Abstract
Kelps are vital for marine ecosystems, yet the genetic diversity underlying their capacity to adapt to climate change remains unknown. In this study, we focused on the kelp Macrocystis pyrifera a species critical to coastal habitats. We developed a protocol to evaluate heat stress response in 204 Macrocystis pyrifera genotypes subjected to heat stress treatments ranging from 21 °C to 27 °C. Here we show that haploid gametophytes exhibiting a heat-stress tolerant (HST) phenotype also produced greater biomass as genetically similar diploid sporophytes in a warm-water ocean farm. HST was measured as chlorophyll autofluorescence per genotype, presented here as fluorescent intensity values. This correlation suggests a predictive relationship between the growth performance of the early microscopic gametophyte stage HST and the later macroscopic sporophyte stage, indicating the potential for selecting resilient kelp strains under warmer ocean temperatures. However, HST kelps showed reduced genetic variation, underscoring the importance of integrating heat tolerance genes into a broader genetic pool to maintain the adaptability of kelp populations in the face of climate change.
Subject terms: Plant sciences, Zoology, Bioinformatics, Imaging, DNA sequencing
204 Macrocystis pyrifera genotypes were screened and a panel of heat-stress tolerant (HST) gametophytes were identified. A subsequent analysis revealed a correlation between the HST strains and increased sporophyte biomass under warmer ocean conditions.
Introduction
The increasing frequency and duration of marine heat waves, likely exacerbated by climate change, present a growing threat to kelp forests, an ecologically important habitat formed by large brown algae (i.e., kelps) in shallow coastal waters that provide a myriad of goods and services to society (as reviewed in refs. 1,2). There is a pressing need to understand how kelps respond to heat stress and to explore their potential for heat-stress tolerance (HST) or adaptation3,4. Extreme climatic events like marine heatwaves often exceed the physiological limits of individual organisms within a population, leading to selective mortality that can drive evolutionary change5. Studies show many species of kelp are highly susceptible to local extinctions and range contractions caused by marine heatwaves6–8. Consequently, decreases in genetic biodiversity caused by temperature extremes may hinder the capacity of kelp populations to adapt to future climate change and other challenges9,10. However, recent findings by Klingbeil et al.11 suggest that kelp populations in southern California have maintained stable genetic diversity despite prolonged warming events, indicating potential HST. Additionally, Mohring et al.12 emphasize the importance of research on the under-explored microscopic gametophyte stage of kelps, particularly their response to abiotic stressors, to fully understand kelps resilience. Building upon this, we hypothesized a predictive relationship between HST traits across the biphasic life stages of kelps13.
The biphasic haplodiplontic life history of kelps (Order Laminariales), characterized by a microscopic haploid gametophyte and macroscopic diploid sporophyte, likely plays a key role in their HST. Veenhof et al.14 underscore that traits associated with the microscopic stages of kelp’s complex life history are central to its adaptive capacity, yet research at this life-cycle stage in this area is sparse. Research by Wernberg et al.8 shed light on the need to monitor the response of marine algae to warming anomalies with the understanding that extreme climate events will shift and damage critical kelp ecosystems. Further, Veenhof et al.14, found that survival, relative growth rate (RGR) and sex ratio of the gametophytes the kelp Ecklonia radiata from different latitudes (high, mid, and low) tended toward adaptation to their local temperatures, with a heat stress maximum of 2–3 °C above in situ temperatures14.
In this study, we examined how genetic differences in HST of the gametophyte stage of Macrocystis pyrifera (M. pyrifera) are associated with the growth of the sporophyte stage. Using microscopy-derived chlorophyll fluorescent intensity (FI) values from a 3D tomography system, we determined the HST of 204 M. pyrifera genotypes obtained from a germplasm collection derived from southern California, USA15,16 and compared this to the biomass yield of related sporophytes from an independent experiment conducted in 2019, in which sporophytes were cultivated in an in situ ocean setting during the warm summer season.
Finally, macroalgal germplasm banking has been introduced in recent years to preserve the biodiversity of marine algal species with the potential to aid in kelp restoration initiatives and regenerative ocean farming efforts16. Here we search for HST tolerant gametophyte strains in a large giant kelp germplasm collection that can assist future restoration efforts. Our findings suggest a predictive relationship between gametophyte-stage HST and sporophyte-stage growth performance, underscoring the potential for selecting resilient kelp strains under warmer ocean temperatures.
Results
Genetic variability and temperature effects on gametophyte fluorescent intensity
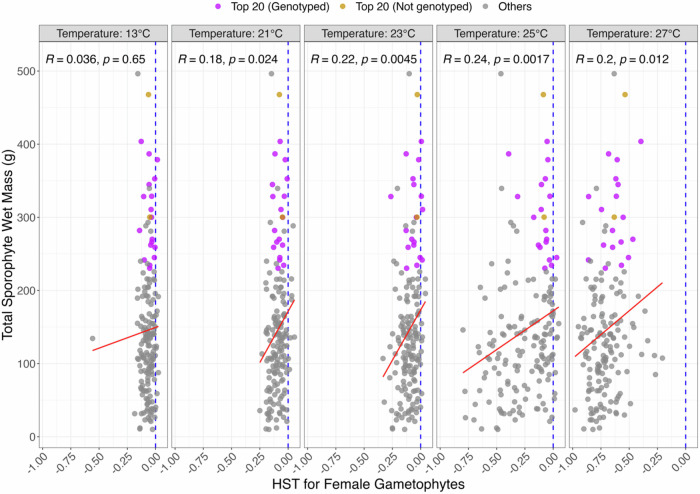
For gametophytes treated at temperatures 13 °C (Control) and 21 °C, we observed homogeneity in the response across genotypes, indicating that the existing genetic variability did not affect gametophyte growth or survival within this temperature range. The onset of phenotypic variation became noticeable at 23 °C and substantial at 25 °C, with certain genotypes exhibiting markedly lesser tolerance to temperatures than others. Notably, at the upper-temperature limit of 27 °C, fluorescent intensity (FI) for all genotypes significantly decreased and remained consistently low during the treatment period, leading to remarkably negative HST values, and suggesting that 27 °C represents a critical threshold beyond which gametophyte viability sharply declines (Fig. 1).
Fig. 1. Correlation analysis reveals a correspondence between sporophytes out-planted in warm water conditions offshore Santa Barbara, CA, and their parental female gametophytes heat-stressed in ex situ conditions.
