Abstract
Cdk9 is the catalytic subunit of TAK (cyclinT1/P-TEFb), a cellular protein kinase that mediates human immunodeficiency virus type 1 (HIV-1) Tat transcriptional activation function. To examine Cdk9 function in cells relevant to HIV-1 infection, we used a murine leukemia virus retrovirus vector to transduce and overexpress the cDNA of a dominant negative mutant Cdk9 protein (Cdk9-dn) in Jurkat T cells and U937 promonocytic cells. In Jurkat cells, overexpression of Cdk9-dn specifically inhibited Tat transactivation and HIV-1 replication but had no inhibitory effect on induction of CD69, CD25, and interleukin-2 following T-cell activation. In U937 cells, overexpression of Cdk9-dn sensitized cells to apoptosis, especially after phorbol myristate acetate (PMA) treatment to induce differentiation to macrophage-like cells. Because Cdk9 function is induced in PMA-treated U937 cells, Cdk9 may play an antiapoptotic role during monocyte differentiation.
Human immunodeficiency virus type 1 (HIV-1) requires the viral transactivator protein Tat for efficient elongation of the integrated proviral genome by RNA polymerase II. Tat acts by recruitment of a cellular protein kinase, TAK (Tat-associated kinase), to the TAR RNA element in nascent viral transcripts (reviewed in references 8 and 37). TAK is composed of Cdk9 as the catalytic subunit, cyclin T1 (cycT1) as a regulatory subunit, and possibly other subunits that remain to be identified (30, 39, 42, 44). TAK is closely related to the general elongation factor P-TEFb (reviewed in reference 31). Multiple P-TEFb complexes exist in human cells that contain Cdk9 and differ according to their regulatory cyclin subunit: cyclins T1, T2a, T2b, and possibly cyclin K (6, 30, 39). P-TEFb/TAK is thought to activate elongation by hyperphosphorylating the carboxyl-terminal domain of RNA polymerase II, thereby relieving repression by negative factors that limit processivity (38, 41). It has been well established that TAK, in the cycT1/P-TEFb complex, mediates Tat function (2, 7, 9, 12, 17, 24, 28, 32, 39, 43, 44).
P-TEFb was originally identified through in vitro transcription studies in Drosophila melanogaster nuclear extracts (25–27). From these in vitro studies, P-TEFb appears to be required for efficient elongation of many cellular promoters. A recent immunofluorescence analysis of Drosophila polytene chromosomes with antibodies against cycT1 indicates that P-TEFb is likely to be involved in regulation of many, but not all, Drosophila genes (23). In human cells, Cdk9 in the cycT1/P-TEFb complex is required for transcription of major histocompatibility class II (MHC II) genes (19). Other than MHC II genes, there is currently little information as to which human genes require P-TEFb function in vivo.
The major target cells for HIV-1 infection are CD4+ lymphocytes and macrophages. Regulation of TAK activity in these cell types is potentially important to HIV-1 pathogenesis, since TAK may be a limiting factor for HIV-1 replication under some conditions. TAK is induced by activation of either peripheral blood lymphocytes (PBL) or purified primary CD4+ T lymphocytes (11, 14, 42). TAK is also induced following differentiation of the promonocytic cell lines U937 and HL-60 by phorbol myristate acetate (PMA) treatment (14, 42). These observations suggest that TAK may have distinct functions during T-cell activation and monocyte differentiation. Activation of TAK in PBLs involves an increase in levels of mRNA and protein for both Cdk9 and cycT1 (11, 14). In contrast, PMA-induced differentiation of promonocytic cell lines involves a large increase in cycT1 protein through a posttranscriptional mechanism, whereas Cdk9 protein levels are constant before and after treatment with PMA (14). The difference in expression of Cdk9 and cycT1 between PBLs and promonocytic cell lines may highlight a difference in function for the proteins between the two cell types.
In this study, we examined potential roles of Cdk9 in the activation of T cells and the differentiation of monocytes. We report here that the overexpression of a dominant negative Cdk9 protein in activated Jurkat T cells has no apparent effect on induction of CD25, CD69, or interleukin-2 (IL-2), three molecules known to be important for T-cell function. By contrast, overexpression of a dominant negative Cdk9 protein in the promonocytic cell line U937 caused the cells to become sensitive to apoptosis, especially following PMA treatment to induce differentiation, suggesting that Cdk9 may have an antiapoptotic function during monocyte differentiation.
MATERIALS AND METHODS
Cells.
Human promonocytic U937 and Jurkat T cell lines were grown in RPMI-1640 (Gibco BRL) supplemented with 10% fetal bovine serum and antibiotics. Densities of cultures were maintained between 2 × 105 and 8 × 105 cells/ml. 293T cells were cultured in Dulbecco modified Eagle medium with high glucose (Gibco BRL) supplemented with 10% fetal bovine serum and antibiotics.
