Abstract
The Multispecies Ovary Tissue Histology Electronic Repository (MOTHER) is a publicly accessible repository of ovary histology images. MOTHER includes hundreds of images from nonhuman primates, as well as ovary histology images from an expanding range of other species. Along with an image, MOTHER provides metadata about the image, and for selected species, follicle identification annotations. Ongoing work includes assisting scientists with contributing their histology images, creation of manual and automated (via machine learning) processing pipelines to identify and count ovarian follicles in different stages of development, and the incorporation of that data into the MOTHER database (MOTHER-DB). MOTHER will be a critical data repository storing and disseminating high-value histology images that are essential for research into ovarian function, fertility, and intra-species variability.
Keywords: ovary morphology, ovary histology, microscopy, follicle identification, oocyte, database, machine learning
The Multispecies Ovary Tissue Histology Electronic Repository (MOTHER) is a public database of nonhuman ovary histology images and associated metadata that is useful for meta-analyses, computational modeling, and education.
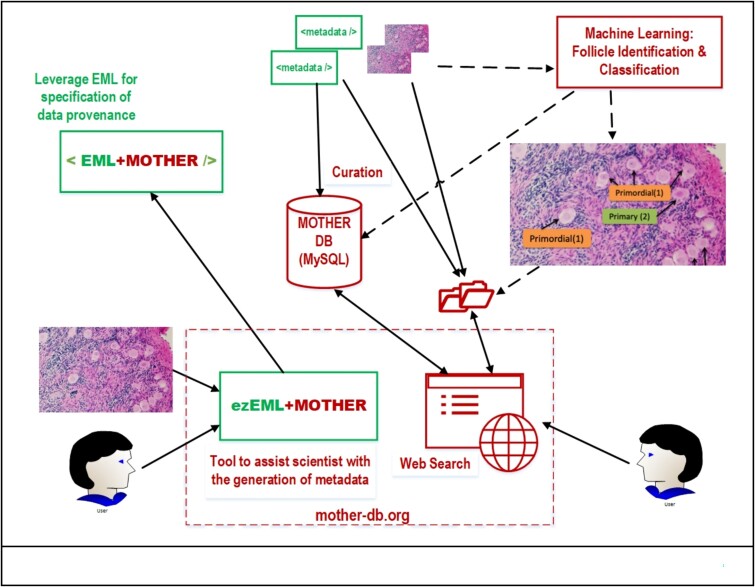
Graphical Abstract
Graphical Abstract.
The Multispecies Ovary Tissue Histology Electronic Repository (MOTHER) is a digital resource for sharing ovary histology image data from a wide range of species. MOTHER supports the archiving, documentation, analysis, dissemination, and reuse of valuable histology data currently stored as physical slides on a shelf, or as individual images on a researcher’s computer. Research publications often only publish a few of the samples that were collected while most samples remain unshared and underutilized. By sharing these histology images in MOTHER [1], cross-species comparisons and meta-analyses will be possible. MOTHER increases the value and impact of these expensive data resources and will enable future studies to promote, protect or control fertility, provide data for the development of predictive computational and machine learning (ML) models, and serve as a biological and educational resource.
Providing high quality image data and metadata is key, and MOTHER complies with the FAIR (Findable, Accessible, Interoperable, Reusable) guiding principles for scientific data management and stewardship [2]. Metadata, which is data about the histology images, are collected with each slide and include information about: the project and personnel; investigator-selected licensing options; the animal donor species and its reproductive stage; image data such as the microscope used, magnification, section thickness, and staining; and if appropriate, e.g. for field-collected specimens, sampling location and date. In cases where histology slides are sent to the MOTHER team for scanning to create a digital image, a workflow log is used to track the slide and its image from the time we receive the slide, through scanning and quality control checks [3]. This ensures that the publicly available image can be traced back to a tissue section on a given slide derived from an identified donor animal. A guiding goal for MOTHER is to maximize the value and re-use of ovary histology images so they will not be lost when an investigator or team’s focus changes.
MOTHER leverages existing infrastructure and technology to develop an adaptable framework for sharing histology images and related metadata in a publicly available searchable database. Specifically, the project leverages the Ecological Metadata Language [4] for collecting metadata that ensures data provenance [5], and includes a purpose-built metadata component to define the structure of the metadata. We developed a Web-based tool, ezEML+MOTHER [1], which extends ezEML [6] to help collaborating scientists share their images and associated metadata. MOTHER curates the submissions, which may require contacting the contributor if there are any issues, before including the image and metadata as part of MOTHER’s collection. MOTHER’s metadata database is searchable from the Web and includes filters to find an image(s) that can then be downloaded along with its metadata. Figure 1 shows an overview of the MOTHER pipeline from submission through curation, search, and retrieval.