Sporophytes were harvested in September 2019. The blue dotted lines show neutral response to heat stress (HST = 0), the red lines are linear regression between y- and x- axes. Gametophyte HST values plotted are averaged, normalized FI values from week 0-week 4 and correlated to sporophyte biomass. Correlations with P-values are placed on the top right corner (R and p, respectively).
Correlation of gametophyte HST with sporophyte biomass
The strongest association between gametophyte HST and sporophyte biomass was observed when HST was estimated at 25 °C (R = 0.24, P = 0.0017), followed by the associations at 23 °C (R = 0.22, P = 0.0045) and 27 °C (R = 0.20, P = 0.012) (Fig. 1). The latter case reinforces the relevance of HST in early-stage gametophytes to the later-stage growth performance of sporophytes. M. pyrifera, a perennial kelp, begin their growing season in the early spring when temperatures are below what one can expect in July and August. Therefore, because we grew sporophytes during warmer summer months, disparate from their regular growing season, we are considering the temperatures at which they grew from juvenile sporophytes to adult sporophytes warmer ocean conditions. It is notable in our research that at 25 °C in ex situ conditions, gametophytes had the highest degree of variability. Examining the genotypes that thrived at 25 °C, we found that these same genotypes also exhibited the highest biomass during warmer summer months.
Identification of high-performing genotypes
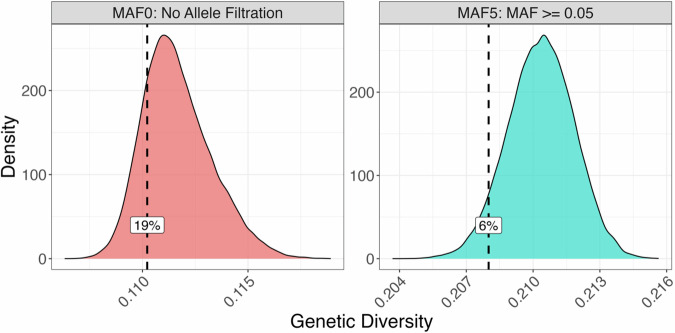
By comparing the HST of female gametophytes treated in the laboratory with the biomass of sporophytes grown in the ocean, we identified 20 gametophytes performing best in both HST at 25 °C and related sporophytes’ biomass across three of the original four populations studied (Table 1). Among these 20 female gametophytes, 18 were genotyped, meaning we could derive haploid single nucleotide polymorphisms for them. Next, we ran a subsequent analysis based on average pairwise Identity-by-State statistics to determine their overall genetic diversity. We chose this approach over conventional heterozygosity estimations because our gametophytes are not diploid organisms. We found that the 18 HST genotypes exhibited remarkably less genetic variability than 18 random individuals sub-sampled with the same population distribution (Fig. 2).
Table 1.
Top 20 performing female gametophytes based on HST and progeny’s biomass; AQ – Arroyo Quemado, CI – Catalina Island, CP – Camp Pendleton. Rank is in order of the top performing genotype for both HST and biomass to the 20th top performing
| Female Gametophyte | Population | HST | Sporophyte’s Biomass | Genotyped | Rank |
|---|---|---|---|---|---|
| CP.46.F.A1 | CP | −0.087 | 467.80 | No | 1 |
| CP.04.F.A3 | CP | −0.054 | 403.75 | Yes | 2 |
| CP.70.F.D3 | CP | −0.047 | 378.75 | Yes | 3 |
| AQ.59 A.F.C1 | AQ | −0.069 | 352.67 | Yes | 4 |
| AQ.62.F.A5 | AQ | −0.029 | 328.75 | Yes | 5 |
| CI.17.F.C3 | CI | −0.104 | 344.80 | Yes | 6 |
| AQ.28.F.A5 | AQ | −0.106 | 310.67 | Yes | 7 |
| CP.45.F.D4 | CP | −0.080 | 300.25 | No | 8 |
| CI.43.F.D3 | CI | −0.065 | 282.00 | Yes | 9 |
| CP.73.F.B2 | CP | −0.172 | 300.00 | Yes | 10 |
| CP.01.F.D3 | CP | −0.057 | 270.00 | Yes | 11 |
| CP.67.F.C4 | CP | −0.062 | 266.25 | Yes | 12 |
| CI.36.F.B4 | CI | 0.031 | 245.00 | Yes | 13 |
| CP.05.F.D2 | CP | −0.116 | 259.00 | Yes | 14 |
| CI.22.F.D2 | CI | −0.130 | 262.00 | Yes | 15 |
| CI.01.F.A4 | CI | −0.032 | 241.67 | Yes | 16 |
| CP.41.F.D1 | CP | −0.015 | 234.40 | Yes | 17 |
| AQ.21.F.C1 | AQ | −0.315 | 328.40 | Yes | 18 |
| CP.66.F.B1 | CP | −0.074 | 230.40 | Yes | 19 |
| CI.58.F.C2 | CI | −0.394 | 386.75 | Yes | 20 |
Fig. 2. Genetic diversity analysis reveals lower genetic variability in 18 heat stress-tolerant (HST) gametophytes in the context of 18 random gametophytes with the same population distribution.
Each panel represents the sampling distribution of genetic diversity (average pairwise Identity-by-State, GenDiv, (Eq. 5)) from 50,000 permutations. The observed GenDiv in the selected 18 HST female gametophytes is indicated by the dashed lines and corresponding percentile values. The left panel is based on the MAF0 SNP dataset, which includes all SNPs, while the right panel uses the MAF5 SNP dataset, where SNPs with minor allele frequencies less than 0.05 were removed.
Discussion
Our results support that gametophyte genotypes exhibiting greater heat-stress tolerance at 25 °C in ex situ laboratory environments also correspond to genetically similar adult sporophytes grown in situ warmer (18 °C–20 °C) summer months exhibiting higher biomass phenotypes. Research by Hollarsmith et al.17 treated M. pyrifera gametophytes from higher-latitude populations in California and found significant reproductive failure at elevated temperatures, whereas lower latitude strains from San Diego populations exhibited greater reproduced success under the same treatment conditions. Such predictive relationships suggest that even at the earliest stages of development, the genetic tolerance or susceptibility to heat stress can be gauged. This finding may have significant ramifications for kelp forest management aimed at mitigating the escalating effects of climate change. For example, Buschmann et al.18, demonstrated that M. pyrifera gametophyte cultivation strategies focused on optimized cultivation practices through selective breeding could help to “future-proof” kelp in regenerative ocean farming conditions under threat of warming ocean conditions. Similarly, the use of HST genotypes in kelp forest restoration has been posited to improve the success and long-term resilience of these initiatives19,20.
