence analysis, the FASTA and BLAST programs from the Genetics Computer Group (GCG) package (11, 15) were used and ORFs were compared to the GenBank and SwissProt databases. Amino acid sequence comparison between potential BoHV-4 genes and HHV-8 and HVS was carried out with GAP from the GCG package.
Phylogenetic analysis.
Phylogenetic analysis was based on multiple sequence alignments performed with the ClustalW module of MacVector. Gaps or insertions unique to a certain species were removed, and the remaining conserved regions were concatenated for each individual protein (30). Trees were then constructed with the PHYLIP program package, using the programs Protdist (Dayhoff PAM matrix) and Neighbor or, alternatively, the program Protpars with randomized input of sequences (20, 21). The trees were statistically evaluated by bootstrap analysis, using the programs Seqboot and Consense.
ori mapping: plasmid constructs.
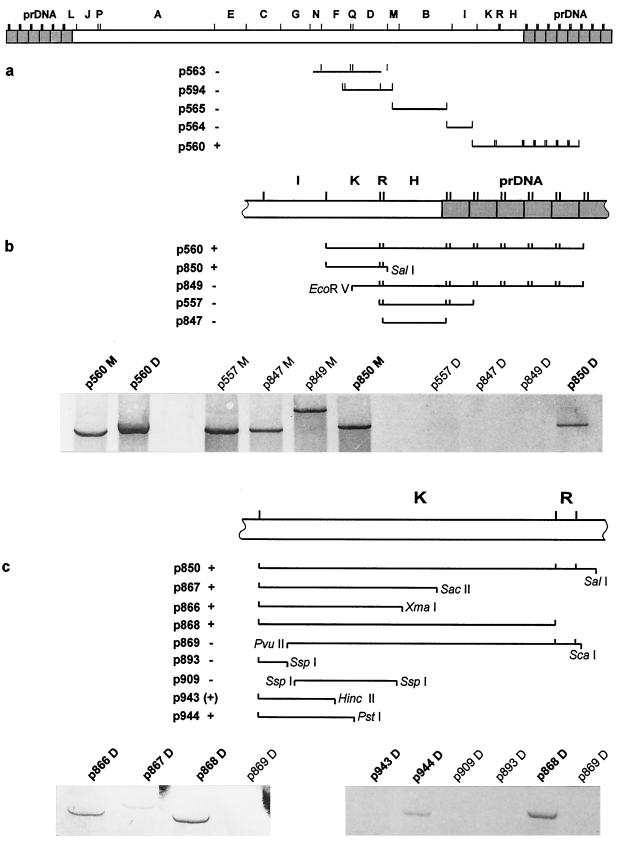
The restriction fragments used for the replication studies are shown in Fig. 2. Plasmid constructs p557, p560, p563, p564, p565, and p594 were generated by cloning BoHV-4 restriction fragments from partial BamHI digestion into the BamHI site of a pBR322-derived vector, p141.31 (W. Hammerschmidt [GSF-Forschungszentrum für Umwelt und Gesundheit, Munich, Germany], unpublished data). Construct p847 contains a single BamHI fragment derived from clone p560. Constructs p849 and p850 were generated by deleting an EcoRV and SalI fragment, respectively, from clone p560 and subsequent religation of the remaining constructs. Constructs p866 and p867 were derived from p850 by deletion of an XbaI or SacII fragment and subsequent religation of the remaining plasmid. Constructs p868 and p869 were derived from p850 by subcloning of a BamHI or a PvuII-ScaI fragment. Constructs p893, p909, p943, and p944 were generated by subcloning BoHV-4 fragments from p866 into pBluescript SK(+) (Stratagene). Construct p893 contains the BamHI-SspI subfragment of p866. Construct p909 contains an SspI subfragment derived from p866. Finally, constructs p943 and p944 were generated by subcloning of BamHI-HincII and BamHI-PstI fragments derived from p866 into the corresponding restriction sites of pBluescript SK(+).
FIG. 2.
Mapping of an ori. (a) Schematic drawing of the complete genome of BoHV-4 is shown at the top. The LUR is given as an open box. BamHI sites are indicated by vertical lines above the genome, and the restriction fragments are labeled in alphabetical order depending on their size, with A as the largest fragment. The positions of the inserts of the first set of plasmids tested are shown below the genome. The corresponding plasmid names are given to the left of the inserts. Replication activity as explained in Results is indicated by “+” or “−” after the plasmid names. (b) Magnification of the region which has been shown to contain an ori in the first set of experiments. Inserts of the second set of plasmids, plasmid names, and results are shown as explained for panel a. Below the drawings, an example of the results of the transient replication assay after visualization of the Southern blots is given. The signals of the BamHI- and MboI-digested Hirt extracts, labeled with “M” behind the plasmid name, are used as size references. Bands resulting from MboI cleavage are not shown because of the weak signals due to the small fragment sizes. Lanes containing the BamHI- and DpnI-digested Hirt extracts are labeled with “D” following the plasmid names. (c) A further magnification step and the inserts of the plasmids used in the second and third round of experiments. Again examples of the Southern blot results are given, showing the BamHI- and DpnI-digested Hirt extracts. For plasmid p943, a very weak signal could be demonstrated which is clearly visible only after long exposition to the alkaline phosphatase substrate, leading to high background.
ori mapping: transient replication assays.
For the localization of a viral origin of replication in the BoHV-4 genome, DpnI assays were performed as described earlier (12, 23). In brief, plasmid DNA was purified from the clones described above, utilizing the plasmid Midi and Maxi Kits (Qiagen), and used for transfection of Georgia bovine kidney cells by electroporation. For complementation of a viral ori present on some of the plasmid constructs with BoHV-4 gene products necessary for viral DNA replication, 18 to 22 h after electroporation the transfected cells were infected with BoHV-4 for 45 min at a multiplicity of infection of 1. Virus suspension was removed, and the cells were provided with fresh medium (Dulbecco's modified Eagle's medium, 5% fetal calf serum). With the beginning of plaque formation 48 to 72 h postinfection, cells were harvested and plasmid DNA recovered (25). DNA was purified by phenol-chloroform-isopropanol extractions and subsequently digested overnight with DpnI and BamHI or, as a control, with MboI and BamHI. While BamHI digestion was used to cleave plasmid and cellular DNA, DpnI and MboI were used to distinguish plasmid fractions replicated in the eucaryotic cells from bacterial input DNA. The DNA was separated by agarose gel electrophoresis, transferred to nylon membranes (Boehringer Mannheim-Roche Diagnostics Corp.), UV cross-linked, and hybridized with digoxigenin-labeled plasmid p141.31. Visualization was performed according to a modified manufacturer's protocol for the digoxigenin system (Boehringer Mannheim-Roche Diagnostics), using naphthol-AS-phosphate and fast blue B.