Figure 1.
Overview of the MOTHER data flow.
The MOTHER project contributed over 300 macaque histology slides for three species: rhesus (Macaca mulatta), Japanese (Macaca fuscata), and cynomolgus (Macaca fascicularis) provided by Dr Zelinski [7]. The images are of ovary tissue cross-sections. Life stages span fetal to adult (post-menopausal) collected over 10 years of research. MOTHER continues to build its collection with histology images from collaborators who study rodents, fish, and birds.
Written project protocols [3, 8, 9] are used to train students to create digital images of ovary histology slides (slide scanning) and to manually segment macaque ovary histology slides (follicle identification and annotation). The training protocols include step-by-step instructions and are available through our Open Science Framework project [10] and the MOTHER web site [1].
We have begun to analyze the nonhuman primate slides using a combination of manual (human) annotators and ML. Training data for the ML algorithms must be accurate and comprehensive in coverage across a variety of histology images. Thus, we developed tools (e.g. a user-friendly Python-based Jupyter notebook) to assist with generating consensus annotations that require at least two human annotators’ versions for each image. To date, we have fully annotated 18 histology images from nonhuman primates including more than 8000 identified follicles. The final consensus annotations will, in the future, be available as additional metadata for the respective slides in MOTHER.
The images and annotations in MOTHER will be a resource for developing ML tools that can identify and classify follicles in histology images, which is an emerging technology of importance in ovarian biology [11, 12]. We have developed a preliminary ML algorithm using transfer learning that can accurately classify macaque follicles into one of six classes ranging from primordial to multilayer follicles. As training data, we used the 18 manually annotated macaque histology images described above. We are developing an automated data segmentation pipeline to generate follicle counts by type for primates, and ultimately, we plan to include an automatic data segmentation process that will annotate and count follicles in submitted images. A significant challenge, and scientific opportunity, is determining the extent to which an ML algorithm that identifies ovarian follicles in different stages of development trained on nonhuman primate data is transferable across species, especially across significantly different species ranging from rodents to non-mammalian species. Innovation in this area will save countless hours of labor-intensive follicle identifications. We are excited to explore the feasibility of using the same ML implementation to identify not only the follicle types in primates, but to also be able to accurately classify and provide follicle-type counts across a variety of species available in MOTHER.
MOTHER is a growing, publicly accessible, and supported resource to collect, archive, and analyze ovarian histology slides from a wide range of species. MOTHER will facilitate the sharing and reuse of ovary histology data for broader applications such as comparative analyses across species, development of predictive, biologically based models, and as an educational resource. Recently, the MOTHER team launched a user-friendly search tool that will allow biologists, mathematical/computational model developers, and educators to find and retrieve the histology images, annotations and any automated analysis results. This will facilitate analysis across species and make contributors’ images widely accessible. We are working with multiple collaborators who are contributing ovary histology images for a variety of species of nonhuman primates, rodents, exotic species, birds, reptiles, fish, and mollusks. Furthermore, we invite scientists to share their ovary histology slides in MOTHER as a permanent data repository to enable its reuse and to enhance education, thereby maximizing the impact of their research.
Acknowledgment
We thank the students and collaborators that have contributed to the MOTHER project.
Conflict of Interest: The authors have declared that no conflict of interest exists.
Footnotes
† Grant Support: Funded by the National Science Foundation award # DBI-2054061; P51 OD011092 (DPCPSI, ORIP, NIH to Oregon National Primate Research Center).
Contributor Information
Karen H Watanabe, School of Mathematical and Natural Sciences, Arizona State University, Glendale, Arizona, United States.
Suzanne W Dietrich, School of Mathematical and Natural Sciences, Arizona State University, Glendale, Arizona, United States.
Yian Ding, School of Mathematical and Natural Sciences, Arizona State University, Glendale, Arizona, United States.
Wenli Ma, School of Mathematical and Natural Sciences, Arizona State University, Glendale, Arizona, United States.
James P Sluka, Biocomplexity Institute, Indiana University, Bloomington, Indiana, United States.
Mary B Zelinski, Oregon National Primate Research Center, Beaverton, Oregon, United States.
Data availability
The data underlying this article are available in the Multispecies Ovary Tissue Histology Electronic Repository accessible at https://mother-db.org.
Author contributions
KHW led the manuscript preparation. She supervised the slide scanning and follicle annotation components of the MOTHER project, engaged collaborators to contribute slides, and contributed to the development of the metadata requirements and machine learning algorithm.
SWD contributed to the manuscript preparation, including the creation of the figures. She is the computational lead of the MOTHER project, responsible for the database, metadata design, metadata tool, and curation, as well as the supervision of the students who contributed to the computational products for the project.