It is important to note that we cannot definitively conclude HST strains will produce HST sporophytes without further investigation. Building upon the HST screening data, future studies should examine the heat tolerance of sporophytes derived from both non-HST and HST strains. As suggest by Umanzor et al.21, functional validation steps in this field could provide valuable insights into the potential for transgenerational inheritance of HST traits in M. pyrifera.
Other environmental factors including light and nutrient availability may contribute to the observed differences in HST between genotypes in both the heat stress screen and in situ sporophyte experiment. Umanzor et al.21 revealed complex interactions between the combined effects of temperature and nitrate availability on juvenile sporophytes. Further studies utilizing the gametophyte data set should include a more comprehensive understanding of interactions of light, temperature, and nutrient availability as our focus solely on temperature manipulations lead to a more conservative estimate of HST differences between genotypes. Finally, in vascular plant systems tissue and organ-dependent HST is variable22; a consideration of future M. pyrifera HST research should seek to understand the translation of HST across life stages as well as tissue types.
In the ex situ gametophyte screening panel for HST, the use of kelp genotypes originating from populations with an approximately 1–2 °C average annual temperature difference and resultant pattern of HST genotypes ranging across populations suggests that HST in M. pyrifera may be influence by other factors such as phenotypic plasticity and genetic diversity. Populations of M. pyrifera along the Chilean coast were determined to have significant variation in their genetic and phenotypic diversity, underscoring the importance of considering genetic and phenotypic diversity with developing breeding programs for this species23. Because our findings shed light on HST from populations exhibiting slight differences in temperature climes, it’s important to consider that heat tolerance is potentially influenced by both genetic and environmental factors.
Our identification of HST genotypes provides insight into the genetic structure and diversity of heat-stress adaptability in M. pyrifera populations in southern California. However, we also conclude here that there is a lower, yet not significant, genetic variation among HST strains. This may be confounded by sample size, however, the need to determine genetic variation rests on the supposition that lower genetic variation in HST genotypes underscores the potential for certain alleles to confer heat tolerance. Therefore, this trait is likely to be selected under the increasing threat of climate change. Management strategies aimed at using resilient genotypes to mitigate the effects of climate change should consider integrating thermal tolerance genes into a wider range of genetic backgrounds. Such introgression would likely help sustain a broad genetic base, which is essential for the long-adaptability and resilience of kelp populations.
This approach could help balance the benefits of HST genotypes with the need to preserve genetic variation that might be critical for other aspects of kelp survival and adaptability. These results may have significant implications for conservation strategies regarding vulnerability to climate change, breeding and restoration programs and monitoring genetic health. Lower genetic diversity in HST strains used in restoration initiatives would likely lack the variability necessary to adapt to changing conditions and may prompt conservationists to preserve a broader genetic base. Finally, regular monitoring of genetic health and diversity for natural and restored populations may be necessitated when incorporating a mix of genotypes that include HST strains and those with higher genetic variability.
Methods
Ex-situ gametophyte heat stress treatments
A phenotyping protocol was developed to evaluate heat stress response in M. pyrifera gametophytes. Chlorophyll autofluorescence of each gametophyte, indicated by fluorescence intensity (FI) values, was used as an indicator of a genotype’s heat stress response. Utilizing In Vivo Imaging System (IVIS) intravital 3D tomography, we analyzed FI values to record heat stress phenotypes in 204 (165 female and 39 male) M. pyrifera gametophytes with 3-fold replication per genotype. Gametophytes were subjected to four heat stress treatments, the independent treatment variables, at 21 °C, 23 °C, 25 °C, and 27 °C, with a control of 13 °C for four weeks.
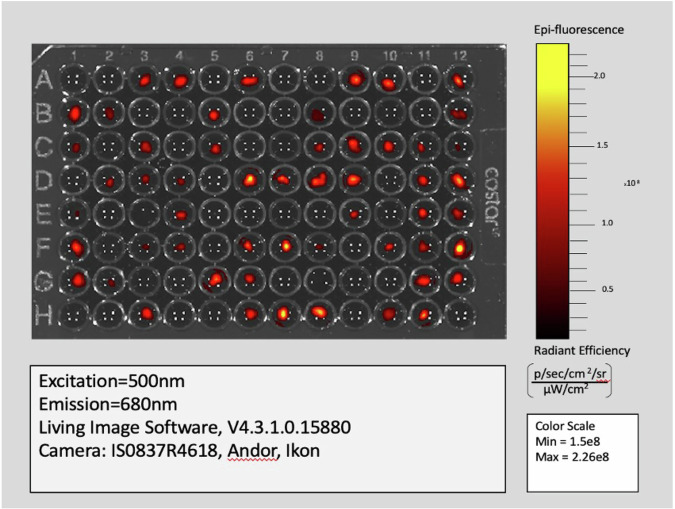
FI values are generated by Living Image Software as an output of the IVIS imaging system (Fig. 3) and are employed in this study to quantify the measure of chlorophyll fluorescent signal present in kelp gametophyte chloroplasts, allowing for relative comparison of signal between living kelp gametophytes over time. Chloroplasts contain a mix of different chlorophyll, the main photosynthetic pigment. We estimate that multiple chlorophyll types are fluorescing at 680 nm emission (Fig. 3); therefore, we broadly refer to “chlorophyll” without specifying the exact type. This non-invasive screening approach is a widely-used tool to study plant physiology, often to understand abiotic stress response as chlorophyll fluorescence is sensitive to abiotic stress that affects photosynthesis24,25 Further, Harris et al.26 developed a high-throughput method to determine photosynthetic thermal tolerance in species of kelp using chlorophyll fluorometry via temperature-dependent fluorescence (T-F0) curve method for three kelp species at the sporophyte stage. The study found that this metric could efficiently detect thermal tolerance differences among three kelp species26.
Fig. 3. Fluorescence imaging of giant kelp gametophytes at Week 0 using the IVIS 3D tomography system.
Only wells with visible red or yellow pixels contains a gametophyte. A 96-well plate containing the samples was positioned within the system with the imaging parameters set to emission 680 nm, excitation 500 nm, Epi-Illumination, Bin: (HR)4, FOV:13.2, F2, 2 s. Fluorescent intensity (FI) values were captured every 2 weeks over a period of four weeks, starting from Week 0, using LivingImage® software with a 12 × 8 grid. Data are presented in continuous fluorescent intensity units of [p/s] / [µW/cm²], allowing for the monitoring of changes in FI values per replicate throughout the thermal stress treatments.