Generation of retroviral vectors.
The cDNA of a dominant negative mutant Cdk9, Cdk9-dn (12), was inserted into a murine leukemia virus (MLV)-based retroviral vector, pBABEMN IRES GFP (derived from pBABE-puro; Mike Rothenberg, Stanford University), upstream to an internal ribosome entry site (IRES) followed by the cDNA for green fluorescent protein (GFP). Pseudotyped MLV particles were produced in 293T cells by cotransfection of the pBabe/Cdk9-dn plasmid, pHit60, which expresses the gag and pol genes of MLV, and a plasmid that expresses the vesicular stomatitis virus G protein (35, 36). One day prior to transfections, a confluent 10-cm culture dish of 293T cells was split 1:5, and on the day of transfection, 25 μg of each of three plasmids (pBABE-Cdk9-dn or parental pBABE/GFP, pVSV-G, and pHit60) was cotransfected by a standard calcium phosphate procedure. After 8 h, the calcium phosphate precipitate was washed from cells and the cells were treated with 10% dimethyl sulfoxide in phosphate-buffered saline (PBS) for 2 min. Cells were cultured in 10 ml of complete media for 72 h, and culture supernatants containing retroviruses were collected. Retrovirus preparations were stored at −70°C until use.
Generation of cell lines expressing Cdk9-dn.
Jurkat T cells or U937 cells were infected at a multiplicity of infection of 0.4 with retroviral vectors, and at 3 days postinfection, GFP+ cells with the top 10% fluorescence intensity were sorted by flow cytometry (EPICS-Altra; Beckman-Coulter). Postsort Jurkat T-cell pools and U937 pools were approximately 85 and 93% GFP+, respectively. These GFP+ populations are referred to as sorted pools. Limiting dilution of pool populations was used to generate clonal cell lines.
Immunoblots.
Cells were washed with PBS and lysed in EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40, 5 mM dithiothreitol) containing protease inhibitors as described previously (16). Protein concentrations were determined by the Bio-Rad protein assay, and 15 μg of total protein was analyzed on sodium dodecyl sulfate–9% polyacrylamide gels. Immunoblotting was preformed by standard procedures by using enhanced chemiluminescence for detection as described previously (15). Antibodies for detection of Cdk9, cyclin T1, and TATA-binding protein (TBP) were purchased from Santa Cruz Biotechnology.
Plasmid transfection assays and HIV-1 infections.
For HIV-1 Tat transactivation assays, 106 Jurkat Cdk9-dn clonal cells were cotransfected with 1 μg of an HIV-1 long terminal repeat (LTR) luciferase reporter plasmid, 3 μg of a simian virus 40 early promoter β-galactosidase (β-Gal) reporter plasmid (pCH110; Pharmacia), and either 1 μg of pCMV-Tat-1 or 1 μg of pCMV parental vector (33). Transfections were performed with Superfect Transfection Reagent (Qiagen) using the manufacturer's suggested protocol. Lysates were prepared 48 h posttransfection. Luciferase assays were performed with the Luciferase Assay System (Promega), and light units were measured using a TD20-e luminometer (Turner). For β-Gal assays, 100 μl of lysates was used in a standard enzyme assay. For human T-cell leukemia virus type 1 (HTLV-1) Tax transactivation assays, 106 cells were cotransfected with 1 μg of an HTLV-1 LTR luciferase reporter (provided by Susan Marriott, Baylor College of Medicine) and either 1 μg of GW1-Tax (provided by Ron Javier, Baylor College of Medicine) or parental vector. Cells were harvested 48 h posttransfection, and luciferase and β-Gal assays were performed as above.
For HIV-1 infections, 6 × 106 Jurkat Cdk9-dn and control vector clonal cell lines were infected with pretitered HIV-1 NL4-3 at a final concentration of 200 ng/ml of p24 antigen. At 4 h postinfection, cells were washed three times with PBS plus 2% fetal bovine serum. Culture media were collected, and p24 concentrations were determined by enzyme-linked immunosorbent assay (ELISA) (Beckman-Coulter, Hialeah, Fla.). Cells remained more than 90% viable throughout infections as determined by trypan blue exclusion.
Measurement of T-cell activation markers.
Jurkat cells were activated with PMA (1 ng/ml) and ionomycin (1 μM) for 18 h. Alternatively, 4.4 μg of anti-CD3 (Pharmingen) in 0.2 M sodium bicarbonate was bound to 6-well plates overnight at 4°C. Binding buffer was removed, and 106 cells plus 4.4 μg of soluble anti-CD28 (Pharmingen) in 3 ml of media were added for 24 h. Cells were collected and washed twice with PBS. Cells were stained with either phycoerythrin-conjugated anti-CD69 or anti-CD25 antibodies (Becton Dickinson) on ice for 30 min. Samples were analyzed by flow cytometry using a Beckman-Coulter XL-MCL cytometer. A total of 10,000 events were collected.
