Abstract
The objective of this study was to identify the active form of the feline calicivirus (FCV) RNA-dependent RNA polymerase (RdRP). Multiple active forms of the FCV RdRP were identified. The most active enzyme was the full-length proteinase-polymerase (Pro-Pol) precursor protein, corresponding to amino acids 1072 to 1763 of the FCV polyprotein encoded by open reading frame 1 of the genome. Deletion of 163 amino acids from the amino terminus of Pro-Pol (the Val-1235 amino terminus) caused a threefold reduction in polymerase activity. Deletion of an additional one (the Thr-1236 amino terminus) or two (the Ala-1237 amino terminus) amino acids produced derivatives that were 7- and 175-fold, respectively, less active than Pro-Pol. FCV proteinase-dependent processing of Pro-Pol in the interdomain region preceding Val-1235 was not observed in the presence of a catalytically active proteinase; however, processing within the polymerase domain was observed. Inactivation of proteinase activity by changing the catalytic cysteine-1193 to glycine permitted the production and purification of intact Pro-Pol. Biochemical analysis of Pro-Pol showed that this enzyme has properties expected of a replicative polymerase, suggesting that Pro-Pol is an active form of the FCV RdRP.
The family Caliciviridae, positive-strand RNA viruses, comprises four genera: Vesivirus, Lagovirus, “Norwalk-like viruses,” and “Sapporo-like viruses.” The last two genera contain human pathogens associated with acute gastroenteritis (5, 8, 15). As a result of the development of molecular tools to identify human caliciviruses, it has become clear that these viruses are responsible for the vast majority of food-borne and waterborne outbreaks of viral gastroenteritis in many communities worldwide (8). The development of strategies to treat human calicivirus infection is compromised by the absence of tissue or organ culture systems and animal models to study the multiplication of the viruses.
In order to understand the molecular mechanisms governing genome replication of viruses in this family and to identify host factors essential for virus multiplication, we have focused on the study of feline calicivirus (FCV), a member of the genus Vesivirus. This virus grows well in cell culture, and an infectious molecular clone exists that can be used to study the virus life cycle in cell culture (21). The FCV RNA genome is 7.7 kb in length. The genome has a protein (VPg) linked to the 5′ end and is polyadenylated at the 3′ end (13a). The genome includes three open reading frames (ORFs). ORF1 encodes nonstructural proteins, some of which are homologous to picornavirus nonstructural proteins (24). For example, 2C-like NTPase, 3C-like proteinase, and 3D-like polymerase motifs have been identified. ORF2 encodes the capsid protein. ORF3 encodes a protein of unknown function, but recent studies suggest that this protein is a minor component of the virion (13, 23). ORF2- and ORF3-encoded proteins are produced by translation of a subgenomic RNA. This RNA is also VPg linked and polyadenylated (13a).
Central to the genome replication process is the RNA-dependent RNA polymerase (RdRP). The only calicivirus RdRP that has been purified and characterized to date is the rabbit hemorrhagic disease virus (RHDV) enzyme (26). In this system, the active polymerase is thought to be a 58-kDa protein derived from processing of the ORF1 polyprotein between the proteinase and polymerase domains by the viral proteinase (26). Our present study was designed to identify the site of processing within the FCV ORF1 polyprotein used by the viral proteinase to produce the active form of the FCV polymerase and to characterize the biochemical properties of this enzyme. In contrast to the RHDV system, we find that the active form of the FCV polymerase is the bifunctional proteinase-polymerase (Pro-Pol) protein. Processing between these domains is not required for RdRP activity. Biochemical data support the conclusion that Pro-Pol is the active form of the FCV RdRP. We discuss the possibility that other calicivirus polymerases may exist as a bifunctional polypeptide.
MATERIALS AND METHODS
Materials.
[α-32P]UTP (>6,000 Ci/mmol) and [α-32P]GTP (>3,000 Ci/mmol) were from NEN Life Science Products; [γ-32P]ATP (>7,000 Ci/mmol) was from ICN. RNA oligonucleotides were from Dharmacon Research; heparin 6000 and poly(rC) were from Sigma; nucleoside 5′-triphosphates, poly(rA), and Q-Sepharose were from Amersham Pharmacia Biotech; 2.5-cm-diameter DE81 filter paper disks and phosphocellulose (P11) resin were from Whatman; polyethyleneimine (PEI)-cellulose thin-layer chromatography (TLC) plates were from EM Science; Ecoscint scintillation fluid was from National Diagnostics; T4 polynucleotide kinase and Deep Vent DNA polymerase were from New England Biolabs; a single-stranded 10-nucleotide (nt) ladder was from Life Technologies; and Ni-nitrilotriacetic acid (NTA) agarose resin was from Qiagen. All other reagents were of the highest grade available from Fisher or Sigma.
Construction of expression vectors.
Expression vectors were prepared by using standard recombinant DNA methods as described previously (9). Briefly, PCR was used to amplify polymerase-coding sequence. Forward primers were designed such that each primer fused coding sequence for the carboxy-terminal three amino acids of ubiquitin to coding sequence for polymerase derivatives containing different amino termini: Val-1235, Thr-1236, or Ala-1237 (Table 1, oligonucleotides 1 to 3). These sequences were located downstream of a SacII site that could be used for cloning. The reverse primer (Table 1, oligonucleotide 4) encoded the carboxy-terminal residues of the polymerase domain followed by (i) a Gly-Ser-Ser-Gly linker, (ii) a hexahistidine tag, (iii) stop codons, and (iv) a SacI site for cloning. The infectious FCV cDNA clone (pQ14) was used as a template (21). Gel-purified PCR products were digested with SacII and SacI and cloned into pET26-Ub (9) to yield pET26-Ub-FCV-PolVal-1235-his, pET26-Ub-FCV-PolThr-1236-his, and pET26-Ub-FCV-Pol-Ala-1237-his. Vectors directing the expression of full-length Pro-Pol derivatives were also constructed. These vectors were prepared as described above. The forward primer (Table 1, oligonucleotide 5) has the standard design for fusion to ubiquitin, and the coding sequence begins with Ser-1072 of FCV ORF1, the amino terminus of the proteinase domain (24). In addition, an oligonucleotide encoding an authentic carboxyl terminus was also designed (Table 1, oligonucleotide 6). The following constructs were prepared: pET26-Ub-FCV-Pro-Pol-his, pET26-Ub-FCV-ProM-Pol-his, and pET26-Ub-FCV-ProM-Pol. The designation “ProM” refers to proteinase-coding sequence in which the codon encoding catalytic Cys-1193 was changed to one encoding a glycine, thereby producing an inactive proteinase. This particular mutation and the corresponding template used for PCR has been described previously (24).