The choice of the control temperature at 13 °C is based on established protocols for maintaining healthy M. pyrifera gametophytes, which have shown optimal growth and development at this temperature27,28. The gradient of heat stress treatments from 21 °C to 27 °C was selected to represent a range of temperatures above the average sea surface temperature along the Southern California Bight, where this species is commonly found. This range is consistent with observed temperature anomalies during marine heatwaves and is relevant for understanding the thermal tolerance of kelp gametophytes in the context of climate change29,30. Our study investigated the effects of these elevated temperatures at the gametophyte stage, as previous research has indicated that temperatures exceeding 23 °C can induce heat stress in kelp species (Deihl et al.31). This information is crucial for predicting the health and adaptability of kelp populations in the face of rising ocean temperatures and for informing conservation and management strategies for kelp ecosystems (Wernberg et al.4).
Two-hundred and four kelp gametophytes originating from a large germplasm collection were randomized across black Eppendorf ™, PCR clean, u-bottomed microplates using the Well-Plate Maker (WPM) package in RStudio32. Gametophytes in the germplasm collection were first released as zoospores and preserved under specific light, nutrient, and temperature conditions from 2018 as described by Osborne et al.15. Per genotype, a single gametophyte was extracted from the germplasm collection and fragmented using Millipore Sigma blue polypropylene pellet pestles (catalog number: Z359947). Mechanically-induced parthogenesis then occurs, allowing for the growth of new gametophyte filaments. Gametophyte filaments were allowed to mature for approximately 1 month. Post fragmentation, three replicates per genotype were chosen. There were no selection criteria utilized other than determining that gametophytes were pigmented and therefore healthy. Sizes of gametophytes varied (between ~100 μM to 1 mm) as did their total number of cells. To control for this variation, fluorescent intensity values were normalized (see FI Normalization and HST Estimation below for details).
Gametophytes were plated in full-strength Provasoli Enriched Seawater media (PES) as per the guidelines in Redmond et al.33. After plating, gametophytes were first conditioned in a Conviron™ Incubator set at 13 °C. Regular white LED lights, covered with red plastic film, were used to facilitate low-light conditions at approximately 8 μMol photons m−2 s−1, for a 12:12-hr light: dark cycle, ensuring that red wavelengths were maintained at minimal levels.
Following the initial conditioning period of one week, gametophytes were exposed to four heat stress treatments within Thermo Scientific™ Heratherm IMC18 Benchtop Incubators. Heat stress temperatures were set at 21 °C, 23 °C, 25 °C, and 27 °C. Inside each incubator, Marreal brand Red LED Strip Lights were installed to ensure that red wavelengths were maintained at low levels, supporting the optimal growth conditions for the gametophytes. Additionally, a control group was maintained at 13 °C.
We recorded FI values at 0, 2, and 4 weeks after initiating heat stress. We estimated HST as the slope in regressions of normalized FI values (dependent variable) against time (independent variable) with a fixed 0 intercept. Negative and positive values of HST represent rates of autofluorescence loss versus increase, respectively. To test whether HST at the microscopic gametophyte stage is associate with HST at the macroscopic sporophyte stage, we estimated the correlation between female gametophytes HST and the biomass of sporophytes phenotyped in an ocean kelp farm experiment (2018–2019). Sporophytes growing in this experiment were bred by crossing a single male gametophyte originating from the kelp germplasm collection, from one population (Leo Carrillo) with 500 female gametophytes from four populations and including the 165 female gametophytes characterized for HST here (Fig. 4). Prior to out planting, sporophytes were cultivated at 13 °C. The sporophyte crossing schema in the ocean farm experiment used the same genetic lines as the female gametophytes screened for HST. Due to this genetic relatedness, we were able to directly compared the HST of the female gametophytes and the biomass of their offspring (sporophytes) under ex situ warmer ocean conditions.
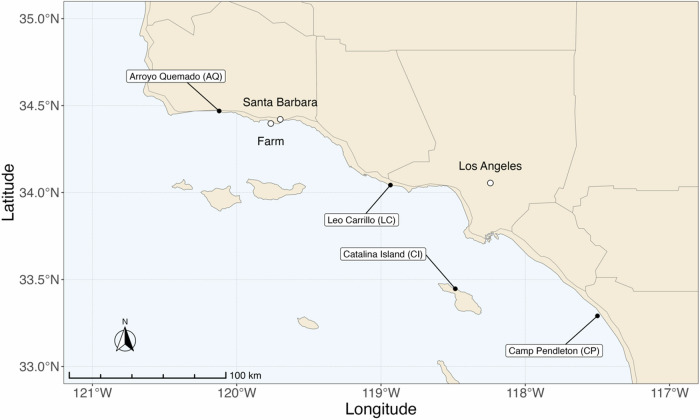
Fig. 4. Map of M. pyrifera sampling locations across the Southern California Bight and the farm location.
This map illustrates each of the four sampling sites. The northernmost population of Arroyo Quemado has a 1–2 °C colder average SST compared to the southernmost population of Camp Pendleton, CA39. The farm location is proximal to the Santa Barbara coastline and demarcated by a white circle. This distribution highlights the distinct sampling regions. Geographic distribution of the farm and sampling locations were marked using latitude and longitude coordinates. The R package “rnaturalearth” was employed to retrieve the base map, the R packages, “maps”, “sf”, “ggspatial”, and “tidyverse” were used to manipulate the spatial data and to plot the map.
Embryonic sporophytes of the 500 genotypes in replicates of 5 per genotype for a total of 2500 sporophytes were out-planted to a coastal ocean farm near Santa Barbara in early May 2019. For the crossing protocol, the Leo Carrillo male was mixed with female gametophytes to allow for fertilization; fertilization can take anywhere from one to two weeks. Successful crosses can produce embryonic sporophytes within 1 to 2 weeks.
Sixty-seven percent of the out planted sporophytes (1686), represented by 491 genotypes, survived until mid-September 2019, a period of approximately four and a half months with an increasing pattern of ocean temperatures (Fig. 5). These sporophytes were harvested and weighted wet, and average biomass was calculated for each of the 491 genotypes. Of the 165 female gametophytes participating in the ex-situ heat stress treatments, 164 were represented among the survived genotypes from the farm biomass data.
