Abstract
Over the past 30 years, the survival rate for osteosarcoma (OS) has remained stagnant, indicating persistent challenges in diagnosis and treatment. Photodynamic therapy (PDT) has emerged as a novel and promising treatment modality for OS. Despite apoptosis being the primary mechanism attributed to PDT, it fails to overcome issues such as low efficacy and resistance. Ferroptosis, a Fe2+-dependent cell death process, has the potential to enhance PDT's efficacy by increasing reactive oxygen species (ROS) through the Fenton reaction. In this study, we investigated the anti-tumor mechanism of PDT and introduced an innovative therapeutic strategy that synergistically induces apoptosis and ferroptosis. Furthermore, we have identified HERC1 as a pivotal protein involved in the ubiquitination and degradation of NCOA4, while also uncovering a potential regulatory factor involving NRF2. Ultimately, by targeting the HERC1-NCOA4 axis during PDT, we successfully achieved full activation of ferroptosis, which significantly enhanced the anti-tumor efficacy of PDT. In conclusion, these findings provide new theoretical evidence for further characterizing mechanism of PDT and offer new molecular targets for the treatment of OS.
Keywords: Osteosarcoma, MPPα-PDT, Ferroptosis, Protein interaction, Ubiquitination
1. Introduction
Osteosarcoma (OS) is a highly malignant bone tumor that predominantly affects males, with a bimodal incidence peaking at ages 18 and 60 [1]. The occurrence of OS is commonly observed at the epiphysis of long bones and in locations characterized by rapid bone growth, including the distal femur (43 %), proximal tibia (23 %), proximal humerus (10 %), as well as sites such as the skull and mandible [2,3]. Currently, the standard treatment for OS involves a combination of surgical intervention and neoadjuvant chemotherapy. While this therapeutic approach markedly improves the prognosis for patients with localized disease, with Event-Free Survival (EFS) exceeding 60 %, approximately 20 % of individuals with secondary metastasis still face a poor prognosis [4]. Therefore, it is essential to continually explore novel therapeutic modalities and identify key targets to enhance the effectiveness of OS treatment.
Photodynamic therapy (PDT), as a new treatment method with non-invasive, low toxicity, and good selectivity, has been confirmed in our previous studies to be an effective way of treating OS [[5], [6], [7], [8]]. By absorbing the radiation energy of light, PDT causes excessive accumulation of reactive oxygen species (ROS) in cells, thereby disrupting the intracellular homeostasis and ultimately leading to cell death [9]. As the key mechanism of PDT, apoptosis involves direct damage to organelles, plasma membranes, proteins, and other components of tumor cells by ROS. The damaged mitochondria trigger a series of signal cascades. Specifically, the reduction in mitochondrial membrane potential (ΔΨm) promotes the release of cytochrome c into the cytoplasm, which activates the expression of caspase 9 and caspase 3, ultimately leading to apoptosis through the mitochondrial pathway [10,11]. The imbalance in the redox system and the activation of programmed cell death can trigger various self-protective mechanisms in tumor cells. In response to damage induced by PDT, these mechanisms can lead to the gradual development of resistance, significantly impeding the efficacy of PDT and potentially resulting in evasion, recurrence, and metastasis [7,12].
Ferroptosis is a form of regulated cell death (RCD) that occurs primarily as a result of damage caused by iron-catalyzed lipid peroxidation [13,14]. The potential synergistic effect between ferroptosis and PDT can be observed: PDT induces a rapid intracellular increase in H2O2 levels during oxidation, which can then serve as a substrate for the “Fenton reaction” that is involved in ferroptosis. Conversely, when ferroptosis occurs, H2O2 can be utilized by Fe2+ to generate highly oxidizing hydroxyl radicals, thereby indirectly enhancing the therapeutic efficacy of PDT [15]. Ferritinophagy is a selective form of autophagy that targets intracellular ferritin for degradation, promoting the breakdown and recycling of iron stores within cells. This process helps maintain an overall balance in the internal environment and reduces oxidative damage caused by excessive accumulation of iron [16]. Nuclear Receptor Coactivator 4 (NCOA4) acts as a cargo receptor in the autophagic degradation of ferritin, facilitating its degradation by transporting it to the autophagolysosome and maintaining cellular iron homeostasis [17]. Moreover, the dysregulation of ferritinophagy has been implicated in the pathogenesis and progression of several diseases, including tumors, neurodegenerative disorders, and hemochromatosis. The regulation of NCOA4 mediated ferritinophagy is intricately controlled by intracellular iron ion levels [18], and is modulated by the supply-demand dynamics of iron ions. Furthermore, research has shown that NCOA4 plays a crucial role in inducing ferroptosis and is also strongly associated with lysosomal function, natural killer cell-mediated cytotoxicity, chemokines, and toll-like receptor genes in various tumors. Moreover, solid tumor patients with elevated NCOA4 expression tend to have prolonged survival [19].
The process of protein ubiquitination is tightly regulated by three key enzymes: namely the E1 ubiquitin activase, E2 ubiquitin conjugase, E3 ubiquitin ligase. HERC1, a member of the larger HERC family, is considered a potential E3 ubiquitin ligase due to its HECT domain. Initially identified in breast cancer, it has been demonstrated that HERC1 has regulatory effects on ARF6, TSC2, and BAK [20]. Inhibition of HERC1 has been reported to suppress C-Raf ubiquitination while enhancing ERK phosphorylation, playing a pivotal role in tumor development [21]. Meanwhile, HERC1 overexpression has been demonstrated to contribute to the development of breast cancer, as well as increase the incidence of intellectual disability, kyphosis, and epilepsy [22].
In this study, we have discovered that HERC1 plays a crucial role in the degradation process of the NCOA4 protein during MPPα-PDT, thereby exerting an inhibitory effect on ferritinophagy. Targeting HERC1 significantly impedes the degradation of the NCOA4 protein and enhances the anti-tumor efficacy of MPPα-PDT. Moreover, we also demonstrated the mechanism by which HERC1 is activated through the NRF2 signaling cascade, particularly under conditions of oxidative stress induced by MPPα-PDT. Finally, through a series of investigations, we have comprehensively elucidated both the signal cascade and the enhanced potential for inducing ferroptosis in OS cells using MPPα-PDT by targeting the NCOA4-HERC1 axis.
2. Materials and methods
2.1. Materials
Pyropheophorbide-a methyl ester (MPPα) was purchased from Sigma-Aldrich Chemical Co. (USA). The CCK-8 assay kit, MG132, and cycloheximide (CHX) were obtained from MCE Co. (USA). The mitochondrial fluorescent probe, Golgi fluorescent probe, endoplasmic reticulum fluorescent probe, and DCFH-DA assay kit were purchased from Beyotime Bio Co. (China). The lysosomal fluorescent probe and the SOSG assay kit were purchased from MeilunBio Co. (China). The JC-1 assay kit was purchased from Solarbio Co. (China). The MDA assay kit, FerroOrange fluorescent probe, Lipid Peroxide assay kit, and C11 Bodipy 581/591 assay reagent were purchased from Dojindo Co. (Japan). DFO and Ferrostatin-1 were purchased from MCE Co. (USA). The primary antibody (Ab) for NCOA4 was purchased from CST Co. (USA), and the HERC1 primary Ab was purchased from Santa Cruz Co. (USA). The primary Abs for PARP, BCL-2, Caspase 9, Caspase 7, Caspase 3, SLC7A11, LC3 II, Atg5, Atg7, and Ubiquitin were all bought from ABMART Co. (China). The primary Abs for BAX, Cytochrome c, GAPDH, GPX4, TFR1, COX2, FTH1, FTL, and NRF2 were all bought from ZenBio Co. (China). Protein A/G beads were bought from Selleck Biotech Co. (USA). The osteosarcoma HOS and 143B cell lines were purchased from Wuhan PriCell Biotechnology Co. (China).
2.2. Cell culture
Osteosarcoma HOS and U2OS cells were cultured in DMEM medium supplemented with 10 % FBS (fetal bovine serum). Osteosarcoma 143B and MG63 cells were cultured in MEM supplemented with 10 % FBS. hFOB1.19 cells were cultured in DMEM/F12 medium supplemented with 10 % FBS. The culture conditions for all the aforementioned cell lines included an incubator set to 37 °C, with moderate humidity and a CO2 concentration of 5 %.
2.3. CCK-8 assay and treatment
Osteosarcoma cells exhibiting good growth were trypsinized and inoculated into 96-well plates at a density of 5,000 cells per well. They were then incubated overnight to ensure adhesion. Following various treatments, 10 μL of the CCK-8 working solution was added to each well and incubated at 37 °C away from light for 1 h. Subsequently, the plate was read using a microplate reader, and the absorbance (OD value) at 450 nm was measured. The formula for calculating cell viability is as follows: Cell viability (%) = (OD value of experimental group - OD value of blank control group)/(OD value of control group - OD value of blank control group) × 100 %. HOS and 143B cells were treated with MPPα for 20 h at a range of concentrations while protected from light, after which they were washed twice with PBS. Culture media was then added to appropriate wells, and cells were exposed to red light (630 nm, 40 mW/cm2) for 120 s in continuous output mode. For further details regarding this application of MPPα-PDT, refer to our previous study [[6], [7], [8]].
2.4. Organelle probe detection
After various treatments, osteosarcoma cells were incubated with diluted organelle-specific fluorescent probes in complete medium (Mitochondria-Tracker Green: 1:1000; Lysosome-Tracker Green: 1:20,000; Endoplasmic Reticulum-Tracker Green: 1:1000; Golgi-Tracker Green: 1:1000). The cells were then incubated for 30 min in a CO2 incubator protected from light, followed by an additional 30-min incubation in the dark. After washing three times with PBS (Phosphate-Buffered Saline), the cells were replenished with fresh complete medium and transferred to a confocal petri dish for observation under a confocal laser scanning microscope (CLSM).
2.5. Western blot analysis
After different treatments, osteosarcoma cells were lysed using RIPA buffer (1 % PMSF and 1 % phosphatase inhibitors) and then incubated on ice for 30 min. Following centrifugation, the supernatant was collected. The protein concentration was determined using the BCA assay. For protein separation, 30 μg of each sample was loaded onto an SDS-PAGE gel and run at a constant voltage of 80–100 V. The proteins were transferred to PVDF films at a constant current of 250 mA, with the duration modified according to the molecular weight of the proteins of interest. The films were blocked with a 5 % skim milk solution in TBST for 2 h at room temperature. Primary antibodies were applied overnight at 4 °C with shaking, followed by a thorough TBST wash. Then, secondary antibodies were added at a 1:5000 dilution and incubated for 2 h at room temperature with shaking. Protein detection was achieved using chemiluminescence, and the resulting images were captured. The Fusion software was utilized to analyze the grayscale values of the bands, allowing for the determination of the relative protein expression levels, with GAPDH serving as an internal control.
2.6. Measurement of ROS
Osteosarcoma cells in the logarithmic growth phase were seeded into culture dishes at a density of 2 × 104 cells per well and incubated overnight for adherence. After various treatments, a fluorescent probe of DCFH-DA diluted in serum-free medium (1:1000) was added. Following a 30-min incubation protected from light, the medium was aspirated and the cells were thoroughly washed with serum-free medium. Fluorescence microscopy or flow cytometry was then used for detection [23].
2.7. SOSG detection
Osteosarcoma cells in the logarithmic phase of growth were pre-inoculated into confocal dishes at a density of 5 × 104 cells per well and incubated overnight for adhesion. After various treatments, a solution of SOSG (10 μM) was added to each confocal dish and incubated away from light for 30 min. After thorough washing, the samples were observed under CLSM.
2.8. MMP assay
The mitochondrial membrane potential (Δψm) was measured using a JC-1 assay kit. Osteosarcoma cells were inoculated into 6-well plates at a density of 1 × 105 cells per well and incubated overnight to allow for adhesion. Following different treatments, JC-1 staining solution was added to each well and protected from light for 20 min. After thorough washing, the cells were observed using fluorescence microscopy or analyzed by flow cytometry.
2.9. MDA assay
Osteosarcoma cells were seeded into culture dishes at a density of 1 × 106 cells per well and incubated overnight to allow for adhesion. Following various treatments, the cells were centrifuged at 300 g for 5 min. Then, 100 μL of Lysis Buffer was added, and the mixture was thoroughly mixed using a vortex shaker. The mixture was left to stand at room temperature for 5 min. Next, the working solution was added and mixed thoroughly again using the vortex shaker. The sample was incubated in a water bath at 95 °C for 15 min, cooled in an ice bath for 5 min, and then centrifuged at 10,000 rpm for 10 min. Subsequently, 100 μL of the supernatant was transferred to a 96-well plate. The absorbance at 590 nm was measured using a microplate reader, and the concentration of MDA in the samples was calculated from the standard curve of MDA.
2.10. LPO, Lipid-ROS, and Fe2+ measurements
Osteosarcoma cells were seeded into 6-well plates at a density of 1 × 105 cells per well and incubated overnight to promote adhesion. After various treatments, an appropriate volume of Lipid Peroxide (Liperfluo) working solution (1:1000) or C11-Bodipy working solution (1:500) or FerroOrange working solution (1:1000) was added and incubated for 30 min at 37 °C in the dark. After the incubation period, the cells were observed under a fluorescence microscope or analyzed using flow cytometry.
2.11. Lentiviral transduction
Osteosarcoma cells in the logarithmic growth phase are plated at 5 × 104 cells/well and transfected with lentivirus once they reach 30 % confluence, using MOI values of 20 for HOS cells and 40 for 143B cells, along with viral stock and Polybrene. After transfection, cells are incubated for 16 h before the medium is replaced with fresh complete medium. Puromycin selection at 2 μg/mL is initiated to isolate stably transfected cell lines, with PBS washes to remove dead cells every 48 h. This process continues with weekly medium changes for one week.