For IL-2 induction, 5 × 106 Jurkat cells were activated with PMA plus ionomycin as described above. Aliquots of culture medium were removed over a time course, and the IL-2 concentration was measured by ELISA according to the manufacturer's protocol (Endogen). Samples were stored at −70°C prior to assaying.
Apoptosis measurements in U937 cells.
U937 Cdk9-dn sorted pool and clones, control vector pool and clones, and parental U937 cells were treated with PMA (1 ng/ml) for 24 or 48 h as described for each experiment. Cells were collected from the plate and washed twice with PBS. Cells were then ethanol fixed and stained with propidium iodide (PI) (Sigma) (50 μg/ml); cells were treated with RNase A (Sigma) (1 mg/ml) for 30 min at 37°C prior to analysis. Cells were analyzed as described above for CD69 and CD25 markers. Annexin V (Pharmingen) staining was performed according to the manufacturer's recommendations.
Caspase-3 activity in cell lysates was determined using a caspase-3 assay kit following the manufacturer's protocol (Pharmingen). Cdk9-dn pool, vector control pool, and parental U937 cells were treated with PMA as described above for 12, 24, or 48 h. Cells were harvested, and frozen pellets were stored at −70°C until lysates were prepared. Fluorescence was measured using a spectrofluorometer (Perkin-Elmer LS50B) with excitation at 380 nm and the emission wavelength at 420 to 460 nm.
RESULTS
Generation of Jurkat T-cell clonal cell lines overexpressing dominant negative Cdk9 protein.
To investigate potential roles of Cdk9 in T-cell activation events, we used an MLV-based retroviral vector to transduce the cDNA of a dominant negative Cdk9 mutant protein (Cdk9-dn) into Jurkat T cells. The dominant negative Cdk9 protein contains a substitution of asparagine for aspartic acid at residue 167, rendering the protein catalytically inactive (10). Upon overexpression, the Cdk9-dn protein inhibits wild-type (wt) Cdk9 function as demonstrated by specific inhibition of Tat transactivation of the HIV-1 LTR (12, 24). The mRNA expressing the Cdk9-dn protein from the MLV LTR contains an IRES element at its 3′ end followed by the coding sequence of the GFP, allowing GFP to be used as a marker for transduction of the Cdk9-dn cDNA. Cdk9-dn expression can be distinguished from endogenous Cdk9 expression by incorporation of the FLAG-epitope tag at the amino terminus of Cdk9-dn.
Jurkat T cells were infected with MLV vectors expressing both Cdk9-dn and GFP or the parental MLV vector expressing only GFP. Three days postinfection, transduced cells that expressed GFP were sorted by flow cytometry. Clonal cell lines were generated from the sorted pool by limiting dilution. Forty clones transduced with the Cdk9-dn cDNA were evaluated in immunoblots for levels of expression of the catalytic mutant protein, and all clones were found to express the Cdk9-dn protein (data not shown). From these clones, three cell lines expressing the highest levels of Cdk9-dn were chosen for subsequent experiments. Five clones from infection with the parental MLV vector expressing GFP alone were also generated, and two of these control cell lines were used for experiments. An immunoblot analysis of the three Cdk9-dn and two control cell lines is shown in Fig. 1. By quantitative Western blot analysis using twofold serial dilutions of cell extracts, we estimate that Cdk9-dn levels in clones 3, 30, and 43 were greater than fourfold more than those of endogenous Cdk9 (data not shown). We observed no evidence during the course of this study that expression of the Cdk9-dn protein had a negative effect on Jurkat T-cell growth, since the percentages of GFP-positive cells in the sorted pool and clonal cell lines were stable over a 3-month period.
FIG. 1.
Jurkat T-cell clonal lines overexpress FLAG-Cdk9-dn. Immunoblots to detect Cdk9 proteins were performed on lysates from the indicated cell lines. FLAG-tagged Cdk9-dn and endogenous Cdk9 proteins are indicated.
In the immunoblots shown in Fig. 1 and 7, both the endogenous wt and epitope-tagged Cdk9-dn proteins often resolved into a pair of bands. We have observed that resolution of Cdk9 proteins into a doublet is variable, and in some immunoblots only a single form of Cdk9 was observed. The explanation for Cdk9 proteins with different electrophoretic mobilities is not known at this time, although it is possible that these forms represent different phosphorylation states of Cdk9.
FIG. 7.
Expression of wt and Cdk9-dn, cyclin T1, and TBP proteins in U937 sorted pools and Jurkat clonal lines. Immunoblots were performed to measure Cdk9, cyclin T1, and TBP expression in lysates of U937 parental cells or sorted U937 cell pools transduced with Cdk9-dn cDNA or vector control and Jurkat Cdk9-dn cell lines 3 and 30 and Jurkat control lines 7 and 10. FLAG-tagged Cdk9-dn and endogenous proteins are indicated.