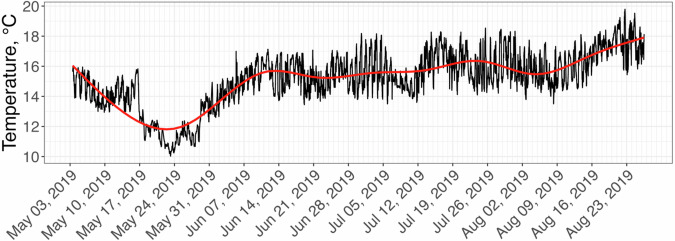
Fig. 5. Averaged temperature pattern on the farm between 05/03/2019 and 08/27/19.
The red line is a smooth approximation representing the temperature trend. Temperature data, provided by HOBO data loggers, reveal average SST for the time that the juvenile sporophytes were grown at the farm from May through August 2019.
IVIS fluorescent imaging analysis
To conduct a fluorescence imaging experiment using the IVIS 3D tomography system, a 96-well plate was placed within the system, ensuring it perfectly aligned to facilitate accurate downstream data analysis. Next, the imaging parameters on the system controls were set at emission 680 nm, excitation 500 nm, Epi-Illumination, Bin: (HR)4, FOV:13.2, F2, 2 s. Starting at Week 0, FI values of each gametophyte were captured every 2 weeks for a total of three time-points, using LivingImage® software Region of Interest (ROI) tool and 12 × 8 grid (Caliper Life Sciences34). Data was extracted as continuous fluorescent intensity units of [p/s] / [µW/cm²]. The imaging data were used to monitor the changes in FI values per replicate throughout the thermal stress treatments (Fig. 3)34.
Sporophyll sample collection and In-Situ experimental setup
Sporophylls (i.e., reproductive tissue) of M. pyrifera sporophytes containing zoospores, the progenitors of gametophytes, were collected from four natural spanning regions in southern California (Arroyo Quemado 34. 468783° N -120.121417 ° W, Leo Carrillo 34.042933° N -118.934500° W, Catalina Island 33.446882° N, -118.485067 ° W, and Camp Pendleton 33.29091° N -117.499969° W).
Genetic groups were first distinguished in these regions in 2015 and are characterized by their rates of genetic divergence15,35. Using 2023-2024 sea surface temperature (SST) data reported by NOAA’s Environmental Research Division Data Access Program, the average annual SST for Arroyo Quemado was 15.59 °C, Leo Carrillo 17.11 °C, Catalina Island 17.73 °C and the southern-most population of Camp Pendleton was 18.05 °C38. These patterns present a notable, albeit non-linear, sea surface temperature gradient from north to south across these marine environments.
While detailed descriptions of the 2019 farm design can be found in Osborne et al.15, the main features will be briefly summarized here. Single genotype male and female gametophyte cell cultures were fragmented into filaments approximately 5–10 cells long15 and “seeded” on polyvinyl strings with a 6 cm long and 2 mm diameter size and exposed to increasing white light levels over four days (15, 22, 35, to 60 μmol photons. m − 2. s − 1), with the resulting crosses growing for a month until sporophytes developed and were shipped to a marine laboratory at the University of California Santa Barbara. Prior to out-planting, the sporophyte strings were attached to a seeding line, and ten seedling lines with 250 genotypes each were fastened to longlines by divers, with one set of 500 genotypes out-planted to two adjacent longlines on the farm in May 2019. Ocean temperatures (Fig. 4) at the experimental farm increased throughout the summer following a brief period of upwelling in May with temperature peaking in August, which is typically the warmest month of the year. Temperature data was collected via 5 HOBO Pendant Temperature and Light Loggers (model # UA-002-64), arrayed across alternating lines and recording data at 10-min intervals. Daily averaged temperatures were recorded as 15.5 ± 1.6 °C (mean ± SD, range = 9.9–21.8 °C). Approximately 125 days after out-planting, the 1686 surviving sporophytes were harvested between September 7–12, 2019, and returned to the laboratory, where they were weighed wet. The wet mass of the 491 (out of 500) surviving genotypes (averaged across replicates) ranged from 5 g to 467.8 g.
Gametophyte isolation and DNA Extraction
Fertile sporophylls, originating from distinct southern California populations35, were shipped overnight to the University of Wisconsin-Milwaukee where we induced spore release via the Oppliger method approximately 24 h. post-collection (ref. 15; Oppliger et al.36). Sporophylls were first cleaned by being dipped into 10% iodine solutions for no more than 30 s to remove excess epiphytes and bacteria. Zoospore release was induced by submerging whole sporophylls containing the reproductive tissue (sori) into sterile Provasoli-Enriched Seawater (PES) (Provasoli37) made with Instant Ocean Sea Salt and ultrapure water (Simplicity water system) at a salinity of 34 parts per thousand (PPT)15. Zoospores were inoculated in 60 × 15 mm Petri dishes with 10 mL of PES. Zoospore release and sporophyll cleaning was carried out at ambient temperatures of approximately 20 °C.
Each treated blade corresponded to an adult kelp sporophyte donor. For each donor, two final densities of 10 and 100 zoospores. mm−2 15. Zoospores were then incubated in growth chambers at 12 °C with a 12:12 h light:dark photoperiod at 6 ± 3 photons m−2 s−1 red light. PES media was replaced every 5 weeks33. After zoospore germination, gametophytes were allowed to grow vegetatively to approximately 100 μm size before being sexed and isolated15. Before DNA extraction, individual gametophyte samples were mildly centrifuged, and the supernatant discarded, yielding 50–100 mg of gametophyte biomass. Gametophyte tissue was ground using liquid nitrogen. The NucleoSpin 96 Plant Kit (Macherey-Nagel) obtained high-quality genomic DNA from 500 female and 100 male gametophyte lines.
Gametophyte Sequencing, SNP Calling and Preprocessing
Extracted gametophyte genomic DNA was prepared for DNA library preparation (BGI North America NGS labs) and whole-genome re-sequencing. Post-library preparation, an Illumina S4 Novaseq platform was used to sequence the samples, yielding 11.2 GB of 150 base pair reads for each sample. A total of 559 samples underwent sequencing. Raw 150 base pair Illumina reads were trimmed of adapters, low-quality reads, and tails using a fast set with standard parameters39 and then aligned to the M. pyrifera nuclear genome.
Trimmed reads were aligned to the nuclear genome of M. pyrifera40 using hisat2 v2.1 with standard parameters41. Bam files had their duplicates marked using the GATK4 v4.1.2 command “MarkDuplicates”, and then multiple bam files for a single individual genotype were collapsed into a single bam file using samtools42,43. Genetic variants were called using the GATK4 v4.1.2 with ploidy set to 143. Individual GVCF files were then merged and converted into a raw VCF file containing variant information and used for downstream applications using GATK v4.1.243.