2.12. ADV-eGFP-LC3 probe transduction
Osteosarcoma cells are seeded onto confocal culture dishes at a density of 5 × 104 cells/dish and incubated overnight to allow for adherence. Once an appropriate growth density is reached, the adenovirus-packaged eGFP-LC3 fluorescent probe is transfected into the cells using the recommended transfection index for osteosarcoma cells (MOI for HOS and 143B cells = 10). The transfected cells are then cultured in complete medium and returned to the incubator for over 24 h before the expression of autophagosomes is observed using CLSM.
2.13. iRNA transduction
Osteosarcoma cells in the logarithmic growth phase are selected, and transfection can begin once the cell density is approximately 50 %. Following the protocol, siRNA and Lipofectamine-2000 transfection reagent are separately diluted in serum-free medium, then mixed to form a complex. This siRNA-transfection reagent complex is slowly added to the cells, and the culture plate is gently rocked to ensure an even distribution of the mixture. After a 6-h incubation, the cell supernatant is aspirated, and the cells are replaced with complete medium and further cultured for 72 h before conducting the relevant assays.
2.14. qPCR analysis
Total RNA was isolated from cells using TRIzol reagent, following the manufacturer's protocol. Subsequently, the PrimeScript RT Reagent Kit was utilized to synthesize complementary DNA (cDNA) from the extracted RNA. Quantitative PCR (qPCR) was then conducted using a SYBR PrimeScript RT-PCR Kit, according to the manufacturer's instructions. The thermocycler program was as follows: an initial denaturation step at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as an endogenous control for normalization.
2.15. RNA sequencing
Following digestion, washing, and centrifugation of osteosarcoma cells that had received different treatments, TRIzol reagent was slowly added to the cells at a ratio of 5 × 106 cells/mL. The mixture was then carefully transferred to RNase-free cryogenic tubes. Subsequently, the tubes were rapidly immersed in liquid nitrogen for 30 min to flash-freeze the samples. After freezing, the samples were removed from the liquid nitrogen, placed into self-sealing bags, and stored at −80 °C for long-term preservation. The samples were then sent to Majorbio Co. (China) for transcriptome sequencing. During the analysis, differentially expressed genes and enriched pathways were identified.
2.16. Bioinformatics analysis
Single-cell RNA sequencing data from human osteosarcoma (GSE162454) were obtained from the Gene Expression Omnibus (GEO) database, with three samples (GSM5155198, GSM5155199, and GSM5155200) selected for detailed analysis. Utilizing the Seurat package in R (version 4.2.2), data processing involved initial integration with the ‘merge’ function, followed by normalization using ‘Normalize Data’. Principal component analysis (PCA) was conducted on the 2000 most variable genes for dimensionality reduction, with the first 40 principal components selected for further study. Cell clustering was executed with ‘Find Neighbors' and ‘Find Clusters', and the ‘Run UMAP’ and ‘Dim Plot’ functions were applied for visualizing the clusters. Marker genes were identified using the CellMarker2.0 database and literature, guiding the naming of cell subgroups. The expression of HERC1 and NCOA4 across subgroups was analyzed and plotted. A Spearman correlation analysis determined the correlation between HERC1 and NCOA4, with correlation coefficients indicating strength. Using the Target osteosarcoma dataset, patients were categorized into high and low HERC1 expression groups based on median expression levels. Finally, a heatmap was generated to examine the relationship between HERC1 expression and the NRF2 signaling pathway.
2.17. Co-immunoprecipitation
Osteosarcoma cells were lysed in IP Lysis Buffer for 30 min on ice. After centrifugation, the part of supernatant was partially reserved as the input control. The remaining supernatant was mixed with Protein A/G beads and rotated at 4 °C to reduce non-specific binding, followed by bead adsorption using a magnetic rack. The supernatant, enriched for the protein of interest, was collected. The protein sample was split into two: one aliquot was incubated with a specific antibody (2.5 μg), and the other with an equivalent amount of IgG as a negative control. Both were then rotated on a mixer at 4 °C for 12 h to form immune complexes. Afterward, the beads were washed to remove unbound proteins and incubated again with the protein samples for an additional 4 h at 4 °C. The proteins were subsequently denatured by heating, and the immunoprecipitated samples were stored at −80 °C for subsequent analysis.
2.18. Immunofluorescent staining
After undergoing various treatments, osteosarcoma cells were fixed with 4 % paraformaldehyde for 15 min. Following fixation, cells were permeabilized with a 100 × dilution of Permeabilization Buffer for an additional 15 min. The permeabilization buffer was then removed, and blocking serum, corresponding to the species of the secondary antibody, was added to the cells. The dishes were sealed and incubated for 2 h at room temperature. After removing the blocking serum, cells were incubated with an immunofluorescence primary antibody diluted at 1:500 overnight in a 4 °C refrigerator or freezer. Unbound primary antibody was removed by washing the cells three times with PBS. A fluorescently labeled secondary antibody was applied, and cells were incubated at room temperature for 2 h to facilitate antibody binding. Excess antibody was washed away with PBS. The cells were finally examined under a fluorescence microscope.
2.19. Xenograft tumor model
Four-week-old nude mice were selected and acclimated to their environment for 1 week. Following various treatments, osteosarcoma cells were prepared as a cell suspension containing 1 × 107 cells, diluted in 200 μL of PBS buffer. After local disinfection with 75 % alcohol, a 200 μL aliquot of the cell suspension was injected subcutaneously into the nude mice. Tumor formation was monitored every two days, and tumor volume was measured every three days using the formula: volume = 1/2 × (short diameter)2 × (long diameter). When tumors in the control group reached approximately 70 mm3, corresponding treatments were initiated for all groups.
2.20. In vivo fluorescence imaging
Once the tumors reached a volume of approximately 100 mm3, in vivo fluorescence imaging was performed. The nude mice were anesthetized using 4 % isoflurane, and their vital signs were closely monitored throughout the procedure. Upon achieving deep anesthesia, photosensitizer MPPα was administered intravenously via the tail vein at a dose of 15 mg/kg. Following the injection, the mice were placed in a light-protected enclosure. Subsequently, in vivo fluorescence imaging was conducted using a specific wavelength of light to excite the photosensitizers within the mice, and emitted fluorescence signals at a wavelength of 630 nm were captured.
2.21. Immunohistochemical staining
Tumor tissue sections were marked and baked for 2 h. After deparaffinization using a gradient of xylene and ethanol, the sections were subjected to antigen retrieval in EDTA solution and heated in a microwave oven at high power for 5 min, followed by medium-low power for 15 min. Once the sections reached room temperature, they were treated with a 0.3 % hydrogen peroxide solution to quench endogenous peroxidase activity and then blocked for 12 min using a blocking solution. The diluted primary antibody (1:500) was applied and incubated at 4 °C overnight in a humidified chamber. After incubation, the sections were washed, and an appropriate amount of secondary antibody was applied and incubated at 37 °C for 2 h. Following DAB colorimetric development, the tissue sections were counterstained with hematoxylin, dehydrated, cleared, and mounted for microscopic examination.
2.22. TUNEL staining
After undergoing deparaffinization with a graded series of xylene and ethanol, tissue sections were treated with DNase-free protease K at a concentration of 20 μg/mL and incubated at 37 °C for 30 min. Following proteolytic treatment, the TUNEL detection solution was applied to the tissues and incubated at 37 °C for 60 min in the dark to label apoptotic cells. Once the TUNEL reaction was complete, the stained tissue sections were counterstained with hematoxylin to visualize the nuclear morphology. Afterward, the sections were processed through a series of graded alcohols for dehydration, cleared with xylene, and then mounted with a coverslip using a suitable mounting medium. The prepared tissue sections were subsequently examined under a fluorescence microscope to assess the TUNEL staining.
2.23. Statistical analysis
All data are expressed as the mean ± SD values and were analyzed with SPSS 22.0 software. Single Student's t-test and one-way ANOVA were used to determine statistical significance between pairs of groups and three or more groups, respectively. Significance levels are shown as *P < 0.05, **P < 0.01, and ***P < 0.001.
3. Results
3.1. Properties of photosensitizer MPPα and killing mechanism of PDT
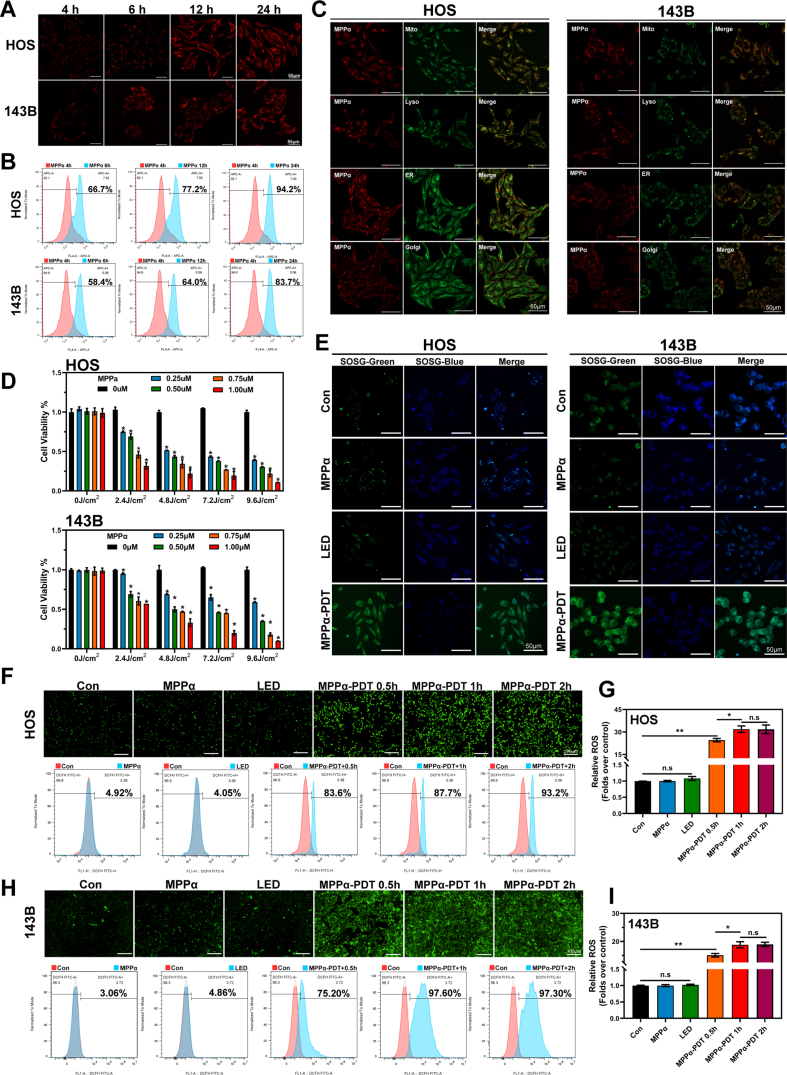
As a second-generation photosensitizer, pyropheophorbide-α methyl ester (MPPα), demonstrates enhanced stability and absorption capacity compared to first-generation photosensitizers. It exhibits improved absorbance and heightened photoelectric sensitivity [7,24,25]. To evaluate the phagocytic capacity of osteosarcoma cells for MPPα, we co-cultured cells with MPPα and observed the emergence of red fluorescence particles within the cells after 4 h using CLSM. Cellular phagocytosis of MPPα increased over time, peaking after 24 h of co-culture (Fig. 1A). Flow cytometry analysis revealed that the percentage of cells with fluorescence markers increased gradually with extended uptake time, reaching a peak of 94.2 % (HOS)/83.7 % (143B) after 24 h of phagocytosis, as depicted in Fig. 1B. Fluorescence co-localization analysis was conducted using organelle fluorescence probes in conjunction with MPPα′s intrinsic fluorescence properties. The yellow fluorescent fusion indicated that MPPα is predominantly localized within mitochondria and lysosomes in osteosarcoma cells, with minimal localization in the endoplasmic reticulum and Golgi apparatus (Fig. 1C). To assess the impact of MPPα-PDT on OS cells activity, we conducted further investigations utilizing a continuous output LED light source with a wavelength of 630 nm and an intensity of 40 mW/cm2. Various concentrations (0 μM, 0.25 μM, 0.5 μM, 0.75 μM, 1 μM) of the MPPα were used in combination with different levels of light energy density (0 J/cm2, 2.4 J/cm2, 4.8 J/cm2, 7.2 J/cm2, 9.6 J/cm2), aiming to determine optimal conditions for further study. The CCK-8 assay indicated that increasing concentrations of MPPα led to a dose-dependent decrease in cell activity, suggesting a stronger inhibitory effect on OS cells. Additionally, as the intensity of light energy increased, cell activity declined gradually with a specific MPPα concentration. Notably, at a light energy density of 4.8 J/cm2, the MPPα concentration required to achieve IC50 in HOS cells was 0.25 μM, with an inhibition rate of 48.4 ± 1.4 %. Similarly, for 143B cells under the same light energy density, a MPPα concentration of 0.5 μM resulted in an inhibition rate of 50.0 ± 6.71 % (Fig. 1D). Singlet oxygen, a highly reactive form of reactive oxygen species (ROS), is an excited state of the oxygen molecule with higher energy than its ground state. We found that MPPα-PDT significantly increased singlet oxygen levels in cells, indicating substantial activation of highly reactive ROS (Fig. 1E). Then ROS was further detected using fluorescence microscopy and flow cytometry, and the results showed a marked increase after 0.5 h of MPPα-PDT treatment, with the intensity rapidly peaking over time (Fig. 1F–I).
Fig. 1.