Nucleotide sequence accession numbers.
The complete LUR for BoHV-4 is accessible from GenBank as AF318573. Accession numbers for the sequences used for comparison in Table 1 and in the phylogenetic analysis are as follows: alcelaphine herpesvirus 1 (AlHV-1), accession no. AF005370; Epstein-Barr virus (EBV), accession no. X00784; equine herpesvirus 2, accession no. U20824; human herpesvirus 8 (HHV-8), accession no. U75698; herpesvirus saimiri (= saimiriine herpesvirus 2) (HVS), accession no. X64346; herpesvirus ateles (= ateline herpesvirus 3) (HVA), accession no. AF083424; murine gammaherpesvirus 68 (MHV-68), accession no. U97553; and rhesus monkey rhadinovirus (RRV), accession no. AF029302.
TABLE 1.
Potential BoHV-4 ORFs and homologues to HHV-8 and HVS
| BoHV-4 ORF | Strand | Position ofa:
|
Size (aa) | HHV-8
|
HVS
|
Description (Fig. 1 abbreviation) | |||
|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | %IDb | ORF | %IDb | ORF | ||||
| Bo1 | − | 633 | 445 | 62 | |||||
| 3 | + | 614 | 4480 | 1,288 | 36.73 | 3 | Tegument protein or v-FGAM-synthase (BORFA1c) | ||
| Bo2 | + | 4608 | 4811 | 67 | BORFA2c | ||||
| Bo3 | + | 5286 | 5513 | 75 | |||||
| 6 | + | 5943 | 9347 | 1,134 | 56.33 | 6 | 58.65 | 6 | Single-stranded DNA binding protein (MDBP) |
| 7 | + | 9352 | 11430 | 692 | 28.00 | 7 | 21.43 | 7 | Transport protein |
| 8 | + | 11420 | 14044 | 874 | 54.30 | 8 | 51.13 | 8 | gB |
| 9 | + | 14184 | 17201 | 1,005 | 62.96 | 9 | 62.49 | 9 | DNA polymerase (DPOL) |
| 10 | + | 17297 | 18577 | 426 | 22.74 | 10 | 28.78 | 10 | BORFB1c |
| Bo4 | − | 19345 | 18848 | 165 | 25.97 | Short ORF of IE1 RNA | |||
| Bo5 | − | 285 | 27.85 | K5 | IE1 RNA | ||||
| Bo5d | − | 19918 | 19414 | 167 | Long ORF of IE1 RNA, exon IV | ||||
| Bo5c | − | 20200 | 20015 | 62 | Long ORF of IE1 RNA, exon III | ||||
| Bo5b | − | 20438 | 20288 | 51 | Long ORF of IE1 RNA, exon II | ||||
| Bo5a | − | 20553 | 20538 | 5 | Long ORF of IE1 RNA, exon I | ||||
| Bo6 | + | 20495 | 20788 | 97 | |||||
| Bo7 | − | 21194 | 20843 | 116 | |||||
| B08 | + | 21519 | 21722 | 67 | Overlapping with 1.7-kb late RNA | ||||
| 16 | + | 22971 | 23651 | 226 | 26.59 | 16 | 26.45 | 16 | v-Bcl-2 homologue (BORFB2c) |
| 17 | − | 25261 | 23714 | 515 | 43.82 | 17 | 49.36 | 17 | Capsid protein |
| 18 | + | 25260 | 26078 | 272 | 47.84 | 18 | 50.79 | 18 | |
| 19 | − | 27706 | 26030 | 558 | 47.22 | 19 | 50.00 | 19 | Tegument protein |
| 20 | − | 28220 | 27408 | 270 | 37.84 | 20 | 40.98 | 20 | |
| 21 | + | 28204 | 29541 | 445 | 32.50 | 21 | 34.72 | 21 | Thymidine kinase (TK) |
| 22 | + | 29552 | 31675 | 707 | 32.47 | 22 | 36.18 | 22 | Glycoprotein H (gH) |
| 23 | − | 32874 | 31672 | 400 | 34.84 | 23 | 40.97 | 23 | |
| 24 | − | 35110 | 32852 | 752 | 48.58 | 24 | 50.90 | 24 | |
| 25 | + | 35100 | 39221 | 1,373 | 66.54 | 25 | 68.03 | 25 | Major capsid protein (MCP) |
| 26 | + | 39257 | 40171 | 304 | 54.60 | 26 | 60.07 | 26 | Capsid protein |
| 27 | + | 40185 | 40823 | 212 | 22.22 | 27 | 24.75 | 27 | |
| Bo9 | + | 40833 | 41132 | 99 | |||||
| 29 | − | 682 | 60.65 | 29 | 61.73 | 29 | Cleavage and/or packaging protein | ||
| 29b | − | 42295 | 41156 | 379 | 29b | 29b | Cleavage and/or packaging protein, exon II | ||
| 30 | + | 42308 | 42550 | 80 | 49.29 | 30 | 36.49 | 30 | |
| 31 | + | 42484 | 43125 | 213 | 41.18 | 31 | 52.68 | 31 | |
| 32 | + | 43071 | 44441 | 456 | 31.92 | 32 | 36.05 | 32 | Viral DNA cleavage or packaging protein |
| 33 | + | 44434 | 45432 | 332 | 36.48 | 33 | 50.76 | 33 | Dispensable for viral growth of HSV-1 |
| 29a | − | 46330 | 45422 | 303 | 29a | 29a | Cleavage and/or packaging protein, exon 1 | ||
| 34 | + | 46329 | 47315 | 328 | 42.81 | 34 | 44.76 | 34 | |
| 35 | + | 47287 | 47760 | 157 | 31.76 | 35 | 37.33 | 35 | |
| 36 | + | 47648 | 48952 | 434 | 33.57 | 36 | 37.44 | 36 | Kinase (PK) |
| 37 | + | 48960 | 50429 | 489 | 45.98 | 37 | 52.08 | 37 | Alkaline exonuclease |