TABLE 1.
Oligonucleotides used in this study
| No. | Name | Sequence |
|---|---|---|
| 1 | FCV-3D-V1235-SacII-for | GCGGTCGACCCGCGGTGGAGTTACTGCTCAAAAATAT |
| 2 | FCV-3D-T1236-SacII-for | GCGGTCGACCCGCGGTGGAACTGCTCAAAAATATGAT |
| 3 | FCV-3D-A1237-SacII-for | GCGGTCGACCCGCGGTGGAGCTCAAAAATATGATGTAACC |
| 4 | FCV-3CD-his-SacI-rev | GCGAAGCTTGAGCTCTTACTAATGGTGGTGATGGTGGTGACCGGAAGAACCAACTTCGAACACATC |
| 5 | FCV-3C-SacII-for3 | GCGGTCGACCCGCGGTGGATCTGGACCCGGCACCAAATTTC |
| 6 | FCV-3D-SacI-rev | GCGAAGCTTGAGCTCTTATCAAACTTCGAACACATC |
Expression and purification of histidine-tagged polymerase derivatives.
To express each His-tagged polymerase derivative, the appropriate plasmid was transformed into Escherichia coli strain BL21(DE3)(pCG1) (9) and grown at 37°C to an A600 of 0.8 to 2 in NZCYM medium that contained 25 μg of kanamycin/ml (K25) and 20 μg of chloramphenicol/ml (C20). The cultures were cooled to 25°C, and expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 500 μM. Cells were harvested by centrifugation after 4 h. The harvested cells were suspended in lysis buffer (100 mM potassium phosphate) [pH 8.0] 20% glycerol, 0.5 mM EDTA, 1 mM 2-mercaptoethanol, 5.6 μg of pepstatin A/ml, 4.0 μg of leupeptin/ml) at 4 ml/g of cells. The suspended cells were lysed by passing them through a French press. Phenylmethylsulfonyl fluoride was added to the cell extract to a final concentration of 2 mM, and nucleic acid was precipitated by the addition of PEI to a final concentration of 0.25% (vol/vol). The extract was stirred slowly at 4°C for 20 min and then centrifuged at 100,000 × g for 35 min. After the PEI supernatant was decanted, solid ammonium sulfate was added slowly to 60% saturation and stirred for 20 min at 4°C. The ammonium sulfate suspension was centrifuged at 100,000 × g for 35 min. The supernatant was decanted, and the pellet was suspended in buffer A (50 mM Tris [pH 8.0], 20% glycerol, 1 mM 2-mercaptoethanol, 0.1% NP-40, and 60 μM ZnCl2) to a final salt concentration of 35 mM based upon the conductivity of the sample.
The suspended sample was loaded onto a 1-mL Ni-NTA column at a flow rate of 0.10 ml/min. The column was washed to baseline with buffer A containing 50 mM NaCl. Protein was eluted with 3 column volumes each of 5, 50, and 500 mM imidazole. The 50 and 500 mM fractions were pooled and diluted in buffer A to a final salt concentration of 35 mM.
The Ni-NTA pool was loaded onto a 1-ml phosphocellulose column at a flow rate of 0.12 ml/min. The passthrough was collected and loaded onto a 0.5-ml Q-Sepharose column at a flow rate of 0.12 ml/min. The column was washed to baseline with 50 mM NaCl in buffer B (50 mM HEPES [pH 7.5], 20% glycerol, 0.1% NP-40, 60 μM ZnCl2, and 1 mM dithiothreitol). The His-tagged polymerase derivatives were eluted from the column by using buffer B containing 500 mM NaCl. The purity of the eluted fractions was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The concentration of the final pool was determined by absorbance at 280 nm in phosphate-buffered (25 mM; pH 7.0) guanidine (6 M) using a calculated extinction coefficient, 61,020 M−1cm−1, for all polymerase derivatives (7).
ProM-Pol-His was expressed and purified as described above with the following modifications. Ammonium sulfate was added slowly to 40% saturation. The protein was eluted from the Q-Sepharose column by using buffer B containing 2 M NaCl. The calculated extinction coefficient used for all Pro-Pol enzymes was 78,800 M−1cm−1 (7).
Expression and purification of ProM-Pol.
Expression and purification of ProM-Pol was performed as described above for the ProM-Pol-His enzyme through the ammonium sulfate precipitation step. The ammonium sulfate-precipitated protein was suspended in buffer A and dialyzed against 10 mM NaCl overnight. The dialyzed sample was adjusted to 50 mM NaCl and loaded onto a 25-ml phosphocellulose column at a flow rate of 1 ml/min. The column was washed to baseline with buffer A containing 50 mM NaCl. Protein was eluted with a linear gradient (6 column volumes) from 50 to 400 mM NaCl in buffer A, and 2.5-ml fractions were collected. The purity of the eluted fractions was assessed by SDS-PAGE.
The pool from the phosphocellulose column was adjusted to 50 mM NaCl and loaded onto a Q-Sepharose column. The subsequent steps were identical to those for the phosphocellulose column.
RdRP activity assays.
RdRP assays were performed in a reaction mixture of 50 mM HEPES buffer (pH 7.5), 10 mM 2-mercaptoethanol, 5 mM MgCl2, 60 μM ZnCl2, 0.2 μCi of α-32P-nucleoside triphosphate (NTP)/μl, and 500 μM NTP. The concentrations of primers and templates used, along with any deviations from the above mentioned reaction conditions, are listed in the appropriate figure legends. The reactions were quenched by the addition of EDTA to a final concentration of 250 mM, unless otherwise specified, and spotted onto DE81 filter paper disks. The DE81 disks were dried completely and then washed in 5% dibasic sodium phosphate for 2 min followed by two 5-min washes. The disks were then rinsed in absolute ethanol. Bound radioactivity was quantitated by liquid scintillation counting in 5 ml of Ecoscint scintillation fluid.
Purity of α-32P-NTPs.
Dilutions (0.1 μCi/μl) of α-32P-NTPs were made in distilled deionized water, and 1 μl was spotted in triplicate onto PE1-cellulose TLC plates. The TLC plates were developed in 0.3 M potassium phosphate buffer, pH 7.0. The plates were dried and exposed to a PhosphorImager screen and viewed and quantitated by using ImageQuant software from Molecular Dynamics. The purity served as a correction factor for the specific activity of α-32P-NTPs used in reactions in order to accurately calculate concentrations of products.