Properties of photosensitizer MPPα and killing mechanism of PDT. (A) Phagocytic activity of OS cells on the photosensitizer MPPα as measured by CLSM. Scale bar: 50 μm. (B) Fluorescence quantitative analysis of OS cells by flow cytometry. (C) Fluorescence co-localization of organelle probes with photosensitizer MPPα as measured by CLSM. Scale bar: 50 μm. (D) Effect of MPPα-PDT on OS cells activity was tested using the CCK-8 assay. (E) The singlet oxygen fluorescent images of OS cells after MPPα-PDT treatment detected by SOSG. Scale bar: 50 μm. (F–I) DCFH-DA fluorescence images and flow quantitative analysis of OS cells after MPPα-PDT treatment. Scale bar: 100 μm. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
3.2. The effect of MPPα-PDT on promoting apoptosis and ferroptosis
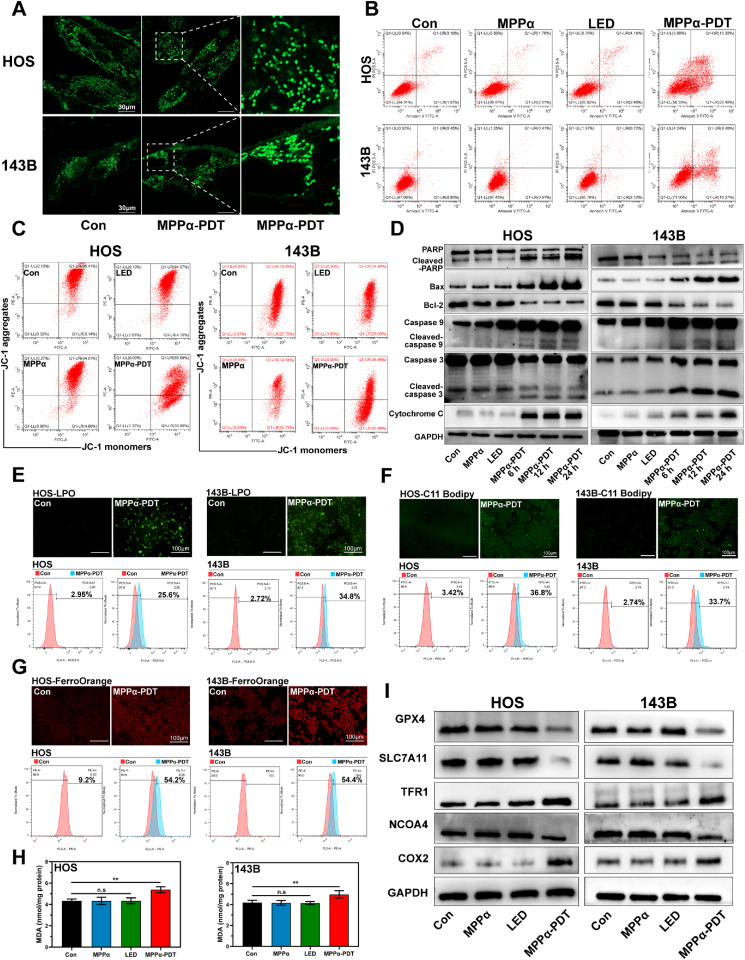
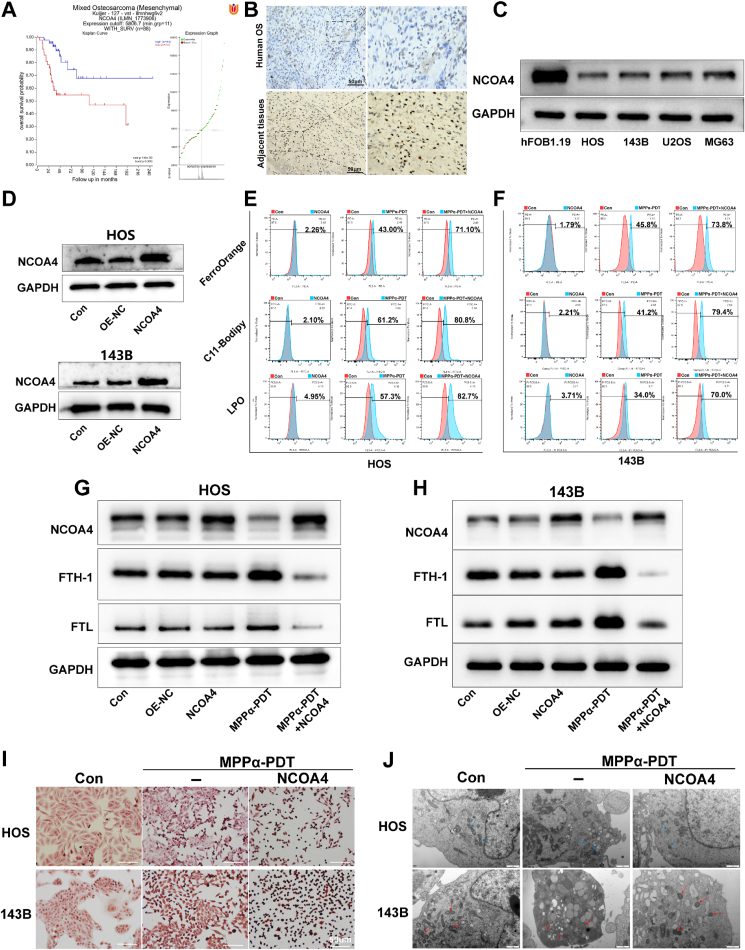
Mitochondria play a crucial role in cellular functions, encompassing energy production, regulation of metabolic processes, and determination of cell fate. Given the predominant localization of the photosensitizer MPPα within mitochondria, we initially examined whether MPPα-PDT could induce morphological alterations in OS cells. Our findings demonstrated that mitochondrial fragmentation could be observed after MPPα-PDT treatment, providing direct evidence for mitochondrial dysfunction (Fig. 2A). Then, it was also revealed a great increase in the apoptosis rate of OS cells following MPPα-PDT treatment, accompanied by a notable reduction in ΔΨm (Fig. 2B–S1A, S1B, and 2C). To further validate the involvement of the mitochondrial pathway in apoptosis, Western Blot (WB) analysis was employed to detect key proteins. The results revealed an increase in cytochrome c, caspase family proteins (caspase 3 and caspase 9), PARP, and Bax proteins with prolonged MPPα-PDT treatment. Concurrently, the expression of the anti-apoptotic protein Bcl-2 was found to be suppressed (Fig. 2D and S2). To determine whether MPPα-PDT can induce ferroptosis, we initially assessed the levels of lipid peroxidation in OS cells using the lipid peroxidation probe C11-Bodipy and a lipid peroxide (LPO) assay. Our findings revealed that MPPα-PDT significantly increased the percentage of fluorescent C11-Bodipy-positive cells and elevated LPO content, indicating induction of ferroptosis (Fig. 2E and F, and S3). To further validate the impact of MPPα-PDT on iron homeostasis in OS cells, we employed the FerroOrange fluorescence probe to measure Fe2+ levels. Our investigation showed that MPPα-PDT significantly induced a substantial increase in Fe2+ content within cells, as observed under fluorescence microscopy. Consistent results were obtained through quantitative analysis using flow cytometry (Fig. 2G and S3). Malondialdehyde (MDA) is an organic compound, which is one of the products of lipid peroxidation reaction and is often used as a biomarker to measure the level of oxidative stress and lipid peroxidation in organisms [14]. The results indicated that the MDA contents in OS cells could be induced by MPPα-PDT (Fig. 2H). Concurrently, WB analysis was also used to assess alterations in ferroptosis-related proteins. Subsequent analysis revealed that MPPα-PDT markedly induced GPX4-mediated ferroptosis in OS cells, as evidenced by a specific reduction in the expression of protein GPX4 and SLC7A11, along with an increase in TFR1 protein expression. Furthermore, we also observed inhibition of NCOA4 expression, a pivotal protein involved in ferritin degradation, which may impede ferritin degradation to some extent and subsequently hinder ferritinophagy occurrence. Therefore, elucidating the underlying cause for decreased NCOA4 could potentially activate ferritinophagy and enhance ferroptosis (Fig. 2I and S4).
Fig. 2.
The effect of MPPα-PDT on promoting apoptosis and ferroptosis. (A) The mitochondrial fragmentation of OS cells as observed by CLSM after MPPα-PDT. Scale bar: 30 μm. (B) The apoptosis rate of OS cells after MPPα-PDT treatment by flow cytometry analysis. (C) The ΔΨm of OS cells after MPPα-PDT treatment by flow cytometry analysis. (D) The expression of mitochondrial apoptotic proteins was detected by Western blot analysis after MPPα-PDT treatment. (E) Fluorescent images and flow cytometry analysis of lipid peroxides in OS cells. Scale bar: 100 μm. (F) Fluorescent images and flow cytometry analysis of lipid-ROS in OS cells. Scale bar: 100 μm. (G) Fluorescent images and flow cytometry analysis of Fe2+ in OS cells. Scale bar: 100 μm. (H) MDA contents in OS cells after various treatments. (I) The expression of ferroptosis related proteins in OS cells was detected by Western blot analysis. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
3.3. Synergistic effect of apoptosis and ferroptosis
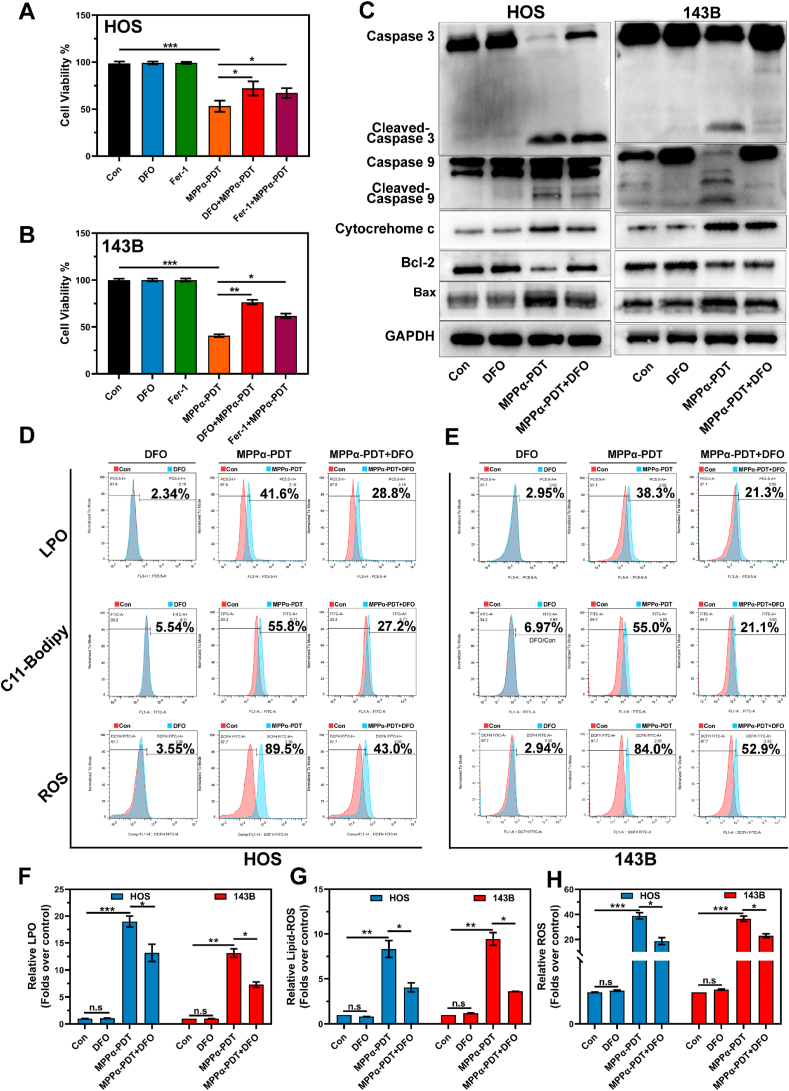
To preliminarily validate the synergistic effect between ferroptosis and apoptosis mediated by MPPα-PDT, we suppressed the iron chelation ability and lipid peroxidation function using ferroptosis inhibitors Deferoxamine (DFO) and Ferrostatin-1 (Fer-1). Initially, the impact of various treatments on the cell viability using the CCK-8 assay. It was observed that while MPPα-PDT combined with ferroptosis inhibitors (DFO or Fer-1) still exhibited some inhibition on OS cell activity compared to MPPα-PDT alone, there was a noticeable recovery could be observed (Fig. 3A and B). Additionally, WB analysis was conducted to detect the apoptotic proteins. It was observed that the combined treatment of OS cells with the ferroptosis inhibitor DFO and MPPα-PDT resulted in inhibited expression of pro-apoptotic proteins. Furthermore, an increased expression of the anti-apoptotic protein Bcl-2 could also be observed, which further supports the validation of a synergistic effect between ferroptosis and apoptosis (Fig. 3C and S5). To further investigate the potential mechanism underlying this synergistic effect, lipid peroxidation in OS cells was measured. Flow cytometry analysis revealed that MPPα-PDT noticeably induced lipid peroxidation in OS cells, whereas combined treatment effectively inhibited this process (Fig. 3D–H). In summary, the concurrent administration of ferroptosis inhibitors can significantly counteract the pro-apoptotic and pro-ferroptotic effects of MPPα-PDT. Hence, we propose that there is a synergistic effect between apoptosis and ferroptosis as mediated by MPPα-PDT.
Fig. 3.