Cdk9-dn overexpression in Jurkat T cells inhibits HIV-1 LTR transactivation by Tat.
To examine the effect of overexpression of the catalytic mutant on Tat function, the Cdk9-dn clonal lines 3 and 30 and control clonal lines 7 and 10 were used in plasmid cotransfection assays. The results shown in Fig. 2 indicate that Tat transactivation of the HIV-1 LTR was reduced more than threefold in both Cdk9-dn lines relative to both control lines.
FIG. 2.
Effect of Cdk9-dn overexpression on transactivation by HIV-1 Tat and HTLV-1 Tax proteins. The indicated Jurkat Cdk9-dn or vector control clonal lines were transfected with an HIV-1 LTR luciferase reporter plasmid plus either a Tat expression or vector control plasmid and a β-Gal internal reference plasmid. Extracts were prepared at 48 h posttransfection, luciferase expression was normalized to β-Gal expression, and Tat transactivation was calculated. Jurkat Cdk9-dn or vector control clones were also transfected with an HTLV-1 LTR luciferase reporter plasmid plus either a Tax expression or vector control plasmid and a β-Gal internal reference plasmid. Tax transactivation was determined as described for Tat transactivation.
To examine the specificity of inhibition by Cdk9-dn overexpression, we examined transactivation of the HTLV-1 LTR by its Tax transactivator protein. The Jurkat clonal cell lines were cotransfected with an HTLV-1 luciferase reporter plasmid and a Tax expression plasmid, and luciferase expression was determined as described above (Fig. 2). Consistent with previous work (12), overexpression of the Cdk9-dn protein did not inhibit Tax transactivation, since activation of the HTLVI LTR by Tax was similar between the Cdk9-dn and control cell lines.
We also examined replication of HIV-1 NL4-3 over a 12-day time course in Cdk9-dn cell lines 3 and 30 and vector control cell lines 7 and 10. As determined by p24 content in culture supernatants, HIV-1 replication in the Cdk9-dn cell lines was significantly below that in the control cell lines (Fig. 3). Cdk9-dn clone 3 showed the lowest levels of HIV-1 replication, more than 185-fold and 25-fold below those of control lines 7 and 10, respectively, at days 8 and 12. Cdk9-dn clone 30 showed HIV-1 replication levels that were at least 14-fold less than those for control clone 7 at days 8 and 12 and 5-fold and approximately 3-fold less than those for control clone 10 at days 8 and 12, respectively. These results confirm those of a previous study in which the Cdk9-dn protein was shown to inhibit HIV-1 replication in Jurkat T cells (5). We used flow cytometry to examine CD4 expression levels in the Cdk9-dn and control cell lines used in the experiment presented in Fig. 3. The percentages of cells in each clonal line expressing CD4 were similar, and the mean fluorescent intensities of the phycoerythrin-conjugated anti-CD4 antibody bound to cells indicated that all clonal lines expressed comparable amounts of CD4 on the cell surface (data not shown). It is therefore unlikely that reduced replication of HIV-1 in the Cdk9-dn cell lines is the consequence of reduced levels of CD4 on the cell surface. Taken together, the results shown in Fig. 2 and 3 indicate that the level of the Cdk9-dn protein in these clonal cell lines is sufficient to inhibit Tat transactivation, thereby indicating that the function of the wt Cdk9 protein is inhibited.
FIG. 3.
HIV-1 NL4-3 replication in Jurkat Cdk9-dn and vector control clonal cell lines. The indicated cell lines were infected with HIV-1 pNL4-3, and p24 levels in culture media were measured by ELISA at the indicated time points.
Cdk9-dn overexpression does not inhibit induction of CD69, CD25, or IL-2 in activated Jurkat cell lines.
To determine whether Cdk9-dn overexpression affects events associated with Jurkat T-cell activation, Jurkat clonal cell lines were activated with PMA plus ionomycin or anti-CD3 plus anti-CD28 antibodies, and induction of the activation cell surface markers CD69 and CD25 was evaluated by flow cytometry (Fig. 4). The results demonstrated no significant effect of overexpression of the Cdk9-dn protein on induction of CD25 or CD69 by either activation protocol. Although there was some clonal variability, each Cdk9-dn clonal line demonstrated induction of both markers to levels that are equivalent to those seen for the vector control clonal line 7. In other experiments, control clonal line 10 was found to express CD25 and CD69 after activation to levels similar to those shown for the cell lines used for Fig. 4 (data not shown).
FIG. 4.