PAGE.
The quenched reaction mixtures were mixed with equal volumes of loading buffer (80% formamide, 100 mM EDTA, 50 mM Tris-borate, 0.15% xylene cyanol, and 0.15% bromophenol blue). Samples were heated at 65°C for 3 min before being loaded on a 1× Tris-borate-EDTA denaturing 10% polyacrylamide gel. A single-stranded 10-nt ladder was also loaded onto the gel. The DNA ladder was labeled by using [γ-32P]ATP and T4 polynucleotide kinase as specified by Life Technologies, Inc. Electrophoresis was carried out in 1× Tris-borate-EDTA at 90 W. Resolved products were visualized by using a PhosphorImager.
Construction of transcription plasmid.
Plasmid pGLT7 was constructed by subcloning the 732-bp NotI fragment of plasmid pGreenLantern-1 (Gibco BRL) into NotI-digested pSPORT1 vector (Gibco BRL). The resulting plasmid contained the green fluorescent protein gene under control of the T7 RNA polymerase promoter.
RNA synthesis.
Plasmid pGLT7 was utilized to prepare heteropolymeric RNA template. The 830-nt-long RNA transcript (GLT7 RNA) was produced from MluI-linearized plasmid by using Ribomax, the large-scale RNA production system from Promega. RNA was analyzed by electrophoresis on 1% agarose gels containing formaldehyde, as described by Sambrook et al. (19).
RNase T1 digestion.
The product synthesized by FCV ProM-Pol by using GLT7 RNA as a template was treated with 5 U of RNase T1 (Gibco BRL) for 1 h at 37°C.
Probe preparation.
5′ 32P-end-labeled RNA was used as a probe for Northern blot analysis. To prepare the RNA probe, 25 pmol of GLT7 RNA was dephosphorylated by using 0.25 U of calf intestinal alkaline phosphatase (Gibco BRL) for 10 min at 37°C. The reaction was stopped by heating the mixture to 70°C for 10 min, and RNA was extracted with phenol and then precipitated with ethanol. For radiolabeling of the RNA 5′ end, the RNA pellet was dissolved in 50 μl of buffer containing 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 100 mM KCl, 1 mM β-mercaptoethanol, and 100 μCi of [γ-32P]ATP (5,000 Ci/mmol; Amersham) and incubated for 15 min at 37°C with 10 U of T4 polynucleotide kinase (Gibco BRL). The reaction was stopped by heat inactivation (10 min at 70°C), and the reaction mixture was extracted twice with phenol prior to ethanol precipitation of RNA.
Northern blot analysis.
Northern blot analysis was performed under conditions similar to those described previously (19). Briefly, RNA was separated on a formaldehyde denaturing 1% agarose gel, transferred to nylon membrane (Schleicher & Schuell), and fixed by UV cross-linking (Stratagene). The blot was hybridized with 32P-labeled RNA probe, and the hybridization conditions were 6× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.1% SDS, and 100 μg of sheared DNA per ml at 65°C overnight. The blot was washed in 2× SSC–0.1% SDS and exposed to film.
RESULTS
Construction, expression, purification, and characterization of FCV polymerase derivatives with different amino termini.
The calicivirus RdRP belongs to the supergroup I family of RdRPs, and this supergroup also includes picornavirus RdRPs, such as poliovirus (PV) 3Dpol. In the case of PV polymerase, an authentic amino terminus is essential for maximal catalytic activity of the enzyme (9), and precursor forms of the polymerase, such as the Pro-Pol precursor (3CDpro), lack polymerase activity (12). Therefore, it seemed reasonable that processing of the FCV Pro-Pol precursor would be required to release the active form of the viral polymerase. However, inspection of the amino acid sequence between defined regions of the FCV proteinase and polymerase domains failed to reveal any obvious cleavage site—that is, an EG, EA, or ET dipeptide that conforms to known calicivirus proteinase cleavage sites (Fig. 1) (6). Furthermore, previous proteolytic mapping studies of the FCV ORF1-encoded polyprotein failed to identify an efficient processing site in this region (24).
FIG. 1.
Comparison of the Pro-Pol interdomain linker of FCV to those from RHDV, SV, Norwalk virus (NV), and Manchester virus (MV). The sequence alignment was performed with ClustalW; conserved residues are denoted by asterisks. Established proteinase and polymerase domains, based upon conserved sequence motifs, are labeled. Processing sites reported for RHDV (25) and SV (18), as well as residues evaluated as the amino terminus of the FCV polymerase domain, are shown in boldface type. The position of each sequence in the ORF1-encoded polyprotein is indicated. The GenBank accession numbers are as follows: FCV, L40021; RHDV, M67473; SV, L07418; NV, M87661; and MV, X86560.
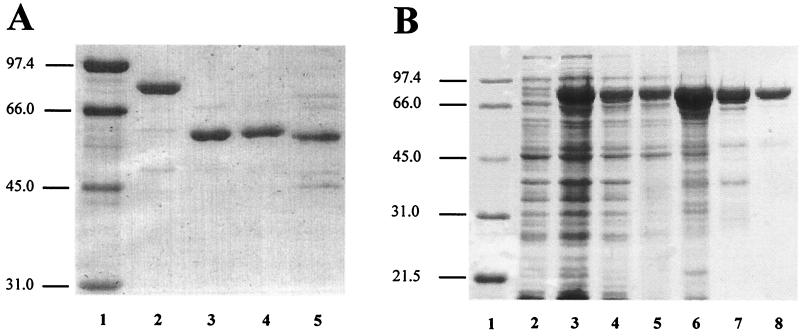
In order to identify the amino terminus of FCV polymerase, we compared the FCV Pro-Pol interdomain linker to those from RHDV and Southampton virus (SV). This analysis helped to further delimit the region in which the cleavage site might be located (Fig. 1). We set out to construct a panel of polymerase derivatives beginning with Val-1235, a residue conserved among all calicivirus polymerases and located prior to the established amino termini of RHDV and SV polymerases (Fig. 1), and continuing inwards until the most active polymerase was produced. The initial set of constructs produced polymerase derivatives with the following amino termini: Val-1235, Thr-1236, and Ala-1237. The derivatives were produced in E. coli by using a pET expression system to produce a ubiquitin-polymerase fusion protein containing a hexahistidine tag on the carboxy-terminal end of the polymerase domain. Only polymerase with the desired amino terminus accumulated intracellularly because the ubiquitin monomer was removed from the fusion protein by a ubiquitin protease that was expressed from a second plasmid (9). Each polymerase derivative was purified to greater-than-90% purity (Fig. 2A) from the soluble fraction of E. coli by using a combination of metal chelate affinity and ion-exchange chromatographies (see Materials and Methods). The authenticity of the amino terminus of each derivative was verified by amino-terminal sequencing (data not shown).