Synergistic effect of apoptosis and ferroptosis. (A–B) Effect of various treatments on OS cells activity was tested using CCK-8 assay. (C) The expression of apoptotic proteins after various treatment were detected by Western blot analysis. (D–E) Analysis of LPO, Lipid-ROS and ROS by flow cytometry after different treatments. (F– H) Quantitative analysis of LPO, Lipid-ROS and ROS by flow cytometry results after different treatments. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
3.4. Ferrintinophagy is inhibited by MPPα-PDT
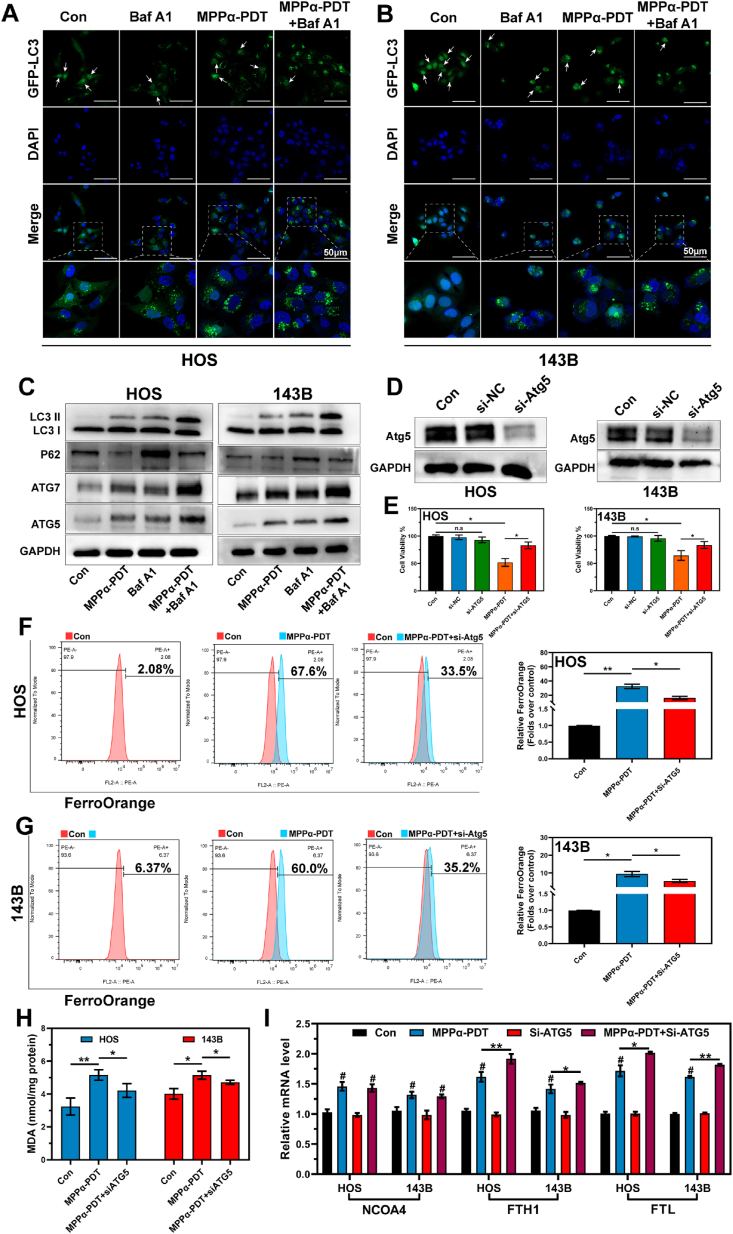
Autophagy is a crucial cellular process involved in the degradation and recycling of intracellular components, playing a vital role in maintaining cellular homeostasis and responding to various physiological processes and stressors. Autophagic flux, which refers to the dynamic sequence of autophagosome formation, fusion with lysosomes, and subsequent cargo degradation, is a key measurement of autophagy activity. Bafilomycin A1 (Baf A1) serves as a commonly tool for monitoring autophagic flux by impeding the fusion between autophagosomes and lysosomes through inhibition of V-ATPase activity, thereby preventing acidification within lysosomal compartments [26]. In this study, we initially used Baf A1 to inhibit autophagy flux in OS cells. Then, a notable increase in the expression of green-fluorescent eGFP-LC3 autophagosomes could be observed using CLSM, confirming the successful blockade of autophagy flux. LC3 is involved in a nucleocytoplasmic transport mechanism that relies on acetylation and deacetylation regulated by autophagy signals, which play a crucial role in the formation of autophagosomes [27]. Under stress conditions, nuclear-localized Sirt1 becomes activated and deacetylates LC3 within the nucleus. The resulting deacetylated LC3 molecules then translocate to the cytoplasm, where they interact with autophagy effectors such as Atg7 [28]. In this study, noticeable fluorescence transfer from the nucleus to the cytoplasm was observed following MPPα-PDT treatment, indicating induction of autophagy. Furthermore, combined treatment further augmented expression of autophagosomes (Fig. 4A and B). Similarly, through WB analysis, we monitored alterations in the expression of autophagy-related proteins following MPPα-PDT treatment and found a significant upregulation of LC3II, Atg3, and Atg5 proteins. Concurrently, there was a substantial inhibition in P62 protein expression (Fig. 4C and S6). As shown in Fig. 2I, we preliminarily confirmed that MPPα-PDT inhibits the expression of NCOA4 and limits the degradation of ferritin, thereby suppressing ferritinophagy. However, we also found that mega-autophagy in OS cells could be induced by MPPα-PDT. To further clarify the interaction between autophagy and ferroptosis mediated by MPPα-PDT, we first used small interfering RNA (siRNA-Atg5) to inhibit the expression of Atg5 protein (Fig. 4D and S7). It was found that silencing Atg5 significantly inhibits the antitumor efficacy of MPPα-PDT (Fig. 4E). The Fe2+ content was monitored using the FerroOrange fluorescence probe, revealing that inhibition of autophagy could attenuate the elevation in Fe2+ mediated by MPPα-PDT (Fig. 4F and G). Furthermore, the assessment of MDA revealed that the inhibition of Atg5 also effectively suppressed lipid peroxidation induced by MPPα-PDT (Fig. 4H). Hence, we tentatively propose that silencing Atg5-mediated autophagy inhibition can partially suppress ferroptosis triggered by MPPα-PDT. Additionally, we observed the transcriptional activity of iron metabolism genes. Interestingly, the expression of NCOA4 was activated following MPPα-PDT treatment, suggesting that ferritinophagy was activated at the transcriptional level (Fig. 4I). Therefore, we hypothesize that NCOA4 may be activated in transcription following MPPα-PDT treatment, but there appears to be some form of inhibition or degradation in translation. Thus, actively investigating the potential mechanisms underlying the inhibition NCOA4 protein could be a crucial factor in overcoming ferritinophagy inhibition and enhancing the efficiency of ferroptosis.
Fig. 4.
Ferrintinophagy is inhibited by MPPα-PDT. (A–B) Fluorescence images of ADV-eGFP-LC3 in OS cells after various treatments. The white arrow indicates LC3 fluorescence within the nucleus. Scale bar: 50 μm. (C) The expression of autophagy related proteins after various treatments was detected by Western blot analysis. (D) The expression of Atg5 protein after siRNA treated was detected by Western blot analysis. (E) Effect of silencing Atg5 on OS cells activity was tested using CCK-8 assay. (F–G) Analysis of Fe2+ by flow cytometry after different treatments. (H) MDA contents in OS cells after various treatments. (I) mRNA expression of NCOA4, FTH1, FTL after different treatments as measured by qPCR. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
3.5. Targeting NCOA4 enhances ferroptosis mediated by MPPα-PDT
NCOA4 functions as a pivotal cargo receptor implicated in ferritin degradation and plays a crucial role in ferritinophagy. To obtain clinical prognosis analysis of the NCOA4 gene in OS patients, we scrutinized the mRNA microarray data from the GEO database (GSE42352) using an online bioinformatics visualization platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). The result revealed that individuals with elevated NCOA4 expression exhibited more favorable clinical prognoses (Fig. 5A). Furthermore, immunohistochemical (IHC) staining revealed a higher expression of NCOA4 in the adjacent tissue compared to the tumor site in human OS tissue (Fig. 5B). Based on these findings, we can indirectly conclude that NCOA4, which acts as an inhibitory protein, has the potential to be a therapeutic target for the treatment of OS. The expression of NCOA4 was simultaneously assessed in HOS, 143B, MG63, U2OS cell lines, and osteoblast Hfob1.19 cells by WB analysis. Notably, the levels of NCOA4 were relatively low in HOS and 143B cell lines (Fig. 5C and S8A). Therefore, we chose HOS and 143B cells as models for subsequent investigations. To ascertain the biological function of NCOA4, the gene was overexpressed by lentiviral vector. The efficiency of viral transduction was then confirmed by WB (Fig. 5D and S8B). The present study confirms the induction of autophagy by MPPα-PDT, which prompts us to explore the synergistic treatment by combining MPPα-PDT with NCOA4 overexpression to fully activate ferritinophagy. The impact of MPPα-PDT combined with NCOA4 overexpression on ferroptosis was investigated by initially assessing the lipid peroxidation in OS cells. Through flow cytometry analysis, we observed an obviously induction of lipid peroxidation upon the combination treatment, as demonstrated by FerroOrange, LPO, and C11-Bodipy fluorescent probes (Fig. 5E and F, S8C-S8E). Afterward, the expression changes in ferritin were also detected via WB. The results revealed that NCOA4 overexpression could not induce alterations in ferritin levels. This may be attributed to the incomplete activation of autophagy under basal conditions, which hinders the degradation pathway of ferritin. However, when combined with MPPα-PDT, NCOA4 overexpression resulted in complete activation of ferritinophagy and subsequently facilitated the degradation of ferritin (Fig. 5G and H, and S8F-S8H). Additionally, we employed Perls staining to assess the iron deposition after treatments in OS cells. It was observed that the combination of MPPα-PDT and NCOA4 overexpression led to the complete activation of ferritinophagy, resulting in the increased degradation of ferritin, gradual elevation of iron ion expression, and subsequent accumulation of numerous iron particles under microscopy (Fig. 5I). Subsequently, we examined the mitochondria after virous treatments using transmission electron microscopy (TEM). Notably, MPPα-PDT induced characteristic apoptotic changes in mitochondria, including significant swelling, irregular arrangement, or breakage of mitochondrial cristae. Simultaneously, when combined with NCOA4 overexpression, MPPα-PDT caused a distinct ferroptosis-like mitochondrial appearance characterized by a reduction in volume and diminished cristae (Fig. 5J).
Fig. 5.
Targeting NCOA4 enhances ferroptosis mediated by MPPα-PDT. (A) The impact of NCOA4 on the clinical prognosis of OS patients was analyzed using data from GSE42352 database. (B) Expression of NCOA4 in human OS and adjacent tissues was detected using IHC. (C) Basic expression of NCOA4 protein in OS cells was tested by Western blot analysis. (D) Expression of the protein in OS cells was tested by Western blot analysis after NCOA4 overexpression. (E–F) Analysis of LPO, Lipid-ROS and Fe2+ by flow cytometry after different treatments. (G–H) The expression of protein NCOA4, FTH1, FTL in OS cells after various treatments were detected by Western blot analysis. (I) Detection of iron ion deposition images in OS cells after different treatments using Prussian blue staining. Scale bar: 50 μm. (J) Observation of mitochondria in OS cells by Transmission Electron Microscopy. The red or blue arrow refers to the mitochondria in OS cells. Scale bar: 1 μm. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Transcriptome sequencing and bioinformatics analysis
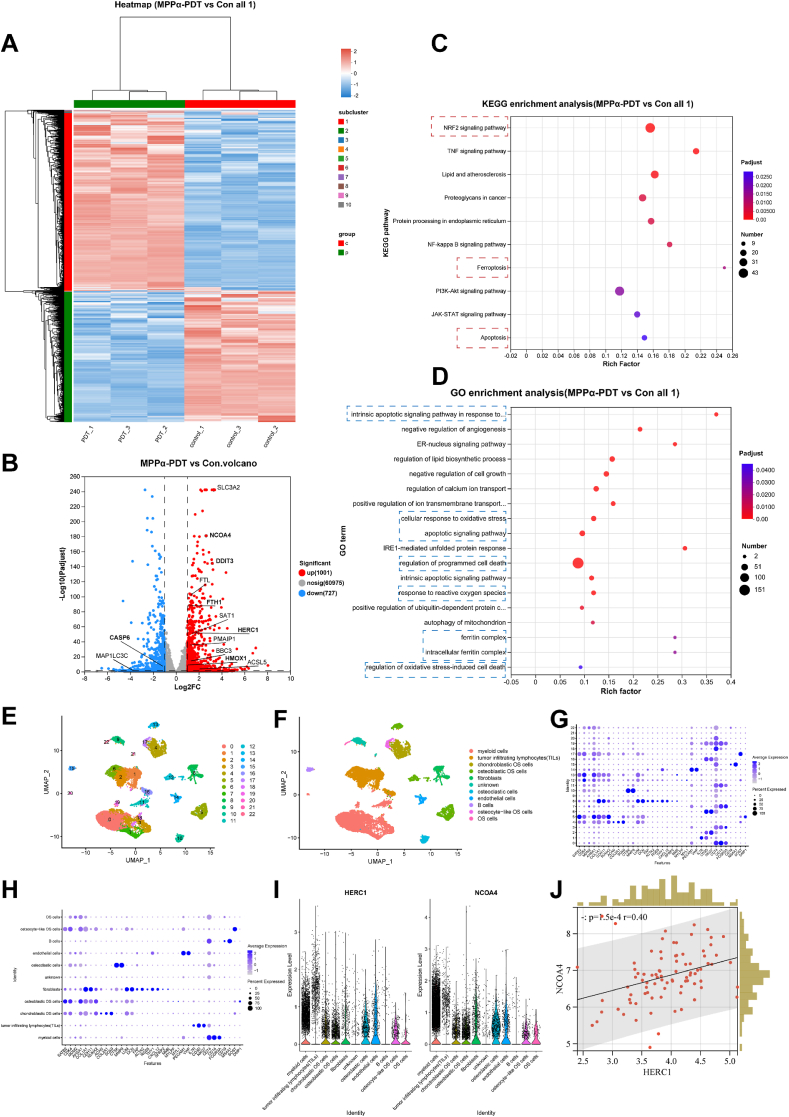
To further elucidate the mechanisms underlying degradation in translation, we initially examined the transcriptomic changes induced by MPPα-PDT using high-throughput transcriptomic sequencing. The findings revealed that MPPα-PDT treatment resulted in the upregulation of 1001 genes and the downregulation of 727 genes (Fig. 6A and B). Importantly, HERC1, which belongs to the extensive HERC family and is recognized as a potential E3 ubiquitin protein ligase due to its HECT domain [29,30], showed a noticeable increase in transcription. Given the existence of multiple ubiquitination regulatory sites on NCOA4 [17], it is hypothesized that HERC1 functions in facilitating the ubiquitin-mediated degradation of NCOA4. Additionally, enrichment analysis of differentially expressed genes revealed marked enrichment of NRF2, TNF, NF-κB, PI3K-Akt, and others in the KEGG analysis. Furthermore, we also observed enriched functional signals related to apoptosis and ferroptosis, providing further evidence for the effective regulation by MPPα-PDT. GO enrichment analysis also demonstrated noticeable enrichment of biological processes such as “apoptosis-related pathways,” “cell response to oxidative stress,” and “ferritin complex” (Fig. 6C and D). To investigate the distribution of HERC1 and NCOA4 genes in OS tissues and their correlation, we performed bioinformatics analysis on single-cell sequencing data, including data integration, quality control, standardization, dimensionality reduction, clustering, and expression analysis of HERC1 and NCOA4. The results identified 23 distinct cell subgroups within the UMAP clusters. The CellMarker2.0 database was utilized to identify marker genes for each cell subgroup based on their expression patterns, and each cell subgroup was subsequently named accordingly. Importantly, our findings demonstrated that both HERC1 and NCOA4 were expressed in OS cells (Fig. 6E–I). Furthermore, Spearman correlation analysis indicated a significant association between HERC1 and NCOA4 (P < 0.001) (Fig. 6J).