| 38 | + | 50381 | 50587 | 68 | 32.73 | 38 | 29.03 | 38 | |
| 39 | − | 51763 | 50654 | 369 | 54.08 | 39 | 50.82 | 39 | Glycoprotein M (gM) |
| 40 | + | 51879 | 53252 | 457 | 27.58 | 40 | 33.64 | 40 | Helicase-primase complex component |
| 41 | + | 53365 | 53866 | 173 | 26.06 | 41 | 31.82 | 41 | Helicase-primase complex component |
| 42 | − | 54753 | 53878 | 291 | 36.59 | 42 | 43.56 | 42 | |
| 43 | − | 56380 | 54530 | 616 | 53.28 | 43 | 59.18 | 43 | Capsid protein |
| 44 | − | 56328 | 58706 | 792 | 61.12 | 44 | 67.73 | 44 | Helicase |
| 45 | − | 59535 | 58810 | 241 | 28.96 | 45 | 33.19 | 45 | |
| 46 | − | 60296 | 59535 | 253 | 57.31 | 46 | 61.51 | 46 | Uracil-DNA-glycosidase |
| 47 | − | 60736 | 60305 | 143 | 29.69 | 47 | 33.33 | 47 | Glycoprotein L (gL) |
| 48 | − | 62387 | 60843 | 514 | 28.61 | 48 | 29.34 | 48 | |
| 49 | − | 63504 | 62605 | 299 | 31.86 | 49 | 24.49 | 49 | |
| 50 | + | 552 | 26.08 | 50 | 28.92 | 50 | R transactivator homologue | ||
| 50a | + | 62591 | 62593 | 1 | IE2, R transactivator homologue, exon I | ||||
| 50b | + | 63529 | 65184 | 551 | IE2, R transactivator homologue, exon II | ||||
| Bo10 | + | 237 | Glycoprotein gp80 | ||||||
| Bo10a | + | 65714 | 66242 | 176 | Glycoprotein gp80, exon I (BORFD1c) | ||||
| Bo10b | + | 66325 | 66617 | 97 | Glycoprotein gp80, exon II (BORFD1c) | ||||
| 52 | − | 67029 | 66643 | 128 | 39.02 | 52 | 38.05 | 52 | |
| 53 | − | 67367 | 67095 | 90 | 36.78 | 53 | 32.56 | 53 | |
| 54 | + | 67436 | 68284 | 282 | 32.86 | 54 | 35.97 | 54 | dUPTase |
| 55 | − | 68945 | 68343 | 200 | 46.70 | 55 | 51.30 | 55 | |
| 56 | + | 68909 | 71440 | 843 | 46.03 | 56 | 48.79 | 56 | DNA replication protein |
| 57 | + | 420 | 32.23 | 57 | 32.42 | 57 | |||
| 57a | + | 71534 | 71561 | 9 | 57 | 57 | Posttranscriptional transactivator? exon I | ||
| 57b | + | 71651 | 72888 | 411 | 57 | 57 | Posttranscriptional transactivator? exon II | ||
| 58 | − | 74364 | 73312 | 350 | 29.27 | 58 | 27.53 | 58 | |
| 59 | − | 75553 | 74378 | 391 | 42.97 | 59 | 37.19 | 59 | DNA replication protein |
| 60 | − | 76623 | 75706 | 305 | 63.93 | 60 | 66.23 | 60 | Ribonucleotide reductase (RR), small subunit |
| 61 | − | 79038 | 76657 | 793 | 53.91 | 61 | 58.25 | 61 | Ribonucleotide reductase (RR), large subunit |
| 62 | − | 80039 | 79020 | 339 | 62 | 62 | Assembly or DNA maturation protein | ||
| 63 | + | 79996 | 82815 | 939 | 33.30 | 63 | 35.06 | 63 | Tegument protein |
| 64 | + | 82830 | 90539 | 2,569 | 33.02 | 64 | 35.44 | 64 | Tegument protein |
| 65 | − | 90947 | 90555 | 130 | 40.83 | 65 | 44.88 | 65 | Capsid protein |
| 66 | − | 92218 | 90944 | 424 | 36.98 | 66 | 39.08 | 66 | |
| 67 | − | 92928 | 92158 | 256 | 47.24 | 67 | 58.12 | 67 | Tegument protein |
| 67.5 | − | 93324 | 92932 | 130 | 39.74 | 67.5 | 55.29 | 67.5 | |
| 68 | + | 93206 | 94555 | 449 | 46.43 | 68 | 48.85 | 68 | Probable glycoprotein |
| 69 | + | 94563 | 95456 | 297 | 48.00 | 69 | 52.53 | 69 | |
| Bo11 | 57 | 1.1-kb late RNA | |||||||
| Bo11b | − | 96220 | 96209 | 3 | Long ORF of spliced 1.1-kb late RNA, exon II | ||||
| Bo11a | − | 97218 | 97057 | 54 | Long ORF of spliced 1.1-kb late RNA, exon I | ||||
| Bo12 | + | 97972 | 98175 | 67 | |||||
| Bo13 | + | 99393 | 99617 | 74 | |||||
| 71 | − | 99431 | 98883 | 182 | 27.41 | K13/71 | 23.42 | 71 | v-FLIP (BORFE2c) |
| 73 | − | 100337 | 99576 | 253 | 15.00 | 73 | 24.79 | 73 | BORFE3c |
| 75 | − | 104745 | 100828 | 1,305 | 25.29 | 75 | 38.85 | 75 | Tegument protein or v-FGAM-synthetase |
| Bo14 | − | 105340 | 104828 | 170 | Proline rich (BORFF1c) | ||||
| Bo15 | − | 106592 | 106278 | 104 | |||||
| Bo16 | + | 106779 | 107048 | 89 | BORFF2c | ||||
| Bo17 | + | 107313 | 108635 | 440 | v-β-1, 6GnT (BORFF3-4c) | ||||
Positions of the respective ORFs of BoHV-4 are given from the first nucleotide of the start codon ATG to the last nucleotide of the stop codon.