FIG. 2.
SDS-PAGE analysis. (A) Purified His-tagged polymerase derivatives. Lanes: 1, molecular mass markers (in kilodaltons); 2, ProM-Pol-His; 3, PolVal-1235-His; 4, PolThr-1236-His; 5, PolAla-1237-His. (B) Purification of FCV ProM-Pol. Lanes: 1, molecular mass markers; 2, uninduced cells; 3, induced cells; 4, lysate; 5, clarified lysate; 6, 40% ammonium sulfate pellet; 7, phosphocellulose pool; 8, Q-Sepharose pool.
The polymerase derivatives were assayed in vitro for poly(rU) and poly(rG) polymerase activity. Each derivative exhibited detectable levels of poly(rU) and poly(rG) polymerase activity, and in each case, poly(rU) polymerase activity was reduced threefold relative to poly(rG) polymerase activity (Table 2). The Val-1235 derivative was most active (30 to 100 pmol of nucleotide incorporated/min/μg of protein). Deletion of the valine (the Thr-1236 derivative) caused a threefold reduction in polymerase activity relative to the Val-1235 enzyme; deletion of the threonine (the Ala-1237 derivative) caused a 90-fold reduction in polymerase activity relative to the Val-1235 enzyme (Table 2). These data suggested that the boundary for the active form of the polymerase was located either at Val-1235 or at a position amino terminal to this residue.
TABLE 2.
Activities of FCV polymerase derivatives determined by using homopolymeric primer-template duplexesa
| Enzyme | Activity (pmol/min/μg) with nucleic acid substrate
|
|
|---|---|---|
| poly(rA) | poly(rC) | |
| PolVal-1235-His | 34 ± 5 | 110 ± 10 |
| PolThr-1236-His | 14 ± 2 | 38 ± 2 |
| PolAla-1237-His | 0.57 ± 0.55 | 1.27 ± 0.61 |
| ProM-Pol-His | 100 ± 18 | 300 ± 37 |
| ProM-Pol | 76 ± 6 | 340 ± 58 |
1 μM enzyme was assayed for RdRP activity at 30°C for 1 min in the presence of poly(rA) or poly(rC) (93.4 μM AMP or CMP), dT15 (1.87 μM; 28 μM TMP), or dG6 (4.67 μM; 28 μM GMP) and 500 μM UTP or GTP.
Identification of the FCV Pro-Pol precursor as an active form of the RdRP.
Instead of systematically adding residues to the amino terminus of the Val-1235 derivative, we constructed an expression vector that produced the authentically processed, wild-type Pro-Pol precursor (see Materials and Methods). When this protein was expressed in E. coli, proteolytic processing was observed. While some of the proteolytic processing was due to bacterial proteinases, some could be attributed to the viral proteinase as well (reference 24 and data not shown). Consistent with previous studies of FCV polyprotein processing, the sites in the Pro-Pol precursor recognized by the viral proteinase were located in regions of the polymerase essential for catalytic function (24). In order to preclude processing within the polymerase domain, a mutation was introduced into the proteinase-coding sequence that changed the active-site Cys-1193 to glycine. This derivative is referred to here as ProM-Pol. In addition, we constructed a derivative containing a hexahistidine tag on the carboxyl terminus of the polymerase domain (ProM-Pol-His) to permit a direct comparison to the truncated polymerase derivatives. The ProM-Pol-His derivative was expressed and purified as described for the truncated enzymes (Fig. 2A). This derivative was threefold more active than the Val-1235 enzyme when either poly(rU) or poly(rG) polymerase activity was evaluated, indicating that the Pro-Pol precursor was an active RdRP.
Purification of ProM-Pol.
It remained possible that the presence of the hexahistidine tag on the carboxyl terminus of the polymerase altered the activity of the enzyme. In order to test this possibility, we developed a purification procedure for the ProM-Pol enzyme. ProM-Pol was produced in E. coli by using the pET-ubiquitin-ubiquitin protease system (9). ProM-Pol accumulated in the soluble fraction to levels on the order of 5% of the total cellular protein (Fig. 2B, lanes 2 to 5, and Table 3). The enzyme was purified to at least 95% purity in three steps: (i) ammonium sulfate precipitation (Fig. 2B, lane 6, and Table 3); (ii) phosphocellulose chromatography (Fig. 2B, lane 7, and Table 3); and (iii) Q-Sepharose chromatography (Fig. 2B, lane 8, and Table 3). The final yield from 30 g (wet weight) of cells was 75 mg of protein (Table 3).
TABLE 3.
Purification of FCV ProM-Pol
| Stepa | Protein concn (mg/ml) | Protein (mg) | Sp act (pmol/min/μg) | Total unitsb 106 | Fold purification | Yield (%) |
|---|---|---|---|---|---|---|
| Lysate | 26 | 4,000 | 12.6 | 50.5 | 100 | |
| 40% AmSO4 | 2.3 | 870 | 134 | 117 | 11 | 232 |
| PC pool | 5.6 | 180 | 260 | 42.3 | 21 | 84 |
| Q pool | 11 | 75 | 334 | 24.9 | 26 | 49 |
Lysate from a 3-liter culture (∼30 g [wet weight] of cells). PC, phosphocellulose; Q, Q-Sepharose.
A unit is defined as 1 pmol of GMP incorporated per min.
Characterization of ProM-Pol by using homopolymeric primer-template duplexes.