Induction of CD69 and CD25 in Jurkat T-cell lines. The indicated Jurkat clones were activated either with anti-CD3 plus anti-CD28 for 24 h or with PMA plus ionomycin for 18 h. Expression of CD25 and CD69 were measured by flow cytometry. Black areas represent marker expression before activation, and white areas represent marker expression after activation. Fold induction values, as determined by comparison of mean fluorescence intensities, of each marker are the following: 6.4 (A), 3.7 (B), 37.6 (C), 204.3 (D), 2.4 (E), 2.1 (F), 21.3 (G), 192.0 (H), 4.8 (I), 10.5 (J), 10.4 (K), 459.9 (L), 10.6 (M), 12.2 (N), 43.2 (O), and 33.1 (P).
Induction of IL-2, a hallmark of T-cell activation, was also measured in the Cdk9-dn clonal cell lines. Cells were activated with PMA plus ionomycin, culture supernatants were removed over a 24-h time course, and IL-2 levels were determined by ELISA (Fig. 5). Each of the Cdk9-dn clonal lines and the vector controls showed strong induction of IL-2, indicating that overexpression of the Cdk9-dn protein had no significant inhibitory consequence for IL-2 induction. Taken together, these results suggest that Cdk9 function is not required for induction of CD25, CD69, or IL-2 following activation of Jurkat T cells.
FIG. 5.
Induction of IL-2 in Jurkat T cell lines. The indicated Cdk9-dn and vector control Jurkat cell lines were activated with PMA plus ionomycin. The concentration of IL-2 in the culture medium was measured for duplicate samples by ELISA.
Cdk9-dn expression renders U937 cells sensitive to apoptosis.
We were also interested in investigating whether Cdk9 may have a role in monocyte differentiation. We therefore used the MLV retroviral vector to transduce the Cdk9-dn cDNA into U937 promonocytic cells. U937 clonal lines expressing the Cdk9-dn protein were then generated by limiting dilution. We observed under light microscopy that many of these clonal cell lines spontaneously contained irregularly shaped cells and the culture media contained high amounts of membranous debris, indicative of apoptosis. When treated with PMA to induce differentiation, many of the U937 clonal lines transduced with the Cdk9-dn cDNA produced massive membrane blebs and cell debris in the culture medium. To examine apoptosis in these clonal cell lines, PI staining and flow cytometry were used to monitor cells with a subdiploid DNA content (Fig. 6). Without PMA treatment, for the parental U937 cells and the vector control clone 2, less than 1% of cells were in the apoptotic subdiploid DNA region, whereas for Cdk9-dn clones 6 and 11, approximately 7% of cells were in the subdiploid DNA region. Upon PMA treatment for 24 h, this percentage increased to 1.7 and 1.3 for the parental U937 cells and the vector control clone 2, respectively, whereas the Cdk9-dn clonal lines 6 and 11 increased to 26.2 and 18.4%, respectively. By 48 h of PMA treatment, the Cdk9-dn clonal lines contained massive cellular debris and too few cells for analysis.
FIG. 6.
PI staining of U937 clonal cell lines. Parental U937, MLV vector control, and Cdk9-dn clonal lines 6 and 11 were treated with PMA (1 ng/ml) for 24 h. DNA content was analyzed by PI staining and flow cytometry. The subdiploid DNA regions are gated, and the percentages of apoptotic cells are indicated.
We chose not to further analyze these U937 clonal lines expressing the Cdk9-dn protein because of concern over selection pressures that might result in second site mutations to allow survival of the cell lines. Therefore, we examined pools of U937 cells expressing the Cdk9-dn protein that were obtained by cell sorting. An immunoblot analysis of a sorted U937 pool demonstrates that Cdk9-dn protein is expressed at a relatively high level in these cells (Fig. 7). We also evaluated expression levels of cycT1 and TBP (20) in U937 pools and Jurkat clonal lines that overexpress the Cdk9-dn protein. Although there was some variation in the levels of cycT1 (lane 3) and TBP (lane 6) in some cells, there was no consistent effect of Cdk9-dn expression on the level of cycT1 or TBP.
Using PI staining and flow cytometry to measure subdiploid DNA content, we observed very little difference among non-PMA-treated parental U937, the Cdk9-dn pool, and the MLV vector control pool (Fig. 8). After 48 h of PMA treatment, however, the Cdk9-dn pool contained approximately threefold more cells in the subdiploid DNA region than either the vector control pool or the parental U937 cells. No significant difference was observed between the PMA-treated vector control pool and the parental U937 cells.
FIG. 8.
PI staining of U937 pool expressing Cdk9-dn protein. Parental U937 or sorted pools of vector control U937 or Cdk9-dn U937 cells were treated with PMA for 48 h. DNA content was analyzed with PI and flow cytometry. The subdiploid DNA regions are gated, and the percentages of apoptotic cells are indicated.