%ID, percent identity of the HHV-8 and HVS ORFs to the indicated BHV-4 ORF was calculated with the GAP program of the GCG package.
ORF designations as used by Lomonte et al. (27).
RESULTS
Cloning and sequencing: primary structure.
In previous studies it was clearly shown that the BoHV-4 genome consists of an LUR which is flanked by prDNA, also designated terminal tandem repeats or H-DNA (Fig. 1) (9, 17). Therefore, BoHV-4 has the same overall genome structure as gammaherpesviruses like HVS, HHV-8, MHV-68, AlHV-1, and others. The borders of the LUR DNA were recently determined in a project where a single prDNA element as well as the genomic termini and the junctions between the prDNA and LUR were characterized (6).
FIG. 1.
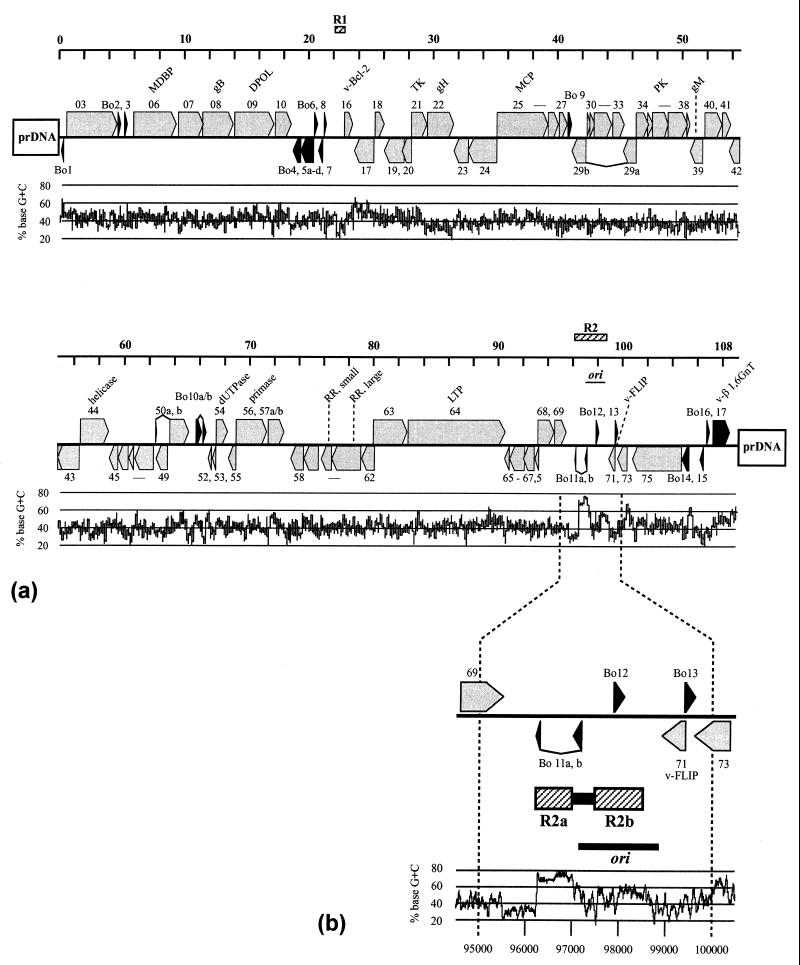
Map of the BoHV-4 genome. (a) The LUR is symbolized by a solid line. ORFs are given as arrows indicating the orientation. ORFs homologous to genes of other gammaherpesviruses are shown in grey, and ORFs unique to BoHV-4 are shown in black. ORFs described in the paper are labeled by protein name. Abbreviations are explained in Table 1. Above the genome, a kilobase scale is shown. The plot below the genome shows the G+C content of the corresponding genomic region. The plot was calculated with MacVector, using a window size of 50 bp. (b) The genome stretch containing the repeat region R2 and the ori is shown in magnification. The kilobase scale is shown below the G+C plot.
The DNA sequence of the LUR was determined by sequencing shotgun-cloned viral DNA fragments. The readings were assembled to a consensus sequence of 108,873 bp with an average G+C content of 41.4%. As described for other members of the Gammaherpesvirinae (1, 19, 41), the CpG dinucleotide frequency in the unique region of the BoHV-4 genome was found to be very low (1%).
Two regions of multiple direct repeats have been identified within the BoHV-4 LUR (Fig. 1). The first region (R1) consists of complex separate stretches of several complete and incomplete direct repeats which are 23, 25, and 65 bp in length, located at positions 21858 to 22631 in the LUR. The length and sequence as well as the number of repeats differ between BoHV-4 strains (4). The second repeat region (R2) consists of two different repeat stretches, R2a and R2b (Fig. 1b). The first repeat stretch, R2a, comprises LUR positions 96250 to 97034, consisting of 28 perfect and 3 imperfect direct repeats of 22 to 23 bp in length. Some of these direct repeats are separated by a GGAAG(2–3) motif, 15 in total. The second repeat stretch, R2b, comprises LUR positions 97500 to 98500. R2b contains several different direct repeats from 8 to 68 bp in length as well as one inverted repeat (LUR positions 97565 to 97616), leading to a predicted hairpin-loop structure. R2 is connected with a remarkable change in the G+C content (Fig. 1b): whereas R2a has a high G+C content of 71%, a 750-bp sequence stretch directly upstream of R2 has a low G+C content (30%).
Coding capacity and gene arrangement.
The following criteria were chosen for the ORF analysis: (i) the presence of a translation start and stop signal, (ii) a minimum length of more than 180 bp (60 amino acids [aa]), and (iii) no more than 50% overlap with another ORF. By applying these criteria and considering the published data on BoHV-4 genes, 79 ORFs have been predicted, 62 of them with homology to HVS and HHV-8 ORFs and 17 unique to BoHV-4 (Table 1). The BoHV-4 sequence and ORF map orientation has been adapted to other published gammaherpesvirus genomes (Fig. 1). In accordance with other rhadinovirus genomes, the nomenclature of the conserved BoHV-4 ORFs follows the homologous HVS genes. The remaining unique BoHV-4 ORFs were designated by the prefix “Bo” (for BoHV-4) and numbered in consecutive order beginning with the left end of the LUR.