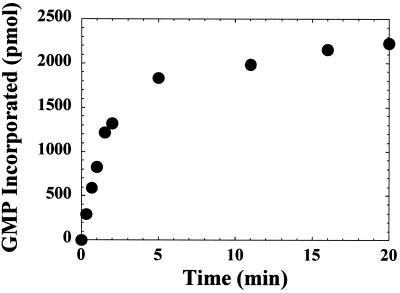
The poly(rU) and poly(rG) polymerase activity of ProM-Pol was evaluated and shown to be within experimental error of the values determined for the ProM-Pol-His derivative (Table 2). DNA [T(pT)14] and RNA [U(pU)14] oligonucleotides served equally well as primers in this assay (data not shown). The kinetics of RNA synthesis were biphasic with both homopolymeric primer-template duplexes analyzed (Fig. 3 and data not shown). Evaluation of the product formed after 3 min no longer provided useful information on the true initial rate of the reaction (Fig. 3). The reduction in the observed rate of the reaction was not due to substrate depletion, as only 10% of the nucleotide was consumed after a 20-min incubation (Fig. 3). The most reasonable explanation for the observed reduction in activity was thermal inactivation of ProM-Pol. This conclusion was based upon the finding that ProM-Pol has a half-life of approximately 1 min in reaction buffer at 30°C (Table 4). The linearity of the reactions shown in Fig. 3 for periods longer than 1 min likely reflects stabilization of the enzyme by nucleotide and/or nucleic acid (3). Polymerase activity was also sensitive to the presence of salt in the reaction; 30 mM NaCl caused a 50% reduction in activity (Table 4).
FIG. 3.
Kinetics of GMP incorporation. The reaction mixtures contained ProM-Pol (0.66 μM), poly(rC) (93.4 μM CMP), and 4.70 μM oligo(dG)6 (28 μM GMP). The reaction mixtures were incubated at 30°C for 5 min, followed by initiation with ProM-Pol. The reactions were quenched at the indicated times by addition of EDTA. The reaction volume was 50 μl.
TABLE 4.
Dependence of ProM-Pol poly(rU) polymerase activity on temperature and NaCl concentration
| Thermal inactivation (min)a | UMP incorporated (pmol) | NaCl concn (mM)b | UMP incorporated (pmol) |
|---|---|---|---|
| 0 | 600 | 0 | 500 |
| 0.33 | 400 | 15 | 320 |
| 0.67 | 320 | 25 | 220 |
| 1 | 290 | 35 | 140 |
| 2 | 160 | 45 | 80 |
| 5 | 110 | 75 | 40 |
| 10 | 70 | 100 | 30 |
| 20 | 50 | 150 | 3 |
Enzyme was incubated in 1× reaction buffer (50 mM HEPES, [pH 7.5], 10 mM 2-mercaptoethanol, 60 μM ZnCl2, and 5 mM MgCl2) at 22°C for the indicated amount of time prior to addition to the complete reaction mixture. Reactions were quenched after 2 min by addition of EDTA. Reaction mixtures contained ProM-Pol (1 μM), 0.2 μM poly(rA) (93.4 μM AMP), and 1.87 μM oligo(dT)15 (28 μM TMP). The reaction volume was 30 μl.
Reaction mixtures contained ProM-Pol (0.83 μM), 0.2 μM poly(rA) (93.4 μM AMP), and 2 μM oligo(dT)15 (30 μM TMP). The reaction volume was 40 μl. Reaction mixtures were incubated at 30°C for 5 min prior to initiation by addition of ProM-Pol. Reactions were quenched after 2 min by addition of EDTA. The NaCl concentration of the reaction mixture before any additional NaCl was added was 1.38 mM due to NaCl in the enzyme preparation.
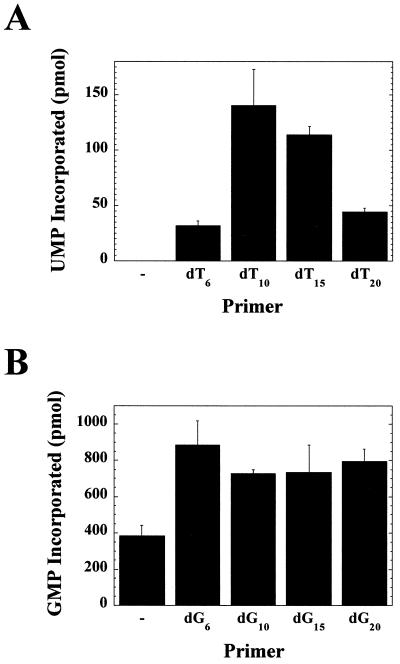
A template-specific primer length dependence was observed (Fig. 4). When poly(rA) was employed as a template, a 10-nt primer was optimal (Fig. 4A). However, when poly(rC) was employed as a template, a 6-nt primer was optimal (Fig. 4B). Increasing the length of the oligo(dT) primers above 10 nt decreased polymerase activity (Fig. 4A). With each primer, the fraction of nucleic acid template devoid of primer remained constant. Therefore, the amount of nucleotide incorporated per bound primer should be independent of primer length. The observed decrease in the product formed as primer length increased may reflect an increase in the number of unproductive complexes formed as a result of the enzyme active site being located at internal positions of the duplex rather than at the 3′ end of the primer. Increasing the length of the oligo(dG) primers was not inhibitory (Fig. 4B). Perhaps the tertiary structures adopted by deoxyguanylate repeats prevented complete annealing of these primers to the template (20). The 6-nt oligo(dT) primer was not as good as the 10-nt primer (Fig. 4A), suggesting that the thermal stability of the primer-template duplex was more important than length for maximal activity. With the poly(rC) template, primer-independent RNA synthesis was also noted (Fig. 4B).
FIG. 4.
Effect of primer length on ProM-Pol-catalyzed RNA synthesis. (A) Synthesis of poly(rU). ProM-Pol (1 μM) was assayed at 30°C for poly(rU) polymerase activity as a function of oligo(dT) primer length. The reaction mixtures contained 0.2 μM poly(rA) (93.4 μM AMP) and oligo(dT)n (where n = 6, 10, 15, or 20). Concentrations of primer sufficient to coat 30% of the homopolymeric template were used. The reactions were initiated by the addition of ProM-Pol and quenched after 1 min by the addition of EDTA. The reaction volume was 30 μl. (B) Synthesis of poly(rG). Reactions were performed as described above, except poly(rC), oligo(dG)n, and GTP were used.