The obtained raw VCF file was utilized for the extraction of biallelic SNPs. Then, we performed hard filtering with the thresholds suggested by GATK for filtering germline short variants44. Next, we kept SNPs present in scaffolds and had a mean depth between 2 and 10 and a percentage of missing data of less than 7.5%. Finally, gametophytes with a mean depth of less than 2 were excluded from the final dataset. Additionally, we removed all monomorphic SNPs generated after removing the gametophytes. The filtered VCF file was comprised of 504 gametophytes and 1,515,399 biallelic SNPs. All extractions were performed using vcftools v0.1.1445 and R46.
To impute missing data, we developed the following pipeline. We recorded the filtered VCF file into the PLINK binary format (*.bed, *.bim, and *.fam) and divided data based on scaffolds. Each scaffold was imputed 100 times using LinkImpute v1.1.547 with the parameter –nummask = 500000 if the number of non-missing entries was more than 500000; otherwise, we used the number of non-missing entries itself as the parameter value. Between two alleles, the final imputed one is that have a bigger score (the sum of two scores equals 100). Then, the imputed scaffolds were merged back. All manipulations with SNP data at this stage, except the imputation, were performed using PLINK v1.948,49 and R.
The resulting dataset was MAF0 (imputed; 504 gametophytes and 1,515,399 SNPs), indicating no allele filtration. Additionally, we prepared a dataset with filtrated alleles based on minor allele frequency. SNPs with minor allele frequency less than 0.05 were excluded resulting in 504 gametophytes and 602,493 SNPs.
FI Normalization and HST Estimation
The extracted FI data was preprocessed and used to estimate HST for the screened 204 gametophytes. FI values reflect the size of gametophytes given that the larger the gametophyte, the more cells or chloroplasts are likely to be present thus increasing the over chlorophyll fluorescence. FI normalization adjusted for initial size differences between gametophytes at week0. Post normalization, HST are a single slope value per genotype; this slope is from the regression of normalized FI on weeks 0, 2 and 4 (see below). A neutral HST of 0 represents a genotype that did not lose significant amounts of FI over the four-week heat stress treatment period. For those with negative HST, there was a loss of FI over the four-week treatment period (Fig. 1). Each replicate had three FI values, FI.0, FI.2, and FI.4, for weeks 0, 2, and 4, respectively, for 5 different treatment temperatures (9180 recorded FI values in total). These values were transformed for each replicate under a particular treatment as follows:
| 1 |
Next, for each replicate, we used a linear regression with a zero intercept to model HST based on the normalized FI values:
| 2 |
where and , respectively. To estimate HST, we used the R function lm() with the formula Norm.FI ~ Week + 046. The HST for a treatment temperature for a genotype was calculated as an average of the replicates’ HST values (1020 values as a result; genotypes had HST value for each of the five temperature treatments). Overall, this approach allows us to treat FI as if they obey an exponential decay/growth model where HST serves as a decay/growth constant depending on its sign.
Correlation analysis
Upon merging the heat stress experiment data (165 female gametophytes) and the farm experiment data (491 parental female gametophytes), we obtained an intersection in 164 female gametophytes, with 145 having genotype information. These 145 genotyped gametophytes are therefore present in the MAF0 and MAF5 datasets. We graphed these 164 gametophytes in Fig. 1, placing the HST data on the x-axis (five facets for the five treatment temperatures) and the sporophytes’ biomass on the y-axis. Depicted regression lines were estimated using the geom_smooth() function with parameters method = “lm” and formula = “y ~ x” from the ggplot2 package50, while correlations with p-values were calculated using the stat_cor() function from the ggpubr package51.
To define the top 20 performing gametophytes, we scaled both the HST at 25 °C and the biomass data (BM) to the range using the following formulas:
| 3 |
A Euclidian distance was calculated between and for each gametophyte. The 20 gametophytes closest to the point were then selected based on these distances. Among these 20 gametophytes, 18 had genotypes, which we emphasized in Fig. 1 by different colors. The population distribution was the following: 4 from Arroyo Quemado, 6 from Catalina Island, and 8 from Camp Pendleton.
Genetic diversity analysis
To assess how genetically distant two gametophytes are, we applied the following formula:
| 4 |
Where g1 and g2 are the genotypes of two gametophytes based on SNPs, and IBS (g1, g2) is Identity-by-State between two genotypes showing the fraction of shared alleles. The proposed metric ranges between 0 and 1, with 0 representing identical genotypes and 1 being the opposite case. To estimate the genetic diversity of a set of gametophytes, we averaged the genetic distances of all unique gametophyte pairs in the set, i.e.,:
| 5 |
where S is the set of gametophytes, P is the set of the unique pairs, and is the number of the unique pairs.
We aimed to investigate the genetic diversity of our 18 top-performing gametophytes to understand if there was a lower genetic diversity for HST genotypes. We created a sampling distribution of genetic diversity with 50,000 random sets of gametophytes drawn from our genotype data. Each set had the same population distribution consisting of 4 from Arroyo Quemado, 6 from Catalina Island, and 8 from Camp Pendleton. Each set was generated to ensure that population-specific subsets weren’t duplicated. This means that if gametophytes from one population were present in a set, then they would not be repeated in other sets. Additionally, we estimated what the percentile of the modeled distribution is and the genetic diversity of the top 18 performing gametophytes. This analysis was performed for both SNP datasets, MAF0 and MAF5.
Statistics and reproducibility
Statistical analysis was performed using R (version 4.2.046). FI data was normalized using Formula 1, where w represents weeks 0, 2 and 4. HST was estimated as the slope from a linear regression analysis, using the R function “lm()”, of normalized FI on weeks 0, 2 and 4 with a zero intercept (Formula 2). Next, HST values were averaged across replicates for each genotype at each treatment temperature (21 °C, 23 °C, 25 °C, and 27 °C).
Sample size for this study included 204 genotyped strains of M. pyrifera gametophytes, each in replicates of 3 per genotype, resulting in a total of 9180 recorded FI values. For each replicate, FI values were measured at weeks 0, 2, and 4 across the five different temperature treatments: 13 °C (Control), and 21 °C, 23 °C, 25 °C, and 27 °C (heat stress treatments). Each replicate comprised FI values (FI.0, FI.2, FI.4) for each of the specified timepoints. Reproducibility was ensured via randomizing gametophytes using the Well-Plate Maker (WPM) package in RStudio32. The control temperature of 13 °C was based on established protocols and previously published standards for M. pyrifera gametophyte cultivation and maintenance15. Further, Normalization of FI allowed for the adjustment of initial size differences between gametophytes at week 0.