The conserved genes of gammaherpesviruses are arranged in a common block organization, but there are individual differences in number, position, and orientation of subgroup-specific or individual ORFs (33, 35). Based on these data, the gene arrangement of BoHV-4 is most closely related to that of HVS. The central parts of the LUR of both viruses contain a stretch of 54 genes with the same orientation (ORFs 16 to 69). Within this stretch there are only two genes without genetic but with positional and orientational homology (Bo9 and Bo10) to HVS genes. In all other gammaherpesviruses sequenced to completion, differences in the presence and/or position of ORF 16 (v-Bcl-2) and the number of individual or subgroup-specific genes between ORFs 50 and 69 were observed. Furthermore, BoHV-4 and HVS have similar numbers of individual genes between the first and the second conserved gene blocks (four versus five), and both viruses have no long noncoding sequence stretches within the LUR.
Comparison of BoHV-4 sequences of different strains.
The previously published sequence data of BoHV-4 are derived from three different strains, 66-p-347, DN599, and V. test. As expected, no differences were seen between the strain 66-p-347 sequence data presented here and the sequences of the gB gene (22), the ORF 29 locus (7), and the prDNA-LUR borders (6) published earlier for the same strain. Very few differences (<1‰) were found between the strain 66-p-347 and the strain DN599 sequences published for immediate-early transcript 1 (IE1) (Bo4-Bo5), IE2 (ORF 50), and the 1.1-kb late RNA transcripts (Bo11 loci) (3, 44, 45). Considerable differences were found in comparison to the V. test sequences of the LUR regions outside the conserved gene blocks (27). The DNA identity values range from 88% (V. test BORFB2 region) to 99% (BORFD region). The sequences with the lowest identities, V. test BORFA2 and BORFB2, not only show numerous bp exchanges, but also deletions of up to 125 bp when aligned to strain 66-p-347.
Notes on individual and subgroup-specific ORFs.
BoHV-4 lacks homologues to HVS ORFs 1, 2, 4, 5, 11 to 15, 28, 51, 70, 72, and 74. With the exception of ORF 11, which was found in all other genomes of the genus Rhadinovirus, all members of this genus lack some or all of these ORFs. Among these ORFs are several homologues of cellular genes. No cytokine- or cytokine receptor-coding genes, G protein-coupled (interleukin) receptor genes, or viral macrophage inflammatory protein α/β gene is present within the LUR of BoHV-4. In this respect, BoHV-4 is most comparable to AlHV-1, another gammaherpesvirus of ruminants.
BoHV-4 contains 17 ORFs (Bo1 to Bo17), which have no obvious counterparts in HVS and, with the exception of Bo5, in other herpesviruses. Among these ORFs are seven which were not described in previous studies (Bo1, -3, -6, -7, -12, -13, and -15). ORF Bo4 and the spliced ORF Bo5 have been described by van Santen to be part of the major immediate-early transcript IE1 (45). van Santen speculated that Bo4 was either a separate ORF or an exon of a late transcript of the IE1 region. Interestingly, Bo4 and Bo5 exhibit partial amino acid homology and may be involved in differential splicing.
ORF Bo8 partially overlaps with the 1.7-kb late RNA which was described by Bermudez-Cruz et al. (4). Bo5 exhibits a 27.85% homology at the amino acid level with the HHV-8 ORF K5 (24). Preliminary data for Bo10 showed that this spliced gene encodes a glycoprotein with a molecular mass of 80 kDa (GenBank accession no. Z84818 and P. Lomonte, personal communication). Bo11 is encoded by a previously described IE2-transactivated, spliced, 1.1-kb late RNA (3), forcing us to make an exception to the ORF product length rule (>60 aa).
Bo17 encodes a viral β-1,6-N-acetylglucosaminyltransferase (β-1,6GnT) that is 81.1% identical on the amino acid level with the human β-1,6GnT-M. The BoHV-4 β-1,6GnT is transcribed during viral replication and is functionally active (43). No other virus is known to have a β-1,6GnT gene.
Notes on further genes.
Until now, RNAs of six genes of BoHV-4 were demonstrated experimentally to be spliced. Besides Bo5, Bo10, and Bo11, these are ORFs 29, 50, and 57 (7, 44). ORF 29 encodes terminase, a protein essential for the viral cleavage and packaging process of herpesviruses after lytic replication of the genomes (2). This gene is present and spliced in all herpesviruses that have so far been analyzed in detail. The ORF 50 product was shown earlier to be a putative R transactivator (EBV BRLF1) homologue, encoded by the spliced IE2 (44). Transactivation activity has been demonstrated for the corresponding gene product (44). For HHV-8 it was demonstrated that the ORF 50 product is involved in reactivation of the virus from latency (29). In our BoHV-4 strain 66-p-347, the intron of the IE2 transcript contains the complete ORF 49 encoding a product with a length of 299 aa, which corresponds to other gammaherpesviruses (i.e., the HVS ORF 49 product has 303 aa). In comparison to strain 66-p-347, the DN 599 isolate published earlier has a single base pair deletion in the ORF 49 codon corresponding to aa 41 which leads to a frameshift and a 52-aa short predicted protein. Due to homology studies, van Santen showed the carboxy-terminal 258 aa of the ORF 49 product without an initial methionine (44). ORF 57 is a spliced gene coding for a posttranscriptional regulator (V. L. van Santen, personal communication, and GenBank accession no. U30519). For HVS it has been shown that the ORF 57 product is responsible for redistribution of the spliceosome complex, thereby modulating mRNA processing in the infected cell (13). As described previously by Lomonte et al. (27), two copies of the putative viral phosphoribosylformylglycinamidine synthase (v-FGAM) are present in reverse orientation near the left and right ends of the BoHV-4 LUR (ORFs 3 and 75). Both conserved positions are also present in HVS and AlHV-1, but only one copy is found in HHV-8 and EBV.
Multiple sequence and phylogenetic analysis.