Fig. 6.
Transcriptome sequencing and bioinformatics analysis. (A) Heat map analysis of differential genes after MPPα-PDT treatment. (B) Volcano map of differential genes after MPPα-PDT treatment. (C) KEGG enrichment analysis of differential genes after MPPα-PDT treatment. (D) GO enrichment analysis of differential genes after MPPα-PDT treatment. (E–F) Cellular umap grouping. (G–H) The expression of marker genes in various cell subpopulations. (I) Expression of HERC1 gene and NCOA4 gene in various cell subpopulations. (J) Spearman correlation analysis between HERC1 gene and NCOA4 gene.
3.7. HERC1 interacts with NCOA4
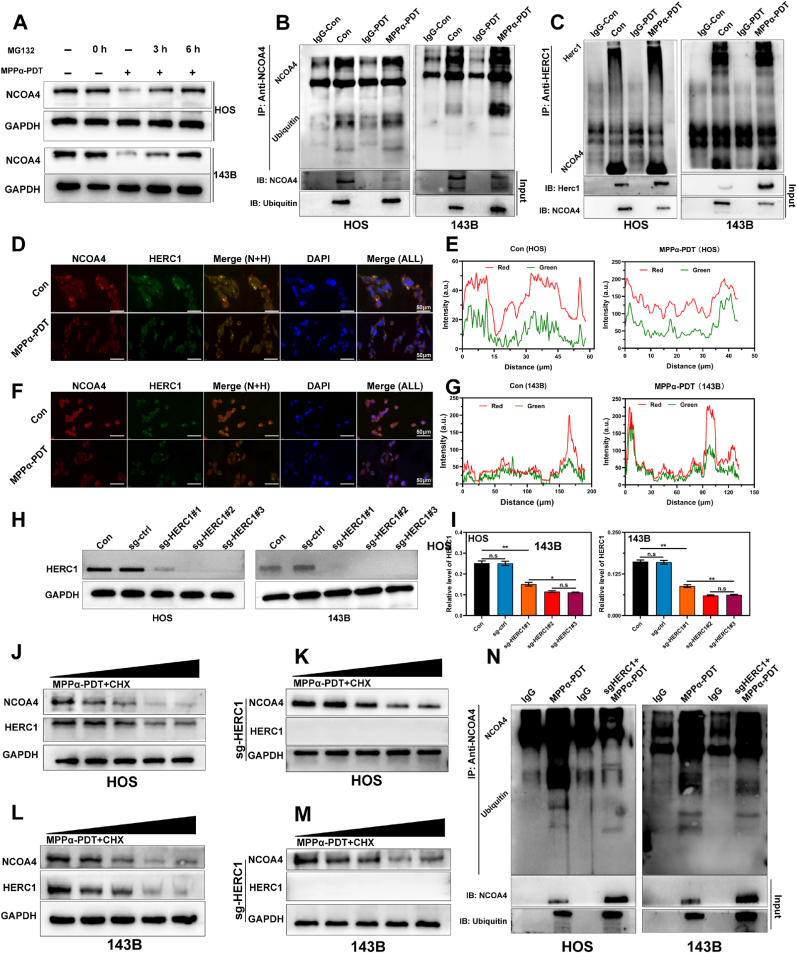
To investigate the potential degradation of NCOA4 protein through the ubiquitination pathway, we initially treated OS cells with a proteasome inhibitor MG132 (15 μM). WB analysis revealed that the combined treatment of MG132 and MPPα-PDT in OS cells led to a gradual recovery in the expression of the NCOA4 protein over time (Fig. 7A and S9). These findings provide preliminary evidence suggesting that the NCOA4 protein may undergo degradation through ubiquitination. To further validate the ubiquitination of NCOA4, we employed Co-immunoprecipitation (co-IP) to ascertain its interaction with ubiquitin. The results demonstrated that the immunoprecipitation antibody specific to NCOA4 effectively captured ubiquitin, providing evidence for the interaction between NCOA4 and ubiquitin (Fig. 7B). This supports the scientific hypothesis that NCOA4 undergoes degradation via the ubiquitination process. Next, to confirm the involvement of HERC1 in the ubiquitination degradation of NCOA4, we employed co-IP to evaluate the interaction between HERC1 and NCOA4 proteins. The results demonstrated successful capture of NCOA4 protein by immunoprecipitation using HERC1 antibody, providing evidence for their potential interaction (Fig. 7C). Subsequently, we utilized immunofluorescence co-localization staining in OS cells to validate the shared subcellular localization of HERC1 and NCOA4 proteins. Our study revealed an obvious increase in subcellular localization of both proteins especially after MPPα-PDT treatment. In conclusion, our findings indicate an interaction between HERC1 and NCOA4 proteins, along with a notable degree of subcellular colocalization (Fig. 7D–G). To investigate the regulatory role of HERC1 on NCOA4, we knocked out the HERC1 gene using CRISPR/CAS9 single vector lentivirus (sg-HERC1). We then detected the knockout efficiency of HERC1 by WB (Fig. 7H and I). Following this, to investigate the impact of HERC1 knockout on NCOA4 protein stability, we initially suppressed protein synthesis using cycloheximide (CHX) and subsequently assessed NCOA4 protein stability via WB analysis. It was observed that the degradation rate of NCOA4 protein dramatically decreased compared to the control when MPPα-PDT was combined with CHX treatment in sg-HERC1 cells. These findings demonstrated that HERC1 knockout substantially enhances NCOA4 protein stability and indirectly suggests the involvement of HERC1 in NCOA4 ubiquitination (Fig. 7J–M and S10). To further verify the effect of HERC1 knockout on the ubiquitination of NCOA4 protein, we conducted co-IP to detect the binding between NCOA4 protein and ubiquitin due to HERC1 was knockout. The results revealed an obvious inhibition in the binding between NCOA4 protein and ubiquitin molecules due to HERC1 knockout. These results demonstrate that HERC1 knockout can effectively inhibit the ubiquitination of NCOA4 protein (Fig. 7N).
Fig. 7.
HERC1 interacts with NCOA4. (A) The expression of NCOA4 protein in OS cells after various treatments was detected by Western blot analysis. (B) co-IP images of NCOA4 protein and ubiquitin protein in OS cells. (C) co-IP images of NCOA4 protein and HERC1 protein in OS cells. (D–G) Images and analysis of immunofluorescence double staining in OS cells after MPPα-PDT treatment. Scale bar: 50 μm. (H) The expression of HERC1 in OS cells was detected by Western blot analysis. (I) Quantitative analysis of HERC1 protein expression in OS cells, with the value of GAPDH as the loading control. (J–K) The expression of proteins in HOS cells after various treatments was detected by Western blot analysis. (L–M) The expression of proteins in 143B cells after various treatments was detected by Western blot analysis. (N) co-IP images of NCOA4 protein and ubiquitin protein in OS cells. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
3.8. Targeting HERC1 enhances the anti-tumor efficiency of MPPα-PDT
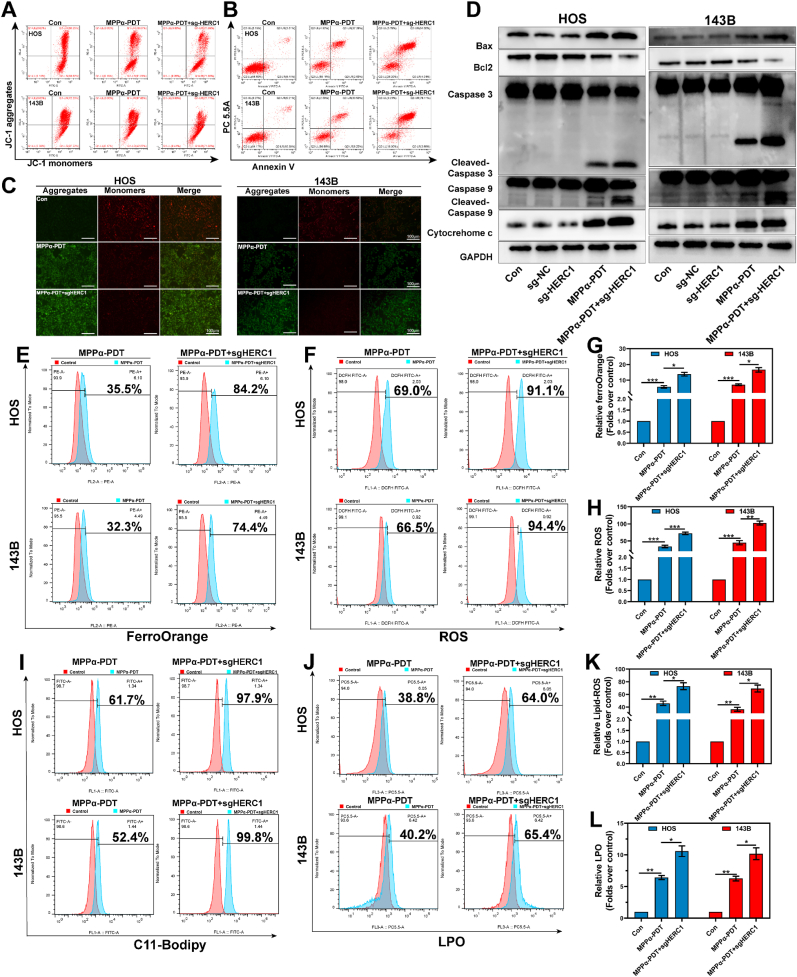
To validate the impact of HERC1 knockout on the anti-tumor efficacy of MPPα-PDT, we initially assessed the apoptosis rate and ΔΨm in OS cells. The results revealed a higher apoptosis rate in OS cells under combined treatment of HERC1 knockout and MPPα-PDT compared to MPPα-PDT alone, accompanied by a notable reduction in ΔΨm (Fig. 8A–C). To further investigate the pro-apoptotic effect of combined treatment on OS cells, we performed WB analysis to examine the expression of pro-apoptotic proteins. The results revealed that a more pronounced pro-apoptotic effect was observed compared to MPPα-PDT treatment when HERC1 was knocked out (Fig. 8D). Similarly, the degradation of NCOA4 protein through ubiquitination is also inhibited, which prevents autophagy-mediated degradation of ferritin and subsequently reduces lipid peroxidation significantly. To further confirm the impact of HERC1 knockout on iron homeostasis in OS cells, we utilized flow cytometry to assess the level of lipid peroxidation. The results demonstrated that combining HERC1 knockout with MPPα-PDT treatment markedly increased lipid peroxidation. Specifically, there was a notable elevation in Fe2+ content as well as level of ROS, LPO, and C11-Bodipy (Fig. 8E–L). In conclusion, our findings indicated that combining HERC1 knockout with MPPα-PDT treatment effectively elevate Fe2+ contents and induce lipid peroxidation in OS cells, thereby promoting ferroptosis.
Fig. 8.