Data validation was done using the LinkImpute package for missing data and genetic diversity analysis was conducted using Identity-by-State (IBS) statistics to assess genetic variability among top-performing genotypes.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank all team members from University of Wisconsin-Milwaukee, University of Southern California, and UCSB who were involved in the harvest and phenotyping of the kelp farm. We also thank our collaborators Gevorg Karapetyan and Seda Mkhitaryan of Children’s Hospital Los Angeles for their assistance in gathering fluorescent intensity data and utilizing the IVIS imaging system. This work is funded by: Advanced Research Project Agency - Energy (Grant Number: GR1022773) and the Builders Initiative, Research and restoration of Giant kelp at Catalina (Grant Number: GR1063448).
Author contributions
Maddelyn Harden: Conceptualization (equal); Data collection (lead); Writing – original draft (lead); Writing – review & editing (lead). Maxim Kovalev: Statistical analysis (lead); Modeling (lead); Writing – review & editing (supporting). Gary Molano: SNP data provision (lead). Christie Yorke: Data collection – sporophyte data (equal); Writing – review & editing (supporting). Robert Miller: Data collection – sporophyte data (equal); Writing – review & editing (supporting). Daniel Reed: Data collection – sporophyte data (equal); Writing – review & editing (supporting). Filipe Alberto: Germplasm collection (lead); Writing – review & editing (supporting). Rusty Lansford: Imaging protocols (equal); Staining protocols (equal); Fixation protocols (equal); Writing – review & editing (supporting). David S. Koos: Imaging protocols (equal); Staining protocols (equal); Fixation protocols (equal); Writing – review & editing (supporting). Sergey Nuzhdin: Conceptualization (lead); Project administration (lead); Supervision (lead); Writing – review & editing (equal).
Peer review
Peer review information
Communications Biology thanks Pablo P. Leal, José Sandoval-Gil and Katja Schweikert for their contribution to the peer review of this work. Primary Handling Editors: Linn Hoffmann and David Favero.
Data availability
The reference genome of Macrocystis pyrifera is available at the Joint Genome Institute portal https://phycocosm.jgi.doe.gov/Macpyr2. The WGS data of 559 gametophytes used to produce the VCF files are available in the NCBI repository under the accession code PRJNA1050779. The correspondence between the gametophytes’ names and the SRA runs in the NCBI repository, as well as the raw fluorescence intensity values, can be found in Supplementary Data 1 and Supplementary Data 2. Numerical source data for the graph presented in Figs. 1–3, and Fig. 5 can be found in the Zenodo repository here: 10.5281/zenodo.13315681. Other data and sources that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
All scripts used in this study are available in a Zenodo repository at 10.5281/zenodo.13315681.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/18/2024
A Correction to this paper has been published: 10.1038/s42003-024-07366-0
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06800-7.
References
- 1.Smale, D. A. Impacts of ocean warming on kelp forest ecosystems. N. Phytol.225, 1447–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 2.United Nations Environment Programme and Norwegian Blue Forests Network. Into the Blue: Securing a Sustainable Future for Kelp Forests. [online]. Available: https://wedocs.unep.org/20.500.11822/42255. [Accessed: Sept. 6, 2024].
- 3.Coleman, M. A. et al. Restore or redefine: future trajectories for restoration. Front. Mar. Sci.7, 237 (2020). [Google Scholar]
- 4.Wernberg, T. et al. Genetic diversity and kelp forest vulnerability to climatic stress. Sci. Rep.8, 1–8. 10.1038/s41598-018-20009-9 (2018). toolkit best practices pipeline. Curr. Protoc. Bioinformatics, 43, 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed]
- 5.Alsuwaiyan, N. A. et al. Genotypic variation in response to extreme events may facilitate kelp adaptation under future climates. Mar. Ecol. Prog. Ser.672, 111–121 (2021). [Google Scholar]
- 6.Cavanaugh, K. C., Reed, D. C., Bell, T. W., Castorani, M. C. & Beas-Luna, R. Spatial variability in the resistance and resilience of giant kelp in southern and Baja California to a multiyear heatwave. Front. Mar. Sci.6, 413 (2019). [Google Scholar]
- 7.Filbee-Dexter, K., Feehan, C. J. & Scheibling, R. E. Large-scale degradation of a kelp ecosystem in an ocean warming hotspot. Mar. Ecol. Prog. Ser.543, 141–152 (2016). [Google Scholar]
- 8.Wernberg, T. et al. An extreme climatic event alters marine ecosystem structure in a global biodiversity hotspot. Nat. Clim. Change3, 78–82 (2013). [Google Scholar]
- 9.Assis, J. et al. Past climate-driven range shifts structuring intraspecific biodiversity levels of the giant kelp (Macrocystis pyrifera) at global scales. Sci. Rep.13, 12046 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurgel, C. F. D., Camacho, O., Minne, A. J. P., Wernberg, T. & Coleman, M. A. Marine heatwave drives cryptic loss of genetic diversity in underwater forests. Curr. Biol.30, 1199–1206.e2 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Klingbeil, G., Montecinos, G. J. & Alberto, F. Giant kelp genetic monitoring before and after disturbance reveals stable genetic diversity in Southern California. Front. Mar. Sci.910.3389/fmars.2022.947393 (2022).
- 12.Mohring, M. B., Wernberg, T., Wright, J. T., Connell, S. D. & Russell, B. D. Biogeographic variation in temperature drives performance of kelp gametophytes during warming. Mar. Ecol. Prog. Ser.513, 85–96 (2014). [Google Scholar]
- 13.Fales, R. J., Weigel, B. L., Carrington, E., Berry, H. D. & Dethier, M. N. Interactive effects of temperature and nitrogen on the physiology of kelps (Nereocystis luetkeana and Saccharina latissima). Front. Mar. Sci.10, 1281104 (2023). [Google Scholar]
- 14.Veenhof, R. J., et al. Projecting kelp (Ecklonia radiata) gametophyte thermal adaptation and persistence under climate change. Ann. Botany133, 153–168 (2024). [DOI] [PMC free article] [PubMed]
- 15.Osborne, M. G. et al. Natural variation of Macrocystis pyrifera gametophyte germplasm culture microbiomes and applications for improving yield in offshore farms. J. Phycol.59, 402–417 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Wade, R. et al. Macroalgal germplasm banking for conservation, food security, and industry. PLoS Biol.18, e3000641 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollarsmith, J. A., Buschmann, A. H., Camus, C. & Grosholz, E. D. Varying reproductive success under ocean warming and acidification across giant kelp (Macrocystis pyrifera) populations. J. Exp. Mar. Biol. Ecol.522, 151247 (2020). [Google Scholar]
- 18.Buschmann, A. H. et al. Enhancing yield on Macrocystis pyrifera (Ochrophyta): the effect of gametophytic developmental strategy. Algal Res.52, 102124 (2020). [Google Scholar]
- 19.Eger, A. et al. The Kelp Forest Challenge: A collaborative global movement to protect and restore 4 million hectares of kelp forests. J. Appl. Phycol. 36, 951–964 (2024).