A phylogenetic analysis of the relationship of BoHV-4 to other herpesviruses using ORF 29, encoding the putative terminase, has previously been reported. In this analysis, BoHV-4 clustered with the gammaherpesviruses, in particular HVS, although with a low bootstrap value (7). To extend this analysis and to determine more precisely the relationship of BoHV-4 to the other gammaherpesvirus species, we analyzed six different ORFs (ORFs 6, 8, 9, 44, 46, and 61) of BoHV-4 which have well-conserved counterparts among all completely sequenced herpesviruses and were already used previously in phylogenetic analysis of Alpha-, Beta-, and Gammaherpesvirinae (except ORF 61) (30). Amino acid sequences of BoHV-4 and eight other completely sequenced gammaherpesvirus species (HVS, HVA, HHV-8, RRV, EHV-2, AlHV-1, MHV-68, and EBV) were aligned by ClustalW and used for phylogenetic tree construction by neighbor joining or the parsimony analysis. Among the nine viruses examined, HVS and HVA, HHV-8 and RRV, and EBV and AlHV-1 always segregated as pairs. However, BoHV-4 (as well as MHV-68 and EHV-2) had different branching patterns with mostly insufficient bootstrap values when individual proteins were examined. This was because the branch point of BoHV-4 was always in the center of the tree, close to the branching point which segregates the HVS/HVA pair from the HHV-8/RRV pair. Although in the majority of the analyses a branching of BoHV-4 with the HVS/HVA pair was seen, the overall picture was inconsistent with both analysis methods (data not shown). Therefore, a phylogenetic locus of BoHV-4 could not be firmly determined.
Mapping of an ori.
In order to map a BoHV-4 ori, a 60-kb genome stretch representing 45% of the LUR and five prDNA elements was first analyzed. The central part of this genome stretch was chosen for analysis by positional homology to lytic origins of other gammaherpesviruses and flanked with additional stretches of the genome which served as negative controls in the replication assays. Restriction fragments covering this 60-kb region were cloned in bacterial plasmid vectors and subsequently tested in a DpnI assay as described in Materials and Methods. Plasmid DNA derived from dam+ Escherichia coli strains is cleaved by DpnI and is resistant to MboI, while plasmids newly replicated in eucaryotic cells are resistant to DpnI and are cleaved by MboI. The positions of the tested restriction fragments are shown in Fig. 2. In the first set of experiments, plasmids which were part of a BamHI library, representing the complete genome of BoHV-4, were used. Plasmid constructs p560, p563, p564, p565, and p594, spanning a region from LUR position 59731 to the right end of the LUR as well as five prDNA elements, have been investigated. Among these plasmids, only construct p560 reacted positively in the replication assay. The insert of this plasmid contains the last three BamHI fragments of the LUR and five prDNA elements. To define the position of cis elements for ori function more precisely, a set of subclones and deletion constructs were made and tested in the DpnI assay. The 5,884-bp insert of plasmid p850 (LUR positions 97143 to 103027) was sufficient for replication. Subsequently, three additional deletion constructs were tested (p867 to p869 [Fig. 2]). Deletion of the 5′ end of the insert of p850 led to plasmid p869, which failed to replicate. In contrast, all plasmids deleted at the 3′ end of the genome region represented in p850 replicated in the DpnI assay. The smallest genomic region, in plasmids p866 and p867, had decreased replication efficiency compared to constructs p850 and p868. For further fine mapping of the ori, five pBluescript-based subclones were tested (p892, p893, p909, p843, and p944). Plasmid p892 contained the same insert as p866; the other plasmids contained this insert with 3′ and 5′ deletions. A 647-bp 5′ deletion of the insert of p892 and p866 resulted in construct p909, which was replication negative. Among the constructs with 3′-deleted inserts, only p944 was clearly able to replicate while the plasmids with shorter inserts failed. In summary, the smallest insert in this study which led to a replication-competent plasmid represents a 1,687-bp BamHI-PstI fragment spanning LUR positions 97143 to 98850. This region contains ORF Bo12 and is partially overlapping with the repeat region R2: the mapped ori is positioned downstream (3′) of the G+C-rich R2a stretch and comprises the second repeat stretch (R2b) with the predicted hairpin-loop structure (Fig. 1b).
DISCUSSION
In this study we present the sequence of the entire LUR of BoHV-4 and localized an origin of replication.
The genome structure, the gene arrangement, and the biological properties confirm that BoHV-4 belongs to the genus Rhadinovirus, and HVS appears to be its closest relative, as proposed in earlier studies (9, 27). AlHV-1, another ruminant gammaherpesvirus which has structural and biological rhadinovirus features (19), is clearly more distantly related. In phylogenetic analyses of individual conserved BoHV-4 proteins, however, the relationship to HVS was not unequivocally seen. This is in accord with recently published phylogenetic analyses of herpesviruses which also do not show a firm locus for BoHV-4 (31).
The comparison of the strain 66-p-347 LUR sequence to available sequence data of BoHV-4 revealed no differences within this strain and only minor ones compared to strain DN599. The most significant difference in the latter strain is a single base pair deletion leading to an incomplete ORF 49. Whether this deletion in DN599 is a sequencing artifact or a mutation has to be proven. More divergence was found between our strain and the V. test strain sequences published by Lomonte et al. (27) with the analysis of the LUR regions outside the conserved gene blocks. The identity values ranging from 88 to 99% and the existence of DNA insertions or deletions in homologous sequences of both strains give evidence for significant strain differences. While some of the ORFs described by this group are found in our sequence with identical lengths, others are different in length due to single base pair exchanges, deletions, and ambiguous base calls in the submitted sequence files. For final judgment of the strain differences and their consequences on the ORFs, sequence data of higher quality are necessary for the V. test strain. As an example, the initial description of the β-1,6GnT (Bo17) gene locus contained a 10-bp deletion within the reading frame, resulting in two ORFs, designated BORFF3 and BORFF4 (27). The corrected version of this V. test strain sequence was published recently in conjunction with the β-1,6GnT gene description (43). This sequence now contains the entire, uninterrupted ORF and is identical to our 66-p-347 strain sequence.