Targeting HERC1 enhances the anti-tumor efficiency of MPPα-PDT. (A) The ΔΨm of OS cells after various treatments by flow cytometry analysis. (B) The apoptosis rate of OS cells after various treatments by flow cytometry analysis. (C) JC1 fluorescence images of OS cells after different treatments. Scale bar: 100 μm. (D) The expression of pro-apoptotic proteins after HERC1 knockout combined with MPPα-PDT was detected by Western blot analysis. (E–F) Detection of Fe2+ and ROS in OS cells after different treatments using flow cytometry. (G–H) Quantitative analysis of flow cytometry data. (I–J) Detection of lipid-ROS and LPO in OS cells after different treatments using flow cytometry. (K–L) Quantitative analysis of flow cytometry data. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
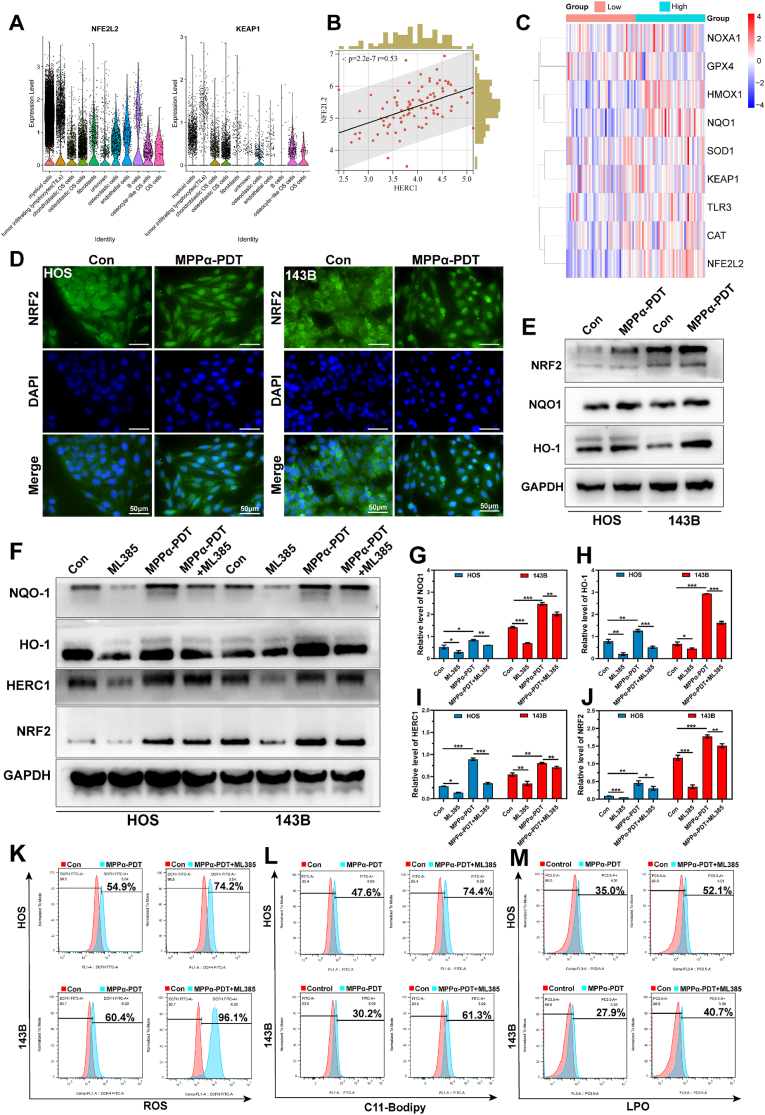
3.9. NRF2 is a potential regulator of HERC1
Nuclear factor erythroid 2-related factor 2 (NRF2) is a pivotal transcription factor involved in regulating the antioxidant response system in cells. Numerous studies have demonstrated that NRF2 exerts direct or indirect control over GPX4, Fe2+, mitochondrial function, and nicotinamide adenine dinucleotide phosphate (NADPH) regeneration among others, thereby governing the process of ferroptosis [31,32]. We have performed KEGG enrichment analysis on differentially expressed genes using RNA sequencing and identified a significant enrichment of the NRF2 signaling pathway. To preliminarily confirm the potential correlation between HERC1 and NRF2, bioinformatics analysis was conducted to examine their association. We found considerable enrichment of NRF2 and KEAP1 genes in OS cells (particularly in osteoblast OS and chondroblast OS), indirectly supporting their involvement in OS cells (Fig. 9A). At the same time, we observed a significant correlation (P < 0.001, R = 0.53) between the NRF2 and HERC1 through Spearman correlation analysis (Fig. 9B). Furthermore, we categorized individuals into high or low group based on the median expression of HERC1 as the threshold value using Target osteosarcoma data. Subsequently, we examined the relationship between the HERC1 and NRF2 signaling pathway in a heatmap format. The findings revealed a greatly positive correlation between the gene HERC1 and genes such as NRF2, HMOX1, NQO1, CAT among others (Fig. 9C). To preliminarily investigate the impact of MPPα-PDT on the NRF2, we initially assessed the nuclear translocation of NRF2 through immunofluorescence staining. Our findings revealed a marked induction of NRF2 nuclear translocation by MPPα-PDT. Additionally, WB analysis was performed to examine protein expression associated with the NRF2 signaling pathway. The results demonstrated that MPPα-PDT effectively activated the NRF2 signaling pathway and induced expression of proteins related to this pathway (Fig. 9D and E, and S11). Then, with the intention of further investigating the regulatory role of NRF2 in HERC1, we pretreated OS cells with the NRF2 inhibitor ML385 and observed a noticeable inhibition of the NRF2 signaling pathway. Concurrently, there was also a varying degree of reduction in HERC1 expression. Furthermore, when combined with MPPα-PDT, ML385 exhibited a certain extent of inhibition on both the NRF2 and HERC1 expression. Therefore, we tentatively suggest that NRF2 may potentially act as a regulator for the HERC1 gene (Fig. 9F–J). NRF2, as a protective factor in tumor cells, plays a crucial role in maintaining the oxidation-antioxidant homeostasis. Therefore, inhibiting NRF2 may be a potential therapeutic strategy. In this study, we found that the lipid peroxidation in OS cells was further increased when treated with an NRF2 inhibitor combined with MPPα-PDT. Considering the potential targeting of HERC1 by NRF2 inhibition, suppression of NRF2 can significantly regulate ferritinophagy and ferroptosis (Fig. 9K–M).
Fig. 9.
NRF2 is a potential regulator of HERC1. (A) The distribution of NRF2 and KEAP1 in OS tissue analyzed through bioinformatics analysis. (B) Spearman correlation analysis between HERC1 gene and NRF2 gene analyzed through bioinformatics analysis. (C) Heat map of the correlation between NRF2 signaling pathway and HERC1 analyzed through bioinformatics analysis. (D) Fluorescence images of OS cells nuclear translocation. Scale bar: 50 μm. (E–F) The expression of NRF2 signaling pathway proteins after MPPα-PDT treatment was detected by Western blot analysis. (G–J) Quantitative analysis of the relative expression level of NRF2, NOQ1, HO1, and HERC1 protein, with the value of GAPDH as the loading control. (L–N) Effect of different treatments on ROS and Lipid-ROS in OS cells by flow cytometry. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
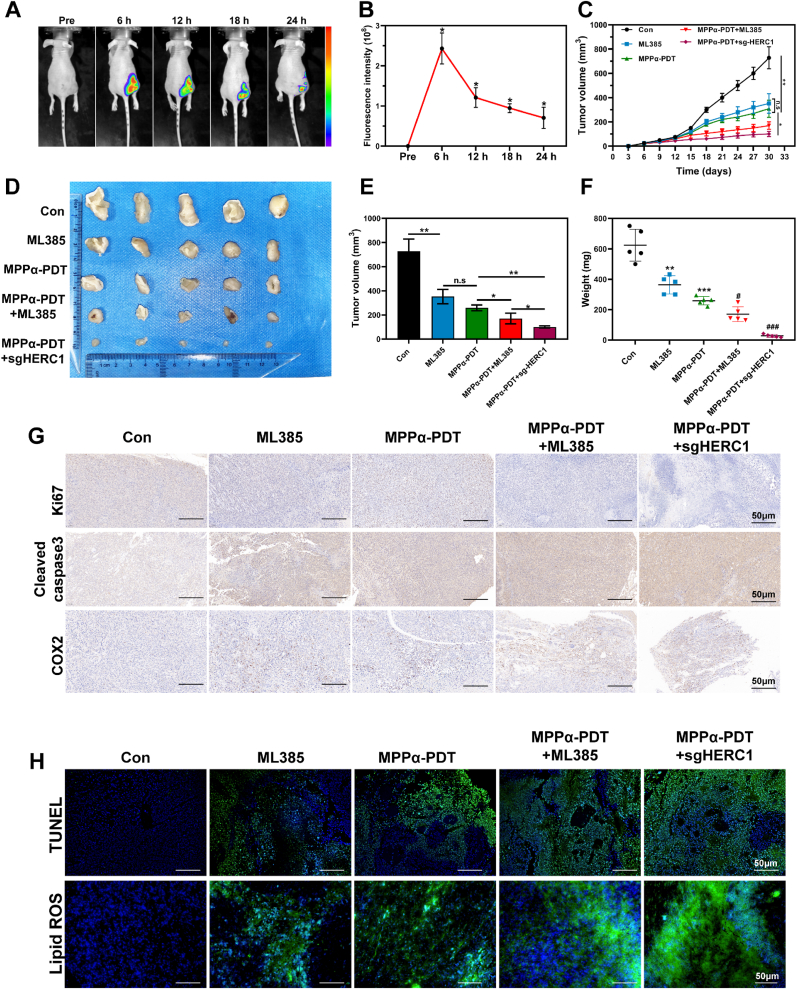
3.10. Targeting HERC1-NCOA4 axis enhances the antitumor effect of MPPα-PDT in vivo
As a second-generation photosensitizer, MPPα is a kind of chlorophyll derivative. Compared to the first-generation photosensitizer, it is more stable and easier to absorb, with better absorbance and stronger photoelectric sensitivity [7,24,25]. These fluorescence characteristics make it suitable for biomarkers and photodynamic diagnosis [33]. In this study, we utilized the biomarker functionality of MPPα to investigate in vivo distribution and tumor targeting capabilities through fluorescence imaging. The results showed that MPPα exhibited noticeable fluorescence enrichment 6 h after injection, particularly in tumor tissue, which further confirmed the specific tumor-targeting capability of MPPα and provided favorable conditions for subsequent PDT treatment (Fig. 10A and B). To further investigate the therapeutic efficacy of targeting the HERC1-NCOA4 axis in combination with MPPα-PDT in vivo, we evaluated the growth inhibition of different treatments on xenograft tumor model. It was observed that treatment with either ML385 or MPPα-PDT alone exhibited limited inhibition on the tumor growth. However, the combined treatment or HERC1 knockout together with MPPα-PDT markedly suppress tumor growth, with the latter one showing more pronounced results. These findings suggest that targeting the HERC1-NCOA4 axis in combination with MPPα-PDT can effectively suppress subcutaneous tumor growth in tumor-bearing nude mice (Fig. 10C–F). Subsequently, we performed IHC staining to detect marker protein associated with proliferation, such as Ki67, and marker related to apoptosis, such as Cleaved-caspase 3. The results revealed that treatment with NRF2 inhibition or MPPα-PDT alone let to some inhibition of tumor growth, as evidenced by the suppression of Ki67 and an increase in Cleaved-caspase 3. Furthermore, when targeting the HERC1-NCOA4 axis in combination with MPPα-PDT, this inhibitory effect was significantly enhanced (Fig. 10G). Consistent results were obtained using the TUNEL staining. (Fig. 10H). To further investigate the potential mechanism of targeting the HERC1-NCOA4 axis on lipid peroxidation induced by MPPα-PDT, we performed IHC staining to assess COX2 expression, a protein closely associated with oxidative stress in tumors. Concurrently, the C11-Bodipy fluorescent probe was employed to assess the lipid peroxidation. It was observed that treatment with ML385 or MPPα-PDT induced a certain degree of oxidative stress and lipid peroxidation. However, when targeting the HERC1-NCOA4 axis in combination with MPPα-PDT, there was a great increase in both oxidative stress and lipid peroxidation within the tumor, as evidenced by notable COX2 protein expression and pronounced enrichment of C11-Bodipy green fluorescence. These findings suggest that by targeting the HERC1-NCOA4 axis in combination with MPPα-PDT, it is possible to effectively increase oxidative stress and lipid peroxidation within the tumor. This indirectly indicates a significant induction of ferroptosis in tumors (Fig. 10G and H).
Fig. 10.
Targeting HERC1-NCOA4 axis enhances the antitumor effect of MPPα-PDT in vivo. (A) Fluorescence images in vivo after MPPα processing. (B) Quantitative analysis of fluorescence intensity at different time points. (C) Growth curves of tumors after different treatments. (D) Photographs of tumors harvested from mice after various treatments. (E) Gross tumor volume of tumors after different treatments. (F) Weight of tumors after different treatments. (G) The expression of Ki67, Cleaved caspase 3, and COX2 proteins detected by IHC. Scale bar: 50 μm. (H) The fluorescent images of TUNEL and lipid-ROS. Scale bar: 50 μm. Analyses were repeated in triplicate (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
4. Discussion
Osteosarcoma (OS) is a primary malignant bone tumor that originates from osteoid cells, which are mesenchymal cells. Approximately 4.4 cases of OS are reported annually per million children [34]. The current standard treatment for OS involves radical tumor resection combined with neoadjuvant chemotherapy, specifically high-dose methotrexate (MTX), doxorubicin (DOX), ifosfamide (IFO), and cisplatin (CIS) [35]. Despite advancements in radiotherapy and immunotherapy, patients with OS still face a challenging prognosis due to the persistent risk of metastasis as indicated by the 5-year survival rates ranging from 60 to 70 % for localized OS [35]. Furthermore, patients who experience metastasis or recurrence after treatment continue to have a poor prognosis [36]. Therefore, it is imperative to explore novel therapeutic targets and develop innovative treatment strategies for OS.