While the activity of ProM-Pol reported here was ∼300,000-fold greater than the activity of the polymerase derivative reported for RHDV (26), this enzyme was still significantly less active (∼10-fold) than PV 3Dpol under the same reaction conditions (1). One possible explanation for this difference was that the divalent cation specificity of ProM-Pol was different than that of the PV polymerase. We performed experiments with ProM-Pol in which poly(rU) and poly(rG) polymerase activity was measured by using a dT15-rA30 or dG15-rC30 primer-template duplex, respectively; Mn2+ was employed as the divalent cation cofactor. These experiments required the use of a 30-nt oligo(rA) template rather than the longer poly(rA) template owing to the insolubility of high concentrations of poly(rA) in the presence of Mn2+ (2). While poly(rC) does not suffer from the same complication, a 30-nt oligo(rC) template was employed in order to facilitate comparison. Interestingly, the response of ProM-Pol to Mn2+ was substrate dependent. Poly(rU) polymerase activity was inhibited (Table 5) while poly(rG) polymerase activity was stimulated (Table 5) in the presence of Mn2+. This substrate-dependent difference likely reflects changes in nucleic acid structure induced by Mn2+ that in turn modulate the ability of ProM-Pol to bind primer-template duplexes and/or extend primers. The 10-fold increase in poly(rG) polymerase activity observed in the presence of Mn2+ was still 10-fold less than that observed for the PV polymerase (2). Therefore, a change in divalent cation specificity was not sufficient to explain the reduced activity of ProM-Pol compared to that of 3Dpol.
TABLE 5.
Effect of Mn2+ on ProM-Pol-catalyzed RNA synthesisa
| Time (min) | NMP incorporated (pmol)
|
|||
|---|---|---|---|---|
| rA30
|
rC30
|
|||
| Mg2+ | Mn2+ | Mg2+ | Mn2+ | |
| 0.5 | 0.20 | 0.11 | 6.5 | 48 |
| 1 | 1.9 | 2.3 | 8.6 | 60 |
| 1.5 | 3.1 | 2.0 | 13 | 71 |
| 2 | 4.0 | 1.9 | 15 | 70 |
| 5 | 9.9 | 4.9 | 25 | 86 |
| 10 | 14 | 7.1 | 33 | 89 |
| 20 | 16 | 9.7 | 42 | 96 |
Reaction mixtures were incubated for 5 min at 30°C, and reactions were initiated by addition of enzyme and quenched at the indicated times by addition of EDTA. Reaction mixtures contained ProM-Pol (0.5 μM), dT15-rA30 or dG15-rC30 (1 μM), UTP or GTP (500 μM), and either 5 mM MgCl2 or MnCl2. The reaction volume was 30 μl.
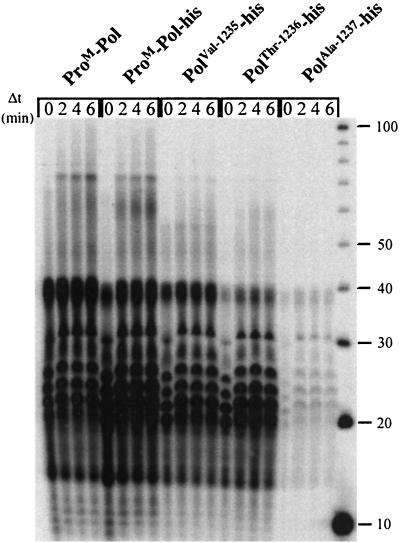
The activity of PV polymerase observed by using homopolymeric primer-template duplexes derives from a few elongating enzymes incorporating nucleotides not only by extending primers to the ends of templates but also by using slippage and template-switching mechanisms (1). Therefore, another possible explanation for the difference in activity between ProM-Pol and 3Dpol was that ProM-Pol was less efficient at slippage and/or template switching. In order to test this possibility, the following experiment was performed. ProM-Pol (5 μM) was incubated with dG15-rC30 (10 μM) and a limiting concentration of radiolabeled GTP (0.25 μM; the total concentration of GTP was 1 μM) to incorporate label into bound primers. After a 1-min labeling period, GTP (500 μM) and heparin (10 μM) were added to chase the initiated primers into final products without reinitiation. Heparin prevents reinitiation (reference 1 and data not shown). During the initial labeling period, products ranging from 16 to 40 nt in length were observed by using ProM-Pol (Fig. 5). This product distribution was consistent with the 15-nt primer being annealed randomly over the entire length of the 30-nt template. A similar pattern was observed by using PV 3Dpol (1). During the chase, products ranging in length from 40 to 100 nt were produced (Fig. 5). Both the efficiency and length distribution of long-product formation were reduced significantly relative to those observed for PV 3Dpol (1). This result suggested that the primary difference between the FCV and PV polymerases was that the FCV enzyme was less efficient at template switching.
FIG. 5.
Template switching catalyzed by FCV polymerase derivatives. The initial reaction mixtures contained dG15-rC30 (10 μM), [α-32P]GTP (0.25 μM), and GTP (0.75 μM) and were initiated by the addition of enzyme (5 μM) followed by incubation at 30°C for 1 min. Heparin (10 μM) and GTP (500 μM) were added, and at the specified times (Δt), the reactions were quenched by the addition of EDTA to a final concentration of 50 mM. The final concentrations of dG15-rC30, [α-32P]GTP, and enzyme were 1, 0.025, and 0.5 μM, respectively. The labeled products were resolved by electrophoresis on a denaturing 10% polyacrylamide gel.
As expected, the outcome of the experiment was unchanged by using the ProM-Pol-His derivative (Fig. 5). However, the ability of the truncated polymerase derivatives to undergo template switching was compromised still further relative to the ProM-Pol and ProM-Pol-His enzymes. The processivity of each polymerase derivative was also decreased based upon the accumulation of products in the 40-nt range (Fig. 5). The processivity of the derivatives followed the order of activity reported in Table 2. The dramatic reduction in activity of the Ala-1237 derivative must be related to a decrease in the efficiency of assembly of this enzyme on the primer-template because the overall labeling of primers was reduced significantly relative to the other polymerase derivatives without a substantial change in the product distribution.
In general, the biochemical properties of FCV ProM-Pol are very similar to those reported for PV 3Dpol (1, 2).
Characterization of ProM-Pol by using a heteropolymeric template.
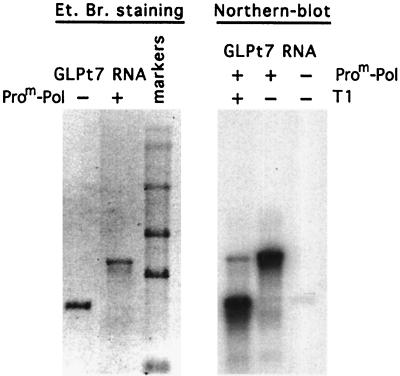
While the analysis of ProM-Pol on homopolymeric primer-template duplexes was quite useful for comparison of the activity of this enzyme to those of others, the biologically relevant nucleic acid substrate is heteropolymeric. We evaluated the ability of ProM-Pol to utilize a heteropolymeric RNA template. This RNA encodes the green fluorescent protein and was produced by in vitro transcription (see Materials and Methods). The RNA is 830 nt in length (Fig. 6, left). Incubation of ProM-Pol with this template and nucleotides followed by denaturing agarose gel electrophoresis showed that the enzyme was capable of copying the entire template (Fig. 6, left). However, the product RNA was twice the size of the input RNA as previously observed with RHDV polymerase (26), suggesting that the product RNA was covalently linked to the template. Template RNA can form a hairpin loop at the 3′ end due to the restriction enzyme used to linearize the template for in vitro transcription. Utilization of this 3′ end as a primer would produce a covalently linked, dimer length RNA product that is sensitive to the single-strand-specific RNase T1. After RNase T1 digestion of the product, electrophoresis under denaturing conditions, and Northern blot analysis with labeled template, a single-stranded RNA product was observed (Fig. 6, right), thus confirming that template and product RNAs were connected by a single-stranded RNA loop.