Several gammaherpesviruses are associated with lymphoproliferative diseases and tumor development. These diseases seem to be associated with immunosuppression or a late-in-life infection of the natural host as well as with infection of a related nonnatural host. For BoHV-4, no link to such diseases has been identified with evidence (32). They may either have been overlooked or be absent due to the lack of transforming genes and cellular homologues in the BoHV-4 genome. Therefore, one aim of this study was to elucidate the coding capacity of this virus to gain further knowledge about the presence of such genes. Gammaherpesvirus genes homologous to cellular genes have often been demonstrated to be involved in cell growth or cell survival, in nucleotide metabolism, and in immune escape. The set of such genes in BoHV-4 is reduced compared to other gammaherpesviruses with known transforming capacity. No cytokine- or cytokine receptor-coding genes like those for the viral interleukins of HVS, HHV-8, and EHV-2, G protein-coupled or interleukin receptors of HVS, HHV-8, MHV-68, and EHV-2, or viral macrophage inflammatory protein α/β of HHV-8 are present within the LUR of BoHV-4.
Additional groups of common cellular homologues in rhadinoviruses are genes involved in cell growth or cell survival and in nucleotide metabolism. BoHV-4 has been shown earlier to include two potential cell cycle regulators, v-bcl-2 (viral B-cell lymphoma gene [ORF 16]) and the death effector domain-containing v-FLIP gene (viral FLICE inhibitory protein gene [ORF 71]) (46). In contrast to HVS and HHV-8, no cyclin D or complement control protein gene homologue is present in BoHV-4. Furthermore, besides the common thymidine kinase, two copies of v-FGAM-synthetase are encoded in BoHV-4 (ORF 3 and ORF 75), but neither viral dehydrofolate reductase as is present in HHV-8, RRV, and HVS nor viral thymidilate synthetase as is present in HHV-8, RRV, HVS, and HVA was found (for an overview, see references 33 and 35). Gene products which can increase the amount of available nucleotides in the infected cell influence not only viral replication but also cell proliferation (33). The recently published β-1,6GnT gene (Bo17) of BoHVH-4 represents a new category of cellular homologue in viruses (43). No other virus is known to have this gene. It was suggested that the Bo17 gene product may be necessary for replication of BoHV-4 in mononuclear blood cells or may be involved in viral immune escape. In summary, it cannot be excluded that BoHV-4 has a lymphoproliferative or transforming capacity under certain conditions, but there is no evidence that this virus has the genes required to cause such diseases.
We mapped an origin of replication in BoHV-4 with positional homology to oriLyt of EBV (23). In both viruses this origin is located downstream of ORF 69 and in the immediate vicinity of a G+C-rich stretch. The BoHV-4 ori core region overlaps by approximately 75 bp with ORF Bo11, the 1.1-kb late RNA (3), and contains ORF Bo12. Whether the transcripts or products of one or both of these ORFs are involved in DNA replication has to be shown in future studies.
Several features make BoHV-4 attractive as a backbone for use as a viral vector: (i) its genome structure is less complex compared to those of several other herpesviruses; (ii) it allows the stable insertion of additional genetic information in the viral genome up to at least 6 kb (M. Goltz et al., unpublished data); (iii) in contrast to several other rhadinoviruses, it is easily productively propagated in cell culture; (iv) to the present knowledge it is safe for the human experimenter; (v) severe, fatal diseases in the natural host seem to be nonexistent or rare events; and (vi) a small animal model (rabbits) is available. BoHV-4 recombinants may be useful as viral vaccines for cattle. Another, more likely application may be as a model system for the use in cell culture and animals to investigate viral and cellular genes. For example, the role of gammaherpesvirus-mediated oncogenesis could be examined by insertion of genes in the BoHV-4 genome. With the knowledge of the complete sequence and the position of the cis element necessary for viral replication (ori), more prerequisites are given for construction and usage of BoHV-4 recombinants.
ACKNOWLEDGMENTS
We thank Jaqueline Weber and Uwe Menzel for help with the different computer programs. We are grateful to Geoff Letchworth for critical reading and to Ursula Erikli for copyediting of the manuscript.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Poon A P, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermudez-Cruz R, Zhang L, van Santen V L. Characterization of a bovine herpesvirus 4 (BHV-4) 1.1-kb RNA and its transactivation by BHV-4 immediate-early 2 gene product. Arch Virol. 1998;143:2391–2412. doi: 10.1007/s007050050469. [DOI] [PubMed] [Google Scholar]
- 4.Bermudez-Cruz R, Zhang L, van Santen V L. Characterization of an abundant, unique 1.7-kilobase bovine herpesvirus 4 (BHV-4) late RNA and mapping of a BHV-4 IE2 transactivator-binding site in its promoter-regulatory region. J Virol. 1997;71:527–538. doi: 10.1128/jvi.71.1.527-538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borodovsky M, McIninch J. GenMark: parallel gene recognition for both DNA strands. Computers Chem. 1993;17:123–133. [Google Scholar]
- 6.Broll H, Buhk H-J, Zimmermann W, Goltz M. Structure and function of the prDNA and the genomic termini of the γ2-herpesvirus bovine herpesvirus type 4. J Gen Virol. 1999;80:979–986. doi: 10.1099/0022-1317-80-4-979. [DOI] [PubMed] [Google Scholar]
- 7.Broll H, Finsterbusch T, Buhk H-J, Goltz M. Genetic analysis of the bovine herpesvirus type 4 gene locus for the putative terminase. Virus Genes. 1999;19:243–250. doi: 10.1023/a:1008145015954. [DOI] [PubMed] [Google Scholar]
- 8.Bublot M, Lomonte P, Lequarre A-S, Albrecht J-C, Nicholas J, Fleckenstein B, Pastoret P-P, Thiry E. Genetic relationship between bovine herpesvirus 4 and the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. Virology. 1992;190:654–665. doi: 10.1016/0042-6822(92)90903-3. [DOI] [PubMed] [Google Scholar]
- 9.Bublot M, Van Bressem M F, Thiry E, Dubuisson J, Pastoret P P. Bovine herpesvirus 4 genome: cloning, mapping and strain variation analysis. J Gen Virol. 1990;71:133–142. doi: 10.1099/0022-1317-71-1-133. [DOI] [PubMed] [Google Scholar]