To confirm that the increase in subdiploid DNA content in the U937 Cdk9-dn pool was the result of apoptosis, PMA-treated cells were examined for PI and Annexin V binding. Annexin V staining is indicative of apoptosis, since it binds to phospholipid phosphatidylserine that is translocated to the cell surface at an early stage of apoptosis. As shown in Table 1, PMA treatment resulted in an increase in both PI and Annexin V staining for the Cdk9-dn pool relative to the vector control pool. After 48 h of PMA treatment, the Cdk9-dn pool contained 4.5-fold more cells with a subdiploid DNA content and 4-fold more Annexin V-positive cells than the vector control pool.
TABLE 1.
Examination of PMA-treated cells for PI and Annexin V binding
| Cell type | PMA treatment time (h) | % Cells
|
|
|---|---|---|---|
| Subdiploid DNA present | Annexin V positive | ||
| Cdk9-dn pool | 0 | 4.0 | 1.2 |
| 24 | 10.9 | 4.1 | |
| 48 | 25.6 | 13.2 | |
| Vector pool | 0 | 2.1 | 1.2 |
| 24 | 6.8 | 1.6 | |
| 48 | 5.7 | 3.4 | |
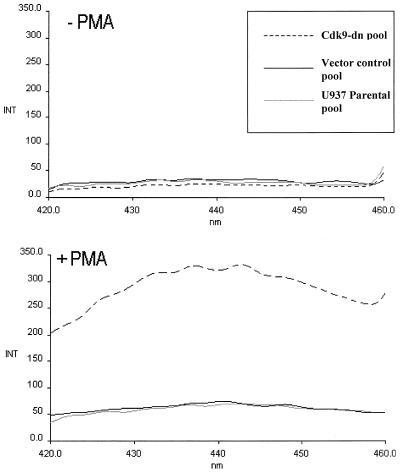
To further confirm that expression of the Cdk9-dn protein renders U937 cells sensitive to apoptosis upon treatment with PMA, we measured activation of caspase-3. Caspase-3 is a protease which is an early marker of apoptosis and cleaves other caspases and additional protein targets in the cytoplasm and nucleus. We observed a sixfold increase in caspase-3 activity in extracts from the PMA-treated Cdk9-dn pool relative to extracts from the vector control pool or parental U937 cells (Fig. 9). No difference in caspase-3 activity was seen between the vector control pool and the parental U937 cells. The results shown in Fig. 6, 8, and 9 and Table 1 indicate that expression of the Cdk9-dn protein renders U937 cells sensitive to apoptosis after PMA treatment.
FIG. 9.
Caspase-3 activity in extracts of U937 cells. Parental U937 cells, sorted pools of vector control U937 cells, or Cdk9-dn U937 cells were either untreated (top) or treated with PMA. Lysates were prepared at 48 h posttreatment, and caspase-3 activity was determined by the ability to cleave a synthetic tetrapeptide fluorogenic substrate, Ac-DEVD-AMC, to release fluorescent 7-amino-4-methylcoumarin, which is measured by a spectrofluorometer. Excitation wavelength was at 380 nm, and the emission was read between 420 and 460 nm.
To examine whether expression of the Cdk9-dn protein might render Jurkat T cells sensitive to apoptosis, we also analyzed the Jurkat Cdk9-dn and control clonal lines used in the experiments presented in Fig. 1 to 5 and described above. To induce apoptosis in Jurkat cells, cultures were treated with anti-Fas antibodies or PMA plus ionomycin, and apoptosis was evaluated by both PI and Annexin V staining. We found no significant difference in the levels of spontaneous or induced apoptosis between the Cdk9-dn and control clonal cell lines (data not shown).
DISCUSSION
Because small molecular inhibitors of Cdk9 can inhibit HIV-1 replication in vitro at concentrations that are not toxic to cells, it has been suggested that the development of selective inhibitors of Cdk9 might lead to novel chemotherapeutic agents against HIV-1 (5). The elucidation of normal cellular functions of Cdk9 in CD4+ T cells and monocytes/macrophages is therefore important for evaluating the feasibility of Cdk9 as a therapeutic target. Additionally, elucidation of Cdk9 function in T cells and monocytes/macrophages may be important for understanding HIV-1 pathogenesis, since infection may perturb these functions. In this study, we utilized a dominant negative Cdk9 mutant protein to investigate roles of Cdk9 in Jurkat T and U937 promonocytic cell lines. We found that overexpression of the Cdk9-dn protein in U937 cells renders cells sensitive to apoptosis, especially following PMA treatment to induce differentiation. This finding suggests that Cdk9 may have an antiapoptotic function during monocyte differentiation. Jurkat cell lines expressing the Cdk9-dn protein were found not to be more sensitive to apoptosis than control Jurkat cell lines (data not shown). However, before conclusions can be drawn about the cell-type specificity of regulation of apoptosis by Cdk9, additional lymphoid cell lines and other cell types will have to be examined.