FIG. 6.
Analysis of RNA product synthesized by FCV ProM-Pol on heteropolymeric RNA template. GLT7 RNA (3 pmol) was employed in a transcription reaction with FCV ProM-Pol (30 pmol). The RNA template and the transcription reaction product corresponding to an equivalent amount of the template were electrophoresed on a formaldehyde 1% agarose gel and visualized by ethidium bromide (Et. Br.) staining (left). The RNA ladder shown in the gel consists of RNA molecules with sizes of 7.46, 4.40, 2.37, 1.35, and 0.24 kb (from top to bottom of the gel). The same amount of transcription product was treated with 5 U of RNase T1 for 1 h at 37°C and analyzed by Northern blotting together with the untreated product and template (right). The RNA bands were visualized by hybridization with the 5′-end-labeled GLT7 RNA (see Materials and Methods).
DISCUSSION
This study was designed to identify the active form of the FCV RdRP. In contrast to a previous report (26), our data were most consistent with the Pro-Pol precursor being the primary, active form of the polymerase. Proteolytic processing between the proteinase and polymerase domains of the Pro-Pol precursor was not required for activity. In fact, the most active enzyme was a precursor (ProM-Pol) in which proteinase activity had been inactivated by site-directed mutagenesis. Biological studies also support our conclusion that Pro-Pol is the active form of the FCV RdRP; only the Pro-Pol precursor accumulates to significant levels when FCV is grown in cell culture (24).
The only other active calicivirus RdRP reported to date is that from RHDV (26). The enzyme characterized in that system lacked the proteinase domain and did not contain an authentic amino terminus owing to the expression system used. The specific poly(rU) polymerase activity of this enzyme was ∼0.3 fmol/min/μg, a value that is 105- to 106-fold lower than that reported here for FCV ProM-Pol. Several possible reasons exist for the difference between the activity of the RHDV and FCV polymerases; none of these explanations are mutually exclusive. First, the additional three residues (Gly-Ser-Met) linked to the amino terminus of the RHDV polymerase derivative might be deleterious to enzyme activity. Indeed, extension of the amino terminus of PV 3Dpol by addition of a single glycine caused a 50-fold reduction in polymerase activity (9). Second, the enzyme may have been partially inactivated by the 3-h incubation at room temperature that was required for thrombin to cleave the glutathione S-transferase domain from the glutathione S-transferase–RHDV polymerase fusion protein and release RHDV polymerase (26). As indicated in Table 4, the activity of the FCV ProM-Pol enzyme was reduced ∼10-fold by a mere 5-min incubation at 30°C in the absence of nucleotide and nucleic acid. Finally, it is possible that the active form of the RHDV polymerase is the Pro-Pol precursor. This possibility was not explored in the previous study (26). The reduced activity of the RHDV enzyme would then be explained by the loss of residues essential for function. The amino terminus of RHDV polymerase has been mapped to threonine-1252 of the RHDV ORF1-encoded polyprotein (Fig. 1) (25). Based upon our alignment of the FCV, RHDV, and SV Pro-Pol proteins, this threonine is located carboxy terminal to FCV Ala-1237 (Fig. 1). Therefore, the RHDV enzyme might be expected to have less activity than the FCV Ala-1237 derivative. However, our data are not sufficient to predict the magnitude of the reduction. The FCV Ala-1237 derivative is 250-fold less active than the ProM-Pol enzyme (Table 2).
We hypothesize that the primary, active form of RHDV polymerase is the Pro-Pol protein. RHDV Pro-Pol accumulates in virus-infected hepatocytes, based upon experiments reported by Konig and colleagues (16). In addition, expression of a truncated precursor containing residues amino terminal and carboxy terminal to the proteinase domain in E. coli showed that only cleavage to release the amino terminus of the proteinase is efficient; cleavage between the proteinase and polymerase domains does not occur readily (25). Recently, a more quantitative analysis of the proposed RHDV Pro-Pol cleavage site was performed which predicts that less than 8% of Pro-Pol molecules should be cleaved to release the polymerase domain (14). Initial studies of SV polyprotein processing showed that precursors larger than Pro-Pol accumulate when translation in vitro occurs in the presence of a full-length genome (17). Expression of truncated SV precursors in E. coli permits the observation of additional processing to liberate a polymerase domain (18). In light of the observations made here, when Pro-Pol processing is evaluated in the future, a quantitative analysis of the products of proteolysis is warranted.
In the PV system, it has been shown that the Pro-Pol precursor, 3CDpro, accumulates in virus-infected cells (12). However, 3CDpro lacks polymerase activity in vitro. The necessity for processing of 3CDpro to yield 3Dpol, the active form of the PV polymerase, was clearly the primary reason that investigators studying caliciviruses searched for processing of the Pro-Pol protein (14, 17, 18, 24, 25). We are therefore forced to ask the question: why is FCV Pro-Pol an active polymerase but PV 3CDpro an inactive polymerase? It is likely that the difference in activity observed for FCV and PV Pro-Pol proteins reflects differences in the conformation of the proteinase domain relative to the polymerase domain in each full-length protein. In the FCV Pro-Pol protein, there is a longer linker between the end of the proteinase domain and the beginning of the polymerase domain than in PV 3CDpro (Fig. 1 and data not shown). Therefore, in the case of FCV Pro-Pol, each domain may fold and function independently of the other. In addition, the absence of an efficient proteinase cleavage site between the two domains in FCV could diminish intra- and/or intermolecular interactions between the proteinase domain and the interdomain linker of Pro-Pol that might occur with PV 3CDpro. This interaction could perturb the structure of the 3CDpro polymerase domain, causing the observed loss of polymerase activity. An unobstructed amino-terminal subdomain of the polymerase is required for activity because the first eight amino acids are predicted to form a β-strand that interacts with two other strands from the finger domain to create a β-sheet that helps to maintain part of the template-binding site in an open conformation (4, 10, 11).