- 10.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 11.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper M, Goodwin D J, Hall K T, Stevenson A J, Meredith D M, Mackham A F, Whitehouse A. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J Gen Virol. 1999;80:1311–1316. doi: 10.1099/0022-1317-80-5-1311. [DOI] [PubMed] [Google Scholar]
- 14.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egyed L. Replication of bovine herpesvirus type 4 in human cells in vitro. J Clin Microbiol. 1998;36:2109–2111. doi: 10.1128/jcm.36.7.2109-2111.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehlers B, Buhk H-J, Ludwig H. Analysis of bovine cytomegalovirus genome structure: cloning and mapping of the monomeric polyrepetitive DNA unit, and comparison of European and American strains. J Gen Virol. 1985;66:55–68. doi: 10.1099/0022-1317-66-1-55. [DOI] [PubMed] [Google Scholar]
- 18.Ehlers B, Goltz M, Ejercito M P, Dasika G K, Letchworth G J. Bovine herpesvirus type 2 is closely related to the primate alphaherpesviruses. Virus Genes. 1999;19:197–203. doi: 10.1023/a:1008184630066. [DOI] [PubMed] [Google Scholar]
- 19.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelaphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein J. Confidence limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP (phylogeny inference package) version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 22.Goltz M, Broll H, Mankertz A, Weigelt W, Ludwig H, Buhk H-J, Borchers K. Glycoprotein B of bovine herpesvirus type 4: its phylogenetic relationship to gB equivalents of the herpesviruses. Virus Genes. 1994;9:53–60. doi: 10.1007/BF01703435. [DOI] [PubMed] [Google Scholar]
- 23.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 24.Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74:2867–2875. doi: 10.1128/jvi.74.6.2867-2875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 26.Kit S, Kit M, Ichimura H, Crandell R, McConnell S. Induction of thymidine kinase activity by viruses with group B DNA genomes: bovine cytomegalovirus (bovine herpesvirus 4) Virus Res. 1986;4:197–212. doi: 10.1016/0168-1702(86)90041-9. [DOI] [PubMed] [Google Scholar]
- 27.Lomonte P, Bublot M, van Santen V, Keil G M, Pastoret P-P, Thiry E. Analysis of bovine herpesvirus 4 genomic regions located outside the conserved gammaherpesvirus gene blocks. J Gen Virol. 1995;76:1835–1841. doi: 10.1099/0022-1317-76-7-1835. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig H. Bovine herpesviruses. In: Roizman B, editor. The herpesviruses. Vol. 2. New York, N.Y: Plenum Press; 1983. pp. 135–214. [Google Scholar]
- 29.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGeoch D J, Cook S, Dolan A, Jamieson F E, Telford E A R. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 31.McGeoch D J, Dolan A, Ralph A C. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol. 2000;74:10401–10406. doi: 10.1128/jvi.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno-Lopez J, Goltz M, Rehbinder C, Valsala K V, Ludwig H. A bovine herpesvirus (BHV-4) as passenger virus in ethmoidal tumours in Indian cattle. J Vet Med B. 1989;36:481–486. doi: 10.1111/j.1439-0450.1989.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 33.Neipel F, Albrecht J-C, Fleckenstein B. Human herpesvirus 8—the first human Rhadinovirus. J Natl Cancer Inst Monogr. 1998;23:73–77. doi: 10.1093/oxfordjournals.jncimonographs.a024178. [DOI] [PubMed] [Google Scholar]
- 34.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2221–2230. [Google Scholar]
- 35.Simas J P, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 36.Storz J, Ehlers B, Todd W J, Ludwig H. Bovine cytomegaloviruses: identification and differential properties. J Gen Virol. 1984;65:697–706. doi: 10.1099/0022-1317-65-4-697. [DOI] [PubMed] [Google Scholar]
- 37.Thomas A, Skolnick M H. A probabilistic model for detecting coding regions in DNA sequences. IMA J Math Appl Med Biol. 1994;11:149–160. doi: 10.1093/imammb/11.3.149. [DOI] [PubMed] [Google Scholar]
- 38.Todd W J, Storz J. Morphogenesis of a cytomegalovirus from an American bison affected with malignant catarrhal fever. J Gen Virol. 1982;64:1025–1030. doi: 10.1099/0022-1317-64-5-1025. [DOI] [PubMed] [Google Scholar]
- 39.Truman D, Ludwig H, Storz J. Bovines herpesvirus type 4 (BHV-4): Untersuchungen zur Biologie und Verbreitung in Rinderbeständen und bei Besamungsbullen. J Vet Med B. 1986;33:485–501. [PubMed] [Google Scholar]
- 40.Uberbacher E C, Mural R J. GRAIL2. Proc Natl Acad Sci USA. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulrich S, Goltz M, Ehlers B. Characterization of the DNA polymerase loci of the novel porcine lymphotropic herpesviruses 1 and 2 in domestic and feral pigs. J Gen Virol. 1999;80:3199–3205. doi: 10.1099/0022-1317-80-12-3199. [DOI] [PubMed] [Google Scholar]
- 42.Vanderplasschen A, Goltz M, Lyaku J, Benarafa C, Buhk H-J, Thiry E, Pastoret P-P. The replication in vitro of the gammaherpesvirus bovine herpesvirus 4 is restricted by its DNA synthesis dependence on the S phase of the cell cycle. Virology. 1995;213:328–340. doi: 10.1006/viro.1995.0006. [DOI] [PubMed] [Google Scholar]
- 43.Vanderplasschen A, Markine-Goriaynoff N, Lomonte P, Suzuki M, Hiraoka N, Yeh J-C, Bureau F, Willems L, Thiry E, Fukuda M, Pastoret P-P. A multipotential beta-1,6-N-acetylglucosaminyltransferase is encoded by bovine herpesvirus type 4. Proc Natl Acad Sci USA. 2000;97:5756–5761. doi: 10.1073/pnas.100058897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Santen V L. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J Virol. 1993;67:773–784. doi: 10.1128/jvi.67.2.773-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Santen V L. Characterization of the bovine herpesvirus 4 major immediate-early transcript. J Virol. 1991;65:5211–5224. doi: 10.1128/jvi.65.10.5211-5224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G-H, Bertin J, Wang Y, Martin D A, Wang J, Tomaselli K J, Armstrong R C, Cohen J I. Bovine herpesvirus 4 BORFE2 protein inhibits Fas- and tumor necrosis factor receptor 1-induced apoptosis and contains death effector domains shared with other gamma-2 herpesviruses. J Virol. 1997;71:8928–8932. doi: 10.1128/jvi.71.11.8928-8932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]