YD reviewed the manuscript. She assisted with and trained students to scan ovary histology slides, performed follicle identifications to create training data sets for the machine learning algorithms, and contributed to a machine learning algorithm for the identification of antral follicles. She established the initial auto-curation pipeline for validating metadata, building upon Wenli Ma’s collection of Python programs.
WM reviewed the manuscript. She contributed to the design and implementation of the database for storing metadata and Python programs for validating the metadata. She was also a metadata provider for the macaque donor animals from Mary Zelinski’s lab.
JPS contributed to the manuscript preparation. He led the machine learning work on image segmentation and follicle identification, and contributed to the metadata specification.
MBZ contributed to the manuscript preparation, donated macaque slides for imaging and assisted with creating metadata, trained students in follicle identification for various projects including machine learning, and enlisted collaborators.
Funding
The MOTHER project is supported by the National Science Foundation award #DBI-2054061, “CIBR Multispecies Ovary Tissue Histology Electronic Repository (MOTHER)”. The macaque ovary histology slides were funded by award #P51 OD011092 from the National Institutes of Health, Division of Program Coordination, Planning, and Strategic Initiatives (DPCPSI), Office of Research Infrastructure Programs (ORIP) to the Oregon National Primate Research Center. Disclaimer: Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
References
- 1. Dietrich SW, Sluka J, Zelinski MB, Watanabe KH. Multispecies Ovary Tissue Histology Electronic Repository. Phoenix, AZ, Arizona State University; 2024. https://mother-db.org/: Accessed 7 June 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva SantosLB, Bourne PE, Bouwman J, Brookes AJ, et al. Comment: the FAIR guiding principles for scientific data management and stewardship. Sci Data 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding Y, Shah G, Ma W, Chu T-Y, Zelinski MB, Watanabe KH. Multispecies Ovary Tissue Histology Electronic Repository (MOTHER) Slide Scanning Protocol. Geneva, Switzerland, Zenodo; 2024. https://doi.org/ 10.5281/zenodo.10636869 Accessed 15 January 2023. [DOI] [Google Scholar]
- 4. Jones MB, O’Brien M, Mecum B, Boettiger C, Schildhauer M, Maier M, Whiteaker T, Earl S, Chong S. Ecological Metadata Language version 2.2.0. KNB Data Repository. 2019. https://eml.ecoinformatics.org: Accessed 13 August 2020.
- 5. Wittner R, Holub P, Mascia C, Frexia F, Müller H, Plass M, Allocca C, Betsou F, Burdett T, Cancio I, Chapman A, Chapman M, et al. Toward a common standard for data and specimen provenance in life sciences. Learn Health Syst 2024; 8:e10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vanderbilt K, Ide J, Gries C, Grossman-Clarke S, Hanson P, O'Brien M, Servilla M, Smith C, Waide R, Zollo-Venecek K. Publishing ecological data in a repository: an easy workflow for everyone. Bull Ecol Soc Am 2022; 103:e2018. [Google Scholar]
- 7. Multispecies Ovary Tissue Histology Electronic Repository (MOTHER) . Zelinski Lab: Macaque Ovary Datasets. In: ASU Library Research Data Repository; 2022: https://dataverse.asu.edu/dataverse/MOTHER. [Google Scholar]
- 8. Sluka J, Watanabe K, Ding Y. MOTHER Ovarian Follicle Annotation using QuPath. Bloomington, IN, IUScholar Works, Indiana University; 2023. https://hdl.handle.net/2022/29016: Accessed 3 July 2023. [Google Scholar]
- 9. Sluka J, Watanabe K, Ding Y, Zelinski M, Dietrich S. MOTHER Step-by-Step Supplementary Files. Bloomington, IN, IUScholar Works, Indiana University; 2023. https://hdl.handle.net/2022/29015: Accessed 3 July 2023. [Google Scholar]
- 10. Dietrich S, Watanabe KH, Sluka JP, Zelinski MB. MOTHER: Multispecies Ovary Tissue Histology Electronic Repository. In Open Science Framework (OSF.io). 2022: 10.17605/OSF.IO/W8D65. [DOI] [PMC free article] [PubMed]
- 11. Blevins GM, Flanagan CL, Kallakuri SS, Meyer OM, Nimmagadda L, Hatch JD, Shea SA, Padmanabhan V, Shikanov A. Quantification of follicles in human ovarian tissue using image processing software and trained artificial intelligence. Biol Reprod 2024; 110:1086–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. İnik Ö, Ceyhan A, Balcıoğlu E, Ülker E. A new method for automatic counting of ovarian follicles on whole slide histological images based on convolutional neural network. Comput Biol Med 2019; 112:103350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the Multispecies Ovary Tissue Histology Electronic Repository accessible at https://mother-db.org.