The activity of FCV ProM-Pol was decreased 10-fold relative to that of PV 3Dpol on homopolymeric primer-templates duplexes owing to an apparent reduction in the efficiency of ProM-Pol-catalyzed template switching (Fig. 5). Template switching is thought to be the primary mechanism for recombination between viral genomes (1). Therefore, if FCV ProM-Pol is, in fact, the active form of the polymerase, then the recombination frequency of FCV should be lower than that of PV. More importantly, because our experiments with FCV ProM-Pol were performed with the same substrates and under the same reaction conditions employed for PV 3Dpol (1), it is possible to conclude for the first time that polymerase sequence and structure can be as much of a determinant of recombination efficiency as nucleic acid sequence and structure.
Our data do not rule out the possibility that active forms of the polymerase shorter than full-length Pro-Pol exist and have some function. For example, it is possible that some caliciviruses have evolved to have one form of the polymerase for synthesis of genomic RNA and another form for synthesis of subgenomic RNA. Alternatively, one form may be used for initiation of RNA synthesis (e.g., with VPg priming) and the other for elongation. A final possibility is that Pro-Pol–Pol hetero-oligomers may have a unique function.
The cleavages within the FCV polymerase domain by the proteinase (24) should inactivate the catalytic and RNA-binding activities of the protein based upon a comparison of the FCV polymerase domain to the PV and hepatitis C virus polymerases (4, 10). This inactivation may not be an artifact of overexpression of Pro-Pol precursors in systems such as E. coli but may be relevant biologically. For example, functional polymerase not actively engaged in RNA synthesis may inhibit elongating enzymes by binding randomly to template RNA. A mechanism to inactivate these idle enzymes would preclude this problem. In addition, processing within the polymerase domain should prevent “spatial” restriction of the proteinase resulting from interaction of polymerase with RNA and/or modulate the substrate specificity of the proteinase. In the PV system, 3CDpro cleaves capsid proteins more efficiently than 3Cpro (12). Finally, it is possible that these proteinase cleavage sites are only exposed in misfolded Pro-Pol molecules. If this is the case, then the presence of these sites may provide a quality control mechanism that ensures the removal of misfolded Pro-Pol, which could interfere with efficient virus multiplication.
In conclusion, this study confirms the functional similarity among supergroup I RdRPs and illuminates the differences that can exist among polymerases within a supergroup. The most striking difference among virus systems that warrants further investigation is the variable nature of the polymerase polypeptide—that is, mono- versus bifunctional. Did one form arise before the other? Is one form more advantageous than the other? The ability to apply reverse genetics to the study of FCV multiplication should permit us to address these and related questions directly. Analysis of other calicivirus systems will be necessary to determine whether the observations reported here for FCV reflect a general rule for this virus family.
ACKNOWLEDGMENT
C.E.C. is the recipient of a Howard Temin Award (CA75118) from the National Cancer Institute, NIH.
REFERENCES
- 1.Arnold J J, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J Biol Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- 2.Arnold J J, Ghosh S K B, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol): divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 3.Arnold J J, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol): assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale R L, Mathieu M, De Francesco R, Rey F A. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba S, Nakata S, Numata-Kinoshita K, Honma S. Sapporo virus: history and recent findings. J Infect Dis. 2000;181:S303–S308. doi: 10.1086/315574. [DOI] [PubMed] [Google Scholar]
- 6.Clarke I N, Lambden P R. The molecular biology of caliciviruses. J Gen Virol. 1997;78:291–301. doi: 10.1099/0022-1317-78-2-291. [DOI] [PubMed] [Google Scholar]
- 7.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 8.Glass R I, Noel J, Ando T, Fankhauser R, Belliot G, Mounts A, Parashar U D, Bresee J S, Monroe S S. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J Infect Dis. 2000;181:S254–S261. doi: 10.1086/315588. [DOI] [PubMed] [Google Scholar]
- 9.Gohara D W, Ha C S, Ghosh S K B, Arnold J J, Wisniewski T J, Cameron C E. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- 10.Gohara D W, Crotty S, Arnold J J, Yoder J D, Andino R, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem. 2000;275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 11.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 12.Harris K S, Reddigari S R, Nicklin M J, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert T P, Brierley I, Brown T D. Detection of the ORF3 polypeptide of feline calicivirus and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J Gen Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- 13a.Herbert T P, Brierley I, Brown T D. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- 14.Joubert P, Pautigny C, Madelaine M, Rasschaert D. Identification of a new cleavage site of the 3C-like protease of rabbit haemorrhagic disease virus. J Gen Virol. 2000;81:481–488. doi: 10.1099/0022-1317-81-2-481. [DOI] [PubMed] [Google Scholar]
- 15.Kapikian A Z, Estes M K, Chanock R M. Norwalk group of viruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 783–810. [Google Scholar]
- 16.Konig M, Thiel H J, Meyers G. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J Virol. 1998;72:4492–4497. doi: 10.1128/jvi.72.5.4492-4497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Clarke I N, Lambden P R. Polyprotein processing in southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J Virol. 1996;70:2605–2610. doi: 10.1128/jvi.70.4.2605-2610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B L, Viljoen G J, Clarke I N, Lambden P R. Identification of further proteolytic cleavage sites in the southampton calicivirus polyprotein by expression of the viral protease in E. coli. J Gen Virol. 1999;80:291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Smith F W, Feigon J. Quadruplex structure of oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 21.Sosnovtsev S, Green K Y. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 22.Sosnovtsev S V, Sosnovtseva S A, Green K Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sosnovtsev S V, Green K Y. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology. 2000;277:193–203. doi: 10.1006/viro.2000.0579. [DOI] [PubMed] [Google Scholar]
- 24.Sosnovtseva S A, Sosnovtsev S V, Green K Y. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J Virol. 1999;73:6626–6633. doi: 10.1128/jvi.73.8.6626-6633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirblich C, Sibilia M, Boniotti M B, Rossi C, Thiel H J, Meyers G. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J Virol. 1995;69:7159–7168. doi: 10.1128/jvi.69.11.7159-7168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez A L, Alonso J M M, Casais R, Boga J A, Parra F. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J Virol. 1998;72:2999–3004. doi: 10.1128/jvi.72.4.2999-3004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]