Photodynamic therapy (PDT) is an emerging minimally invasive treatment modality expected to be utilized for OS treatment. The three fundamental components of PDT include a photosensitizer (PS), specific wavelength light, and oxygen [37], which need to be integrated. As the pivotal component among these, the photosensitizer plays a decisive role in determining therapeutic efficacy throughout the entire PDT process due to its distinctive physicochemical properties and biological effects. Importantly, subcellular localization of the photosensitizer holds paramount importance in the PDT procedure and specificity as it markedly influences treatment efficiency, cell death mechanisms, and potential side effects [38]. In this study, we observed that the photosensitizer MPPα predominantly localizes within the mitochondria, which are vital organelles responsible for ATP production and play crucial roles in cellular processes such as cell growth, death, division, and signal transduction. Consequently, we initially hypothesized that MPPα-PDT could induce damage to mitochondria of OS cells. Subsequent investigations revealed mitochondrial fragmentation, providing direct evidence of impairment caused by MPPα-PDT and serving as one of the initiating factors leading to cell death. Mitochondrial damage leads to depolarization of the inner membrane and subsequent dysfunction, resulting in the release of cytochrome c, mitochondrial DNA (mtDNA), and other contents. The electron transport chain on the inner membrane is a primary site for generating reactive oxygen species (ROS) within cells. When mitochondrial function is compromised, excessive ROS production exceeds the body's ability to clear them, activating specific signal transduction pathways that trigger cellular damage or other reactions [39]. This process involves multiple sequential steps and crucial protein interactions, including sensing and responding to cellular stress and injury signals, regulating the activity of bcl-2 proteins, inducing mitochondrial membrane permeabilization (MOMP), forming apoptotic bodies, activating the Caspase cascade, executing apoptosis, and clearing cellular debris [40,41]. The present study demonstrates that MPPα-PDT effectively triggers mitochondrial impairment, consequence causing apoptosis through the mitochondrial pathway. PARP is a nuclear enzyme crucial for recognizing and repairing DNA damage, thereby preserving genomic integrity and cell survival, especially in response to strong genotoxic stresses or therapeutic interventions like chemotherapy, radiation, or photodynamic therapy [42,43]. Caspase-3 and Caspase-7 act as primary effector proteases, cleaving various proteins involved in apoptosis, including PARP [44]. Under conditions that induce apoptosis, an increase in PARP levels can occur, driven by heightened DNA damage response and stress signaling preceding cell death [45,46]. Differences in apoptotic signaling pathways, p53 status, ROS response, and metabolic traits may explain the varying responses to apoptotic signals across different cell types [4,47,48]. These factors collectively contribute to distinct sensitivities towards apoptosis and observed response patterns in experimental settings.
Ferroptosis is a novel form of programmed cell death that distinguishes itself from other cell death in terms of morphology, genetics, and biochemistry, which is characterized by the accumulation of iron-dependent lipid peroxides and oxidative damage [49]. Our findings demonstrated that ferroptosis could be significantly induced by MPPα-PDT in OS cells, as evidenced by notable lipid peroxidation and iron overload. Additionally, the expression of proteins such as GPX4, SLC7A11, and TFR1 were also found to be altered. However, it is noteworthy that the inhibition of NCOA4, a crucial protein involved in ferritin degradation, was observed after MPPα-PDT treated. This indirectly suggests a limited degradation of ferritin, potentially leading to impaired ferritinophagy. Ferroptosis and apoptosis are distinct modes of cell death, however, they can influence cell fate in pathological conditions through complex interactions and cross-regulation of signaling pathways. Iron ions play a pivotal role in ferroptosis, as their excessive accumulation can induce the expression of the pro-apoptotic polyunsaturated fatty acids (PUMA) via the ERS-mediated PERK-eIF2α-ATF4-CHOP pathway, thereby triggering apoptotic signaling. Moreover, oxidative stress serves as a critical link between apoptosis and ferroptosis: ROS accumulation not only activates mitochondrial pathway-mediated apoptosis but also plays a key role in lipid peroxidation during ferroptosis [50,51]. In this study, we treated OS cells with a ferroptosis inhibitor in combination with MPPα-PDT. We discovered that inhibiting ferroptosis could restrain the pro-apoptotic effect mediated by MPPα-PDT. Therefore, the synergistic effect of ferroptosis and apoptosis reveals the complexity and dynamics in the regulation of cell death and offers a new perspective for understanding how cells respond to different death signals.
Autophagy is a highly conserved catabolic process that plays a crucial role in cellular waste clearance. It primarily facilitates the recycling or degradation of cellular components through lysosomes or vacuoles, thereby providing cells with energy and metabolites [52]. Excessive or dysfunctional autophagy can result in cell death, known as “autophagy-dependent cell death,” which represents a unique regulated cell death (RCD) [53]. Although the specific cascade signals remain to be elucidated, this process mainly involves the progressive depletion or degradation of cytoprotective proteins. In our previous research [11,54], we have demonstrated preliminary evidence that MPPα-PDT can effectively induce autophagy in MG-63 cells. Furthermore, targeting autophagy as a therapeutic approach significantly enhances the therapeutic efficacy of MPPα-PDT. In this study, we further observed that MPPα-PDT prominently stimulates autophagy in OS cells, establishing a close association between autophagy and ferroptosis.
Ferritinophagy is an autophagic process intimately associated with ferroptosis. Its activation is entirely dependent on NCOA4 and regulated by intracellular Fe2+ levels [55]. NCOA4 functions as a selective cargo receptor that plays a role in the autophagic degradation of ferritin [55]. Upon activation of ferritinophagy, NCOA4 binds to FTH1, MAP1, and LC3-PE on the autophagosomal membrane, sequestering the ferritin complex into the autophagosome [56]. This ultimately leads to the fusion of the autophagosome with the lysosome, resulting in the complete degradation of ferritin. In this study, we have demonstrated that ferroptosis induced by MPPα-PDT depends on autophagy and contributes to the anti-tumor mechanism. Additionally, we observed an upregulation of NCOA4 in transcription following MPPα-PDT treatment. However, there appears to be an inhibition or degradation of the NCOA4 protein, leading to limited ferritin degradation and suppressed ferroptosis. Studies have demonstrated that the expression of NCOA4 is regulated by various genes and influences the occurrence of ferritinophagy. Autophagy-related genes (ATG) can activate NCOA4, while overexpression of NCOA4 can enhance intracellular iron transport and ferritinophagy, thereby inducing ferroptosis [57,58]. Conversely, knockdown of NCOA4 or ATG (such as ATG3, ATG5, ATG7, and ATG13) significantly inhibits Erastin-induced ferritin degradation, iron accumulation, and lipid peroxidation [59]. Therefore, targeting ferritinophagy to promote ferritin degradation is of great significance in enhancing PDT-mediated ferroptosis. In this study, we employed lentiviral-packaged plasmids to overexpress the NCOA4 in combination with MPPα-PDT for a synergistic treatment approach to OS. The results revealed that the combined treatment markedly activated the process of ferritinophagy in OS cells, leading to enhanced ferritin degradation and considerable ferroptosis. Specifically, notable iron overload and lipid peroxidation were observed along with characteristic features indicative of ferroptosis within mitochondria including reduced size, decreased or absent mitochondrial cristae, and rupture on the outer membrane.
The ubiquitin-proteasome system (UPS) is a crucial post-translational modification mechanism responsible for degrading approximately 80 % of cellular proteins [60]. This intricate system comprises various components, including ubiquitin (Ub), E1 ubiquitin activase, E2 ubiquitin conjugase, E3 ubiquitin ligase, E4 ubiquitin chain elongation factor, 26S proteasome, and deubiquitinating enzyme (DUB). Its primary function is to eliminate misfolded or surplus proteins as well as damaged ones to maintain protein homeostasis. In this study, through a series of experiments, we have demonstrated that MPPα-PDT can effectively facilitate the ubiquitination and subsequent degradation of the NCOA4 protein in OS cells. As a result, this impacts the subsequent activation of ferritinophagy. Therefore, actively identifying key factors involved in NCOA4 degradation becomes an essential approach towards activation of ferritinophagy and ferroptosis.
The E3 ubiquitin ligase acts as a mediator in the UPS, transferring ubiquitin from the E2 conjugase to substrate proteins. It ensures the specificity of the ubiquitination process by interacting with specific sequences or domains of substrate proteins. This specific recognition mechanism enables selective labeling of target proteins within the cell, thereby regulating their stability and activity [61]. HERC1 is an E3 ubiquitin ligase that contains a HECT domain and has been well-established for over two decades. The HERC1 gene is located on the 15q22 locus of the human chromosome [62] and exhibits widespread expression across various mammalian tissues, with higher levels observed in the brain and testis, and minimal expression in the liver. Importantly, HERC1 demonstrates significant upregulation in diverse human tumor cell lines compared to those derived from normal fibroblasts [63]. Extensive research has revealed frequent mutations within the HERC family across different cancer types, including leukemia and solid tumors [30]. Mutations identified in HERC1 have underscored its pivotal role in oncogenesis among patients diagnosed with acute T-Cell Acute Lymphoblastic Leukemia (T-ALL) [64]. In this study, we initially screened the key protein HERC1 which potentially participates in the ubiquitination degradation of NCOA4, through transcriptome sequencing combined with bioinformatics analysis. Subsequently, a series of investigations substantiated the interaction between HERC1 and NCOA4. Moreover, by employing CRISPR/CAS9 technology to knock out the HERC1 gene, we observed a significant inhibition of NCOA4 ubiquitination and an enhancement in its stability. Finally, to comprehensively demonstrate the impact of targeting HERC1 on MPPα-PDT's efficacy, we combined HERC1 knockout with MPPα-PDT for OS treatment. The results unequivocally indicated that targeting HERC1 substantially augmented the therapeutic effect of MPPα-PDT, as evidenced by a notable increase in OS cell apoptosis and lipid peroxidation.
NRF2, a transcription factor and member of the leucine zipper family, is involved in transcribing genes related to antioxidant and detoxification responses [65]. While its role in maintaining redox homeostasis has been well established, it also plays a crucial role in mediating various metabolic pathways such as protein stability, exogenous drug metabolism, iron/heme metabolism, lipid metabolism, and programmed cell death. Dysregulation of the NRF2 signaling pathway contributes to tumor occurrence and progression [66,67]. Studies have shown that NRF2 exhibits pronounced oncogenic activity in certain tumors, specifically characterized by elevated NRF2 expression. This heightened expression is primarily attributed to somatic mutations occurring in KEAP1, thereby facilitating tumor progression, metastasis, and conferring resistance against anti-tumor therapies [68,69]. Additionally, clinical patients with elevated NRF2 expression demonstrate a poor long-term prognosis [70]. Therefore, targeting NRF2 holds promise as a potential approach for anti-tumor treatment. Furthermore, several studies have revealed that multiple proteins involved in ferroptosis are direct target genes of NRF2, such as SLC7A11 and the catalytic and modified subunit of GCLC/GCLM. These proteins synergistically interact to regulate the levels of GSH. In this study, we preliminarily confirmed through a series of experiments that HERC1 is a potential target gene of NRF2. The NRF2 signaling pathway is fully activated after MPPα-PDT treatment, subsequently regulating the downstream target gene HERC1. It has been demonstrated that HERC1 functions as an E3 ubiquitin ligase for NCOA4, participating in its ubiquitination and degradation process. Ultimately, by targeting HERC1, we successfully inhibited the ubiquitination and degradation of NCOA4 protein, leading to the activation of ferritinophagy and ferroptosis.
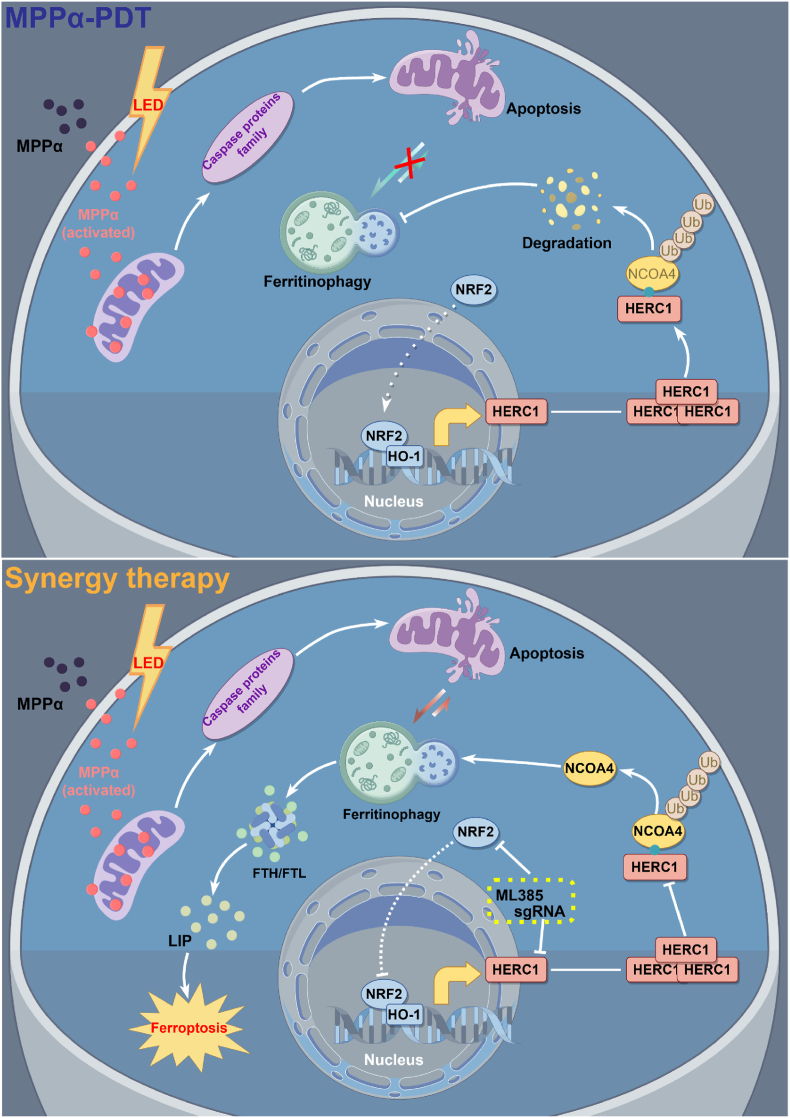
In conclusion, this study comprehensively elucidated the anti-tumor mechanism of MPPα-PDT through a series of investigations combined with transcriptomics, bioinformatics analysis, and in vivo tests. Furthermore, we proposed a novel approach titled “Apoptosis induced by MPPα-PDT combined with ferroptosis in osteosarcoma treatment” (Scheme 1). Additionally, we identified HERC1 as a crucial factor involved in the ubiquitination and degradation of NCOA4, along with the potential upstream signaling pathway NRF2 associated with HERC1. This provided a comprehensive demonstration of the signaling logic chain. Finally, by targeting the HERC1-NCOA4 axis, we successfully achieved complete activation of ferritinophagy after MPPα-PDT treatment, which greatly facilitated ferroptosis and enhanced the anti-tumor efficacy of MPPα-PDT.