- 20.Eger, A. et al. A roadmap for protecting and restoring 4 million hectares of kelp forests by 2040. 10.13140/RG.2.2.26800.12809 (2023b).
- 21.Umanzor, S. et al. Short‐term stress responses and recovery of giant kelp (Macrocystis pyrifera, Laminariales, Phaeophyceae) juvenile sporophytes to a simulated marine heatwave and nitrate scarcity1. J. Phycol.57, 1604–1618 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Wahid, A., Gelani, S., Ashraf, M. & Foolad, M. R. Heat tolerance in plants: an overview. Environ. Exp. Bot.61, 199–223 (2007). [Google Scholar]
- 23.Camus, C., Faugeron, S. & Buschmann, A. H. Assessment of genetic and phenotypic diversity of the giant kelp, Macrocystis pyrifera, to support breeding programs. Algal Res.30, 101–112 (2018). [Google Scholar]
- 24.Song, Y., Wang, J. & Wang, L. Satellite solar-induced chlorophyll fluorescence reveals heat stress impacts on wheat yield in India. Remote Sens.12, 3277 (2020). [Google Scholar]
- 25.Van der Westhuizen, M., Oosterhuis, D., Berner, J. & Boogaers, N. Chlorophyll a fluorescence as an indicator of heat stress in cotton (Gossypium hirsutum L). South African J. Plant Soil37, 116–119 (2020).
- 26.Harris, R. J. et al. A novel and high throughput approach to assess photosynthetic thermal tolerance of kelp using chlorophyll-a fluorometry. J. Phycol59, 179–192 (2022). [DOI] [PubMed]
- 27.Graham, M. H., Vasquez, J. A. & Buschmann, A. H. Global ecology of the giant kelp Macrocystis: from ecotypes to ecosystems. Oceanogr. Mar. Biol.45, 39 (2007). [Google Scholar]
- 28.Hsiao, S. I. & Druehl, L. D. Environmental control of gametogenesis in Laminaria saccharina. J. Phycol.9, 128–132 (1973). [Google Scholar]
- 29.Di Lorenzo, E. & Mantua, N. Multi-year persistence of the 2014/15 North Pacific marine heatwave. Nat. Clim. Change6, 1042–1047 (2016). [Google Scholar]
- 30.Edwards, M. S. Estimating scale-dependency in disturbance impacts: El Niños and giant kelp forests in the northeast Pacific. Oecologia138, 436–447 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Diehl, N., Roleda, M. Y., Bartsch, I., Karsten, U. & Bischof, K. Summer heatwave impacts on the European kelp Saccharina latissima across Its latitudinal distribution gradient. Front. Mar. Sci.8, 695821 (2021).
- 32.Borges, H. et al. Well Plate Maker: a user-friendly randomized block design application to limit batch effects in large-scale biomedical studies. Bioinformatics37, 2770–2771 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Redmond, S., Green, L., Yarish, C., Kim, J., & Neefus, C. New England Seaweed Culture Handbook. Seaweed Cultivation. 1. https://digitalcommons.lib.uconn.edu/seagrant_weedcult/1 (2014).
- 34.Caliper Life Sciences. Living Image® Software User's Manual (Version 3.2). Xenogen Corporation (2009).
- 35.Johansson, M. L. et al. Seascape drivers of Macrocystis pyrifera population genetic structure in the northeast Pacific. Mol. Ecol.24, 4866–4885 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Oppliger, L. V. et al. Sex ratio variation in the lessonia nigrescens complex (Laminariales, Paeophyceae): Effect of Latitude, temperature, and marginality. J Phycol. 47, 5–12 (2011). [DOI] [PubMed]
- 37.Provasoli L. Media and prospects for the cultivation of marine algae. Cultures and collections of algae. Proc US Japan Conference, Hakone (eds Watanabe, A. & Hattori, R.) (1968).
- 38.Huang, B. et al. “Improvements of the daily optimum interpolation sea surface temperature (DOISST) version 2.1.”. J. Clim.34, 2923–2939 (2021). [Google Scholar]
- 39.Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diesel, J. et al. A scaffolded and annotated reference genome of giant kelp (Macrocystis pyrifera). BMC Genomics24, 543 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, H. et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Auwera, G. A. et al. From FastQ data to high‐confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinforma.43, 11–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hard-filtering germline short variants. Retrieved from https://gatk.broadinstitute.org/hc/en-us/articles/360035890471-Hard-filtering-germline-short-variants (2024).
- 45.Danecek, P., 1000 Genomes Project Analysis Group. et al. The variant call format and VCFtools. Bioinformatics27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Core Team. (2023). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Retrieved from https://www.R-project.org/.
- 47.Money, D. et al. LinkImpute: fast and accurate genotype imputation for nonmodel organisms. G3 Genes|Genomes|Genet.5, 2383–2390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience4, s13742-015-0047-8 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed]
- 49.Purcell, S., Chang, C. PLINK v 1.9. (n.d.). Retrieved from www.cog-genomics.org/plink/1.9/.
- 50.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. Retrieved from https://ggplot2.tidyverse.org (2023).
- 51.Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.6.0. Retrieved from https://rpkgs.datanovia.com/ggpubr/ (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The reference genome of Macrocystis pyrifera is available at the Joint Genome Institute portal https://phycocosm.jgi.doe.gov/Macpyr2. The WGS data of 559 gametophytes used to produce the VCF files are available in the NCBI repository under the accession code PRJNA1050779. The correspondence between the gametophytes’ names and the SRA runs in the NCBI repository, as well as the raw fluorescence intensity values, can be found in Supplementary Data 1 and Supplementary Data 2. Numerical source data for the graph presented in Figs. 1–3, and Fig. 5 can be found in the Zenodo repository here: 10.5281/zenodo.13315681. Other data and sources that support the findings of this study are available from the corresponding author upon reasonable request.
All scripts used in this study are available in a Zenodo repository at 10.5281/zenodo.13315681.