The ability to readily undergo apoptosis is important to monocyte homeostasis, since monocytes normally circulate in the blood for a period of only a few days, during which time they either emigrate to tissues and differentiate to macrophages or die through apoptosis (13, 21). Previous studies of U937 and HL-60 cells showed that Cdk9 catalytic activity is low in promonocytic cells due to limiting amounts of the cycT1 regulatory subunit (14, 42). The findings in this study suggest that a low level of cycT1 protein in monocytes, and therefore a low level of Cdk9 function, may be important for their requirement to readily undergo apoptosis in the absence of differentiation. The mechanisms responsible for the increased rate of apoptosis in U937 cells expressing the Cdk9-dn protein remain to be established.
The antiapoptotic function of Cdk9 could be due to a direct role in an apoptosis pathway or to a block in the differentiation program of monocytes by the Cdk9-dn protein. Previous work on monocyte differentiation indicates that the cells have an intrinsic program to differentiate when apoptosis is blocked by enforced expression of Bcl-2 (21). This might suggest that Cdk9 functions in the P-TEFb complex to regulate transcription of genes, such as the gene for p21WAF1, whose expression confers resistance to apoptosis prior to monocyte differentiation (1, 40). An additional possibility is that Cdk9 function is necessary for the differentiation program of monocytes.
Our experiments with Jurkat T cells indicate that overexpression of a Cdk9-dn protein to levels that inhibit Tat function does not inhibit induction of CD69, CD25, and IL-2 following T-cell activation. Because induction of each of these corresponding genes occurs at the transcriptional level (3, 4, 18, 22, 34, 45), our rather surprising results suggest that Cdk9 in the P-TEFb complex is not required for their transcriptional induction. Since Tat has been shown to sequester P-TEFb activity from MHC II genes (19), a requirement for P-TEFb in the transcription of IL-2 would be expected to have been detectable in our experiments. The lack of a requirement for Cdk9 for IL-2 production in this study is consistent with the observation that HIV-1 infection and Tat expression can actually superinduce IL-2, suggesting that sequestration of Cdk9 and cyclin T1 in a complex with Tat does not inhibit some basic T-cell functions (29).
Cdk9 in the P-TEFb complex is thought to be a general elongation factor required for expression of many genes. We were therefore somewhat surprised to observe that in Jurkat T cells transduced with the Cdk9-dn cDNA, the percentages of GFP+ cells and therefore Cdk-9dn expression in sorted pools and clonal lines were stable over a 3-month period. It may be informative to utilize DNA microarray technology to investigate how overexpression of Cdk9-dn affects global gene expression in both activated and nonactivated Jurkat T cells.
The results of this study suggest that Cdk9 function is likely to be crucial to the monocyte/macrophage life cycle. This raises the possibility that HIV-1 infection might perturb normal monocyte homeostasis through sequestering Cdk9 for Tat function, leading to pathogenic consequences. Because Cdk9 appears to play an important role in monocyte apoptosis and differentiation, it may not be a feasible therapeutic target in treatment of HIV-1 infection.
ACKNOWLEDGMENTS
We thank Richard Sutton for advice on retroviral vectors and Jeff Scott for cell sorting and flow cytometry analysis.
The work was supported by grant AI35381 (A.P.R.) and the Center of AIDS Research at BCM (AI 36211) from the National Institutes of Health. S.M.F. was supported by Training Grant T32 AI07471 from the National Institutes of Health.
REFERENCES
- 1.Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci USA. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castellanos M C, Munoz C, Montoya M C, Lara-Pezzi E, Lopez-Cabrera M, de Landazuri M O. Expression of the leukocyte early activation antigen CD69 is regulated by the transcription factor AP-1. J Immunol. 1997;159:5463–5473. [PubMed] [Google Scholar]
- 4.Durand D B, Bush M R, Morgan J G, Weiss A, Crabtree G R. A 275 basepair fragment at the 5′ end of the interleukin 2 gene enhances expression from a heterologous promoter in response to signals from the T cell antigen receptor. J Exp Med. 1987;165:395–407. doi: 10.1084/jem.165.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores O, Lee G, Kessler J, Miller M, Schlief W, Tomassini J, Hazuda D. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc Natl Acad Sci USA. 1999;96:7208–7213. doi: 10.1073/pnas.96.13.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu T J, Peng J, Lee G, Price D H, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem. 1999;274:34527–34530. doi: 10.1074/jbc.274.49.34527. [DOI] [PubMed] [Google Scholar]
- 7.Fujinaga K, Cujec T P, Peng J, Garriga J, Price D H, Grana X, Peterlin B M. The ability of positive transcription elongation factor b to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garber M E, Jones K A. HIV-1 Tat: coping with negative elongation factors. Curr Opin Immunol. 1999;11:460–465. doi: 10.1016/S0952-7915(99)80077-6. [DOI] [PubMed] [Google Scholar]