Scheme 1.
Mechanism diagram for synergistically enhancing photodynamic sensitivity of osteosarcoma by targeting the HERC1-NCOA4 axis to induce ferroptosis.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82172682 and No. 82373221), Natural Science Foundation of Chongqing, China (No. CSTB2023NSCQ-MSX0472), First-class Discipline Construction Project of the First Affiliated Hospital of Chongqing Medical University (No. CYYY-BSYJSCXXM-202207, No. CYYY-BSYJSCXXM-202304).
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
CRediT authorship contribution statement
Ye Zhang: Writing – original draft, Visualization, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation. Yuxing Chen: Methodology, Formal analysis. Hai Mou: Resources, Project administration. Qiu Huang: Software, Resources. Changchun Jian: Writing – original draft, Software. Yong Tao: Visualization. Fuqiang Tan: Software. Yunsheng Ou: Funding acquisition, Conceptualization.
Declaration of competing interest
We declare that we have no financial or personal relationships with any individuals or organizations that could potentially influence our work. There are no professional or personal interests of any nature in any product, service, or company that could be perceived as exerting an influence on the content or the review of the manuscript entitle“Synergistic induction of ferroptosis by targeting HERC1-NCOA4 axis to enhance the photodynamic sensitivity of osteosarcoma”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103328.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Shoaib Z., Fan T.M., Irudayaraj J.M.K. Osteosarcoma mechanobiology and therapeutic targets. Br. J. Pharmacol. 2022;179:201–217. doi: 10.1111/bph.15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheng G., Gao Y., Yang Y., Wu H. Osteosarcoma and metastasis. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.780264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson M.E. Update on survival in osteosarcoma. Orthop. Clin. N. Am. 2016;47:283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Gill J., Gorlick R. Advancing therapy for osteosarcoma. Nat. Rev. Clin. Oncol. 2021;18:609–624. doi: 10.1038/s41571-021-00519-8. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., et al. Homologous targeting nanoparticles for enhanced PDT against osteosarcoma HOS cells and the related molecular mechanisms. J. Nanobiotechnol. 2022;20:83. doi: 10.1186/s12951-021-01201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H., et al. Targeting X box-binding protein-1 (XBP1) enhances the sensitivity of HOS osteosarcoma cells to pyropheophorbide- α methyl ester-mediated photodynamic therapy. Photodiagnosis Photodyn. Ther. 2022;37 doi: 10.1016/j.pdpdt.2021.102646. [DOI] [PubMed] [Google Scholar]
- 7.Zhan F., et al. RhoA enhances osteosarcoma resistance to MPPa-PDT via the Hippo/YAP signaling pathway. Cell Biosci. 2021;11:179. doi: 10.1186/s13578-021-00690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo Q., et al. Targeting GRP78 enhances the sensitivity of HOS osteosarcoma cells to pyropheophorbide-α methyl ester-mediated photodynamic therapy via the Wnt/β-catenin signaling pathway. Acta Biochim Biophys Sin (Shanghai) 2021;53:1387–1397. doi: 10.1093/abbs/gmab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji B., Wei M., Yang B. Recent advances in nanomedicines for photodynamic therapy (PDT)-driven cancer immunotherapy. Theranostics. 2022;12:434–458. doi: 10.7150/thno.67300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessel D., Reiners J.J. Photodynamic therapy: autophagy and mitophagy, apoptosis and paraptosis. Autophagy. 2020;16:2098–2101. doi: 10.1080/15548627.2020.1783823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q., Ou Y.S., Tao Y., Yin H., Tu P.H. Apoptosis and autophagy induced by pyropheophorbide-α methyl ester-mediated photodynamic therapy in human osteosarcoma MG-63 cells. Apoptosis. 2016;21:749–760. doi: 10.1007/s10495-016-1243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen C.E., et al. Development of resistance to photodynamic therapy (PDT) in human breast cancer cells is photosensitizer-dependent: possible mechanisms and approaches for overcoming PDT-resistance. Biochem. Pharmacol. 2017;144:63–77. doi: 10.1016/j.bcp.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon S.J., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z., et al. Oxygen-boosted biomimetic nanoplatform for synergetic phototherapy/ferroptosis activation and reversal of immune-suppressed tumor microenvironment. Biomaterials. 2022;290 doi: 10.1016/j.biomaterials.2022.121832. [DOI] [PubMed] [Google Scholar]
- 16.Jin X., et al. Ferritinophagy in the etiopathogenic mechanism of related diseases. J. Nutr. Biochem. 2023;117 doi: 10.1016/j.jnutbio.2023.109339. [DOI] [PubMed] [Google Scholar]
- 17.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santana-Codina N., Mancias J.D. The role of NCOA4-mediated ferritinophagy in health and disease. Pharmaceuticals (Basel) 2018;11 doi: 10.3390/ph11040114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology. 2017;66:449–465. doi: 10.1002/hep.29117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renault L., Kuhlmann J., Henkel A., Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 21.Pedrazza L., Schneider T., Bartrons R., Ventura F., Rosa J.L. The ubiquitin ligase HERC1 regulates cell migration via RAF-dependent regulation of MKK3/p38 signaling. Sci. Rep. 2020;10:824. doi: 10.1038/s41598-020-57756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utine G.E., et al. HERC1 mutations in idiopathic intellectual disability. Eur. J. Med. Genet. 2017;60:279–283. doi: 10.1016/j.ejmg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. A Versatile Chitosan-Based Hydrogel Accelerates Infected Wound Healing via Bacterial Elimination, Antioxidation, Immunoregulation, and Angiogenesis. Adv. Healthc Mater. 2024;13 doi: 10.1002/adhm.202400318. [DOI] [PubMed] [Google Scholar]
- 24.Zhong S., et al. Targeting PERK-ATF4-P21 axis enhances the sensitivity of osteosarcoma HOS cells to Mppα-PDT. Aging (Albany NY) 2024;16 doi: 10.18632/aging.205511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu P.H., et al. Induction of cell death by pyropheophorbide-α methyl ester-mediated photodynamic therapy in lung cancer A549 cells. Cancer Med. 2017;6:631–639. doi: 10.1002/cam4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R., et al. Molecular basis of V-ATPase inhibition by bafilomycin A1. Nat. Commun. 2021;12:1782. doi: 10.1038/s41467-021-22111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang R., Liu W. Identifying an essential role of nuclear LC3 for autophagy. Autophagy. 2015;11:852–853. doi: 10.1080/15548627.2015.1038016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R., et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell. 2015;57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Rossi F.A., et al. HERC1 regulates breast cancer cells migration and invasion. Cancers (Basel) 2021;13 doi: 10.3390/cancers13061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider T., et al. Large HERCs function as tumor suppressors. Front. Oncol. 2019;9:524. doi: 10.3389/fonc.2019.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Koppula P., Gan B. Regulation of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell Cycle. 2019;18:773–783. doi: 10.1080/15384101.2019.1597506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan Z., et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6:e371. doi: 10.1038/oncsis.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Refaat A., et al. In vivo fluorescence imaging: success in preclinical imaging paves the way for clinical applications. J. Nanobiotechnol. 2022;20:450. doi: 10.1186/s12951-022-01648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirabello L., Troisi R.J., Savage S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison D.C., et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012 doi: 10.1155/2012/704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues J.A., Correia J.H. Enhanced photodynamic therapy: a review of combined energy sources. Cells. 2022;11 doi: 10.3390/cells11243995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwiatkowski S., et al. Photodynamic therapy - mechanisms, photosensitizers and combinations. Biomed. Pharmacother. 2018;106:1098–1107. doi: 10.1016/j.biopha.2018.07.049. [DOI] [PubMed] [Google Scholar]
- 39.Hamanaka R.B., Chandel N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youle R.J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 41.Wei M.C., et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H., et al. PARP inhibitors in pancreatic cancer: molecular mechanisms and clinical applications. Mol. Cancer. 2020;19:49. doi: 10.1186/s12943-020-01167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zall H., Weber A., Besch R., Zantl N., Häcker G. Chemotherapeutic drugs sensitize human renal cell carcinoma cells to ABT-737 by a mechanism involving the Noxa-dependent inactivation of Mcl-1 or A1. Mol. Cancer. 2010;9:164. doi: 10.1186/1476-4598-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schossler Garcia C., et al. Effect of m-Trifluoromethyl-diphenyl diselenide on the pain-depression dyad induced by reserpine: insights on oxidative stress, apoptotic, and glucocorticoid receptor modulation. Mol. Neurobiol. 2021;58:5078–5089. doi: 10.1007/s12035-021-02483-x. [DOI] [PubMed] [Google Scholar]
- 45.Chaitanya G.V., Steven A.J., Babu P.P. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicoletti V.G., Stella A.M. Role of PARP under stress conditions: cell death or protection? Neurochem. Res. 2003;28:187–194. doi: 10.1023/a:1022316914492. [DOI] [PubMed] [Google Scholar]
- 47.Vousden K.H., Lane D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 48.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le J., et al. Targeting ferroptosis in gastric cancer: strategies and opportunities. Immunol. Rev. 2024;321:228–245. doi: 10.1111/imr.13280. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y.S., Lee D.H., Choudry H.A., Bartlett D.L., Lee Y.J. Ferroptosis-induced endoplasmic reticulum stress: cross-talk between ferroptosis and apoptosis. Mol. Cancer Res. 2018;16:1073–1076. doi: 10.1158/1541-7786.MCR-18-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh A.P., Klocke B.J., Ballestas M.E., Roth K.A. CHOP potentially co-operates with FOXO3a in neuronal cells to regulate PUMA and BIM expression in response to ER stress. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruyama T., et al. Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis. Nat. Struct. Mol. Biol. 2021;28:583–593. doi: 10.1038/s41594-021-00614-5. [DOI] [PubMed] [Google Scholar]
- 53.Galluzzi L., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y., et al. Antitumor effects and mechanisms of pyropheophorbide-α methyl ester-mediated photodynamic therapy on the human osteosarcoma cell line MG-63. Int. J. Mol. Med. 2020;45:971–982. doi: 10.3892/ijmm.2020.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mancias J.D., et al. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife. 2015;4 doi: 10.7554/eLife.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 57.Li J., et al. d-Borneol enhances cisplatin sensitivity via autophagy dependent EMT signaling and NCOA4-mediated ferritinophagy. Phytomedicine. 2022;106 doi: 10.1016/j.phymed.2022.154411. [DOI] [PubMed] [Google Scholar]
- 58.Nai A., et al. NCOA4-mediated ferritinophagy in macrophages is crucial to sustain erythropoiesis in mice. Haematologica. 2021;106:795–805. doi: 10.3324/haematol.2019.241232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou W., et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z., Li H., Zhu J., Wang H., Jin X. The roles of E3 ligases in Hepatocellular carcinoma. Am. J. Cancer Res. 2022;12:1179–1214. [PMC free article] [PubMed] [Google Scholar]
- 61.Sosič I., Bricelj A., Steinebach C. E3 ligase ligand chemistries: from building blocks to protein degraders. Chem. Soc. Rev. 2022;51:3487–3534. doi: 10.1039/d2cs00148a. [DOI] [PubMed] [Google Scholar]
- 62.Cruz C., et al. Assignment of the human P532 gene (HERC1) to chromosome 15q22 by fluorescence in situ hybridization. Cytogenet. Cell Genet. 1999;86:68–69. doi: 10.1159/000015414. [DOI] [PubMed] [Google Scholar]
- 63.Sánchez-Tena S., Cubillos-Rojas M., Schneider T., Rosa J.L. Functional and pathological relevance of HERC family proteins: a decade later. Cell. Mol. Life Sci. 2016;73:1955–1968. doi: 10.1007/s00018-016-2139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neumann M., et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–2766. doi: 10.18632/oncotarget.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J., Roh J.L. Targeting Nrf2 for ferroptosis-based therapy: implications for overcoming ferroptosis evasion and therapy resistance in cancer. Biochim. Biophys. Acta, Mol. Basis Dis. 2023;1869 doi: 10.1016/j.bbadis.2023.166788. [DOI] [PubMed] [Google Scholar]
- 66.Dodson M., et al. Modulating NRF2 in disease: timing is everything. Annu. Rev. Pharmacol. Toxicol. 2019;59:555–575. doi: 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dodson M., Castro-Portuguez R., Zhang D.D. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23 doi: 10.1016/j.redox.2019.101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denicola G.M., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H., et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl. Med. 2016;8:334ra51. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- 70.Kitamura H., Motohashi H. NRF2 addiction in cancer cells. Cancer Sci. 2018;109:900–911. doi: 10.1111/cas.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Data will be made available on request.