- 9.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garriga J, Mayol X, Grana X. The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem J. 1996;319:293–298. doi: 10.1042/bj3190293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garriga J, Peng J, Parreno M, Price D H, Henderson E E, Grana X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene. 1998;17:3093–3102. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- 12.Gold M O, Yang X, Herrmann C H, Rice A P. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidenreich S. Monocyte CD14: a multifunctional receptor engaged in apoptosis from both sides. J Leukoc Biol. 1999;65:737–743. doi: 10.1002/jlb.65.6.737. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann C H, Carroll R G, Wei P, Jones K A, Rice A P. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann C H, Gold M O, Rice A P. Viral transactivators specifically target distinct cellular protein kinases that phosphorylate the RNA polymerase II C-terminal domain. Nucleic Acids Res. 1996;24:501–509. doi: 10.1093/nar/24.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov D, Kwak Y T, Nee E, Guo J, Garcia-Martinez L F, Gaynor R B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for Tat-activation. J Mol Biol. 1999;288:41–56. doi: 10.1006/jmbi.1999.2663. [DOI] [PubMed] [Google Scholar]
- 18.John S, Robbins C M, Leonard W J. An IL-2 response element in the human IL-2 receptor alpha chain promoter is a composite element that binds Stat5, Elf-1, HMG-I(Y) and a GATA family protein. EMBO J. 1996;15:5627–5635. [PMC free article] [PubMed] [Google Scholar]
- 19.Kanazawa S, Okamoto T, Peterlin B M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 20.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator Tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 21.Lagasse E, Weissman I L. Enforced expression of Bcl-2 in monocytes rescues macrophages and partially reverses osteopetrosis in op/op mice. Cell. 1997;89:1021–1031. doi: 10.1016/s0092-8674(00)80290-1. [DOI] [PubMed] [Google Scholar]
- 22.Lecine P, Algarte M, Rameil P, Beadling C, Bucher P, Nabholz M, Imbert J. Elf-1 and Stat5 bind to a critical element in a new enhancer of the human interleukin-2 receptor alpha gene. Mol Cell Biol. 1996;16:6829–6840. doi: 10.1128/mcb.16.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lis J T, Mason P, Peng J, Price D H, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 24.Mancebo H S Y, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D H, Flores O. P-TEFb kinase is required for HIV Tat transactivation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 26.Marshall N F, Price D H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 28.Napolitano G, Licciardo P, Gallo P, Majello B, Giordano A, Lania L. The CDK9-associated cyclins T1 and T2 exert opposite effects on HIV-1 Tat activity. AIDS. 1999;13:1453–1459. doi: 10.1097/00002030-199908200-00003. [DOI] [PubMed] [Google Scholar]
- 29.Ott M, Emiliani S, Van Lint C, Herbein H, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science. 1997;275:1481–1485. doi: 10.1126/science.275.5305.1481. [DOI] [PubMed] [Google Scholar]
- 30.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price D H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanathan Y, Reza S M, Young T M, Mathews M B, Pe'ery T. Human and rodent transcription elongation factor P-TEFb: interactions with human immunodeficiency virus type 1 Tat and carboxy-terminal domain substrate. J Virol. 1999;73:5448–5458. doi: 10.1128/jvi.73.7.5448-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhim H, Rice A P. Exon2 of HIV-2 Tat contributes to transactivation of the HIV-2 LTR by increasing binding affinity to HIV-2 TAR RNA. Nucleic Acids Res. 1994;22:4405–4413. doi: 10.1093/nar/22.21.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serfling E, Avots A, Neumann M. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim Biophys Acta. 1995;1263:181–200. doi: 10.1016/0167-4781(95)00112-t. [DOI] [PubMed] [Google Scholar]
- 35.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutton R E, Wu H T, Rigg R, Bohnlein E, Brown P O. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J Virol. 1998;72:5781–5788. doi: 10.1128/jvi.72.7.5781-5788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taube R, Fujinaga K, Wimmer J, Barboric M, Peterlin B M. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology. 1999;264:245–253. doi: 10.1006/viro.1999.9944. [DOI] [PubMed] [Google Scholar]
- 38.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 40.Xaus J, Cardo M, Valledor A F, Soler C, Lloberas J, Celada A. Interferon gamma induces the expression of p21waf-1 and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity. 1999;11:103–113. doi: 10.1016/s1074-7613(00)80085-0. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 42.Yang X, Gold M O, Tang D N, Lewis D E, Aguilar-Cordova E, Rice A P, Herrmann C H. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N F, Marshall T A B, Amendt B, Mathews M B, Price D H. Transcriptional elongation factor p-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler S F, Levin S D, Johnson L, Copeland N G, Gilbert D J, Jenkins N A, Baker E, Sutherland G R, Feldhaus A L, Ramsdell F. The mouse CD69 gene. Structure, expression, and mapping to the NK gene complex. J Immunol. 1994;152:1228–1236. [PubMed] [Google Scholar]